Abstract
Accumulation and deposition of ß-amyloid peptide, a major constituent in neuritic plaques are hallmarks of Alzheimer’s disease (AD) and AD-related neurodegenerative diseases. ß-Amyloid (Aß) is derived from the proteolytic cleavage of amyloid precursor protein (APP), a transmembrane protein present in three major isoforms in brain comprising 695, 751 and 770 amino acids, respectively. Among other post-translational modifications, APP is modified during maturation by N- and O- glycosylation, which are thought to be responsible for its expression and secretion. Unlike N-glycosylation, no sites of O-glycosylation of APP have previously been reported. We report here the identification of three specific O-glycosylation sites of the secreted APP695 (sAPP695) produced in CHO cells, using a combination of high performance liquid chromatography and electrospray - tandem mass spectrometry. With the use of electron transfer dissociation and collision induced dissociation (ETD and CID), we identified type, composition and structures of the Core 1 type O-linked glycans attached at the residues: Thr 291, Thr 292 and Thr 576 of the full length APP695. The glycosylations comprise multiple short glycans, containing N-acetyl galactosamine (GalNAc), Gal-GalNAc and sialic acid terminated structures. The presence of the glycopeptides in the tryptic mixture was identified using the CID-generated sugar oxonium ions. ETD proved to be valuable for the unambiguous identification of the modified sites as ETD fragmentation occurred along the peptide backbone with little or no cleavage of the glycans. Thus, the combination of the CID and ETD techniques in LC-MS is shown here, as a powerful tool for de novo identification of O-glycosylations at unknown modification sites in proteins.
Keywords: ß-Amyloid, amyloid-precursor protein, Alzheimer’s disease, O-glycosylation, tandem-mass spectrometry, electron transfer-dissociation, collision-induced dissociation
Introduction
The amyloid precursor protein (APP) is a transmembrane protein that plays major roles in the nervous system where it is involved in synaptogenesis and synaptic plasticity. There are three major isoforms of APP, containing 695-, 751- and 770 amino acid residues (APP695, APP751 and APP770, respectively) 1, derived from alternative splicing of the mRNA of a single gene located on chromosome 21 2. In nerve cells, the predominantly expressed isoform is APP695, while isoforms APP751 and APP770 predominate in the other cell types 3. The secreted extracellular domain of APP derived from the non-amyloidogenic processing pathway, sAPPα 4, 5, acts as a growth factor for many cell types and promotes neuritogenesis in post-mitotic neurons 1. Interestingly, APP was not initially discovered for its role in the healthy brain, but rather because of the characteristic deposition of ß-amyloid containing senile plaques in AD. ß-amyloid, the major component of amyloid plaques, is a neurotoxic product released enzymatically from APP by ß- and γ-secretases during amyloidogenic processing 6, 7. The deposition of neuritic plaques containing the transmembrane Aß fragment is a hallmark of Alzheimer’s disease (AD) 8, 9.
During transit through the intracellular protein secretory pathway, APP is modified by N- and O-glycosylation, phosphorylation and tyrosine sulfation 10-12. After loss of the signal sequence and cleavage by α - secretase, the extracellular portion of the neuronal isoform of APP (sAPPα) contains 594 amino acid residues and the non-posttranslationally modified protein has an expected molecular weight of 68 kDa. There are two consensus sites for N-glycosylation, N467 and N496, (numbering as in full length APP695), of which only N467 has been shown to be modified with complex type glycans 13, 14 (see Figure 1). It has been reported that N- and O- glycosylation of the extracellular portion of APP are prerequisites for phosphorylation of Thr 668 of the cytoplasmic domain during neuronal differentiation 15. Mutants defective in O-glycosylation also have altered cellular metabolism compared to wild type APP695 16. To our knowledge, no O-glycosylation sites have been reported for APP, although the types of glycans from the total O-glycosylation pool of APP695 expressed in CHO cells have been previously reported 14. Identification of sites of O-glycosylation for APP may lead to a better understanding of the role this post-translational event plays in the cellular metabolism of the amyloid precursor.
Figure 1.
Schematic representation of the non-amyloidogenic cleavage pathway of the transmembrane human APP695 generating the secreted fragment sAPPα and the C-terminal fragment CTFα; the amino acid sequence of sAPPα, residues 18 - 612, is depicted at the bottom. The N-glycosylation sites are highlighted in grey. The identified O-glycosylation sites are indicated in purple. The peptides derived from the use of different enzymes observed by LC-MS/MS are underlined.
Mass spectrometry (MS) using matrix-assisted laser desorption/ionization (MALDI) and/or electrospray ionization (ESI) 17, 18, is increasingly used in glycobiology for oligosaccharide and glycopeptide analysis. Furthermore, the complementary mass spectrometric fragmentation techniques, collision - induced dissociation (CID) and electron capture dissociation (ECD) 19, 20, or electron transfer dissociation (ETD) 21, 22, have tremendously expanded the analytical options in glycoproteomics. CID tandem mass spectra (MS/MS) of glycopeptides contain mainly fragments arising from glycosydic bond cleavage, and, thus, the structure of the glycan is directly attainable 23, 24. In the ETD process, an electron from a singly charged radical anion (e.g. gaseous fluoranthene anion) is transferred to a multiply charged peptide/glycopeptide ion. The resulting charge reduced odd electron species undergo, in a manner similar to ECD, predominant cleavage of the N - Cα bonds generating c’ and z· fragment ions (nomenclature of Zubarev and co-workers 25, 26). The intact oligosaccharide moieties are retained in the fragment ions containing the site of glycosylation. Consequently, ECD and ETD represent excellent tools for localization of sites of modification in glycopeptides and other post-translationally modified proteins 27-33. Despite the advantages provided by ETD, there have been only a few reports in which this technique has been used in the characterization of N- 34, 35 and O-glycopeptides 28, 33.
In the present study the O-glycopeptides of APP695 secreted from CHO cells were analyzed by liquid chromatography in combination with data dependent ETD and CID using an ion trap instrument. Three threonine residues, Thr 291, Thr 292 and Thr 576, were found to be modified with Core 1 type glycans, containing N-acetyl galactosamine (GalNAc), Gal - GalNAc and sialic acid terminated structures. Whereas information about the composition of the attached glycan(s) was extracted from the CID experiments, the identity of the glycosylated sites was unambiguously determined from ETD, which revealed characteristic c’ and z· fragment ions retaining the intact O-glycan. In addition, radical loss of 219 Da, corresponding to ·OGalNAc, from the glycosylated threonine side chain was observed. Here, we show that ETD and CID represent a powerful combination for the identification of unknown O-glycosylation sites in proteins as demonstrated by the results of our study of APP695 glycosylation.
Experimental Procedures
Materials
Mouse anti-human β-amyloid precursor protein (aa695) monoclonal antibody (clone 6A6) was purchased from US Biological (Swampscott, MA). Dithiothreitol, ammonium bicarbonate, and 96% formic acid were purchased from Sigma-Aldrich (St. Louis, MO). Sequencing grade-modified porcine trypsin was obtained from Promega (Madison, WI) and sequencing grade bovine α-chymotrypsin was obtained from Roche (Penzberg, Germany). WesternBreeze® chemiluminescent Western blot immunodetection kit, NuPage 4 - 12 % Bis-Tris pre-cast gels, sample and running buffers and Coomasie SimplyBlue were purchased from Invitrogen (Carlsbad, CA). Acetonitrile was purchased from Caledon Laboratories, Ltd. (Georgetown, Ontario). Purified water (17.8 MΩ) was obtained from an in-house Hydro Picopure 2 system. All chemicals were used without further purification unless otherwise specified.
Methods
Cell Culture
CHO-s cells were obtained from Invitrogen. The cells were grown serum free in suspension culture using CD-CHO (GIBCO) containing HT supplement (Gibco) and 8 mM glutamine.
Protein Production
The APP695 protein was produced using the pHD-APP695 plasmid, containing the APP695 gene under the control of the HSV promotor with the SV40 enhancer sequence 36. CHO-s cells were transiently transfected with the pHD-APP695 plasmid using Fugene HD (Roche). A ratio of 4 uL of Fugene HD per 1 ug of plasmid gave the best results. The APP695 protein was allowed to accumulate in the media for 48 to 72 hrs post-transfection before the media was harvested.
Q-Sepharose chromatography
The media containing the APP695 protein was centrifuged at 350 x g for 10 min to remove all of the cells. The media was further clarified by passage through a 0.2 um Acrodisk syringe filter before being flow loaded onto a 10 mm by 20 cm Q sepharose FF (Sigma) column at 1mL/min. The column had been pre-equilibrated with 25 mM HEPES pH 7.5 buffer containing 150 mM NaCl. After the sample was loaded, the column was washed with 3 column volumes of the pre-equilibration buffer. APP695 was eluted from the Q sepharose column using a 10 column volume linear gradient from 150 mM NaCl to 1 M NaCl in 25 mM HEPES pH 7.5. The load flow through and column wash were collected in batch, while the elution was collected in 2 mL fractions. The fractions that contained APP695 were identified by SDS-PAGE and western blot. The chromatography was performed at 4°C using an AKTA purifier (GE Healthcare).
SDS-PAGE and Western blot analysis
The protein peak eluted from the Q-sepharose column was separated on 4-12% Bis-Tris pre-cast gels as follows: 30 μL of each fraction were incubated for 10 minutes at 90° C with 10 μL sample buffer containing 100 mM dithiothreitol. Electrophoresis was performed at 200 V and a maximum of 100 mA, for 90 minutes. The bands were stained over night with Coomasie Simply Blue. For Western blot analysis, the proteins separated by SDS-PAGE were transferred onto nitrocellulose membranes for 90 minutes at 25 V and 125 mA. The membrane was rinsed with deionized water and blocked for 30 minutes in the blocking solution prepared from the reagents provided with the WesternBreeze® kit. Subsequently, the membrane was submersed for one hour in a solution containing 1 μg/mL of the mouse anti-ß-APP695 monoclonal antibody (primary antibody). Following washing, the membrane was incubated for 30 minutes with a solution containing the anti-mouse, alkaline phosphatase - conjugated, secondary antibody. The chemiluminiscent substrate was added to the membrane surface and the reaction was allowed to develop for 5 minutes before the membrane was exposed to the X-ray film for 10 seconds.
In-gel digestion
The protein band corresponding to secreted APP695 was manually cut and digested with trypsin for 8 hours at 37° C in an automated fashion with a Progest robotic digester (Genomic Solutions). In-gel digestion with α-chymotrypsin was performed manually; 4 μL of a 0.25 μg/μL stock solution of α- chymotrypsin in 1 mM HCl were diluted to 50 μL with 25 mM ammonium bicarbonate and subsequently added to the destained protein band. Digestion was performed over night at 25° C Samples were lyophilized to dryness and resuspended in 0.1% formic acid.
Mass Spectrometry
LC/ESI/MS/MS analyses were performed using an Agilent 6340 (Santa Clara, CA) Ion Trap equipped with an HPLC Chip Cube MS interface, an Agilent 1200 nanoHPLC and an electron transfer dissociation module. The solvents used for chromatography were 0.1% formic acid in deionized water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). Ion trap-MS/MS analyses were performed as follows: 20 μL injections of the tryptic or chymotryptic digests dissolved in 0.1% formic acid were loaded onto a 40 nL enrichment column followed by a 43 mm × 75 μm analytical column, packed with ZORBAX 300SB C18 particles. Linear gradients of 3-50% solvent B were performed over 50 min at a flow rate of 500 nL/min. The parameter settings for positive ion ESI-MS were as follows: capillary voltage - 2000 V, end plate offset - 500 V, capillary exit - 180 V, nebulizer - 2 psi, dry gas - 4 L/min, dry gas temperature - 325 °C. For MS/MS (ETD or CID), automated data dependent acquisitions of the four most abundant ions were employed. For CID, the fragmentation amplitude was 0.80 V, which was scanned from 30% to 200% of this preset value (SmartFrag parameter on the instrument tune page). For ETD analyses, the accumulation time of gaseous fluoranthene anions was 10 - 12 msec, and the reaction time was typically 100 or 150 ms. The ETD/CID voltage was set at 0.07 - 0.10 V when supplemental ion activation of the charged reduced molecule radical ions was employed.
Database search
Prior to database searching, data were processed (including ions with a S/N ratio greater than 3:1) using Mascot Distiller software (Matrix Science, UK) and saved as an *.mgf file. These data were searched against the NCBInr protein data base by means of the Mascot MS/MS Ion Search engine, using a precursor tolerance of 0.2 Da and an MS/MS tolerance of 0.1 Da. The sequences determined from the MS/MS data obtained for the identified peptides were validated manually.
Results and Discussion
Expression and purification of human APP695
The amyloid precursor protein APP695 was expressed in CHO cells following the procedure described by Sato et al. 14; however, no fetal calf serum was supplemented to the cell culture medium, which greatly simplified the purification procedure. The cell culture medium was subjected to Q-sepharose chromatography, using a linear NaCl gradient 14 and a single peak containing the APP695 eluted from the column. We observed an identical binding efficiency of APP695 to the Q-sepharose column as previously reported by Sato et al. 14. APP695 was detected by Western blot analysis in the cell culture medium and in the fraction eluted from the Q-sepharose column as a band migrating at approximately 75 to 105 kDa on SDS-PAGE. The identity of the protein was confirmed by LC-MS/MS analysis of the tryptic digest and database search using the NCBInr protein database. Despite the co-elution of multiple proteins from the Q-sepharose column, the SDS-PAGE separation of the protein band of interest was satisfactory, so that no additional purification step was required. The secreted (unmodified) APP695 sequence has an expected molecular weight of approximately 68 kDa, and the difference between the theoretical and the apparent molecular weight observed on SDS-PAGE is largely ascribed to glycosylation.
Identification and characterization of O-linked glycosylation
The O-glycopeptides from the secreted human APP695 (sAPP695) were identified by nano-LC-ESI-MS/MS analysis of a tryptic digest of the corresponding protein band separated by SDS-PAGE, using data dependent CID acquisition. The glycopeptides containing O-linked sugars were characterized at elution times between 19.5 and 24 minutes by the presence of characteristic sugar-oxonium ions under CID-MS/MS conditions (m/z 292.1, protonated neuraminic acid (SA+); m/z 274.1, protonated dehydrated neuraminic acid (SA+ - H2O); m/z 204.1, protonated N-acetyl hexosamine (HexNAc+); m/z 366.1, protonated hexose-N-acetyl hexosamine (HexHexNAc+)). These ions arise during CID of the protonated glycopeptides, by glycosydic bond cleavages of the sugar moiety from its non-reducing end (Y/B-type fragmentation 37), including complete loss of the oligosaccharide 38. The appearance of ions corresponding to the non-glycosylated tryptic peptides provided initial information of the peptide’s identity. Two independent MS/MS analyses of the tryptic peptides were carried out under identical chromatographic conditions: (i), first, data dependent ETD was performed and, (ii) second, data dependent CID spectra were acquired. In this manner, the amino acid sequences of the peptides containing the glycans, and the location of the O-linked sites were elucidated. No consensus sequence for O-glycosylations has been defined at present. In addition, a significant challenge for the “de novo” identification of the O-linked glycopeptides and their glycosylation sites was due to the primary structure of full length human APP695 which contains 30 serine and 45 threonine residues, of which 27 serine and 39 threonine residues are contained in the secreted form, sAPPα (see Figure 1). A total of 15 serine and 26 threonine residues were observed in the tryptic and chymotryptic digests of sAPP695 by LC-MS/MS analysis. Sequence coverage obtained by trypsin and α-chymotrypsin digestion, and the combination of both proteases is highlighted in Figure 1.
ETD and CID tandem mass spectrometric identification of O-glycosylations
Two peptides with potential O-glycosylation were identified in the LC/MS analyses at the partial sequences, (289-302) and (574-587) (see Figure 1). Three threonine residues, Thr 291, Thr 292 and Thr 296, and one serine, Ser 295, are located in the sequence (289 - 302), while three threonine residues, Thr 576, Thr 577 and Thr 584, and one serine residue, Ser 581, are located in peptide (574 - 587) (numbering as in the full length APP695). The CID and ETD fragmentation of these peptides were interrogated in detail to identify the sites and composition of the glycans.
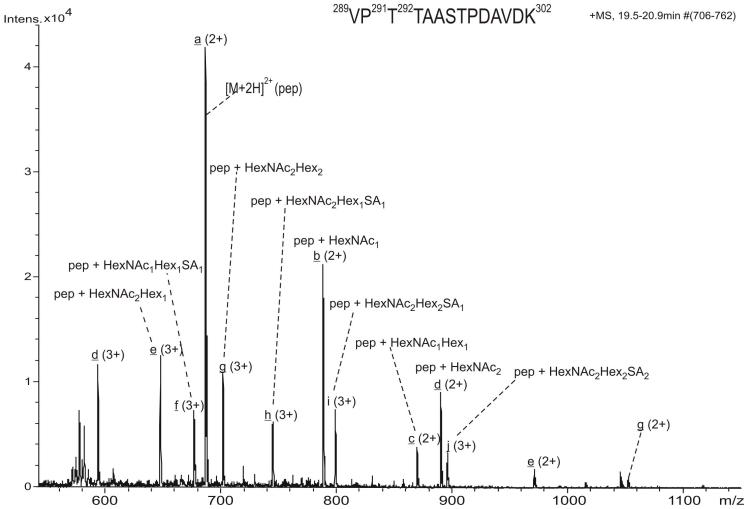
The averaged ESI-MS spectrum over the chromatographic retention time 19.5 - 20.9 min (Figure 2) showed the population of the glycopeptides within the sequence (289 - 302), and the heterogeneity resulting from the attachment of distinct glycans. We can not exclude that a low extent of in source fragmentation may occur under our MS conditions. However, the extracted ion currents of each individual glycoform are centered at distinct chromatographic elution times, suggesting that the observed heterogeneity is of biological nature, and not artifactual. In the discussion of our results, we refer to the observed relative abundance of each individual glycoform, not corrected for differences in ionization efficiencies resulting from the attachment of distinct glycans. In addition to the glycosylated forms, the unmodified peptide (289 - 302) was detected as the doubly-protonated molecule of m/z 686.6 and represented the most abundant species. The sugar compositions of the glycopeptides, determined from the molecular weight difference of each individual glycopeptide compared to the non-glycosylated peptide, are indicated in Figure 2. The subsequent analysis of the CID and ETD spectra of the ions provided (i), the determination of the composition of the oligosaccharide modification for each glycopeptide from the CID tandem mass spectra, and (ii) the identification of the site(s) of modification from the ETD MS/MS.
Figure 2.
Positive ion scan ESI mass spectrum summed over the chromatographic retention time 19.5 - 20.9 min, indicating the major glycoforms of the peptide 289-302 of the full length APP695. The individual glycopeptides are highlighted with letters from a through j. The charge states and the composition of the glycans determined for every glycopeptide are indicated for each glycoform.
The most abundant glycoform of peptide (289 - 302), observed as both doubly (m/z 788.2) and triply charged (m/z 525.7) ions, was identified as containing a single N-acetyl hexosamine unit. This monosaccharide is most likely assigned to N-acetyl galactosamine (GalNAc), because the biosynthesis of O-glycans in vertebrates is initiated by the attachment of α-linked GalNAc to Ser/Thr residues 39. In the CID spectrum of the ion of m/z 788.2 (2+), loss of the GalNAc residue and charge reduction of the precursor give rise to the base peak, m/z 1372.3, representing the singly charged unmodified peptide ion (data not shown). The GalNAc attachment site was unambiguously determined from the ETD of the parent ion m/z 788.2 (2+) (Figure 3A). The base peak in this spectrum corresponds to the charge reduced species of m/z 1575.4, the singly protonated, even-electron glycopeptide (Figure 3B). This species may arise either from the transfer of a proton from the 2+ precursor to the fluoranthene (radical) anion or by neutral loss of a hydrogen radical from the charge-reduced radical molecular ion [M+2H]+·. The 1 Da heavier isotopomer (peak) (m/z 1576.4), having a similar abundance as the 12C isotopomer, represents a mixture of the 13C isotope of the [M+H]+ species and of the 12C isotope of the [M+2H]+· odd-electron species arising from the transfer of a single electron to the precursor ion.
Figure 3.
(A.) ETD of the precursor ion m/z 788.2 (2+), corresponding to the peptide 289-302 of the full length APP695 containing the monosaccharide N-acetyl galactosamine (yellow rectangle) attached at Thr 292. The red label indicates the radical loss of 219Da (·OGalNAc) from the radical species [M+2H]+· and z11·, respectively. The fragment ions relevant for determination of the glycosylation site are indicated with black boxes. The inset in this spectrum shows the peak distribution of the species z7, z10 and z12 (see discussion in text). The spectrum was obtained using data dependent acquisition without supplemental ion activation. (B) Inset over the mass range m/z 1510 - 1580, showing the small neutral losses derived from the molecule radical ion of m/z 1576.4.
Abundant fragmentation along the peptide backbone resulted in formation of z· and c’ species, which covered almost the complete sequence of the peptide (289 - 302) (Figure 3A). Based on the observed c’ and z· ions, in particular z10· (m/z 958.3) and z11· (m/z 1262.4), which are separated by the mass increment of the glycosylated threonine residue, it was possible to unambiguously assign Thr 292 as the O-glycosylation site, and to rule out other possible modification sites. A number of ions was observed to result from hydrogen rearrangement to and from z· fragments, as demonstrated previously in ECD experiments 40, 41. Specifically, the fragment z7· (m/z 729.2), resulting from N-Cα cleavage between the Ser and Thr residues, showed pronounced hydrogen rearrangement with formation of both species z7′ (1 Da heavier than z7·) and [z7· - 1H·]. In contrast, radical migration to the fragment z10· (m/z 958.3) results exclusively in formation of the abundant even-electron species z10′ at m/z 959.3 (see insert in Figure 3A). Similarly, one hydrogen radical is transferred to the fragment z12· (m/z 1363.3) to form the abundant z12′. According to Savitski et al., the extent of H· rearrangement is dependent on the nature of the residues adjacent to the N-Cα bond 42, with Thr adjacent to the radical site promoting both H· addition and/or H· loss. In agreement with this observation, pronounced H· transfer to z7·, z10· and z12· was observed, whereas H· loss was abundant only for the z7· species.
The presence of N-acetyl hexosamine at Thr 292 was found to have a significant effect on H· transfer/abstraction to/from the adjacent z· radical ions and this effect is currently under investigation. A comparison of the ETD spectra of the precursor ions of m/z 788.2 (2+) and m/z 686.7 (2+), corresponding to the peptide modified with HexNAc at Thr 292, and to the non-modified peptide (289-302) respectively, are shown in the Supplemental material. The presence of HexNAc at Thr 292 inhibits H· transfer to the radical ion z9· (m/z 887.2) (cleavage between Ala 293 and Ala 294) compared to that observed in the ETD spectrum of the non-modified peptide. For the fragmentation leading to formation of the z10· radical ion (m/z 958.2), by cleavage C-terminal to the glycosylation site, the H· transfer appears to be promoted when Thr 292 is glycosylated. In addition, a species corresponding to the abstraction of one H· was observed at m/z 957.2 in the ETD spectrum of the non-glycosylated peptide; however, in the absence of the accurate mass, this species may be assigned as H· abstraction from z10· and/or c10, as these species are isobaric (m/z 958.1) (see Supplemental material). The major cleavage C-terminal to Pro 290, in the non-glycosylated peptide ion, is the z12· fragment (m/z 1160.2), but enhanced H· subtraction was also observed leading to the ion of m/z 1159.2. The latter process was not significant for the z12· species in the ETD spectrum of the glycosylated peptide (m/z 1363.3).
Small neutral losses were observed from the diprotonated molecular radical cation [M+2H]+·, m/z 1576.4 (see Figure 3B), with loss of water (18 Da) and ·OH (17 Da) probably from the Ser/Thr side chain representing the most abundant processes. The loss of 43 Da may be explained either by loss of CH3HC·CH3 from the side chain of valine or by loss of CH3C·O from the N-acetyl galactosamine unit, while the losses of 44 and 45 Da may arise from neutral loss of CO2 and radical loss ·CHO2, respectively, from the Asp side chain. Loss of 60 Da, giving rise to the ion m/z 1516.4, may be explained as simultaneous loss of 43 and 17 Da or as loss of CH3COOH from the aspartic acid side chain. As a result of the limited mass accuracy of the ion trap, it was not possible to unambiguously assign the identity of these neutral losses. Such side chain losses have recently been reported in the ETD spectra of N-glycopeptides 43, and previously for the ECD of O-glycopeptides 30.
Interestingly, radical losses of ·O-GalNAc (219 Da) from the radical species [M+2H]+· and z11· were observed (see Figure 3A and the bottom spectrum in the insert in Figure 3A). Similar observations have been reported by Catalina, et al., who described the loss of the complete glycosylated Asn-side chain from a tryptic N-glycosylated peptide under ETD conditions43, and by Mormann, et al., for O-glycosylated peptides under ECD conditions 30. In ECD, side chain losses result from radical transfer from the initially formed C-terminal z· fragment, followed by elimination of radical species from the side chain and subsequent formation of a double bond between the Cα and Cß atoms of the residue 44. Most likely, a similar mechanism based on radical migration or ·H transfer is responsible for loss of ·O-GalNAc observed from the charge reduced molecular radical cation. In addition, neutral loss of GalNAc (203 Da) from both [M+H]+ and [M+2H]+· ions was observed (see insert in Figure 3A).
Figure 4 shows the ETD spectrum of the precursor ion m/z 676.8 (3+), corresponding to the glycopeptide (289 - 302) modified at Thr 292 with a Core 1 type glycan terminated by sialic acid (SA), having the structure: GalNAc - Gal - SA. The linear structure of this O-glycan was derived from the CID spectrum of the precursor ion (not shown) where the sugar oxonium ion, m/z 454.1 (Gal - SA+), indicates that SA and Gal are interconnected. This glycoform was less abundant than that modified with O-GalNAc (see Figure 2). As described above, neutral losses of 17, 43 and 60 Da were observed from the charge reduced odd-electron species m/z 1015.3, assigned as [M+3H]2+·. The series of c’ and z· ions, in particular c3 (m/z 315.0) and c4 (m/z 1072.22), demonstrated that the O-glycosylation site is located at Thr 292. The fragment ions containing the modified Thr 292 were found to retain the intact glycan, despite the instability of sialic acid containing sugars. Loss of sialic acid by glycosydic bond cleavage as a result of ETD was minimal and was observed as the ion of m/z 1737.5, corresponding to [M+H]+-SA. Although the spectrum shown in Figure 4 was obtained only by ETD acquisition, pronounced loss of sialic acid was observed upon additional ion activation (typically 0.07 - 0.1 V for these experiments), which induced dissociation of the charge reduced odd-electron species indicating the labile character of this monosaccharide.
Figure 4.
ETD of the precursor ion m/z 676.8 (3+), glycopeptide 289-302 of the full length APP695, showing the Core 1 type trisaccharide attached at Thr 292. The spectrum was obtained performing targeted MS/MS of the ion m/z 676.8 (3+), without supplemental ion activation. Color code: yellow - N-acetyl galactosamine, blue - galactose, purple - sialic acid. The fragment ions relevant for determination of the glycosylation site are indicated with black boxes.
An additional glycoform of the peptide (289 - 302) containing GalNAc - Gal at Thr 292 was observed as a doubly protonated molecule of m/z 869.1 (ion c, Figure 2). The location of the disaccharide on this peptide was established using ETD (data not shown). The abundances of this glycoform and of the sialic acid-terminated form were comparable and significantly lower than the O-GalNAc modified peptide (ion b, Figure 2).
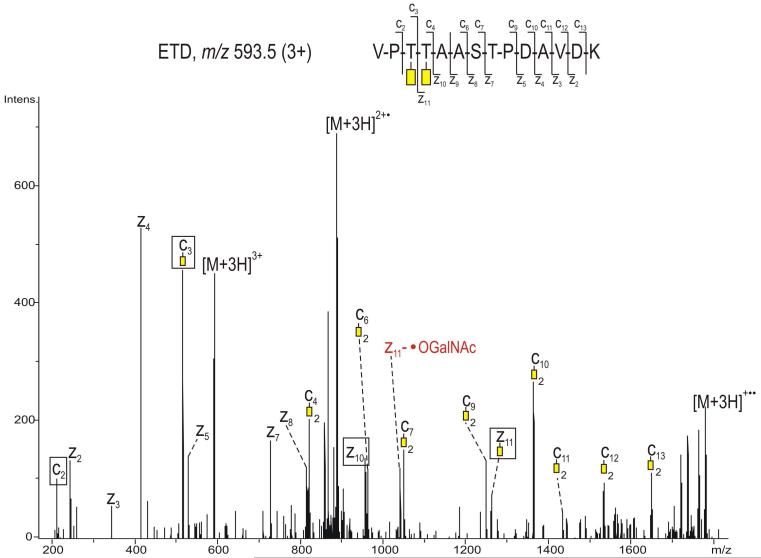
Furthermore, an additional set of six different glycopeptides, with the amino acid sequence (289 - 302), were found glycosylated at both Thr 291 and Thr 292. Among these, the glycoform containing one GalNAc at each site was the most abundant, albeit its abundance includes both the doubly (m/z 889.7) and the triply protonated species (m/z 593.5). The composition of the glycan attached to the peptide was derived from the CID data as GalNAc2 (not shown), in which the base peak ion (m/z 788.2, 2+) arises from the loss of a single GalNAc unit. One could argue that the N-acetyl hexosamine units are interconnected and attached to a single side chain; however from these data no information upon the connectivity of the sugar moieties was obtained. The ETD spectrum of the ion of m/z 593.5 (3+) is shown in Figure 5. A nearly complete series of c’ and z· ions were observed in addition to the small mass neutral losses from the charged reduced molecule ion. The fragment ions c2 (m/z 214.0), c3 (m/z 518.1) and c4 (m/z 822.2), as well as z10· (m/z 958.1) and z11· (m/z 1262.1) indicate that Thr 291 and Thr 292 are each modified by GalNAc, because the mass difference between each of these corresponds to the mass of a threonine residue modified with N-acetyl hexosamine. In addition, the ion of m/z 1043.1 was assigned as arising from the loss of the ·O-GalNAc radical (219 Da) from the z11· radical ion (m/z 1262.1). In this spectrum, the loss of ·O-GalNAc from the charge reduced radical species was minimal compared to that observed in the ETD spectrum of the precursor ion m/z 788.2 (2+) (Figure 3).
Figure 5.
ETD spectrum of the precursor ion m/z 593.5 (3+), corresponding to glycopeptide 289-302 of the full length APP695, showing each of the amino acids Thr 291 and Thr 292 occupied with N-acetyl galactosamine (yellow rectangle). The radical loss of 219 Da (·OGalNAc) from the z11 ion is indicated in red. The spectrum was obtained using data dependent acquisition without supplemental ion activation. The fragment ions relevant for determination of the glycosylation site(s) are indicated with black boxes.
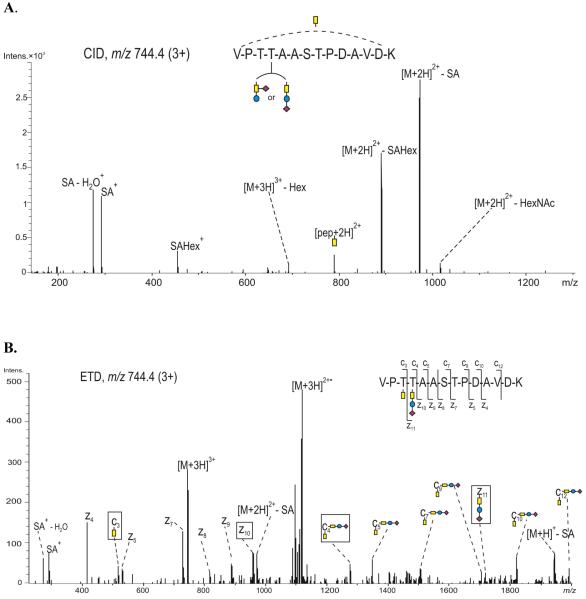
The CID and ETD mass spectra of the triply protonated precursor ion m/z 744.4, assigned to the glycopeptide (289 - 302) are presented in Figure 6A and 6B, respectively. The fragment ions observed in the CID spectrum (Figure 6A) are largely derived from processing of the glycan from the non-reducing end; from this, the composition of the glycan was derived as HexNAc2Hex1SA1, which may be assigned to either a Core 2 type glycan, containing GalNAc, GlcNAc, Gal and SA 39, or to two distinct glycans, GalNAc1, and GalNAc1Gal1SA1, respectively. The loss of HexNAc from the precursor ion with retention of the remaining monosaccharides indicates that the HexNAc is terminally linked, which may result from decomposition of either a Core 2 type glycan or a glycan containing a single GalNAc. In addition, primary losses of terminal sialic acid and hexose from the precursor ion indicate that these moieties represent non-reducing ends. This rules out the Core 2 type glycan, because elongation of the GalNAc with three branches has not been reported in vertebrates. This might also indicate a branched Core 1 type glycan, possibly GalNAc (SA) - Gal. However, the oxonium ion m/z 454.1 (SAHex+) indicates a linear structure for the glycan GalNAc - Gal - SA. In summary, the CID data are only consistent with two residues in the peptide (289 - 302) being modified by the O-glycans GalNAc and a mixture of the isomers GalNAc (SA) - Gal and GalNAc - Gal - SA. The glycosylated residues in this sequence (289 - 302) were unambiguously identified from the ETD spectrum (Figure 6B). The ion c3 (m/z 518.1) indicates that the GalNAc unit is attached at Thr 291, while c4 (m/z 1275.3) and z11· (m/z 1715.3) indicate that Thr 292 is occupied with the glycan GalNAc1Gal1SA1. The ions z7· (m/z 729.1) and z8· (m/z 816.2) suggest that Ser 295 and Thr 296 are not modified. The loss of terminal sialic acid was also observed (Figure 6B) as a result of the additional ion activation (0.07 V) applied to improve the efficiency of ETD.
Figure 6.
(A) CID, and (B) ETD spectrum, of the precursor ion m/z 744.4 (3+), corresponding to glycopeptide 289-302 of the full length APP695, showing each of the amino acids Thr 291 and Thr 292 occupied with N-acetyl galactosamine and a Core 1 type trisaccharide, respectively. The spectrum was obtained using data dependent acquisition with a supplemental ion activation of 0.07 V. Color code: yellow - N-acetyl galactosamine, blue - galactose and purple - sialic acid. The fragment ions relevant for determination of the glycosylation site(s) are indicated with black boxes.
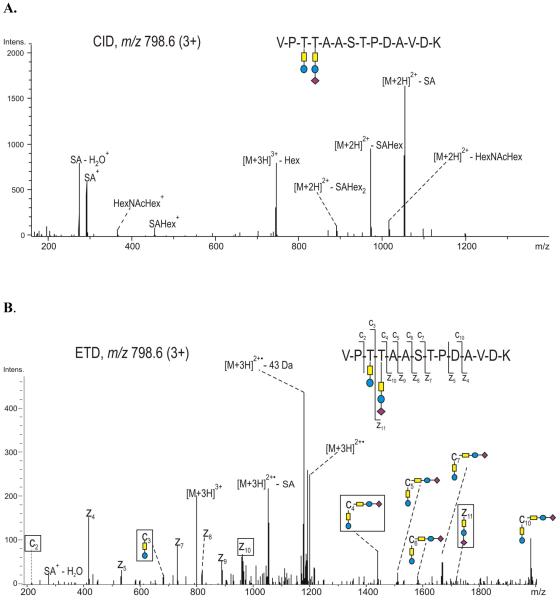
An additional example of the glycopeptide (289 - 302), modified at Thr 291 and Thr 292 with GalNAc - Gal and GalNAc1Gal1SA1 respectively, is presented in Figures 7A and 7B. As described above, the composition of the oligosaccharide contained on the peptide (289 - 302), determined as HexNAc2Hex2SA, was derived from the CID spectrum (Figure 7A). The spectrum indicates terminal sialic acid and terminal hexose residues, which may be assigned either to two distinct Core 1 type glycans (GalNAc1Gal1 and GalNAc1Gal1SA1), or to a Core 2 glycan (GalNAc (GlcNAc-Gal) Gal - SA). On the basis of the sequence ions observed in the ETD spectrum (Figure 7B), the modification sites were assigned to Thr 291 modified with GalNAc - Gal, and to Thr 292 modified with GalNAc - Gal - SA.
Figure 7.
(A) CID and (B) ETD spectra of the precursor ion m/z 798.6 (3+) corresponding to glycopeptide 289-302 of the full length APP695, showing the amino acids Thr 291 and Thr 292 occupied with two distinct Core 1 type glycans. The spectrum was obtained using data dependent acquisition and the activation energy was 0.07 V. Color code: yellow - N-acetyl galactosamine, blue - galactose, purple - sialic acid. The fragment ions relevant for determination of the glycosylation site(s) are indicated with black boxes.
In summary, the peptide 289VPTTAASTPDAVDK302 contains four potential O-glycosylation sites, Thr 291, Thr 292, Ser 295 and Thr 296, of which only Thr 292 was observed to be O-glycosylated in all glycopeptides identified. In addition, Thr 291 and 292 were both found modified by multiple short Core 1 type glycans. The minimal O-glycan decorating these sites was N-acetyl galactosamine (GalNAc), while elongated structures contain the GalNAc - Gal core terminated with sialic acid, attached either in a linear fashion to galactose, or branched with attachment to GalNAc (see proposed glycosylated structures in Figure 9). The observed molecular weights and the retention time of each glycoform are summarized in Table 1. Peptides containing both glycosylated sites Thr 291 and Thr 292 were found to elute earlier than those containing a single glycosylation (Table 1). Within each glycopeptide form, the sialylated glycopeptides were found with slightly longer retention times than the non-sialylated glycoforms. The distinct chromatographic retention times of each glycoform (see Table 1) indicates that the observed heterogeneity results primarily from the biochemistry of O-glycosylation and to a lesser extent from in source decomposition of the glycan chain.
Figure 9.
Proposed structure types and linkages of identified O-glycosylations at Thr-291, Thr-292 and Thr-576.
Table 1.
Composition of the Core 1 type glycans determined for each glycosylation site in glycopeptide 289 - 302
| Glycoform | Experimental MW (Da) | Theoretical MW (Da) | Retention time (min) | Occupied sites observed in peptide 289 - 302 |
|
|---|---|---|---|---|---|
| Thr 291 | Thr 292 | ||||
| a | 1371.3 | 1371.69 | 20.8 | - | - |
| b | 1574.3 | 1574.77 | 20.6 | - | GalNAc |
| c | 1736.2 | 1736.82 | 20.4 | - | GalNAcGal |
| d | 1777.5 | 1777.85 | 20.4 | GalNAc | GalNAc |
| e | 1939.4 | 1939.90 | 19.8 | GalNAc | GalNAcGal |
| f | 2027.3 | 2027.92 | 20.7 | - | GalNAcGalSA |
| g | 2101.4 | 2101.95 | 19.5 | GalNAcGal | GalNAcGal |
| h | 2230.3 | 2230.99 | 20.3 | GalNAc | GalNAcGalSA |
| i | 2392.3 | 2393.05 | 20.1 | GalNAcGal | GalNAcGalSA |
| j | 2683.3 | 2684.14 | 20.2 | GalNAcGalSA | GalNAcGalSA |
ETD and CID tandem mass spectrometric identification of the O-glycosylation in the APP domain (574 - 587)
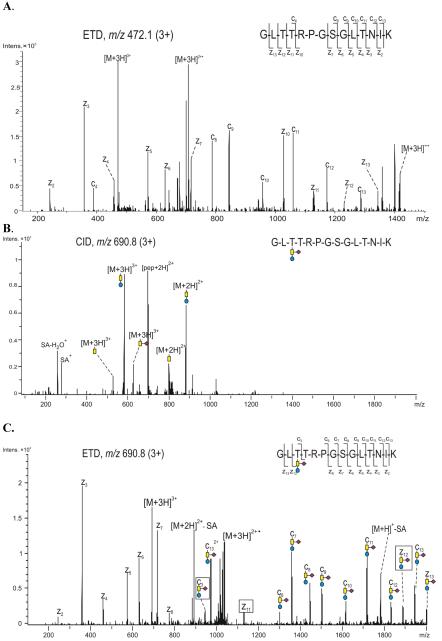
A further O-glycosylation site was identified in the APP sequence (574-587) (574GLTTRPGSGLTNIK587), containing three threonine residues and one serine residue as potential glycosylation sites. The ETD spectrum of the unmodified peptide (Figure 8A) contains a nearly complete z· ion series, identifying the amino acid sequence. The CID and ETD spectra of the corresponding precursor ion of m/z 690.8 (3+), eluting at 23.2 min, are shown in Figures 8B and 8C, respectively. The composition of the glycan was identified as a Core 1 type trisaccharide GalNAc (SA) - Gal, based on the mass difference between the unmodified and the glycosylated peptide, and the glycoforms identified in the CID spectrum (Figure 8B). The presence of sialic acid is clearly shown by the sugar oxonium ions m/z 292.1 (SA+) and m/z 274.1 (SA - H2O+). Primary losses of hexose and sialic acid from the parent ion indicate a branched structure for the glycan: GalNAc (SA) - Gal. Complete decomposition of the glycan resulted in the formation of the doubly protonated peptide of m/z 707.7, which is consistent with the amino acid sequence (574-587) for the O-glycopeptide.
Figure 8.
(A) ETD spectrum of the precursor ion m/z 472.1 (3+), showing the non-modified peptide 574-587 of the full length APP695 containing four potential O-glycosylation sites. (B) CID and (C) ETD of the precursor ion m/z 690.8 (3+), corresponding to glycopeptide 574-587 of the full length APP695, showing amino acid Thr 576 occupied with the indicated Core 1 type trisaccharide. The ETD spectrum was obtained using data dependent acquisition and the activation energy was 0.10 V. Color code: yellow - N-acetyl galactosamine, blue - galactose, purple - sialic acid. The fragment ions relevant for determination of the glycosylation site are indicated with black boxes.
The amino acid sequence of the peptide was confirmed and the location of the modification site, Thr 576, was identified on the basis of the ETD spectrum of the precursor ion of m/z 690.8 (Figure 8C). The observed z· ions (z2 - m/z 244.0, z3 - m/z 358.1, z4 - m/z 459.1, z5 - m/z 572.1, z6 - m/z 629.2, z7 - m/z 716.2, z11 - m/z 1127.3,) are identical to the z· ions observed in the spectrum of the non-modified peptide, indicating that the residues Thr 584, Ser 581 and Thr 577 are not modified. The remaining candidate for O-glycosylation, Thr 576, was confirmed as glycosylated by the observation of the ions c3 (m/z 945.3), z11· (m/z 1127.4) and z12· (m/z 1884.6), of which z11· and z12· are separated by the incremental mass corresponding to the glycosylated threonine residue. Consequently, the c ions indicated in Figure 8B have a mass shift of 656.1 compared to those c ions observed in the ETD spectrum of the non-modified peptide. Pronounced loss of sialic acid from the precursor ion was observed, which may have been induced by the additional CID voltage (0.10 V) employed to increase the fragmentation efficiency of the charge reduced molecule ions. However, no such neutral loss from any of the fragment ions was observed.
The proposed structure types and linkages of all glycosylations are illustrated in Figure 9. The heterogeneity of the oligosaccharides attached at Thr 576 was significantly lower than that observed for the glycopeptide (289 - 302), with only two glycoforms observed at this residue: GalNAc (SA) - Gal (the most abundant glycoform) and GalNAc (SA) - Gal - SA (observed as the triply protonated molecule of m/z 787.7). The abundance of the non-glycosylated peptide was considerably larger compared to those of the two glycoforms. Interestingly, the residue Thr 576 is only 21 amino acid residues away from the N-terminus of the Aß-sequence (Asp 597), suggesting that this glycosylation may play a role in the ß-secretase proteolytic processing of the amyloid precursor protein.
Conclusions
Using a combination of nanoHPLC and the fragmentation techniques ETD and CID, we identified two distinct O-glycopeptide structures and three glycosylation sites from the secreted amyloid precursor protein (sAPP695) expressed in CHO cells. The challenge of this approach for “de novo” characterization of unknown sites of O-glycosylation is primarily due to: (i) the large number of serine and threonine residues contained in the protein sequence (27 serine and 39 threonine residues in the secreted APP fragment); (ii) the absence of a defined consensus sequence for O-glycosylation; (iii) heterogeneity of O-glycosylation; and decreased ionization efficiency of O-glycopeptides compared to the unmodified peptides. While characteristic sugar oxonium ions in the CID spectra indicated the presence of glycopeptides in the LC separation, no information about the glycosylation site or peptide sequence could be extracted from that data. Moreover, CID data alone may lead to false interpretation of the data, when multiple O-glycosylation sites are present in the same peptide sequence. In contrast, N-glycosylated peptides with only one consensus glycosylation sequence may be readily interpreted on the basis of the CID spectra, if the protein sequence is known, due to the knowledge of the consensus sequence for N-glycosylation 38. In these cases ETD proved to be highly useful, as it contained fragment ions derived from cleavage across the peptide backbone, while retaining the intact glycan modification. Thus, not only the sequence of the peptide was revealed, but also the glycosylation site and the exact composition of the glycan at each site. The peptide (289 - 302), containing both modified Thr 291 and Thr 292, represented such a challenging example, which could not be confidently assigned from CID alone; here, the specificity of the electron transfer to the peptide backbone and the radical driven decomposition of the odd-electron species into c and z· fragment ions readily provided the information on the modification sites and glycan attached. The single instance when neutral loss of a sialic acid was observed, occurred when supplemental ion activation of the charge reduced species was performed in order to improve the efficiency of the ETD process. From the structures examined here, we conclude that the necessity of this additional CID excitation parameter (typically 0.07 - 0.10 V) is probably dependent on the charge state and mass of the precursor ion and needs to be determined for each individual experiment. A large number of the fragment ion types observed in ETD have been reported for the ECD process, such as neutral losses from the radical molecular ion or amino acid side chains 30, 43, 44. Previously, loss of the entire glycosylated Asn-side chain was observed in ETD 43. Similarly, we observed neutral loss of the N-acetyl galactosylated threonine side chain, either from the odd-electron molecular ion, or from C-terminal z· radical fragments. It is noteworthy that the glycans determined at the three O-linked sites from the LC-MS/MS analyses of the sAPP695 glycopeptides are in agreement with the data reported by Sato et al. 14, who analyzed the total O-glycan pool released by hydrazinolysis from the secreted APP695.
In conclusion, we show in this study that ETD and CID represent highly useful and reliable complementary tools for mapping unknown O-glycosylation sites in proteins.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. This work has been supported in part by the University of Konstanz, and by the Deutsche Forschungsgemeinschaft (Biopolymer-MS and FG-753). The authors would like to thank Ms. Katina Johnson and Dr. Jason Williams for the help provided with the Agilent software, as well as Dr. Lake Paul for the help provided with the Western blot experiments.
Glossary
Abbreviations
- APP
amyloid precursor protein
- ETD
electron transfer dissociation
- CID
collision induced dissociation
- MS/MS
tandem mass spectrometry
References
- 1.Gralle M, Ferreira ST. Structure and functions of the human amyloid precursor protein: the whole is more than the sum of its parts. Prog Neurobiol. 2007;82(1):11–32. doi: 10.1016/j.pneurobio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Goldgaber D, Lerman MI, McBride OW, Saffiotti U, Gajdusek DC. Characterization and chromosomal localization of a cDNA encoding brain amyloid of Alzheimer’s disease. Science. 1987;235(4791):877–80. doi: 10.1126/science.3810169. [DOI] [PubMed] [Google Scholar]
- 3.Palmert MR, Podlisny MB, Witker DS, Oltersdorf T, Younkin LH, Selkoe DJ, Younkin SG. The beta-amyloid protein precursor of Alzheimer disease has soluble derivatives found in human brain and cerebrospinal fluid. Proc Natl Acad Sci U S A. 1989;86(16):6338–42. doi: 10.1073/pnas.86.16.6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci U S A. 1999;96(7):3922–7. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allinson TM, Parkin ET, Turner AJ, Hooper NM. ADAMs family members as amyloid precursor protein alpha-secretases. J Neurosci Res. 2003;74(3):342–52. doi: 10.1002/jnr.10737. [DOI] [PubMed] [Google Scholar]
- 6.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–41. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 7.Wilquet V, De Strooper B. Amyloid-beta precursor protein processing in neurodegeneration. Curr Opin Neurobiol. 2004;14(5):582–8. doi: 10.1016/j.conb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Selkoe DJ. Normal and abnormal biology of the beta-amyloid precursor protein. Annu Rev Neurosci. 1994;17:489–517. doi: 10.1146/annurev.ne.17.030194.002421. [DOI] [PubMed] [Google Scholar]
- 9.Selkoe DJ. Cell biology of the amyloid beta-protein precursor and the mechanism of Alzheimer’s disease. Annu Rev Cell Biol. 1994;10:373–403. doi: 10.1146/annurev.cb.10.110194.002105. [DOI] [PubMed] [Google Scholar]
- 10.Weidemann A, Konig G, Bunke D, Fischer P, Salbaum JM, Masters CL, Beyreuther K. Identification, biogenesis, and localization of precursors of Alzheimer’s disease A4 amyloid protein. Cell. 1989;57(1):115–26. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]
- 11.Pahlsson P, Spitalnik SL. The role of glycosylation in synthesis and secretion of beta-amyloid precursor protein by Chinese hamster ovary cells. Arch Biochem Biophys. 1996;331(2):177–86. doi: 10.1006/abbi.1996.0296. [DOI] [PubMed] [Google Scholar]
- 12.Sodhi CP, Perez RG, Gottardi-Littell NR. Phosphorylation of beta-amyloid precursor protein (APP) cytoplasmic tail facilitates amyloidogenic processing during apoptosis. Brain Res. 2008;1198:204–12. doi: 10.1016/j.brainres.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 13.Pahlsson P, Shakin-Eshleman SH, Spitalnik SL. N-linked glycosylation of beta-amyloid precursor protein. Biochem Biophys Res Commun. 1992;189(3):1667–73. doi: 10.1016/0006-291x(92)90269-q. [DOI] [PubMed] [Google Scholar]
- 14.Sato Y, Liu C, Wojczyk BS, Kobata A, Spitalnik SL, Endo T. Study of the sugar chains of recombinant human amyloid precursor protein produced by Chinese hamster ovary cells. Biochim Biophys Acta. 1999;1472(12):344–58. doi: 10.1016/s0304-4165(99)00140-3. [DOI] [PubMed] [Google Scholar]
- 15.Ando K, Oishi M, Takeda S, Iijima K, Isohara T, Nairn AC, Kirino Y, Greengard P, Suzuki T. Role of phosphorylation of Alzheimer’s amyloid precursor protein during neuronal differentiation. J Neurosci. 1999;19(11):4421–7. doi: 10.1523/JNEUROSCI.19-11-04421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomita S, Kirino Y, Suzuki T. Cleavage of Alzheimer’s amyloid precursor protein (APP) by secretases occurs after O-glycosylation of APP in the protein secretory pathway. Identification of intracellular compartments in which APP cleavage occurs without using toxic agents that interfere with protein metabolism. J Biol Chem. 1998;273(11):6277–84. doi: 10.1074/jbc.273.11.6277. [DOI] [PubMed] [Google Scholar]
- 17.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246(4926):64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 18.Hillenkamp F, Karas M, Beavis RC, Chait BT. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal Chem. 1991;63(24):1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 19.Zubarev RA, Kelleher NL, McLafferty FW. Electron Capture Dissociation of Multiply Charged Protein Cations - a Nonergodic Process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 20.Beu SC, Senko MW, Quinn JP, Wampler FM, McLafferty FW. Fourier-transform electrospray instrumentation for tandem high-resolution mass spectrometry of large molecules. J Am Soc Mass Spectrom. 1993;4:557–565. doi: 10.1016/1044-0305(93)85017-R. [DOI] [PubMed] [Google Scholar]
- 21.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(26):9528–33. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitteri SJ, Chrisman PA, Hogan JM, McLuckey SA. Electron transfer ion/ion reactions in a three-dimensional quadrupole ion trap: reactions of doubly and triply protonated peptides with SO2*. Anal Chem. 2005;77(6):1831–9. doi: 10.1021/ac0483872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demelbauer UM, Zehl M, Plematl A, Allmaier G, Rizzi A. Determination of glycopeptide structures by multistage mass spectrometry with low-energy collision-induced dissociation: comparison of electrospray ionization quadrupole ion trap and matrix-assisted laser desorption/ionization quadrupole ion trap reflectron time-of-flight approaches. Rapid Commun Mass Spectrom. 2004;18(14):1575–82. doi: 10.1002/rcm.1521. [DOI] [PubMed] [Google Scholar]
- 24.Carr SA, Huddleston MJ, Bean MF. Selective identification and differentiation of N- and O-linked oligosaccharides in glycoproteins by liquid chromatography-mass spectrometry. Protein Sci. 1993;2(2):183–96. doi: 10.1002/pro.5560020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kjeldsen F, Haselmann KF, Budnik BA, Jensen F, Zubarev RA. Dissociative capture of hot (3-13 eV) electrons by polypeptide polycations: An efficient process accompanied by secondary fragmentation. Chem. Phys. Lett. 2002;356:201–206. [Google Scholar]
- 26.Haselmann KF, Jorgensen TJ, Budnik BA, Jensen F, Zubarev RA. Electron capture dissociation of weakly bound polypeptide polycationic complexes. Rapid Commun Mass Spectrom. 2002;16(24):2260–5. doi: 10.1002/rcm.853. [DOI] [PubMed] [Google Scholar]
- 27.Kjeldsen F, Haselmann KF, Budnik BA, Sorensen ES, Zubarev RA. Complete characterization of posttranslational modification sites in the bovine milk protein PP3 by tandem mass spectrometry with electron capture dissociation as the last stage. Anal Chem. 2003;75(10):2355–61. doi: 10.1021/ac026295b. [DOI] [PubMed] [Google Scholar]
- 28.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764(12):1811–22. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirgorodskaya E, Roepstorff P, Zubarev RA. Localization of O-glycosylation sites in peptides by electron capture dissociation in a Fourier transform mass spectrometer. Anal Chem. 1999;71(20):4431–6. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]
- 30.Mormann M, Paulsen H, Peter-Katalinic J. Electron capture dissociation of O-glycosylated peptides: radical site-induced fragmentation of glycosidic bonds. Eur J Mass Spectrom (Chichester, Eng) 2005;11(5):497–511. doi: 10.1255/ejms.738. [DOI] [PubMed] [Google Scholar]
- 31.Renfrow MB, Mackay CL, Chalmers MJ, Julian BA, Mestecky J, Kilian M, Poulsen K, Emmett MR, Marshall AG, Novak J. Analysis of O-glycan heterogeneity in IgA1 myeloma proteins by Fourier transform ion cyclotron resonance mass spectrometry: implications for IgA nephropathy. Anal Bioanal Chem. 2007;389(5):1397–407. doi: 10.1007/s00216-007-1500-z. [DOI] [PubMed] [Google Scholar]
- 32.Wu SL, Huhmer AF, Hao Z, Karger BL. On-line LC-MS approach combining collision-induced dissociation (CID), electron-transfer dissociation (ETD), and CID of an isolated charge-reduced species for the trace-level characterization of proteins with post-translational modifications. J Proteome Res. 2007;6(11):4230–44. doi: 10.1021/pr070313u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroeder MJ, Webb DJ, Shabanowitz J, Horwitz AF, Hunt DF. Methods for the detection of paxillin post-translational modifications and interacting proteins by mass spectrometry. J Proteome Res. 2005;4(5):1832–41. doi: 10.1021/pr0502020. [DOI] [PubMed] [Google Scholar]
- 34.Wuhrer M, Catalina MI, Deelder AM, Hokke CH. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;849(12):115–28. doi: 10.1016/j.jchromb.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 35.Hogan JM, Pitteri SJ, Chrisman PA, McLuckey SA. Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. J Proteome Res. 2005;4(2):628–32. doi: 10.1021/pr049770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller G, Ruppert S, Schmid E, Schutz G. Functional analysis of alternatively spliced tyrosinase gene transcripts. Embo J. 1988;7(9):2723–30. doi: 10.1002/j.1460-2075.1988.tb03126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Domon B, Costello CE. A Systematic Nomenclature for Carbohydrate Fragmentations in FAB-MS/MS Spectra of Glycoconjugates. Glycoconjugate J. 1988;5:397–409. [Google Scholar]
- 38.Zaia J. Mass spectrometry of oligosaccharides. Mass Spectrom Rev. 2004;23(3):161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- 39.Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J. Essentials of Glycobiology. 1999:101–113. [Google Scholar]
- 40.Hakansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptic to yield complementary sequence information. Anal Chem. 2001;73(18):4530–6. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 41.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72(3):563–73. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 42.Savitski MM, Kjeldsen F, Nielsen ML, Zubarev RA. Hydrogen rearrangement to and from radical z fragments in electron capture dissociation of peptides. J Am Soc Mass Spectrom. 2007;18(1):113–20. doi: 10.1016/j.jasms.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 43.Catalina MI, Koeleman CA, Deelder AM, Wuhrer M. Electron transfer dissociation of N-glycopeptides: loss of the entire N-glycosylated asparagine side chain. Rapid Commun Mass Spectrom. 2007;21(6):1053–61. doi: 10.1002/rcm.2929. [DOI] [PubMed] [Google Scholar]
- 44.Savitski MM, Nielsen ML, Zubarev RA. Side-chain losses in electron capture dissociation to improve peptide identification. Anal Chem. 2007;79(6):2296–302. doi: 10.1021/ac0619332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.