Abstract
This report demonstrates that introduction of physiologically relevant miRNAs can enhance somatic cell reprogramming. The mouse embryonic stem (ES) cell specific microRNAs (miRNA) miR-291-3p, miR-294, and miR-295 enhanced the efficiency of Klf4, Oct4 and Sox2 induced pluripotency. These miRNAs did not further enhance reprogramming in the presence of cMyc. cMyc binds the promoter of these miRNAs, suggesting that they are downstream effectors of cMyc promoted pluripotency. However, unlike exogenous cMyc, these miRNAs induced a homogeneous population of reprogrammed colonies suggesting overlapping and independent functions of cMyc and the miRNAs.
The miR-290 cluster constitutes over 70% of the entire miRNA population in mouse ES cells1. Its expression is rapidly down-regulated upon ES cell differentiation2. A subset of the miR-290 cluster, called the embryonic stem cell cycle (ESCC) regulating miRNAs, enhances the unique stem cell cell cycle3. This subset includes miR-291-3p, miR-294, and miR-295. To test whether ESCC miRNAs could promote the induction of pluripotency, we introduced these miRNAs along with retroviruses4 expressing Oct4, Sox2, and Klf4 (OSK) into mouse embryonic fibroblasts (MEFs). The MEFs carried two reporters: an Oct4-GFP reporter that activates GFP with the induction of pluripotency and ubiquitous expression of a β-galactosidase/neo fusion from the Rosa26 locus5. MiRNAs were introduced on days 0 and 6 post-infection by transfection of synthesized double-stranded RNAs that mimic their mature endogenous counterparts. This method transiently recapitulates ES-like levels of the miR-290 cluster miRNAs (Supplementary Fig. 1).
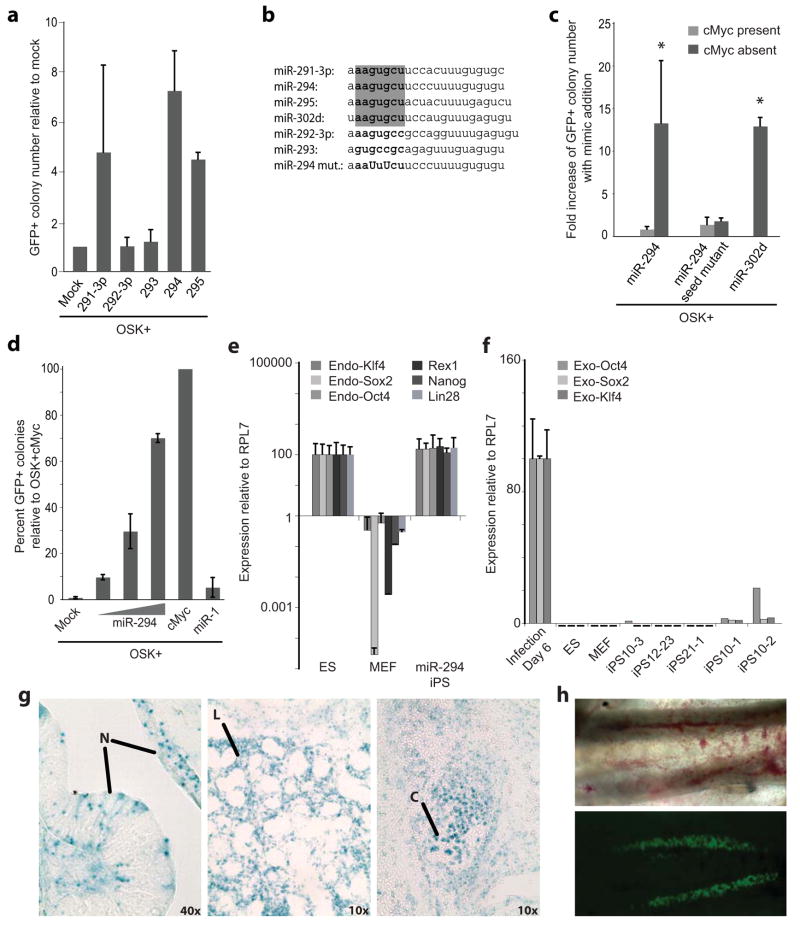
OSK plus miR-291-3p, miR-294, or miR-295 consistently increased the number of Oct4-GFP+ colonies as compared to controls transduced with OSK plus transfection reagent (Fig. 1a). The miR-294 mimic showed the greatest effects, increasing efficiency from 0.01–0.05% to 0.1–0.3% of transduced MEFs. Introduction of a chemically synthesized miR-294 pre-miRNA similarly enhanced reprogramming (Supplementary Figure 2). Two other members of the miR-290 cluster that are not ESCC miRNAs, miR-292-3p and miR-293, did not increase colony number (Fig. 1a). The ESCC miRNAs share a conserved seed sequence, which largely specifies target mRNAs (Fig. 1b). MiR-302d, a member of another miRNA cluster that has the same seed sequence also enhanced reprogramming (Fig. 1b&c). Mutation of the seed sequence in miR-294 blocked the increase in colony number (Fig. 1b&c). In summary, together with Oct4, Sox2, and Klf4, the ESCC miRNAs and related miRNAs with a common seed sequence promote the de-differentiation of fibroblasts into Oct4-GFP+ ES cell-like colonies.
Figure 1.
The ESCC miRNAs promote three-factor, but not four-factor induced pluripotency. (a) Fold increase of day 10 Oct-GFP+ colonies with retroviruses expressing Oct4, Sox2, and Klf4 (OSK) together with 16nM miRNA mimic relative to transfection reagent only (Mock). N=3. Raw data in Supplementary Table 1. (b) Sequence of miR-290 cluster, miR-302d, and miR-294 seed sequence mutant. Bold indicates seed sequence. Capitals indicate point mutations. Grey box highlights ESCC seed-sequence. (c) Fold increase in day 10 Oct4-GFP+ colonies with addition of mimic to OSK in the presence (light grey) or absence (dark grey) of cMyc retrovirus. Bars represent the number of GFP+ colonies after mimic transfection divided by the number of GFP+ colonies after mock transfection. N= 6, 26, 2, 5, & 3 left to right. Asterisk indicates p-value ≤ 0.0001. Raw data for bars 1&2 in Supplementary Table 2. (d) Percent day 10 Oct4-GFP colonies for OSK plus 1.6, 16 and 160nM transfected miR-294 mimic or 160nM miR-1 relative to OSKM. (e) Quantitative RT-PCR for endogenous pluripotency markers in control (V6.5) ES cells, MEFs, and miR-294-iPS lines. N=3, 3, & 5. RPL7 was used as input control. Data was normalized to ES expression. (f) Quantitative RT-PCR for exogenous Oct4, Sox2, and Klf4 in MEFs 6 days after viral infection, control (V6.5) ES cells, and MEFs (each N=3) and 5 individual miR-294-iPS lines. Horizontal black bars indicate Ct > 40. RPL7 was used as input control. Data was normalized to MEF expression 6 days after viral infection. (g) X-gal staining demonstrates miR-294-iPS chimeric contribution to ectoderm (neural tissue, N), endoderm (lung, L), and mesoderm (cartilage, C). (h) GFP expression in genital ridges of E12.5 chimera demonstrates Oct4-GFP miR-294-iPS contribution to germline. All error bars indicate standard deviation.
Consistent with previous observations that ESCC miRNAs act redundantly3, mixes of the different ESCC miRNAs did not further enhance reprogramming efficiency (Supplementary Fig. 3). Therefore, further studies focused on miR-294. Increasing doses of miR-294 further enhanced Oct4-GFP+ colony formation and the Oct4-GFP+ cellular fraction (Fig. 1d & Supplementary Fig. 4). At the highest doses, miR-294 increased the number of colonies to approximately 75 percent of that achieved with OSK and cMyc (OSKM) (0.4–0.7% of starting MEFs) (Fig. 1d). Addition of miR-294 mimic increased the kinetics of OSK reprogramming to rates comparable to OSKM reprogramming (Supplementary Fig. 5a). Transfection of miR-294 did not further enhance the reprogramming efficiency of any other three-factor combination or OSKM (Fig. 1c & Supplementary Fig. 5b). Therefore, miR-294 substituted for, but did not further enhance, cMyc’s contribution to reprogramming efficiency.
ES-like Oct4-GFP+ colonies induced by OSK and miR-294 (miR-294-iPS) were expanded and verified as induced pluripotent stem (iPS) cells. MiR-294-iPS lines expressed endogenous Oct4, Sox2, and Klf4, while retrovirus expression was silenced (Fig. 1e&f). Colonies showed an ES-like morphology and stained positively for the ES cell markers, Nanog and SSEA-1 (Supplementary Fig. 6a). The cell lines had normal karyotypes and efficiently induced teratoma formation with differentiation down all three germ layers (Supplementary Fig. 6b & Supplementary Fig. 7a–c). Injection of miR-294-iPS cells into blastocysts resulted in high-grade chimeras, with contribution of donor iPS cells to all three germ layers, and to germ line (Fig. 1g–h & Supplementary Fig. 6c).
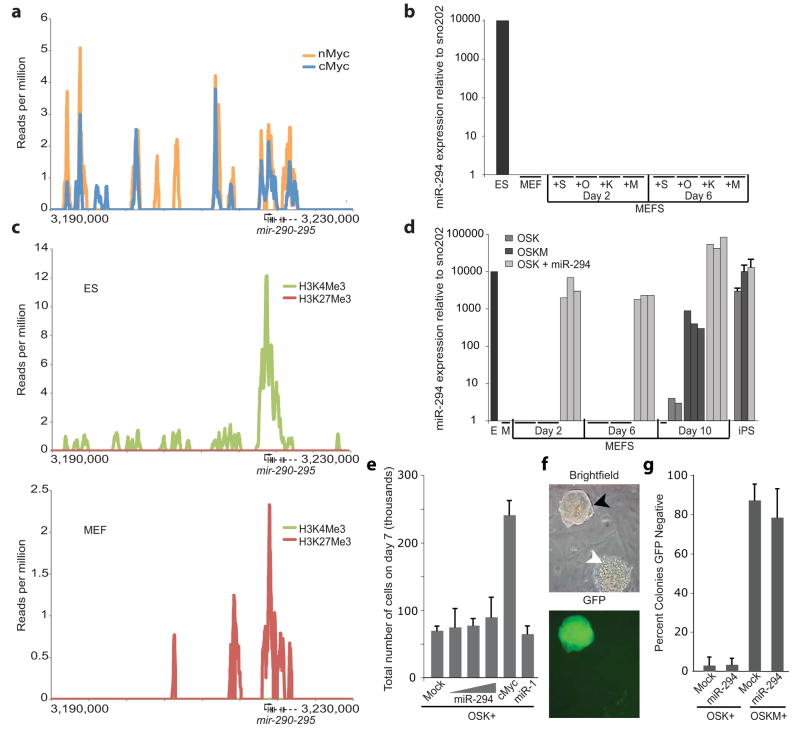
The mechanism for how ESCC miRNAs substitutes for cMyc in reprogramming is not entirely clear. However, bioinformatic analysis of ES ChIP-seq data6 showed that both c-Myc and n-Myc bind to the promoter region of the miR-290 cluster (Fig. 2a). Oct4, Sox2 and Nanog have also been reported to bind the promoter of the miR-290 cluster1. Transduction of cMyc, Oct4, Sox2, or Klf4 expressing retrovirus individually failed to induce expression of the miR-290 cluster in fibroblasts (Fig. 2b). Analysis of ChIP-seq data7 for different histone modifications (Fig. 2c) showed that the miR-290 promoter is H3K27 methylated in MEFs, a modification associated with transcriptional silencing. In contrast, the promoter is H3K4 methylated in ES cells, a modification associated with transcriptional activity. Therefore, these transcription factors likely can only induce the expression of the miR-290 cluster as cells replace promoter-associated H3K27 with H3K4 methylation during the reprogramming process. Indeed, with OSKM transduction, miR-294 was robustly activated late in the reprogramming process, similar to the reported timing for expression of endogenous Oct4, and other critical members of the core ES machinery (Fig. 2d)8, 9. These data suggest that miR-294 is downstream of cMyc, but requires epigenetic remodeling for expression.
Figure 2.
Characterization of the relationship between Myc and miR-294. (a) cMyc (blue) and nMyc (yellow) bind the miR-290 cluster promoter. ChIP-seq data reads6 were aligned to the mm9 assembly of the genome and peaks were generated with Findpeaks16. Vertical hash marks denote the positions of the miR-290 cluster miRNAs. (b) Quantitative RT-PCR for total mature miR-294 expression in control (V6.5) ES cells, MEFs, and MEFs infected with viruses expressing Sox2 (S), Oct4 (O), Klf4 (K) or cMyc (M). RNA was collected on days 2 and 6. N=3. Horizontal black bars indicate Ct > 40. Sno202 was used as input control. Data was normalized to ES cells. (c) H3K4me3 (green) and H3K27me3 (red) surrounding the miR-290 cluster in MEFs. Chip-seq data7 were analyzed as described in a. (d) Quantitative RT-PCR for total mature miR-294 expression in control (V6.5) ES cells (E), MEFs (M), MEFs infected with either Oct4, Sox2, Klf4 (OSK); Oct4, Sox2, Klf4, and cMyc (OSKM); or OSK+miR-294, and established iPS lines resulting from these conditions (iPS). RNA was collected on days 2, 6, and 10 of reprogramming. Three independent experiments are shown. Horizontal black bars indicate Ct > 40. Sno202 was used as input control. Data was normalized to ES cells. (e) Total cell number during reprogramming. Cells were counted on day 7 after infection with OSKM or OSK +/− miRNA mimic. Concentrations of miR-294 mimic: 1.6, 16 and 160nM. Concentration of miR-1 mimic: 160nM. (f) GFP negative colonies in presence of cMyc. Oct4-GFP+, ES-like colonies (black arrow) and GFP-negative, nonES-like colonies (white arrow). (g) Quantification of number of day 10 GFP-negative colonies after infection with OSKM or OSK +/− miR-294 mimic. All error bars indicate standard deviation of N=3.
The downstream effects of the ESCC miRNAs versus cMyc on the reprogramming process were not identical. Unlike cMyc, miR-294 did not promote proliferation of MEFs early in the reprogramming process (Fig. 2e). Furthermore, as previously reported, approximately 80% of the OSKM colonies failed to express GFP and lacked ES-like morphology10 (Fig. 2f&g). In contrast, OSK+miR-294 produced a predominantly uniform population of ES-like GFP+ colonies. The Oct4-GFP− colonies were induced by cMyc, not inhibited by miR-294, as the introduction of both produced a similar number of GFP−, non-ES-like colonies as cMyc alone (Fig. 2g). Finally, when cells were injected into immunodeficient mice to produce teratomas, more than a third of the teratomas resulting from cMyc-iPS cells invaded into the underlying body wall, while none of teratomas resulting from miR-294-iPS cells did so (Supplementary Fig. 7b&c). These findings show that while miR-294 can substitute for cMyc to enhance reprogramming, its effects on the cell population are not identical.
In summary, our data show that miRNAs can replace cMyc in promoting the de-differentiation of somatic cells into induced pluripotent stem cells. An exciting possibility is that other small RNAs could replace additional factors, which together with other approaches may eventually substitute for the use of introduced DNA elements. Additionally, further analysis of the targets of these miRNAs may offer valuable insights into the mechanism of reprogramming. The ESCC miRNAs are highly expressed in ES cells where they promote progression through the ES cell cell cycle, by accelerating the transition through the G1/S restriction point3. Their expression is downregulated with ES cell differentiation as the G1 phase of the cell cycle extends2, 11. ESCC miRNAs have also been shown to induce the expression of the de novo methyltransferases in ES cells, although how this may promote self-renewal is unclear12, 13. As a target of Myc, the miR-290 cluster likely acts downstream, but only after erasure of silencing histone modifications within its promoter. cMyc certainly has additional targets, reflected in the differences in outcomes between the introduction of cMyc and miR-294.
The ESCC miRNAs share a common seed sequence with a larger family of small RNAs known to promote cellular proliferation3. This family includes “onco-miRs”, such as members of the miR-17 cluster, miR-106, and miR-302 miRNAs14,15. These miRNAs, like the ESCC miRNAs, may be acting by enhancing cell cycle progression and promoting de-differentiation of the cells. Such parallels between induced de-differentiation and cancer will be an exciting area of future pursuit.
Supplementary Material
Acknowledgments
We would like to thank Deepa Subramanyam, Yangming Wang, and Kathryn Blaschke for technical advice and experimental suggestions as well as Marco Conti, Diana Laird, and members of the Blelloch Laboratory for critical reading of the manuscript. This work was supported by funds to RB from CIRM (RS1-00161) and NIH (K08 NS48118 and R01 NS057221). RJ is supported by a NSF Graduate Research Fellowship. MV is supported by a NIH training fellowship (F32NS058042 from the National Institutes of Neurological Disorders and Stroke). RB is a Pew Scholar.
Footnotes
Author Contributions: RLJ performed all experiments. JB analyzed ChIP-seq data. MV processed chimera tissue. RLJ and RB designed all experiments, analyzed data and wrote the manuscript.
Methods: See Supplemental Information.
References
- 1.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Blelloch R, Venere M, Yen J, Ramalho-Santos M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell. 2007;1:245–247. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 7.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brambrink T, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 11.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 12.Sinkkonen L, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 13.Benetti R, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 14.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voorhoeve PM, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 16.Fejes AP, et al. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.