Abstract
Proteomics, the large-scale study of protein expression in organisms, offers the potential to evaluate global changes in protein expression and their post-translational modifications that take place in response to normal or pathological stimuli. One challenge has been the requirement for substantial amounts of tissue in order to perform comprehensive proteomic characterization. In heterogeneous tissues, such as brain, this has limited the application of proteomic methodologies. Efforts to adapt standard methods of tissue sampling, protein extraction, arraying, and identification are reviewed, with an emphasis on those appropriate to smaller samples ranging in size from several microliters down to single cells. The effects of miniaturization on these analyses are highlighted using neuroscience-related examples, as are statistical issues unique to the high-dimensional datasets generated by proteomic experiments.
Keywords: 2D gel electrophoresis, imaging mass spectrometry, capillary electrophoresis, microdissection
I. INTRODUCTION
The last decade has seen a resurgence of interest in the study of the proteome—the set of proteins that can be expressed by the genetic material of an organism. Advancements in protein extraction, purification, and identification have been driven by improvements in sensitivity, throughput, and resolution of separations and mass analyzers. However, proteomic approaches have not advanced to the extent of genomic technologies. Over the past 30 years, major technological innovations have improved the throughput of genomic sequencing and analysis techniques to the point where sequencing the complete genome of organisms is commonplace. In addition, transcriptomic analysis is feasible in samples as small as single cells. Unfortunately, the analytical capabilities of proteomic technologies lack the speed and robust characteristics of current microarray methods. Given these limitations, why should one pursue proteomic studies? Clearly, the promise implied in proteomics is acquiring global information on changes in protein expression, including post-translational modifications (PTMs) from specific biological tissues under unique physiological conditions. Such information certainly is relevant to biological function, because proteins play an integral role in cell function.
It should not be surprising that changes in gene expression might not correlate well with changes in protein expression (Anderson & Seilhammer, 1997; Futcher et al., 1999); proteins are activated by mechanisms such as precursor cleavages, PTMs, and unique localization, which are not direct consequences of changes in protein production. Furthermore, many physiological responses of organisms are not always genetically mediated. For example, homeostatic responses are relatively uniform despite the genetic variability between specific individuals. One final, but perhaps most important point, the lifetime of proteins can vary by orders of magnitude so that the amounts of two proteins can be surprisingly different even with similar translation rates. Thus, proteomic characterization complements transcriptomic studies, and might be more relevant for the characterization of complex genetic and homeostatic phenomena.
Complexity presents a fundamental challenge in proteomics. Although the genetic code is composed of four bases, the “protein code” consists of over 20 amino acids plus many possible PTMs. The genome for many organisms is known, and does not vary between cells. Although each organism has only one genome, it has many transcriptomes and even more proteomes. Protein expression patterns differ among cell types, and physiological changes alter these expression patterns; these alterations also differ between specific cells of the same type.
Also of concern, tissues of interest can be microscopically small and surprisingly heterogeneous, requiring the dissection and characterization of specific cells from a region of interest. Whereas recent technological developments have made micro-dissection feasible (see below), extracting, separating, and detecting the wide range of proteins present in extremely small samples present significant issues. Geneticists have overcome these obstacles with the development of the polymerase chain reaction to amplify DNA. However, analogous methods do not exist for protein amplification.
In the classical proteomic workflow, one obtains the tissue of interest, extracts proteins from the tissue, separates and/or arrays the proteins, identifies proteins of interest, and quantifies changes in protein expression and PTMs. Each step requires greater care and expertise as the sample size decreases. Obligate losses during the extraction and fractionation procedures become more of an issue as one draws closer to the limits of protein detection. Protein diversity decreases in small samples, and fewer proteins will be abundant enough to exceed the sensitivity limit of specific technologies. It also becomes more difficult to evaluate differences between conditions with small sample sizes, because variability increases as one operates closer to the limits of the analytical techniques. Although information content is decreased in smaller samples so that only higher abundance compounds are detected, useful knowledge can still be gained, even from a single cell (Rubakhin, Greenough, & Sweedler, 2003; Hummon, Amare, & Sweedler, 2006; Rubakhin et al., 2006; Neupert et al., 2007; Predel et al., 2007; Rubakhin & Sweedler, 2007).
The main focus of this review is to describe recent developments that have made “microproteomics” a reality. Sampling issues are covered first, followed by a discussion of new methods that have been developed to improve the sensitivity and selectivity of conventional sampling techniques, such as laser capture microdissection (LCM). Approaches to protein separation, including multidimensional separations and smaller gels, are described. Improvements in mass spectrometric resolution and sensitivity are also presented, as well as innovative statistical methods for analyzing and cataloging the complex data generated by these experiments. Approaches to characterize preselected proteins such as with immunoaffinity approaches or GFP-labeling are not covered. Protein arrays based on antibodies or other selective capture agents are an exciting area undergoing rapid development, and have been the focus of several recent reviews (Borrebaeck & Wingren, 2007; Lu & Liu, 2007; von Eggeling, Melle, & Ernst, 2007). In this review, mass-spectrometric-based identification methods are highlighted. We briefly discuss recent developments, such as mass spectrometric imaging (MSI), which bypass steps in the conventional proteomic workflow and allow small samples to be characterized. As the dynamic range of these cutting-edge technologies improves, these “shorter” approaches may provide attractive alternatives for the proteomic analysis of small samples.
II. TISSUE SAMPLING
The first and most critical issue facing anyone trying to analyze small samples is how to acquire the optimal specimen of interest. If a poor quality sample is obtained, or if proteins are degraded or contaminated with other unwanted tissue types, then even the best protein identification techniques will not rescue the experiment. There are many possible techniques that can be employed; choosing the appropriate one depends on experimental requirements. For example, if anatomical precision is not required, then freehand microdissection can be used. Flow cytometry is useful for identifying cells that express specific biomarkers, but anatomic orientation and resolution is lost. These limitations are also shared by some emerging technologies, such as dielectrophoresis-activated cell sorting (Hu et al., 2005c), use of optical tweezers (Grier, 2003), and immunomagnetic separation (Safarik & Safarikova, 1999). Several approaches have been developed to reproducibly obtain small samples of defined anatomical regions. An excellent method for microscope-aided microdissection in the CNS is described by Cuello and colleagues (Cuello & Carson, 1983), which relies on the differential light transmittance of myelinated and unmyelinated tissues in fresh brain slices. Because myelination decreases light transmittance, myelinated tissues appear dark when transilluminated and unmyelinated tissues appear lighter. With 2–300 µm sections are used, this method can provide a surprising degree of anatomical detail and precision. It is important to maintain temperatures as close to freezing as possible while ensuring that ice crystals do not develop during the microdissection process. If they do, much of the anatomical resolution inherent to transillumination is lost. Further, if temperatures rise, then significant protein degradation can occur. The addition of protease and phosphatase inhibitors to bath solutions should also be considered to help improve the consistency of results.
Another useful sampling technique, the micropunch method, was developed by Palkovits and Brownstein (1983). Punch microdissection entails making small punches to remove tissue from anatomically defined regions of fresh or frozen samples, and can also be aided by the use of a loupe or dissecting microscope. Although anatomical delineation is not as precise as with transillumination, the ability to immediately snap-freeze specimens and keep them frozen throughout the dissection process is a huge advantage for proteomic studies. Micropunch and transillumination both require substantial amounts of technical skill and practice to obtain the requisite precision and reproducibility.
When examining invertebrate neurons, individual cells can be in the range of 20–500 µm in diameter, a size that allows direct sampling of these cells, from microdissection to subsequent protease treatments in preparation for matrix-assisted laser desorption/ionization mass spectrometry (MALDI MS) (Li, Garden, & Sweedler, 2000). As shown in Figure 1, individual neurons can be divided into subsections such as soma and neurites, and each assayed for their neuropeptide content (Rubakhin, Greenough, & Sweedler, 2003). Similarly, micro-dissection and/or isolation of mammalian cells (e.g., red blood cells, pituitary cells) have been demonstrated to be effective for direct analysis using mass spectrometry (MS). Single cell analysis of pituitary cells using MALDI MS has allowed positive identification of 14 peptides derived from pro-opiomelanocortin prohormone (Rubakhin et al., 2006). Similarly, using capillary electrophoresis (CE) coupled to an electrospray ionization-Fourier transform ion cyclotron mass spectrometer, Hofstadler et al. (1995) were able to identify α- and β-chains of hemoglobin from a few red blood cells. Furthermore, MALDI MS has been shown to be robust in analyzing peptides in samples as small as 2 µm dense core vesicles (Rubakhin et al., 2000). Analysis at the single cell or organelle level can provide a comprehensive view of how an individual cell processes protein products with fewer confounding factors.
Figure 1.
Direct profiling of the subcompartments of an individual neuron using MALDI TOF. A: Transmission light micrograph of a cultured bag cell neuron after 30 min of 4% paraformaldehyde fixation, extracellular solution removal, neuron dehydration, and MALDI matrix application. B: MALDI mass spectral profile of the cultured bag cell obtained from the cell soma and neurites, postparaformaldehyde fixation. Peaks are labeled based on the accurate mass match to a single known neuropeptide prohormone: ε-BCP, epsilon bag cell peptide; AP, acidic peptide; ELH, egg laying hormone. Reprinted with permission from Rubakhin, Greenough, and Sweedler (2003), copyright 2003 American Chemical Society.
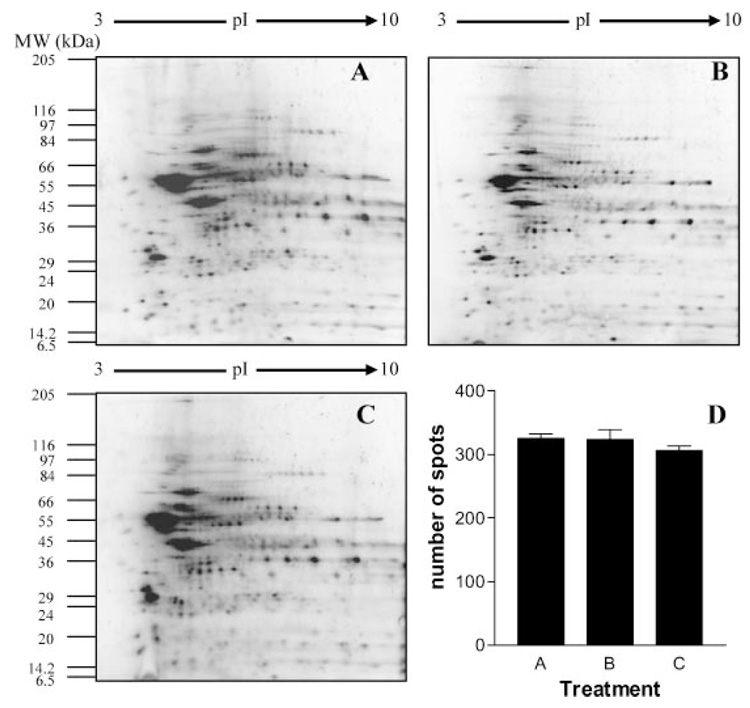
A relatively recent improvement in tissue sampling is the development of LCM by Emmert-Buck and colleagues (Emmert-Buck et al., 1996). This technique uses a finely-focused laser to either cut around cells of interest or adhere them to a plastic film, depending on the instrument. Slices of ethanol-post-fixed tissues from 8–40 µm thick can be dissected, depending on the instrument and exact technique employed. Several investigators have adapted LCM to proteomic studies. As shown in Figure 2, we demonstrated that LCM and the required ethanol postfixation (30 sec in 70% ethanol) does not interfere with two-dimensional gel electrophoresis (2D)E-based protein separation (Moulédous et al., 2002). However, we found that conventional histological staining methods in brain (cresyl violet, hematoxylin/eosin, and toluidine blue) as well as some more unusual stains (chlorazol black E, Sudan black B) did interfere with 2DE-based protein separation (Moulédous et al., 2002). In some tissues, such as kidney and esophagus, conventional tissue-staining techniques can be used to provide anatomic detail without interfering with protein recovery and identification (Banks et al., 1999; Emmert-Buck et al., 2000). However, we have found that in brain, higher stain concentrations are required and interfere with protein recovery.
Figure 2.
The effect of tissue fixation and LCM on 2D gel-based protein separation. Samples of the rat brain region called the striatum were used to determine whether fixation or dissection method affected protein separation using 2-D gel electrophoresis. Three conditions were tested: samples were either manually dissected without fixation, fixed in ethanol and then manually dissected, or fixed and dissected using LCM. Twenty-five microgram of total protein was loaded onto immobilized pH gradient (IPG) strips and separated by 2-D gel electrophoresis as described. A: Unfixed, unstained, manually dissected sample. B: Ethanol-fixed, manually dissected sample. C: Ethanol-fixed, LCM sample. D: Number of spots detected on each gel. Results are expressed as mean±SEM of three independent experiments. No statistically significant difference was observed between any of the treatment groups. Reprinted with permission from Moulédous et al. (2002), copyright 2002 Association of Biomolecular Resource Facilities.
An alternative to direct sample staining is the technique of “navigated” LCM (Wong et al., 2000; Moulédous et al., 2003a). In this method, a stained section is used to guide the dissection of an adjacent, unstained section of tissue. Accurate image registration is necessary to ensure the success of this technique, which can be accomplished by the use of a variety of image analysis programs. Such fine dissections permit detailed questions to be addressed; for example, what are the proteomic changes seen in a specific type of cell expressing a specific receptor? This issue can be elucidated by using “immuno-LCM” to harvest cells expressing specific markers (Moulédous et al., 2003b). However, conventional avidin-biotin chemistry interferes with protein extraction and 2DE. Craven and colleagues also showed that immunogold staining in renal tissue resulted in the loss of 60% of total protein (Craven et al., 2002). Fortunately, fluorescently tagged antibodies do not significantly reduce protein recovery, nor do they interfere with subsequent analyses (Moulédous et al., 2003b). Moreover, immunostaining does not significantly change the percentage of protein coverage for proteins identified with peptide mass fingerprinting (PMF) after 2DE. Thus, with the aid of a fluorescent-capable LCM, more detailed and precise studies can be undertaken. The tradeoff is that much more starting tissue is required to perform proteomic studies on specific subsets of cells.
Several groups have combined LCM with the technique of differential in-gel electrophoresis (DIGE; also see section VII), a technique to reduce measurement variability in 2D gels (Zhou et al., 2002; Lee et al., 2003; Greengauz-Roberts et al., 2005; Wilson et al., 2005;Kondo&Hirohashi, 2006; Sitek et al., 2006). One recent study demonstrated the ability to detect 900 protein spots on 2D gels generated from less than 1 µg of kidney glomeruli using LCM followed by DIGE (Sitek et al., 2006). Other modes of protein separation have also been effectively coupled with LCM. One recent study captured approximately 1500 cells using LCM, then subjected the sample to nano-LC followed by FT-MS identifications (Mustafa et al., 2007). Using this approach, 4 proteins that were uniquely expressed in the blood vessels of brain tumors were identified. Several investigators have also shown that proteins can also be identified by direct mass spectrometric analysis of laser-captured tissues using MALDI-MS and SELDI-MS (Xu et al., 2002; Bhattacharya, Gal, & Murray, 2003; Kwapiszewska et al., 2004; de Groot et al., 2005; Guo et al., 2005; Krieg et al., 2005). Caprioli and colleagues demonstrated that meaningful spectra could be obtained from samples containing as few as 10 laser-captured cells (Xu et al., 2002).
III. PROTEIN EXTRACTION
Once tissue samples have been obtained, the next step is to extract the proteins from the tissue. Although this step is perhaps the most critical one in performing a successful proteomics experiment, it has been given the least attention by current investigators. All extractions are aimed toward an elusive and as yet unachieved goal, the complete extraction of all proteins from a sample. The wide range of physicochemical properties of cellular proteins and their differing solubilities make it difficult to efficiently extract all proteins, which range in size from small cytoplasmic signaling molecules to lipophilic, megadalton membrane receptors. However, the introduction of new techniques and optimization of classical methods have improved extraction efficiencies for small samples. Extractions have been categorized from “gentle”, which consists of osmotic, chemical, or enzymatic treatments or mild grinding, to “vigorous”, which are active mechanical cellular disruption methods (Scopes, 1994). In order to achieve as complete an extraction as possible, most investigators use a combination of cellular disruption and treatments with strong detergents and chaotropes. Although mechanical cell disruption might improve extraction efficiencies over simple chemical extraction, there is no method of cell disruption that is clearly superior for small samples. The only limitation is the size of the probes or vessels available for sample processing. Ultrasonication, homogenization, freeze-thawing, pressure-cycling, and bead mills have all been used (Schutte & Kula, 1990a,b; Hopkins, 1991; Melendres et al., 1991; Belo et al., 1996; Raynie, 2004; Butt & Coorssen, 2006; Flanagan et al., 2006; Smejkal et al., 2006). We have had success in using ultrasonication with small-diameter probes. Methods such as bead mills and pressure cycling, an extension of the old “french press” technology, are currently difficult to use with the smallest samples. However, they do have potential advantages for certain types of tissues and extractions that warrant re-examination when the equipment has been appropriately miniaturized. Care must be taken to avoid any heating of the sample during cell disruption, which can degrade proteins prior to subsequent analyses.
Chemical solubilization, the second phase of the process, has also improved markedly over the past several years. Solubilization has three major components: (a) minimize interactions between proteins as well as between proteins and other substances (e.g., nucleic acids, lipids); (b) remove contaminants; and (c) prevent protein precipitation during the process of protein separation (Rabilloud, 1996). The chemical strategies employed are dependent on (a)whether the proteins are needed in their native or a denatured conformation; (b) which nonprotein substances need to be removed from the sample; and (c) the subsequent separation technology to be used. The goal of denaturation is to eliminate protein interactions by disrupting noncovalent bonds as well as covalent disulfide bonds. This will disrupt secondary and tertiary protein structure and so is not desirable when examining protein complexes. However, denaturation improves the ease and effectiveness of extraction. A wider variety of solvents and cosolvents can be used within the constraints of the subsequent separation technology. There are excellent reviews available on this subject (Deutscher, 1990; Rabilloud, 1996; Molloy et al., 1998;Herbert, 1999; Santoni et al., 1999).However, it is important to be aware that any protocol that involves precipitation or phase-partitioning of proteins will be more difficult to use with smaller samples. Manipulations result in some protein losses, which in the case of small samples, might mean the loss of an unacceptably high percentage of the total protein available.
Recently, Wang and colleagues developed a microscale, high-througput extraction and sample preparation method for LC-MS using the organic solvent triflouroethanol (Wang et al., 2005). These promising findings suggest a new direction for the extraction and separation of extremely small samples, minimizing protein losses due to sample handling and multiple extraction procedures. Also, application of the emerging field of nanotechnology to the extraction and separation of proteins suggests that more thorough coverage of the proteome from samples as small as a single cell may soon be possible (Hellmich et al., 2005; Ivanov et al., 2006). Recently, the detection of single proteins, some of low abundance, from individual cells has been demonstrated (Cai, Friedman, & Xie, 2006; Huang et al., 2007).
IV. SEPARATION OF PROTEIN MIXTURES
There are limits to the number of proteins that can be detected and identified from a single protein mixture. It has been estimated that the average cell expresses 10–15,000 unique proteins (Hastie & Bishop, 1976), and has perhaps an order of magnitude higher chemical diversity due to chemical modifications; however, the highest resolution separation methods currently available can only distinguish a few thousand distinct species (Anderson, 1995). Therefore, sample complexity should be reduced or many constituents will remain unidentified. A commonly suggested first step is fractionation of cells to focus on a specific organelle or region (e.g., synaptic cleft) of interest. Taken to an extreme, a tissue can be fractionated so that only a single compound will be isolated via immunohistochemical approaches. However, Subcellular fractionation methods require relatively large amounts of starting material, and it can be difficult to obtain a completely pure subcellular fraction (Pasquali, Fialka, & Huber, 1999). Nevertheless, these techniques could be used to obtain small amounts of a specific organelle needed for study (e.g., mitochondria, synaptosomes) when enough tissue is initially available. Several recent examples highlight the ability to perform proteomic measurements on synaptic fractions (Collins et al., 2006; Burre, Zimmermann, & Volknandt, 2007; Moron et al., 2007). Other specialized capillary-scale separations can be used for high sensitivity characterization of small-volume organelle sampling (Olson, Ahmadzadeh, & Arriaga, 2005). Another approach to decrease sample complexity is sequential extraction. For example, a hypotonic lysis buffer with mild detergent can be used to extract cytoplasmic proteins followed by a buffer that contains strong detergents and chaotropes to extract less soluble (membrane) proteins. Although each step results in some sample loss (Molloy et al., 1998; Herbert, 1999), sequential extraction provides a straightforward approach to fractionate smaller tissue samples (100–250 µg) with less protein loss. We have found that with samples under 50 µg total protein content, wide-ranging single-stage protein extractions appear to provide the best compromise between detectable protein diversity and sample loss.
Given these issues, what are the choices available to separate these protein/peptide mixtures? Most available methods are based on chromatographic or electrophoretic techniques, or some combination of the two. Liquid chromatography (LC) has become one of the most common methods to separate proteins, allowing their identification via MS. With a reverse phase column, peptides, normally in the presence of an ion-pairing reagent, separate based on hydrophobic interactions with the stationary phase. Similarly, ion-exchange columns separate analytes based on charge, and size-exclusion columns based on molecular size. The peak capacity of a separation is an indication of how many different analytes can be separated in a single separation (Giddings, 1991). Normally, the peak capacity (maximum number of theoretical resolution planes) of LC is several hundred, so that an LC separation interfaced to a mass spectrometer can be used to identify several hundred peptides (some degree of peak overlap is acceptable with MS detection). Because many samples have greater complexity, multi-dimensional chromatographic separation methods have been developed. These combined systems have vastly improved peak capacities (Giddings, 1991; Davis & Blumberg, 2005) and allow much more complex samples to be studied; thus, they are well suited to proteomic experiments. In these methods, the mixture is separated in stages, with subsequent separations based on differing properties. For example, the analytes can be separated by charge, pH, hydrophobicity, size, and ability to bind to specific stationary phases, to name a few. The number of different combinations of techniques that have been used is quite large (Evans&Jorgenson, 2004).One specific method of analysis that involves several stages of separation followed by MS analysis has been termed “MudPIT” (multidimensional peptide identification technology) (Washburn, Wolters, & Yates, 2001). MudPIT involves the combination of two chromatographic approaches (strong cation exchange and reverse phase) directly coupled to a mass spectrometer. Protein samples are first digested into component peptides, applied to the chromatographic system, and then eluted in cycles before being introduced to the second stage. Each cycle involves eluting peptides from the ion exchange (SCX resin) with a specific salt concentration, followed by a solvent gradient of increasing hydrophobicity to separate peptides on the reverse phase column; every subsequent cycle begins with an increased salt concentration. Often, the separation by cation exchange resin is performed offline, and the fractions are individually processed through reverse-phase liquid chromatography coupled to the mass spectrometer. Although this offline method is time consuming, there is an increase in the number of peptides identified compared to the direct SCX-reverse phase-high performance LC; for example, more than 5000 peptides were characterized in a single experiment by Li et al. (2005b). In general, such methods require large amounts of strating material as there tend to be protein losses with each fractionation step. Hence, MudPIT is not currently useful for the analysis of smaller samples.
Electrophoretic separations for proteins are among the older separation modes (Tiselius, 1937), and are often used to simplify complex mixtures. Electrophoretic techniques have usually been associated with the use of a gel matrix to assist with the separation (Andrews, 1989). Simple one-dimensional separations by molecular weight can be used, as well as 2DE, as pioneered by O’Farrell and Klose in the 1970s (Klose, 1975; O’Farrell, 1975). Conventional 2DE first separates proteins by their isoelectric points and second by molecular weights. Under ideal circumstances, electrophoresis can be used to resolve hundreds to thousands of individual proteins. However, scaling laws favor miniaturization, so that small 2DE separations have long been known to have advantages for volume-limited samples and for high separation efficiencies (Ruchel, 1977; Poehling & Neuhoff, 1980). We have had good success with “minigel” formats (first dimension isoelectric focusing strips 7–11 cm wide, and second dimension sodium dodecyl sulfate polyacrylamide gel electrophoresis, or SDS-PAGE gels) for micro-proteomic applications.
The multiple steps in the 2DE process can lead to protein losses (Zhou et al., 2005). Although 2DE has had a reputation for being slow, difficult, and poorly reproducible, these issues have been largely resolved with the development of immobilized pH gradient strips that provide more consistent separations in the first dimension, as well as electrophoretic devices capable of running multiple gels quickly and consistently (Gorg et al., 2000) A renewed interest in radiolabeling strategies (Vuong et al., 2000) and the development of more sensitive fluorescent protein stains (Patton, 2002) have markedly improved detection sensitivity. There have been efforts to directly analyze samples from gel-based separations or through a intermediate transfer to a polyvinylidene fluoride membrane using MALDI time-of-flight MS; however, this approach has yet to be widely applied (Gusev, 2000).
Capillary electrophoresis (CE) is a method that combines several features of column chromatography and electrophoresis. In this method, analytes are introduced into a buffer (conducting electrolyte) and injected onto an open capillary. When a potential is applied, the analytes migrate along the capillary with their velocity depending on their electrophoretic mobility and electroosmotic (buffer) flow(Jorgenson&Lukacs, 1983). During electrophoretic separations one applies a current across a solution, which generates heat. This heat tends to disturb the separation process. In CE, heat dissipation is more effective as the capillary diameter is reduced, so that smaller capillaries can be used with higher voltages to improve and speed up separations. Like LC, CE can be directly interfaced with mass spectrometers. However, combining CE with an MS instrument presents several challenges, because the mass of the material in a nanoliter-volume (or smaller) analyte band is difficult to characterize with MS. Reviews of how these challenges have been overcome include Simpson and Smith (2005), Banks (1997), and others (Ohnesorge, Neususs, & Watzig, 2005).
There are several operational modes of separation byCEthat have been interfaced with a mass spectrometer, notably, capillary zone electrophoresis (CZE) and capillary isoelectrofocusing (CIEF). CZE is similar to LC in that proteins are injected onto the column, and the components are separated into discrete bands that elute from the capillary and are subsequently introduced into the mass spectrometer. CIEF is similar to the gel-based approach because the separation is based on the pI (Shen&Smith, 2002).A direct comparison of LC, CZE, and CIEF for analyses of yeast cytosol digest showed that the CE methods have a high resolving power, separation efficiency, and shorter run times compared to LC(Shen et al., 2000). However, CIEF allows more material to be introduced into the capillary (Cooper, Wang, & Lee, 2004). Smaller sample volumes suggest that CE would be an ideal tool for microproteomics. However, these small sample bands mean that a smaller dynamic range of proteins can be detected. Taken together, these characteristics suggest that CE separation would be well-suited for the last stage of a comprehensive multistage separation, or as a sole technique for the smallest samples.
V. IDENTIFICATION OF PROTEINS USING MASS SPECTROMETRY (MS)
As mentioned earlier, one challenge in sequencing proteins arises from the complexities inherent in having 20 amino acid building blocks and a variety of PTMs. This complexity is even greater because protein translation levels differ both temporally and spatially. Gel- and immunohistochemical-based methods to identify proteins have been used successfully in a wide variety of samples. Edman degradation has also been a powerful approach. However, the throughput of these methods is low. Although MS technologies have been utilized for decades, it was only with the development of ionization methods that allow large protein molecules to be introduced into mass analyzers has MS been widely applied to proteomics. Specifically, ESI and MALDI revolutionized protein characterization (Tanaka et al., 1988; Hillenkamp et al., 1991; Karas, Bahr, & Gieβmann, 1991). In MS-based methods, much smaller sample amounts are required to sequence a peptide. Amounts can be in the attomole or even zeptomole range under ideal circumstances.
All mass analyzers require the ionization of a protein molecule in the gas phase; therefore, inefficiencies in the vaporization and ionization processes can often limit overall system performance. Because it is easier to ionize and characterize peptides, proteins are often cleaved into complex peptide mixtures prior to measurements. In this so-called “bottom-up” approach, a protein is digested into its component peptides with enzymes such as trypsin or chymotrypsin, and the peptides characterized with MS. Once the peptide sequences are confirmed, they are matched to precursor proteins using various databases, in an approach called peptide mass fingerprinting (PMF) (James et al., 1993). This “bottom up” approach has been widely used.
Global proteomics by means of PMF normally requires the detection of multiple peptides to unambiguously identify the protein; measuring the peptides with sufficiently high mass accuracy can permit proteins to be identified based on fewer peptides. This method also uses accurate mass information and the retention times from a list of previously identified peptides to decrease the sequencing time spent on those peptides by the mass spectrometer. In a report that demonstrated the utility of this global approach to proteomics, 61% of the predicted peptides in the Deinococcus radiodurans proteome were identified (Lipton et al., 2002).
A newer protein identification strategy is the “top-down” approach, which uses extremely accurate masses of intact proteins obtained from a Fourier transform mass spectrometer as well as masses obtained from their subsequent CID fragmentation to identify proteins (Kelleher, 2004; Han et al., 2006). A major advantage of the “top-down” approach is its robustness for analyzing modified proteins (Pesavento et al., 2004). The “bottom-up” approach will miss PTMs located on peptides that are not identified via MS.
VI. INTEGRATING TISSUE SAMPLING AND CHARACTERIZATION IN A SINGLE STEP: MASS SPECTROMETRIC IMAGING (MSI)
Usually, scientists utilize immunohistochemical techniques to determine protein localization in a tissue, and MS to identify proteins of interest. However, over the last 30 years several forms of probe-based MS have been adapted to protein localization studies. Recently, Caprioli and others have pioneered the adaptation of MALDI MS to image proteins in their anatomical context in tissue samples (Spengler, Hubert, & Kaufmann, 1994; Stoeckli, Farmer, & Caprioli, 1999; Stoeckli et al., 2001; Todd et al., 2001; Altelaar et al., 2005; Caldwell & Caprioli, 2005; Crecelius et al., 2005; Jurchen, Rubakhin, & Sweedler, 2005; McDonnell et al., 2005; Rubakhin et al., 2005; Monroe et al., 2006). In MALDI MSI, a tissue section is prepared and matrix is applied. The laser is focused onto a discrete point, and a mass spectrum acquired. At each point, the amount of tissue sampled is in the nanoliter-volume range; thus, this approach is inherently microproteomic. The laser is rastered across the sample to obtain spatially resolved mass spectra, creating a “spectral image” of the tissue sample. Because imaging requires the acquisition of thousands of spectra, MSI is heavily dependent on instrumental and data-storage capabilities. One advantage of this approach is that because minimal sample handling is required, protein losses are reduced. However, the depth of proteomic coverage is less than when conventional separations are performed. Also, the procedures used to collect and prepare samples for MSI are critical, especially when analyzing phosphoproteins or neuropeptides, both of which can be quickly modified or degraded if sample preparation is suboptimal. New, innovative ionization methods such as desorption electrospray ionization (DESI) are also being applied to MSI. In contrast to MALDI, DESI permits images to be created from tissue slices under ambient conditions (Cooks et al., 2006). This method performs well, but its upper mass range must be extended before its application to biomedical proteomics is practical. Because MSI is a rapidly changing field, readers are encouraged to examine recent publications devoted to this technology, such as (Heeren & Sweedler, 2007).
VII. QUANTIFICATION OF PROTEIN EXPRESSION DIFFERENCES
Often, some of the most important information gained from a proteomic study is the determination of differences in protein expression between experimental conditions. Although one frequently uses the term quantification to indicate the absolute amount of a material (in concentration or mass), in most proteomic measurements, the term implies relative abundance, such as the percent change between two treatment groups. Quantification is usually done after the sample is fractionated, but can be performed either before or after protein identification. The advantage of quantifying before identification is that efforts can then be focused on identifying only those proteins that are differentially expressed. Quantification can follow either a gel-based separation, by comparing differences in the intensity of corresponding spots, or after MS measurement, by evaluating differences in the intensities/areas of corresponding mass spectral peaks. Because of the expense and effort required for many proteomic measurements, care must be taken to ensure that the experimental design balances the running of samples from the different groups to avoid any systematic bias. There are numerous case studies that illustrate the dangers of design flaws that can lead to systematic bias in proteomic studies (e.g., see Baggerly et al., 2004a; Baggerly, Morris, & Coombes, 2004b; Baggerly, Coombes, & Morris, 2005a; Baggerly et al., 2005b; Hu et al., 2005a). This problem can be avoided by applying standard experimental design principles such as blocking and randomization, as described in standard textbooks (e.g., Box, Hunter, & Hunter, 2005), within the laboratory setting.
How is quantification performed? Gel-based methods usually involve fluorescent and some visible stains, as well as radiolabeling. The relationship between spot intensity and protein amount is linear for these stains over several orders of magnitude. Silver stains do not have as linear of a relationship, and are applied less frequently for quantification. DIGE is a method that can reduce variability by running control and treatment samples together on the same gel, as well as a pooled sample from the two groups. Dyes with different spectral characteristics are used to detect and quantify spots from the different groups. The most common set of dyes are the Cy dyes (Friedman, 2006). Although these approaches are well validated, areas that can be improved when working with the smallest samples include the use of more efficient (saturation) protein labeling, use of more sensitive dyes, and reducing the effect that dyes have on changing protein migration.
After scanning the gels for a given experiment, a key quantitative challenge is to detect spots on the gels while filtering out artifacts, and to match the spots across gels and quantify them. Assuming N gels and p protein spots in the set, the goal of this step is to construct an N-by-p matrix that contains the protein expression values for each spot for each gel, which can be surveyed to identify those proteins related to the factor of interest. Various commercial and noncommercial methods have been developed to accomplish this goal, but unfortunately, these methods encounter numerous difficulties that limit their effectiveness, especially for larger studies. Various errors, such as spot detection, spot matching, and spot boundary estimation errors, are typical and reduce the probability of finding protein expression differences in the data. These problems tend to accumulate with larger studies, and encourage some investigators to perform smaller studies that are underpowered to find realistic-sized differences (Fig. 3).
Figure 3.
Comparison of yeast cytosol tryptic digest (at 5 mg/mL) using three different CE separation modes. A: Capillary isoelectric focusing (CIEF);B: capillary zone electrophoresis (CZE); andC: reverse phase LC. The concentration used for CIEF was 16-fold lower than the samples used for CZE and LC. Reprinted with permission from Shen et al. (2000), copyright 2000 American Chemical Society.
In response to these problems, we have developed an approach for preprocessing 2DE data called Pinnacle (Morris, Clark, & Gutstein, 2008). The name derives from the fact that, unlike most existing methods, this method performs spot quantification using pixel intensities at peak spot intensity values in the horizontal and vertical dimensions (pinnacles) rather than spot volumes. The underlying philosophy is to keep the preprocessing as simple as possible in order to avoid any extra bias and variance that can result from the propagation of errors that characterize more complex methods. The steps of the Pinnacle method are as follows:
Align the 2DE gel images.
Compute the average gel, averaging the staining intensities across gels for each pixel in the image.
Denoise the average gel using the two-dimensional undecimated discrete wavelet transform (UDWT).
Detect pinnacles on the denoised average gel, which are pixel locations that are local maxima in the horizontal and vertical directions with an intensity above some minimum threshold (e.g., the 75th percentile for the average gel).
Perform spot quantification on the individual gels by taking the maximum intensity within a stated tolerance of each pinnacle location.
Assuming p pinnacles are detected on the average gel, one obtains an N-by-p matrix of spot intensities that can be surveyed for potential biomarkers.
This method is significantly quicker and simpler than the typical spot detection algorithms used in 2DE, which perform spot detection on individual gels with complex spot definitions, quantifying spot volumes, and matching spots across gels. This usual approach is time-consuming and error-prone, and is especially problematic for larger studies, because these errors propagate as the number of gels increases. It takes a great deal of time-consuming hand-editing to try to fix these errors. Further, the process of estimating spot boundaries is also difficult and error-prone, and leads to increased coefficients of variance when spot volumes are used to quantify the proteins. Simulation studies have demonstrated that the Pinnacle method leads to more reliable and precise quantifications (Morris, Clark, & Gutstein, 2008). Statistical principles suggest that performing spot detection on the average gel leads to greater sensitivity and specificity, because true protein spots are reinforced across gels whereas artifacts and noise average out. As a result, this method has the potential to reliably detect and quantify fainter spots, thus increasing the realized dynamic range of the technology (Fig. 4).
Figure 4.

Typical average gel with detected pinnacles. The average gel was created by taking the pixel-wise average over 28 gels in a dilution series created from Escherichia coli lysates by Nishihara and Champion (2002) (gels provided courtesy of Dr. Kathleen Champion-Francissen). “Hotter” colors indicate regions of higher intensity, whereas “cooler” colors indicate lower intensities. Intensities above 350 were removed from the scale to improve contrast. The units of the x and y axes are pixel distance from the origin (upper left corner of the image). White “x’s” mark the 1,380 pinnacles detected using Pinnacle, which represent local maxima in the x- and y-directions with intensities greater than 47.2, the 75th percentile intensity on the average gel. Note that a wide range of spot (pinnacle) intensities are detected, whereas artifacts are minimized by the averaging process. Reprinted with permission from Jeffrey, Walla, and Gutstein (2008), copyright 2008 Oxford Journals.
VIII. MASS SPECTROMETRIC-BASED QUANTIFICATION
The ability to detect a protein via MS depends on multiple variables, including vaporization/desorption and ionization efficiency (which depend on sample constituents besides proteins), and losses during separation and sample handling. Thus, it is difficult to directly compare peaks between treatment groups unless these variables are constant. Although a direct correlation between peak intensities from two samples is challenging, evolving methods use parallel MS experiments for a direct comparison of the peptide peaks(Wiener et al., 2004).Perhaps the most common method for quantification involves tagging the proteins with labels that differ only by stable isotopes (Fricker et al., 2006). Using such methods, proteins in two or more proteome samples are tagged with different isotopic tags that vary in their molecular weight but are chemically nearly identical; thus, any sample or ionization differences between the two can be corrected, and their peak intensities directly correlated to their concentrations. Several different chemical tags using H/2H, 12C/13C, 16O/18O, or 14N/15N isotopes have been reported (Leitner & Lindner, 2004; Righetti et al., 2004).
One can avoid the requirement for derivatizing peptides/proteins with the isotopic label by using in situ labeling. For example, isotopic labeling of proteins in cell cultures is achieved by using stable isotope-enriched amino acids in the culture media (known as SILAC) (Ong, Foster, & Mann, 2003). This method is used for cell cultures under different conditions and allows the quantification of the expression levels of hundreds of proteins, thus promising insights into cell signaling pathways and also several interconnected signaling pathways.
Methods similar to Pinnacle discussed above have also been developed to detect and quantify peaks in one-dimensional MS data, such as MALDI MS and surface-enhanced laser desorption-ionization (SELDI) MS (Coombes et al., 2005; Morris et al., 2005). Stand-alone software is freely available for applying the method to MS analysis (Karpievitch et al., 2007). The steps are as follows:
Align the spectra on the time scale by choosing a linear change of variables for each spectrum in order to maximize the correlation between pairs. We have found that alignment can be done much more simply and efficiently on the time scale rather than the m/z scale.
Compute the mean of the aligned raw spectra.
Denoise the mean spectrum using the UDWT.
Locate intervals that contain peaks by finding local maxima and minima in the denoised mean spectrum.
Quantify peaks in individual raw spectra by recording the maximum and minimum height in each interval, which should contain a peak. This quantification method implicitly removes the baseline artifact.
Calibrate all spectra using the mean of the full set of calibration experiments.
Assuming that a total of p peaks are detected on the average gel, an N-by-p matrix of peak intensities is obtained for the N spectra in the study that can be surveyed for potential biomarkers.
A key component of these approaches as described for two-and one-dimensional data is that we perform peak (or spot) detection on the average spectrum (gel), rather than on individual spectra (gel); a number of advantages are obtained (Morris et al., 2005). First, it avoids the difficult and error-prone peak-matching step that is necessary when using individual spectra. Eliminating peak-matching improves the accuracy of the data, and results in no missing data. Second, it tends to result in greater sensitivity and specificity for peak detection, because averaging across N spectra reinforces the true signal while weakening the noise by a factor of . Averaging enables us to better detect real peaks down near the noise region of the spectra, and decreases the chance of flagging spurious peaks that are noise. The simulation study presented in Morris et al. (2005) demonstrates that using the average spectrum results in improved peak detection, with the largest improvement being for low-abundance peaks of high prevalence. Third, it speeds preprocessing time considerably, because peak matching is by far the most time-consuming preprocessing step.
IX. DETERMINATION OF STATISTICAL SIGNIFICANCE
Given an N×p matrix that contains relative quantifications for p features (peak/spot quantifications) for each of N spectra/gels, the next analysis step is to identify which features are significantly associated with the factor of interest, and that might indicate potential biomarkers. Significance should be determined with appropriate statistical tests, not simply ad hoc criteria such as fold-change, because statistical tests appropriately account for the variability of the measurements when making the determinations, whereas simple use of fold-change does not. The particular test to use depends on the number of groups and the experimental design of the study. For example, if the goal is to find proteins that are differentially expressed between two experimental conditions, then a t-test (or nonparametric rank-sum test if normality cannot be assumed) can be done for each p feature, and then all features with sufficiently small P values flagged as significant.
In this setting, the selection of this P-value cutoff must be done with care. Because these assays survey hundreds or even thousands of proteins simultaneously, we expect a certain number of them to have small P values even if none are truly related to the factor of interest. In the absence of true differences, we expect ca. 50 P values of <0.05 for every 1,000 protein spots. In statistics, this is called the multiplicity or multiple testing problem. One classical approach to deal with multiplicities is Bonferroni, which would use a cutoff of 0.05/p to determining significance, where p is the number of features considered. This cutoff would control the experiment-wise error rate at 0.05, and mean that we expect that the probability of at least one false positive result is 0.05.
Because this criterion strongly controls the false positive rate, but results in a large number of false negatives, it is widely considered too conservative for exploratory analyses like these. Other methods control the false discovery rate, or FDR (Benjamini & Hochberg, 1995). Controlling the FDR at 0.05 means that of the features declared significant, we expect5% or fewer of them to be false positives. There are a number of procedures for controlling FDR (Benjamini & Hochberg, 1995; Benjamini& Liu, 1999; Yekutieli & Benjamini, 1999; Genovese & Wasserman, 2002; Storey, 2002; Ishwaran & Rao, 2003; Pounds & Morris, 2003; Storey, 2003; Efron, 2004; Newton et al., 2004; Pounds & Cheng, 2004; Datta & Datta, 2005). Many of these methods take advantage of the property that the P values that correspond to features that are not associated with the factor of interest should follow a uniform distribution, whereas the distribution of P values for predictive features should be characterized by an overabundance of small P values. In most of these methods, one inputs the list of P values and desired FDR, and receives as output a P-value cutoff that preserves the desired FDR. Typically, this cutoff is smaller than 0.05, but quite a bit larger than the corresponding Bonferroni bound 0.05/p. Using the FDR of 0.05, we still expect that one out of every 20 flagged proteins are false positives. We must be willing to accept some false positives so that we have sufficient power to capture relevant markers. Thus, it is important to validate any flagged markers (e.g., by Western blotting or multiple reaction monitoring MS).
Instead of performing peak/spot detection and applying statistical tests to each feature, another alternative is to model the spectra or gel images using functional data techniques. One flexible method that has been developed for this purpose is the wavelet-based functional mixed model (Morris, Clark, & Gutstein, 2008). This approach does not depend on peak or spot detection at all, and can search for differentially expressed regions of the spectra or gel images in a way that takes statistical considerations and fold-change into account, and accounts for the multiplicity problem. These methods are more complex and computationally intensive, and just under development, but show considerable promise for the analysis of proteomic data.
Power calculations can be done to assess how large a sample size (number of gels, number of subjects) is needed for a particular proteomics study. In recent years, various methods have been developed to perform sample size calculations for settings where FDR is used to deal with the multiplicity problem(Hu, Zou, & Wright, 2005b; Jung, 2005; Li et al., 2005a; Pounds & Cheng, 2005; Liu & Hwang, 2007). In each case, the calculations depend on the reproducibility and assumed subject-to-subject variability in the data, as well as the magnitude of the effect sizes for the differentially expressed proteins. If preliminary studies in the tissue of interest are available, then these data can be used to estimate the reproducibility (variance across replicate gels) and subject-to-subject variability (variance across gels for different subjects), and the effect size (difference between group means on the log scale) for the set of spots detected in the preliminary data. Absent these preliminary studies, it is difficult to perform a sample size/power calculation.
The use of small biological samples in proteomic analyses is likely to affect the statistical properties of these studies. As we operate closer to the detection limit of the available technology, it is likely that technical variability will increase, potentially reducing the statistical power of the research. This effect can be mitigated by performing larger studies with more replicates and/or subjects. Another possibility is to pool samples. By increasing the amount of total protein per assay, this pooling can help mitigate the effects of analyzing samples near the detection limit, but at the cost of losing protein information for individual samples. Pooling may be an acceptable tradeoff in studies attempt to find proteins differentially expressed across groups, but this pooling prevents the possibility of finding proteins correlated with the subjects’ individual outcomes.
X. CONCLUSIONS
Proteomics is a rapidly evolving field. Technological advances permit us to examine a greater fraction of the proteome from less tissue. Statistical and analytical advances also permit us to determine significant changes in protein expression and PTMs from smaller samples with greater certainty and specificity.
However, more progress must be made. For example, improvements in protein extraction methods as well as the specificity of protein identification techniques to allow us to evaluate lower-concentration proteins, or a greater fraction of the membrane proteins are certainly needed, as are better methods to characterize more complex PTMs such as carbohydrates. Of course, such advances should also be coupled to greater sensitivity so that smaller samples can be assayed.
What options await us on the horizon? Methods such as MSI or DESI that bypass much of the conventional proteomics workflow are intriguing, because they minimize the number of steps needed to identify proteins from tissue. These systems could minimize obligate protein losses from extraction and separation techniques, and so perhaps will provide more efficient protein identification from small groups of cells. Nanotechnology, another rapidly developing area, is poised to make significant contributions to microproteomics as novel miniaturized platforms are applied to protein extraction, separation, and identification. Further evaluation and development of these and other promising and exciting methods are needed to allow researchers to take advantage of the information offered by proteomics. As we move toward an ultimate goal of examining the complete proteome from single cells or small cell groups, the keys to achieving this end will be creativity, flexibility, and a willingness to take risks and innovate past the ultimate limitations of conventional approaches.
ACKNOWLEDGMENTS
The support of the National Institute on Drug Abuse to the UIUC Neuroproteomics Center on Cell to Cell Signaling through Award No. DA018310, is gratefully acknowledged. Work in the authors’ laboratories is also supported by additional NIH grants from NIDA, NIAAA, and NIGMS. We thank Marlen Banda for manuscript preparation assistance.
Biographies
Howard B. Gutstein is a practicing Pediatric Anesthesiologist with a clinical interest in treating children with cancer pain. Clinical experiences fueled his research interests in developing more effective treatments for pain and two devastating side effects of pain treatment, narcotic tolerance and drug addiction. His laboratory focuses on the development and use of novel proteomics technologies in conjunction with state of the art behavioral and neuroanatomical approaches to identify new targets for treating pain and addiction. Gutstein received his M.D. from Johns Hopkins University, and did postdoctoral training with Huda Akil at the University of Michigan. He is currently Associate Professor in the departments of Anesthesiology and Molecular Genetics at the MD Anderson Cancer Center.

Jeffrey S. Morris received his Ph.D. in Statistics in 2000 from Texas A&M University, working with Raymond J. Carroll to develop innovative statistical methods for colon carcinogenesis data, after which he joined the Department of Biostatistics at The University of Texas MD Anderson Cancer Center, where he is currently an Associate Professor. His research interests include bioinformatics, proteomics, and developing new methodology for functional and image data.

Suresh P. Annangudi, Ph.D. Dr. Annangudi received his Master of Science in organic chemistry from Indian Institute of Technology-Bombay, India and received his PhD under the direction of Prof. Robert G. Salomon in chemistry from Case Western Reserve University. Currently as a post-doctoral associate, he is interested in developing methods to discover, characterize and quantitate cell-to-cell signaling molecules in discrete biological tissues using mass spectrometry with Prof. Jonathan Sweedler at the University of Illinois.

Jonathan V. Sweedler received his Ph.D. in 1988 from the University of Arizona, and has been at the University of Illinois since 1991. He is currently the Lycan Professor of Chemistry at the University of Illinois and is associated with the Beckman Institute, is the director of the Carver Biotechnology Center and head of the Neuroproteomics Center on Cell to Cell Signaling. He has appointments in the Neuroscience Program, the Department of Physiology and the Bioengineering Program. His research interests are in bioanalytical chemistry, and focus on developing new methods for assaying small volume samples, and applying these methods to study novel neurochemistry. He and his group are developing new sampling methods interfaced to capillary scale separations, nanoliter volume NMR, single-cell mass spectrometry, information rich spectroscopic detectors for capillary-scale separations, and hybrid nanofluidic/microfluidic devices for neuronal sampling. Using this suite of technology, he is investigating the roles that peptide hormones, neurotransmitters and neuromodulators play in behavior, learning and memory. He has received numerous awards including the PittCon Award, the Merck Prize, the Instrumentation Award from the Analytical Division of the American Chemical Society, the Gill Prize and the Benedetti-Pichler Award for Microanalysis.

REFERENCES
- Altelaar AF, van Minnen J, Jimenez CR, Heeren RM, Piersma SR. Direct molecular imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal Chem. 2005;77:735–741. doi: 10.1021/ac048329g. [DOI] [PubMed] [Google Scholar]
- Anderson L. From genome to proteome: Looking at a cell’s proteins. Science. 1995;270:369–371. doi: 10.1126/science.270.5235.369. [DOI] [PubMed] [Google Scholar]
- Anderson L, Seilhammer J. A comparsion of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- Andrews AT. Electrophoresis: Theory, techniques, and biochemical and clinical applications. 2nd edition. Oxford: Clarendon Press; 1989. [Google Scholar]
- Baggerly KA, Edmonson SR, Morris JS, Coombes KR. High-resolution serum proteomic patterns for ovarian cancer detection. Endocr Relat Cancer. 2004a;11:583–584. doi: 10.1677/erc.1.00868. author reply 585–587. [DOI] [PubMed] [Google Scholar]
- Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: Comparing datasets from different experiments. Bioinformatics. 2004b;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- Baggerly KA, Coombes KR, Morris JS. Are the NCI/FDA ovarian proteomic data biased? A reply to producers and consumers. Cancer Inform. 2005a;1:9–14. [PMC free article] [PubMed] [Google Scholar]
- Baggerly KA, Morris JS, Edmonson SR, Coombes KR. Signal in noise: Evaluating reported reproducibility of serum proteomic tests for ovarian cancer. J Natl Cancer Inst. 2005b;97:307–309. doi: 10.1093/jnci/dji008. [DOI] [PubMed] [Google Scholar]
- Banks JF. Recent advances in capillary electrophoresis/electrospray/mass spectrometry. Electrophoresis. 1997;18:2255–2266. doi: 10.1002/elps.1150181216. [DOI] [PubMed] [Google Scholar]
- Banks R, Dunn M, Forbes M, Stanley A, Pappin D, Naven T, Gough M, Harnden P, Selby P. The potential use of laser capture microdissection to selectively obtain distinct populations of cells for proteomic analysis—Preliminary findings. Electrophoresis. 1999;20:689–700. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<689::AID-ELPS689>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Belo I, Santos JA, Cabral JM, Mota M. Optimization study of Escherichia coli TB1 cell disruption for cytochrome b5 recovery in a small-scale bead mill. Biotechnol Progress. 1996;12:201–204. doi: 10.1021/bp950085l. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- Benjamini Y, Liu W. A step-down multiple hypotheses testing procedure that controls the false discovery rate under independence. J Stat Plann Inference. 1999;82:163–170. [Google Scholar]
- Bhattacharya SH, Gal AA, Murray KK. Laser capture microdissection MALDI for direct analysis of archival tissue. J Proteome Res. 2003;2:95–98. doi: 10.1021/pr025547m. [DOI] [PubMed] [Google Scholar]
- Borrebaeck CA, Wingren C. High-throughput proteomics using antibody microarrays: An update. Expert Rev Mol Diagn. 2007;7:673–686. doi: 10.1586/14737159.7.5.673. [DOI] [PubMed] [Google Scholar]
- Box GEP, Hunter WG, Hunter JS. Statistics for experimenters: An introduction to design, data analysis, and model building. 2nd edition. New York: Wiley; 2005. [Google Scholar]
- Burre J, Zimmermann H, Volknandt W. Immunoisolation and subfractionation of synaptic vesicle proteins. Anal Biochem. 2007;362:172–181. doi: 10.1016/j.ab.2006.12.045. [DOI] [PubMed] [Google Scholar]
- Butt RH, Coorssen JR. Pre-extraction sample handling by automated frozen disruption significantly improves subsequent proteomic analyses. J Proteome Res. 2006;5:437–448. doi: 10.1021/pr0503634. [DOI] [PubMed] [Google Scholar]
- Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- Caldwell RL, Caprioli RM. Tissue profiling by mass spectrometry: A review of methodology and applications. Mol Cell Proteomics. 2005;4:394–401. doi: 10.1074/mcp.R500006-MCP200. [DOI] [PubMed] [Google Scholar]
- Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97 Suppl 1:16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Detection technologies. Ambient Mass Spectrom Sci. 2006;311:1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- Coombes KR, Tsavachidis S, Morris JS, Baggerly KA, Hung MC, Kuerer HM. Improved peak detection and quantification of mass spectrometry data acquired from surface-enhanced laser desorption and ionization by denoising spectra with the undecimated discrete wavelet transform. Proteomics. 2005;5:4107–4117. doi: 10.1002/pmic.200401261. [DOI] [PubMed] [Google Scholar]
- Cooper JW, Wang Y, Lee CS. Recent advances in capillary separations for proteomics. Electrophoresis. 2004;25:3913–3926. doi: 10.1002/elps.200406154. [DOI] [PubMed] [Google Scholar]
- Craven RA, Totty N, Harnden P, Selby PJ, Banks RE. Laser capture microdissection and two-dimensional polyacrylamide gel electrophoresis: Evaluation of tissue preparation and sample limitations. Am J Pathol. 2002;160:815–822. doi: 10.1016/S0002-9440(10)64904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crecelius AC, Cornett DS, Caprioli RM, Williams B, Dawant BM, Bodenheimer B. Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. J Am Soc Mass Spectrom. 2005;16:1093–1099. doi: 10.1016/j.jasms.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Cuello AC, Carson S. Microdissection of fresh rat brain tissue slices. In: Cuello AC, editor. Brain microdissection techniques. Chichester: John Wiley and Sons; 1983. pp. 37–126. [Google Scholar]
- Datta S, Datta S. Empirical Bayes screening of many P-values with applications to microarray studies. Bioinformatics. 2005;21:1987–1994. doi: 10.1093/bioinformatics/bti301. [DOI] [PubMed] [Google Scholar]
- Davis JM, Blumberg LM. Probability theory for number of mixture components resolved by n independent columns. J Chromatogr A. 2005;1096:28–39. doi: 10.1016/j.chroma.2005.03.137. [DOI] [PubMed] [Google Scholar]
- de Groot CJM, Steegers-Theunissen RP, Guzel C, Steegers EAP, Luider TM. Peptide patterns of laser dissected human trophoblasts analyzed by matrix-assisted laser desorption/ionisation-time of flight mass spectrometry. Proteomics. 2005;5:597–607. doi: 10.1002/pmic.200400974. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. Maintaining protein stability. Methods Enzymol. 1990;182:83–89. doi: 10.1016/0076-6879(90)82010-y. [DOI] [PubMed] [Google Scholar]
- Efron B. Large-scale simultaneous hypothesis testing: The choice of a null hypothesis. Journal of the American Statistical Association. 2004;99:96–104. [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Emmert-Buck MR, Gillespie JW, Paweletz CP, Ornstein DK, Basrur V, Appella E, Wang QH, Huang J, Hu N, Taylor P, Petricoin EFr. An approach to proteomic analysis of human tumors. Molecular Carcinogenesis. 2000;27:158–165. [PubMed] [Google Scholar]
- Evans CR, Jorgenson JW. Multidimensional LC-LC and LC-CE for high-resolution separations of biological molecules. Anal Bioanal Chem. 2004;378:1952–1961. doi: 10.1007/s00216-004-2516-2. [DOI] [PubMed] [Google Scholar]
- Flanagan RJ, Morgan PE, Spencer EP, Whelpton R. Micro-extraction techniques in analytical toxicology: Short review. Biomedical Chromatography. 2006;20:530–538. doi: 10.1002/bmc.671. [DOI] [PubMed] [Google Scholar]
- Fricker LD, Lim J, Pan H, Che FY. Peptidomics: Identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom Rev. 2006;25:327–344. doi: 10.1002/mas.20079. [DOI] [PubMed] [Google Scholar]
- Friedman DB. Quantitative proteomics for two-dimensional gels using difference gel electrophoresis. Methods Mol Biol. 2006;367:219–240. doi: 10.1385/1-59745-275-0:219. [DOI] [PubMed] [Google Scholar]
- Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. A sampling of the yeast proteome. Mol Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese C, Wasserman L. Operating characteristics and extensions of the false discovery rate procedure. J R Stat Soc Ser B Stat Methodol. 2002;64:499–517. [Google Scholar]
- Giddings JC. Unified separation science. New York: Wiley; 1991. [Google Scholar]
- Gorg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000;21:1037–1053. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Greengauz-Roberts O, Stoppler H, Nomura S, Yamaguchi H, Goldenring JR, Podolsky RH, Lee JR, Dynan WS. Saturation labeling with cysteine-reactive cyanine fluorescent dyes provides increased sensitivity for protein expression profiling of laser-microdissected clinical specimens. Proteomics. 2005;5:1746–1757. doi: 10.1002/pmic.200401068. [DOI] [PubMed] [Google Scholar]
- Grier DG. A revolution in optical manipulation. Nature. 2003;424:810–816. doi: 10.1038/nature01935. [DOI] [PubMed] [Google Scholar]
- Guo J, Colgan TJ, DeSouza LV, Rodrigues MJ, Romaschin AD, Siu KWM. Direct analysis of laser capture microdissected endometrial carcinoma and epithelium by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:2762–2766. doi: 10.1002/rcm.2119. [DOI] [PubMed] [Google Scholar]
- Gusev AI. Interfacing matrix-assisted laser desorption/ionization mass spectrometry with column and planar separations. Fresenius J Anal Chem. 2000;366:691–700. doi: 10.1007/s002160051563. [DOI] [PubMed] [Google Scholar]
- Han X, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- Hastie ND, Bishop JO. The expression of three abundance classes of messenger RNA in mouse tissues. Cell. 1976;9:761–774. doi: 10.1016/0092-8674(76)90139-2. [DOI] [PubMed] [Google Scholar]
- Heeren RMA, Sweedler JV. Imaging mass spectrometry special issue. Int J Mass Spectrom. 2007;260:89–252. [Google Scholar]
- Hellmich W, Pelargus C, Leffhalm K, Ros A, Anselmetti D. Single cell manipulation, analytics, and label-free protein detection in microfluidic devices for systems nanobiology. Electrophoresis. 2005;26:3689–3696. doi: 10.1002/elps.200500185. [DOI] [PubMed] [Google Scholar]
- Herbert B. Advances in protein solubilsation for two-dimensional electrophoresis. Electrophoresis. 1999;20:660–663. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<660::AID-ELPS660>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Hillenkamp F, Karas M, Beavis RC, Chait BT. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal Chem. 1991;63:1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- Hofstadler SA, Swanek FD, Gale DC, Ewing AG, Smith RD. Capillary electrophoresis-electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry for direct analysis of cellular proteins. Anal Chem. 1995;67:1477–1480. doi: 10.1021/ac00104a028. [DOI] [PubMed] [Google Scholar]
- Hopkins TR. Physical and chemical cell disruption for the recovery of intracellular proteins. Bioprocess Technol. 1991;12:57–83. [PubMed] [Google Scholar]
- Hu J, Coombes KR, Morris JS, Baggerly KA. The importance of experimental design in proteomic mass spectrometry experiments: Some cautionary tales. Brief Funct Genomic Proteomic. 2005a;3:322–331. doi: 10.1093/bfgp/3.4.322. [DOI] [PubMed] [Google Scholar]
- Hu J, Zou F, Wright FA. Practical FDR-based sample size calculations in microarray experiments. Bioinformatics. 2005b;21:3264–3272. doi: 10.1093/bioinformatics/bti519. [DOI] [PubMed] [Google Scholar]
- Hu X, Bessette PH, Qian J, Meinhart CD, Daugherty PS, Soh HT. Marker-specific sorting of rare cells using dielectrophoresis. PNAS. 2005c;102:15757–15761. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wu H, Bhaya D, Grossman A, Granier S, Kobilka BK, Zare RN. Counting low-copy number proteins in a single cell. Science. 2007;315:81–84. doi: 10.1126/science.1133992. [DOI] [PubMed] [Google Scholar]
- Hummon AB, Amare A, Sweedler JV. Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom Rev. 2006;25:77–98. doi: 10.1002/mas.20055. [DOI] [PubMed] [Google Scholar]
- Ishwaran H, Rao JS. Detecting differentially expressed genes in microarrays using Bayesian model selection. JAm Stat Assoc. 2003;98:438–455. [Google Scholar]
- Ivanov YD, Govorun VM, Bykov VB, Archakov AI. Nanotechnologies in proteomics. Proteomics. 2006;6:1399–1414. doi: 10.1002/pmic.200402087. [DOI] [PubMed] [Google Scholar]
- James P, Quadroni M, Carafoli E, Gonnet G. Protein identification by mass profile fingerprinting. Biochem Biophys Res Commun. 1993;195:58–64. doi: 10.1006/bbrc.1993.2009. [DOI] [PubMed] [Google Scholar]
- Jorgenson JW, Lukacs KD. Capillary zone electrophoresis. Science. 1983;222:266–272. doi: 10.1126/science.6623076. [DOI] [PubMed] [Google Scholar]
- Jung SH. Sample size for FDR-control in microarray data analysis. Bioinformatics. 2005;21:3097–3104. doi: 10.1093/bioinformatics/bti456. [DOI] [PubMed] [Google Scholar]
- Jurchen JC, Rubakhin SS, Sweedler JV. MALDI-MS imaging of features smaller than the size of the laser beam. J Am Soc Mass Spectrom. 2005;16:1654–1659. doi: 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Karas M, Bahr U, Gieβmann U. Matrix-assisted laser desorption ionization mass spectrometry. Mass Spectrom Rev. 1991;10(5):335–357. [Google Scholar]
- Karpievitch YV, Hill EG, Smolka AJ, Morris JS, Coombes KR, Baggerly KA, Almeida JS. PrepMS: TOFMS data graphical preprocessing tool. Bioinformatics. 2007;23:264–265. doi: 10.1093/bioinformatics/btl583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher NL. Top-down proteomics. Anal Chem. 2004;76:197A–203A. [PubMed] [Google Scholar]
- Klose J. Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik. 1975;26:231–243. doi: 10.1007/BF00281458. [DOI] [PubMed] [Google Scholar]
- Kondo T, Hirohashi S. Application of highly sensitive fluorescent dyes (CyDye DIGE Fluor saturation dyes) to laser microdissection and two-dimensional difference gel electrophoresis (2D-DIGE) for cancer proteomics. Nat Protocols. 2006;1:2940–2956. doi: 10.1038/nprot.2006.421. [DOI] [PubMed] [Google Scholar]
- Krieg RC, Gaisa NT, Paweletz CP, Knuechel R. Proteomic analysis of human bladder tissue using SELDI approach following microdissection techniques. Methods Mol Biol. 2005;293:255–267. doi: 10.1385/1-59259-853-6:255. [DOI] [PubMed] [Google Scholar]
- Kwapiszewska G, Meyer M, Bogumil R, Bohle RM, Seeger W, Weissmann N, Fink L. Identification of proteins in laser-microdissected small cell numbers by SELDI-TOF and Tandem MS. BMC Biotechnol. 2004;4:30. doi: 10.1186/1472-6750-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Baxter TM, Yamaguchi H, Wang TC, Goldenring JR, Anderson MG. Differential protein analysis of spasomolytic polypeptide expressing metaplasia using laser capture microdissection and two-dimensional difference gel electrophoresis. Appl Immunohistochem Mol Morphol. 2003;11:188–193. doi: 10.1097/00129039-200306000-00018. [DOI] [PubMed] [Google Scholar]
- Leitner A, Lindner W. Current chemical tagging strategies for proteome analysis by mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;813:1–26. doi: 10.1016/j.jchromb.2004.09.057. [DOI] [PubMed] [Google Scholar]
- Li L, Garden RW, Sweedler JV. Single-cell MALDI: A new tool for direct peptide profiling. Trends Biotechnol. 2000;18:151–160. doi: 10.1016/s0167-7799(00)01427-x. [DOI] [PubMed] [Google Scholar]
- Li SS, Bigler J, Lampe JW, Potter JD, Feng Z. FDR-controlling testing procedures and sample size determination for microarrays. Stat Med. 2005a;24:2267–2280. doi: 10.1002/sim.2119. [DOI] [PubMed] [Google Scholar]
- Li X, Gong Y, Wang Y, Wu S, Cai Y, He P, Lu Z, Ying W, Zhang Y, Jiao L, He H, Zhang Z, He F, Zhao X, Qian X. Comparison of alternative analytical techniques for the characterisation of the human serum proteome in HUPO Plasma Proteome Project. Proteomics. 2005b;5:3423–3441. doi: 10.1002/pmic.200401226. [DOI] [PubMed] [Google Scholar]
- Lipton MS, Pasa-Tolic L, Anderson GA, Anderson DJ, Auberry DL, Battista JR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, Masselon C, Markillie LM, Moore RJ, Romine MF, Shen Y, Stritmatter E, Tolic N, Udseth HR, Venkateswaran A, Wong KK, Zhao R, Smith RD. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc Natl Acad Sci USA. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Hwang JT. Quick calculation for sample size while controlling false discovery rate with application to microarray analysis. Bioinformatics. 2007;23:739–746. doi: 10.1093/bioinformatics/btl664. [DOI] [PubMed] [Google Scholar]
- Lu LL, Liu BC. High-throughput antibody microarrays for quantitative proteomic analysis. Expert Rev Proteomics. 2007;4:505–513. doi: 10.1586/14789450.4.4.505. [DOI] [PubMed] [Google Scholar]
- McDonnell LA, Piersma SR, Maarten Altelaar AF, Mize TH, Luxembourg SL, Verhaert PD, van Minnen J, Heeren RM. Subcellular imaging mass spectrometry of brain tissue. J Mass Spectrom. 2005;40:160–168. doi: 10.1002/jms.735. [DOI] [PubMed] [Google Scholar]
- Melendres AV, Honda H, Shiragami N, Unno H. A kinetic analysis of cell disruption by bead mill. The influence of bead loading, bead size and agitator speed. Bioseparation. 1991;2:231–236. [PubMed] [Google Scholar]
- Molloy MP, Herbert BR, Walsh BJ, Tyler MI, Traini M, Sanchez JC, Hochstrasser DF, Williams KL, Gooley AA. Extraction of membrane proteins by differential solubilization for separation using two-dimensional gel electrophoresis. Electrophoresis. 1998;19:837–844. doi: 10.1002/elps.1150190539. [DOI] [PubMed] [Google Scholar]
- Monroe EB, Jurchen JC, Koszczuk BA, Losh JL, Rubakhin SS, Sweedler JV. Massively parallel sample preparation for the MALDI MS analyses of tissues. Anal Chem. 2006;78:6826–6832. doi: 10.1021/ac060652r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moron JA, Abul-Husn NS, Rozenfeld R, Dolios G, Wang R, Devi LA. Morphine administration alters the profile of hippocampal postsynaptic density-associated proteins: A proteomics study focusing on endocytic proteins. Mol Cell Proteomics. 2007;6:29–42. doi: 10.1074/mcp.M600184-MCP200. [DOI] [PubMed] [Google Scholar]
- Morris JS, Coombes KR, Koomen J, Baggerly KA, Kobayashi R. Feature extraction and quantification for mass spectrometry in biomedical applications using the mean spectrum. Bioinformatics. 2005;21:1764–1775. doi: 10.1093/bioinformatics/bti254. [DOI] [PubMed] [Google Scholar]
- Morris JS, Clark BN, Gutstein HB. Pinnacle: A fast, automatic and accurate method for detecting and quantifying protein spots in 2-dimensional gel electrophoresis data. Bioinformtics. 2008 doi: 10.1093/bioinformatics/btm590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Brown PS, Herrick RC, Baggerly KA, Coombs KR. Bayesign analysis of mass spectrometry data using wavelet-based functional mixed models. Biometrics. 2008 doi: 10.1111/j.1541-0420.2007.00895.x. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulédous L, Hunt S, Harcourt R, Harry J, Williams KL, Gutstein HB. Lack of compatibility of histological staining methods with proteomic analysis of laser-capture microdissected brain samples. J Biomol Tech. 2002;13:258–264. [PMC free article] [PubMed] [Google Scholar]
- Moulédous L, Hunt S, Hopwood F, Harcourt R, Harry J, Williams KL, Gutstein HB. Navigated laser-capture microdissection as an alternative to direct histological staining for proteomic analysis of brain samples. Proteomics. 2003a;3:610–615. doi: 10.1002/pmic.200300398. [DOI] [PubMed] [Google Scholar]
- Moulédous L, Hunt S, Hopwood F, Harcourt R, Harry J, Williams KL, Gutstein HB. Proteomic analysis of immunostained, laser-capture microdissected samples. Electrophoresis. 2003b;24:296–302. doi: 10.1002/elps.200390026. [DOI] [PubMed] [Google Scholar]
- Mustafa DAN, Burgers PC, Dekker LJ, Charif H, Titulaer MK, Smitt PAES, Luider TM, Kros JM. Identification of glioma neovascularization-related proteins by using MALDI-FTMS and nano-LC fractionation to microdissected tumor vessels. Mol Cell Proteomics. 2007;6:1147–1157. doi: 10.1074/mcp.M600295-MCP200. [DOI] [PubMed] [Google Scholar]
- Neupert S, Johard HA, Nassel DR, Predel R. Single-cell peptidomics of drosophila melanogaster neurons identified by Gal4-driven fluorescence. Anal Chem. 2007;79:3690–3694. doi: 10.1021/ac062411p. [DOI] [PubMed] [Google Scholar]
- Newton MA, Noueiry A, Sarkar D, Ahlquist P. Detecting differential gene expression with a semiparametric hierarchical mixture method. Biostatistics. 2004;5:155–176. doi: 10.1093/biostatistics/5.2.155. [DOI] [PubMed] [Google Scholar]
- Nishihara JC, Champion KM. Quantitative evaluation of proteins in one- and two-dimensional polyacrylamide gels using fluorescent stain. Electrophoresis. 2002;23:2203–2215. doi: 10.1002/1522-2683(200207)23:14<2203::AID-ELPS2203>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ohnesorge J, Neususs C, Watzig H. Quantitation in capillary electrophoresis-mass spectrometry. Electrophoresis. 2005;26:3973–3987. doi: 10.1002/elps.200500398. [DOI] [PubMed] [Google Scholar]
- Olson KJ, Ahmadzadeh H, Arriaga EA. Within the cell: Analytical techniques for subcellular analysis. Anal Bioanal Chem. 2005;382:906–917. doi: 10.1007/s00216-005-3135-2. [DOI] [PubMed] [Google Scholar]
- Ong S-E, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- Palkovits M, Brownstein MJ. Microdissection of brain areas by the punch technique. In: Cuello AC, editor. Brain microdissection techniques. Chichester: John Wiley and Sons; 1983. pp. 1–36. [Google Scholar]
- Pasquali C, Fialka I, Huber HA. Subcellular fractionation, electro-migration analysis and mapping of organelles. J Chromatogr. 1999;722:89–102. doi: 10.1016/s0378-4347(98)00314-4. [DOI] [PubMed] [Google Scholar]
- Patton WF. Detection technologies in proteome analysis. J Chromatogr B Anal Technol Biomed Life Sci. 2002;771:3–31. doi: 10.1016/s1570-0232(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Pesavento JJ, Kim YB, Taylor GK, Kelleher NL. Shotgun annotation of histone modifications: A new approach for streamlined characterization of proteins by top down mass spectrometry. J Am Chem Soc. 2004;126:3386–3387. doi: 10.1021/ja039748i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehling HM, Neuhoff V. One- and two-dimensional eletrophoresis in micro-slab gels. Electrophoresis. 1980;1:90–102. [Google Scholar]
- Pounds S, Cheng C. Improving false discovery rate estimation. Bioinformatics. 2004;20:1737–1745. doi: 10.1093/bioinformatics/bth160. [DOI] [PubMed] [Google Scholar]
- Pounds S, Cheng C. Sample size determination for the false discovery rate. Bioinformatics. 2005;21:4263–4271. doi: 10.1093/bioinformatics/bti699. [DOI] [PubMed] [Google Scholar]
- Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236–1242. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- Predel R, Eckert M, Pollak E, Molnar L, Scheibner O, Neupert S. Peptidomics of identified neurons demonstrates a highly differentiated expression pattern of FXPRLamides in the neuroendocrine system of an insect. J Comp Neurol. 2007;500:498–512. doi: 10.1002/cne.21183. [DOI] [PubMed] [Google Scholar]
- Rabilloud T. Solubilization of proteins for electrophoretic analyses. Electrophoresis. 1996;17:813–829. doi: 10.1002/elps.1150170503. [DOI] [PubMed] [Google Scholar]
- Raynie DE. Modern extraction techniques. Anal Chem. 2004;76:4659–4664. doi: 10.1021/ac040117w. [DOI] [PubMed] [Google Scholar]
- Righetti PG, Campostrini N, Pascali J, Hamdan M, Astner H. Quantitative proteomics: A review of different methodologies. Eur J Mass Spectrom (Chichester, Eng) 2004;10:335–348. doi: 10.1255/ejms.600. [DOI] [PubMed] [Google Scholar]
- Rubakhin SS, Sweedler JV. Characterizing peptides in individual mammalian cells using mass spectrometry. Nat Protoc. 2007;2:1987–1997. doi: 10.1038/nprot.2007.277. [DOI] [PubMed] [Google Scholar]
- Rubakhin SS, Garden RW, Fuller RR, Sweedler JV. Measuring the peptides in individual organelles with mass spectrometry. Nat Biotechnol. 2000;18:172–175. doi: 10.1038/72622. [DOI] [PubMed] [Google Scholar]
- Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI MS: Distribution of neuropeptides within single neurons. Anal Chem. 2003;75:5374–5380. doi: 10.1021/ac034498+. [DOI] [PubMed] [Google Scholar]
- Rubakhin SS, Jurchen JC, Monroe EB, Sweedler JV. Imaging mass spectrometry: Fundamentals and applications to drug discovery. Drug Discov Today. 2005;10:823–837. doi: 10.1016/S1359-6446(05)03458-6. [DOI] [PubMed] [Google Scholar]
- Rubakhin SS, Churchill JD, Greenough WT, Sweedler JV. Profiling signaling peptides in single mammalian cells using mass spectrometry. Anal Chem. 2006;78:7267–7272. doi: 10.1021/ac0607010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchel R. Two-dimensional micro-separation technique for proteins and peptides, combining isoelectric focusing and gel gradient electrophoresis. J Chromatogr. 1977;132:451–468. doi: 10.1016/s0021-9673(00)82909-x. [DOI] [PubMed] [Google Scholar]
- Safarik I, Safarikova M. Use of magnetic techniques for the isolation of cells. J Chromatogr B Biomed Sci Appl. 1999;722:33–53. [PubMed] [Google Scholar]
- Santoni V, Rabilloud T, Doumas P, Rouquie D, Mansion M, Kieffer S, Garin J, Rossignol M. Solubilization of proteins in 2-D electrophoresis. An outline. Methods Mol Biol. 1999;112:9–19. doi: 10.1385/1-59259-584-7:9. [DOI] [PubMed] [Google Scholar]
- Schutte H, Kula MR. Pilot- and process-scale techniques for cell disruption. Biotechnol Appl Biochem. 1990a;12:599–620. [PubMed] [Google Scholar]
- Schutte H, Kula MR. Separation processes in biotechnology. Bead mill disruption. Bioprocess Technol. 1990b;9:107–141. [PubMed] [Google Scholar]
- Scopes RK. Protein purification: Principles and practice. New York: Springer-Verlag; 1994. [Google Scholar]
- Shen Y, Smith RD. Proteomics based on high-efficiency capillary separations. Electrophoresis. 2002;23:3106–3124. doi: 10.1002/1522-2683(200209)23:18<3106::AID-ELPS3106>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Shen Y, Berger SJ, Anderson GA, Smith RD. High-efficiency capillary isoelectric focusing of peptides. Anal Chem. 2000;72:2154–2159. doi: 10.1021/ac991367t. [DOI] [PubMed] [Google Scholar]
- Simpson DC, Smith RD. Combining capillary electrophoresis with mass spectrometry for applications in proteomics. Electrophoresis. 2005;26:1291–1305. doi: 10.1002/elps.200410132. [DOI] [PubMed] [Google Scholar]
- Sitek B, Potthoff S, Schulenborg T, Stegbauer J, Vinke T, Rump L-C, Meyer HE, Vonend O, Stuhler K. Novel approaches to analyse glomerular proteins from smallest scale murine and human samples using DIGE saturation labelling. Proteomics. 2006;6:4337–4345. doi: 10.1002/pmic.200500739. [DOI] [PubMed] [Google Scholar]
- Smejkal GB, Robinson MH, Lawrence NP, Tao F, Saravis CA, Schumacher RT. Increased protein yields from Escherichia coli using pressure-cycling technology. J Biomol Tech. 2006;17:173–175. [PMC free article] [PubMed] [Google Scholar]
- Spengler B, Hubert M, Kaufmann R. 42nd ASMS conference on mass spectrometry and allied topics. Chicago, IL, USA: 1994. p. 1041. [Google Scholar]
- Stoeckli M, Farmer TB, Caprioli RM. Automated mass spectrometry imaging with a matrix-assisted laser desorption ionization time-of-flight instrument. J Am Soc Mass Spectrom. 1999;10:67–71. doi: 10.1016/S1044-0305(98)00126-3. [DOI] [PubMed] [Google Scholar]
- Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: A new technology for the analysis of protein expression in mammalian tissues. Nat Med. 2001;7:493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J R Stat Soci Ser B Stat Methodol. 2002;64:479–498. [Google Scholar]
- Storey JD. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann Stat. 2003;31:2013–2035. [Google Scholar]
- Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T, Matsuo T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1988;2(8):151–153. [Google Scholar]
- Tiselius A. A new apparatus for electrophoretic analysis of colloidal mixtures. Trans Faraday Soc. 1937;33:524–531. [Google Scholar]
- Todd PJ, Schaaff TG, Chaurand P, Caprioli RM. Organic ion imaging of biological tissue with secondary ion mass spectrometry and matrix-assisted laser desorption/ionization. J Mass Spectrom. 2001;36:355–369. doi: 10.1002/jms.153. [DOI] [PubMed] [Google Scholar]
- von Eggeling F, Melle C, Ernst G. Microdissecting the proteome. Proteomics. 2007;7:2729–2737. doi: 10.1002/pmic.200700079. [DOI] [PubMed] [Google Scholar]
- Vuong GL, Weiss SM, Kammer W, Priemer M, Vingron M, Nordheim A, Cahill MA. Improved sensitivity proteomics by postharvest alkylation and radioactive labelling of proteins. Electrophoresis. 2000;21:2594–2605. doi: 10.1002/1522-2683(20000701)21:13<2594::AID-ELPS2594>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wang H, Qian W-J, Mottaz HM, Clauss TRW, Anderson DJ, Moore RJ, Camp DG, II, Khan AH, Sforza DM, Pallavicini M, Smith DJ, Smith RD. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005;4:2397–2403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wiener MC, Sachs JR, Deyanova EG, Yates NA. Differential mass spectrometry: A label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal Chem. 2004;76:6085–6096. doi: 10.1021/ac0493875. [DOI] [PubMed] [Google Scholar]
- Wilson KE, Marouga R, Prime JE, Pashby DP, Orange PR, Crosier S, Keith AB, Lathe R, Mullins J, Estibeiro P, Bergling H, Hawkins E, Morris CM. Comparative proteomic analysis using samples obtained with laser microdissection and saturation dye labelling. Proteomics. 2005;5:3851–3858. doi: 10.1002/pmic.200401255. [DOI] [PubMed] [Google Scholar]
- Wong MH, Saam JR, Stappenbeck TS, Rexer CH, Gordon JI. Genetic mosaic analysis based on cre recombinase and navigated laser capture microdissection. Proc Natl Acad Sci USA. 2000;97:12601–12606. doi: 10.1073/pnas.230237997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu BJ, Caprioli RM, Sanders ME, Jensen RA. Direct analysis of laser capture microdissected cells by MALDI mass spectrometry. J Am Soc Mass Spectrom. 2002;13:1292–1297. doi: 10.1016/S1044-0305(02)00644-X. [DOI] [PubMed] [Google Scholar]
- Yekutieli D, Benjamini Y. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J Stat Plann Inference. 1999;82:171–196. [Google Scholar]
- Zhou G, Li H, DeCamp D, Chen S, Shu H, Gong Y, Flaig M, Gillespie JW, Hu N, Taylor PR, Emmert-Buck MR, Liotta LA, Petricoin EF, III, Zhao Y. 2D differential in-gel electrophoresis for the identification of esophageal scans cell cancer-specific protein markers. Mol Cell Proteomics. 2002;1:117–124. doi: 10.1074/mcp.m100015-mcp200. [DOI] [PubMed] [Google Scholar]
- Zhou S, Bailey MJ, Dunn MJ, Preedy VR, Emery PW. A quantitative investigation into the losses of proteins at different stages of a two-dimensional gel electrophoresis procedure. Proteomics. 2005;5:2739–2747. doi: 10.1002/pmic.200401178. [DOI] [PubMed] [Google Scholar]