Abstract
Hepcidin, the principal regulator of the iron metabolism, is up-regulated in response to inflammatory stimuli, bone morphogenic proteins (BMPs), and iron excess. There are two murine hepcidin genes: hepcidin-1 (Hamp1) and hepcidin-2 (Hamp2). Hamp1 gene responds to both IL-6 and BMPs while Hamp2 responds to neither. We replaced the putative functional regulatory motifs of the Hamp1 promoter with the corresponding putative “non-functional” Hamp2 motifs and vice versa in reporter constructs. Conversion of the Hamp1 STAT site into the Hamp2 site reduced the basal level of reporter expression but did not affect IL-6 and BMP responsiveness; replacing Hamp2 site with the Hamp1 site only resulted in partial responsiveness. These data are in contrast to the role of the STAT site in the human hepcidin promoter which is important in both basal level and IL-6 inducible promoter activity. The murine AP1, E-box and TIEG motifs were found to neither influence the basal level of expression of Hamp1 and HAMP promoters nor play a critical role in the IL-6 and BMP-9 induced response. Our data suggest that the STAT site (nt −148 to −130) is important for the regulation of basal level expression of Hamp1 but there are additional regions that are responsible for the IL-6 and BMP-9 responsiveness within the Hamp1 promoter.
Keywords: promoter, transcription
Introduction
Hepcidin is a principal regulator of iron metabolism affecting iron uptake from intestine and release of iron from body stores in liver and macrophages [1]. Its transcription is increased by diverse stimuli such as the cytokines IL-6, IL-1α and IL-1β, the bone morphogenetic proteins -2, -4 and -9 (BMP-2, BMP-4 and BMP-9) and is decreased by hypoxia and anemia [1–6].
There is one hepcidin gene in humans (HAMP) while there are two murine genes, Hamp1 and Hamp2. The Hamp1 and HAMP are functionally equivalent since both gene products bind ferroportin and modulate iron metabolism [7,8]. In contrast, the product of Hamp2, although up-regulated by iron excess, does not seem to affect iron metabolism in transgenic mice over-expressing Hamp2 [9]. Moreover, in contrast to Hamp1, which is responsive to IL-6 and BMPs, Hamp2 is responsive to neither [10,11].
The transcriptional regulation of hepcidin appears to be complex. Courselaud et al. [12] found that CCAAT/enhancer-binding protein alpha (C/EBP-α) and C/EBP-β were very potent and weak activators, respectively. Bayele et al. [13] concluded that members of the basic helix-loop-helix leucine zipper (bHLH-ZIP) family of transcriptional regulators control hepcidin expression through binding to the canonical E-box sequences. The bHLH-ZIP family of transcription factors include upstream stimulatory factor 1 and 2 (USF1 and USF2). USF2 appeared to exert a polar or cis-acting effect, while USF1 was thought possibly to act in trans to control hepcidin expression. Interestingly, co-expression of USF1/USF2 with hepcidin promoter reporter constructs demonstrated significant up-regulation of Hamp1 but not Hamp2. The SMAD4 transcriptional regulator, which is a part of TGF and BMP signaling pathway, affects hepcidin expression since lack of SMAD4 reduces basal expression of Hamp1 by 100 fold and reduces hepcidin response to IL-6, BMP-4 and TGF-β1 [5]. SMAD4 might regulate hepcidin directly via SMAD consensus motifs or possibly through TGFβ-inducible early gene (TIEG) responsive elements within the hepcidin promoter [5,14–16]. Recently, it has been shown that STAT-3 plays a role in the inflammatory regulation of HAMP and that a crucial binding site is located at nt −97 to−75 from start of transcription (nt −148 to −130 from start of translation) and that a change of the STAT core binding residues TTC into GGA led to a loss of responsiveness of HAMP promoter to IL-6 [17–19].
BMP-9 was selected as a molecule representing the BMP signaling pathway because of its high potency to stimulate hepcidin [4] and its predominantly liver expression [20] and IL-6 was selected as a molecule representing the inflammatory pathway in order to see whether these signaling pathways share responsive elements and whether the regulation by STAT-3 is conserved between human and mice..
In the present report we demonstrate that conversion of the Hamp1 STAT site to the putative non-functional Hamp2 STAT site does not abolish its responsiveness to IL-6, while replacing the Hamp2 non-functional STAT site with the Hamp1 STAT site increases responsiveness of this promoter to IL-6 but not to the extent present in native Hamp1 promoter. Moreover, the AP1 site, TIEG box as well as E box sequences within 650 bp of the proximal promoter are not required for the response of hepcidin to BMP-9 and IL-6.
Materials and methods
Materials
Human recombinant BMP-9 and IL-6 were obtained from R&D Systems (Minneapolis, MN). Minimal Essential Medium, L- glutamine, penicillin/streptomycin solution, fetal bovine serum and polymyxin B sulfate were from Invitrogen (Carlsbad, CA).
Cell lines
The human hepatoma cell line HepG2 was obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in Minimal Essential Medium supplemented with 5% FBS, 100U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate and 2 mM L-glutamine.
Cloning of Hamp1, Hamp2 and HAMP promoter fragments into pGL3 basic
All nucleotides are numbered from start of translation, the nucleotide immediately 5′ to the start ATG begin designated −1. The proximal 1.0 kb, 260 bp, 200 bp and 140 bp fragments of the Hamp1 promoter, 1.2 Kb fragment of the Hamp2 gene were amplified by PCR and cloned into the Promega (Madison, WI) pGL3 basic vector containing the firefly luciferase reporter (luc) gene. Each promoter fragment was amplified by PCR using unique forward primers and a common reverse primer, that ends 5 nucleotides after the start of transcription (nt −40) and was inserted into the pGL3 basic vector or appropriately modified pGL3 basic vector using the primers and restriction enzymes listed in Table 1. All cloned fragments were originally amplified from genomic C57BL/6J DNA and sequences of all promoter constructs were verified by direct sequencing using the ABI Prism 3100 Genetic Analyzer (AME Bioscience A/S, Toroed, Norway). Using the primers listed in Table 1 we also made a human hepcidin promoter reporter construct by amplifying a 2.0 Kb promoter fragment using the primers described in Table 1, digesting with HindIII and inserting the resulting 1.3 Kb fragment into pGL3 basic.
Table 1. Primers, restriction enzymes and vector backbones used for cloning of the reporter vectors.
Desired inserts were amplified by PCR and cut with selected restriction enzymes and cloned into pGL3 basic or appropriately modified reporter vector based on pGL3 basic.
| Name | Forward primer | Reverse primer | Nt from ATG |
Vector backbone |
|---|---|---|---|---|
| Hamp1 140 bp | gagctcttacgcgtgttcttggaaatgagtcagag ca (MluI) | cttagatcgcagatctctgtgtggtggct-gtctaggagc (BglII) | −143 to −35 | pGL3 basic |
| Hamp1 200 bp | acgcgtgctagcaccctgtcccctgtt(NheI) | gatcgcagatctcatgccttctgttctgctgtgc (BglII) | −200 to −4 | pGL3 basic |
| Hamp1 260 bp | ccgagctcttacgcgtgcttgtgtccctggttctgtctgcccca (MluI) | cttagatcgcagatctctgtgtggtggct-gtctaggagc (BglII) | −264 to −35 | pGL3 basic |
| Hamp1 1.0 Kb | tgtaggctgggtaccaatctc (KpnI) | tgccttcagatctgctgtgc (BglII) | −983 to −2 | pGL3 basic |
| Hamp2 1.2 Kb | gagctcttacgcgtgtcttgagaaccctgtctttgg(MluI) | atcgcagatctcgagcttgtgtggtggctgtctaggt (XhoI) | −1267 to −34 | pGL3 basic |
| HAMP 1.3 Kb | tgcagccttgaccaactaca | ggaagcttgtgacagtcgcttttatggggcctgc (HinDIII) | −1269 to −62 | pGL3 basic |
Mutagenesis of STAT/AP-1 sites in murine hepcidin 1, 2 and human hepcidin promoter
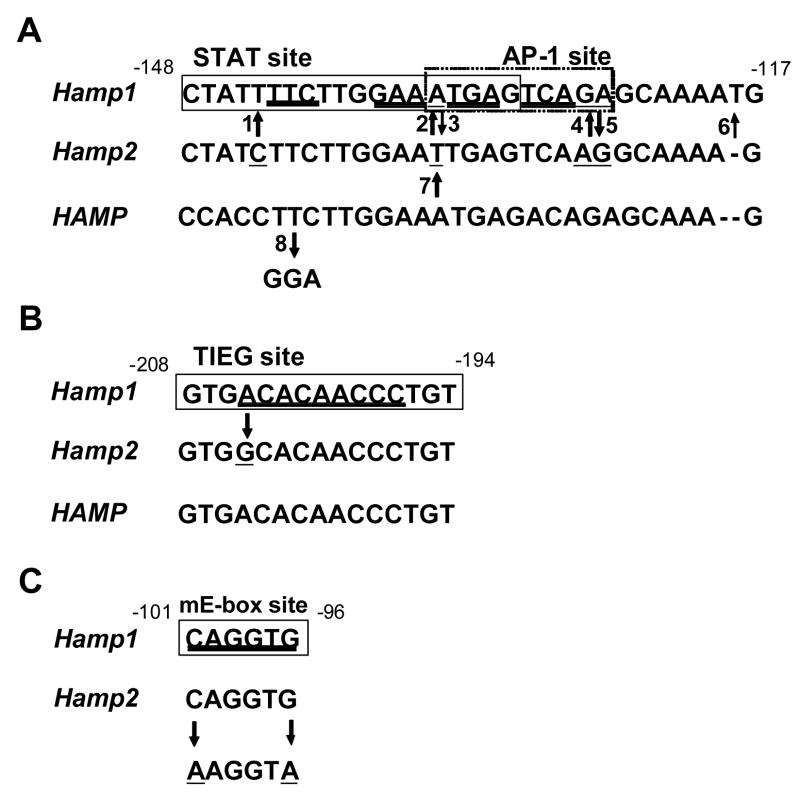
All nucleotide changes depicted in Figure 1 were carried out by using the Quikchange mutagenesis kit (Stratagene, La Jolla, CA) and were verified by sequencing and mutant sequences are shown in Figure 2.
Fig. 1. Scheme of mutagenesis of the STAT (A), TIEG (B) and E-box sites (C).
Black rectangles mark the binding sites. The critical binding residues are double underlined. The mutagenized residues are underlined and arrows show to what nucleotides they have been changed. The number accompanying the arrows is used to refer to this particular mutation throughout the text. Numbering is from start of translation and according to the murine Hamp1 promoter.
Fig. 2. Scheme of the mutagenized constructs.
The mutagenized residues are underlined and the names which are derived from Figure 1 are used to refer to the particular mutation throughout the text.
Mutagenesis of TIEG and E-box site in murine hepcidin 1 promoter
In order to define the role of the TIEG (nt −208 to −194) and E-box (nt −101 to −96) site located within 260 bp promoter fragment of the Hamp1 gene, we created reporter vectors designated as mTIEG and mE-box by mutagenizing them with Quikchange mutagenesis kit (Stratagene, La Jolla, CA). The TIEG 5′-gtgacacaaccctgt-3′ site was changed to 5′-gtggcacaaccctgt-3′ so that it was identical to the sequence present in the Hamp2 promoter and E-box caggtg site was changed to aaggta resulting in complete disruption of the canonical sequence (Figure 1).
Transfection and IL-6 treatment
Cells were plated onto 24-well plates (Corning, NY) at a density of 5×104 cells per well. The next day 200 ng of selected plasmid constructs containing the firefly luciferase and 10 ng of normalization plasmid pRL-TK (Promega, Madison, WI) containing the renilla luciferase were co-transfected into the cells using FuGene6 transfection reagent (Roche Applied Science, Indianapolis, IN) according to manufacturer’s instructions. Four to 6 hours post-transfection, the cells were treated with 10 ng/ml of human IL-6 or BMP-9 for 12–16 hours in the presence of 3 μg/ml of polymyxin to inhibit any possible contribution by cytokines generated by contaminating lipopolysaccharide (LPS).
Dual Luciferase Assay (Promega)
BMP-9 or IL-6 induced cells were lysed using 150 μl of the passive lysis buffer (Promega, Madison, WI). Thirty-five μl of LARII solution (Promega) were added rapidly to ten μl of the cell lysate and the light output was measured using a LB 96V microplate luminometer (Berthold Technologies GmbH&Co.KG, Bad Wildbad, Germany). Next, 35 μl of Stop&Glow solution (Promega) were added and the renilla luciferase luminescence was measured to permit normalization. Results are expressed as fold induction of firefly (luc)/renilla luminescence ratio of treated cells over the ratio obtained from non-treated cells expressing the same construct. Basal level of expression is expressed as fold induction of firefly/renilla luminescence ratio of reporter expressing cells over negative control represented by cells transfected with pGL3 only.
Results
In order to examine the role of the STAT (nt −148 to −130 nt), AP-1 (nt −134 to −125), E-box (nt −101 to −96) and TIEG (nt −208 to −194) binding motifs in responsiveness of the murine hepcidin promoter to IL-6 and BMP-9, we took advantage of the fact that murine Hamp1 responds to IL-6 and BMP-9 while murine Hamp2 does not. We hypothesized that some of the nucleotide differences in the proximal promoter region between Hamp1 and Hamp2 might be responsible for the lack of responsiveness of Hamp2 toward these stimuli. By interconverting Hamp1 and Hamp2 at these sites, we hoped to delineate which of these motifs were required for responsiveness to IL-6 and BMP-9.
The Effect of STAT, AP-1, TIEG and E-box mutagenesis on basal level of reporter expression
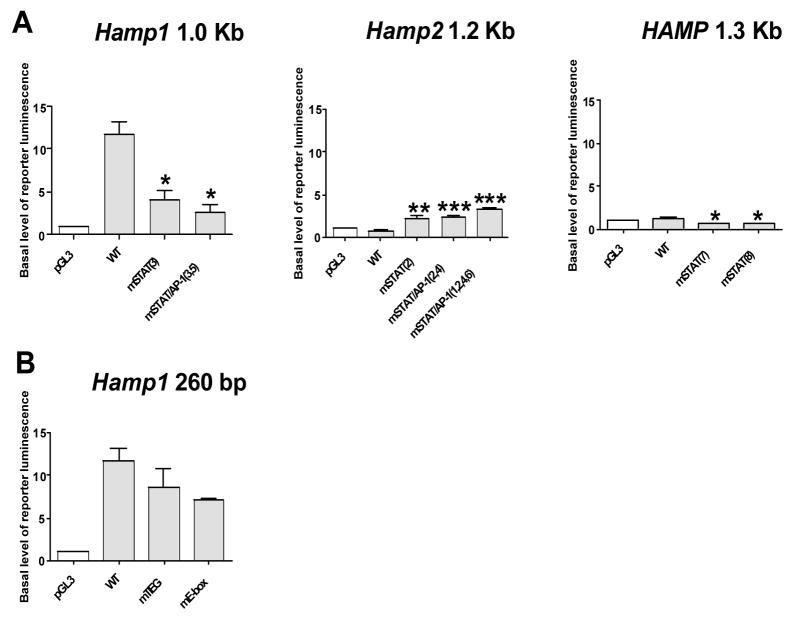
The murine Hamp1 promoter-containing constructs showed markedly higher basal level of reporter expression than constructs driven by murine Hamp2 and human HAMP. We found that changing the STAT site in the Hamp1 promoter into the corresponding supposedly non-functional STAT site present in Hamp2 [mSTAT(3)] resulted in a significant decrease in basal level of the luciferase reporter expression. Mutagenesis of the adjacent Hamp1 AP-1 site to the corresponding Hamp2 AP-1 site resulting in a STAT/AP-1 double mutant [mSTAT/AP-1(3,5)] did not have an additional effect on the basal level expression (Fig. 3A).
Fig. 3. Effect of mutagenesis of the STAT (A) and TIEG and E-box sites (B) on basal level of luciferase reporter expression.
HepG2 cells were transfected with a pGL3 promoterless firefly luciferase reporter vector (pGL3) or wild type (WT) hepcidin promoter construct containing murine Hamp1 260 bp, murine Hamp2 1.2 Kb or human HAMP 1.3 Kb promoter fragment with native sequence as well as with promoter fragments containing the desired mutation in STAT (mSTAT), STAT+AP-1 (mSTAT/AP-1), TIEG (mTIEG) and E-box (mE-box) sites with a number that corresponds to each mutation shown in Figure 1 and 2. The basal level of reporter luminescence is expressed as fold increase of firefly/renilla luminescence ratio of reporter expressing cells over the ratio of the negative control represented by cells transfected with promoterless pGL3 only. All values represent the mean ± SEM from at least two independent experiments. Results were analysed by the Graphpad software using t-test analysis. The p value of 0.05-0.01 is marked as *(significant), 0.01-0.001 as ** (very significant) and <0.001 as *** (extremely significant).
Inversely, changing the Hamp2 STAT site to the corresponding STAT site present in the Hamp1 promoter [mSTAT(2)] led to a significant increase in the basal reporter levels, however, not to the extent seen in the native Hamp1 promoter; the mutant promoter basal level was approximately 3 times while native Hamp1 promoter approximately 12 times higher than the promoterless negative pGL3 control. Mutagenesis of the adjacent Hamp2 AP-1 site as well as mutagenesis of two other nucleotides that differ between Hamp1 and Hamp2 into the sequence present in Hamp1 [mSTAT/AP-1(2,4) and mSTAT/AP-1(1,2,4,6)] did not have an additional effect on basal level expression of the luciferase reporter (Fig. 3A).
Mutation of the STAT site [mSTAT(7), mSTAT(8)] in the human HAMP promoter also led to decrease in the basal level of reporter expression although the effect was less dramatic than that observed with the murine promoter (Fig 3A). Mutation of the murine E-box (mE-box) as well as the TIEG mutation (mTIEG) did not significantly change the basal level of reporter expression (Fig. 3B).
The effect of STAT mutagenesis on IL-6 and BMP-9 responsiveness of the hepcidin reporter
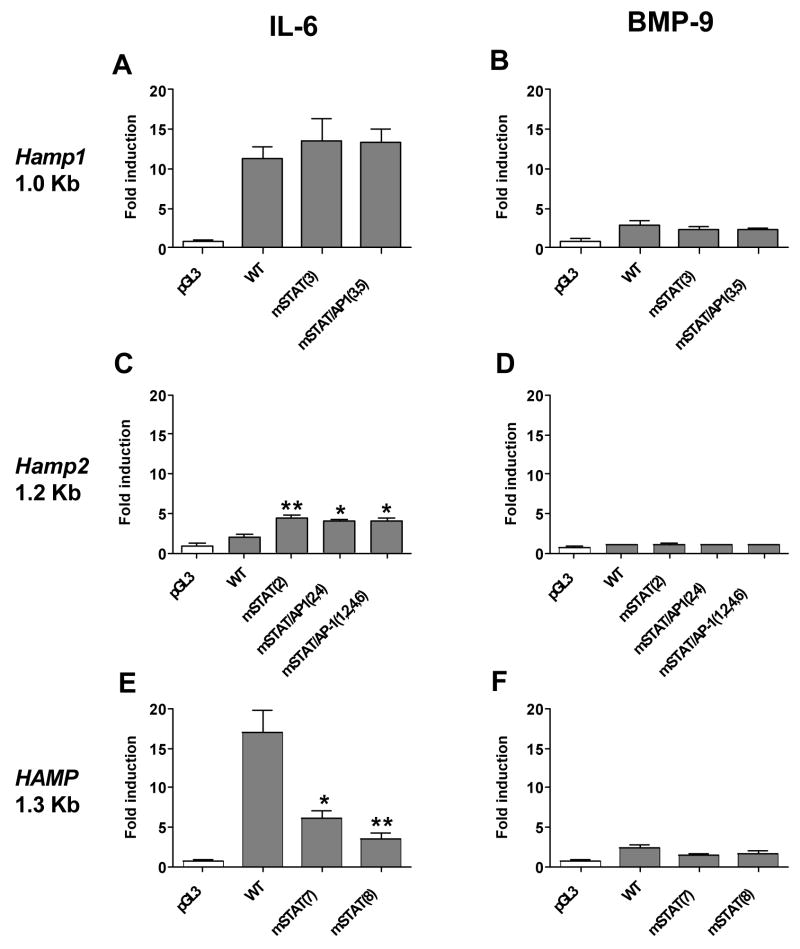
Conversion of either the STAT site [mSTAT (3)] or STAT and AP-1 site [mSTAT/AP-1 (3,5)] of Hamp1 to the corresponding Hamp2 sites, did not affect the responsiveness of the promoter reporter constructs to IL-6 and BMP-9 (Fig. 4A,B). In contrast, conversion of the supposedly non-functional Hamp2 promoter STAT site [mSTAT(2)], STAT+AP-1 sites [mSTAT/AP-1(2,4)] and conversion of two other nucleotides different between the two murine promoters [mSTAT/AP-1(1,2,4,6)] to the corresponding functional Hamp1 STAT and AP1 sites increased the responsiveness to IL-6 although not to the extent seen in the native Hamp1 promoter (mutant promoters are induced ~5 fold while native Hamp1 promoter ~15 fold) and had a negligible effect on responsiveness to BMP-9 (Fig. 4C,D). Importantly, the stimulating effect was observed with the STAT site mutation [mSTAT(2)] alone and none of the other mutations [ mSTAT/AP-1(2,4) and mSTAT/AP-1(1,2,4,6)] had a significant additional effect (Fig. 4C,D).
Fig. 4. Effect of STAT mutagenesis on IL-6 and BMP-9 responsiveness of the luciferase reporter driven by murine Hamp1 (A,B), murine Hamp2 (C,D) and human HAMP (E,F).
HepG2 cells were transfected with a pGL3 promoterless firefly luciferase reporter vector (pGL3) or wild type (WT) hepcidin promoter construct containing murine Hamp1 1.0 Kb, murine Hamp2 1.2 Kb or human HAMP 1.3 Kb promoter fragment with native sequence as well as with promoter fragments containing the desired mutation in STAT (mSTAT) or STAT+AP-1 (mSTAT/AP-1) sites with a number that corresponds to each mutation shown in Figure 1 and 2. The results are expressed as fold induction of firefly/renilla luminescence ratio of treated cells over the ratio of non-treated cells expressing the same construct. All values represent the mean ± SEM from at least three independent experiments. Results were analysed by the Graphpad software using t-test analysis. The p value of 0.05-0.01 is marked as *(significant), 0.01-0.001 as ** (very significant) and <0.001 as *** (extremely significant).
Since these data were not consistent with the observations made with the human hepcidin promoter by Wrighting et al. and Verga Falzacappa et al. [18,19] which clearly demonstrated the importance of the STAT site in IL-6 responsiveness, we compared the human HAMP mutant described by Writing et al (19) changing the core STAT binding nucleotides TTC to GGA [mSTAT(7)], with the HAMP mutant changing the STAT site to the murine Hamp2 STAT site (−134A>T). Our data clearly demonstrate that mutagenesis of the human STAT site by either mutation resulted in significantly decreased responsiveness to IL-6 (Fig. 4E,F), confirming the published results and demonstrating a difference in importance of the STAT site between human and mouse hepcidin promoters.
The effect of TIEG and E-box sites mutagenesis on IL-6 and BMP-9 responsiveness of hepcidin reporter
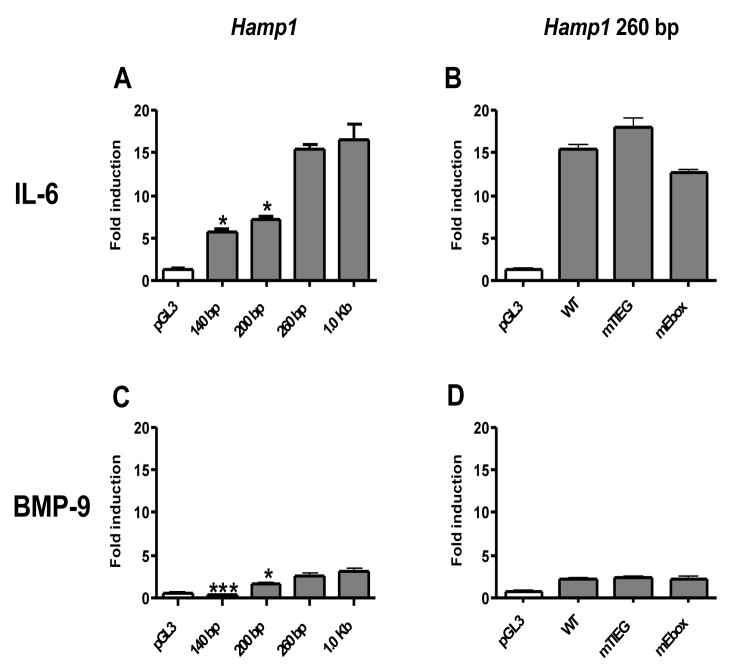
The response to BMP-9 and IL-6 was identical between the reporter constructs driven by the 1.0 Kb Hamp1 promoter fragment and the 260 bp Hamp1 promoter fragment (Fig. 5A,C). This suggests that the minimal promoter required for IL-6 and BMP responsiveness lies within the proximal 260 bp region and that there were no additional elements for IL-6 and BMP responsiveness between the −260 bp and −1.0 kb of the Hamp1 promoter region.
Fig. 5. Effect of murine Hamp1 promoter length (A,C) and TIEG and E-box mutagenesis (B,D) on IL-6 and BMP-9 responsiveness of the luciferase reporter.
HepG2 cells were transfected with a pGL3 promoterless firefly luciferase reporter vector (pGL3), 140 bp, 200 bp, 260 bp and 1.0 Kb promoter fragment of murine Hamp1 promoter construct or wild type (WT) construct (Hamp1 260 bp) with native sequence as well as with promoter containing the desired mutation in TIEG (mTIEG) and E-box sites (mE-box). The results are expressed as fold induction of firefly/renilla luminescence ratio of treated cells over the ratio of non-treated cells expressing the same construct. All values represent the mean ± SEM from at least two independent experiments. Results were analysed by the Graphpad software using t-test analysis. The p value of 0.05-0.01 is marked as *(significant), 0.01-0.001 as ** (very significant) and <0.001 as *** (extremely significant).
MatInspector software (Genomatix Software GmbH, München, Germany) identified two TIEG sites within the 260 bp proximal promoter of Hamp1. The distal TIEG site (nt −208 to −194) is conserved between murine Hepc1 and human hepcidin but the proximal TIEG site is not conserved. In addition, there is a single nucleotide difference between Hamp1 and Hamp2 in the distal TIEG site. Since IL-6 and BMP-9 stimulates Hamp1 and not Hamp2 expression, we mutated this TIEG site in the WT Hamp1 construct (260 bp Hamp1 promoter) to the corresponding sequence present in Hamp2. HepG2 cells transfected with the mTIEG construct did not show any loss in responsiveness to BMP-9 or IL-6 as compared to the WT Hamp1 promoter construct (Fig. 5B,D).
Although there are four E-box motifs within 650 bp of the proximal promoter of Hamp1, the most distal three E-box motifs (nt −250 to −650) are not likely to contribute to responsiveness to IL-6 and BMP-9 since there is no difference in responsiveness between the 1.0 kb and 260 bp Hamp1 promoter constructs (Fig. 5A,C). There is one E-box motif (nt −101 to −96) within the 260 bp proximal promoter of Hamp1 that is also conserved in Hamp2. This Hamp1 E-box motif was mutated so that the canonical sequence was disrupted. We found that the mutant E-box (mE-box) construct was as responsive to IL-6 and BMP-9 as the non-mutated WT hepcidin promoter construct containing 260 bp of the Hamp1 promoter (Fig. 5B,D).
Discussion
We tested the role of the STAT site (nt −148 to −130) as a critical regulator of murine hepcidin expression and responsiveness to IL-6 and BMP, by taking advantage of highly homologous murine Hamp1 and Hamp2 promoter sequences which differ in their responsiveness to IL-6 and BMP [10,11]. We show that murine Hamp1 promoter-containing constructs showed markedly higher basal levels of reporter expression compared to constructs driven by murine Hamp2 and human HAMP. The differences in the basal level of expression between the different promoter constructs might reflect differences in transcriptional elements between the constructs and/or species-specific transcription factors, since the studies were performed in the human HepG2 hepatoma cell line. Nevertheless, all reporter constructs respond to IL-6 and BMP in a manner consistent with their counterpart endogenous hepcidins.
Our data support the critical role of the STAT site (nt −148 to −130) in the regulation of basal level expression of hepcidin since mutagenesis of the STAT site in both murine Hamp1 as well as human HAMP to the “inactive” Hamp2 STAT site led to a decrease in basal level of reporter expression. In agreement, “correction” of the STAT site in the weak murine Hamp2 promoter resulted in significant up-regulation of basal level of a luciferase reporter, although not to the level observed with the native murine Hamp1 promoter. Our data complement the work by Wrighting et al [19] which demonstrated that mutagenesis of three nucleotides (nt −143 to −141) in the STAT reduced the basal level of hepcidin expression and its responsiveness to IL-6.
Our data demonstrated that mutagenesis of the murine Hamp1 STAT had no effect on IL-6 responsiveness. This appeared to be specific for the murine Hamp1 promoter, since mutagenesis of human STAT site in the human HAMP resulted in a significant reduction in IL-6 responsiveness. This suggests that the murine Hamp1 promoter might have additional elements that support IL-6 responsiveness. Mutagenesis of murine Hamp2 to “restore” an active STAT site was ineffective in restoring complete responsiveness to IL-6.
The STAT site in both human HAMP and murine Hamp1 did not play an important role in BMP responsiveness since mutagenesis of the STAT site had no significant effect on BMP responsiveness. This suggests that the IL-6 responsive region is distinct from the BMP responsive region.
Additionally, mutagenesis of the region surrounding the STAT site including the adjacent AP-1 site did not affect the responsiveness of such constructs to IL-6 and BMP-9; showing that the sequence surrounding the STAT site neither affects the basal level of expression nor the responsiveness of murine Hamp1 to IL-6/BMP-9.
We also demonstrated that the proximal 260 bp promoter of murine Hamp1 was as responsive to BMP-9 as the 1.0 kb promoter of Hamp1 and we speculated that BMP-9 might bind/activate the TGFβ-inducible early gene (TIEG) responsive element (nt −208 to −194) conserved between murine Hamp1 and human HAMP. Nevertheless, we found that mutagenizing this Hamp1 TIEG site into the corresponding supposedly “non-functional” sequence found in the Hamp2 site had no effect on BMP-9 responsiveness, thus showing that the BMP-9 response is probably not mediated through this TIEG site.
Studies by Bayele et al. [13] suggested that the canonical E-box sequences could regulate transcription of hepcidin by binding the USF1 and USF2 bHLH transcription factors. There are four E-box sites located in the proximal 650 pb of murine Hamp1, similar to the four human HAMP E-box motifs. Only one of these E-box sequences is located within the 260 bp proximal promoter of Hamp1, the region that permits a maximal response to IL-6 as well as a response to BMP-9. We found that disruption of the canonical sequence of the potentially critical E-box in the 260 bp segment did not alter the responsiveness to IL-6 or BMP-9. This finding suggest that the 4 E-box sequences within 650 bp of the Hamp1 promoter region are probably not key regulatory elements involved in BMP-9 and IL-6 responsiveness, however, it does not exclude the role of such sites located further upstream.
The regulation of hepcidin seems to be a complex signaling network and our findings indicate that although the STAT site definitely regulates the basal level of hepcidin expression it seems that the responsiveness to IL-6 and BMP-9 might require other, not yet identified element(s) and also suggest that regulation of murine Hamp1 and human HAMP promoters may not be identical.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jaroslav Truksa, Department of Molecular and Experimental Medicine, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA, 92037, USA, truksa@scripps.edu.
Pauline Lee, Department of Molecular and Experimental Medicine, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA, 92037, USA, plee@scripps.edu.
Ernest Beutler, Department of Molecular and Experimental Medicine, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA, 92037, USA, e-mail: beutler@scripps.edu.
References
- 1.Nemeth E, Ganz T. Regulation of iron metabolism by hepcidin. Annu Rev Nutr. 2006;26:323–342. doi: 10.1146/annurev.nutr.26.061505.111303. [DOI] [PubMed] [Google Scholar]
- 2.Lee P, Peng H, Gelbart T, et al. Regulation of hepcidin transcription by interleukin-1 and interleukin -6. Proc Natl Acad Sci USA. 2005;102:1906–1910. doi: 10.1073/pnas.0409808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babitt JL, Huang FW, Wrighting DM, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 4.Truksa J, Peng H, Gelbart T, et al. Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1 expression independently of Hfe, transferrin receptor 2 (Tfr2), and IL-6. Proc Natl Acad Sci USA. 2006;103:10289–10293. doi: 10.1073/pnas.0603124103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang RH, Li CL, Xu XL, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metabolism. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Nicolas G, Chauvet C, Viatte L, et al. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002;110:1037–1044. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Domenico I, Ward DM, Langelier C, et al. The Molecular Mechanism of Hepcidin-mediated Ferroportin Down-Regulation. Mol Biol Cell. 2007 doi: 10.1091/mbc.E07-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemeth E, Tuttle MS, Powelson J, et al. Hepcidin Regulates Iron Efflux by Binding to Ferroportin and Inducing Its Internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 9.Lou DQ, Nicolas G, Lesbordes JC, et al. Functional differences between hepcidin-1 and -2 in transgenic mice. Blood. 2004;103:2816–2821. doi: 10.1182/blood-2003-07-2524. [DOI] [PubMed] [Google Scholar]
- 10.Krijt J, Cmejla R, Sykora V, et al. Different expression pattern of hepcidin genes in the liver and pancreas of C57BL/6N and DBA/2N mice. J Hepatol. 2004;40:891–896. doi: 10.1016/j.jhep.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 11.Truksa J, Peng H, Lee P, Beutler E. Different regulatory elements are required for response of hepcidin to IL-6 and bone morphogenetic proteins BMP 4 and 9. Br J Haematol. 2007 doi: 10.1111/j.1365-2141.2007.06728.x. In Press. [DOI] [PubMed] [Google Scholar]
- 12.Courselaud B, Pigeon C, Inoue Y, et al. C/EBPalpha regulates hepatic transcription of hepcidin, an antimicrobial peptide and regulator of iron metabolism. Cross-talk between C/EBP pathway and iron metabolism. J Biol Chem. 2002;277:41163–41170. doi: 10.1074/jbc.M202653200. [DOI] [PubMed] [Google Scholar]
- 13.Bayele HK, McArdle H, Srai SKS. Cis and trans regulation of hepcidin expression by upstream stimulatory factor. Blood. 2006;108:4237–4245. doi: 10.1182/blood-2005-07-027037. [DOI] [PubMed] [Google Scholar]
- 14.Johnsen SA, Subramaniam M, Katagiri T, et al. Transcriptional regulation of Smad2 is required for enhancement of TGFbeta/Smad signaling by TGFbeta inducible early gene. J Cell Biochem. 2002;87:233–241. doi: 10.1002/jcb.10299. [DOI] [PubMed] [Google Scholar]
- 15.Johnsen SA, Subramaniam M, Janknecht R, Spelsberg TC. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene. 2002;21:5783–5790. doi: 10.1038/sj.onc.1205681. [DOI] [PubMed] [Google Scholar]
- 16.Subramaniam M, Harris SA, Oursler MJ, et al. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pietrangelo A, Dierssen U, Valli L, et al. STAT3 Is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterology. 2007;132:294–300. doi: 10.1053/j.gastro.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Verga Falzacappa MV, Vujic SM, Kessler R, et al. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109:353–358. doi: 10.1182/blood-2006-07-033969. [DOI] [PubMed] [Google Scholar]
- 19.Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood. 2006;108:3204–3209. doi: 10.1182/blood-2006-06-027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller AF, Harvey SA, Thies RS, Olson MS. Bone morphogenetic protein-9. An autocrine/paracrine cytokine in the liver. J Biol Chem. 2000;275:17937–17945. doi: 10.1074/jbc.275.24.17937. [DOI] [PubMed] [Google Scholar]