Abstract
Ejection of DNA from the capsid is an early step in infection by all herpesviruses. Ejection or DNA uncoating occurs after a parental capsid has entered the host cell cytoplasm, migrated to the nucleus and bound to a nuclear pore. DNA exits the capsid through the portal vertex and proceeds by way of the nuclear pore complex into the nucleoplasm where it is transcribed and replicated. Here we describe use of an in vitro uncoating system to determine which genome end exits first from the herpes simplex virus (HSV-1) capsid. Purified DNA-containing capsids were bound to a solid surface and warmed under conditions in which some, but not all, of the DNA was ejected. Restriction endonuclease digestion was then used to identify the genomic origin of the ejected DNA. The results support the view that the S segment end exits the capsid first. Preferential release at the S end demonstrates that herpesvirus DNA uncoating conforms to the paradigm in dsDNA bacteriophage where the last end packaged is the first to be ejected. Release of HSV-1 DNA beginning at the S end causes the first gene to enter the host cell nucleus to be α4, a transcription factor required for expression of early genes.
Keywords: herpes simplex virus, DNA uncoating, DNA ends, polarity of DNA ejection, flow chamber
Introduction
The nucleic acid in mature, extracellular viruses is highly condensed and coated with protein. The two properties together serve to protect the genome from chemical and physical damage as the virus traffics from one host cell to the next. Once associated with a new host cell, however, the genome needs to be uncoated and expanded so it can be replicated and the virus genes expressed.
In herpesviruses including herpes simplex virus (HSV-1), uncoating the dsDNA genome takes place at the nuclear membrane.1–3 A DNA-containing capsid enters the peripheral cytoplasm as a result of fusion between the virus and host cell membranes. The capsid then migrates to the nucleus, docks at a nuclear pore and releases its DNA through the pore and into the nucleoplasm.2,4,54 DNA exits the capsid as a single double helix with release presumed to occur through the portal, a cylindrical structure located at unique capsid vertex and specialized for function in DNA entry and exit.5–7 The parental capsid does not enter the nucleus; after its role in the docking process, it appears to play no further role in infection.1,3
The HSV-1 genome is a single molecule of dsDNA 152,261 bp in length. It encodes a total of 75 genes and is divided into two major segments, L and S.8–11 Each segment contains repeated DNA sequences at the ends and a longer region of unique sequence in the middle. Replication of HSV-1 DNA takes place in the infected cell nucleus and results in formation of a multi-genome concatemer in which individual genomes occur in a linear array.12–15 As DNA is replicated, the L and S segments invert with respect to each other creating a total of four genome isomers, called P, IS, IL and ISL, which occur in equal proportions in wild-type HSV-1 populations.16,17
The DNA concatemer has the interesting property that exposed DNA ends are only observed from the L segment. All or nearly all S segment ends are found in junctions with L.13,15 Preferential exposure of one genome end in the concatemer is observed in other herpesviruses including human cytomegalovirus, equine herpesvirus 1, murine cytomegalovirus, guinea pig cytomegalovirus and bovine herpesvirus 1.18–21 Although more complicated explanations are possible, selective exposure of L ends in HSV-1 has led to the assumption that DNA encapsidation begins at the L end and proceeds toward S. Further, since S is the last end packaged it is thought that uncoating must begin at S, as this is the case in dsDNA bacteriophage with the same mechanism of DNA packaging. 22,23
Here we describe the results of experiments designed to test the idea that HSV-1 DNA uncoating begins at the S end. Using an in vitro uncoating system, purified DNA-containing capsids were induced to eject some, but not all, of their DNA. Restriction enzyme analysis was then used to identify the genomic origin of the extruded fraction.
RESULTS
DNA extrusion on a solid surface: effect of formaldehyde
In vitro DNA uncoating was carried out by the method we have described recently.5 DNA-containing capsids (C capsids) were isolated from the nuclei of infected cells, bound to a plastic surface and induced to release their DNA by brief warming at 37°C. Under these conditions DNA is extruded as a single double helix which arises from one vertex assumed to contain the portal. All DNA is ejected, and DNA is ejected only when capsids are both bound to a solid surface and warmed to 37°C. 5 In the current studies, the length of DNA extruded was limited by pre-treatment of capsids with formaldehyde, fixative able to introduce cross-links between DNA and the capsid protein shell.24
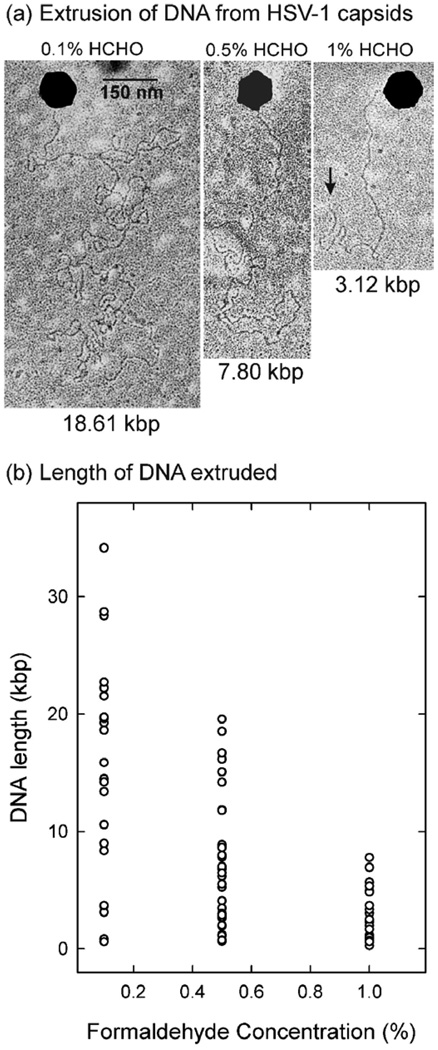
A study of the effect of formaldehyde concentration was performed by pre-treating capsids with concentrations in the range of 0.1%–1%. After treatment, capsids were bound to the surface of a Formvar-coated surface (electron microscope grid), and warmed at 37°C to induce DNA release. The length of DNA released was determined by electron microscopy. Fig. 1a shows representative micrographs of DNA extruded from capsids treated with 0.1%, 0.5% and 1.0% formaldehyde. As noted previously, in all cases DNA was released as a single double helix with ejection occurring at a single capsid vertex (Fig. 1a).5 In some cases a DNA end could be identified as shown in Fig. 1a (arrow in right panel).
Figure 1.
Effect of formaldehyde treatment on DNA ejection from HSV-1 C capsids. (a) Electron micrographs showing formaldehyde-treated C capsids induced to eject their DNA on a plastic surface (a coated electron microscope grid). Left, center and right panels show DNA extruded from capsids pre-treated with 0.1%, 0.5% and 1.0% formaldehyde, respectively. The length of extruded DNA (in kbp) is indicated beneath each image. The extruded DNA end is indicated by an arrow in the right panel. Note that DNA is released as a single double helix from one capsid vertex (assumed to be the portal vertex). (b) Plot showing the effect of formaldehyde on the length of DNA extruded from individual C capsids. Measurements were made from micrographs similar to those shown in (a). Each data point indicates the length of DNA extruded from a separate capsid. The average value in each distribution is marked. Note that the length of extruded DNA is greater at lower formaldehyde concentration.
Examination of a larger number of capsids demonstrated that the length of DNA extruded was dependent on the formaldehyde concentration. Less DNA was released at higher formaldehyde concentration (Fig. 1b). It is suggested that a greater number of DNA-capsid cross-links were introduced at higher formaldehyde concentration causing less DNA to be ejected. A wide range of extruded DNA lengths was observed at each formaldehyde concentration tested (Fig. 1b). In the case of 0.1% formaldehyde experiments, for instance, the standard deviation of extruded DNA lengths was approximately 2/3 of the mean (15.9 ± 10.0 kb [n=23]). Mean and standard deviations for 0.5% and 1.0% formaldehyde experiments were 7.5 ± 5.6 kb (n=33) and 2.6 ± 2.2 kb (n=25), respectively.
Genome origin of ejected DNA
Experiments to identify the genomic origin of extruded DNA were carried out with capsids treated with 0.1% formaldehyde, a concentration found to be permissive for DNA release from a high proportion of capsids (data not shown). Formaldehyde-treated capsids were adsorbed to a plastic surface, warmed to induce DNA release and treated with BamHI to digest extruded DNA. Agarose gel electrophoresis and Southern blotting were then used to compare the digestion products derived from formaldehyde-treated and control capsids. It was expected that the genomic region ejected first would be enriched in the BamHI digestion product from that end. In contrast, a more equal distribution of end fragments was expected from control capsids not treated with formaldehyde as the entire genome is extruded in that case.5
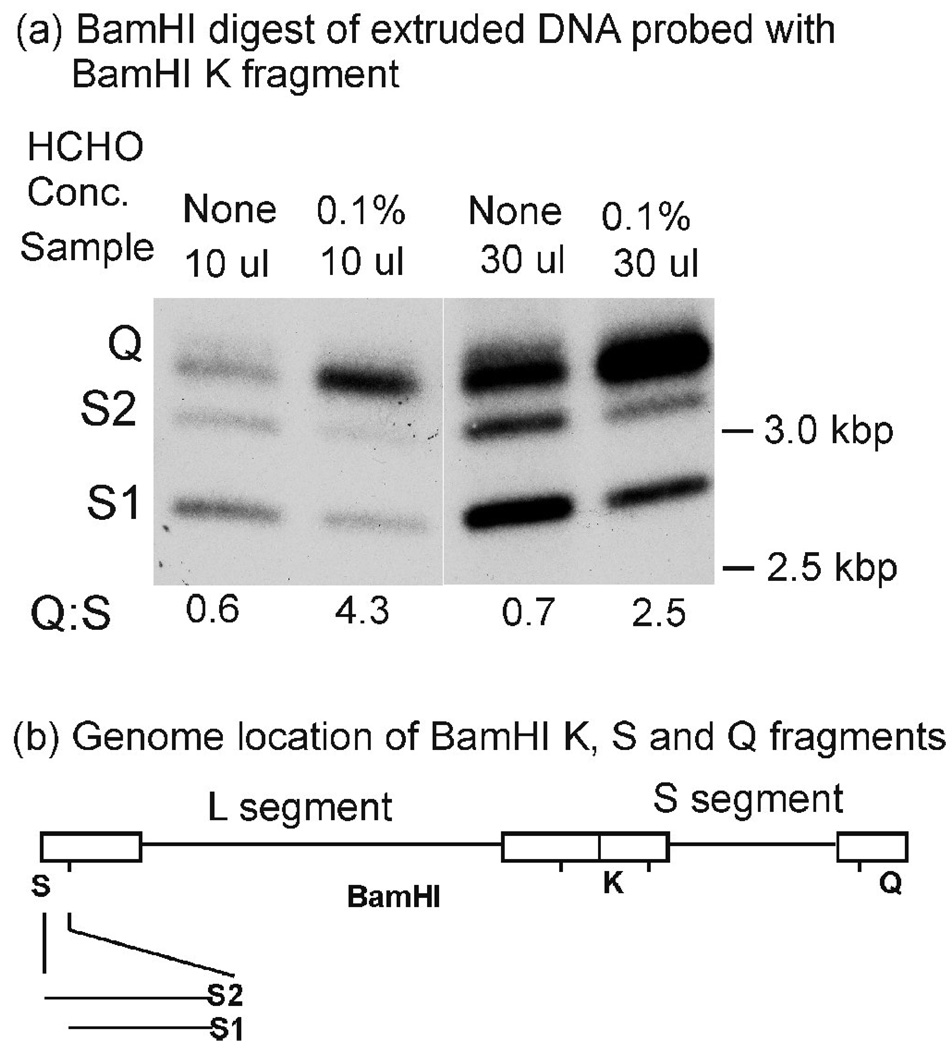
Experiments involved analysis of the BamHI Q fragment (3.4 kbp) derived from the S segment terminus and the BamHI S fragment derived from the L segment end (see Fig. 2b). HSV-1 populations include virions with one of two BamHI S fragments, S1 (2.9 kbp) or S2 (3.3 kbp) that differ in the number of 250–400 bp “a” sequences present, S1 has one and S2 two. The Q fragment contains one a sequence. 53
Figure 2.
Southern blot analysis of DNA extruded from control and formaldehyde-treated C capsids. As described in Materials and Methods, BamHI digests of the extruded DNA were subjected to agarose gel electrophoresis, blotting and probing with the BamHI K fragment (containing both Q and S fragments). The Q and S regions of the gel are shown in (a). Quantitative determination of the Q: S1+S2 ratio is shown below the blot. Note that Q fragments are enriched in the digests of DNA extruded from formaldehyde-treated compared to control capsids. The genomic locations of BamHI Q (3.4 kbp), S (S1: 2.9 kbp; S2 3.3 kbp) and K (6.1 kbp) fragments are shown in (b). S1 and S2 differ in the number of a sequences present; S1 has one and S2 has two.53
The result of a representative Southern blot is shown in Fig. 2a. It indicates that the Q fragment (right end) was enriched compared to S2 and S1 (left end) in digests of DNA extruded from capsids treated with 0.1% formaldehyde (compare the first and second lanes or the third and fourth lanes). Quantitative determination of the band intensities demonstrated a 7-fold (Q:S 4.3/Q:S 0.6= 7.2) or 3-fold (Q:S 2.5/Q:S 0.7=3.6) preference for DNA extrusion from the S segment end. The difference in the two ratios is suggested to be due to saturation of the signal when the Q fragment is present in high amounts (Fig. 2a, lane 4). Qualitatively similar results were obtained when the formaldehyde concentration was 0.3% instead of 0.1% (data not shown).
DNA uncoating in a flow chamber
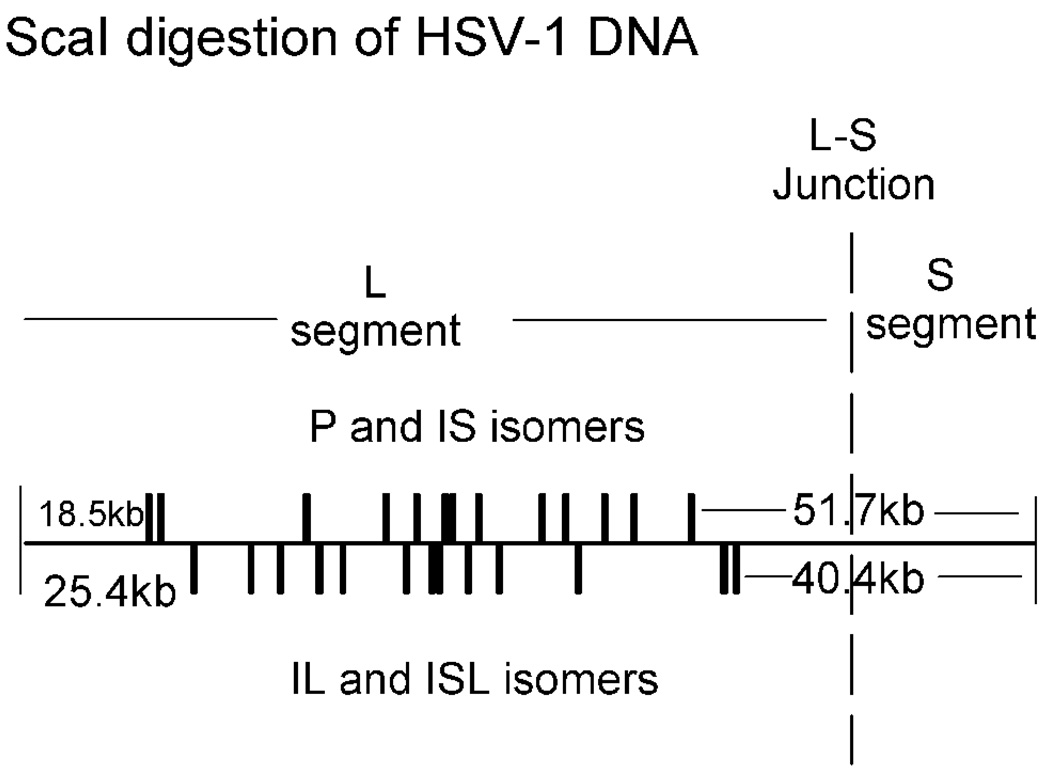
The genomic origin of the first DNA ejected was also examined by a flow chamber methodology.25,26 Formaldehyde-treated C capsids were bound and induced to release their DNA in a chamber in which the extruded DNA was extended by a stream of flowing buffer. Restriction endonuclease digestion was then carried out and the length of DNA remaining associated with the capsid was measured by fluorescence microscopy. The results were interpreted with reference to the known sites for digestion by the restriction enzyme used, ScaI (Fig. 3).
Figure 3.
Linear map of the HSV-1 genome showing the positions of ScaI cleavage sites. Note that cut sites are the same for the P and IS isomers and also for IL and ISL. The position of the junction between the L and S segments is indicated.
It was considered that digestions with ScaI would be revealing because long, un-cut fragments are expected if DNA exits the capsid beginning at the right (S segment) end, but not the left (L segment). Cleavages nearest the S end occur 51.7 kbp and 40.4 kbp away depending on the genome isomer as shown in Fig. 3. Extruded, un-cut DNA molecules up to 51.7 kbp and 40.4 kbp in length are therefore expected only if S end DNA is ejected first. In contrast, if ejection begins at the L end, then the longest exposed, un-cut fragments are expected to be 18.5 kbp and 25.4 kbp (Fig. 3).
Effects of formaldehyde
As in the case of the non-flow experiments described above, control flow chamber studies were carried out to test the effect of formaldehyde concentration on the length of DNA extruded. C capsids were treated in solution with formaldehyde, bound inside the flow cell and induced to release their DNA under conditions where ejected DNA was extended by flow of stain-containing buffer. Images of such preparations were recorded by fluorescence microscopy and DNA lengths were measured from the images.
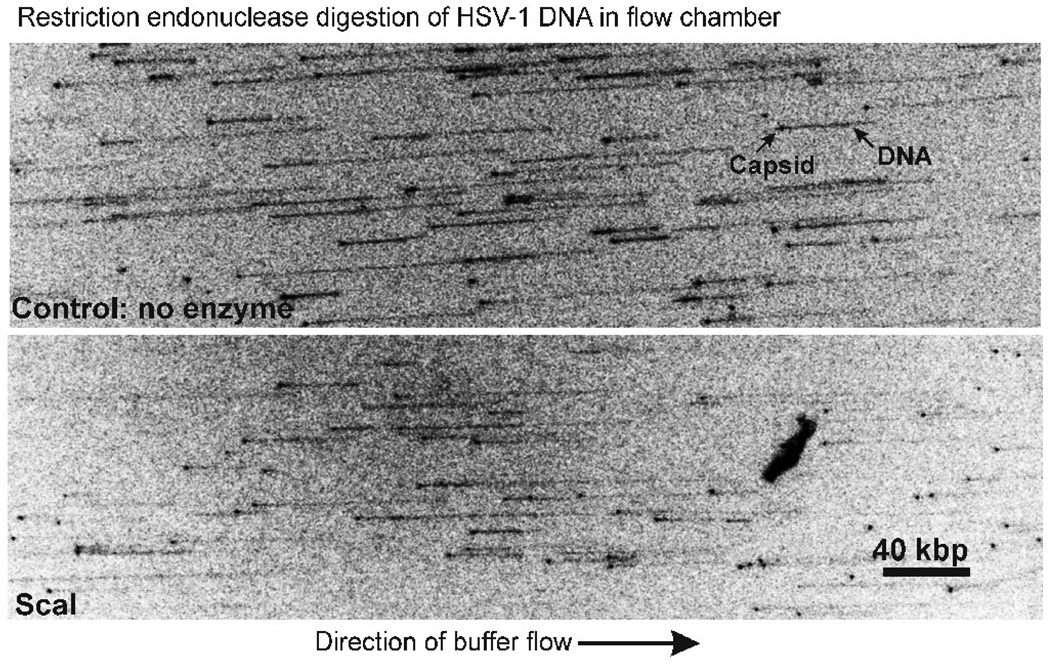
A representative micrograph is shown in the top panel of Fig. 4. In this experiment capsids were treated with 0.1% formaldehyde and buffer flow was from left to right. Spots and thinner lines extending from the spots correspond to capsids and DNA molecules, respectively.25,26 When the direction of buffer flow was reversed, the positions of spots were not affected, but the direction of DNA chains was reversed indicating that while capsids were attached to the flow cell surface, DNA chains were free to rotate (micrographs not shown). All DNA was released from capsids not treated with formaldehyde.
Figure 4.
Fluorescence micrographs showing the effects of ScaI digestion on DNA extruded from C capsids. The Materials and Methods section describes procedures by which capsids were treated with 0.1% formaldehyde, bound to the surface of a flow chamber, and induced to extrude DNA by heating to 37°C. Extruded DNA was then extended from the capsid by a flow of buffer, digested with ScaI and visualized by fluorescence microscopy (GelRed stain). Top: DNA extruded from control capsids not treated with ScaI; bottom: extruded DNA after cleavage with ScaI. Arrows in the top panel indicate the capsid and extruded DNA in a representative image. Note that the length of DNA is decreased after ScaI digestion.
As in the case of static, non-flow experiments, the length of DNA extruded in flow chambers was found to be inversely related to the formaldehyde concentration as shown in Table 1. The high degree of variability in DNA lengths seen with the non-flow studies was also observed in the flow experiments. For instance, the standard deviation of the extruded lengths was greater than 50% of the mean for all four formaldehyde concentrations tested under buffer flow conditions (Table 1). It was consistently observed that the length of extruded DNA was greater in flow cell compared to non-flow, grid studies. For instance, at a formaldehyde concentration of 0.5%, the mean extruded DNA lengths were 19.3 kb and 7.5 kb in flow and grid experiments, respectively. We suggest this may be related to an effect of surface-associated DNA to resist the pressure driving DNA extrusion, a force that would not be present under conditions of DNA extension by buffer flow. In flow chamber studies buffer movement would be expected to potentiate DNA extrusion.
Table 1.
Effect of formaldehyde concentration on the length of DNA extruded in flow chamber experiments
| HCHO | Mean DNA length | |
|---|---|---|
| Concentration |
(kb ± S.D.) |
Measurements |
| None | 95.4 ± 37.6 | 19 |
| 0.1% | 60.7 ± 38.5 | 82 |
| 0.2% | 37.6 ± 28.9 | 90 |
| 0.5% | 19.3 ± 16.4 | 27 |
| 1.0% | 15.4 ± 11.6 | 37 |
ScaI digestion
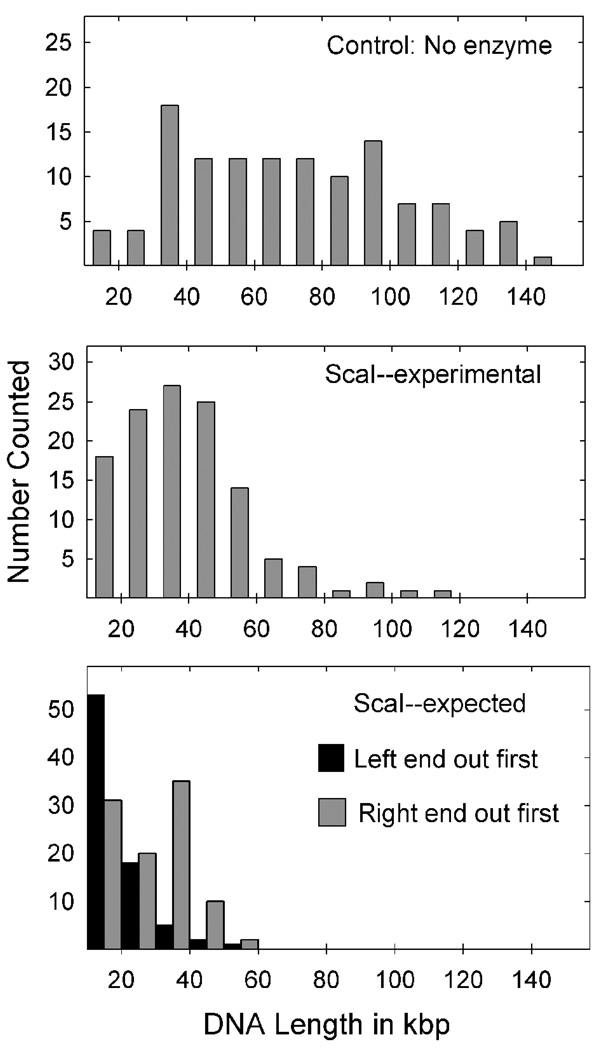
The effect of ScaI treatment on extruded DNA length is shown in Fig. 4 and Fig 5. From a visual comparison of DNA lengths in control and ScaI-treated preparations it was seen that control lengths were generally longer than those seen after ScaI digestion (Fig. 4). Measurement of DNA lengths supported the same conclusion (Fig. 5). While the measured lengths in control preparations extended from less than 20 kbp to greater than 130 kbp (i.e. nearly the full genome length), after ScaI digestion, extruded fragments were found in a reasonably uniform distribution centered about a mean of ~35 kbp (Fig. 5, middle panel). There was an abundance of capsids with extruded DNA lengths of 30–50 kbp supporting the view that the S end is released first. The distribution was not truncated at 18.5 or 25.4 kbp as expected if release begins at the L end.
Figure 5.
Histograms showing the extruded DNA lengths measured in flow chamber experiments from capsids in control (top) and ScaI-treated preparations (middle panel). Measurements were made from micrographs recorded in experiments identical to those shown in Fig. 4. Measured DNA lengths were converted to number of base pairs as described in Materials and Methods. The bottom panel shows the predicted distribution of DNA lengths after ScaI digestion assuming DNA was ejected beginning at the left (black) and right (gray) genome ends. Note that the experimentally-determined length distribution (middle panel) corresponds better to the distribution expected from extrusion beginning at the right end.
Comparison of the ScaI and control distributions demonstrated that ScaI effectively cleaved extruded DNA greater than ~50–60 kbp in length (compare the center and top panels in Fig. 5). This observation provides evidence that ScaI was enzymatically active under the conditions of the experiment and most exposed ScaI sites were cleaved. The small number of un-cut fragments (Fig. 5, middle panel) is interpreted to result from rare sites resistant to ScaI cleavage or cut with slower kinetics.
For comparison with the experimentally-determined fragment distribution, two theoretically expected distributions were calculated assuming DNA is ejected beginning at the left and right ends, respectively. Calculations were made beginning with the control, un-cut fragment data set (121 capsids; Fig. 5, top panel), and all four genome isomers were included in the calculation; each was assumed to constitute 25% of the capsid population. The two calculated distributions differed in that a large number of long (30–50 kbp), un-cut fragments were predicted if the right end is ejected first, while fewer are predicted in the left end first model (Fig. 5, bottom panel). As an abundance of 30–50 kbp fragments are found in the experimental distribution, we conclude that the results fit better with DNA exit beginning at the right end.
Discussion
Experimental design
The experiments described here were designed in such a way that the results could be interpreted despite the existence of genome segment inversion in HSV-1. The two possible L segment isomers occur in equal numbers in wt HSV-1 populations and the same is true of S. Restriction enzyme digestion of DNA from such populations can therefore result in junctional fragments expected in 1/2 or 1/4 the amount present in non-junctional ones. In the case where analysis was performed with BamHI digests of released DNA, it was felt that the BamHI Q and S fragments would be particularly appropriate as they occur uniquely at the right and left ends, respectively, of the HSV-1 genome regardless of the segment orientation. Q and S occur together in a junctional fragment (K; 6.1 kbp) at the L-S interface, but this fragment is longer than Q (3.4 kbp) or S (2.9 or 3.3 kbp) and cannot be confused with either.
Use of formaldehyde treatment of C capsids was central to the design of the experiments. It had previously been demonstrated that treatment of C capsids with 1% formaldehyde blocked DNA extrusion in experiments with attached capsids.5 It is novel to the experiments described here that lower doses of formaldehyde were found to permit limited DNA extrusion in a dose-dependent manner (Fig. 1, Table 1). Stable extrusion of HSV-1 DNA from formaldehyde-treated C capsids, as observed in flow chambers, may be useful for other types of studies such as DNA-protein interactions and nuclease specificity.
Interpretation of results
Experiments with BamHI digests of extruded DNA demonstrated enrichment in BamHI Q compared to S digestion products in formaldehyde-treated in contrast to untreated capsids (Fig. 2). Since DNA extruded from treated capsids is enriched in the end ejected first (compared to untreated capsids in which all or nearly all DNA is extruded 5), we conclude the Q end is ejected first in the majority of capsids. The preference for Q (right) end ejection was found to be 3–7-fold in the experiments shown Fig. 2. Although DNA extrusion beginning at the right end was shown to predominate, the presence of a low level of S fragments (Fig. 2a) raises the possibility that the right end may not be ejected first in every capsid. On the other hand, the low level of L segment ends (S1 and S2) in BamHI digests may be due to DNA released from rare damaged or leaky capsids that result from attachment to the plastic surface or to handling during the experimental operations.
Experiments with flow chamber methodology are also interpreted to indicate that the right end of the genome is ejected first. The most telling observation is the existence in the capsid-associated, extruded DNA population of long (30–50 kbp) ScaI-resistant regions expected in abundance if DNA is released beginning at the right genome end, but rare if release begins at the left. For a protected 30–50 kbp fragment to arise from DNA exit beginning at the left end, nearly all the capsid DNA would need to be extruded first, an event that is rare in the formaldehyde-treated capsids examined here (see Fig. 3 and Fig. 5, top panel). The same interpretation is derived from comparison of the observed fragment length distribution with distributions calculated to arise from DNA exit beginning at the right and left genome ends. A better match is seen in the case of right end exit (compare Fig. 5, middle and lower panels).
Observation of polarized DNA release from the HSV-1 capsid as described here strengthens the idea that the in vitro system employed is a reliable one for examining the uncoating process. Prior studies demonstrated that DNA release in the in vitro system occurs as a single double helix with release taking place from a single capsid vertex as expected for uncoating in infected cells.5 Preferential DNA release at the right genome end further supports the authenticity of observations with the in vitro system. Studies with this system are the first we are aware of where polarized nucleic acid uncoating is demonstrated in an animal virus.
Polarized DNA ejection in bacteriophage
Polarized DNA ejection as described here for HSV-1 is found to be the rule in dsDNA bacteriophage where appropriate studies have been done. Polarized release has been documented, for example, in phages lambda, T7/T3, φ29, SPP1 and T5.22,23,27–31,55 T4 is the only phage we are aware of where the potential exists for DNA to be extruded beginning at either genome end.32 The results reported here therefore support the view that DNA release in HSV-1 occurs by the same, end-specific mechanism observed in the majority of dsDNA phage.
In the case of phages lambda, T7/T3, SPP1 and φ29, experimental studies have demonstrated a polarity to both DNA ejection and to the DNA encapsidation process.22,23,27–30,32–37,55 For example, lambda phage DNA is packaged beginning at the left genome end and ejected beginning at the right. A similar situation is observed in T7/T3, SPP1 and φ29. The first DNA end to enter the capsid as the virus is assembled is at the opposite end of the genome from the first extruded in a new infection. It is suggested that this paradigm may apply broadly among dsDNA phage.38,39,40,55
HSV-1 DNA uncoating
If the paradigm described above applies to HSV-1 as well as to phage, then it is suggested that DNA packaging begins at the L segment end. Since the S end is ejected first as shown by the results described here, then the phage model indicates that encapsidation should begin at L. The idea that HSV-1 DNA packaging begins at an L segment end is consistent with studies demonstrating that only L ends are exposed in the concatemer, the primary HSV-1 DNA replication product.13,15 S segment ends are present only in junctions with L. It is suggested that packaging must begin at L ends as these are the only ones exposed in newly replicated DNA (although sequence-specific endonuclease cutting as observed in some phage is also a possibility).
Preferential exposure of one genome end in the DNA concatemer has been observed in other herpesviruses, including equine herpesvirus 1, human cytomegalovirus, guinea pig cytomegalovirus, mouse cytomegalovirus and bovine herpesvirus 1.18–21 By the reasoning described above, it is expected that DNA encapsidation will begin at the selectively exposed terminus with ejection beginning at the opposite end. A similar polarity may exist in other herpesviruses.
Structural studies with phage lambda have added information about the location of specific DNA regions in the capsid. Mature wt phage were etched in an Ar+ plasma under conditions in which the capsid and the outermost DNA were eroded by sputtering while internal DNA remained intact.41 Restriction enzyme analysis of the DNA remaining demonstrated that it arose selectively from the left genome end, the end that enters the capsid first and exits last.22,23,32,34,37 Applied to HSV-1, this result suggests that the first DNA ejected, the S end, will be located at the exterior of the packaged DNA mass in the mature capsid. Studies are currently underway to test this expectation.
HSV-1 α4 gene
Polarized ejection of HSV-1 DNA beginning at the S end requires that α4 is the first virus gene to enter the nucleus. Two copies of α4 (also called RS1) are present in the HSV-1 genome with one copy at each end of the S segment.10,11,42 α4 is therefore the first gene to enter the nucleus in both possible S segment orientations. It is intriguing to speculate that early entry of α4 may be important for HSV-1 replication or pathogenesis. α4 is required for HSV-1 replication.11 The encoded protein, ICP4, binds DNA directly in a sequence-specific fashion,43,44 and ICP4 positively regulates transcription of most HSV-1 β and γ genes.42,45 α4 expression is regulated by a complex promoter whose activation does not require the function of any HSV-1-encoded protein indicating that the α4 gene could be expressed promptly if it were the first to enter an infected cell nucleus.47,48 It might be revealing, therefore, to determine whether virus replication would be affected by moving the α4 genes to a different position in the HSV-1 genome.
An analogous situation is observed in the case of phage T7 where the first gene to enter the host (E. coli) cell encodes RNA-dependent RNA polymerase.28 The gene is expressed promptly after it enters the cell, and the enzyme is involved in promoting uptake of the remaining T7 DNA and in transcription of early virus genes.
Materials and Methods
Cell and virus growth; C capsid isolation
All experiments were carried out with the KOS strain of HSV-1 which was grown on monolayer cultures of Vero cells as previously described.49 C capsids were isolated from cells that had been infected for 20 h at 37°C, harvested by scraping in PBS, pelleted and frozen at −20°C. Newcomb et al. 50 have described the procedure used for isolation of capsids from infected cell nuclei. Once A, B and C capsids were separated by sucrose density gradient centrifugation (for 40 min at 24,000 rpm in an SW55Ti rotor), C capsids were removed from the gradient, pelleted and further purified by centrifugation at 23,000 rpm (SW50.1 rotor) for 1 hr at 4°C on a 600 µl gradient of 20%–50% sucrose in TNE (0.01M Tris-HCl, 0.5M NaCl, 1 mM EDTA, pH 7.5). Capsids were harvested from the gradient, adjusted with TNE to a concentration of 0.5 mg protein/ml and used in the experiments described below. Preparations contained 85% C capsids with the remainder A’s and B’s.
Effect of formaldehyde treatment on DNA ejection
Studies were carried out beginning with 50 µl aliquots of C capsids at a concentration of 5 µg/ml in TNE. Solutions were adjusted to the desired formaldehyde concentration (by addition from a 20% stock solution), incubated for 10 sec at room temperature and the formaldehyde quenched by adding 1/10 volume of 2 M glycine, pH 8.0. Previously-described procedures were used for adsorbing capsids to Formvar-carbon-coated electron microscope grids, warming for 2 h at 37°C to induce DNA release and staining with 1% uranyl acetate.5 Stained specimens were then rotary shadowed with Pt-carbon in a Bal-Tec Med-020 vacuum evaporator operated at 1.90 keV and 70 mA (15° angle of evaporation). Electron microscopy was performed on a Philips EM400T transmission electron microscope operated at 80 keV. Images were recorded on film and converted to digital form in a flatbed scanner. DNA lengths were measured with ImageJ.
BamHI digestion of DNA extruded on coverslips
The strategy of coverslip experiments was to cause DNA release from C capsids by warming under conditions where the extent of release was limited by pre-treatment of capsids with formaldehyde. After capsids were exposed to formaldehyde, they were bound to a solid surface (a microscope coverslip), warmed to cause DNA extrusion and treated with a restriction enzyme (BamHI) to digest extruded DNA. DNA remaining inside the capsid was not digested. Southern hybridization was then used to determine the relative proportion of L compared to S segment genome ends in the digest. It was expected that the result would be revealing about the identity of the first DNA end extruded from the capsid.
Experiments were performed beginning with 125 µl of C capsids at a concentration of 50 µg protein/ml in TNE. The procedure described above was used to treat capsids (as necessary) with 0.1% formaldehyde for 10 sec at room temperature and quench the formaldehyde with 2 M glycine. The entire 125 µl specimen was then sandwiched between two glow discharged, square, plastic coverslips (22 mm × 22 mm) and the capsids were allowed to attach by incubation for 10 min at 4°C. Two such sandwiches were used for each formaldehyde concentration tested. Sandwiches containing capsids treated with the same formaldehyde concentration were then dropped into 1 l buffer (0.1 M NaCl, 0.1% Triton X-100, 20 mM Tris-HCl, pH 8.0), a treatment that caused the sandwich to come apart into single coverslips.
Each of the resulting coverslips was washed (3X) with BamHI restriction enzyme buffer (0.1 M NaCl, 10 mM MgCl2, 1 mM DTT, 50 mM Tris-HCl, pH 7.9) containing 0.1% Triton X-100. 50 µl BamHI (40U/µl; no Triton X-100) was then added to the capsid side of one coverslip and a second was positioned to create a sandwich with 50 µl BamHI separating the capsid sides of two coverslips. After incubation for 45 min at 37°C to allow the enzyme to digest exposed DNA, coverslips were aligned radially in a 50 ml conical tube so that centrifugation would extract the BamHI digest from between the coverslips. Centrifugation was for 5 min at low speed. The digest (~100 µl) was transferred to a fresh tube, centrifuged at 30,000 rpm (Beckman Coulter TLX ultracentrifuge; TLA-55 rotor) and used for Southern blot experiments as described below.
The L and S segment ends of the HSV-1 genome can be monitored by Southern blot analysis of virus DNA digested with BamHI and probed with the HSV-1 BamHI K fragment (6.1 kbp).51 Viral DNA with free chromosomal ends yields the terminal Q (3.4 kbp) and S fragments (S1: 2.9 kbp; S2: 3.3 kbp) from the right and left genome ends, respectively. DNA extruded from capsids and digested with BamHI as described above was subjected to agarose gel electrophoresis, transferred to a nylon membrane and hybridized with 32P-labeled K fragment as described previously.51,52 Blots were scanned with a Storm 840 PhosporImager and the bands determined quantitatively with ImageQuant software.
Flow chamber experiments
Methods used were similar to those described by Mangenot et al.26 and by Grayson et al.25 C capsids at a concentration of 0.5 µg protein/ml in TNE were treated with 0.1% formaldehyde as described above and 100 µl was introduced into a microscope slide flow chamber (ibidi, Verona, WI, µ-Slide I, cat. no. 80111). Capsids were allowed to attach during incubation for 15 min at 4°C and unattached capsids were removed by washing in 1 ml of TNE (twice). TNE was then removed by two washes with ScaI buffer (50 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, pH 7.9). Next, 100 µl of ScaI (0.5 units/ µl) was introduced into the chamber, incubated for 1 h at 37°C to digest exposed DNA and removed by washing with 2 ml TNE containing GelRed stain (1:10,000 dilution). A flow of buffer (TNE plus GelRed) was produced in the chamber by adding more buffer to one side than the other. The resulting gravity-induced stream was sufficient to cause the DNA to extend in a straight line while remaining tethered to the capsid due to the formaldehyde cross-linking.5 Images of capsids with extruded DNA were recorded in a Nikon Eclipse TE2000-E inverted, epifluorescence microscope operated with Improvision Inc. (Waltham, MA) Openlab software. The magnification was determined by imaging a 400 mesh electron microscope grid. DNA lengths were measured with Photoshop and converted to base pairs assuming an inter-nucleotide spacing of 0.34 nm. Data were plotted with SigmaPlot 10. ScaI (10,000 units/ml) was obtained from New England Biolabs.
Calculation of expected DNA fragment lengths
Calculation of the expected distribution of extruded, capsid-associated, ScaI-resistant DNA segments was carried out for flow chamber experiments. Calculations began with the measured segment lengths in the control, undigested population shown in Fig. 5 (top panel; 121 capsids with extruded DNA). DNA segments were assigned to specific ScaI restriction fragments assuming the smallest were derived from left genome end or the right. Separate computations were made for all four genome isomers which were assumed to occur in equal numbers in the capsid population. Expected segment lengths from left-end-first and right-end-first results were merged separately, binned and plotted as a histogram (Fig. 5, bottom panel).
Other methods
The HSV-1 DNA sequence (NC_001806) was extracted from public databases and manipulated with GCG (version 11.1.3; Accelrys).
Acknowledgements
We thank Dr. Paul Grayson for advice about flow chamber methodology. For comments on the manuscript we gratefully acknowledge Ann Beyer, Anna Maria Copeland and Sarah French. This work was supported by NIH awards AI041644 (JCB) and AI060836 (FLH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell Biol. 2000;20:4922–4931. doi: 10.1128/mcb.20.13.4922-4931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tognon M, Furlong D, Conley AJ, Roizman B. Molecular genetics of herpes simplex virus. V. Characterization of a mutant defective in ability to form plaques at low temperatures and in a viral fraction which prevents accumulation of coreless capsids at nuclear pores late in infection. J Virol. 1981;40:870–880. doi: 10.1128/jvi.40.3.870-880.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sodeik B, Ebersold MW, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newcomb WW, Booy FP, Brown JC. Uncoating the herpes simplex virus genome. J Mol. Biol. 2007;370:633–642. doi: 10.1016/j.jmb.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 2001;75:10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. Journal of Virology. 2004;78(22):12668–12671. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison AJ, Dargan DJ, Stow ND. Fundamental and accessory systems in herpesviruses. Antiviral Res. 2002;56:1–11. doi: 10.1016/s0166-3542(02)00107-9. [DOI] [PubMed] [Google Scholar]
- 9.McGeoch DJ, Dalrymple MA, Davison AJ, Dolan A, Frame MC, McNab D, Perry LJ, Scott JE, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen. Virol. 1988;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 10.McGeoch DJ, Dolan A, Donald S, Rixon FJ. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 1985;181:1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- 11.Roizman B. The function of herpes simplex virus genes: A primer for genetic engineering of novel vectors. Proc. Natl. Acad. Sci. USA. 1996;93:11307–11312. doi: 10.1073/pnas.93.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob RJ, Morse LS, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J. Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez R, Sarisky RT, Weber PC, Weller SK. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severini A, Morgan AR, Tovell DR, Tyrrell DL. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virol. 1994;200:428–435. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virol. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]
- 16.Delius H, Clements JB. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J. Gen. Virol. 1976;33:125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- 17.Hayward GS, Jacob RJ, Wadsworth SC, Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc. Natl. Acad. Sci. U. S. A. 1975;72:4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McVoy MA, Adler SP. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J. Virol. 1994;68:1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McVoy MA, Nixon DE, Hur JK, Adler SP. The ends on herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences. J Virol. 2000;74:1587–1592. doi: 10.1128/jvi.74.3.1587-1592.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schynts F, McVoy MA, Meurens F, Detry B, Epstein AL, Thiry E. The structures of bovine herpesvirus 1 virion and concatemeric DNA: implications for cleavage and packaging of herpesvirus genomes. Virol. 2003;314:326–335. doi: 10.1016/s0042-6822(03)00437-9. [DOI] [PubMed] [Google Scholar]
- 21.Slobedman B, Simmons A. Concatemeric intermediates of equine herpesvirus type 1 DNA replication contain frequent inversions of adjacent long segments of the viral genome. Virol. 1997;229:415–420. doi: 10.1006/viro.1997.8447. [DOI] [PubMed] [Google Scholar]
- 22.Chattoraj DK, Inman RB. Location of DNA ends in P2, 186, P4 and lambda bacteriophage heads. J Mol. Biol. 1974;87:11–22. doi: 10.1016/0022-2836(74)90556-7. [DOI] [PubMed] [Google Scholar]
- 23.Thomas JO. Chemical linkage of the tail to the right-hand end of bacteriophage lambda DNA. J Mol. Biol. 1974;87:1–9. doi: 10.1016/0022-2836(74)90555-5. [DOI] [PubMed] [Google Scholar]
- 24.Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl. Acad. Sci. U. S. A. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grayson P, Han L, Winther T, Phillips R. Real-time observations of single bacteriophage lambda DNA ejections in vitro. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14652–14657. doi: 10.1073/pnas.0703274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangenot S, Hochrein M, Radler J, Letellier L. Real-time imaging of DNA ejection from single phage particles. Curr. Biol. 2005;15:430–435. doi: 10.1016/j.cub.2004.12.080. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Huici V, Salas M, Hermoso JM. The push-pull mechanism of bacteriophage O29 DNA injection. Mol. Microbiol. 2004;52:529–540. doi: 10.1111/j.1365-2958.2004.03993.x. [DOI] [PubMed] [Google Scholar]
- 28.Molineux IJ. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 2001;40:1–8. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- 29.Pao CC, Speyer JF. Order of injection of T7 bacteriophage DNA. J Virol. 1973;11:1024–1026. doi: 10.1128/jvi.11.6.1024-1026.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saigo K. Polar DNA ejection in bacteriophage T7. Virol. 1975;65:120–127. doi: 10.1016/0042-6822(75)90012-4. [DOI] [PubMed] [Google Scholar]
- 31.Shaw AR, Davison J. Polarized injection of the bacteriophage T5 chromosome. J Virol. 1979;30:933–935. doi: 10.1128/jvi.30.3.933-935.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Black LW, Silverman DJ. Model for DNA packaging into bacteriophage T4 heads. J Virol. 1978;28:643–655. doi: 10.1128/jvi.28.2.643-655.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becker A, Murialdo H. Bacteriophage lambda DNA: the beginning of the end. J Bacteriol. 1990;172:2819–2824. doi: 10.1128/jb.172.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bjornsti M, Reilly BE, Anderson DL. Morphogenesis of bacteriophage phi-29 of Bacillus subtilis: Oriented and quantized in vitro packaging of DNA protein gp3. J. Virol. 1983;45:383–396. doi: 10.1128/jvi.45.1.383-396.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Catalano CE, Cue D, Feiss M. Virus DNA packaging: the strategy used by phage lambda. Molec. Microbiol. 1995;16:1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 36.Fujisawa H, Kimura M, Hashimoto C. In vitro cleavage of the concatemer joint of bacteriophage T3 DNA. Virol. 1990;174:26–34. doi: 10.1016/0042-6822(90)90050-2. [DOI] [PubMed] [Google Scholar]
- 37.Son M, Watson RH, Serwer P. The direction and rate of bacteriophage T7 DNA packaging in vitro. Virol. 1993;196:282–289. doi: 10.1006/viro.1993.1476. [DOI] [PubMed] [Google Scholar]
- 38.Yang Q, Catalano CE. Biochemical characterization of bacteriophage lambda genome packaging in vitro. Virol. 2003;305:276–287. doi: 10.1006/viro.2002.1602. [DOI] [PubMed] [Google Scholar]
- 39.Black LW. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 40.Murialdo H, Becker A. Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol. Rev. 1978;42:529–576. doi: 10.1128/mr.42.3.529-576.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown JC, Newcomb WW. Ion etching of bacteriophage lambda: evidence that the right end of the DNA is located at the outside of the phage DNA mass. J. Virol. 1986;60:564–568. doi: 10.1128/jvi.60.2.564-568.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGeoch DJ, Dolan A, Donald S, Brauer DH. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986;14:1727–1745. doi: 10.1093/nar/14.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeLuca NA, McCarthy AM, Schaffer PA. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sampath P, DeLuca NA. Binding of ICP4, TATA-binding protein, and RNA polymerase II to herpes simplex virus type 1 intermediate-early, early and late promoters in virus-infected cells. J. Virol. 2008;82:2339–2349. doi: 10.1128/JVI.02459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faber SW, Wilcox KW. Association of herpes simplex virus regulatory protein ICP4 with sequences spanning the ICP4 gene transcription initiation site. Nucleic Acids Res. 1988;16:555–570. doi: 10.1093/nar/16.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith CA, Schaffer PA. Mutants defective in herpes simplex virus type 2 ICP4: isolation and preliminary characterization. J Virol. 1987;61:1092–1097. doi: 10.1128/jvi.61.4.1092-1097.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mackem S, Roizman B. Differentiation between alpha promoter and regulator regions of herpes simplex virus 1: the functional domains and sequence of a movable alpha regulator. Proc. Natl. Acad. Sci. U. S. A. 1982;79:4917–4921. doi: 10.1073/pnas.79.16.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Post LE, Mackem S, Roizman B. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell. 1981;24:555–565. doi: 10.1016/0092-8674(81)90346-9. [DOI] [PubMed] [Google Scholar]
- 49.Sheaffer AK, Newcomb WW, Brown JC, Gao M, Weller SK, Tenney DJ. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids [In Process Citation] J. Virol. 2000;74:6838–6848. doi: 10.1128/jvi.74.15.6838-6848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newcomb WW, Homa FL, Brown JC. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J Virol. 2006;80:6286–6294. doi: 10.1128/JVI.02648-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Homa FL, Otal TM, Glorioso JC, Levine M. Transcriptional control signals of a herpes simplex virus type 1 late (gamma 2) gene lie within bases −34 to +124 relative to the 5' terminus of the mRNA. Mol. Cell Biol. 1986;6:3652–3666. doi: 10.1128/mcb.6.11.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tengelsen LA, Pederson NE, Shaver PR, Wathen MW, Homa FL. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mocarski ES, Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc. Natl. Acad. Sci. U. S. A. 1982;79:5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Copeland AM, Newcomb WW, Brown JC. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J Virol. 2009;83:1660–1668. doi: 10.1128/JVI.01139-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tavares P, Lurz R, Stiege A, Ruckert B, Trautner TA. Sequential headful packaging and fate of the cleaved DNA ends in bacteriophage SPP1. J. Mol. Biol. 1996;264:954–967. doi: 10.1006/jmbi.1996.0689. [DOI] [PubMed] [Google Scholar]