Summary
Spt4-Spt5 complex is an essential RNA Polymerase II elongation factor found in all eukaryotes and important for gene regulation. We report here the crystal structure of Saccharomyces cerevisiae Spt4 bound to the NGN domain of Spt5. This structure reveals that Spt4-Spt5 binding is governed by an acid-dipole interaction between Spt5 and Spt4. Mutations that disrupt this interaction disrupt the complex. Residues forming this pivotal interaction are conserved in the archaeal homologues of Spt4 and Spt5, which we demonstrate to also form a complex. Even though bacteria lack an Spt4 homologue, the NGN domains of Spt5 and its bacterial homologues are structurally similar. Spt4 is located at a position that may help to maintain the functional conformation of the following KOW domain in Spt5. This structural and evolutionary perspective of the Spt4-Spt5 complex and its homologues suggest that it is an ancient, core component of the transcription elongation machinery.
Introduction
In all living organisms, transcription of protein-encoding genes is mediated by a common multiprotein machinery(Allison et al., 1985). The RNA polymerase (RNAP) is highly conserved across species, and the structure and enzymatic mechanisms of the prokaryotic and eukaryotic RNA polymerases share many similarities(Korzheva and Mustaev, 2001; Vassylyev et al., 2007). Numerous factors regulate transcription by influencing the ability of RNAP to access, bind and transcribe specific genes in response to appropriate signals. Some of the most important mechanisms regulating prokaryotic and eukaryotic gene expression target the movement of template-engaged RNA polymerases(Mooney et al., 2005; Saunders et al., 2006).
In eukaryotes, nucleosomes provide an additional challenge to RNAPs elongating through ORFs. In the case of RNAP II, accessory factors are required for processive elongation through chromatin, and modifiers of chromatin structure and function associate with the elongating polymerase(Saunders et al., 2006). In addition, RNA processing events in eukaryotes are coordinated with elongation(Bentley, 2005; Saunders et al., 2006). Thus, eukaryotic elongation factors appear to be part of an extensively coupled network, linking elongation to RNA processing and regulation of the chromatin template.
Spt4 and Spt5 form a heterodimeric protein complex that functions in the control of RNAP II processivity(Hartzog et al., 1998; Wada et al., 1998). Spt5 shares homology to bacterial NusG (Figure 1), a general transcription factor that suppresses pausing and increases Rho-dependent termination(Ciampi, 2006; Ponting, 2002). The metazoan Spt4-Spt5 complex, DSIF, exerts both positive and negative transcription regulation by acting in concert with NELF to mediate promoter proximal pausing of RNAP II(Cheng and Price, 2007; Wada et al., 1998; Wu et al., 2003; Yamaguchi et al., 1999). Both Spt5 and NusG are essential for life, and Spt4-Spt5 and NusG bind to and travel with elongating RNAP(Andrulis et al., 2000; Burns et al., 1998; Kaplan et al., 2000), suggesting a fundamental role in RNAP elongation.
Figure 1. Linear maps of the eukaryotic Spt4, Spt5 and the prokaryotic NusG, RpoE″.
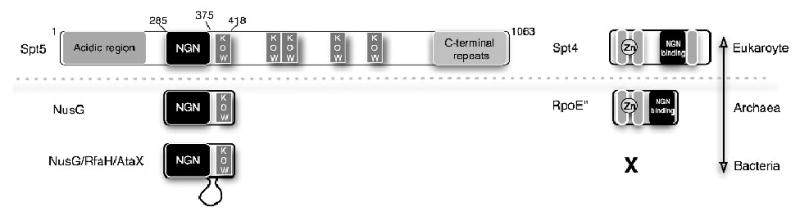
Conserved motifs are shown as boxes. The C-terminal portion of Spt4 is absent in RpoE″. Residue numbers refer to Saccharomyces cerevisiae Spt5.
NusG proteins have an N-terminal NusG (NGN) domain and a C-terminal KOW domain, a nucleic-acid binding domain also found in some ribosomal proteins(Ponting, 2002; Steiner et al., 2002). Spt5 proteins are more complex; they typically have an acidic N-terminus, a single NGN domain, 5-6 KOW domains and a set of simple, C-terminal repeats that are targets of regulatory kinases(Ponting, 2002; Saunders et al., 2006). This more complex structure is most likely a reflection of eukaryote-specific roles for Spt4-Spt5 in chromatin structure and RNA processing; genetic and biochemical assays implicate Spt4-Spt5 in regulation of elongation, chromatin, capping, splicing, RNA export and transcription termination(Bucheli and Buratowski, 2005; Burckin et al., 2005; Cui and Denis, 2003; Kaplan et al., 2000; Lindstrom et al., 2003; Pei and Shuman, 2002; Wen and Shatkin, 1999).
Unlike SPT5, SPT4 is not essential for viability in yeast(Malone et al., 1993). However, there is a near complete overlap between spt4Δ phenotypes and partial loss-of-function mutations in SPT5, and spt4 and spt5 mutations exhibit strong genetic interactions(Hartzog et al., 1998; Swanson et al., 1991; Swanson and Winston, 1992). These data support the idea that Spt4 is critical for normal function of the Spt4-Spt5 complex. However, Spt4 homologues are not found in bacteria and the mechanistic role of Spt4 as an elongation factor in the Spt4-Spt5 complex is still unclear.
Here we determined the crystal structure of Spt4 complexed with the Spt5NGN domain from Saccharomyces cerevisiae and carried out further genetic and biochemical analyses that indicate that Spt4 and NGN domain of Spt5 form the structural core of the Spt4-Spt5 complex in archaea and eukaryotes, and that the NGN domain is likely a conserved regulator of transcription elongation across all forms of life.
Results
Architecture of Spt4-Spt5NGN complex
In our initial efforts to characterize the core of the Saccharomyces cerevisiae Spt4-Spt5 complex, we observed that binding of Spt4 to Spt5NGN increased with decreasing salt concentration. We hypothesized that if these two protein modules were joined together as a fusion protein, they would pack on each other more efficiently as we slowly decreased the salt concentration, and might therefore facilitate crystallization. We therefore created a series of proteins consisting of Spt4 (residues 1-99) fused to the NGN domain (285-375) of Spt5. Fusions with different linker lengths between Spt4 and Spt5NGN were tested for crystallization. A reverse diffusion method was applied to crystallize the fused Spt4/Spt5NGN protein (see experimental procedures for details). One of these fusions yielded crystals with acceptable diffraction and the structure of this protein was determined by single-wavelength anomalous dispersion (SAD) using the zinc atom associated with Spt4's zinc finger domain (Table S1 in the Supplemental data). A map of the electron densities at 2.2 Å resolution was obtained for all amino acids of the fusion protein except for two residues at the N terminus of Spt4 and six His-tag residues at the C-terminus of Spt5NGN (Figure 2A). The asymmetric unit consists of a single monomer of the Spt4/Spt5NGN fusion protein (Figure S1).
Figure 2. Architecture of Spt4-Spt5NGN complex.
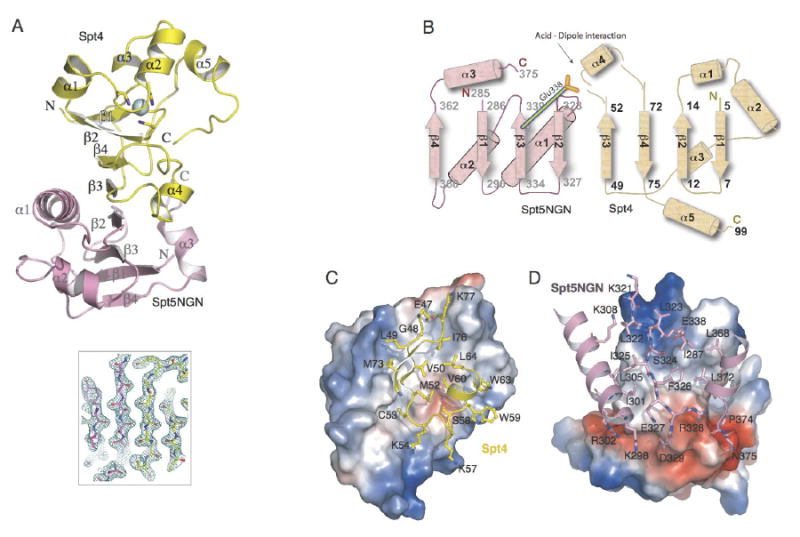
(A) Ribbon diagram of the structure of Spt4-Spt5NGN complex. The Spt4 and Spt5 NGN domains are shown in yellow and pink, respectively; the Zn atom is shown as a blue sphere. The central β-sheet is shown by the 2mfo-Dfc map calculated with refmac coefficients contoured at 2σ level. (B) Topology of the secondary structure elements. Residue numbers and the N and C termini of Spt4 and Spt5NGN are indicated. (C, D) Views of the Spt4-Spt5NGN interface. C, Surface of Spt5NGN is shown and colored according to electrostatic, interacting residues in Spt4 are labeled and shown as yellow sticks; D, Surface of Spt4 and interacting residues in Spt5NGN are shown, interacting residues in Spt5NGN are labeled and shown as pink sticks.
The Spt4 and Spt5NGN domains of the fusion protein fold independently and are connected by an extended loop composed of residues 98-99 of Spt4 (Gly-Ser), the engineered linker of two amino acids (Gly-Ser) and residues 285-286 of Spt5 (Ala-Thr). The Spt5NGN domain consists of a four stranded antiparallel beta sheet flanked by three alpha helices (Figure 2A). Spt4 has four β-strands, β1-β4, in its center, with four α-helices clustered at the top, and a short α-helix motif, α4, appended at its bottom (Figure 2A). Spt4 contains a zinc finger, formed by four conserved cysteine residues (Figure 2A). Zinc binding was confirmed by the atomic absorbance spectrum of the purified protein and the x-ray absorption edge scan of Spt4/Spt5NGN crystals (data not shown). The zinc finger forms the top part of Spt4's hydrophobic core. Mutations altering any of the four finger cysteines of yeast Spt4 cause loss-of-function phenotypes and in several instances, greatly decreased Spt4 levels(Basrai et al., 1996; Malone et al., 1993). Thus, it is likely that the zinc finger is an essential structural element of Spt4.
The Spt4-Spt5 Interface is held by alignment of their beta sheets and an acid-dipole interaction
Monomers of the Spt4/Spt5NGN fusion protein pack on one another in the crystal to form symmetric intermolecular dimers. Spt4 of one monomer binds to the Spt5NGN domain of an adjacent monomer, analogous to domain swaps often seen in crystallization of multidomain proteins(Simader et al., 2006; Yang et al., 2004)(Figure 2A and S1). In contrast, within a monomer of the fusion protein, Spt4 and the Spt5NGN domain do not contact each other (Figure S1). Sequence conservation and mutation data, described below, also strongly support the model that the interface of Spt4 and Spt5NGN domains from distinct monomers represents the normal mode of interaction between Spt4 and Spt5 in vivo.
The central feature of the Spt4-Spt5 interface is an antiparallel β-sheet formed by four β-strands of Spt4 and the four β-strands of the Spt5NGN domain (Figure 2A and 2B). Direct backbone contacts are formed between β3 of Spt4 and β2 of Spt5NGN. These are positioned within a large region of continuous hydrophobic interactions (Figures 2C and 2D). The intermolecular contact area is 1400 Å2, covering over 24% of the surface area of Spt4 (Figure 2C and 2D). Eight α-helices surround this extended β-sheet (Figure 2A) and these mostly display hydrophilic residues on their surfaces, as is typical for protein-protein interaction domains(Jones and Thornton, 1996).
In addition to the beta sheet interface between Spt4 and Spt5NGN, several hydrogen bonds contribute to the specificity of Spt4-Spt5NGN interactions. In particular, Ser58, at the N-terminal -1 position of α4 in Spt4, makes a strictly conserved hydrogen-bonding interaction with Glu338 in Spt5NGN (Figure 3A and 4D). Furthermore, this hydrogen bond fixes the γ-carboxyl of Glu338 towards the N-terminus of α4 of Spt4, which results in a strong acid-α-helix dipole interaction(Nicholson et al., 1988). This interaction may be a pivotal determinant of the specificity and high affinity between Spt4 and the Spt5NGN domain. In summary, Spt4 and Spt5NGN interact via hydrophobic interfaces that appear to be held in register by peripheral polar interactions and alignment of their beta sheets.
Figure 3. Mutations of the acid-dipole interaction disrupt the Spt4-Spt5 complex.
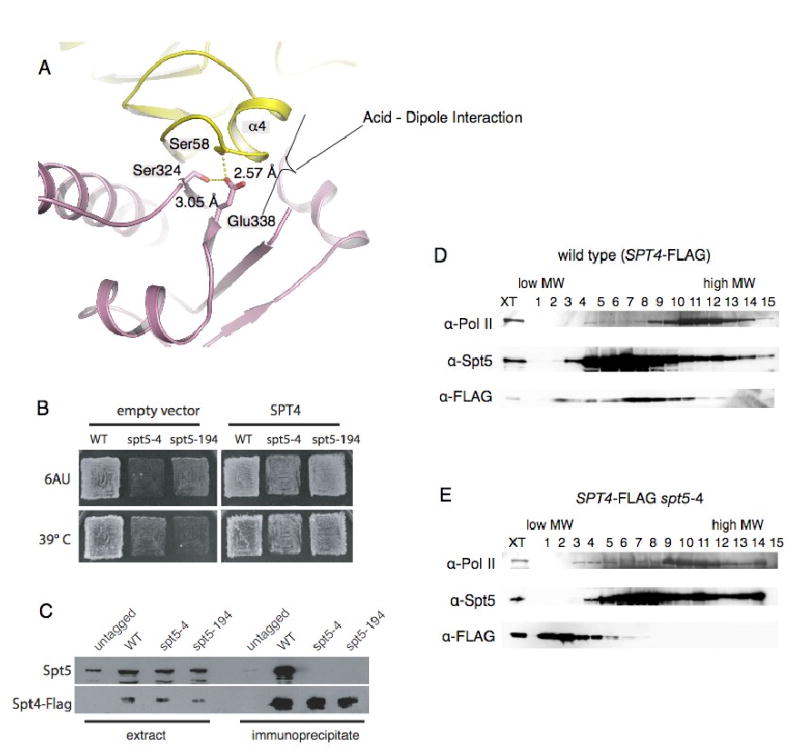
(A) Conserved acid--α-helix dipole interaction between Helix_α4 of Spt4 and Glu338 of Spt5NGN. The carboxyl of Glu338 is also fixed by two hydrogen-bonds with Spt4_Ser58 and Spt5_Ser324. The coloring is the same as in Figure 2. (B) Suppression of spt5-4 and spt5-194 by overexpression of Spt4. The indicated strains were transformed with either empty vector or a high copy Spt4 plasmid and replica plated to YPD media at 39° C or SC-ura media with 50 microgram per ml 6-azauracil at 30° C. (C) Co-immunoprecipitation of Spt4-Spt5 from extracts of Spt5, Spt5-4 or Spt5-194 cells. (D, E) The spt5-4 mutation disrupts the Spt4-Spt5 complex; Extracts of cells expressing wild type Spt5 or Spt5-4 and flag-tagged Spt4 were fractionated on 10-30% sucrose gradients. Fractions were separated on SDS gels and blotted with the indicated anti-sera.
Figure 4. Structural conservation of Spt4 and Spt5NGN domain.
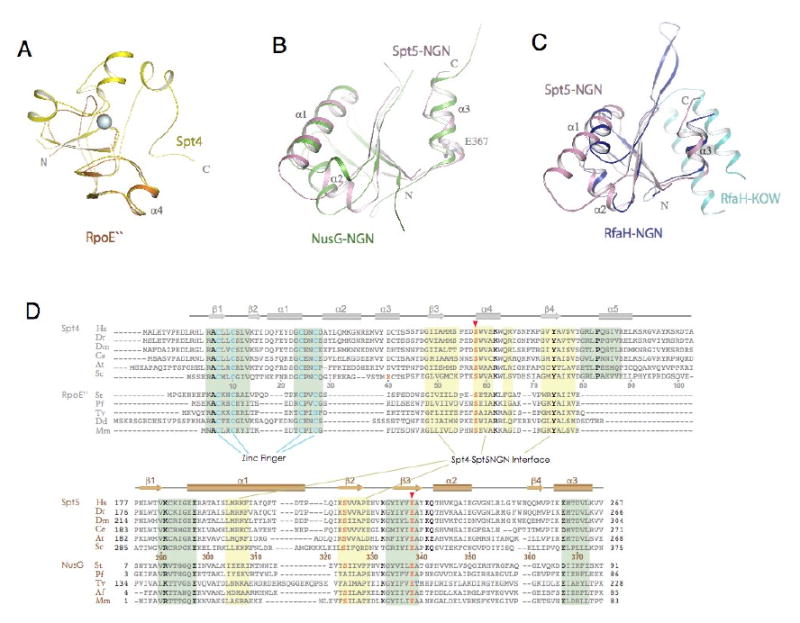
(A) Structural superimposition of Spt4 (yellow) with Pyrococcus furiosus RpoE″ (orange, pdb1ryq). (B) Structural superimposition of Spt5NGN domain with the Aquifex aeolius NusG (green, pdb1npp). (C) structural superimposition of Spt5NGN domain with the e.coli RfaH (pdb2oug, NGN in dark blue and KOW in light blue). (D) Alignment of representative Spt4 and Spt5 homologues. Residues that fall in the binding interface between Spt4 and the Spt5NGN domain are noted with a yellow background; residues that contribute to structural stability are noted by the green background. Highly conserved residues are depicted in bold type; residues in pink are involved in the acid-dipole interaction. Abbreviations: Hs, Homo sapiens; Dr, Danio rerio; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana; Sc, Saccharomyces cerevisiae; St, Sulfolobus tokodaii; Pf, Pyrococcus furiosus; Tv, Thermoplasma volcanium; Dd, Dictyostelium discoideum; Af, Archaeoglobus fulgidus; Mm, Methanococcus maripaludis;
Mutations of the acid-dipole interaction disrupt the Complex
We previously characterized two mutations, spt5-4 and spt5-194, that share several phenotypes with spt4 mutants, including a moderate Ts- growth defect at 39°, sensitivity to 6-azauracil (6AUs) and a strong synthetic Ts- phenotypes when combined with a deletion of DST1, which encodes elongation factor TFIIS(Hartzog et al., 1998). We used a high-copy plasmid to over-express Spt4 in spt5-4 dst1Δ and spt5-4 dst1Δ double mutants and found that overexpression of Spt4 suppressed the strong synthetic Ts-phenotype (data not shown). We also overexpressed Spt4 in spt5-194, spt5-4 and dst1Δ single mutants and observed that elevated levels of Spt4 suppressed the 6AUs and moderate Ts- phenotypes of spt5-194 and spt5-4 but not the 6AUs phenotype of dst1Δ (Figure 3B and data not shown).
One possible explanation for the results described above is that the spt5-4 and spt5-194 mutations weaken the interaction of Spt4 with Spt5, and that this defect can be overcome by increasing the dosage of Spt4 in the cell. To test this model, we performed anti-Flag immunoprecipitations from extracts of wildtype, spt5-4 and spt5-194 strains in which Spt4 was tagged with three copies of the Flag epitope. We observed robust, epitope-dependent co-precipitation of Spt4 and Spt5 in the wild-type extracts, but not in the spt5-4 or spt5-194 extracts (Figure 3C). We observed similar disruption of Spt4-Spt5 complexes when we size fractionated spt5-4 or spt5-194 extracts on sucrose gradients (Figure 3D, 3E and data not shown). These findings demonstrate that the spt5-4 and spt5-194 mutations disrupt Spt4-Spt5 binding.
We rescued and sequenced the spt5-4 and spt5-194 alleles. The spt5-4 mutation causes a single amino acid change, Glu338Lys. This dramatic change of side chain charge may disrupt the hydrogen bond with Ser58 of Spt4, and the acid-α-helix dipole interaction(Nicholson et al., 1988)(Figure 2B and 3A). The spt5-194 allele causes two amino acid changes, Ser324Phe and Gly610Asp; when we separated these mutations, we found that all of the spt5-194 phenotypes were caused by the Ser324Phe mutation. Ser324 is located at the centre of the Spt4-Spt5NGN interface, and it directly contacts Val50, Met52, Ser58 and Val60 of Spt4 and forms a hydrogen bond with Glu338 of Spt5 (Figure 3A). The new, bulky phenylalanine side chain introduced by the Ser324Phe mutation likely leads to steric conflicts that alter and weaken these interactions. These results are consistent with the idea that the Glu338-Helix_α4 acid-dipole interaction plays a central role in formation of the Spt4-Spt5 complex.
Structural conservation of Spt4 and the Spt5NGN domain
A structural similarity search(Holm and Sander, 1996) did not identify proteins similar to Spt4 in archaea. However, Spt4 was previously suggested to share sequence similarity with archaeal RpoE″ (DNA-directed RNA polymerase subunit E″)(Ponting, 2002), and direct comparison of their structures revealed that RpoE″ resembles the bottom part of Spt4, which includes the interface with Spt5 (Figure 4A). A similar zinc finger is also conserved in archaeal RpoE″ proteins (β1-β2-α1, Figure 1 and 4D). The strict conservation of the zinc finger domain suggests it plays a fundamental role in Spt4/RpoE″ function.
Comparison of the sequence and structure of Spt5's NGN domain with those of bacterial NusG and its paralogue RfaH(Artsimovitch and Landick, 2002), reveals conservation between bacteria and eukaryotes (Figure 4B-4D). Remarkably, a glutamate residue at the N-terminus of α3, Glu367 in Spt5, is highly conserved in both Spt5 and NusG proteins suggesting they might have a general functional role at the α3 side of the NGN domain (Figure 4B and 4D). In the bacterial NGN protein RfaH, a cavity beneath this alpha-3 helix is proposed to be an RNAP-binding surface(Belogurov et al., 2007). Consistent with that model, the upper side of α3 in the Spt5NGN domain makes significant contacts with Spt4, leaving the lower site exposed in the Spt4-Spt5NGN complex structure (Figure 2A).
The ancestral Spt4-Spt5 complex: Evidence that RpoE″ and NusG form a complex in archaea
Spt5/NusG proteins are found in all three kingdoms of life and are therefore among the most ancient components of the transcriptional machinery(Ciampi, 2006; Harris et al., 2003; Koonin et al., 2000; Peterlin and Price, 2006; Ponting, 2002). The functional and structural similarities of the Spt5NGN domain and bacterial NGN proteins imply that they also share similar mechanisms of transcription regulation.
In contrast to Spt5, Spt4 sequence homologues are not found in bacteria but exist universally in archaea, in the form of RpoE″(Ponting, 2002). Spt4 residues that contact Spt5NGN are conserved in RpoE″. The acid-dipole interaction between residues Ser58, Helix_α4 of Spt4 and Spt5NGN-Glu338 are strictly conserved in RpoE″ and archaeal NusG (Figure 4D). These observations strongly argue that RpoE″ and archaeal NusG interact in a manner similar to Spt4-Spt5NGN, and this hypothesis is supported by superimposition of the Spt4 and RpoE″ structures (Figure 4A) and by docking RpoE″ on the modeled P. furiosus NusG structure (Figure 5A).
Figure 5. Conservation of the Spt4-Spt5 Interface.
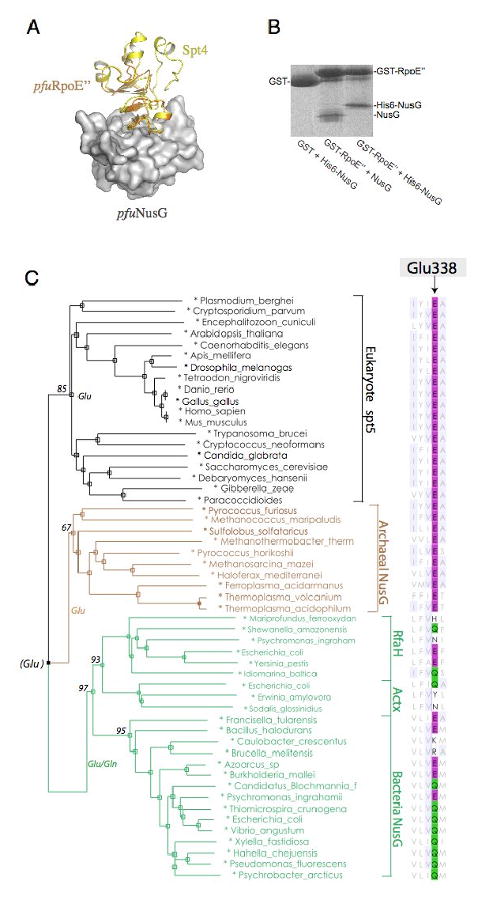
(A) Model of Archaeal NusG bound to RpoE″ : Archaeal RpoE″-NusG complex was modeled by superimposing the pfuRpoE″ structure and pfuNusG model onto the Spt4-Spt5NGN structure. The pfuNusG model is shown as a white surface; yellow, Spt4; orange, pfuRpoE″. (B) GST Pull-down assay of Methanocaldococcus jannaschii RpoE″-NusG interaction. (C) A sequence-based phylogeny derived from an alignment of NGN domain in Spt5/NusG homologues. Organism names are color-coded according to the domain of life. Alignment of Glu338 for the acid-dipole interaction is shown at right. Bootstrap support is indicated for major braches.
To directly examine whether the archaeal homologues of Spt4 and Spt5 can bind to each other, we expressed the Methanocaldococcus jannaschii NusG and RpoE″ proteins. Purified mjaNusG was incubated with glutathione beads bound to either GST-RpoE″ or a GST protein control (Figure 5B). The mjaNusG protein was retained on the GST-RpoE″ beads, even in the presence of 1M NaCl. These two proteins also co-purified on gel filtration and Ni-NTA affinity columns (data not shown). Thus, like Spt4-Spt5, RpoE″ and NusG form a tight complex.
Discussion
By creating an Spt4/Spt5NGN fusion protein we have been able to determine the structure of S. cerevisiae Spt4 bound to the NGN domain of Spt5. The Spt5 NGN domain bears remarkable similarities to its bacterial counterparts, even though bacteria lack an Spt4 homologue. Spt4 is structurally similar to RpoE″, which was previously suggested to be an Spt4 homologue and we further demonstrate that archaeal NusG binds to RpoE″. The homology between Spt4 and RpoE″ is largely limited to the region of Spt4 that contacts Spt5 in the Spt4-Spt5NGN structure, suggesting that the intermolecular contacts we observed are not crystal-packing artifacts and further indicating that the basic architecture of the Spt4-Spt5 complex is conserved in archaea.
What is the function of Spt4? In yeast, SPT4 is not essential for viability(Malone et al., 1993). However, there is a near complete overlap between spt4Δ phenotypes and partial loss-of-function mutations in SPT5, and spt4 and spt5 mutations exhibit strong genetic interactions(Hartzog et al., 1998; Swanson et al., 1991; Swanson and Winston, 1992). Although it is possible that, in the intact Spt4-Spt5/RNAPII elongation complex, the zinc finger contacts DNA or RNA, we have not detected any nucleic acid binding by recombinant Spt4 alone (G.A.H. and Berra Yazar, unpublished data). In both yeast (Figure 3D, 3E and data not shown) and in extracts of human cells (Kim et al., 2003), most, if not all, Spt4 is bound to Spt5. These data support the idea that Spt4 is important for and functions entirely in the context of the Spt4-Spt5 complex.
One potential role for Spt4 is as a regulator of Spt5 conformation. Although our structure does not include the KOW domains of Spt5, several structures of NusG proteins with their single KOW domains have been solved(Knowlton et al., 2003; Reay et al., 2004; Steiner et al., 2002). The KOW domain adopts a variety of orientations in these structures, indicating a highly flexible linkage between the NGN and KOW domains. A functional conformation of the NGN and KOW domains might be achieved by interaction with other transcription factors. For example, NusG only stably binds the E.coli RNAP elongation complex in the presence of rho protein(Nehrke and Platt, 1994). In eukaryotes and archaea it is possible that the binding of Spt4 or RpoE″ restricts the spatial relationships of the NGN and KOW domains. Structural superimposition of Spt4-Spt5NGN and NusG structures suggest the observed conformations of the KOW domain in NusG are compatible with the placement of Spt4 on the Spt5NGN (Figure 6B). Moreover, in one of these superimpositions, the loop between NusG's NGN and KOW domains travels near a deep groove on the surface of Spt4, placing the NusG KOW domain close to the opposite side (α2 helix) of Spt4 (Figure 6B). Thus, it is possible that this groove in Spt4 helps position Spt5's KOW domains relative to the Spt4-Spt5NGN core. Although few sequence changes of spt4 and spt5 mutations have been reported, our structure will provide a framework for designing and interpreting effects of mutations affecting the core of the Spt4-Spt5 complex. Future challenges in studies of Spt4-Spt5 function will be to obtain structures of complexes that include the KOW domains and to understand the dynamic relationships between these domains and the core of the complex.
Figure 6. Evolution of the Spt4-Spt5 Complex.
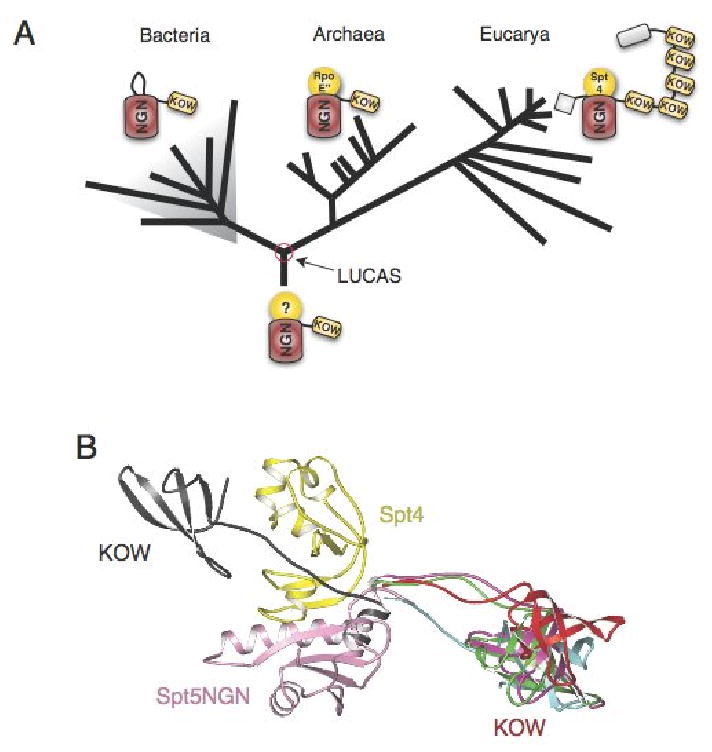
(A) An evolutionary model of Spt4-Spt5 shows the distribution and domain organization of their homologues across evolution. (B) Superimposition of 5 distinct bacterial aaeNusG structures(described in Reay et al., 2004) and the eukaryotic Spt4-Spt5NGN complex. Only the Spt4-Spt5NGN and KOW structures are shown, the superimposed NusG_NGN domains were omitted for clarity. A notable feature of the NusG structure is the flexible linkage between NGN and KOW domains. In one of the 5 NusG structures shown here (in black, pdb1npp, chain D), the orientation of the KOW domain relative to the NGN domain is dramatically different from that of the other 4 structures.
Spt5-Glu338, which is critical for Spt4 binding, is conserved in all eukaryotic Spt5NGN and archaeal NusG proteins (Figure 5C). Curiously, even though bacteria lack an apparent Spt4 homologue, many bacterial NusG and RfaH proteins have a glutamate or glutamine at a position homologous to Glu338 of Spt5 (Figure 5C). The fact that these proteins are widely spread and deeply branched in the bacterial lineage argues against the idea that this conservation is due to convergent evolution, which usually shows a more sporadic pattern(Barton et al., 2007). Thus, the last common ancestor of life likely had a Glu or Gln at the same position (Figure 5C).
Several models may explain the conservation of a Glu/Gln at this position in bacterial proteins where Spt4 is not present. First, it is possible that the evolutionary ancestor of bacteria had an Spt4/RpoE″ progenitor which was subsequently lost; second, this Glu/Gln may be important for the structure of the NGN domain; third, bacterial NusG proteins may have an alternate conformational state in which residue 338 is important. Interestingly, bacterial NGN domains have an insertion in the NGN domain which creates a β sandwich or hairpin that is incompatible with a protein binding in the same fashion as Spt4 does to the Spt5 NGN domain (Figure S2), suggesting a correlation between the presence of this insertion and the absence of Spt4 gene in bacteria (Figure 6A).
Among the RNAP II elongation control co-factors, Spt4-Spt5 is probably the oldest(Peterlin and Price, 2006). Several observations suggest that Spt4-Spt5, archaeal NusG-RpoE″ and bacterial NusG proteins are ancient proteins involved in a core function of transcription. First, NGN proteins are ubiquitous in all branches of life and are conserved at the sequence and structural levels (Figure 4B, 4C and 4D). Second, in both bacteria and eukaryotes, they are implicated in the regulation of transcription elongation and related events, such as RNA processing and transcription termination. Furthermore, Spt4-Spt5 associates with RNA Pol I and rRNA genes and an spt4 mutation causes a modest decrease in growth and rRNA synthesis rates, indicating a potential role for Spt4-Spt5 in transcription elongation by RNA Pol I and rRNA processing(Schneider et al., 2006). Similar observations have been made for the multi-subunit DNA-dependent RNA polymerases: they are found in all organisms, they share sequence, structural homology and employ similar catalytic mechanisms(Vassylyev et al., 2007). It is most reasonable to propose therefore, that Spt4-Spt5, NusG-RpoE″ and bacterial NusG proteins share a core set of functions and mode of interacting with elongating RNA polymerases that were present in the last common ancestor of life, forming the basal transcription elongation apparatus with the primordial RNAP.
Experimental Procedures
Expression plasmids
To facilitate protein expression and crystallization, the S. cerevisiae Spt4 and the Spt5NGN domain were subcloned into a single pET22b vector. The linkage between them was optimized according to the crystallization performance of different constructs. The final construct contained Spt4 (1-99) followed by a 2-residues linker (GS) and the Spt5NGN domain (285-375) with a C-terminal 6 × His tag (LEHHHHHH). The Methanocaldococcus jannaschii NusG and RpoE″ genes were synthesized with optimized codons (sequence available upon request), and subcloned into pHis-tev and pGST-tev vectors (a gift from Dr. DC. Wang).
Expression and purification
Proteins were overexpressed in Escherichia coli strain Rossetta (DE3) (Novagen, San Diego, CA). Cells were grown at 37°C to an optical density of 0.7 at 600 nm, induced with 1 mM IPTG for 8h at 30°C, and disrupted by sonication in 20 mM Tris, pH 8.4, 0.5 M NaCl, 5 mM imidazole and 0.1% Triton X-100. After centrifugation, the pellet was re-dissolved in a similar buffer except with 1 M NaCl, and loaded onto a Ni2+-NTA (Qiagen, Valencia, CA) column. The column was washed with 20mM Tris, pH 8.4, 1.0 M NaCl, 25 mM imidazole, 0.1% Triton X-100 and eluted with 20 mM Tris, pH 8.4, 1.0 M NaCl, 370 mM imidazole. N-terminal His-tagged NusG and GST-RpoE″ was purified using the corresponding affinity columns. To get non-tagged NusG, His-tagged NusG was cleaved by Tev protease and further purified on a Ni2+-NTA column.
Crystallization
Spt4/Spt5NGN purified by Ni2+-NTA chromatography (10 mg ml-1 in 20 mM Tris, pH 8.4, 1.0 M NaCl) was used directly for crystallization. Taking advantage of our observation that the purified Spt4/Spt5NGN fusion protein was only soluble above 700 mM NaCl, we used a modified version of the hanging-drop vapor-diffusion method to obtain crystals. Protein samples (2 μl) were mixed with 1 μl reservoir solution with lower salt concentration (100 mM Tris, pH 8.8, 50 mM NaCl and 36% ethanol). Droplets of the protein sample expanded during equilibration, which decreased the salt concentration and promoted the packing of Spt4/Spt5NGN protein. Crystals, which appeared after 3-4 days, were first equilibrated in a stabilization solution (100 mM Tris, pH 8.8 and 40% ethanol), then transferred to a cryo-protectant buffer containing stabilization solution plus 25% MPD and flash-frozen in liquid nitrogen. The diffraction quality of these crystals was sensitive to subtle changes in growth temperature and equilibration speed.
Data collection and structure determination
Anomalous diffraction data were collected at the BSRF by using wavelength 1.2834 Å corresponding to the peak of a Zn-MAD (multi-wavelength anomalous dispersion) experiment. In addition, a 2.2 Å native data set was collected on a Rigaku RA-Micro7 generator equipped with Mar345 detector. Data were processed with HKL2000(Otwinowski and Minor, 1997) or Mosflm and Scala(1994). The initial structure was determined with 3.0 Å SAD data by SOLVE/RESOLVE(Terwilliger and Berendzen, 1999). Model building and TLS (translation-libration-screw) refinement were performed with the 2.2 Å native dataset by using O(Jones et al., 1991) and REFMAC5(1994). Data collection and refinement statistics are summarized in Supplementary Table 1. The final atomic model includes one fusion protein, two ethanol molecules, one MPD molecule and 104 water molecules in the asymmetric unit.
Yeast genetics
Yeast media was prepared and strain construction, plasmid transformations and other genetic manipulations were performed according to standard protocols (Rose et al., 1990). SC-Ura+6-azauracil plates included 50 μg/ml 6-azauracil. Complete strain genotypes are given in Table S2.
Immunoprecipitation
Immunoprecipitation and western blots were performed as described previously(Lindstrom et al., 2003). Extracts of strains expressing Flag-tagged Spt4 were subjected to anti-Flag immunoprecipitation. Extracts and the immunoprecipitates were separated on an SDS gel, blotted and probed with anti-sera specific to the Flag epitope and Spt5.
GST pull-down experiment
Purified GST or GST fusion RpoE″ was immobilized on glutathione–Sepharose 4B beads and combined with the indicated purified proteins in 0.5 ml binding buffer (20 mM Tris pH 8.0, 150 mM NaCl and 0.1% Triton X-100). The mixtures were incubated at 4°C for 1.5 h with constant rocking. The beads were then washed three times with 1 ml of binding buffer, and 1ml binding buffer with 1M NaCl. Proteins associated with the beads were released by boiling in loading buffer and subjected to SDS–PAGE followed by staining with coomassie blue.
Sequence Alignment and phylogenetic Analyses
Sequences were downloaded from the National Center for Biotechnology Information nonredundant database and the Integrated Microbial Genomes with Microbiome Samples database(Markowitz et al., 2006). Sequence alignments were performed by ClustalW(Larkin et al., 2007) and also edited by using the Jalview alignment editor(Clamp et al., 2004), guided by the structure-based alignments. For constructing the phylogenetic tree for NGN domains, only regions of NGN domains in Spt5/NusG homologues were selected and used for alignment. PHYLIP(Felsenstein, 2005) was used to construct the phylogenetic tree by using Neighbour Joining method and to compute the bootstrap value. Njplot(Perriere and Gouy, 1996) and Jalview were used for displaying and drawing tree.
Accession code
The coordinates and structure factors for the crystal structure of Spt4-Spt5NGN complex have been deposited in the Protein Data Bank under ID code: 2EXU.
Supplementary Material
Acknowledgments
This work was supported by grants to L.N. and M. T from the Chinese National Natural Science Foundation (grants 30121001, 30025012 and 30571066), the Chinese Ministry of Science and Technology (grants 2006CB806500 and 2006AA02A318), and the Chinese Academy of Sciences (grant no. KSCX1-YW-R-60). G.A.H was supported by NIH grant GM060479. We thank Melissa Jurica, Manny Ares and Berra Yazar for their comments on the manuscript. We thank Sarah Mosely for her assistance in the early stages of the high-copy suppressor screen and Pengfei Fang for his assistance in the initial crystallization screen. We also thank Drs. Yuhui Dong and Peng Liu at BSRF for their support for Zn-SAD data collection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes & development. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Barton NH, Briggs DEG, Eisen JA, Goldstein DB, Patel NH. Evolution. Cold Spring Harbor Laboratory Press; 2007. Evolution; pp. 109–128. [Google Scholar]
- Basrai MA, Kingsbury J, Koshland D, Spencer F, Hieter P. Faithful chromosome transmission requires Spt4p a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2838–2847. doi: 10.1128/mcb.16.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Molecular cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Current opinion in cell biology. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bucheli ME, Buratowski S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. The EMBO journal. 2005;24:2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nature structural & molecular biology. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- Burns CM, Richardson LV, Richardson JP. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. Journal of molecular biology. 1998;278:307–316. doi: 10.1006/jmbi.1998.1691. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. The Journal of biological chemistry. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- Ciampi MS. Microbiology (Reading, England) Vol. 152. 2006. Rho-dependent terminators and transcription termination; pp. 2515–2528. [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. Bioinformatics (Oxford, England) Vol. 20. 2004. The Jalview Java alignment editor; pp. 426–427. [DOI] [PubMed] [Google Scholar]
- Cui Y, Denis CL. In vivo evidence that defects in the transcriptional elongation factors RPB2, TFIIS, and SPT5 enhance upstream poly(A) site utilization. Molecular and cellular biology. 2003;23:7887–7901. doi: 10.1128/MCB.23.21.7887-7901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington; Seattle: 2005. Distributed by the author. [Google Scholar]
- Harris JK, Kelley ST, Spiegelman GB, Pace NR. The genetic core of the universal ancestor. Genome research. 2003;13:407–412. doi: 10.1101/gr.652803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & development. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- Jones S, Thornton JM. Prinples of Protein-protein interaction. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. Pt 2. Vol. 47. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models; pp. 110–119. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes & development. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DK, Inukai N, Yamada T, Furuya A, Sato H, Yamaguchi Y, Wada T, Handa H. Structure-function analysis of human Spt4: evidence that hSpt4 and hSpt5 exert their roles in transcriptional elongation as parts of the DSIF complex. Genes Cells. 2003;8:371–378. doi: 10.1046/j.1365-2443.2003.00638.x. [DOI] [PubMed] [Google Scholar]
- Knowlton JR, Bubunenko M, Andrykovitch M, Guo W, Routzahn KM, Waugh DS, Court DL, Ji X. A spring-loaded state of NusG in its functional cycle is suggested by X-ray crystallography and supported by site-directed mutants. Biochemistry. 2003;42:2275–2281. doi: 10.1021/bi0272508. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Aravind L, Kondrashov AS. The impact of comparative genomics on our understanding of evolution. Cell. 2000;101:573–576. doi: 10.1016/s0092-8674(00)80867-3. [DOI] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A. Transcription elongation complex: structure and function. Current opinion in microbiology. 2001;4:119–125. doi: 10.1016/s1369-5274(00)00176-4. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics (Oxford, England) 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR, 3rd, Hartzog GA. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone EA, Fassler JS, Winston F. Molecular and genetic characterization of SPT4, a gene important for transcription initiation in Saccharomyces cerevisiae. Mol Gen Genet. 1993;237:449–459. doi: 10.1007/BF00279450. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, Zhao X, Dubchak I, Hugenholtz P, Anderson I, et al. The integrated microbial genomes (IMG) system. Nucleic acids research. 2006;34:D344–348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: an on-again, off-again relationship? Molecular cell. 2005;20:335–345. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Nehrke KW, Platt T. A quaternary transcription termination complex Reciprocal stabilization by Rho factor and NusG protein. Journal of molecular biology. 1994;243:830–839. doi: 10.1006/jmbi.1994.1685. [DOI] [PubMed] [Google Scholar]
- Nicholson H, Becktel WJ, Matthews BW. Enhanced protein thermostability from designed mutations that interact with alpha-helix dipoles. Nature. 1988;336:651–656. doi: 10.1038/336651a0. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. The Journal of biological chemistry. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Molecular cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Ponting CP. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic acids research. 2002;30:3643–3652. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reay P, Yamasaki K, Terada T, Kuramitsu S, Shirouzu M, Yokoyama S. Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus. Proteins. 2004;56:40–51. doi: 10.1002/prot.20054. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: a laboratory course manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nature reviews. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci U S A. 2006;103:12707–12712. doi: 10.1073/pnas.0605686103. Epub 12006 Aug 12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simader H, Hothorn M, Kohler C, Basquin J, Simos G, Suck D. Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes. Nucleic acids research. 2006;34:3968–3979. doi: 10.1093/nar/gkl560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner T, Kaiser JT, Marinkovic S, Huber R, Wahl MC. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. The EMBO journal. 2002;21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:4286. doi: 10.1128/mcb.11.8.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Winston F. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & development. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes & development. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes & development. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yang S, Cho SS, Levy Y, Cheung MS, Levine H, Wolynes PG, Onuchic JN. Domain swapping is a consequence of minimal frustration. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13786–13791. doi: 10.1073/pnas.0403724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


