Abstract
Describing the “ORFeome” of an organism, including all major isoforms, is essential for a systems understanding of any species; however, conventional cloning and sequencing approaches are prohibitively costly and labor-intensive. We describe a potentially genome-wide methodology for efficiently capturing novel coding isoforms using RT-PCR recombinational cloning, “deep well” pooling, and a “next generation” sequencing platform. This ORFeome discovery pipeline will be applicable to any eukaryotic species with a sequenced genome.
Experimental definition of the complete set of protein-coding transcript sequences (“ORFeome”) is fundamental for complete understanding of any organism, but this has not been achieved to date for any metazoan. Adding to the uncertainty, many eukaryotic genes exhibit alternative splicing, leading to a diversity of ORFs encoded by a single gene. Currently, ~74% of human multi-exon genes and ~13% of Caenorhabditis elegans genes are predicted to undergo alternative splicing1,2. Expansion of the “isoform-space” in more complex organisms may partly explain the paradoxical lack of correlation between organismal complexity and gene number, and underscores the need to efficiently and comprehensively capture the full ORFeome. Historically, determination of intron-exon boundaries in eukaryotes has been addressed mainly by large-scale sequencing of random cDNAs (expressed sequence tags or ESTs) followed by alignment to a reference genomic DNA sequence. Although EST collections are extremely helpful, the human isoform-space remains under-explored. A targeted cloning and full length sequencing strategy could provide the desired information, but is impractically resource-intensive.
Next–generation parallel sequencing technologies, such as the Roche 454 FLX, offer the prospect of sequencing at a much faster pace and lower cost than conventional Sanger-capillary platforms3. Most applications described so far have entailed resequencing of megabase-scale genomic DNA fragments4–7 or of small sequence tags8–11. A disadvantage of the latter approach is that cis-connectivity is lost between the reads; therefore, although the reads can be assembled into contigs, mRNAs can not be assembled unambiguously when splice variants are involved. Sequencing of kilobase-scale DNA fragments from complex pools in which fragments have heterogeneous abundance has not yet been tested, nor has correct assembly of hundreds to thousands of full-length cDNAs in parallel from a complex mixture been proven feasible.
Previous and ongoing full-length cDNA isolation projects aim to discover one isoform per gene, without attempting to investigate the depth of “isoform space”. Here we describe and demonstrate the feasibility of a pipeline for large-scale discovery and cloning of coding isoforms. We tested each individual component of the pipeline and demonstrated overall effectiveness for isolation of novel coding isoforms, successfully sequencing and assembling ~820 ORFs in parallel.
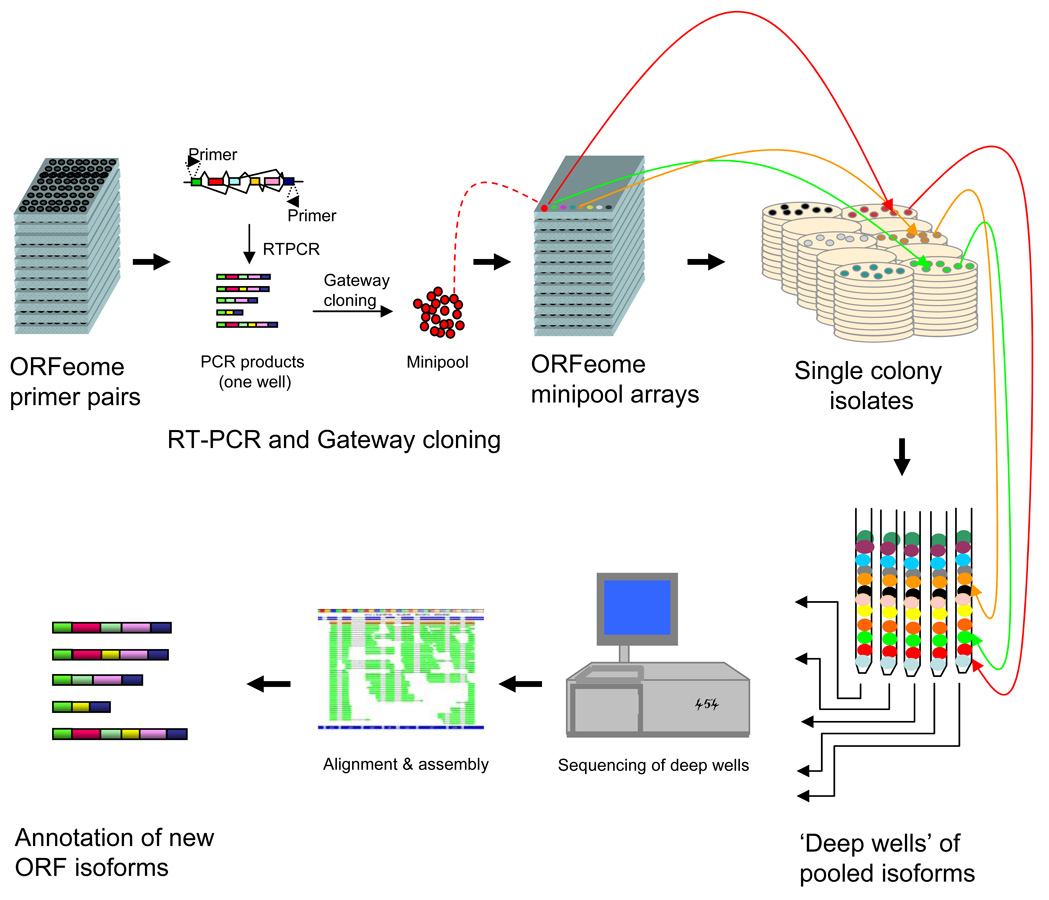
The “deep well” strategy has three elements (Fig 1): 1) efficient capture and cloning of ORF isoforms; 2) “deep well” pooling; and, 3) parallel sequencing and assembly of the obtained fragmentary ORF Sequence Tags (fOSTs) into full-length contigs. The capture of coding isoforms starts with RT-PCR using primers annealing to annotated ORFs. Complex mixtures of RNAs from one or more tissues are reverse transcribed, PCR amplified, and cloned using the Gateway recombination methodology12. As each PCR reaction can generate products containing mixtures of several splice variants, the obtained bacterial transformants represent “minipools” that potentially contain different isoforms of the same gene. Individual colonies are picked from minipools and arrayed across 96 or 384 well plates for archival storage and for subsequent consolidation into equimolar normalized pools of single colony isolates. In “deep well” pooling, aliquots from the same individual well from each plate of a set of arrayed plates are combined such that each pool contains one colony from each of the targeted gene loci (in other words, only one colony from any given minipool is included in each deep well). Deep well pooling creates a library that is perfectly normalized across genes, unlike non-normalized cDNA libraries which may be dominated by a few abundantly transcribed genes. Transcripts are “segregated” in the sense that each deep-well pool contains just one coding variant from each gene locus in the target set. This segregation is critical to ensure that each assembled contig is composed of sequence fragments arising from one specific transcript for any given gene.
Figure 1.
The isoform discovery pipeline. First, ORFs are captured by RT-PCR experiments, recombinationally cloned and transformed into E. coli. “minipools” of transformants for each gene may contain different isoforms. Second, “deep well” pools are constructed by pooling the PCR-amplified ORF sequence from one transformant for each of many genes. This method of pooling ensures normalization of ORFs and avoids concurrent sequencing of multiple isoforms. Third, parallel sequencing is carried out separately on each deep well. The obtained reads are assembled using a “Smart Bridging Assembly” (SBA) algorithm (Supplemental Methods online). Resulting ORF contigs are filtered for the presence of non-canonical splice acceptor/receptor sites and prior presence in sequence databases to identify unique novel isoforms.
The search space along an ORF is established by the choice of primer pairs. To focus on human coding potential, our primer pairs are directed solely against coding regions of the annotated human cDNAs available from the Mammalian Gene Collection (MGC)13,14. This strategy discovers novel coding variants that share the primer sites with the original cDNA used to design the primer pairs for cloning ORFs14, and has the further advantage that the resulting clones are immediately useful for protein expression.
To demonstrate that coding regions of isoforms can be robustly amplified, we carried out a medium-scale RT-PCR experiment on 94 human ORFs (randomly chosen among genes with available primer pairs based on the Human ORFeome 3.1 collection15), using five normal human tissue RNA preparations as template (Supplementary Fig. 1a and Supplementary Methods online). Our PCR success rates were 75% to 88% for all five tissues. We then carried out RT-PCR amplification of ~820 “disease” ORFs (i.e., associated with one or more human disorders in OMIM16), observing success rates of 67%, 66%, 78%, 34% and 53% from testis, brain, heart, liver and placenta RNA, respectively (Supplementary Fig. 1b online).
Subsets of these PCR products were used for subsequent Gateway cloning. To generate Set 1 (“pooled tissue”), RT-PCR reactions for 22 ORFs (chosen from the 94 above) from each of five tissue RNAs were pooled and cloned (Supplementary Fig. 1a online). Set 2 (“brain”) and Set 3 (“testis”) each corresponded to RT-PCR amplifications of a different set of ORFs randomly selected from the OMIM set. These were cloned from brain and testes RNA, respectively (Supplementary Fig. 1b online). As a control, HSD3B7 (hydroxy-delta-5-steroid dehydrogenase, 3-beta- and steroid delta-isomerase 7) was included in Set 1 as well as Sets 2 and 3.
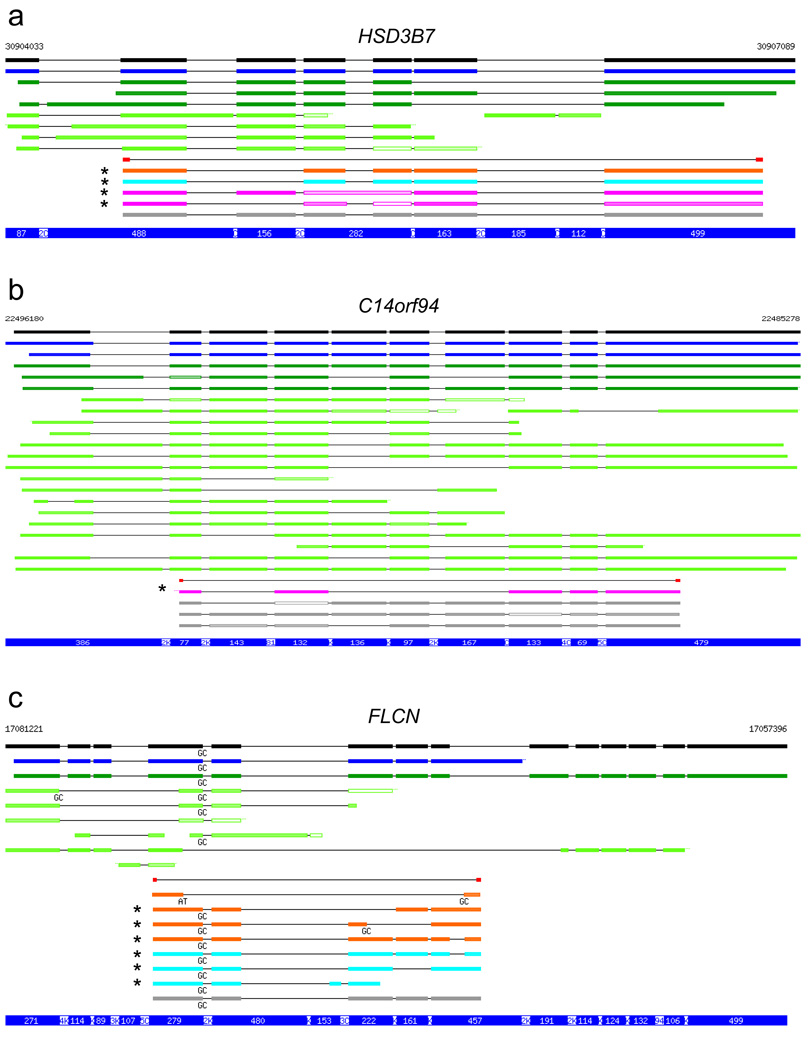
The cloning results are summarized (Table 1, 2), and genomic alignments of three examples are shown (Fig. 2) (Supplementary Fig. 2 shows the complete set of aligned sequences.) A sequence was considered “novel” unless it was recapitulated in its entirety by a single transcript from the MGC, RefSeq, and GenBank resources, including dbEST. For Set 1, in which PCR products were pooled before cloning, we discovered 20 novel variants in the 22 genes tested. These included 16 novel variants in 12 genes with only GY…AG splice signals (i.e., with canonical GT…AG or with GC…AG), and 4 novel variants with at least one unusual splice signal. Of the 22 genes examined in Set 2 and Set 3, we discovered 23 novel splice variants in 9 genes. These included 10 novel GY…AG variants in 6 genes for Set 2, and 4 such variants in 3 genes for Set 3. For HSD37, one novel GY…AG variant occurred in all three sets. In summary, we isolated an average of 18 clones per gene from a small number of tissues, and discovered novel splice variants with canonical or typical alternative splice signals for almost half (19 out of 44) of the genes examined (see also Supplementary Note online).
Table 1.
Splice variants captured from pooled RT-PCRs of brain, testis, heart, liver and placenta.
| No. | Gene ID | Gene symbol | Non- novel | Novel | |
|---|---|---|---|---|---|
| GY-AG | Other | ||||
| 1 | 10449 | ACAA2 | 1 | 1 | 0 |
| 2 | 27237 | ARHGEF16 | 1 | 1 | 0 |
| 3 | 408 | ARRB1 | 2 | 2 | 0 |
| 4 | 7918 | BAT4 | 1 | 1 | 2 |
| 5 | 54930 | C14orf94 | 3 | 1 | 0 |
| 6 | 79411 | GLB1L | 1 | 1 | 0 |
| 7 | 80270 | HSD3B7 | 1 | 2 | 0 |
| 8 | 28981 | IFT81 | 1 | 1 | 0 |
| 9 | 9776 | KIAA0652 | 0 | 3 | 1 |
| 10 | 8569 | MKNK1 | 2 | 1 | 0 |
| 11 | 55471 | PRO1853 | 1 | 0 | 1 |
| 12 | 51100 | SH3GLB1 | 1 | 1 | 0 |
| 13 | 10629 | TAF6L | 1 | 1 | 0 |
| 14 | 95 | ACY1 | 1 | 0 | 0 |
| 15 | 123 | ADFP | 1 | 0 | 0 |
| 16 | 57332 | CBX8 | 1 | 0 | 0 |
| 17 | 1848 | DUSP6 | 2 | 0 | 0 |
| 18 | 7157 | TP53 | 1 | 0 | 0 |
| 19 | 84790 | TUBA6 | 1 | 0 | 0 |
| 20 | 26100 | WIPI-2 | 1 | 0 | 0 |
| 21 | 84287 | ZDHHC16 | 2 | 0 | 0 |
| 22 | 10617 | STAMBP | 0 | 0 | 0 |
| Total | 26 | 16 | 4 | ||
Non-novel: Captured isoforms represented in their entirety by individual transcripts in GenBank (including ESTs), RefSeq, or MGC. Novel, GY-AG: novel isoforms with GT-AG (canonical) or GC-AG splice donor-acceptor signals (Y: pyrimidine). Novel, Other: novel isoforms with at least one splice donor-acceptor pair other than GT-AG or GC-AG. Redundant sequences were not counted.
Table 2.
Splice variants captured by RT-PCR from Set 2 (brain) and Set 3 (testis).
| No. | GeneID | Symbol | Non-novel | Novel, Set 2 | Novel, Set 3 | ||
|---|---|---|---|---|---|---|---|
| GY-AG | Other | GY-AG | Other | ||||
| 1 | 445 | ASS1 | 1 | 1 | 3 | 0 | 0 |
| 2 | 1497 | CTNS | 3 | 1 | 0 | 0 | 0 |
| 3 | 201163 | FLCN1 | 1 | 3 | 1 | 2 | 0 |
| 4 | 3043 | HBB | 1 | 0 | 1 | 0 | 0 |
| 5 | 80270 | HSD3B72 | 1 | 0 | 0 | 0 | 0 |
| 6 | 4953 | ODC1 | 1 | 0 | 1 | 0 | 0 |
| 7 | 80025 | PANK2 | 1 | 0 | 0 | 1 | 0 |
| 8 | 5213 | PFKM3 | 2 | 1 | 3 | 1 | 0 |
| 9 | 5660 | PSAP | 1 | 3 | 0 | 0 | 0 |
| 10 | 6102 | RP2 | 1 | 1 | 0 | 0 | 0 |
| 11 | 8542 | APOL1 | 2 | 0 | 0 | 0 | 0 |
| 12 | 1738 | DLD | 1 | 0 | 0 | 0 | 0 |
| 13 | 55670 | PEX26 | 2 | 0 | 0 | 0 | 0 |
| 14 | 57104 | PNPLA2 | 1 | 0 | 0 | 0 | 0 |
| 15 | 5538 | PPT1 | 2 | 0 | 0 | 0 | 0 |
| 16 | 6403 | SELP | 3 | 0 | 0 | 0 | 0 |
| 17 | 219736 | STOX1 | 1 | 0 | 0 | 0 | 0 |
| 18 | 1861 | TOR1A | 1 | 0 | 0 | 0 | 0 |
| 19 | 7391 | USF1 | 1 | 0 | 0 | 0 | 0 |
| 20 | 7422 | VEGF | 3 | 0 | 0 | 0 | 0 |
| 21 | 8565 | YARS | 3 | 0 | 0 | 0 | 0 |
| 22 | 56652 | PEO1 | 0 | 0 | 0 | 0 | 0 |
| Totall | 33 | 10 | 9 | 4 | 0 | ||
Non-novel: Captured isoforms represented in their entirety by individual transcripts in GenBank (including ESTs), RefSeq, or MGC. Novel, GY-AG: novel isoforms with GT-AG (canonical) or GC-AG splice donor-acceptor signals (Y: pyrimidine). Novel, Other: novel isoforms with at least one splice donor-acceptor pair other than GT-AG or GC-AG. Redundant sequences were not counted.
A novel GY-AG variant was detected in both sets but is reported only for Set 2; see Supplementary Fig. 2.
Figure 2.
Examples of identified transcripts. Genomic alignments of three representative genes from sets 1–3 compared with RefSeq (black), MGC (blue), GenBank (dark green) and dbEST (light green), following removal of redundant alignments. Results are shown for 3 of 44 genes from which ORFs were cloned (the complete set is in Supplementary Fig. 2 online). Transcripts with exon/intron structures that were exactly recapitulated, over the entire length, by individual MGC, Refseq, or GenBank transcripts, including ESTs, are shown in gray, while those that are novel are shown for the pooled tissue (purple), brain (orange), and testis (cyan) cloning experiments. The positions of primers used for RT-PCR are shown in red. Color saturation indicates % identity, ranging from light ( ≤ 90% identity) to dark ( ≥ 99% identity). Splice signals other than the canonical GT donor and AG acceptor are shown for all sequences. Novel isoforms with only canonical or GC…AG signals are indicated by an asterisk ( * ). For simplicity, ESTs with unusual splice signals are not shown, but they were included in the assessment of novelty. Chromosomal coordinates are indicated at the top of each panel. The blue bar at the bottom of each panel indicates the lengths of exonic (white on blue) and intronic (reversed) segments, in bp (C = 100; K = 1000); introns are compressed to highlight exons.
Next generation sequencing technologies provide greatly reduced cost per raw base sequenced but have, to varying degrees, the serious drawback of shorter read lengths. To assess whether our deep well strategy can be coupled to the 454 pyrosequencing technology, we tested the set of cloned OMIM disease ORFs (Supplementary Fig. 1b online). To eliminate ambiguity in assembly of short reads, the set was chosen such that no two ORFs shared any 50 bp subsequence with > 90% identity. The resulting ORF set, taken from the human ORFeome 3.1 collection15, encompassed a broad size range (~0.15 kb to 5.1 kb), and corresponded to ~4% of the protein coding genes annotated in the human RefSeq (currently ~19,200). PCR amplification using vector primers flanking the ORFs successfully amplified ~820 ORFs (Supplementary Fig. 1b online). These PCR products were pooled and sequenced by the 454 platform (Supplementary Methods online). The sequencing run produced 145,318 reads with an average read length of 240 bases (approximately 35 million bases total) and ~25 fold coverage of each base in the set (Supplementary Fig. 3 online).
We assembled contigs from fOSTs using only the human genome sequence as the template for assembly. Knowing the genomic location of ORF-targeted primers allows limiting template sequences to genomic regions between targeting primers. In aligning fOSTs to the genome, bridging of consecutive exons by BLAT requires that a “bridging” read has a sufficient length of exonic sequence on each side of the intervening intron. Although these length requirements may be relaxed when strong hypotheses about the locations of exon ends are available, such information is not currently used by BLAT. After initially employing a pipeline that combines existing software packages (Methods), we developed a more advanced pipeline that better reveals intron-exon structure (“Smart Bridging Assembly” or SBA; Supplementary Fig. 4 online). SBA introduces two features not present in our initial conventional assembly method. First, adjacent contigs with inner termini that correspond approximately to exon ends may be “bridged” by examination of unaligned sequence segments that may have been too short for BLAT to justify the introduction of an intron-sized gap. Second, where two contigs aligned to the genome are separated by a gap that is too small to contain an intron, these contigs are “bridged” by filling the gap with the known genomic sequence.
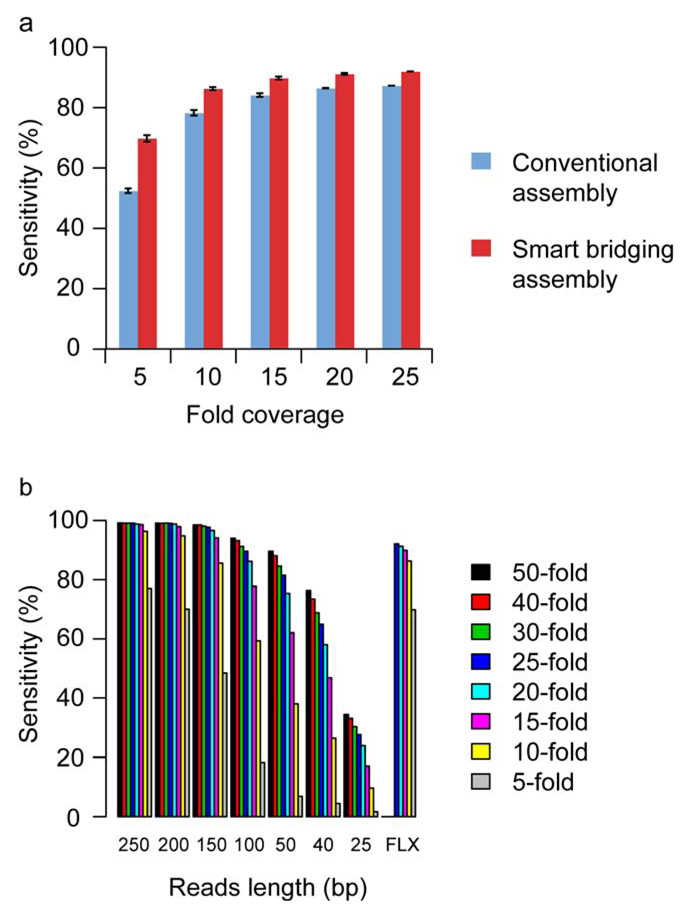
We applied both conventional and SBA assembly methods to all the 454 FLX reads and computed the percentage of ORFs with 100% correctly assembled gene structure (Fig. 3a, 25-fold coverage column). We also simulated the effects of reduced coverage by assembling a randomly chosen subset of 20%, 40%, 60%, and 80% of the 454 FLX reads corresponding to 5, 10, 15, and 20 fold coverage. We repeated this procedure 10 times to compute the average percentage of correctly assembled ORFs. The SBA method clearly outperforms the conventional method, particularly when coverage is low (Fig. 3a). For example, at 5-fold coverage, the SBA method assembles 70% of ORFs correctly, as compared with 52% for the conventional method.
Figure 3.
Sequence assembly results and simulation. (a) Success rates of assembly using conventional and smart bridging assembly methods, at varying fold-coverage (see text for details). The percentage of ORFs with 100% correctly assembled gene structure (exon-intron) was computed (n = 10 repeats). Error bars represent the standard deviation. (b) The set of ORF sequences used in the 454 FLX run were randomly fragmented in silico with average fragment size of 550 base pairs and range of 300–800 bp. Different sequence read lengths and fold coverages were simulated. For each ORF, we assembled contigs based on all available sequence reads that have a corresponding best match in the genomic region of the ORF. The graphs illustrate sensitivity by gene, that is, the percentage of ORFs whose gene structure (all exons) is 100% correctly assembled.
Other next-generation sequencing platforms offer significantly shorter reads than the 454 FLX system, with reduced per-base cost of sequencing. To evaluate compatibility of such platforms with the deep well strategy, we tested in silico the rates of successful assembly at different read lengths and different depths of coverage (Fig. 3b). Not surprisingly, the fraction of genes with completely correct assembly of all exons was strongly affected by read length. As a reference we used sequence reads from the actual 454 FLX reactions, finding that at least 15-fold coverage is needed to achieve 90% per-gene sensitivity. For short read lengths the same level of assembly could be achieved by increased overall coverage. In the simulation, to achieve a similar assembly success rate for read lengths of 200, 150 and 100 bases, fold coverages of 10, 15 and 25 are needed, respectively (Fig. 3b). Even at 50-fold coverage, sequence reads of 25 bases achieved a per-gene sensitivity of only 34%; however, 50-fold coverage allowed 76% and nearly 90% per-gene sensitivity, respectively, for 40 and 50 base read lengths. Taken together, our experiments suggest that transcripts can be assembled accurately with read lengths smaller than those produced by the 454 FLX technology; however, substantial increases in coverage would be needed for read lengths shorter than 40 bp (see also Supplementary Fig. 5–10 and Supplementary Note online for more details).
In summary, a deep well normalized pooling scheme, combined with a tailored assembly algorithm, was used with a cost-efficient parallel sequencing platform to unambiguously define the exon/intron structure of coding sequences. The deep well isoform discovery methodology described and validated here can now be used for genome-wide isoform discovery projects. One potential source of ambiguity in the deep well strategy is the presence of paralogs with high nucleotide sequence similarity. This difficulty is easily addressed by separating clones from paralogs into distinct deep well pools. Assembly of deep well pools is easily done with available robotic liquid handling systems such that “tailored” sets of pools of any size and composition can be generated. For genome-scale implementation of the deep well strategy for humans using 454 FLX technology, the set of targeted genes (currently estimated between ~20,500 (ref. 17) to ~34,000 (ref. 18) might be best separated into deep-well pools of ~4,000 genes each. Each pool would contain ~4 Mbs of unique sequence and 454 FLX sequencing could produce 10x coverage at current capacity. The optimal number of clones in each deep well depends on both the raw quantity of sequence that a sequencing run can generate, as well as the read length (since this determines the required sequence coverage). If we were to limit our search space to the current RefSeq transcripts, we can expect as few as 18 × 4 deep wells (corresponding to 18 colonies sequenced for 19,000 RefSeq ORFs, or the equivalent of ~342,000 sequencing reactions) to yield novel isoforms for about half of the RefSeq genes not found in GenBank, including among 7.8 million EST entries. Given the efficiency of this approach, conjoined with rapidly increasing capacity and read length of emerging sequencing methods, whole genome experiments can be carried out with increasing rapidity and cost effectiveness.
Supplementary Material
Supplementary Figure 1 | Large scale amplification of human ORFs.
Supplementary Figure 2 | Alignments of sequences obtained from cloning of RT-PCR products.
Supplementary Figure 3 | Size distribution of the obtained 454 reads and an example of genomic alignment of the assembled contigs.
Supplementary Figure 4 | Smart Bridging Assembly (SBA).
Supplementary Figure 5 | In silico simulation of contig assembly for different read lengths.
Supplementary Figure 6 | Computer simulation of contig assembly.
Supplementary Figure 7 | Computer simulation of the effect of fragment size on contig assembly.
Supplementary Figure 9 | The effect of exon number on transcript assembly.
Supplementary Figure 10 | The distribution of sequence read coverage of the FLX reads.
Acknowledgments
This work was funded in part by a grant from the Ellison Foundation (awarded to M.V.), and in part by Institute Sponsored Research funds from the Dan Farber Cancer Institute Strategic Initiative in support of Center for Cancer Systems Biology. F.P.R. gratefully acknowledges support from National Institutes of Health grants NS054052, HG003224, HL081341. W.T. was supported in part by National Institutes of Health grant DK070078. We thank the West Quad Computing Group at Harvard Medical School as well as Research Computing at Massachusetts General Hospital for assistance with computational resources. We thank Gary Temple for helpful comments on the manuscript.
Footnotes
Competing interest statement
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 2.Zahler AM. Alternative splicing in C. elegans. WormBook. 2005:1–13. doi: 10.1895/wormbook.1.31.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster SC. Next-generation sequencing transforms today's biology. Nat. Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 4.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 6.Moore MJ, et al. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006;6:17. doi: 10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh JD, et al. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. USA. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres TT, Metta M, Ottenwalder B, Schlotterer C. Gene expression profiling by massively parallel sequencing. Genome Res. 2008;18:172–177. doi: 10.1101/gr.6984908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nat. Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 10.Emrich SJ, Barbazuk WB, Li L, Schnable PS. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007;17:69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wicker T, et al. 454 sequencing put to the test using the complex genome of barley. BMC Genomics. 2006;7:275. doi: 10.1186/1471-2164-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walhout AJ, et al. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 13.The MGC Project Team. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rual JF, et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamesch P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clamp M, et al. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. USA. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamasaki C, et al. The H-Invitational Database (H-InvDB), a comprehensive annotation resource for human genes and transcripts. Nucleic Acids Res. 2008;36:D793–D799. doi: 10.1093/nar/gkm999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 | Large scale amplification of human ORFs.
Supplementary Figure 2 | Alignments of sequences obtained from cloning of RT-PCR products.
Supplementary Figure 3 | Size distribution of the obtained 454 reads and an example of genomic alignment of the assembled contigs.
Supplementary Figure 4 | Smart Bridging Assembly (SBA).
Supplementary Figure 5 | In silico simulation of contig assembly for different read lengths.
Supplementary Figure 6 | Computer simulation of contig assembly.
Supplementary Figure 7 | Computer simulation of the effect of fragment size on contig assembly.
Supplementary Figure 9 | The effect of exon number on transcript assembly.
Supplementary Figure 10 | The distribution of sequence read coverage of the FLX reads.