Abstract
Clinical trials have demonstrated the importance of aromatase inhibitor (AI) therapy in the effective treatment of hormone-dependent breast cancers. In contrast to tamoxifen, an antagonist of the estrogen receptor (ER), AIs have shown to be better tolerated along with decreased recurrence rates of the disease. Currently, three third-generation AIs are being used: exemestane, letrozole and anastrozole. Our laboratory is attempting to understand several aspects of aromatase inhibitor functionality. In this paper, we first review recent findings from our structure-function studies of aromatase as well as the molecular characterization of the interaction between AIs and aromatase. Based on these studies, we propose new evidence for the interaction of letrozole and exemestane with aromatase. In addition, we will discuss recent results generated from our AI-resistant cell lines. Our laboratory has generated MCF-7aro cells that are resistant to letrozole, anastrozole, exemestane and tamoxifen. Basic functional characterization of aromatase and ERα in these resistant cell lines has been done and microarray analysis has been employed in order to better understand the mechanism responsible for AI resistance on a genome-wide scale. The results generated so far suggest the presence of at least four types of resistant cell lines. Overall, the information presented in this paper supplements our understanding of AI function, and such information can be valuable for the development of treatment strategies against AI resistant breast cancers.
Keywords: models, aromatase inhibitor resistance
Introduction
Aromatase, a cytochrome P450, catalyzes three consecutive hydroxylation reactions converting C19 androgens to aromatic C18 estrogens. Upon receiving electrons from NADPH-cytochrome P450 reductase, aromatase converts androstenedione and testosterone to estrone and estradiol, respectively. Estrogens are female hormones involved in the development and growth of breast tumors. Approximately 60% of premenopausal and 75% of postmenopausal breast cancer patients have estrogen-dependent carcinomas [1].
Aromatase inhibitors (AIs), which function to effectively block the synthesis of estrogens, have moved to the forefront of treatment therapies for hormone-dependent breast cancers in post-menopausal women. Based on several major clinical trials, currently available AIs produce greater clinical benefit with near-complete specificity. These drugs were also found to be better tolerated than the commonly used ER antagonist tamoxifen, and were associated with lower incidences of endometrial cancer, vaginal bleeding and discharge, cerebrovascular events, venous thromboembolic events, and hot flashes [2]. In addition, the incidence of contralateral breast cancer occurrence was found to be significantly lower in the aromatase inhibitor group than the tamoxifen group [3–5].
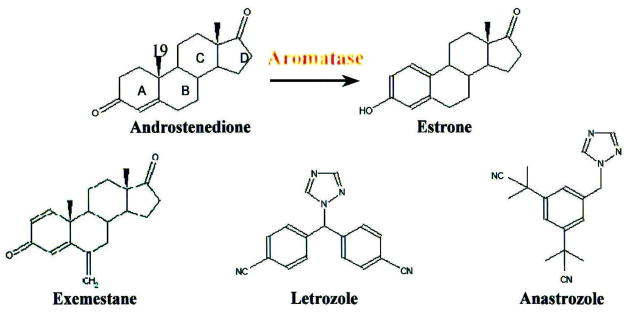
In the past 3 decades, a series of AIs have been produced. Historically, AIs have been grouped into three generations. The ‘first-generation inhibitor’ aminoglutethimide was the first drug to be used as an aromatase inhibitor [6]. Its non-specific inhibition of P450 enzymes, other than aromatase, caused significant side effects. The representative of ‘second-generation inhibitors’, 4-hydroxy-4-androstene-3,17-dione (4-OHA), was the first selective aromatase inhibitor to be used clinically and was effective and well tolerated [7–9]. Yet, due to the extensive first-pass metabolism, 4-OHA needs to be administrated intramuscularly. The three FDA-approved AIs currently available in the US (i.e., exemestane, letrozole and anastrozole) (structure see Fig. 1) are referred to as ‘third-generation inhibitors.’ 4-OHA and current FDA approved agents are all specific, more potent and offer significant safety advantages over their nonselective predecessors [2–5].
Figure 1.
Structures of exemestane, anastrozole and letrozole.
Based on their structures, AIs can be grouped into ‘non-steroidal’ and ‘steroidal’ inhibitors. Non-steroidal inhibitors (e.g., letrozole and anastrozole) have the triazole functional group that interact with the heme prosthetic group of aromatase and act as competitive inhibitors with respect to the androgen substrates. Steroidal inhibitors (e.g., exemestane and 4-OHA) were originally designed as substrate analogs that compete with the substrate of aromatase. These two steroidal inhibitors are also mechanism-based inhibitors which require the catalytic ability of active aromatase to convert them into active intermediates. The intermediates then bind irreversibly to the enzyme and cause its inactivation in a time-dependent manner.
Inhibitory mechanism of AIs
While these third-generation AIs are shown to be very potent and specific, the structural basis of drug recognition by aromatase has remained elusive because the three-dimensional structure of this enzyme is not yet determined, and also because these AIs were developed through extensive structure-activity relationship (SAR) studies. Our laboratory has had a long-term interest in the structure-function relationship of aromatase. We have published extensively and made significant progress in this area during the last ten years [10–14]. We have expressed and purified functionally active recombinant human aromatase from E. coli. The Km and Vmax values of the recombinant enzyme were estimated to be 301 nM and 130 nmol/mg/min for androstenedione [14]. Using this preparation, the three-dimensional folding of aromatase was revealed by proteomic analysis [14]. Combined with site-directed mutagenesis, several critical residues involved in enzymatic catalysis and suicide inhibition by exemestane were evaluated. Based on our results, a new clamping mechanism of steroid substrate/exemestane binding to the active site of aromatase is proposed [14].
Mechanism-based inhibition of aromatase by exemestane has been demonstrated by UV/Vis spectral analysis using our recombinant enzyme preparation [14]. In the oxidized state, ligand-free aromatase exhibited a Soret absorption maximum at approximately 420nm. This is associated with the low-spin state of the heme iron with a water molecule as a sixth proximal axial ligand. When bound to androstenedione, the complex produced a type I binding spectrum, characterized by a reduction in the Soret band at 420 nm and a corresponding absorption maximum at 394 nm. The observation of a type I binding spectrum in the presence of exemestane indicates that exemestane binds to the substrate-binding site during the first step of inhibition. It is well known that mechanism-based inhibitors cause time-dependent inactivation of aromatase only in the presence of cofactors such as NADPH [15]. To better define exemestane as a mechanism-based inhibitor, exemestane was incubated with purified aromatase in the presence of human NADPH-P450 reductase and NADPH [14]. The reaction mixture was kept at 4°C subsequent to the 10 min incubation at 37°C, and the time course spectra were recorded. At zero time, there was no peak at 420nm in the reaction mixture with exemestane or androstenedione. The absorption at 420nm appeared in the reaction mixture with androstenedione after a 30 min incubation on ice. However, the reaction mixture with exemestane failed to recover the 420nm peak even after overnight incubation on ice. These results indicate that after the aromatization of androstenedione, estrone releases from the enzyme, allowing a water molecule to re-ligate to iron, switching it back to a six-fold coordination state. In contrast, acting as a mechanism-based inhibitor, exemestane (or its intermediates) fails to release once it binds to the enzyme.
In contrast to the type I binding spectrum observed for the exemestane-aromatase complex, a type II binding spectrum was observed with the letrozole-aromatase complex [14]. In this spectral analysis of the letrozole-aromatase complex, we identified an increase in the absorption at 422 nm and a decrease at 394nm. This spectral change is associated with the direct interaction of letrozole with the Fe3+ displacing the water molecule as the sixth axial ligand, thus increasing in the low-spin character of the Fe3+.
In order to better understand differences between the functionality of these drugs, we have further studied the effects of AIs on aromatase protein stability. We have investigated the effects of three FDA-approved aromatase inhibitors, exemestane, letrozole and anastrozole, in the aromatase-over expressing breast cancer cell line MCF-7aro [16]. Using western analysis as the major technique, we have found that exemestane treatment significantly reduces aromatase protein level. Exemestane induces aromatase degradation in a dose-responsive manner (25 to 200 nM), and the effect can be seen in as early as 2 hours. Metabolic labeling with 35S-methionine was used to determine the half-life of aromatase protein. In the presence of 200 nM exemestane, the half-life of aromatase was reduced to 12.5 hours, compared to 28.2 hours in the untreated cells. Furthermore, exemestane-induced aromatase degradation can be completely blocked by 10 μM MG132, indicating that the degradation is mediated by proteasome. The two non-steroidal AIs, letrozole and anastrozole, at concentrations as low as 8 nM, caused an increase of aromatase protein levels. These results were expected because letrozole and anastrozole bind to aromatase with high affinities that stabilize the structure of aromatase protein. In addition, androgen substrates, i.e., testosterone and androstenedione, have no effect on the stability of aromatase protein. Therefore, we have found that exemestane, different from letrozole and anastrozole can destabilize the aromatase protein, following the enzyme inactivation step.
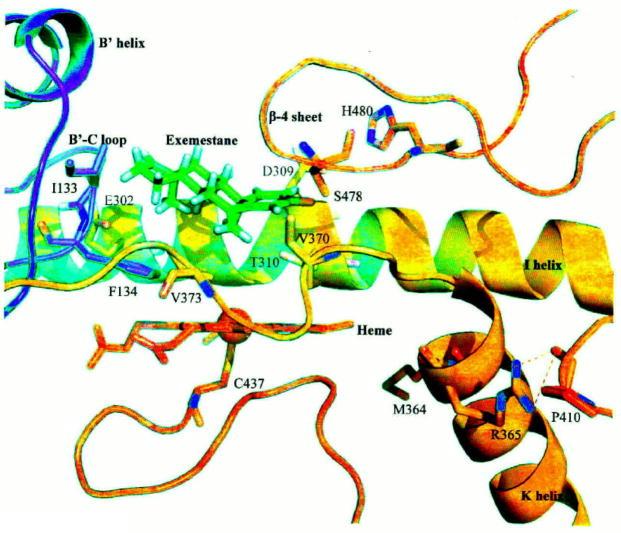
These recent findings from our laboratory help us better understand the inhibitory mechanisms of the steroidal aromatase inhibitor exemestane, and the non-steroidal AIs, letrozole and anastrozole. Taken together, our data is further supported by the 3-D structural aromatase model generated by Favia et al. [17], which is more reliable than those previously generated in this and other laboratories. By carefully examining this new model and the results from our recent structure-function studies of aromatase, a new clamping mechanism of steroidal substrate/inhibitor, binding to the active site, has been proposed (14). The heme iron is ligated by a conserved cysteine (C437) and the propionates of the heme interact with the side chains of R115, W141, R145, R375, and R435. The steroid substrate/inhibitor sits above the heme with its C19 methyl group pointing to the heme iron and is positioned next to the I helix (Fig. 2). Our site-directed mutagenesis data allows us to identify three additional important regions in the active site of aromatase. Together with D309 (in the I helix), S478 and H480 (in the β-4 sheet at the carboxy-terminus) are thought to participate in a charge relay system that leads to the aromatization of the A ring of the androgen substrate. I133 and F134 in the B′-C loop are hypothesized to interact with the D ring of the substrate/inhibitor through Van der Waals forces. The 3′-flanking loop (P368-M374) of the K helix is thought to participate in forming the hydrophobic ligand-binding pocket and hence residue V373 possibly interacts specifically with the B ring of the steroidal substrate/inhibitor. This loop (P368-M374), together with the B′-C loop and β4-including loop, holds the steroid substrate/inhibitor at the correct orientation. We hypothesize that exemestane is converted to reactive intermediates by the heme through the hydroxylation of the C-19 group, helped by D309 and T310. Subsequently, the intermediates bind irreversibly to the enzyme, causing suicide inhibition in which D309 may also be involved.
Figure 2.
Clamping mechanism of exemestane binding provided by the heme, I helix, B′-C loop, β-4 sheet, and the 3′-flanking loop of the K helix.
In a recent study, Ma et al. identified and characterized genetic polymorphisms in the human aromatase gene [18]. There are four single nucleotide polymorphisms (SNPs) in the coding region. These cSNPs alter the following amino acids: W39R, T201M, R264C and M364T. Interestingly, the M364T variant was found to be less stable and to have significantly lower affinities for the androgen substrate and for the inhibitor exemestane. Our laboratory previously generated two mutants, R365A and R365K [19]. These mutants were not active. The immunoprecipitation analysis revealed that these mutants were expressed, but at levels lower than that of the WT enzyme. These results indicate that R365 plays a very critical role during enzyme catalysis because it cannot be replaced with a lysine residue. The computer modeling analysis has revealed that M364 and R365 are situated in the K helix (Fig. 2). It is thought that the side chain of R365 forms a hydrogen bond with the backbone carbonyl oxygen of P410, which is located in the loop between the β-1/β-2 sheets (R375-I395) and L helix (G439-R456) [20]. C437, the heme-binding cysteine residue, is located at the end of this loop. Possibly, R365 stabilizes this loop structure. M364 faces inside the active site, although it is not close to the heme and steroidal ligand. It is likely that M364 helps to form the hydrophobic pocket together with the loop P368-M374 [20]. Overall, our structure-function studies offer new information regarding interactions between aromatase and its inhibitors, and we provide new evidence that further delineates between steroidal and non-steroidal aromatase inhibitor functionality.
AI resistance
Increasing evidence has shown that AIs are superior to the conventional anti-estrogen tamoxifen in treating hormone-dependent breast cancer in postmenopausal women [2–5]. Although AI treatment has shown to be effective in the clinic, resistance to these therapies still occurs and is highly problematic, due to the lack of response to current endocrine therapy. There are two types of endocrine resistance. De novo/intrinsic resistance refers to lack of response at initial exposure to endocrine therapy of aromatase-positive and estrogen receptor (ER)-positive breast cancers. In contrast, acquired resistance is developed during endocrine therapy of patients who respond to the treatment initially. We and other investigators believe that elucidating the mechanisms of resistance to AIs/anti-estrogens, on the molecular level will be extremely valuable for the effective treatment of hormone-dependent breast cancers and for the development of novel approaches to treat patients who fail endocrine therapy.
It is understood that only aromatase-positive and ER-positive breast cancer would respond to the treatment of aromatase inhibitors. Abnormally higher expression of aromatase in breast cancer cells and/or surrounding adipose stromal cells than normal breast tissue, have been demonstrated by a number of laboratories by aromatase activity measurement [21–23], immunohistochemical analysis [24–27] and RT-PCR analysis [28, 29]. The in situ estrogen biosynthesis is thought to have a significant influence on tumor maintenance and growth in breast cancer patients. It is well known that not all ER positive breast cancers respond to endocrine therapy with anti-estrogens or AIs. Typical AI response rates vary from 20 to 50% (as discussed by Dowsett et al. [30]). The exact reason for the lack of response to AIs, or de novo resistance, in some ER+ patients is not known. At this meeting, Dr. W. Miller indicated that non-responsiveness may not be due to the ineffectiveness of AIs because the expression of estrogen-responsive genes such as pS2 is down-regulated following AI treatment. At the present time, we know very little about the mechanisms of de novo resistance.
Acquired resistance
The most obvious mechanism of acquired resistance involves a selection process. All tumors are heterogeneous. Each breast tumor contains ER+/estrogen responsive as well as ER- or estrogen independent cells. During endocrine therapy, the population of ER+/estrogen responsive cells reduces, and with time, ER- or estrogen independent cells become the dominant group of cells in tumors. At this stage, the tumors will stop responding to anti-estrogens or AIs, which is referred to as acquired resistance.
However, clinically, many of the endocrine resistant tumors are still ER positive. The mechanisms of such acquired resistance are probably similar to those discussed for de novo/intrinsic resistance, except that resistance develops during treatment. It is very unlikely that acquired resistance results from aromatase or ER mutation developed during endocrine treatment. Most likely, such resistance results from cross talk between ER and growth factor pathways or other currently unidentified pathways. Almost all the data on acquired resistance are at present derived from laboratory studies. A major hypothesis is that the adaptation to estrogen withdrawal is involved in resistance to both tamoxifen and AIs. Due to the ability of breast cancer cells to be adaptive, these endocrine therapies that function to block hormone-dependent signaling cascades required for breast cancer proliferation, may cause novel signaling mechanisms which circumvent the effects of an AI or anti-estrogen. An attractive hypothesis is that resistance results from estrogen hypersensitivity or estrogen-independent activation of ER. To address this question, studies have been undertaken to investigate long-term estrogen deprivation (LTED), since AIs function to effectively block the synthesis of estrogens. LTED cells have been generated in Dr. R. Santen’s, Dr. M. Dowsett’s, Dr. R. Nicholson’s and Dr. A. Brodie’s laboratories [31–34]. The key findings from these laboratories have been reviewed in a recent report [35]. Briefly, studies from these laboratories have revealed that growth factor pathways are activated in these estrogen withdrawal cell lines, namely HER2 and IGF-1R. These activated growth factor receptors crosstalk with ER and result in increases in ER expression and phosphorylation, which further activates the receptor in a ligand-independent manner, leading to breast cancer proliferation.
In addition to LTED cells, the Brodie laboratory has initiated the first direct study of aromatase inhibitor resistance, using the non-steroidal AI letrozole [36]. It was observed that letrozole-resistance involves HER2 crosstalk with ERα, leading to activation of MAPK and phosphorylation of ERα, resulting in breast cancer cell proliferation [36, 37]. Interestingly, levels of ERα in the letrozole-resistant cells were found to be 50% of those in the wild-type cells.
Characterization of AI-resistant and tamoxifen-resistant cell lines in our laboratory
By reviewing what has been accomplished in other laboratories, we have learned several important lessons:
The estrogen withdrawal cell lines generated from different laboratories are not exactly identical as indicated by the stimulation of different molecular pathways in these lines, i.e. activation of either HER2 or IGF-1R signaling pathways.
The estrogen withdrawal cell lines are not exactly equivalent to AI resistant cell lines, as demonstrated through the comparison of molecular features between the estrogen withdrawal cell lines and letrozole resistant cell line generated in Brodie’s laboratory [33, 36, 37]. As discussed above, exemestane, letrozole and anastrozole inhibit aromatase through different mechanisms. Therefore, it is logical to believe that different AIs may have unique resistance mechanisms.
AIs will only be effective in the aromatase-positive cells when the enzyme is actively converting androgen to estrogen. Therefore, androgen should be present in the resistant breast cancer cells. Recently, Macedo et al. [37] have indicated that AI treatment may suppress estrogen-dependent proliferation as well as unmask the inhibitory effect of androgen.
The information generated thus far on acquired resistance, from the estrogen withdrawal cell lines and Brodie’s letrozole-resistant line, has been very informative, but the possibility exists of other currently unidentified pathways that may further augment resistance. In addition, comparing resistance mechanisms between different AIs has not been addressed to date. Thus, it is important to apply a non-biased method, e.g., cDNA microarray analysis, to identify additional novel genes or pathways that play a role in AI resistance mechanisms.
To investigate AI resistance, our laboratory has generated AI resistant lines (using exemestane, letrozole and anastrozole) and anti-estrogen resistant lines (using tamoxifen) for comparison. In terms of a suitable cell line that can be used for resistance studies, an ER-positive breast cancer line that does express high levels of aromatase is needed, but does not exist. MCF-7aro cells (stably transfected with the aromatase gene [38]) were generated in our lab and are used as a model system to study AI response and were therefore used to produce the drug resistant lines.
In order to generate drug-resistant cell lines, MCF-7aro cells were cultured long-term in the presence of testosterone plus the appropriate inhibitor. Initially, inhibitor treatment of these breast cancer cells induced massive cell death, but after prolonged culture (2–8 months) in the presence of the inhibitor, resistance to these drugs was acquired (i.e., T+LetR, T+AnaR, T+ExeR, and T+TamR). Cell lines were considered to be established or resistant once they proliferated at the similar rate prior to treatment. Six independent sets of each resistant line were generated. As proper controls, three sets of MCF-7aro cells were also cultured in the presence of testosterone only (i.e., AroT), in addition to three sets of cells that were grown in medium alone (long term estrogen deprived (LTEDaro) cell lines). We feel that study of a single resistant line for each inhibitor is not an unbiased approach. Furthermore, our lab has also generated three independent sets of each resistant line that are grown without testosterone (i.e., LetR, AnaR, ExeR and TamR). This was done to determine if differential resistance pathways exist between cells cultured with or without testosterone. In addition, we prepared three independent sets of MCF-7aro cells cultured for 5 days without testosterone or inhibitor. These cells (MCF-7aro) served as reference cells that did not reach resistance status, and were used as reference cells for our AI responsive studies (discussed below).
We feel that it is important to examine the effects of AIs on gene expression in MCF-7aro cells, or AI-responsive studies, before the analysis of AI-resistant cell lines. Given that we are looking for genes involved in resistance pathways, we do want to tease out gene expression changes due solely to inhibitor response. We cultured MCF-7aro cells short-term with four different inhibitors, three aromatase inhibitors (exemestane, letrozole and anastrozole) and one ER antagonist (tamoxifen), in the presence of testosterone. The AI-responsive results generated from treatment of letrozole, anastrozole or tamoxifen were published in 2005 [40].
In contrast to AI/tamoxifen-responsive cells, cell proliferation assays demonstrate that the AI-resistant cell lines all proliferated similarly to the testosterone control, implying that these cells had adapted a mechanism to grow despite the presence of the inhibitor [35]. The time to generate the different resistant cell lines varied among different inhibitors. Cells growing in the presence of testosterone and inhibitor as well as LTEDaro cells had similar generation times and were all established by 3 months. The inhibitor only resistant cells had more variation among them. ExeR and AnaR were established in 2 months while LetR and TamR were established in 8 months and 5 months respectively (Table 1). This large panel of resistant cell lines generated in our study will be useful in determining any heterogeneity that may exist in signaling mechanisms specific to each inhibitor. Also, for microarray and other experiments, multiple biological replicates will allow for a more thorough experimental and statistical analysis.
Table 1.
Generation time of AI resistant cell lines
| Resistant Cell Line | Generation Time |
|---|---|
| T+ExeR | 3 months |
| T+LetR | 3 months |
| T+AnaR | 3 months |
| T+TamR | 3 months |
| LTEDaro | 3 months |
| ExeR | 2 months |
| LetR | 8 months |
| AnaR | 2 months |
| TamR | 5 months |
In addition to cell proliferation assays, aromatase and ER expression and activity levels in the resistant cell lines were examined in order to determine if these proteins play a role in resistance. The aromatase and ERα mRNA levels in the resistant cell lines remained at similar levels as the original MCF-7aro cell line, except for the LTEDaro and AnaR cells in which ERα transcript levels were elevated. Our results indicate that LTEDaro cell lines are similar to LTED/estrogen withdrawal cell lines generated in other laboratories, where ERα expression is elevated. Interestingly, ERα expression only increases in AnaR, but not in the other types of AI-resistant cell lines. The aromatase protein level and activity in the T+LetR, T+AnaR, and T+TamR-resistant cell lines are similar to the control AroT cell lines. In addition, aromatase is still functional in these resistant lines and is responsive to the treatment of AIs, as measured by a commonly used ‘in cell’ aromatase assay [41]. These results indicate that AI resistance is not a result of change in aromatase expression or in its response to AIs. As expected, we detect a low level of aromatase activity and aromatase protein in the T+ExeR and ExeR cell lines because exemestane is a mechanism-based inhibitor and destabilizes the aromatase enzyme. In contrast to aromatase expression and activity, which remain fairly unchanged, we do detect differences in ERα protein levels in our resistant cell lines. These changes in ERα are currently under investigation.
Beside these basic characterizations of the resistant lines, microarray analysis of these resistant cell lines has been performed in order to observe changes in gene expression profiles that could be unique to aromatase inhibitor resistance. Preliminary data suggests that the mechanism of resistance to aromatase inhibitors differs between steroidal and non-steroidal AIs. To date, no study has addressed differences between AI-resistance mechanisms and variations that may exist between these signaling pathways. To address this question, microarray analysis has been employed to elucidate AI-resistance on a genome-wide basis, making this analysis an unbiased approach to study resistance. Previously in other labs, microarray analysis has been employed with tamoxifen-resistant cell lines, in an attempt to better understand the anti-estrogen resistance mechanism. It has been reported that genes involved in the apoptotic response, the growth factor signaling pathway and many estrogen-responsive genes were found to be differentially regulated in tamoxifen-resistant cells [42–44]. Therefore, microarray analysis will be a useful tool to understand differences in drug resistance in steroidal versus non-steroidal aromatase inhibitors, in contrast to the anti-estrogen tamoxifen.
To check the quality of our microarray analysis, a hierarchical clustering analysis of the data has been carried out [35]. As a crucial quality control assessment, we are very pleased with our analysis in which replicates of each type of resistant lines do cluster together. The results demonstrate the high quality of our data where similar genes are modulated in each type of resistant lines although they have different growth rates. As expected, our results indicate that data of T+LetR, T+AnaR and T+ExeR lines are more similar than those of T+TamR and AroT lines. The data of AroT lines are very different from those of T+TamR lines. Furthermore, the data of AroT lines are in a group different from the resistant lines. In addition, clustering analysis has revealed that the testosterone-containing resistant lines (T+LetR and T+AnaR) cluster separately from the inhibitor-only lines (no T) (i.e., LetR and AnaR). This does suggest that for non-steroidal inhibitors, inherent differences do exist between the hormone containing lines versus the inhibitor-only lines. Interestingly, the gene profiles of T+ExeR and ExeR are very similar.
We have started the examination of the expression patterns of a group of estrogen responsive genes from our microarray analysis of resistant cell lines. In our previously published studies on AI/tamoxifen responsive cells, a group of genes were found to be up regulated by androgen/estrogen, whose expression was returned to basal level following the treatment of AI/tamoxifen. Since ER function was found to be up regulated, in the absence of estrogen, in a few resistant cell lines, we would expect that the expression of most estrogen-responsive genes remains high even in the presence of AI/tamoxifen. Interestingly, while genes like CCND1, CTSD and TFF1 were found to be up regulated in all resistant cell lines, PGR was found to be up regulated in AroT, T+AnaR, T+LetR, T+ExeR, ExeR and T+TamR, but not in LTEDaro and AnaR. These results would indicate that the expression of PGR in LTEDaro and AnaR is not regulated through ER. With the results generated so far, we think that we have four types of hormone resistant cell lines. The first type includes LTEDaro and AnaR which are ER over-expressing, with constitutively active ER, and with ER+/PR− phenotype. The second type includes T+AnaR and T+LetR which are with constitutively active ER. The third type includes ExeR and T+ExeR which contain ER that is estrogen-dependent or hormone responsive. The fourth type includes T+TamR which has the gene expression profile that is clearly different from those of LTEDaro and AI resistant cell lines. Using microarray analysis, we believe that our large panel of AI and tamoxifen-resistant lines will provide new insight into mechanistic differences between steroidal versus non-steroidal aromatase inhibitors and how these pathways differ from tamoxifen-resistance.
In summary, resistance to AIs is emerging as a complex phenomenon, based on new experimental information discussed in this paper. Thus far, analysis of acquired resistance pathways has focused primarily on growth factor and nuclear receptor crosstalk. This information has been quite valuable, but may not be complete. Therefore, analysis of a large panel of resistant cell lines by microarray is an unbiased genome-wide examination of signaling pathways responsible for steroidal and non-steroidal AI-resistance. Acquired resistance to AIs is a hindrance in the clinic and better understanding of the molecular mechanisms responsible for such occurrences would be beneficial for effectively treating hormone-dependent breast cancers.
Acknowledgments
This research was supported by NIH grant CA44735 (SC) and a grant from the Flanigan Foundation. Furthermore, Selma Masri is supported by a predoctoral fellowship from NCI (CA123691), and Yanyan Hong is supported by a predoctoral fellowship from the UC Breast Cancer Research Program. Dr. Xin Wang is a Beckman Fellow of the City of Hope.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen S. Aromatase and breast cancer. Front Biosci. 1998;3:d922–d933. doi: 10.2741/a333. [DOI] [PubMed] [Google Scholar]
- 2.Smith IE, Dowsett M. Aromatase inhibitors in breast cancer. N Engl JMed. 2003;348:2431–2442. doi: 10.1056/NEJMra023246. [DOI] [PubMed] [Google Scholar]
- 3.Baum M, Budzar AU, Cuzick J, Forbes J, Houghton JH, Klijn JG, Sahmoud T. Anastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early breast cancer: first results of the ATAC randomised trial. Lancet. 2002;359:2131–2139. doi: 10.1016/s0140-6736(02)09088-8. [DOI] [PubMed] [Google Scholar]
- 4.Coombes RC, Hall E, Gibson LJ, Paridaens R, Jassem J, Delozier T, Jones SE, Alvarez I, Bertelli G, Ortmann O, Coates AS, Bajetta E, Dodwell D, Coleman RE, Fallow field LJ, Mickiewicz E, Andersen J, Lonning PE, Cocconi G, Stewart A, Stuart N, Snowdon CF, Carpentieri M, Massimini G, Bliss JM. A randomized trial of exemestane after two to three years of tamoxifen therapy in postmenopausal women with primary breast cancer. N Engl J Med. 2004;350:1081–1092. doi: 10.1056/NEJMoa040331. [DOI] [PubMed] [Google Scholar]
- 5.Goss PE, Ingle JN, Martino S, Robert NJ, Muss HB, Piccart MJ, Castiglione M, Tu D, Shepherd LE, Pritchard KI, Livingston RB, Davidson NE, Norton L, Perez EA, Abrams JS, Therasse P, Palmer MJ, Pater JL. A randomized trial of letrozole in postmenopausal women after five years of tamoxifen therapy for early-stage breast cancer. N Engl J Med. 2003;349:1793–1802. doi: 10.1056/NEJMoa032312. [DOI] [PubMed] [Google Scholar]
- 6.Santen RJ. Suppression of estrogens with aminoglutethimide and hydrocortisone (medical adrenalectomy) as treatment of advanced breast carcinoma: a review. Breast Cancer Res Treat. 1981;1:183–202. doi: 10.1007/BF01806259. [DOI] [PubMed] [Google Scholar]
- 7.Dowsett M, Cunningham DC, Stein RC, Evans S, Dehennin L, Hedley A, Coombes RC. Dose-related endocrine effects and pharmacokinetics of oral and intramuscular 4- hydroxyandrostenedione in postmenopausal breast cancer patients. Cancer Res. 1989;49:1306–1312. [PubMed] [Google Scholar]
- 8.Brodie AM. Aromatase inhibitors in the treatment of breast cancer. J Steroid Biochem Mol Biol. 1994;49:281–287. doi: 10.1016/0960-0760(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 9.Coombes RC, Goss P, Dowsett M, Gazet JC, Brodie A. 4-Hydroxyandrostenedione in treatment of postmenopausal patients with advanced breast cancer. Lancet. 1984;2:1237–1239. doi: 10.1016/s0140-6736(84)92795-8. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Kao YC, Laughton CA. Binding characteristics of aromatase inhibitors and phytoestrogens to human aromatase. J Steroid Biochem Mol Biol. 1997;61:107–115. [PubMed] [Google Scholar]
- 11.Chen S, Zhang F, Sherman MA, Kijima I, Cho M, Yuan YC, Toma Y, Osawa Y, Zhou D, Eng ET. Structure-function studies of aromatase and its inhibitors: a progress report. J Steroid Biochem Mol Biol. 2003;86:231–237. doi: 10.1016/s0960-0760(03)00361-3. [DOI] [PubMed] [Google Scholar]
- 12.Kao YC, Cam LL, Laughton CA, Zhou D, Chen S. Binding characteristics of seven inhibitors of human aromatase: a site-directed mutagenesis study. Cancer Res. 1996;56:3451–3460. [PubMed] [Google Scholar]
- 13.Kao YC, Korzekwa KR, Laughton CA, Chen S. Evaluation of the mechanism of aromatase cytochrome P450. A site-directed mutagenesis study. Eur J Biochem. 2001;268:243–251. doi: 10.1046/j.1432-1033.2001.01886.x. [DOI] [PubMed] [Google Scholar]
- 14.Hong Y, Yu B, Sherman M, Yuan YC, Zhou D, Chen S. Molecular basis for the aromatization reaction and exemestane-mediated irreversible inhibition of human aromatase. Mol Endocrinol. 2006 doi: 10.1210/me.2006-0281. in press. [DOI] [PubMed] [Google Scholar]
- 15.Brueggemeier RW, Hackett JC, Diaz-Cruz ES. Aromatase inhibitors in the treatment of breast cancer. Endocr Rev. 2006;26:331–345. doi: 10.1210/er.2004-0015. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Chen S. Aromatase destablizer: novel action of exemestane, and FDA approved aromatase inhibitor. Cancer Res. 2006 doi: 10.1158/0008-5472.CAN-06-2134. in press. [DOI] [PubMed] [Google Scholar]
- 17.Favia AD, Cavalli A, Masetti M, Carotti A, Recanatini M. Three-dimensional model of the human aromatase enzyme and density functional parameterization of the iron-containing protoporphyrin IX for a molecular dynamics study of heme-cysteinato cytochromes. Proteins. 2006;62:1074–1087. doi: 10.1002/prot.20829. [DOI] [PubMed] [Google Scholar]
- 18.Ma CX, Adjei AA, Salavaggione OE, Coronel J, Pelleymounter L, Wang L, Eckloff BW, Schaid D, Wieben ED, Adjei AA, Weinshilboum RM. Human aromatase: gene resequencing and functional genomics. Cancer Res. 2005;65:11071–11082. doi: 10.1158/0008-5472.CAN-05-1218. [DOI] [PubMed] [Google Scholar]
- 19.Chen S, Zhou D. Functional domains of aromatase cytochrome P450 inferred from comparative analyses of amino acid sequences and substantiated by site-directed mutagenesis experiments. J Biol Chem. 1992;267:22587–22594. [PubMed] [Google Scholar]
- 20.Hong Y, Chen S. Aromatase inhibitors: Structure features and biochemical characterization. Ann N Y Acad Sci. 2006 doi: 10.1196/annals.1386.022. in press. [DOI] [PubMed] [Google Scholar]
- 21.Miller WR, O’Neill J. The importance of local synthesis of estrogen within the breast. Steroids. 1997;50:537–548. doi: 10.1016/0039-128x(87)90037-7. [DOI] [PubMed] [Google Scholar]
- 22.James VH, McNeill JM, Lai LC, Newton CJ, Ghilchik MW, Reed MJ. Aromatase activity in normal breast and breast tumor tissues: in vivo and in vitro studies. Steroids. 1987;50:269–279. doi: 10.1016/0039-128x(83)90077-6. [DOI] [PubMed] [Google Scholar]
- 23.Vermeulen A, Deslypere JP, Paridaens R, Leclercq G, Roy F, Heuson JC, Aromatase 17 beta-hydroxysteroid dehydrogenase and intratissular sex hormone concentrations in cancerous and normal glandular breast tissue in postmenopausal women. Eur J Cancer Clin Oncol. 1986;22:515–525. doi: 10.1016/0277-5379(86)90121-5. [DOI] [PubMed] [Google Scholar]
- 24.Esteban JM, Warsi Z, Haniu M, Hall P, Shively JE, Chen S. Detection of intratumoral aromatase in breast carcinomas. An immunohistochemical study with clinicopathologic correlation. Am J Pathol. 1992;140:337–343. [PMC free article] [PubMed] [Google Scholar]
- 25.Santen RJ, Martel J, Hoagland M, Naftolin F, Roa L, Harada N, Hafer L, Zaino R, Santner SJ. Stromal spindle cells contain aromatase in human breast tumors. J Clin Endocrinol Metab. 1994;79:627–632. doi: 10.1210/jcem.79.2.8045987. [DOI] [PubMed] [Google Scholar]
- 26.Lu Q, Nakmura J, Savinov A, Yue W, Weisz J, Dabbs DJ, Wolz G, Brodie A. Expression of aromatase protein and messenger ribonucleic acid in tumor epithelial cells and evidence of functional significance of locally produced estrogen in human breast cancers. Endocrinology. 1996;137:3061–3068. doi: 10.1210/endo.137.7.8770932. 1996. [DOI] [PubMed] [Google Scholar]
- 27.Sasano H, Anderson TJ, Silverberg SG, Santen RJ, Conway M, Edwards DP, Krause A, Bhatnagar AS, Evans DB, Miller WR. The validation of new aromatase monoclonal antibodies for immunohistochemistry--a correlation with biochemical activities in 46 cases of breast cancer. J Steroid Biochem Mol Biol. 2005;95:35–39. doi: 10.1016/j.jsbmb.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 28.Harada N. Aberrant expression of aromatase in breast cancer tissues. J Steroid Biochem Mol Biol. 1997;61:175–184. [PubMed] [Google Scholar]
- 29.Bulun SE, Price TM, Aitken J, Mahendroo MS, Simpson ER. A link between breast cancer and local estrogen biosynthesis suggested by quantification of breast adipose tissue aromatase cytochrome P450 transcripts using competitive polymerase chain reaction after reverse transcription. J Clin Endocrinol Metab. 1993;77:1622–1628. doi: 10.1210/jcem.77.6.8117355. [DOI] [PubMed] [Google Scholar]
- 30.Dowsett M, Martin LA, Smith I, Johnston S. Mechanisms of resistance to aromatase inhibitors. J Steroid Biochem Mol Biol. 2005;95:167–172. doi: 10.1016/j.jsbmb.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 31.Martin LA, Farmer I, Johnston SR, Ali S, Dowsett M. Elevated ERK1/ERK2/estrogen receptor cross-talk enhances estrogen-mediated signaling during long-term estrogen deprivation. Endocr Relat Cancer. 2005;12 Suppl 1:S75–84. doi: 10.1677/erc.1.01023. [DOI] [PubMed] [Google Scholar]
- 32.Nicholson RI, Staka C, Boyns F, Hutcheson IR, Gee JM. Growth factor-driven mechanisms associated with resistance to estrogen deprivation in breast cancer: new opportunities for therapy. Endocr Relat Cancer. 2004;11:623–641. doi: 10.1677/erc.1.00778. [DOI] [PubMed] [Google Scholar]
- 33.Sabnis GJ, Jelovac D, Long B, Brodie A. The role of growth factor receptor pathways in human breast cancer cells adapted to long-term estrogen deprivation. Cancer Res. 2005;65:3903–3910. doi: 10.1158/0008-5472.CAN-04-4092. [DOI] [PubMed] [Google Scholar]
- 34.Santen RJ, Song RX, Zhang Z, Kumar R, Jeng MH, Masamura S, Lawrence J, Jr, MacMahon LP, Yue W, Berstein L. Adaptive hypersensitivity to estrogen: mechanisms and clinical relevance to aromatase inhibitor therapy in breast cancer treatment. J Steroid Biochem Mol Biol. 2005;95:155–165. doi: 10.1016/j.jsbmb.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 35.Chen S, Masri S, Wang X, Phung S, Yuan YC, Wu X. What do we know about the mechanisms of aromatase inhibitor resistance? J Steroid Biochem Mol Biol. 2006 doi: 10.1016/j.jsbmb.2006.09.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brodie A, Jelovac D, Sabnis G, Long B, Macedo L, Goloubeva O. Model systems: mechanisms involved in the loss of sensitivity to letrozole. J Steroid Biochem Mol Biol. 2005;95:41–48. doi: 10.1016/j.jsbmb.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 37.Jelovac D, Sabnis G, Long BJ, Macedo L, Goloubeva OG, Brodie AM. Activation of mitogen-activated protein kinase in xenografts and cells during prolonged treatment with aromatase inhibitor letrozole. Cancer Res. 2005;65:5380–5389. doi: 10.1158/0008-5472.CAN-04-4502. [DOI] [PubMed] [Google Scholar]
- 38.Macedo LF, Guo Z, Tilghman SL, Sabnis GJ, Qiu Y, Brodie A. Role of androgens on MCF-7 breast cancer cell growth and on the inhibitory effect of letrozole. Cancer Res. 2005;66:7775–7782. doi: 10.1158/0008-5472.CAN-05-3984. [DOI] [PubMed] [Google Scholar]
- 39.Sun XZ, Zhou D, Chen S. Autocrine and paracrine actions of breast tumor aromatase. A three-dimensional cell culture study involving aromatase transfected MCF-7 and T-47D cells. J Steroid Biochem Mol Biol. 1997;63:29–36. doi: 10.1016/s0960-0760(97)00068-x. [DOI] [PubMed] [Google Scholar]
- 40.Itoh T, Karlsberg K, Kijima I, Yuan YC, Smith D, Ye J, Chen S. Letrozole-, anastrozole-, and tamoxifen-responsive genes in MCF-7aro cells: a microarray approach. Mol Cancer Res. 2005;3:203–218. doi: 10.1158/1541-7786.MCR-04-0122. [DOI] [PubMed] [Google Scholar]
- 41.Thompson EA, Jr, Siiteri PK. Utilization of oxygen and reduced nicotinamide adenine dinucleotide phosphate by human placental microsomes during aromatization of androstenedione. J Biol Chem. 1974;249:5364–5372. [PubMed] [Google Scholar]
- 42.Omoto Y, Hayashi S. A study of estrogen signaling using DNA microarray in human breast cancer. Breast Cancer. 2002;9:308–311. doi: 10.1007/BF02967609. [DOI] [PubMed] [Google Scholar]
- 43.Treeck O, Zhou R, Diedrich K, Ortmann O. Tamoxifen long-term treatment in vitro alters the apoptotic response of MCF-7 breast cancer cells. Anticancer Drugs. 2004;15:787–793. doi: 10.1097/00001813-200409000-00008. [DOI] [PubMed] [Google Scholar]
- 44.Becker M, Sommer A, Kratzschmar JR, Seidel H, Pohlenz HD, Fichtner I. Distinct gene expression patterns in a tamoxifen-sensitive human mammary carcinoma xenograft and its tamoxifen-resistant subline MaCa 3366/TAM. Mol Cancer Ther. 2005;4:151–168. [PubMed] [Google Scholar]