Abstract
Evasion of apoptosis contributes importantly to c-Myc-induced tumorigenesis. The BH3-only Bcl-2 family members Puma and Noxa are critical pro-apoptotic transcriptional targets of p53, a major mediator of Myc-induced apoptosis and suppressor of Myc-induced tumorigenesis. Hence, we have explored the impact of their individual or combined loss on myc-driven lymphomagenesis. Notably, Puma deficiency both increased B-lineage cells and accelerated the development of B lymphoma, accompanied by leukaemia, but not of pre-B lymphoma. Noxa deficiency alone also increased B-lineage cells but did not accelerate lymphomagenesis. However, its deficiency combined with loss of one puma allele produced more rapid onset of both pre-B and B lymphomas than did loss of a single puma allele alone. Nevertheless, the acceleration evoked by loss of both genes was not as marked as that caused by p53 heterozygosity. These results show that Puma imposes a significant, and Noxa a minor barrier to c-Myc-driven lymphomagenesis. They also indicate that additional BH3-only proteins probably also drive Myc-induced apoptosis and that non-apoptotic functions of p53 may contribute substantially to its tumour suppressor role.
Keywords: apoptosis, p53, Puma, Noxa, tumour suppressor
The ability of p53 to induce apoptosis in cells subjected to genotoxic stress or deregulated activation of oncogenes, (e.g., c-myc), is a vital component of its tumour suppressor function.1,2 p53 triggers apoptosis through the ‘Bcl-2- regulated’ pathway, as the response can be inhibited by overexpression of Bcl-2 or its pro-survival homologues.3–5 The Bcl-2 protein family, which regulates developmentally programmed cell death and cytotoxic stress-induced apoptosis,6–8 contains three structurally and functionally distinct subgroups: Bcl-2-like pro-survival proteins, which share up to four Bcl-2 homology (BH) regions; pro-apoptotic Bax/Bak-like proteins, which contain the BH1, BH2 and BH3 regions; and the pro-apoptotic BH3-only proteins, which share only the BH3 domain. The BH3-only proteins initiate apoptosis signalling, whereas Bax/Bak-like proteins act downstream by disrupting the mitochondrial outer membrane.
Experiments with genetically modified mice have shown that just as pro-survival Bcl-2 family members can be oncogenic,9 certain pro-apoptotic BH3-only proteins (e.g., Bim, Puma) can function as tumour suppressors.10,11 Moreover, links between deficiencies in BH3-only proteins – in particular Bim – and human cancer are accumulating.12–14
Two BH3-only genes, noxa and puma, are direct transcriptional targets of p53,15–17 although they can also be induced by p53-independent mechanisms.15,18 Studies with gene-targeted mice have shown that Puma plays a major role in p53-mediated apoptosis of many cell types, including B and T lymphocytes, as well as in some p53-independent apoptotic pathways19–21 and that Noxa participates in the DNA damage response of fibroblasts and keratinocytes.19,22,23 Recently, we showed that the combined absence of Noxa and Puma protected mouse embryo fibroblasts from etoposide-induced apoptosis to a greater extent than loss of either gene alone; remarkably, following whole body γ-irradiation, their concomitant loss protected thymocytes as potently as p53 loss.24
As the pro-apoptotic activity of p53 is considered to be critical for its tumour suppressor function1,2 and Puma and Noxa appear to be the critical pro-apoptotic effectors induced by p53, animals lacking Noxa or Puma might be expected to be abnormally tumour prone. Surprisingly, however, mice lacking either of these apoptotic triggers, or even both, are not tumour prone.19,20,24 Nevertheless, their loss might contribute to tumorigenesis in the context of an oncogenic lesion that activates the p53 pathway.
In Eμ-myc transgenic mice, a model of B-lymphoma development,25 the high c-Myc expression throughout B-cell development provokes an expansion of cycling pre-B cells due to increased proliferation and reduced differentiation from the pre-B to the mature B-cell stage,26 which is to a certain extent counter-balanced by increased apoptosis.27 Although Eμ-myc mice all eventually develop disseminated pre-B and/or B-cell lymphomas, usually with associated leukaemia, somatic mutations to activate additional oncogenes or inactivate tumour suppressors are required.25,28 Transformation by Myc is greatly limited by its tendency to induce apoptosis under stress conditions, such as limiting growth factors.27,29,30 Myc triggers apoptosis, in part, by activating the tumour suppressor p19Arf, which upregulates p53 by suppressing Mdm2 activity.31,32 Accordingly, Eμ-myc lymphomas often contain mutations in the p19Arf/Mdm2/p53 pathway.33
To assess the tumour suppressor potential of Puma and Noxa, we have evaluated the impact of their individual or combined loss on lymphoma development in Eμ-myc mice.
Results
Myc upregulates expression of Puma and Noxa
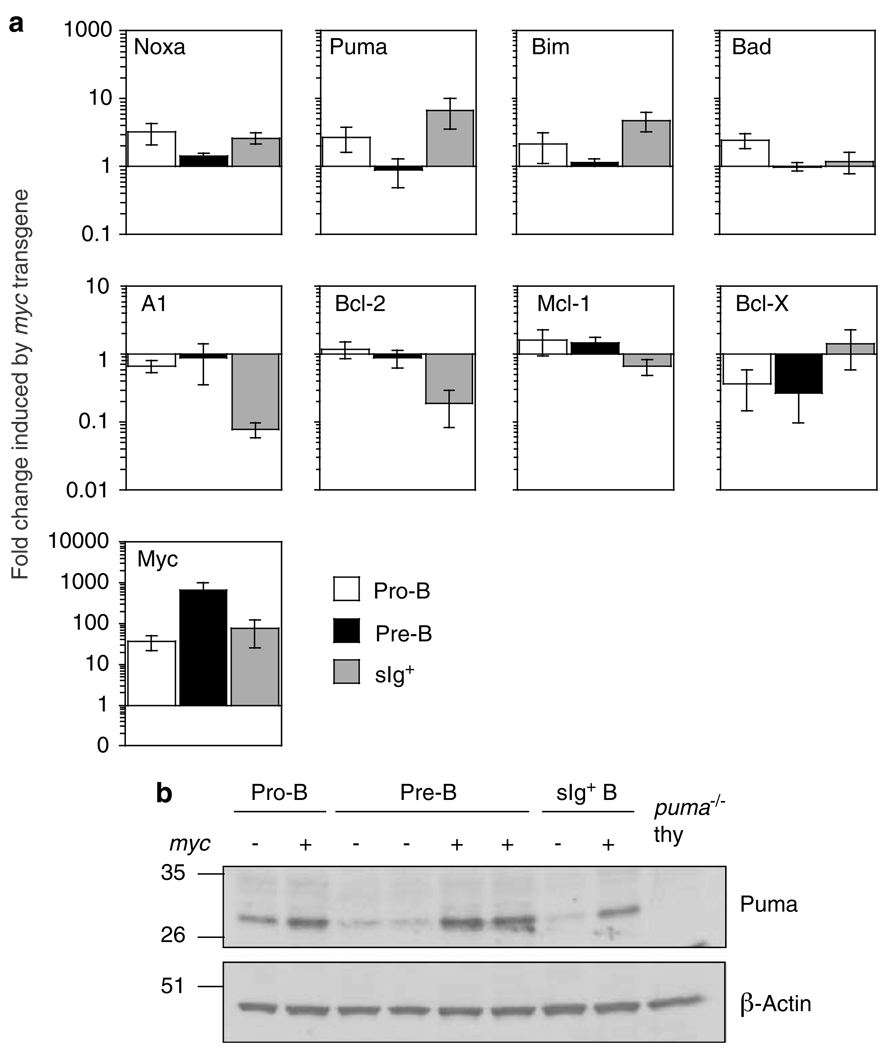
Myc indirectly upregulates p53,31 which would be expected to stimulate the transcription of puma and noxa. To investigate the impact of constitutive c-Myc expression on puma and noxa expression in B-lineage cells, we compared the levels of their mRNAs, and those of several other Bcl-2 family members, by qRT-PCR in pro-B, pre-B and sIg+ B cells sorted from the bone marrow of healthy (pre-malignant) Eμ-myc and non-transgenic mice (Figure 1a). As expected, all three B-cell subsets from Eμ-myc mice had c-myc levels at least 20-fold above those from non-transgenic littermates. The levels of noxa mRNA were ~3-fold higher in pro-B cells from Eμ-myc than non-transgenic mice and ~2.5-fold higher in the sIg+ B-cell subset but only marginally higher in the pre-B cells. Similarly, the constitutive Myc expression increased puma mRNA levels ~2.5-fold in pro-B and ~7-fold in sIg+ B cells but had no impact in pre-B cells. In accord with a previous study,10 bim was also elevated in the Eμ-myc pro-B and sIg+ B cells, whereas bad levels rose only in the pro-B cells. In accordance with other studies,10,34,35 Myc overexpression also elicited stage-specific alterations in the levels of the mRNAs for pro-survival Bcl-2 family members (Figure 1a): it did not change the levels of a1 and bcl-2 in pre- B cells, but decreased both in sIg+ B cells, whereas bcl-x levels were decreased in Eμ-myc pro-B and pre-B cells but not in sIg+ B cells.
Figure 1. Puma and Noxa expression is increased in B cells from Eμ-myc mice.
(a) Differences in the levels of RNA for Bcl-2 family members and Myc between cells from Eμ-myc and non-transgenic mice. Pro-B, pre-B and B-cell subsets were FACS purified from healthy 5- to 6-week-old Eμ-myc or non-transgenic syngeneic (C57BL/6) mice. SYBR green real-time PCR analysis was performed on cDNA. Relative RNA expression levels were calculated by normalising to the signal for β-actin in each sample and then dividing the transgenic by the non-transgenic value. Mean expression is shown ± S.E.M. of cells from 3–4 individual mice of each genotype from at least three separate experiments. (b) Western blot analysis of Puma and β-actin (loading control) on protein isolated from the cell populations described in A. Puma-deficient thymocytes were included as a control for antibody specificity. Protein size standards in kDa are indicated on the left
Western blot analysis confirmed the higher Puma expression in the Eμ-myc pro-B and sIg+ B cells (Figure 1b) and also indicated that Puma protein levels are higher in Eμ-myc pre-B cells. The increase in Puma protein in the pre-B cells might result from translation of the increased mRNA in the pro-B cells. Unfortunately, we have not identified a suitable antibody for mouse Noxa.
Loss of Puma expanded the B-cell compartment of Eμ-myc mice
No abnormalities in haematopoiesis have been observed in unstressed puma−/− mice,19,20 but the B-lineage compartment of pre-malignant Eμ-myc mice is perturbed by increased cell cycling,26 which is partially balanced by increased apoptosis.27 To determine how the absence of Puma affects haematopoiesis in that context, we enumerated leukocytes in the peripheral blood of young tumour-free animals. Eμ-myc/puma−/− mice had ~4-fold more leukocytes than wt non-transgenic littermates and ~3-fold more than Eμ-myc littermates (Figure 2a). This increase was specific to the transgenic mice, as non-transgenic puma−/−, puma+/− and wt mice all had similar numbers. At 5–6 weeks of age, blood leukocyte levels of Eμ-myc mice and Eμ-myc/puma+/− littermates remained higher than in their non-transgenic counterparts, but the Eμ-myc/puma−/− mice had the highest level (Figure 2b). Analysis for B-lymphoid differentiation markers revealed that the leukocyte excess in Puma-deficient Eμ-myc animals reflected an increase in B-lineage cells, as their blood contained several-fold more pro-B/pre-B cells (B220+sIg−), virgin B cells (B220+sIgM+IgDlo) and mature B cells (B220+sIgM+sIgDhi) compared with Eμ-myc wt mice (Figure 2b). A comparable increase was observed for virgin B cells in the lymph nodes and spleen (Supplementary Figure 1). Accordingly, the pre-malignant Eμ-myc/puma−/− mice had marked splenomegaly compared with Eμ-myc mice (P<0.02) (Figure 2c), and this must reflect the excess total B-lineage cells, because, as expected, loss of Puma on the Eμ-myc background did not affect the numbers of CD4+ or CD8+ T cells, macrophages, granulocytes or nucleated erythroid progenitors in any tissue examined (data not shown).
Figure 2. In pre-neoplastic Eμ-myc mice, loss of Puma increases leukocytes and B-lymphoid cells.
(a) White blood cell counts (means ± S.E.M.) from pre-neoplastic 4-week-old mice of the indicated Eμ-myc and puma genotypes. Differences between transgenic genotypes were significant as indicated. (b) Blood cellularity and subset composition in pre-neoplastic 5- to 6-week-old mice of the indicated genotypes. All differences between non-transgenic and Eμ-myc mice were significant for all puma genotypes (P<0.05) except for mature B cells from non-transgenic versus Eμ-myc/puma−/− mice. For comparisons of Eμ-myc transgenic mice of the different puma genotypes, statistically significant differences in addition to those indicated, included all subset comparisons between Eμ-myc/puma+/− and Eμ-myc/puma−/− mice. (c) Spleen weights (means ± S.E.M.) of pre-neoplastic 5- to 6-week-old mice. Differences between non-transgenic and Eμ-myc mice were significant for all puma genotypes (P<0.03), and for those indicated. (d) Bone marrow cellularity (both femora) and subset composition in pre-neoplastic 5- to 6-week-old mice of the indicated genotypes. Differences between non-transgenic and Eμ-myc mice were significant (P<0.05) for all puma genotypes for total B and pre-B cell comparisons and for reduction in mature B cells for wt versus Eμ-myc and wt versus Eμ-myc/puma−/− mice. For comparisons of the Eμ-myc transgenic mice of the different puma genotypes, all statistically significant differences are indicated. Values represent means ± S.E.M. from 4 to 8 mice of each genotype. *P<0.05, **P<0.01, ***P<0.005
We also examined the bone marrow, where B cells develop in adult mice. As reported,26 wt Eμ-myc bone marrow had around two times as many pro-B and pre-B cells as non-transgenic littermates but fewer sIg+ B cells (Figure 2d). Only the sIg+ B cells (but not the pro-B and pre-B cells) of Eμ-myc/puma−/− mice were significantly elevated above the levels in wt Eμ-myc mice (Figure 2d).
Puma contributes to Myc-driven apoptosis of pre-B cells
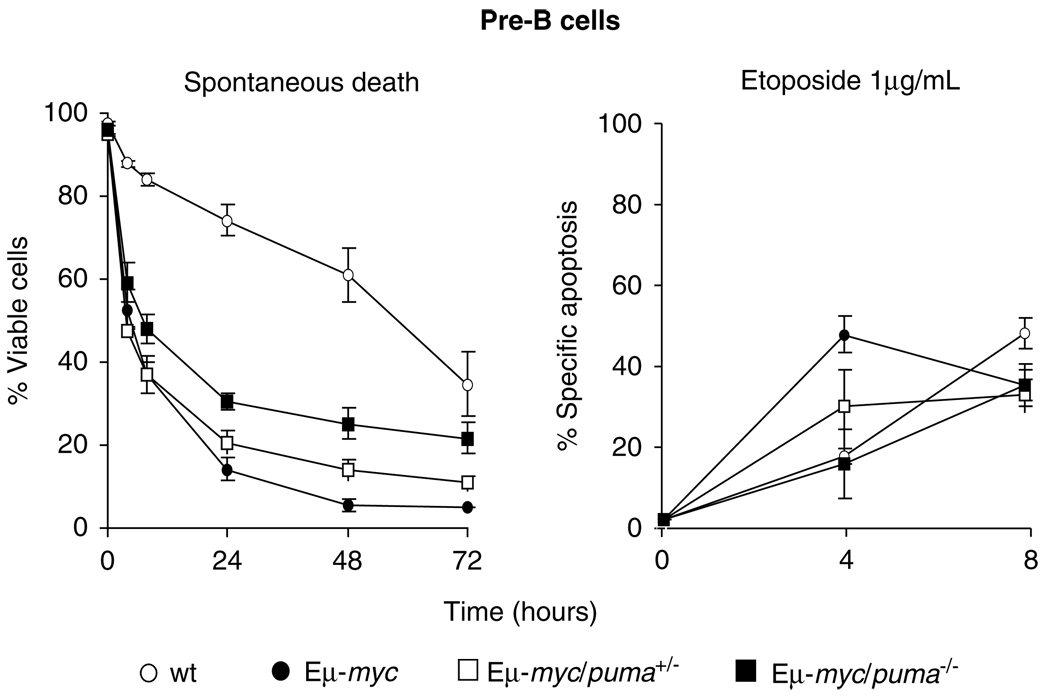
The higher levels of Puma in Eμ-myc B-lymphoid cells (Figure 1) and the modest rise in B-lineage cell numbers in Eμ-myc/puma−/− over wt Eμ-myc animals (Figure 2) indicated that Puma loss might retard the apoptosis normally induced by c-Myc overexpression.29,30 To investigate this, pro-B, pre-B and virgin/mature B cells from young healthy mice were cultured in simple medium (representing cytokine deprivation) or with the DNA-damaging drug etoposide. Eμ-myc/puma+/− and Eμ-myc/puma−/− pro-B cells (Supplementary Figure 2a) and sIg+ B cells (Supplementary Figure 2b) were no more refractory to either stimulus than their Eμ-myc wt counterparts. However, the absence of Puma significantly protected pre-B cells against cytokine deprivation and DNA damage (Figure 3), and the survival of Eμ-myc/puma+/− pre-B cells was intermediate for cytokine deprivation. The virgin/mature B cells were also tested for their response to cross-linking of the B-cell antigen receptor, but the Eμ-myc/puma−/− and wt Eμ-myc B cells died at the same rate (data not shown).
Figure 3. Loss of Puma enhances survival of Eμ-myc pre-B cells in culture.
Pre-B cells sorted from the bone marrow of 5- to 6-week-old pre-neoplastic wt or Eμ-myc mice of the indicated puma genotypes were cultured in simple medium (no added cytokines) or treated with etoposide (1 µg/ml) for the indicated times. The percentages of viable Eμ-myc/puma−/− pre-B cells remaining in culture at 24 and 48 h were significantly greater than for Eμ-myc pre-B cells (P<0.01 and P<0.001, respectively). A similar protection was observed for Eμ-myc/puma−/− pre-B cells after 4 h in culture with etoposide, compared with Eμ-myc pre-B cells (P<0.02). Values represent means ± S.E.M. of cells from 3 to 6 independent experiments for each genotype
Loss of Puma accelerated B-cell lymphomagenesis in Eμ-myc mice
The effect of Puma on cellularity of the B lineage in Eμ-myc mice and on transgenic pre-B-cell survival led us to examine how Puma loss affected lymphomagenesis by monitoring cohorts of Eμ-myc mice of different puma genotypes. Eμ-myc (puma+/+) mice (n=64) became ill between 50 and 470 days of age (median 100 days) as reported earlier,25,28 and the Eμ-myc/puma+/− mice (n=100) succumbed no faster (median 95 days, P=0.46; Figure 4a). In contrast, the Eμ-myc/puma−/− mice (n=33) had a median survival of 66 days, and all were unwell by 110 days of age, when a quarter of the wt Eμ-myc animals remained healthy (P<0.0001) (Figure 4a). Despite this acceleration, the lymphoma development in Eμ-myc/puma−/− mice remained slower than in Eμ-myc/p53+/− animals (n=23, P<0.0001) (Figure 4a).
Figure 4. Loss of Puma accelerates lymphoma development in Eμ-myc transgenic mice.
(a) Kaplan–Meier analysis of tumour-free survival of mice of the indicated genotypes. Lymphomas arose earlier in Eμ-myc/puma−/− mice than in Eμ-myc or Eμ-myc/puma+/− mice (P<0.001). Differences in tumour onset between Eμ-myc and Eμ-myc/puma+/− mice were not significant (P=0.46). (b) B lymphomas arose earlier in Eμ-myc/puma−/− mice than Eμ-myc or Eμ-myc/puma+/− mice (P<0.001). (c) Pre-B lymphoma development was not accelerated in Eμ-myc mice by loss of one or both puma alleles. (d) Proportions of pro-B/pre-B, mixed and sIg+ lymphomas in ill Eμ-myc, Eμ-myc/puma+/−, Eμ-myc/puma−/− (P=0.07, trend analysis) and Eμ-myc/p53+/− mice. (e) Numbers of leukocytes (white blood cells: WBC) in the blood of control (healthy) 102-day-old wt C57BL/6 mice and sick Eμ-myc mice of the indicated puma genotypes. Each circle represents a single animal. Bars represent mean leukocyte counts. Numbers of C57BL/6, Eμ-myc, Eμ-myc/puma+/− and Eμ-myc/puma−/− mice were 10, 33, 54 and 18, respectively. (f) Spleen weights of control 102-day-old (healthy) wt C57BL/6 mice and sick Eμ-myc mice of the indicated puma genotypes. Bars represent means. Numbers of C57BL/6, Eμ-myc, Eμ-myc/puma+/− and Eμ-myc/puma−/− mice were 10, 34, 65 and 21, respectively. **P<0.01, ***P<0.005
Sick Eμ-myc/puma−/− mice presented with enlarged lymph nodes, spleen, and thymus, as in classical Eμ-myc lymphoma. 25,28 To confirm that the lymphomas were malignant, tumour cells (2×106) were injected intra-peritoneally into C57BL/6 recipients. Five of seven control Eμ-myc, six of seven Eμ-myc/puma+/− and seven of eight Eμ-myc/puma−/− lymphomas produced tumours in recipients within 11–54 days, comparable to the 90% transplantability reported earlier for Eμ-myc lymphomas.25,28
Immunophenotyping of the primary lymphomas revealed, as expected28 that nearly all comprised B220+ sIg− pro-B/pre-B lymphomas or B220+ sIgM+ sIgDlo B-cell lymphomas (Supplementary Figures 3a and b), although a small minority contained both sIg− and sIg+ B populations (Supplementary Figure 3c). The puma genotype did not significantly affect the overall proportion of pre-B versus mature B-cell tumours. Pre-B lymphomas comprised 17/26, 27/53, and 7/17 of the tumours arising in wt, puma+/− and puma−/− mice, respectively (Figure 4d). Nevertheless, the accelerated morbidity of the Eμ-myc/puma−/− cohort was due to earlier onset of sIg+ B lymphomas, which arose much sooner than in Eμ-myc/puma+/− or Eμ-myc mice (Figure 4b): median survival was 91 days for Eμ-myc/puma−/− versus 174 days for Eμ-myc/puma+/− mice (P<0.001). Although Eμ-myc/puma+/− mice also developed sIg+ B lymphomas slightly earlier than Eμmycmice (whose median survival was 244 days), their overall survival was not significantly impaired. The accelerated disease in the absence of Puma was entirely ascribable to the faster onset of B lymphomas as pre-B lymphomas did not arise earlier in Eμ-myc/puma−/− mice than control Eμ-myc animals (Figure 4c).
Diminished Puma levels enhance leukaemia
Postmortem analysis of tumour-bearing animals revealed that the absence of Puma, or even loss of one allele, resulted in a higher leukaemic burden than in wt Eμ-myc mice. Although blood leukocyte counts at autopsy varied markedly amongst Eμ-myc/puma+/− and Eμ-myc/puma−/− mice, the mean count was >3-fold higher for Eμ-myc/puma−/− than wt Eμ-myc mice (P<0.005) (Figure 4e). Splenomegaly correlated with the extent of leukaemia, spleen weights being 1.6-fold greater in ill Eμ-myc/puma−/− than ill wt Eμ-myc animals. As reported for Eμ-myc/bim−/− mice,10 the increased leukaemia in Eμ-myc/puma−/− mice occurred exclusively in the setting of sIg+ B-cell lymphomas: their mean leukocyte numbers were four times that seen in animals succumbing to pre-B cell tumours (209±125×106/ml versus 51±32×106/ml; P<0.01). Leukaemia was also several-fold higher in Eμ-myc/puma+/− mice succumbing to sIg+ B-cell lymphomas compared with those succumbing to pre-B lymphomas (data not shown).
Loss of both Puma and Noxa did not enhance the pre-malignant phenotype more than loss of Puma alone
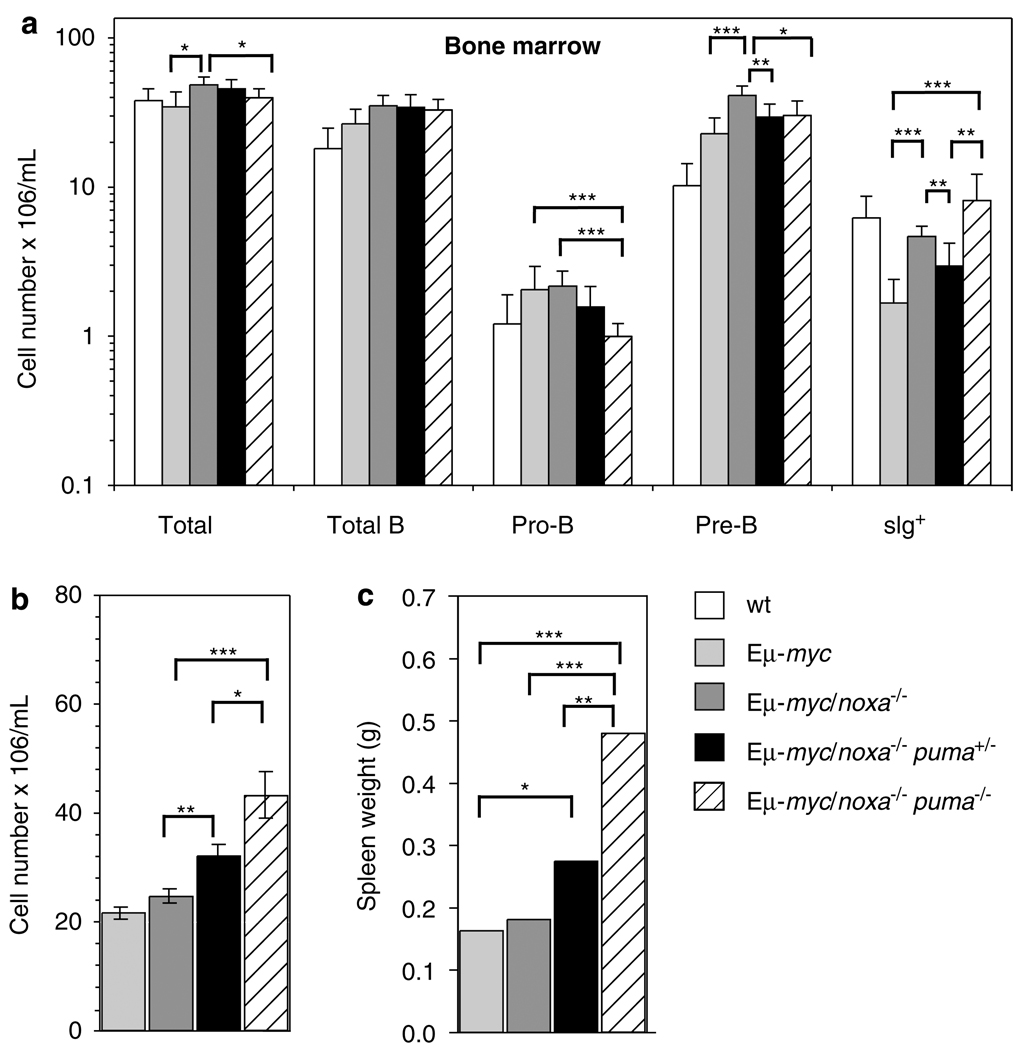
We have previously shown that functional overlap of Noxa and Puma exists in DNA damage-induced apoptosis of certain cell types, including pro-B/pre-B cells.24 We therefore examined whether loss of Noxa, like loss of Puma, had an impact on the Eμ-myc mouse model. To do this, we first investigated whether loss of Noxa, alone or together with loss of one or both puma alleles, affected the haematopoietic system of healthy young Eμ-myc mice. On the Eμ-myc background, Noxa deficiency alone did not elevate blood leukocytes above the level caused by deregulated Myc expression (Supplementary Figure 4a), affect spleen size (Supplementary Figure 4b) or elevate B-lymphoid cells in the lymph nodes, blood or spleen (Supplementary Figure 4c). In the transgenic bone marrow, however, Noxa loss led to excess total B-lineage cells, pre-B cells and virgin/mature B220+ sIg+ B cells over the wt transgenic populations (Figure 5a).
Figure 5. Loss of Noxa did not further augment the elevated leukocytes in pre-malignant Eμ-myc mice lacking one or both puma alleles.
(a) Bone marrow cellularity (both femora) and cell subset composition of pre-neoplastic 5- to 6-week-old mice of the indicated genotypes. Values represent means ± S.E.M. from 5 to 8 mice of each genotype. All differences between non-transgenic and Eμ-myc mice were significant (P<0.05) for total B cells and pre-B cells; as were comparisons of total B cells and pro-B cells for wt versus Eμ-myc/noxa−/− mice and for mature B cells for wt versus Eμ-myc and for wt versus Eμ-myc/noxa−/−puma+/− mice. For comparisons of Eμ-myc transgenic mice of the different noxa and puma genotypes, statistically significant differences are indicated. (b) White blood cell counts (means ± S.E.M.) of pre-neoplastic 4 week-old mice of the indicated genotypes. Numbers of Eμ-myc, Eμ-myc/noxa−/−, Eμ-myc/noxa−/−puma+/− mice and Eμ-myc/noxa−/−puma−/− mice were 31, 31, 32 and 14, respectively. (c) Spleen weights of pre-neoplastic 5- to 6-week-old mice. For B and C, differences between transgenic genotypes were significant as indicated. Values represent means ± S.E.M. from 5 to 8 mice of each genotype. *P<0.05, **P<0.01, ***P<0.005
When loss of Noxa was combined with loss of one allele of Puma, the resulting Eμ-myc/noxa−/−puma+/− mice had higher blood leukocyte numbers (Figure 5b) and greater spleen weights (Figure 5c) than Eμ-myc/noxa−/− animals, and the spleen weights were also significantly higher than those of Eμ-myc/puma+/− mice (P<0.02) (compare Figures 2a and, c with Figures 5b and c). Eμ-myc/noxa−/− puma+/− and Eμ-myc/puma+/− mice had similar B-lymphoid cell numbers in the bone marrow (Figure 2d and Figure 5a) and lymph nodes (Supplementary Figure 1a and Supplementary Figure 4c) although, perhaps surprisingly, the levels of pre-B and sIg+ cells were lower than those in Eμ-myc/noxa−/− animals (Figure 5c). In the bone marrow, Eμ-myc mice deficient for both Noxa and Puma also had fewer immature B cells than in the Eμ-myc/noxa−/− mice and strikingly fewer pro-B cells than either the Eμ-myc/noxa−/− or Eμ-myc/puma−/− mice (P<0.0001) (compare Figure 2d with Figure 5a). The reason for this reduction is not clear.
In contrast, B-cell numbers in the peripheral blood and spleens of Eμ-myc/noxa−/−puma+/− mice were significantly higher than in Eμ-myc/puma+/− mice, but not as high as in Eμ-myc/puma−/− or Eμ-myc/noxa−/−puma−/− mice (compare Figure 2b and Supplementary Figure 1 with Supplementary Figure 4c), due to increased pre-B and immature B cells in the peripheral blood (Eμ-myc/puma+/− versus Eμ-myc/noxa−/−puma+/− P<0.02 and P<0.01) and immature B and mature B cells in the spleen (Eμ-myc/puma+/− versus Eμ-myc/noxa−/−puma+/− P<0.02 and P<0.04). Eμ-myc/noxa−/−puma−/− mice had more mature B cells in peripheral blood (P<0.03) and spleen (P<0.05) than did Eμ-myc/puma−/− and more B cells of all lineages in the peripheral blood than did Eμ-myc/noxa−/− mice (Supplementary Figure 4c).
The elevated levels of noxa mRNA in Eμ-myc B-lymphoid cells (Figure 1a) and the increased cell numbers within bone marrow B-cell subsets provoked by Noxa loss suggested that loss of Noxa, like that of Puma, might retard the apoptosis normally induced by c-Myc. Survival assays with FACS-purified sub-populations, however, showed that the absence of Noxa alone did not protect cells at any stage of B-cell development against either cytokine deprivation or etoposide, compared with cells from Eμ-myc or Eμ-myc/puma+/− mice (Supplementary Figure 5). This is in contrast to loss of Puma, where protection relative to Eμ-myc was significant (Figure 3). B-lymphoid cells from Eμ-myc/noxa−/−puma−/− mice did not survive better in culture than those from Eμ-myc/puma−/− mice.
Combined loss of Noxa and Puma in Eμ-myc mice accelerated pre-B as well as B-lymphomagenesis
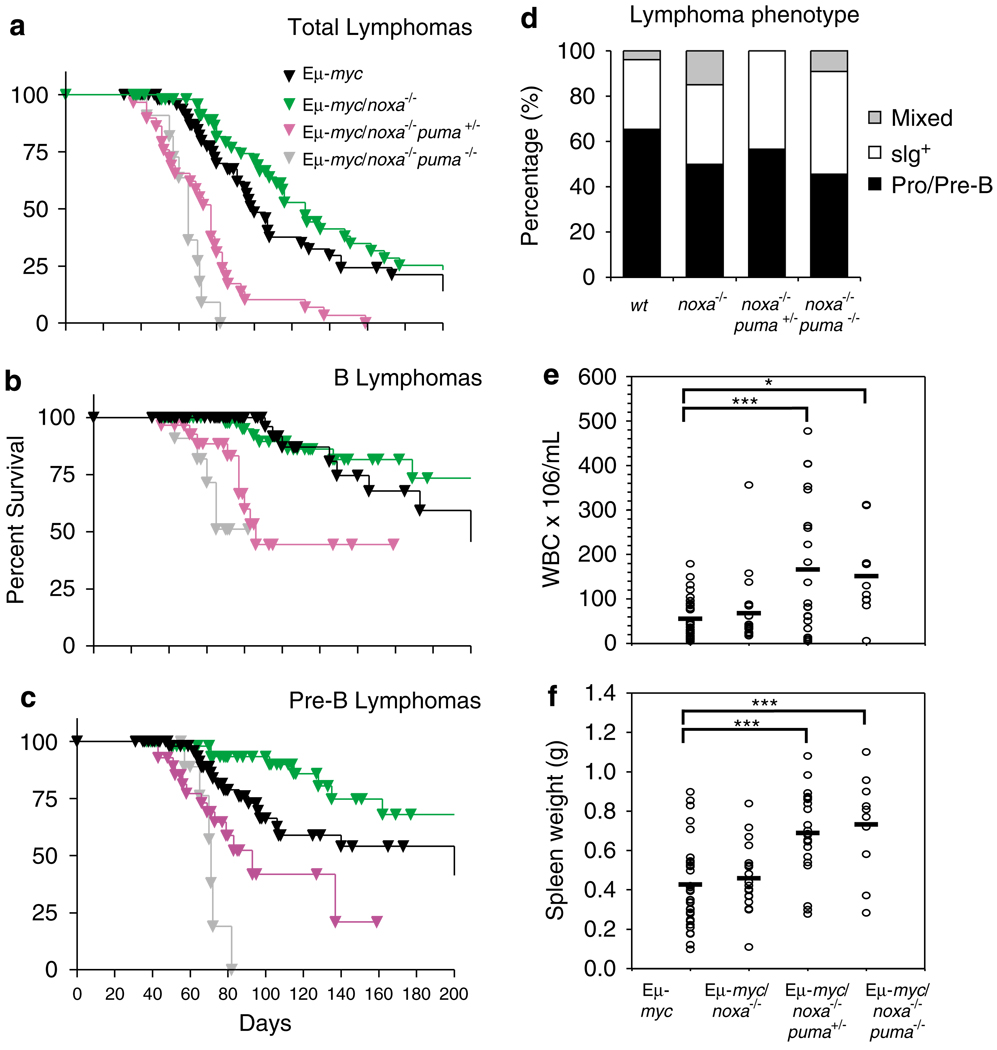
The overlapping roles of Noxa and Puma in DNA damage-induced apoptosis of pro-B/pre-B cells24 indicated to us that Noxa and Puma might cooperate to limit Myc-induced lymphomagenesis. We first investigated whether loss of Noxa alone could promote lymphomagenesis, by monitoring cohorts of Eμ-myc/noxa+/− and Eμ-myc/noxa−/− mice. Loss of Noxa alone had little effect. The median survival of Eμ-myc/noxa+/− (n=76), Eμ-myc/noxa−/− (n=56) and Eμ-myc (n=64) animals was very similar (Supplementary Figure 6). Indeed, tumour development in the absence of Noxa actually delayed onset of pre-B lymphomas (P<0.05, Figure 6c), for unknown reasons.
Figure 6. Loss of Noxa accelerates lymphoma development in Eμ-myc/puma+/− mice.
(a) Kaplan–Meier analysis of tumour-free survival of mice of the indicated genotypes. Lymphomas developed earlier in Eμ-myc/noxa−/−puma−/− (n=11) than Eμ-myc/noxa−/−puma+/− (n=29) mice (P<0.04) and both arose earlier than in Eμ-myc mice (n=64) (P<0.0001). (b) B lymphomas arose earlier in Eμ-myc/noxa−/−puma+/− than Eμ-myc/puma+/− mice (compare with Figure 4; P<0.01). (c) Pre-B lymphomas arose later in Eμ-myc/noxa−/− than Eμ-myc mice (P<0.05) but earlier in Eμ-myc/noxa−/−puma+/− than in Eμ-myc/puma+/− mice (P<0.01). Eμ-myc/noxa−/−puma−/− mice succumbed earlier than Eμ-myc/puma−/− mice (compare to Figure 4c; P<0.02). (d) The proportions of pre-B, sIg+ and mixed pre-B/B-cell lymphomas arising in Eμ-myc/puma+/− and Eμ-myc/puma−/− were not affected by additional loss of Noxa. (e) White blood cell numbers in sick Eμ-myc mice of the indicated noxa and puma genotypes. Each circle represents a single animal. Bars indicate means. Numbers of Eμ-myc, Eμ-myc/noxa−/−, Eμ-myc/noxa−/−puma+/− mice and Eμ-myc/noxa−/−puma−/− mice were 33, 20, 23 and 10, respectively. (f) Spleen weights of sick mice of the indicated genotypes. Bars indicate means. Numbers of Eμ-myc, Eμ-myc/noxa−/−, Eμ-myc/noxa−/−puma+/− mice and Eμ-myc/noxa−/−puma−/− mice were 34, 21, 22 and 10, respectively. *P<0.05, ***P<0.005
Next, cohorts of Noxa-deficient Eμ-myc transgenic mice lacking one or both alleles of puma were monitored, to investigate if a role for Noxa in tumour suppression might become evident when combined with Puma deficiency. Strikingly, Eμ-myc/noxa−/−puma+/− mice (n=29) succumbed to lymphoma with a median survival of 77 days (Figure 6a), a rate significantly faster (P<0.0001) than that seen in Eμ-myc/puma+/− mice (median survival 95 days, Figure 4a). Indeed, the rate approached that observed with Eμ-myc/puma−/− mice (Figure 4a). In contrast, the loss of one allele each of noxa and puma did not appreciably alter survival compared to loss of one puma allele alone (median survival 91 days, n=13, data not shown). Loss of both the puma and the noxa alleles (Eμ-myc/noxa−/−puma−/− mice) further accelerated tumour onset (median survival 65 days, n=11) over that in Eμ-myc/noxa−/−puma+/− mice (P<0.04), albeit not significantly faster than in Eμ-myc/puma−/− mice (P=0.12). The accelerated tumorigenesis in Eμ-myc/noxa−/− puma+/− and Eμ-myc/noxa−/−puma−/− mice involved both the pre-B and B-cell compartments (Figures 6B and C). Thus, Noxa has a role in restraining Myc-induced lymphomagenesis, albeit less substantial than that of Puma.
Although Noxa loss alone did not affect development of B-cell lymphomas, on a puma heterozygous background Noxa loss significantly accelerated B-cell lymphoma onset (median survival 86 days) compared with puma heterozygosity alone (median survival 174 days; P<0.01) (Figure 6b). Curiously, although Noxa loss alone actually delayed pre-B lymphoma development (P<0.05), additional loss of one puma allele accelerated pre-B lymphoma onset (Figure 6c), the Eμ-myc/noxa−/−puma+/− mice succumbing earlier than Eμ-myc/puma+/− mice (median survival 93 versus 140 days; P<0.02). Notably, Eμ-myc/noxa−/−puma−/− mice also succumbed to pre-B lymphomas significantly earlier than Eμ-myc/puma−/− mice (median survival 71 versus 107 days; P<0.02) (compare Figure 6c with Figure 4c). The proportion of pre-B versus sIg+ B-cell tumours arising in Eμ-myc/puma+/− and Eμ-myc/puma−/− was not, however, affected by additional loss of noxa (Figure 6d).
In contrast to Puma loss, Noxa loss did not increase the leukaemic burden in Eμ-myc mice. The mean white blood cell counts and spleen weights at autopsy for ill Eμ-myc/noxa+/− (not shown) and Eμ-myc/noxa−/− mice (Figure 6e and f) were not significantly greater compared with their ill Eμ-myc counterparts. Although killed Eμ-myc/noxa−/−puma+/− mice had spleen weights and blood leukocyte numbers similar to Eμ-myc/puma−/− mice (compare Figures 4e and f with Figures 6e and f), loss of the second puma allele on the Noxa-deficient background provoked no greater increase.
Loss of heterozygosity for puma is not required for Eμ-myc-induced lymphomagenesis
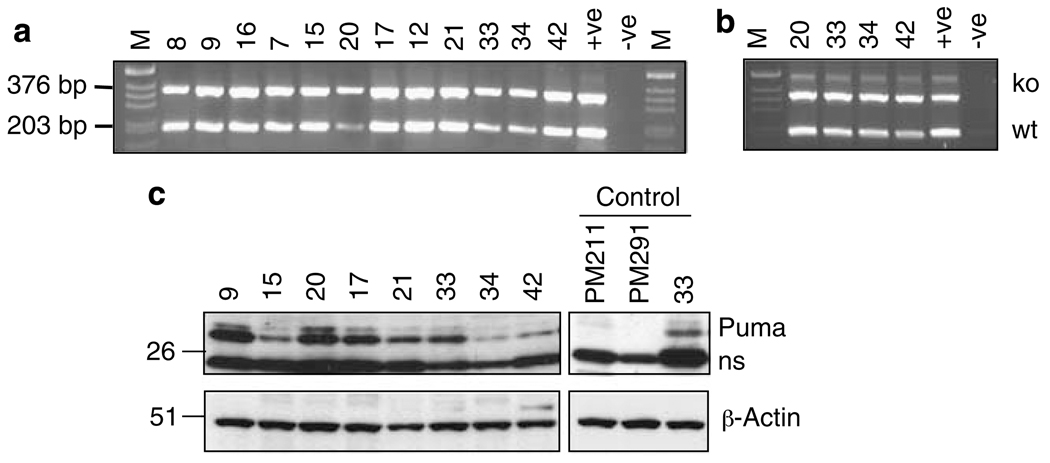
The faster tumour onset in Eμ-myc/noxa−/−puma+/− than Eμ-myc/puma−/− mice could be due simply to the absence of noxa, but it might instead reflect loss of the remaining puma allele. To distinguish between these possibilities, we performed allele-specific PCR for puma on 12 randomly selected Eμ-myc/noxa−/−puma+/− tumours. All retained the wt puma allele (Figure 7a). To rule out the possibility that the wt puma allele had been amplified from normal stromal tissue contaminating the lymphoma specimen, four tumours were FACS-sorted for large B220+ lymphoma cells. The allele-specific PCR yielded the same result (Figure 7b). Western blotting of eight Eμ-myc/noxa−/−puma+/− tumours tested revealed Puma protein (Figure 7c), also arguing against loss of heterozygosity. Interestingly, however, the Puma levels were markedly reduced in several tumours (e.g., no. 15, 34 and 42 in Figure 7c). This suggests that Myc-induced lymphomagenesis sometimes selects for reduced Puma expression, at transcriptional and/or post-transcriptional levels.
Figure 7. Tumours from Eμ-myc/noxa−/−puma+/− mice retain the wt puma allele but some exhibit reduced levels of Puma expression.
Allele-specific PCR for retention of the residual wt puma allele was performed on (a) 12 randomly selected tumours from Eμ-myc/noxa−/−puma+/− mice and (b) lymphoma cells sorted from four Eμ-myc/noxa−/−puma+/− mice based on large forward scatter and staining for B220. The wt and knockout puma alleles are 203 and 376 bp, respectively. A positive control for puma heterozygosity (+ve) and a no-DNA control (−ve) were included. (c) Western blot analysis of Puma and β-actin (loading control) in randomly selected Eμ-myc/noxa−/−puma+/− tumours. One Eμ-myc/noxa−/−puma+/− tumour (no. 33) is repeated as a loading control alongside two tumours (PM211, PM291) from Eμ-myc/puma−/− mice, in which no Puma could be detected. The anti-Puma antibody also detects a non-specific band (ns). Protein size standards in kDa are indicated on the left
Loss of Puma but not loss of Noxa reduced selection for a mutated p53 pathway in Eμ-myc lymphoma
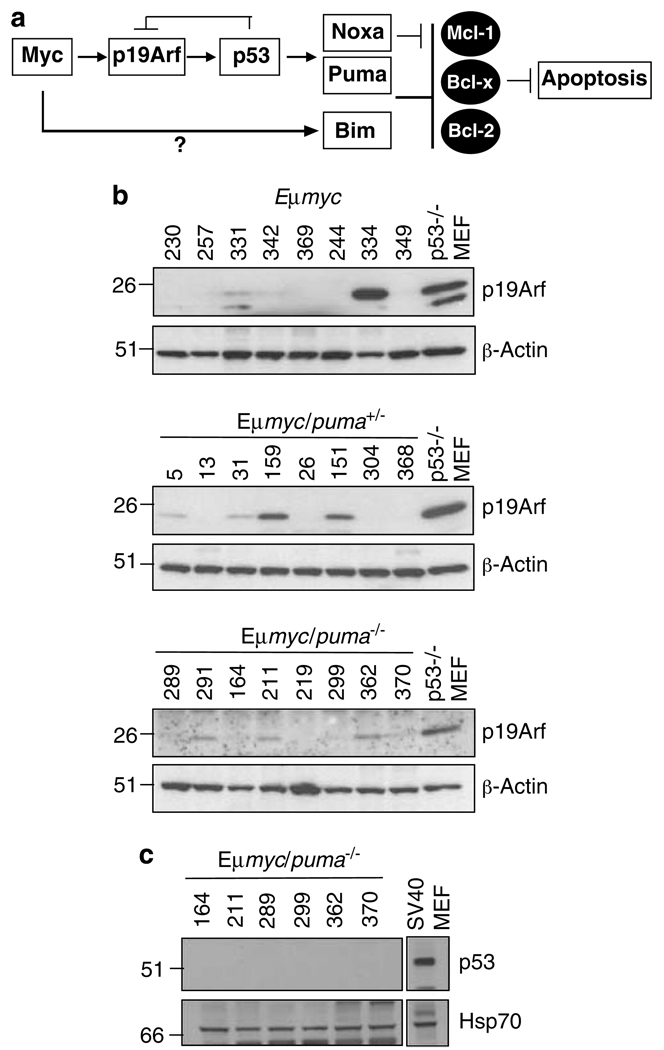
To investigate whether lymphomas from Eμ-myc/puma−/− mice differed from control Eμ-myc lymphomas in their need to select for mutations in the p19Arf/p53 pathway, the p53 pathway status of randomly selected tumours was examined in several ways: from the levels of p19Arf protein (as p53 normally downregulates p19Arf expression by a negative feedback loop33), the presence or deletion of the Ink4a/Arf locus by genomic PCR,36 the level of p53 protein (high levels indicating mutant, stabilised p53 protein) and the sequencing of exons 4–10 of the p53 gene. High levels of p19Arf, indicative of loss of p53 function due to impaired negative regulation37 (Figure 8a), were noted in 3/23 Eμ-myc, 2/12 Eμmyc/puma+/−, 3/10 Eμ-myc/noxa−/− and 2/8 Eμ-myc/noxa−/− puma+/− lymphomas (Figure 8b and Supplementary Figure 7 and 8 and data not shown). Although none of the 15 Eμ-myc/puma−/− lymphomas tested exhibited high p19Arf levels, 1 of 9 Eμ-myc/noxa−/−puma−/− lymphomas (no. 104 in Supplementary Figure 7) had a clear mutant p53 phenotype, as evidenced by high levels of both the Arf and p53 proteins. A lower level of p19Arf was found in a few tumours (e.g., no. 291, 211 and 362 in Figure 8b); we think the sensitivity of the antibody used allowed detection of Arf upregulation by oncogenic stress in the absence of loss of p53 function, 38 as those tumours retained wt p53 function by all other criteria.
Figure 8. Model for Myc-induced apoptosis and detection of alterations in the p53 pathway.
(a) The diagram depicts two pathways by which Myc engages the core apoptotic machinery, one through p19Arf, p53 and Puma and Noxa, and the other through Bim (modified from Egle et al.10). Western blot analysis of (b) p19Arf and β-actin (loading control), or (c) p53, and Hsp70 (loading control) in randomly selected Eμ-myc lymphomas of the indicated puma genotypes. p53−/− mouse embryonic fibroblasts (MEF) or MEF immortalised with SV40 large T antigen, were included as positive controls for p19Arf overexpression and p53 overexpression, respectively. In (c) the MEF control was run on the same gel but a lane not required was spliced out of this gel. Protein size standards in kDa are indicated on the left
Six lymphomas lacking Puma alone were examined in most detail. None of them expressed a high level of p53 protein (Figure 8c), had a p53 mutation detectable by sequencing or had deleted the Ink4A/Arf locus (Supplementary Figure 8 and data not shown). In addition, none of the 11 cell lines derived from Eμ-myc/puma−/− lymphomas were refractory to treatment with etoposide, as was, in contrast, the known p53 mutant lymphoma, Eμ-myc/noxa−/−puma−/− no. 104 (Supplementary Figure 8 and data not shown). Thus, although the absence of Puma does not totally ablate the selection for mutations in the p53 pathway, it appears to substantially reduce it.
Discussion
Although the BH3-only protein Puma and, to a lesser extent, Noxa, are crucial for apoptosis mediated by the tumour suppressor p53, mice deficient for noxa or puma,19,20,22 or even both,24 have not proven prone to tumour development. Nonetheless, we reasoned that Puma and Noxa might constrain tumour development in the context of an oncogenic lesion that promotes apoptosis. As the major barrier to Myc-induced lymphomagenesis is thought to be Myc-induced apoptosis, which acts predominantly through the p19Arf-p53 pathway, regulated by the Bcl-2 protein family,32 we have explored whether loss of Puma and/or Noxa could substitute for p53 loss in accelerating Myc-induced lymphomagenesis.
Consistent with a role in mediating Myc-induced apoptosis, puma and noxa mRNA expression levels were elevated in pro-B and mature B cells from Eμ-myc mice, albeit not in pre-B cells. Two pathways for induction of apoptosis by c-Myc have been identified. In the better-studied path, c-Myc induces p19Arf expression, and p19Arf in turn prevents the degradation of p53 by sequestering its ubiquitin ligase, Mdm2.31,37 The resulting elevated p53 level induces transcription of noxa and puma,15–17 which most likely explains the elevated levels of noxa and puma mRNA and of Puma protein produced by Myc. However, at least in bone marrow-derived B-lymphoid cells35 c-Myc can also induce apoptosis by a p19Arf- and p53-independent mechanism,39 mediated in part by induction of Bim,10 as well as repression of anti-apoptotic Bcl-xL 34 and Bcl-2.10 Theoretically, c-Myc might also enhance Puma and Noxa expression by this second pathway.
Loss of Puma impaired the Myc-induced apoptotic programme
Puma loss protects diverse cell types, including B lymphocytes, against a range of apoptotic stimuli both in vitro19,20 and in vivo.21,24 Pertinently, Puma loss rendered growth factor-deprived myeloid progenitor cells refractory to c-Myc-induced apoptosis in culture.20 Consistent with this, compared with Eμ-myc (wt) cells, Eμ-myc pre-B cells deficient for Puma were partially protected from spontaneous death in vitro (cytokine deprivation) and presumably also in vivo.
In accordance with the enhanced survival of Eμ-myc/puma−/− pre-B cells in vitro, pre-malignant Eμ-myc/puma−/− mice had more pre-B cells in their peripheral blood than did control Eμ-myc animals. In addition, the number of mature B cells in the bone marrow was increased, consistent with the acceleration in B-lymphoma development observed in Eμ-myc/puma−/− mice. Lastly, pre-B cells lacking Puma were partially protected from DNA damage-induced death. Conversely, Noxa-deficient pre-neoplastic Eμ-myc mice exhibited excess pre-B and B cells in the bone marrow, but these cells did not survive better in vitro with the stimuli tested and the increase in the bone marrow was not associated with acceleration in either pre-B or B-cell lymphoma. These findings suggest that the number of B-lymphoid cells present in the pre-malignant state need not correlate with the rate of tumour onset. Consistent with this idea, Eμ-myc mice deficient for Bcl-2 had much fewer B-lymphoid cells but developed lymphomas as rapidly as wt Eμ-myc mice.40 Loss of Puma may accelerate Eμ-myc-induced lymphomagenesis not only by increasing the number of target cells but also by prolonging the survival of small numbers of pre-B cells subjected to limiting cytokine conditions and/or oncogenic stress, thereby increasing their chance acquisition of oncogenic lesions that cooperate with Myc in neoplastic transformation. As more Puma-deficient mice succumbed to sIg+ B lymphomas than pre-B lymphomas, Puma loss may preferentially allow inappropriate cell survival at the pre-B to sIg+ B-cell transition.
puma heterozygosity increased tumour burden but did not hasten tumour onset
Loss of one puma allele was not sufficient to accelerate tumour onset significantly, even for B lymphomas. This was somewhat surprising, given the significant protection from apoptosis afforded by haploinsufficiency of puma in lymphoid cells19–21 and the finding that a shRNA construct, which diminished but did not abolish Puma expression, accelerated lymphomagenesis in lethally irradiated mice reconstituted with fetal liver-derived stem cells from Eμ-myc mice.11 Although the level of puma knockdown achieved may have exceeded 50%, it is also possible that c-Myc-induced lymphomagenesis in irradiated, reconstituted mice is more sensitive to Puma dosage than that in unmanipulated Eμ-myc mice. Puma knockdown might, for example, have enhanced survival of fetal liver stem cells during the in vitro infection period with the shRNA retrovirus, thereby facilitating more rapid lymphomagenesis in the transplant recipients.
Interestingly, ill Eμ-myc/puma−/− mice and even some Eμ-myc/puma+/− mice exhibited higher levels of leukaemia and splenomegaly than those seen in ill wt Eμ-myc mice. As lymphoma/leukaemia cells in wt Eμ-myc mice exhibit spontaneous apoptosis, 36 Puma must play a critical role in the death of these cells in vivo and thereby limit their accumulation. However, the increased leukaemia and spleen size in sick Eμ-myc/puma+/− mice did not lead to significantly earlier deaths compared with Eμ-myc (wt) mice.
Puma delayed the onset and limited the tumour burden of B-cell lymphoma in Eμ-myc mice
Pre-B lymphomas predominate in wt Eμ-myc mice,10,28 but Puma deficiency, like loss of Bim10 or loss of one allele of p53, favoured the development of sIg+ B lymphomas, which arose much earlier in the Eμ-myc/puma−/− mice. Unlike Bim,10 however, Puma proved not to be important for apoptosis at the B-cell receptor checkpoint, as loss of Bim but not of Puma protected against killing by cross-linking of the B-cell antigen receptor.
Thus, despite the marked similarities of loss of Bim and loss of Puma on Myc-induced lymphomagenesis – both yielding more rapid sIg+ B lymphomas with associated leukaemia – slightly different stages of development may be affected. The increased Puma protein levels at the pre-B stage may suggest that Puma plays a role at the pre-B to B checkpoint. At that developmental stage, the process of Ig light chain gene rearrangement, which requires DNA breakage, may both predispose the cells to acquire oncogenic mutations and render them sensitive to Puma-mediated apoptosis. The predominant mature B-cell immunophenotype of Puma-deficient tumours in this study contrasts with the report that tumours arising in mice receiving shRNA-mediated knockdown of either puma or p53 were pre-B lymphomas, but the phenotype of only three lymphomas of each genotype was reported and the pre-B compartment in that system might be more vulnerable to Puma knockdown.11
Noxa and Puma can cooperate to affect tumour onset in both pre-B and B cells
It is not surprising that Puma generally has a greater role in apoptosis than Noxa, because Puma (like Bim) can engage and inactivate all of the pro-survival Bcl-2 family members, whereas Noxa antagonises only Mcl-1 and A1.41,42 Nevertheless, the functional overlap of Noxa and Puma in DNA damage-induced apoptosis of certain cell types, including pro-B/pre-B cells,24 indicated that Noxa and Puma might cooperate in suppressing Myc-induced lymphomagenesis. This did indeed occur to a significant extent, as the loss of Noxa on a puma+/− background accelerated Eμ-myc-induced lymphomagenesis almost as much as loss of both puma alleles. Moreover, loss of both Puma and Noxa accelerated the development of pre-B as well as mature B-cell tumours. As loss of Puma alone did not accelerate development of pre-B lymphoma, Noxa must contribute at least in the pro-B/pre-B compartment to tumour suppression. It is possible that when Puma function is compromised, for example in pre-malignant Eμ-myc/puma−/− B-lineage cells, the levels and/or activation state of Noxa are heightened. Their synergy was illustrated by step-wise increases in B-cell subsets in the peripheral blood observed with loss of one or both alleles of puma on a Noxa-deficient background.
The accelerated tumour onset in Eμ-myc/noxa−/−puma+/− mice was not accompanied by increased leukaemia/lymphoma burden, as spleen size and leukocyte numbers resembled those in Eμ-myc/puma+/− mice. As we found no evidence that the acceleration was associated with loss of the remaining puma allele in Eμ-myc/noxa−/−puma+/− mice, loss of one allele of puma and both noxa alleles may reach a threshold of apoptosis resistance sufficient for tumorigenesis. Nevertheless, the markedly low Puma protein level observed in a number of tumours with a single puma allele may well indicate that tumorigenesis can select for suppression of expression from that allele at the transcriptional and/or post-transcriptional level.43
It is curious that although Noxa deficiency accelerated Myc-driven lymphoma development in a Puma heterozygous background, Noxa deficiency in a Puma wild-type background appeared to delay the onset of at least the pre-B tumours. At present the basis for this difference is unknown, as Noxa and Puma both function at least primarily by engaging pro-survival family members, albeit with overlapping specificity. The fact that Noxa and Puma can be regulated in multiple ways might be relevant. We do not think the differing impact of Noxa deficiency on tumorigenesis reflects some compensation between Noxa and Puma, because we have previously reported24 that loss of Puma had no impact on the expression of Noxa or three other BH3-only proteins (Bim, Bad or Bid) and that, conversely, loss of Noxa has no impact on the expression of Puma, Bad, Bim or Bid, in either unstressed lymphoid cells or those exposed to two apoptotic stimuli. Furthermore, we are aware of no reported evidence for compensating changes in expression of BH3-only proteins in other systems.
Loss of both Noxa and Puma was not equivalent to loss of p53 in lymphomagenesis but loss of Puma did reduce selection for p53 loss
Clearly, neither loss of Puma, nor even loss of both Noxa and Puma, accelerated Eμ-myc-induced lymphomagenesis as much as loss of even one allele of p53. Interestingly, shRNA-mediated knockdown of puma accelerated retroviral c-Myc-induced lymphoma development as potently as did knockdown of p53, 11 perhaps due to differences in that experimental system (see Discussion above). Pertinently, in response to stimuli that activate p53, the puma knockdown cells underwent G1 cell cycle arrest normally,11 consistent with the notion that Puma mediates p53’s apoptotic action but not its cell cycle arrest function.11,16,17 Collectively, these findings indicate that the pro-apoptotic activity of p53 is only one of the ways it safeguards against tumour development. Its roles in cell cycle arrest, DNA repair and cellular senescence are likely to contribute to tumour suppression.1
We assessed the functionality of the p53 pathway in the lymphoma cells from the levels of p19/Arf protein (as p53 normally downregulates p19/Arf expression by a negative feedback loop33), the presence of the Ink4a/Arf locus by genomic PCR36 the level of p53 protein (high levels indicating mutant, stabilised p53 protein) and by sequencing of the p53 gene. We found only a single Puma-deficient lymphoma (which also lacked Noxa) in which the p53 pathway was clearly inactive. Garrison et al.43 have now also reported that the p53 pathway is perturbed in some Puma-deficient lymphomas, in contrast to a prior study involving knockdown of puma.11 Hence, we conclude that the loss of Puma does not entirely supplant the need for loss of p53 function, but it does appear to substantially reduce the selective pressure. Retention of some selection for p53 loss is to be expected, because p53 can prevent tumour development through several non-apoptotic mechanisms, such as DNA repair and senescence.1
Concluding remarks
In the Eμ-myc lymphoma model, Puma appears to have a major tumour suppressor function, particularly in sIg+ B cells, where Bim seems to have a similar role.10 Although the role of Noxa seems more limited, the accelerated lymphomagenesis in both the pre-B and B-cell compartments produced by loss of Noxa on a puma heterozygous background demonstrates that it can contribute. The cell type-specific action of Noxa and Puma demonstrated here underlines the complexities of the apoptotic programme and the process of tumorigenesis and has important implications for the understanding and treatment of human cancer. As around half of all human tumours have lost p53 function, it will be important to learn how to restore this apoptotic pathway by identifying other ways to increase Puma or Noxa expression or to mimic their function, such as with BH3 mimetic agents.8
Materials and Methods
Mice
All experiments with mice conformed to the guidelines of the Melbourne Research Directorate Animal Ethics Committee. Generation and genotyping of mice deficient for Noxa, 19 Puma19 or both24 (all generated on an inbred C57BL/6 background) and those lacking p5344 (generated on a mixed C57BL/6x129SV background but backcrossed with C57BL/6 mice for >20 generations) have been described. Eμ-myc mice (backcrossed onto a C57BL/6 background for >20 generations), which overexpress c-Myc in B-lymphoid cells under control of the immunoglobulin heavy chain gene enhancer (Eμ), have been described.25,28 For this study, Eμ-myc males were crossed with p53−/− females to generate Eμ-myc/p53+/− offspring and with puma−/− and noxa−/− females to produce Eμ-myc/puma+/− or Eμ-myc/noxa+/− males, which were then crossed with puma−/− or noxa−/− females to yield Eμ-myc/puma−/− or Eμ-myc/noxa−/− progeny, respectively. Similarly, Eμ-myc/noxa−/− males were crossed with noxa−/−puma−/− double knockout (DKO) females to generate Eμ-myc/noxa−/−puma+/− males, which were then bred to noxa−/−puma−/− females to yield Eμ-myc/noxa−/−puma−/− mice. Eμ-myc transgenic offspring were identified at 4 weeks of age from the increased size of their leukocyte nuclei using a Coulter particle count and size analyser Z2 (Beckman Coulter).28 Genotyping for p53 alleles was performed by PCR on genomic DNA using primers 5′TTATGAGCCACCCGAGGT, 5′TATACTCAGAGCCGGCCT and 5′TCCTCGTGCTTTACGGTATC. Mice used for mating were censored from the analysis as mated mice have a delayed tumour onset (EM Michalak, A Strasser unpublished observations).
Cell culture and cell viability assays
FACS-sorted pro-B, pre-B and sIg+ B cells were cultured at a starting density of 2–5×105/ml at 37°C in a humidified 10% CO2 incubator in high-glucose Dulbecco’s Modified Eagle’s medium supplemented with 10% fetal calf serum (JRH Biosciences), 50 µM 2-mercaptoethanol (Sigma) and 100 µM asparagine (Sigma). Cell viability was measured by FACS analysis as the fraction of cells not stained by either Annexin V-FITC or PI as described.19 The extent of apoptosis induced specifically by etoposide was calculated using the following equation: [induced apoptosis – (spontaneous apoptosis/100 – spontaneous apoptosis)].
Immunofluorescence staining, flow cytometric analysis and cell sorting
FACS analysis and cell sorting were performed using monoclonal antibodies as described19 and a FACScan (Becton Dickinson) for analysis. For survival assays, pro-B cells (B220+CD43+sIgM−sIgD− or B220+ c-Kit+sIgM−sIgD−), pre-B cells (B220+CD43−sIgM−sIgD− or B220+ c-Kit−sIgM−sIgD−) and immature plus mature B cells (B220+CD43− sIgM+sIgD+ or B220+c-Kit−sIgM+sIgD+) were sorted from bone marrow or spleen using a MoFlo (Cytomation) or a Diva (Becton Dickinson) cell sorter.
Haematopoietic cell analysis
Spleen, lymph nodes (axillary, brachial, inguinal), bone marrow (both femora) and peripheral blood of healthy 4- to 6-weekold mice were analysed as described.24 To confirm that mice lacked transplantable tumour cells at the time of analysis, two C57BL/6 recipients per mouse analysed were injected with 2–6×106 spleen cells and monitored for lymphoma-free survival for at least 50 days. Blood leukocytes for 4-week old mice were enumerated using a Coulter particle count and size analyser Z2 (Beckman Coulter) and those from 5- to 6-week-old pre-malignant mice and tumour-burdened mice using an ADVIA haematology system (Bayer).
Western blot analysis
Western blot analysis was performed by standard procedures using protein extracted from primary lymphomas, and the blots probed with the following antibodies: rabbit polyclonal anti-Puma (directed to the Puma N terminus; ProSci), CM5 rabbit polyclonal anti-mouse p53 (Novacastra), 3F11 hamster monoclonal anti-mouse Bcl-2 (Pharmingen), rabbit polyclonal anti-p19Arf (AbCam) and 5-C3-1 rat monoclonal anti-mouse p19Arf (kind gift of Dr. Roussel), rabbit polyclonal anti-Bim (Stressgen), 3C5 rat monoclonal anti-Bim (Alexis), 2H12 mouse monoclonal anti-Bcl-X (BD Pharmingen), rabbit polyclonal anti-Mcl-1 (Rockland). Antibodies to β-actin (AC-40, Sigma) or HSP70 (rat monoclonal antibody N6, a gift from Dr R Anderson, Peter MacCallum Cancer Centre, Melbourne) provided loading controls.
Real-time qRT-PCR analysis
Bone marrow B-lymphoid subsets were sorted as described above. RNA was prepared using the RNeasy Kit (Qiagen). First strand cDNA was prepared from 0.5–1.5 µg RNA using the Taqman RT system (Roche). Real-time PCR was performed using the ABI Prism 7900 (Applied Biosystems) and the QuantiTect SYBR Green PCR Kit (Qiagen) in 15 µl reaction volumes. Data analyses were performed with the ΔCT method using β-actin as an internal control. Primer sequences are provided in the Supplementary Materials.
Analysis of Ink4A/Arf locus and p53 mutation status in lymphomas
Vials of cryo-preserved lymphomas were thawed, dead cells removed using a Ficoll gradient and genomic DNA generated. Multiplex PCR analysis of genomic DNA for the Ink4A/Arf locus was performed to reveal gross deletions, using exon-specific primers for α-actin, exons 2, 1α and 1β as described earlier.36,45 Sequencing of p53 from genomic DNA was performed using two primer sets designed for each of exons 4–10 (the second set used if required for clarification). PCR products were purified and sequencing was performed using the BigDye Terminator cycle sequencing kit (Applied Biosystems). Primer sequences are available upon request.
Statistical analysis
Prism software (GraphPad Software Inc.) was used for generating Kaplan–Meier plots and for performing statistical analysis (using a log-rank test) of survival of mice and tumour onset. All other statistical analyses used two-tailed t-tests assuming equal variances. Cell counts and spleen weights were tested on the log-scale, whereas percentage data were tested on the arc sine scale.46 Trend test was performed by linear regression. P-values of less than 0.05 were considered to indicate statistical significance.
Supplementary Material
Acknowledgements
We thank Professor S Cory and Drs. A Villunger, DCS Huang, H Puthalakath, P Bouillet, C Vandenberg and A Egle for their input and interesting discussions; K Vella, K Pioch, N Iannarella, G Siciliano and A Naughton for animal care; Dr. F Battye, C Tarlinton, V Milovac, J Garbe and C Young for cell sorting; T Nikolaou and G Thomas for γ-irradiation; J Corbin for automated blood analysis; B Helbert and C Young for genotyping; Dr. S Mihajlovic, A Hasanein, K Weston for histological sections; P Bouillet and the late A W Harris for mice. This work was supported by fellowships and grants from the NHMRC (Canberra; program no. 257502; RD Wright Biomedical CDA 406675); the Leukemia and Lymphoma Society (SCOR grant no. 7015); the NIH (CA043540-18 and CA80188-6); the JDRF/NHMRC; the Leukemia Research Foundation (LRF) (Post-graduate Scholarship to LH); and the Cancer Council Victoria (Postdoctoral Cancer Research Fellowship to EMM). EMM contributed to experimental design, performed experiments, analysed results, made the figures and wrote the paper; EJ, LH, MSC and LT contributed to some experiments; GKS aided the statistical analysis; CLS designed experiments, assisted with interpretation and contributed to writing the article; JMA assisted with interpretation and contributed to writing the article; AS conceived the study, designed the research, assisted with interpretation and contributed to writing the article.
Abbreviations
- BH
Bcl-2-homology
- DKO
double knockout
- DP
double positive
Footnotes
Conflict of interest/disclosure: The authors declare no competing financial interests.
References
- 1.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Kastan MB. Wild-type p53: tumors canșt stand it. Cell. 2007;128:837–840. doi: 10.1016/j.cell.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Szekely L, Okan I, Klein G, Wiman KG. Wild-type p53-triggered apoptosis is inhibited by bcl-2 in a v-myc-induced T-cell lymphoma line. Oncogene. 1993;8:3427–3431. [PubMed] [Google Scholar]
- 4.Strasser A, Harris AW, Cory S. Bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell. 1991;67:889–899. doi: 10.1016/0092-8674(91)90362-3. [DOI] [PubMed] [Google Scholar]
- 5.Strasser A, Harris AW, Jacks T, Cory S. DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell. 1994;79:329–339. doi: 10.1016/0092-8674(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 6.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat rev. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 7.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 8.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 10.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemann MT, Zilfou JT, Zhao Z, Burgess DJ, Hannon GJ, Lowe SW. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci USA. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sturm I, Stephan C, Gillissen B, Siebert R, Janz M, Radetzki S, et al. Loss of the tissue-specific proapoptotic BH3-only protein Nbk/Bik is a unifying feature of renal cell carcinoma. Cell Death Differ. 2006;13:619–627. doi: 10.1038/sj.cdd.4401782. [DOI] [PubMed] [Google Scholar]
- 13.Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A, et al. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene. 2005;24:1348–1358. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- 14.Mestre-Escorihuela C, Rubio-Moscardo F, Richter JA, Siebert R, Climent J, Fresquet V, et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109:271–280. doi: 10.1182/blood-2006-06-026500. [DOI] [PubMed] [Google Scholar]
- 15.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, et al. Noxa, a BH3-only member of the bcl-2 family and candidate mediator of p53-induced apoptosis. Science (New York, NY) 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 16.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Flemington C, Houghton AB, Gu Z, Zambetti GP, Lutz RJ, et al. Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc Natl Acad Sci USA. 2001;98:11318–11323. doi: 10.1073/pnas.201208798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villunger A, Michalak EM, Coultas L, Müllauer F, Böck G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins Puma and Noxa. Science (New York, NY) 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 20.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 21.Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, et al. BH3-only proteins Puma and Bim are rate-limiting for {gamma} -radiation and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood. 2005;106:4131–4138. doi: 10.1182/blood-2005-04-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibue T, Takeda K, Oda E, Tanaka H, Murasawa H, Takaoka A, et al. Integral role of Noxa in p53-mediated apoptotic response. Genes Dev. 2003;17:2233–2238. doi: 10.1101/gad.1103603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naik E, Michalak EM, Villunger A, Adams JM, Strasser A. UV-radiation triggers apoptosis of fibroblasts and skin keratinocytes mainly via the BH3-only protein Noxa. J Cell Biol. 2007;176:415–424. doi: 10.1083/jcb.200608070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michalak EM, Villunger A, Adams JM, Strasser A. In several cell types the tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 2008;15:1019–1029. doi: 10.1038/cdd.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 26.Langdon WY, Harris AW, Cory S, Adams JM. The c-myc oncogene perturbs B lymphocyte development in Eμ-myc transgenic mice. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- 27.Jacobsen KA, Prasad VS, Sidman CL, Osmond DG. Apoptosis and macrophage-mediated deletion of precursor B cells in the bone marrow of Eμ-myc transgenic mice. Blood. 1994;84:2784–2794. [PubMed] [Google Scholar]
- 28.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The Eμ-myc transgenic mouse: a model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167:353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 30.Strasser A, Elefanty AG, Harris AW, Cory S. Progenitor tumours from Em-bcl-2-myc transgenic mice have lymphomyeloid differentiation potential and reveal developmental differences in cell survival. EMBO J. 1996;15:3823–3834. [PMC free article] [PubMed] [Google Scholar]
- 31.Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsson JA, Cleveland JL. Myc pathways provoking cell suicide and cancer. Oncogene. 2003;22:9007–9021. doi: 10.1038/sj.onc.1207261. [DOI] [PubMed] [Google Scholar]
- 33.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARFMdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eischen CM, Woo D, Roussel MF, Cleveland JL. Apoptosis triggered by myc-induced suppression of Bcl-XL or Bcl-2 Is bypassed during lymphomagenesis. Mol cell biol. 2001;21:5063–5070. doi: 10.1128/MCB.21.15.5063-5070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maclean KH, Keller UB, Rodriguez-Galindo C, Nilsson JA, Cleveland JL. c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol cell biol. 2003;23:7256–7270. doi: 10.1128/MCB.23.20.7256-7270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 38.Bertwistle D, Zindy F, Sherr CJ, Roussel MF. Monoclonal antibodies to the mouse p19(Arf) tumor suppressor protein. Hybridoma and hybridomics. 2004;23:293–300. doi: 10.1089/hyb.2004.23.293. [DOI] [PubMed] [Google Scholar]
- 39.Juin P, Hueber A-O, Littlewood T, Evan G. c-Myc-induced sensitization to apoptosis is mediated through cytochrome c release. Genes Dev. 1999;13:1367–1381. doi: 10.1101/gad.13.11.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly PN, Puthalakath H, Adams JM, Strasser A. Endogenous bcl-2 is not required for the development of E{micro}-myc-induced B-cell lymphoma. Blood. 2007;109:4907–4913. doi: 10.1182/blood-2006-10-051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of pro-survival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 42.Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, et al. BH3 domains of BH3-only proteins differentially regulate bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Garrison SP, Jeffers JR, Yang C, Nilsson JA, Hall MA, Rehg JE, et al. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol cell biol. 2008;28:5391–5402. doi: 10.1128/MCB.00907-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 45.Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 46.Zar JH. Biostatistical Analysis. New Jersey: Prentice Hall Int; 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.