Abstract
Alzheimer’s Disease (AD) is characterized by the cerebral accumulation of misfolded and aggregated amyloid-β protein (Aβ). Disease symptoms can be alleviated, in vitro and in vivo, by “β-sheet breaker” pentapeptides that reduce plaque volume. However the peptide nature of these compounds, made them biologically unstable and unable to penetrate membranes with high efficiency. The main goal of this study was to use computational methods to identify small molecule mimetics with better drug-like properties. For this purpose, the docked conformations of the active peptides were used to identify compounds with similar activities. A series of related β-sheet breaker peptides were docked to solid state NMR structures of a fibrillar form of Aβ. The lowest energy conformations of the active peptides were used to design three dimensional (3D)-pharmacophores, suitable for screening the NCI database with Unity. Small molecular weight compounds with physicochemical features in a conformation similar to the active peptides were selected, ranked by docking solubility parameters. Of 16 diverse compounds selected for experimental screening, 2 prevented and reversed Aβ aggregation at 2–3 μM concentration, as measured by Thioflavin T (ThT) fluorescence and ELISA assays. They also prevented the toxic effects of aggregated Aβ on neuroblastoma cells. Their low molecular weight and aqueous solubility makes them promising lead compounds for treating AD.
Keywords: Alzheimer’s disease, amyloid fibrils, aggregation inhibitors, pharmacophore modeling, database screening
1. Introduction
Alzheimer’s Disease (AD) is the most common form of dementia in the elderly, affecting 6–10% of people over the age of 651. A progressive neurodegenerative disease2, AD’s clinical symptoms include personality and behavioral changes, periods of disorientation, difficulty communicating, and gradual loss of memory. Although the molecular basis of AD has been extensively studied, there is still no early diagnosis or cure. The hallmark neuropathological features of the disease are the accumulation of extracellular plaques composed of the amyloid-β protein (Aβ), and intracellular, hyper-phosphorylated tau forming neurofibrillary tangles and dystrophic neurites. The progression of Aβ plaque deposition in humans begins in the temporal cortex, proceeds to the hippocampus and then to the entorhinal and transentorhinal cortexes3, while tau deposits progress in the opposite direction4. Several possible molecular mechanisms may initiate AD. However, considerable genetic and biochemical evidence suggests that the Aβ misfolding, oligomerization and accumulation in the brain is the primary cause of the neuronal dysfunction5.
Amyloid is a generic term used to refer to protein aggregates adopting a cross-β-sheet structure6. Several studies have shown that the aggregation and fibril formation of Aβ involves a change in conformation, from a (soluble) helical or coiled structure to an insoluble oligomer 7. Solid state NMR studies of 6–10 nm in diameter fibrils of purified Aβ have revealed an underlying superstructure of anti-parallel intra-molecular β-strands stabilized predominantly by backbone hydrogen bonds and salt bridges, and parallel, inter-molecular β-sheets stabilized predominantly by backbone hydrogen bonds and hydrophobic interactions8–11. A network of inter- and intra- molecular hydrogen bonds maintains the stability of the fibril once it has formed12–15. Aβ can form amyloid-like fibrils in the absence of other proteins7, indicating that the potential to form amyloid originates within its own sequence. Substitution of hydrophilic for hydrophobic residues in the central hydrophobic region of Aβ (17 to 21) impairs fibril formation12, 16–21, and peptides designed to mimic the structure of this region could inhibit aggregation. A series of “β-sheet breaker peptides” were designed based on Aβ amino acids17–20 (LVFFD), and then chemically modified to obtain more active and stable pentapeptides with the basic sequence LPFFD22–25. Such compounds were demonstrated by us and other groups to be active in destabilizing the pathological Aβ conformation, leading to both inhibition of amyloid formation and disassembly of pre-formed fibrils. The lead β-sheet breaker peptide is able to inhibit and disassemble amyloid fibrils in vitro, to prevent Aβ neurotoxicity in cell culture, and to arrest and dissolve amyloid plaques in several in vivo animals models23, 24, 26. Treatment with this peptide also inhibited neuronal death, brain inflammation and memory impairment in vivo23, 24. In addition, the modified peptide showed low toxicity, low immunogenicity, high solubility and reasonably high brain uptake23, 25. In spite of the good activity in vivo, the major weaknesses of this peptide are that it is rapidly degradable and has low permeability to cross biological barriers25. These are serious limitations because a more frequent administration of large quantities by inconvenient routes (injection) is likely to be necessary. It may also be difficult or impossible to reach the appropriate doses for anti-amyloid activity in the large volume of the human brain.
The major goal of the current study is to use the knowledge accumulated over several years on the structure-activity relationship studies of β-sheet breaker peptides and their 3-dimensional structure to design and identify small molecule peptidemimetic compounds. For this purpose, we used a computational/docking approach to identify features of this peptide series that correlated with their ability to prevent Aβ aggregation. We docked the peptide series to the fibrillar structures of Aβ determined from solid state NMR data. While the docking energies could not be used to discriminate the best inhibitors of aggregation, active peptides preferentially bound to different sites on the fibril from those favored by inactive ones. We used the docked conformations of the peptides to obtain a 3D-molecular pharmacophore, which could be used with the UNITY program to screen the NIH library for compounds with the desired physicochemical properties. These were initially ranked based on number of pharmacophores they matched and then by molecular docking to the experimentally determined structures of fibrillar Aβ. The compounds with the best docking scores at the optimal peptide positions were further selected for solubility (low logP: the coefficient for solvent partitioning between 1-octanol and water), and low molecular weight. Finally, 16 compounds from the NCI library27 (http://dtp.nci.nih.gov/webdata.html) were selected and assayed for their ability to inhibit and reverse the aggregation of Aβ, in three different assays, and the toxic effects of aggregates on neuronal cells. Two compounds inhibited Aβ aggregation and cytotoxicity to neuroblastoma cells. They are thus promising lead compounds for developing novel treatments for AD.
2. Results
2.1 Docking of β-sheet breaker peptides and selected compounds to the Aβ fibril
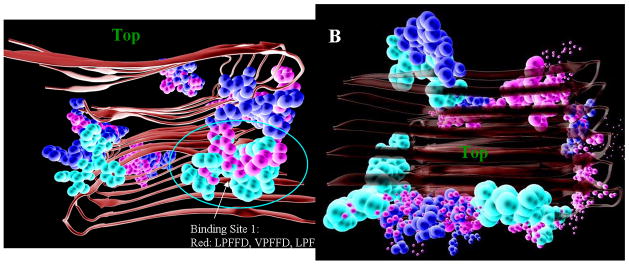
We first docked β-sheet breaker peptides to the fibril structures, to relate binding energies to their activities in disaggregating Aβ. Thirty pentapeptides, with known and varying ability to inhibit Aβ aggregation, were initially docked to the larger grid box (see Experimental 5.2, 5.3), to find the site on the fibril for which they had the highest affinity. While the peptides all had similar Autodock scores (Table S1, supplementary), their preferred docking sites were quite different (Fig. 1). The most active peptides (red; peptides with relative activities > 70 in Supplementary Material Table 1s) bound to a few sites on each fibril structure, while the inactive ones showed less specificity. The first preferred docking site includes residues GLU11-PHE19 and LEU34-VAL40 of 4 outer neighbor monomers. The second preferred docking site includes residues 37–39 from the upper 6 monomers and residues 28–32 from the bottom 6 monomers.
Fig. 1.
Docking positions of 30 β-sheet breaker pentapeptides on two fibril structures of Aβ, Aβ40m2_1.pdb (left), and Aβ40p2_1.pdb (right), colored according to their activities in preventing Aβ aggregation (red, highest relative activities > 70% of LPFFD activity, or ln(Act) > 4.25; green, lowest relative activities <=7% or ln(Act) < 2.0; yellow have intermediate activities (70> Relative activity > 8; or 4.25 > (ln(Act) < 2). The top of the fibril is indicated for orientation purposes.
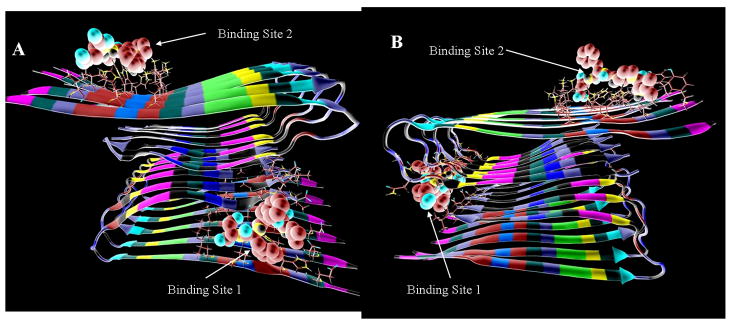
For pharmacophore design and compound docking, two optimal binding (Fig. 2) were selected for each fibril model. The first binding sites, where LPFFD has the best AutoDock score to the Aβ fibril models Aβ40m2_1.pdb and Aβ40p2_1.pdb, are the same as the preferred docking sites shown in Fig. 1. The second binding site chosen is that with the next lowest docking energy that is at least 15Å away from the first site. The second binding sites for both fibril structures, Aβ40m2_1.pdb and Aβ40p2_1.pdb, are close to the docking site of DFPPL to Aβ identified by NMR28, which primarily interacted with residues Lys16-Phe20 on the top surface of several monomers. The most active β-sheet breaker pentapeptides25 bind around site 1 in both structures. For comparison purposes, all 30 compounds were also docked to two binding sites on the Aβ40m2_1.pdb and Aβ40p2_10.pdb models (Fig. 2). Theses sites were also later used to determine the docking energy of compounds selected from NCI database (to match the pharmacophores). Based on the docking results, active compounds, including the lead compound LPFFD which is used for pharmacophore design, can form hydrogen bonds with the backbone hydrogen bond forming atoms. We will discuss the preferred interactions between the active compounds and the fibril later.
Fig. 2.
Fig. 2. A, B) Preferred docking sites for LPFFD (shown with CPK coloring) to the fibril stuctures A) Aβ40m2_1.pdb and B) Aβ40p2_1.pdb.
2.2 Pharmacophore Design
We assumed that the preferred docking sites for the active peptides represented optimal binding positions on the Aβ fibril. The pattern of bonds formed by LPFFD with the fibril in the lowest energy docked conformation were analyzed, to derive features and distance constraints for the design of four 3D-pharmacophores, based on docking to Aβ40m2_1.pdb and Aβ40p2_1.pdb at sites 1 and 2 (Fig. 3).
Fig. 3.
Pharmacophores designed based on the docking model of LPFFD to A, B) docking sites 1, 2 of the fibril structure Aβ40m2_1. C,D) docking sites 1,2 of the fibril structure Aβ40p2_1.
2.3 Database Screening with UNITY
The pharmacophores were used with the UNITY program to search ~250,000 structures from the NCI library. The numbers of compounds that match each of the four 3D-pharmacophores were 980, 1130, 1720 and 1440 for pharmacophore 1, 2 3 and 4 respectively. These were docked to the Aβ fibril structures at sites 1 and 2. The number of compounds with lower docking energies than LPFFD was 68, 26, 61 and 33 for site 1 of Aβ40m2_1.pdb, site 2 of Aβ40m2_1.pdb, site 1 of Aβ40p2_1.pdb and site 2 of Aβ40p2_1.pdb respectively. We further reduced the number of compounds for assay by giving highest priority to compounds selected by UNITY as a potential match for multiple pharmacophores (27 compounds were identified as similar to all 4 pharmacophores, 42 were found in 3 of the 4, 105 in 2, and the remaining 4800 compounds matched only one pharmacophore).
We chose 31 compounds with higher calculated docking affinities than LPFFD from the 69 compounds that matched at least 3 pharmacophores. Although we knew from the peptide series that docking energies alone are not sufficient to distinguish the active peptides from the inactive ones, it was also clear that to reverse the aggregation, an inhibitor should preferentially bind Aβ. Of these, 16 were selected based on their physical properties (logP is not over 5, molecular weight is not over 500, number of hydrogen bond acceptors (HBA) is not over 10, and hydrogen bond donors (HBD) is not over 5). If two or more parameters were out of range, the compounds would not be selected. The structure of the 16 compounds and their NCI number are shown in Figure 4. Table 1 lists their docking scores and physicochemical properties.
Fig. 4.
Structures and NCI number of NCI compounds selected for bioassays.
Table 1.
Autodock scores (“binding energy”) and physical properties of compounds selected for bioassays. The most active compounds are bold, the least active (control in Fig. 4) is underlined.
| Compound | NCI Number | AutoDock Score at each binding site (Binding Energy kcal/mol) | Num. of Pharm. Matched | Num. of HBD | Num. of HBA | cLogP | |||
|---|---|---|---|---|---|---|---|---|---|
| BS1a of Model 1c | BS2b of Model 1c | BS1a of Model 2d | BS2b of Model 2d | ||||||
| 1 | NSC124147 | −16.2 | −16.3 | −15.4 | −14.1 | 3 | 2 | 10 | 2.05 |
| 2 | NSC58883 | −11.8 | −9.6 | −12.4 | −9.9 | 3 | 4 | 6 | −0.05 |
| 3 | NSC20645 | −10.7 | −11.6 | −9.4 | −8.5 | 4 | 4 | 8 | 2.29 |
| 4 | NSC114532 | −10.9 | −11.1 | −9.6 | −8.9 | 4 | 4 | 6 | 1.37 |
| 5 | NSC202510 | −11.4 | −11.3 | −13.5 | −11.6 | 3 | 4 | 7 | 3.59 |
| 6 | NSC123435 | −11.8 | −12.1 | −11.8 | −11.6 | 4 | 4 | 6 | 2.81 |
| 7 | NSC3198 | −12.1 | −11.3 | −10.5 | −11.0 | 4 | 4 | 6 | 1.83 |
| 8 | NSC118474 | −10.6 | −11.2 | −11.1 | −10.9 | 4 | 5 | 8 | −0.83 |
| 9 | NSC319654 | −11.9 | −11.6 | −11.7 | −12.3 | 4 | 5 | 6 | 1.7 |
| 10 | NSC164104 | −11.2 | −11.9 | −10.1 | −9.5 | 4 | 3 | 5 | 3.84 |
| 11 | NSC164103 | −10.9 | −11.9 | −9.8 | −9.6 | 4 | 3 | 6 | 1.56 |
| 12 | NSC178256 | −10.7 | −8.7 | −12.3 | −11.8 | 3 | 5 | 10 | 0.6 |
| 13 | NSC89268 | −11.7 | −12.6 | −9.7 | −8.8 | 3 | 4 | 6 | 3.67 |
| 14 | NSC156276 | −11.8 | −10.8 | −16.3 | −12.7 | 3 | 5 | 12 | −0.35 |
| 15 | NSC343724 | −10.7 | −9.2 | −11.5 | −10.5 | 3 | 6 | 8 | −0.37 |
| 16 | NSC333767 | −11.5 | −12.3 | −9.7 | −8.6 | 4 | 4 | 6 | 2.99 |
| DPFFL | −10.6 | −8.8 | −10.2 | −11.5 | |||||
| LPFFD | −10.1 | −9.0 | −11.7 | −10.8 | |||||
: Binding site 1
: Binding site 2
: the fibril stuctures Aβ40m2_1.pdb
: the fibril stuctures Aβ40p2_1.pdb.
2.4. Bioassays
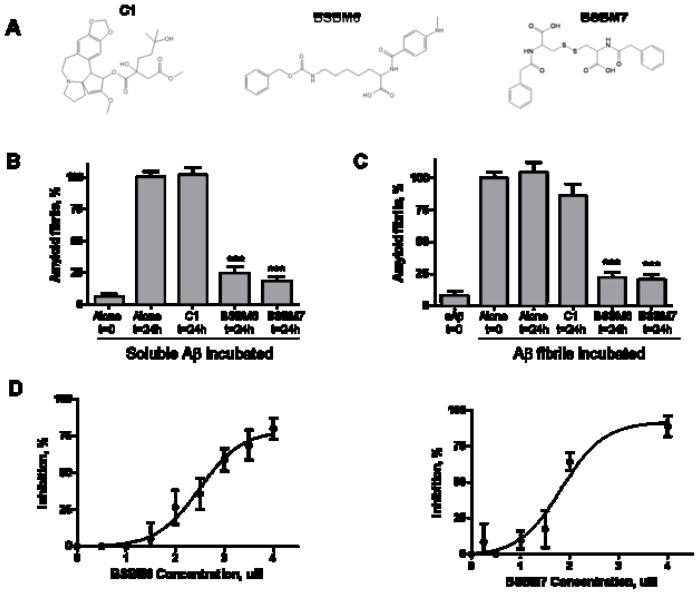
The selected molecules were first screened using a medium throughput in vitro assay based on the specific interaction between amyloid fibrils and ThT. Incubation of soluble Aβ alone for 24h resulted in extensive formation of amyloid fibrils (Fig. 5). However, co-incubation with an equimolar concentration of several of the compounds led to a highly significant inhibition of fibril formation. Two compounds, termed BSBM6 and BSBM7 (beta-sheet breaker mimetic 6 and 7, respectively, Fig. 5a), were selected for further studies, since they showed the highest reproducible inhibition in all assays. Equimolar concentration of these compounds led to >70% inhibition of fibril formation (Fig. 5b). In contrast, an inactive compound (C1 from the initial series) did not alter Aβ amyloidogenesis at the concentration studied. BSBM 6 and 7 were also able to disassemble pre-formed Aβ fibrils (Fig. 5C), decreasing the amount of pre-formed fibrils by >70%. Again, C1 did not alter significantly the amount of fibrils. As controls the compounds alone were added to the ThT assay and the results showed that none of the compounds studied altered ThT fluorescence (data not shown). To confirm the results using an in vitro assay based on a different principle, and to assess the concentration-dependent effect of the compounds in Aβ aggregation, we measured the compounds’ activity using a sedimentation assay, and measured the amount of Aβ using an ELISA assay. Increasing concentrations of BSBM6 or BSBM7 inhibited aggregation, reaching a maximum of around 80% at approximately around equimolarity with the Aβ concentration (4 μM; Fig. 5D). The IC50 values for BSBM6 and BSBM7 in this assay are 2.75 and 1.95 μM, respectively.
Fig. 5.
In vitro activity of selected compounds on Aβ fibrillogenesis. A: Chemical structure of two putative β-sheet breaker mimetics: β-sheet breaker mimetic 6 (BSBM6) and β-sheet breaker mimetic 7 (BSBM7), and the inactive C1 control compound. B: The effect of selected compounds on Aβ amyloid formation was studied by incubation of soluble Aβ1–42 in the absence or the presence of an equimolar concentration of the molecules. Amyloid formation was measured by ThT, as described in Methods. Results are expressed as a percentage of fibrils formed by the peptide incubated alone for 24h. The data was analyzed by student-t test by comparing each result with the control of Aβ incubated alone. ***, P<0.001. C: The ability of the compounds to disassemble pre-form fibrils was assessed by incubation of the molecules with Aβ aggregates made by pre-incubation of Aβ1–42 alone. The amount of fibrils before and after incubation with the compounds was studied by ThT. Results are expressed as a percentage of fibrils remaining after incubation alone for 24h. The data was analyzed by student-t test by comparing with the control of fibrils incubated alone. ***, P<0.001. D: The concentration-dependent effect of BSBM6 and BSBM7 on Aβ aggregation was studied by incubating soluble Aβ1–42 with various quantities of the compounds for 24h at 37°C. Formation of aggregates was quantified by sedimentation assay, followed by ELISA, as described in Methods. The data in panels B, C and D corresponds to the average ± standard error of three different experiments.
2.5. BSBM6 and 7 reduce the neurotoxicity of Aβ aggregates
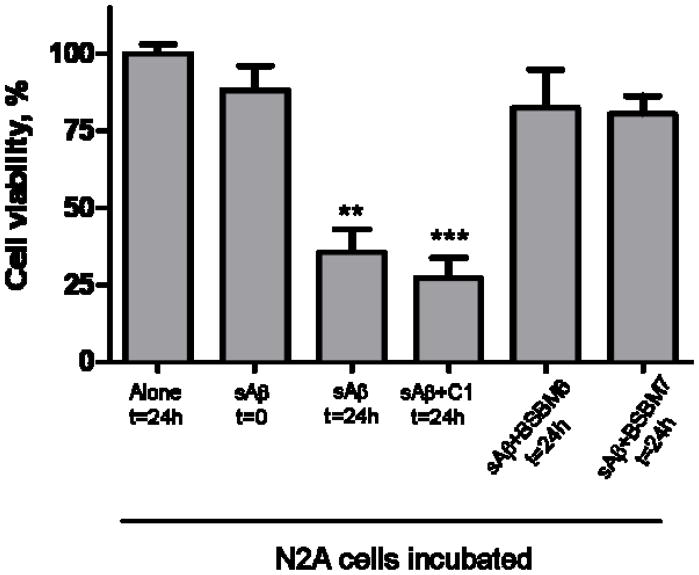
Aβ aggregates decrease the viability of cultured N2A mouse neuroblastoma cells (Fig. 6). Treatment with Aβ pre-incubated for 24h, which contain a mixture of oligomeric and fibrillar species, substantially reduced cell viability. This effect could be prevented if the Aβ was incubated with equimolar concentrations of BSBM6 and 7, indicating that formation of toxic forms of misfolded Aβ was substantially inhibited. The control compound 1 did not prevent Aβ cytotoxicity and indeed, may have increased cell death. None of the compounds tested, on their own, were significantly toxic to cells (as measured by the MTS assay) even at quantities 10 times higher than the active concentration (data not shown).
Fig. 6.
The activity of selected compounds on preventing Aβ neurotoxicity was studied in cell cultures. N2A cells were treated with soluble Aβ1–42 (3.3 μM) which was pre-incubated for 48h either alone or in the presence of 3.3 μM of BSBM6, BSBM7 or C1. After 24h incubation with the mixture peptide/compounds, cell viability was evaluated by the MTS assay. Data represent the average ± standard error of 3 determinations and is expressed as the percentage of viability obtained when cells were incubated alone for 24h. Data for the compounds was statistically compared with cells incubated alone suing the student-t test. ** P<0.01; *** P<0.001.
3. Discussion
There have been extensive studies of the mechanism of Aβ misfolding and aggregation20, 29, 30, and many different compounds have been identified that interfere with this pathway (for reviews, see 31–35). We previously reported that Aβ fibrillogenesis can be prevented and reversed by short peptides designed to bind and inhibit β-sheet misfolding of Aβ (β-sheet breaker peptides). These peptides are active in vitro, in cells and in several in vivo animal models of AD22–26 Moreover, they are well-tolerated in humans, although they exhibited a short half-life and are not orally bioavailable25. Using the knowledge gained about the structure-activity relationships of the peptide series, we used computational modeling and docking techniques to study the interaction with Aβ the peptide series, and relate the interactions of their sidechains to their different activities in preventing aggregation (Figs 1–2, Supplementary data table 1s). The purpose of this study was to identify small molecule peptide mimetics capable to maintain the ability to inhibit and reverse Aβ misfolding and aggregation, but with better drug-like properties. We identified two sites on the fibril structure where the most active peptides bound preferentially. As β-sheet breakers bind to the fibril at different positions and the fibril may have different conformations, we used the pharmacophores (schematic structures of an ideal small molecule inhibitor) from the 3D-quantitative structure-activity relationship (3D-QSAR; the crossvalidated r2 (q2) for the β-sheet breakers is only about 0.5) only as approximate guidance for the relative positioning of reactive groups. To obtain more detailed 3D-pharmacophores, we relied on docked conformations of the active peptides with the fibril structure (Fig. 3). These were used to screen the NCI database, a diverse library which includes many drug like molecules, for potential aggregation inhibitors. A small group of compounds were then selected for testing in a medium throughput assay (Table 1), and two compounds identified as consistently active in preventing and reversing Aβ fibrillogenesis (Fig. 5), measured in three different ways, and blocking Aβ neurotoxicity (Fig. 6). For these initial experiments, we used the smaller NCI database. Now that we have a better idea of what constitutes an active aggregation inhibitor, larger databases such as. ZINC (http://zinc.docking.org), may be used for future investigations.
The basis for this study was previous work, where we used pharmacophore design and Autodock to find small compounds that bound to the Anthrax edema factor36, 37. Autodock was quite accurate in finding the conformation and docking position of substrate analogues (i.e., the docked conformation matched the conformation determined by X-ray crystallography)37. However, docking energies were only approximate indicators of activity36, 37. Here, we faced a more difficult challenge, as we had several possible structures for the Aβ fibril, and only indirect evidence for where aggregation inhibitors should bind. Moreover, we knew from the peptide data that we could not rely on docking/binding energies to indicate which inhibitors would be most active in inhibiting the aggregation of Aβ. We thus decided to select molecules that resembled the docked conformations of the most active peptides, and then test molecules that had good molecular properties, and similar or higher calculated binding affinities for these sites. A summary of our data indicates that the molecule with the highest calculated binding affinity (first in Table 1) was a very poor inhibitor, and indeed was later used as a negative control. Our two best inhibitors had only slightly better binding affinity than the best β-sheet breaker peptides. This illustrates the complexity of using calculated binding energies to identify inhibitors of the aggregation process, and the need for experimental data to direct the selection process.
One explanation for our results would be that our good inhibitors, such as the peptide LPFFD and BSBM6 and BSBM7, interfere with electrostatic interactions during aggregation38 or destabilize the Aβ-fibril internal hydrogen bond network, necessary to maintain the β-sheet structure, by forming strong hydrogen bonds with the Aβ subunits. The lowest energy docked conformations of active compounds, such as the peptide LPFFD, and BMBS6 and BMBS7, show hydrogen bonds (red arrows in Fig 7C for BSBM7 and 7D for LPFFD) that would compete with inter-monomer ones. These hydrogen bonds between the ligand and the fibril would be expected to weaken or break the hydrogen bond network stabilizing the sheets, and thereby inhibit or reverse aggregation of Aβ. While C1 has high affinity for the fibril, according to the docking scores, one reason for inactivity could be that the large multi-cyclic part of C1 (Fig. 5, top right) could also nucleate the formation of even larger aggregates. Continuing with our analysis, inactive compounds such as C1 and the inactive peptide LTicFFD (please see table S1) form hydrogen bonds with atoms of Aβ that do not form network stabilizing bonds with neighboring monomers (blue arrows in Fig 7A, B). Thus, we hypothesize base on docking that our active compounds interfere with bonds needed to maintain the inter-monomer sheets, while the inactive ones do not. Molecular dynamic simulations for C1, LPFFD, LHFFD, BMBS6 and BMBS7 (in preparation) show that the active compounds bind more strongly to the fibril than inactive ones. This result agrees with a study by Bernard Reif’s group28 on the interaction of Aβ fibrils with two peptides, a good inhibitor (LPFFD) and a weak one (DPFFL) which showed clearly that DPFFL had a much higher on/off rate (in both Biacore and NMR studies).
Figure 7.
Comparison, based on the the best ranked conformations (lowest binding scores) from AutoDock, of the type of hydrogen bonds formed between atoms of the bad aggregation inhibitors (A,B) and good ones (C,D) and the fibril. The side chains of the pentapeptides LTicFFD (inactive) and LPFFD (active) are not shown. The hydrogen bonds between ligands and fibril are indicated by red arrows (those that would be expected to break or weaken the β sheet hydrogen bond network of the fibril) and blue arrows (those expected to enhance the stability of the e β sheet hydrogen bond network). Yellow arrows indicate those close interactions that should not affect β sheet hydrogen bond network.
These results indicate that molecular docking can be used to discriminate features of compounds that are important for activity, even when the overall docking energies do not directly correlate with activity. Considering the success of the first screening, we plan to redesign the active compounds to further analyze the role of substituents in aggregation. In future work, we will also analyze the calculated H-bond networks of the docked conformation of the compounds that meet our structural criteria, and use this information to select compounds for experimental testing. This approach should lead to the identification of potent and specific β-sheet breaker mimetics to inhibit and reverse Aβ misfolding and oligomerization and thus offer a new avenue for developing drugs for AD treatment.
4. Conclusions
This work’s major significance is that the two compounds we identified, that inhibit and reverse Aβ aggregation and neurotoxicity, are possible lead compounds for AD treatment, as they are easy to produce and have low toxicity to cells. Two of the 16 compounds inhibited aggregation at less than equimolar concentration with Aβ in the assay, in the range of 2–3 μM. The compounds showed good activity in four different set of experiments, with different methods for measuring aggregates: in preventing aggregation of soluble Aβ, measured by ThT fluorescence or by centrifugation followed by quantitation with ELISA, in disaggregating preformed Aβ fibrils, and in inhibiting the neurotoxic effects of Aβ on cultured neuroblastoma cells. Indeed, as our least active compound (the C1 control in the assay of Figures 5 and 6) had the most favorable (lowest) docking score, we conclude that we must analyze not just how many bonds a compound is capable of forming with the aggregate, a number summarized broadly in docking scores, but also what effect those bonds will have on destabilizing the inter-subunit contact areas.
The next phase of our work will be to better define the mechanism by which our active compounds inhibit and reverse the aggregation of Aβ. One way will be to test variations on our active compounds that, according to their docked structures, form bonds that should destabilize the aggregates. Additional data on the position of the ligands on the fibril, for example from solid state NMR, would also aid in our search for more active derivatives
5. Experimental
5.1. Structural Model Selection for Amyloid Beta Peptides
The docking targets were fibril structures of Aβ1–40, prepared in a fashion similar to the methods for inducing aggregation of Aβ used for the experiments below, determined with solid state NMR8–11. TEM images and MPL data from STEM images of Aβ1–42 are very similar to those of agitated Aβ1–40 fibrils, suggesting that the structure of fibril Aβ1–42 is very similar to Aβ1–4039. PDB-formatted files for these fibril structures were obtained directly from Dr. Robert Tycko at the NIH (Aβ40m2_1.pdb, and Aβ40p2_1.pdb, which differ in the orientation of the strands). The structures of Aβ40m2_1.pdb and Aβ40p2_1.pdb are different in the monomer stacking within the β-sheets11. Conformational data were available for residues from Gly9 to Val40; residues 1–8 were highly disordered. The conformation of a (weak) β sheet breaker peptide, DPFFL, bound to fibrillar Aβ1–40, determined using transferred nuclear Overhauser effect (trNOE) and transferred residual dipolar couplings (trRDC) were used for reference28.
5.2. Inhibitor Peptides for Docking
Activity data for a group of peptides related to LPFFD and DPFFL were previously published25 (supplementary data, table 1, which also includes their docking energies).
5.3. Docking and Scoring
Selection of the optimal docking site for the active peptide series on the NMR fibril structures Aβ40m2_1.pdb and Aβ40p2_1.pdb was done with AutoDock http://www.scripps.edu/mb/olson/doc/autodock) 40, 41(version 3.0.5), using the “Lamarckian” genetic algorithm (LGA). To allow the ligands to rotate freely over the whole structure to determine the approximate region of preferred docking to the fibril, the grid box size was set initially to 120 × 110 × 90 points with grid spacing of 0.60Å and centered on the center of the Aβ fibril. The optimal site on the fibril for each peptide was refined by repeating the docking with a smaller grid box (either 60 × 60 × 60 or 80 × 80 × 80 points with grid spacing of 0.375Å. During docking, the Aβ fibril was kept rigid and 200 conformations of the flexible ligand peptide (LPFFD and related) were searched and docked. Other parameters were default, except the population size (100). AutoDock docking energy, which estimates the intermolecular potential energy between the ligand and Aβ and the torsional free energy of ligand, was minimized.
5.4. 3D-Pharmacophore Design
Our previous work showed that AutoDock is quite accurate in positioning molecules on a given target and determining lowest energy conformations37. Pharmacophores representing an optimal configuration of the active groups of the active peptides were derived, with distance constrainst obtains from the lowest energy docking conformations of LPFFD on the two Aβ models Aβ40m2_1.pdb and Aβ40p2_10.pdb. The pharmacophore features, such as HBA (including negatively charged groups), HBD (including postively charged groups), and hydrophobic (HP), were defined manually based on the interactions of LPFFD with the Aβ fibril. A hydrogen bond interaction was defined if any atom of LPFFD was within (2.8Å) of any atom of the fibril. HP was defined if the hydrophobic groups or atoms of the ligands were close (<4.0Å) to the hydrophobic groups or atoms of Aβ.
5.5 Database Screening and Molecular Docking
The resulting 3D-pharmacophores were used for 3D- database screening with the UNITY program (Tripos, Inc.) in SYBYL7.1. The NCI-2000 database, integrated in SYBYL, with about 250,000 compounds stored as 3D structures converted from their 2D forms by Concord, was screened for compounds matching the pharmacophores, using tolerance for HBA and HBD features of 0.3Å and for HP of 0.8Å..
Selected compounds were docked to fibril sites 1 and 2 of the two Aβ models Aβ40m2_1.pdb and Aβ40p2_10.pdb with AutoDock, using the methods described above, and ranked according to their docking scores. Compounds with cLogP was <5 and molecular weight <500 were considered if their binding energies were lower than the binding energy of LPFFD. Highest priority was given to compounds selected several times in the UNITY searches with the different pharmacophores.
5.6 Preparation of Aβ peptide and compounds for the aggregation assay
Aβ1–42 was synthesized using solid phase chemistry by the protein core at Yale University (W.M. Keck Facility, University of Yale). 0.3 mg of lyophilized powder was dissolved in 200 μl of hexafluoro-2-propanol (Sigma-Aldrich) for 10–20 minutes at room temperature. Soluble Aβ was made by drying the HFIP and adding H2O in a siliconized Eppendorf tube to obtain a final peptide concentration of 60 uM. For the experiments Aβ was diluted to 3.3 μM in 0.1 mM Tris in the absence and presence of selected small molecules at a one to one molar ratio. Compounds were obtained in powder form from the stock collection of the Developmental Therapeutics Program (DTP), NCI, Bethesda, MD (http://dtp.nci.nih.gov) and dissolved in DMSO to 0.4 mM for storage at −20°C. They were diluted 100X in Hepes, pH 7.0, before adding to the assay. The same amount of DMSO was added to the controls.
5.7. Assays for aggregation inhibition
The compounds were screened first with a medium throughput in vitro fluorometric assay based on the fluorescence emission of thioflavin T (ThT) when bound to amyloid fibrils42. Soluble Aβ peptide at a concentration of 3.3μM in 0.1M Tris buffer was incubated alone or with the indicated concentrations of the compounds for 24h at 37°C. Aliquots (20 μl) of samples were incubated 15 minutes at room temperature in 50 mM glycine, pH 9.2 and 2 μM ThT. Fluorescence was measured at excitation wavelength of 435 nm with emission at 485 nm. Compounds 1, 6 and 7 (3.3 μM) were further tested for their ability to dissassemble pre-formed fibrillar Aβ after incubation for 24 hr. As control soluble (non-fibrillar) Aβ and fibrils incubated alone were included. The quantity of fibrils was measured by ThT fluorescence. To rule out that the compounds interfere with ThT fluorescence, the effect of the compounds alone on the assay was measured.
Aβ aggregates were also assayed by sedimentation. After incubation with or without compounds, samples were centrifuged at 16000×g for 10 min at 4°C. Soluble peptide was measured in the supernatant by ELISA using the 4G8 anti-Aβ monoclonal antibody.
5.8. Neuronal toxicity assay
Compounds 1, 6 and 7 (3.3 μM) were incubated with soluble Aβ (3.3 μM) for 48 hr. Controls were soluble Aβ incubated at the same concentration either with addition of the inactive compound 1 or the same amount of DMSO (the final concentration of DMSO in all assays was <0.1%). Aliquots of samples (10 μl) were added to 80% confluent mouse neuroblastoma cells N2a, incubated 24h in a 96 well plate at 37°C in a humidified, 5% CO2 atmosphere. Cell survival was assessed by the MTS assay (Promega). The combined MTS/PMS solution (20 μl) was added to the culture medium and the cells were incubated 1–4 h at 37°C in a humidified, 5% CO2 atmosphere. Absorbance was read at 490 nm.
Supplementary Material
Acknowledgments
We thank Dr. Rakez Kayed (Neurology, UTMB) for his advice and assistance with the assays. This work was supported by a Pilot Award from the George and Cynthia Mitchell Center for Neurodegenerative Diseases to Catherine Schein, grants from the NIH (AG028821) and the Mitchell Foundation to Claudio Soto. The infrastructure of the Sealy Center for Structural Biology and Molecular Biophysics, and of the Sealy Center for Vaccine Design, were also used. Compounds used in this study were obtained from the stock collection of the Developmental Therapeutics Program (DTP), NCI, Bethesda, MD (http://dtp.nci.nih.gov).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hendrie HC. Epidemiology of dementia and Alzheimer’s disease. Am J Geriatr Psychiatry. 1998;6:S3–18. doi: 10.1097/00019442-199821001-00002. [DOI] [PubMed] [Google Scholar]
- 2.Alzheimer A, Stelzmann RA, Schnitzlein HN, Murtagh FR. An English translation of Alzheimer’s 1907 paper, “Uber eine eigenartige Erkankung der Hirnrinde”. Clin Anat. 1995;8(6):429–431. doi: 10.1002/ca.980080612. [DOI] [PubMed] [Google Scholar]
- 3.Thal DR, Rub U, Orantes M, Braak H. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology. 2002;58(12):1791–1800. doi: 10.1212/wnl.58.12.1791. [DOI] [PubMed] [Google Scholar]
- 4.Braak H, Braak E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging. 1995;16(3):271–278. doi: 10.1016/0197-4580(95)00021-6. [DOI] [PubMed] [Google Scholar]
- 5.Selkoe DJ. Toward a comprehensive theory for Alzheimer’s disease. Hypothesis: Alzheimer’s disease is caused by the cerebral accumulation and cytotoxicity of amyloid beta-protein. Ann N Y Acad Sci. 2000;924:17–25. doi: 10.1111/j.1749-6632.2000.tb05554.x. [DOI] [PubMed] [Google Scholar]
- 6.Cohen AS, Jones LA. Amyloidosis. Curr Opin Rheumatol. 1991;3(1):125–138. doi: 10.1097/00002281-199102000-00018. [DOI] [PubMed] [Google Scholar]
- 7.Soto C, Branes MC, Alvarez J, Inestrosa NC. Structural determinants of the Alzheimer’s amyloid beta-peptide. J Neurochem. 1994;63(4):1191–1198. doi: 10.1046/j.1471-4159.1994.63041191.x. [DOI] [PubMed] [Google Scholar]
- 8.Petkova AT, Buntkowsky G, Dyda F, Leapman RD, Yau WM, Tycko R. Solid state NMR reveals a pH-dependent antiparallel beta-sheet registry in fibrils formed by a beta-amyloid peptide. J Mol Biol. 2004;335(1):247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 9.Tycko R. Solid-state NMR as a probe of amyloid fibril structure. Curr Opin Chem Biol. 2000;4(5):500–506. doi: 10.1016/s1367-5931(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 10.Tycko R. Characterization of amyloid structures at the molecular level by solid state nuclear magnetic resonance spectroscopy. Methods Enzymol. 2006;413:103–122. doi: 10.1016/S0076-6879(06)13006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q Rev Biophys. 2006;39(1):1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 12.Kheterpal I, Chen M, Cook KD, Wetzel R. Structural differences in Abeta amyloid protofibrils and fibrils mapped by hydrogen exchange--mass spectrometry with on-line proteolytic fragmentation. J Mol Biol. 2006;361(4):785–795. doi: 10.1016/j.jmb.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 13.Kheterpal I, Wetzel R. Hydrogen/Deuterium exchange mass spectrometry-a window into amyloid structure. Acc Chem Res. 2006;39(9):584–593. doi: 10.1021/ar050057w. [DOI] [PubMed] [Google Scholar]
- 14.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Dobeli H, Schubert D, Riek R. 3D structure of Alzheimer’s amyloid-{beta}(1–42) fibrils. Proc Natl Acad Sci USA. 2005;102(48):17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45(2):498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams AD, Shivaprasad S, Wetzel R. Alanine scanning mutagenesis of A beta(1–40) amyloid fibril stability. Journal of Molecular Biology. 2006;357(4):1283–1294. doi: 10.1016/j.jmb.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 17.Hilbich C, Kisters-Woike B, Reed J, Masters CL, Beyreuther K. Substitutions of hydrophobic amino acids reduce the amyloidogenicity of Alzheimer’s disease beta A4 peptides. J Mol Biol. 1992;228(2):460–473. doi: 10.1016/0022-2836(92)90835-8. [DOI] [PubMed] [Google Scholar]
- 18.Jarrett JT, Berger EP, Lansbury PTJ. The C-terminus of the beta protein is critical in amyloidogenesis. Ann N Y Acad Sci. 1993;695:144–148. doi: 10.1111/j.1749-6632.1993.tb23043.x. [DOI] [PubMed] [Google Scholar]
- 19.Soto C, Castano EM, Frangione B, Inestrosa NC. The alpha-helical to beta-strand transition in the amino-terminal fragment of the amyloid beta-peptide modulates amyloid formation. J Biol Chem. 1995;270(7):3063–3067. doi: 10.1074/jbc.270.7.3063. [DOI] [PubMed] [Google Scholar]
- 20.Wood SJ, Wetzel R, Martin JD, Hurle MR. Prolines and amyloidogenicity in fragments of the Alzheimer’s peptide beta/A4. Biochemistry. 1995;34(3):724–730. doi: 10.1021/bi00003a003. [DOI] [PubMed] [Google Scholar]
- 21.Shivaprasad S, Wetzel R. Scanning cysteine mutagenesis analysis of A beta-(1–40) amyloid fibrils. Journal of Biological Chemistry. 2006;281(2):993–1000. doi: 10.1074/jbc.M505091200. [DOI] [PubMed] [Google Scholar]
- 22.Soto C, Kindy MS, Baumann M, Frangione B. Inhibition of Alzheimer’s amyloidosis by peptides that prevent beta-sheet conformation. Biochem Biophys Res Commun. 1996;226(3):672–680. doi: 10.1006/bbrc.1996.1413. [DOI] [PubMed] [Google Scholar]
- 23.Permanne B, Adessi C, Saborio GP, Fraga S, Frossard MJ, Van Dorpe J, Dewachter I, Banks WA, Van Leuven F, Soto C. Reduction of amyloid load and cerebral damage in a transgenic mouse model of Alzheimer’s disease by treatment with a beta-sheet breaker peptide. FASEB J. 2002;16(8):860–862. doi: 10.1096/fj.01-0841fje. [DOI] [PubMed] [Google Scholar]
- 24.Soto C, Sigurdsson EM, Morelli L, Kumar RA, Castaño EM, Frangione B. Beta-sheet breaker peptides inhibit fibrillogenesis in a rat brain model of amyloidosis: implications for Alzheimer’s therapy. Nat Med. 1998;4(7):822–826. doi: 10.1038/nm0798-822. [DOI] [PubMed] [Google Scholar]
- 25.Adessi C, Frossard MJ, Boissard C, Fraga S, Bieler S, Ruckle T, Vilbois F, Robinson SM, Mutter M, Banks WA, Soto C. Pharmacological profiles of peptide drug candidates for the treatment of Alzheimer’s disease. J Biol Chem. 2003;278(16):13905–13911. doi: 10.1074/jbc.M211976200. [DOI] [PubMed] [Google Scholar]
- 26.Chacón MA, Barría MI, Soto C, Inestrosa NC. Beta-sheet breaker peptide prevents Abeta-induced spatial memory impairments with partial reduction of amyloid deposits. Mol Psychiatry. 2004;9(10):953–961. doi: 10.1038/sj.mp.4001516. [DOI] [PubMed] [Google Scholar]
- 27.Milne GW, Nicklaus MC, Driscoll JS, Wang S, Zaharevitz D. National Cancer Institute Drug Information System 3D database. J Chem Inf Comput Sci. 1994;34(5):1219–1224. doi: 10.1021/ci00021a032. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, Krause G, Reif B. Structure and orientation of peptide inhibitors bound to beta-amyloid fibrils. J Mol Biol. 2005;354(4):760–776. doi: 10.1016/j.jmb.2005.09.055. [DOI] [PubMed] [Google Scholar]
- 29.Etienne MA, Edwin NJ, Aucoin JP, Russo PS, McCarley RL, Hammer RP. Beta-amyloid protein aggregation. Methods Mol Biol. 2007;386:203–225. doi: 10.1007/1-59745-430-3_7. [DOI] [PubMed] [Google Scholar]
- 30.Teplow DB. Structural and kinetic features of amyloid beta-protein fibrillogenesis. Biochemistry. 1998;37(11):3602–3611. doi: 10.3109/13506129808995290. [DOI] [PubMed] [Google Scholar]
- 31.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426(6968):905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 32.De Lorenzi E, Giorgetti S, Grossi S, Merlini G, Caccialanza G, Bellotti V. Pharmaceutical strategies against amyloidosis: old and new drugs in targeting a “protein misfolding disease”. Curr Med Chem. 2004;11(8):1065–1084. doi: 10.2174/0929867043455549. [DOI] [PubMed] [Google Scholar]
- 33.Estrada LD, Soto C. Disrupting beta-amyloid aggregation for Alzheimer disease treatment. Curr Top Med Chem. 2007;7(1):115–126. doi: 10.2174/156802607779318262. [DOI] [PubMed] [Google Scholar]
- 34.LeVine H. The challenge of inhibiting Abeta polymerization. Curr Med Chem. 2002;9(11):1121–1133. doi: 10.2174/0929867023370167. [DOI] [PubMed] [Google Scholar]
- 35.Mason JM, Kokkoni N, Stott K, Doig AJ. Design strategies for anti-amyloid agents. Curr Opin Struct Biol. 2003;13(4):526–532. doi: 10.1016/s0959-440x(03)00100-3. [DOI] [PubMed] [Google Scholar]
- 36.Chen D, Misra M, Sower L, Peterson JW, Kellogg GE, Schein CH. Novel inhibitors of anthrax edema factor. Bioorg Med Chem. 2008;16(15):7225–7233. doi: 10.1016/j.bmc.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen D, Menche G, Power TD, Sower L, Peterson JW, Schein CH. Accounting for ligand-bound metal ions in docking small molecules on adenylyl cyclase toxins. Proteins-Structure Function and Bioinformatics. 2007;67(3):593–605. doi: 10.1002/prot.21249. [DOI] [PubMed] [Google Scholar]
- 38.Yun S, Urbanc B, Cruz L, Bitan G, Teplow DB, Stanley HE. Role of electrostatic interactions in amyloid beta-protein (A beta) oligomer formation: a discrete molecular dynamics study. Biophys J. 2007;92(11):4064–4077. doi: 10.1529/biophysj.106.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer’s beta-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry. 2002;41(51):15436–15450. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- 40.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19(14):1639–1662. [Google Scholar]
- 41.Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des. 1996;10(4):293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 42.LeVine H. Thioflavine T interaction with synthetic Alzheimer’s disease beta-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 1993;2(3):404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.