Abstract
Small interfering RNA (siRNA) and short hairpin RNA (shRNA) targeting different regions of transforming growth factor β1 (TGF-β1) mRNA were designed and the silencing effect was determined after transfection into immortalized rat liver stellate cells (HSC-T6). There was not only significant decrease in TGF-β1, tissue inhibitor of metalloproteinase 1 (TIMP-1), α-smooth muscle actin (α-SMA) and type I collagen after transfection with TGF-β1 siRNAs, but also synergism in gene silencing when siRNAs targeting two different start sites were used as a pool for transfection. The two siRNA sequences which efficiently inhibited TGF-β1 gene expression were converted to shRNAs via cloning into the pSilencer1.0. There was significant decrease in TGF-β1 and TIMP-1 when HSC-T6 cells were transfected with pshRNA targeting the same regions of TGF-β1 mRNA as siRNAs. Furthermore, TGF-β1 gene silencing in HSC-T6 cells significantly decreased the levels of inflammatory cykokines, such as tumor necrosis factor-alpha (TNF-α) and interleukin-1 beta (IL-1β). In conclusion, both siRNA and shRNA showed sequence-specific and dose dependent TGF-β1 gene silencing, and has the potential to treat liver fibrosis.
Keywords: siRNA, shRNA, transforming growth factor β1 (TGF-β1), hepatic stellate cells, liver fibrosis
INTRODUCTION
Liver fibrosis is the excessive accumulation of extracellular matrix (ECM) proteins resulting from chronic liver damage. In general, it is an imbalance between the synthesis and degradation of ECM. In the presence of chronic liver injuries, hepatic stellate cells (HSCs) become activated and transform to proliferative myofibroblast-like cells, which account for the major source of ECM expression.1, 2
There is no standard treatment for liver fibrosis and therefore the effective anti-fibrotic medicines are needed urgently.1, 3 Among many inflammatory cytokines involved in liver fibrosis, TGF-β1 appears to be the most important one (Fig. 1).1, 4 Cytokines of TGF family affect a variety of cellular processes, including differentiation, proliferation, apoptosis and migration. Among them, TGF-β1 is the most potent profibrogenic factor involved in initiation and maintenance of fibrogenesis in the liver.5, 6 Stimulation of activated HSCs by TGF-β1 is believed to be the key fibrogenic response in liver fibrosis because of following evidences: i) higher TGF-β1 expression in activated HSC; ii) potency of TGF-β1 to up-regulate ECM expression; iii) higher expression of TGF-β receptors on HSC; iv) TGF-β1 increases the expression of tissue inhibitor of metalloproteinases 1 (TIMP-1).5–7 TGF-β1 in the liver is secreted by hepatocytes, kupffer cells, stellate cells, endothelial cells and infiltrating mononuclear cells.5
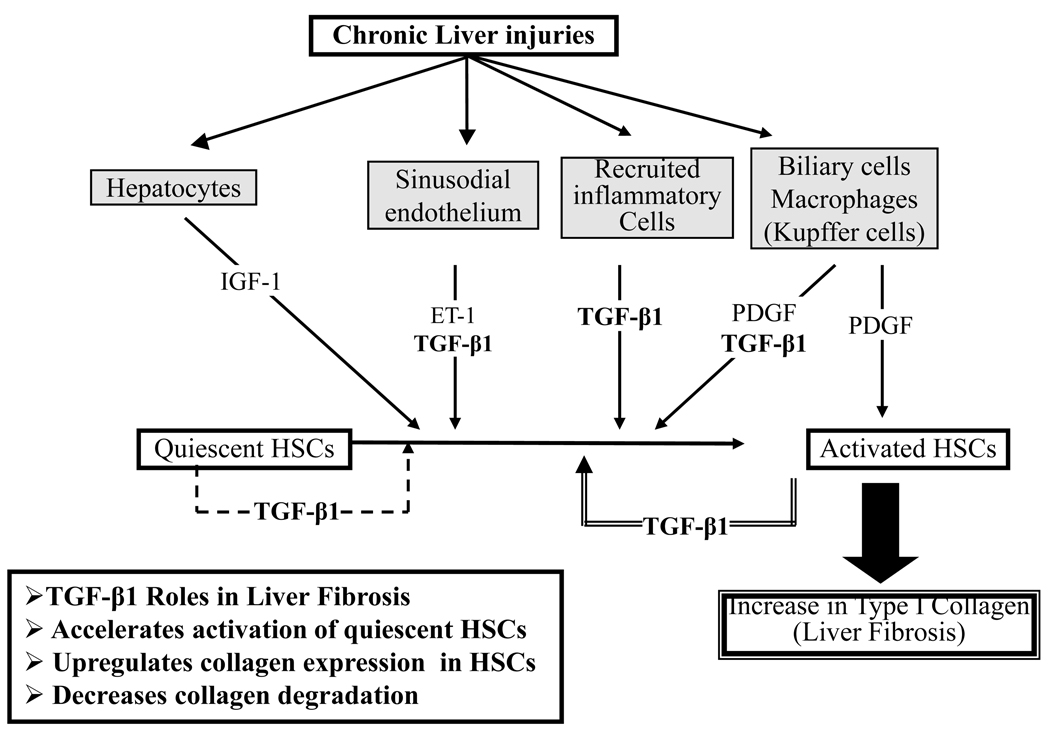
Figure 1.
Role of Transforming Growth Factor-β1 (TGF-β1) in liver fibrosis. TGF-β1 is the most potent single profibrogenic factor involved in initiation and maintenance of fibrogenesis in the liver. TGF-β1 accelerates activation of quiescent Hepatic Stellate Cells (HSCs), upregulates collagen expression, and decreases collagen degradation.
Strategies aimed at disrupting TGF-β1 expression or signaling pathways are extensively being investigated because blocking this cytokine may not only inhibit matrix production, but also accelerate its degradation.8 Animal experiments using different strategies to block TGF-β1 have demonstrated significant anti-fibrotic effect for liver fibrosis.6, 9–14 RNA interference (RNAi) is the phenomenon in which siRNA of 21–23 nt in length silences a target gene by binding to its complementary mRNA and triggering its degradation. Potent knockdown of the target gene with high sequence specificity makes siRNA a promising therapeutic strategy.2 Compared to antisense oligonucleotides, neutralizing antibodies and soluble TGF-β receptors strategies, siRNA targeting TGF-β1 has the potent knockdown of the target gene with high sequence specificity. siRNAs targeting other pathways have been proven effective in treating liver fibrosis15–18 and renal fibrosis.19 There are three ways to deliver siRNA: synthetic duplex, plasmid and viral vectors. While viral vectors give high transduction efficiency, their immune reactions limit their application in therapeutics. On the other hand, plasmid DNA complexes with cationic liposomes may not pass through the sinusoidal gaps, because these fenestrae get lost during liver fibrosis. In contrast, low molecular weight of synthetic duplex siRNA is expected to pass through the sinusoidal gaps in fibrotic liver and thus it may be an ideal candidate for treating fibrosis.
In this study, we designed and screened ten chemically synthesized siRNAs targeting different regions of TGF-β1 mRNA and then converted the most potent siRNA sequences into shRNA via cloning into pSilencer1.0 vector. Both synthetic siRNAs and shRNA expression plasmids were tested in HSC-T6 cells for gene silencing and therapeutic efficacy.
MATERIALS AND METHODS
Materials
Dulbecco’s Modified Eagle’s Medium (DMEM), penicillin G (5000U/ml), Trypsin-EDTA, Trizol, DNase I, Lipofectamine 2000 were purchased from Invitrogen Corporation (Carlsbad, CA). pSilencer1.0 was purchased from GenScript Corporation (Piscataway, NJ). Bovine serum albumin (BSA) (fraction V, purity >98%) was purchased from USB Corporation (Cleveland, OH). Restriction enzymes were purchased from New England Biolabs (Ipswich, MA). SYBR Green-1 dye universal master mix and Multiscript reverse transcriptase were purchased from Applied Biosystems, Inc. (Foster City, CA). TGF-β1 enzyme-linked immunosorbent assay (ELISA) kit was purchased from R&D Systems, Inc. (Minneapol,is, MN). TNF-α and IL-1β ELISA kits were purchased from eBioscience, Inc. (San Diego, CA).
siRNA Design and Synthesis
Ten synthetic siRNAs targeting TGF-β1 mRNA (Accession#: NM_021578) and one control siRNA were designed using BLOCK-iT™ RNAi Designer and purchased from Invitrogen Corporation (Carlsbad, CA) and their sequences are listed in Table 1. These siRNAs are of 19–21 nt with a 2 thymidine deoxnucleotide overhangs at the 3’-end. All designed siRNA sequences were blasted against the rat genome database to eliminate cross-silence phenomenon with non-target genes.
Table 1.
Pre-designed siRNAs for rat TGF-β1 using BLOCK-iT™ RNAi Designer
| Start | Sequence (DNA) |
|---|---|
| 297 | GCCAGATCCTGTCCAAACT |
| 448 | GGACTACTACGCCAAAGAA |
| 499 | CGCAATCTATGACAAAACC |
| 640 | GCAACACGTAGAACTCTAC |
| 769 | GAACCAAGGAGACGGAATA |
| 888 | GCACCATCCATGACATGAA |
| 1033 | GCAGCTGTACATTGACTTT |
| 1036 | GCTGTACATTGACTTTAGG |
| 1167 | CCCTCTACAACCAACACAA |
| 1245 | TCTACTACGTGGGTCGCAA |
Design and Construction of shRNA Expression Plasmids
Following screening of different siRNA sequences, two effective and one control siRNA sequences were converted into shRNA (Table 2). pshRNA-769, pshRNA-1033 and control vectors were constructed using pSilencer 1.0 vector, which carries an shRNA expression cassette under the control of U6 promoter (GenScript Corporation, Piscataway, NJ). As shown in Table 2, these shRNAs contain two complementary oligonucleotides that were annealed to form a double-stranded DNA for ligation into pSilencer1.0 vector at corresponding sites under U6 promoter using T4 DNA ligase for 2h at 25°C. Following transformation in Top 10 supercompetent cells and amplification in broth media, plasmids were purified using QIAGEN Plasmid Mini Kit.
Table 2.
Sequences of shRNA against different target regions of TGF-β1
| Control shRNA | Antisense: 5’-ACTTCATAAGGCGCATGCTTTCAAGAGAAGCATGCGCCTTATGAAGTTTTTTT-3’ |
| Sense: 5’-AATTAAAAAAACTTCATAAGGCGCATGCTTCTCTTGAAAGCATGCGCCTTATGAAGTGGCC-3 | |
| shRNA-769 | Antisense: 5’-GAACCAAGGAGACGGAATATTCAAGAGATATTCCGTCTCCTTGGTTCTTTTTT-3’ |
| Sense: 5’-AATTAAAAAAGAACCAAGGAGACGGAATATCTCTTGAATATTCCGTCTCCTTGGTTCGGCC-3 | |
| shRNA-1033 | Antisense: 5’-GCAGCTGTACATTGACTTTTTCAAGAGAAAAGTCAATGTACAGCTGCTTTTTT-3 |
| Sense: 5’-AATTAAAAAAGCAGCTGTACATTGACTTTTCTCTTGAAAAAGTCAATGTACAGCTGCGGCC-3 |
Transfection of siRNA and shRNA
Immortalized rat hepatic stellate cells (HSC-T6), kindly provided by Dr Scott Friedman (Mount Sinai School of Medicine, New York), were seeded in 6-well plates at a density of 1.1×105 cells 12 h before transfection. siRNA duplexes were mixed with 4 µl Lipofectamine 2000 in 200µl Opti-MEM I medium for 20 min at room temperature to allow complex formation. The transfection mixture was then added to each well with 1.3 ml fetal bovine serum (FBS) free DMEM medium at a concentration of 150nM after washing cells with phosphate buffered saline (PBS). After 24 h of incubation, 0.5ml DMEM containing 10% FBS was added and incubated for another 24 h. For siRNA pool, the equal amounts of two siRNAs were mixed together and the final concentration of total siRNAs is 150 nM.
In case of shRNA expression plasmids, HSC-T6 cells were transfected with shRNA expression plasmids pshRNA-769 and pshRNA-1033 after complex formation in 250 µl Opti-MEM I medium with pyridinium lipid (C16:1, amide linker, transisomer)/ L-α-dioleoyl phosphatidylethanolamine (DOPE) (1:1 mol/mol) cationic liposomes as described by Zhu et al.20 The transfection mixture was then added to each well with 2 ml FBS free DMEM. At 3h post-incubation, 200 µl FBS was added and then incubated for additional 42 h. The supernatant was collected for western blot assay.
Real Time RT-PCR
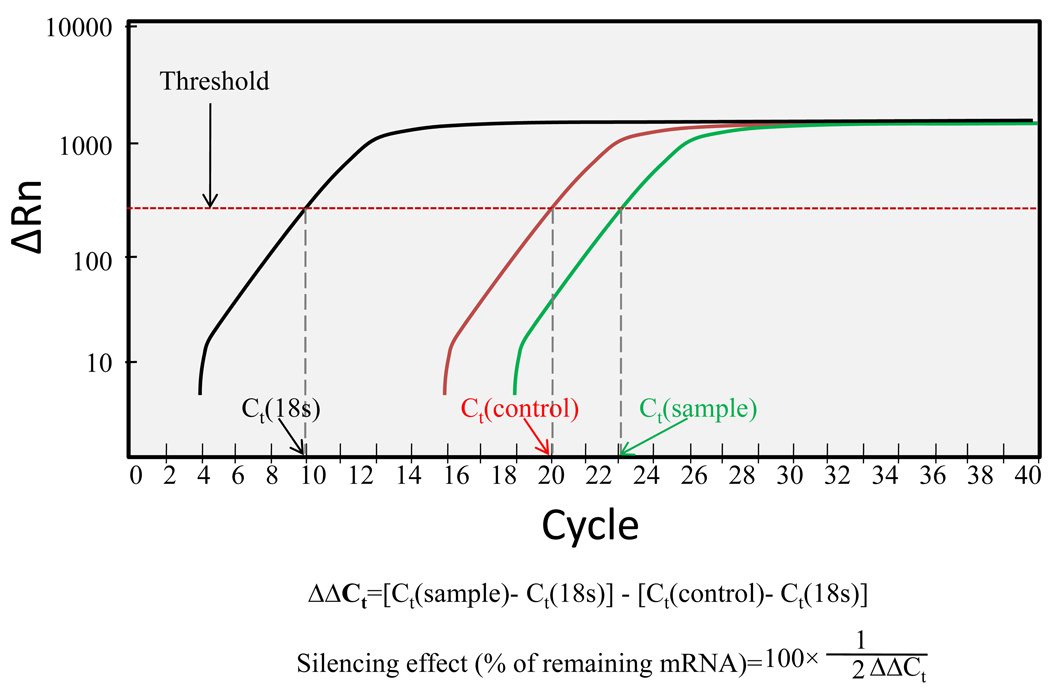
Total RNA was isolated from the cell pellets using Trizol reagent. Total RNA (1 µg) was converted to cDNA using MultiScribe Reverse Transcriptase Reagent and random hexamers (Applied Biosystems, Inc., Branchburg. NJ). One hundred nanograms of the cDNA was amplified by Real Time PCR using SYBR Green-1 dye universal master mix on ABI Prism 7700 Sequence Detection System (Applied Biosystems, Inc., Foster City, CA). We used the following primers: (i) TGF-β1: Forward: 5’-CATCCATGACATGAACCG ACCCTT-3’ and reverse: 5’-ACAGAAGTTGGCATGGTAGCCCTT-3’; (ii) TIMP-1: forward: 5’-CCTCTGGCATCCTCTTGTTGCTAT-3’ and reverse: 5’-CATTTCCCACAG CGTCGAATCCTT-3’; (iii) α1(I) collagen mRNA: Forward: 5’-TGGTCCCAAAGGTTC TCCTGGT-3’ and reverse: 5’-TTAGGTCCAGGGAATCCCATCACA-3’; and (iv) α-SMA: forward: 5’-ACAACGTGCCTATCTATGAGGGCT-3’ and reverse: 5’-AGCGACATA GCACAGCTTCTCCTT-3’. We used 18S ribosomal RNA as an internal control: forward: 5’-GTCTGTGATGCCCTTAGATG-3’ and reverse: 5’-AGCTTATGACCCGCACTTAC-3’. To confirm PCR specificity, the PCR products were subjected to a melting-curve analysis. Comparative threshold (Ct) method was used for calculating the relative amount of mRNA of treated sample compared to control samples.21 The illustrative real-time RT-PCR plot and calculation details were provided in Figure 2.
Figure 2.
The real time PCR plot. Black curve is the intrinsic reference (18s). Red curve is the control group. Green curve is the group with treatment.
Western Blot Assay
Proteins in the cell culture medium were purified using Microcon YM-30 columns (Millipore) and then lysed using 2x Laemmli sodium dodecyl sulfate (SDS) sample buffer containing 100 mM Tris, pH 6.8, 200 mM dithiothreitol (DTT), 4% SDS, 20% glycerol and 0.2% bromophenol blue. To immunodetect cellular proteins, cells were lysed directly with 1x Laemmli SDS sample buffer. The lysate was then boiled for 5min and subjected to 6% or 10% SDS-polyacrylamide (SDS-PAGE) gel electrophoresis and subsequent transfer to Immobilon polyvinylidene fluoride (PVDF) membrane (Millipore). After blocking with 5% non-fat dry milk in 1x PBST (PBS containing 0.05% Tween-20) for 1h at room temperature. The membranes were incubated with rabbit anti-rat type I collagen (Calbiochem), TGF-β1 (Santa Cruz); β-actin (Santa Cruz) primary antibodies for 16h at 4°C. Membrane was then incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Santa Cruz) for 1h at room temperature. Target proteins were detected by enhanced chemiluminescence (ECL) detection kit (GE Healthcare Life Sciences, Pittsburgh, PA).
ELISA of TGF-β1, TNF-α and IL-1β
HSC-T6 cells were transfected with shRNA expression vectors at a dose of 1 µg/well after complex formation with cationic liposomes prepared using pyridinium lipid (C16:1, amide linker, trans isomer) and DOPE at 1:1 molar ratio as described by Zhu et al.20 Forty eight hours after transfection, supernatant of the cultured cells was collected and the concentration of TGF-β1, TNF-α and IL-1β were measured using ELISA kits according to the manufacturer's protocol.
Persistence of TGF-β1 Gene Silencing
HSC-T6 cells were seeded in 6-well plates and transfected with siRNA and shRNA when the cell confluence reached 40%. One microgram siRNA duplexes or shRNA were mixed with 8µl pyridinium lipid (C16:1, amide linker, trans)/DOPE (1:1 mol/mol) cationic liposomes. The transfection mixture was then added to each plate with 2 ml FBS free DMEM at a concentration of 150 nM for siRNA and 1 µg per well for shRNA, both conditions have been optimized for siRNA and shRNA transfection in this case. After 3 h of incubation, 200 µl FBS was added and incubated for another 24 h, 48h and 72h. At different time points, the cells are collected and total RNA was isolated for real time RT-PCR analysis.
Statistical Analysis
Data were expressed as the mean ± standard deviation (SD). Difference between any two groups was determined by ANOVA. P<0.05 was considered statistically significantly.
RESULTS
Effect of siRNA Sequences and Dose on TGF-β1 Gene Silencing
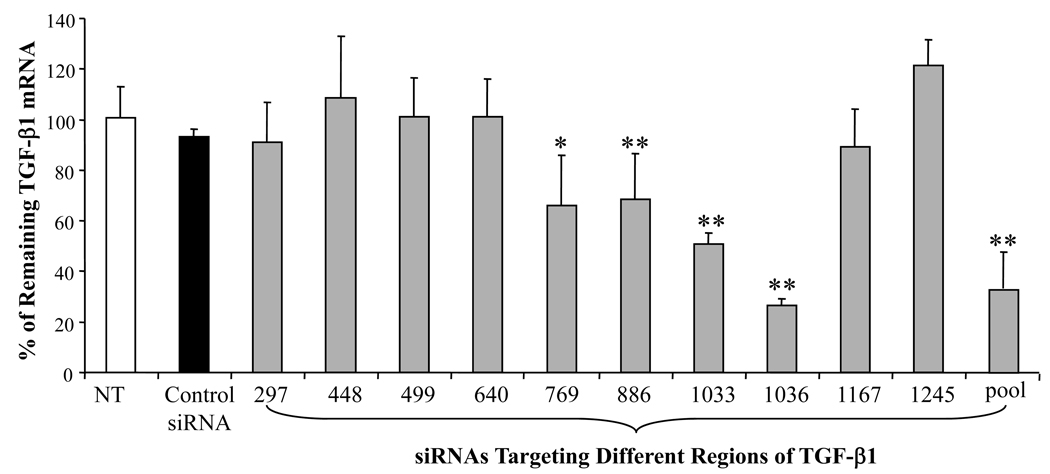
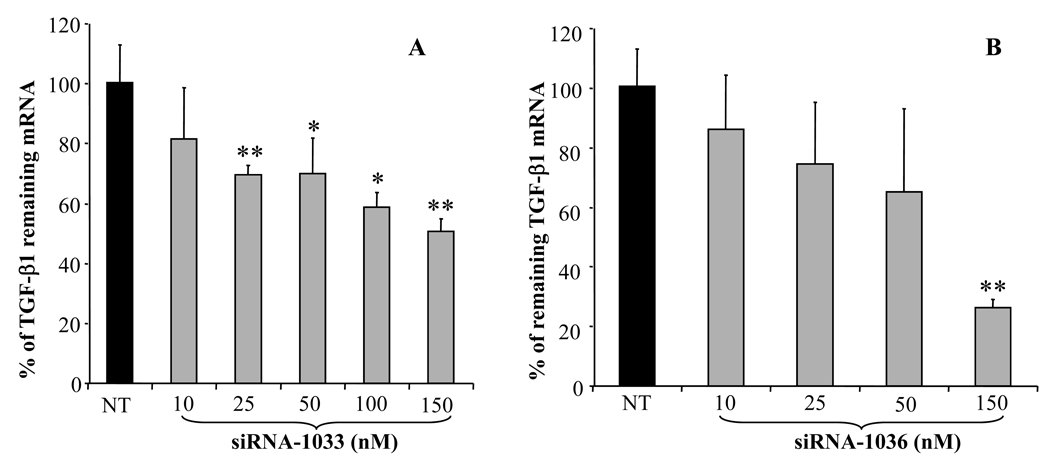
To examine the ability of siRNA to silence gene expression, we selected 19bp synthetic siRNA duplexes targeting different regions of the rat TGF-β1 mRNA (Table 1). These siRNA duplexes were transfected into rat HSC-T6 cells after complex formation with Lipofectamine 2000. As shown in Figure 3, TGF-β1 gene silencing by siRNAs was sequence specific. Three siRNAs targeting TGF-β1 start sites of 769, 1033, and 1036 caused significant inhibition of TGF-β1 expression, while the control siRNA had no effect on TGF-β1 gene expression. Among all the siRNA sequences tested, siRNA-1036 showed the highest silencing effect up to 70%, while siRNA-1033 showed up to 55% of gene silencing. Therefore, we selected siRNA-1036 and siRNA-1033 to determine the effect of siRNA dose on TGF-β1 gene silencing. We also tested the pool of siRNAs targeting 769 and 1033, which showed higher TGF-β gene silencing effect than single siRNA. As shown in Figures 4A and 4B, there was significant increase in the TGF-β1 silencing effect as we increased the doses of siRNA-1033 and siRNA-1036 from 10 nM to 150 nM, with siRNA-1036 showing the highest TGF-β1 gene silencing effect at 150nM.
Figure 3.
Effect of siRNA sequence on TGF-β1 gene silencing. Nine different siRNAs targeting different regions of Transforming Growth Factor-β1(TGF-β1) mRNA were transfected into HSC-T6 cells at a dose of 150nM after complex formation with Lipofectamine 2000. TGF-β1 gene silencing was determined by Real Time RT-PCR at 48h post-transfection. Results were represented as the mean ± SD (n=3). *: p < 0.05, **:p<0.01
Figure 4.
Effect of siRNA concentration on Transforming Growth Factor-β1 (TGF-β1) silencing. Lipofectamine 2000/siRNA complexes were transfected into HSC-T6 cells at doses of 10, 25, 50, 100 and 150 nM. TGF-β1 gene silencing was determined by real time RT-PCR at 48h post-transfection. Results were represented as the mean ± SD (n=3). *: p < 0.05, **:p<0.01
We also determined the level of TGF-β1 secreted into the cell culture medium by Western blot analysis at days 2 post-transfection of HSC-T6 cells with siRNAs targeting TGF-β1 start sites of 769 and 1033 as well as the pool of these two siRNAs. As shown in Figure 5, the level of TGF-β1 protein secreted in the culture medium was significantly lower than that of non-transfected and control-siRNA transfected samples.
Figure 5.
Western blot analysis for Transforming Growth Factor-β1 (TGF- β1) gene silencing after transfection of HSC-T6 cells with Lipofectamine/siRNA complexes. From left to right, no siRNA, control siRNA, siRNA-1033, siRNA-769 and pool of siRNA-1033 and siRNA-769. From top to bottom, TGF-β1, type I collagen and β-actin.
Effect of TGF-β1 Gene Silencing on TIMP-1 Expression
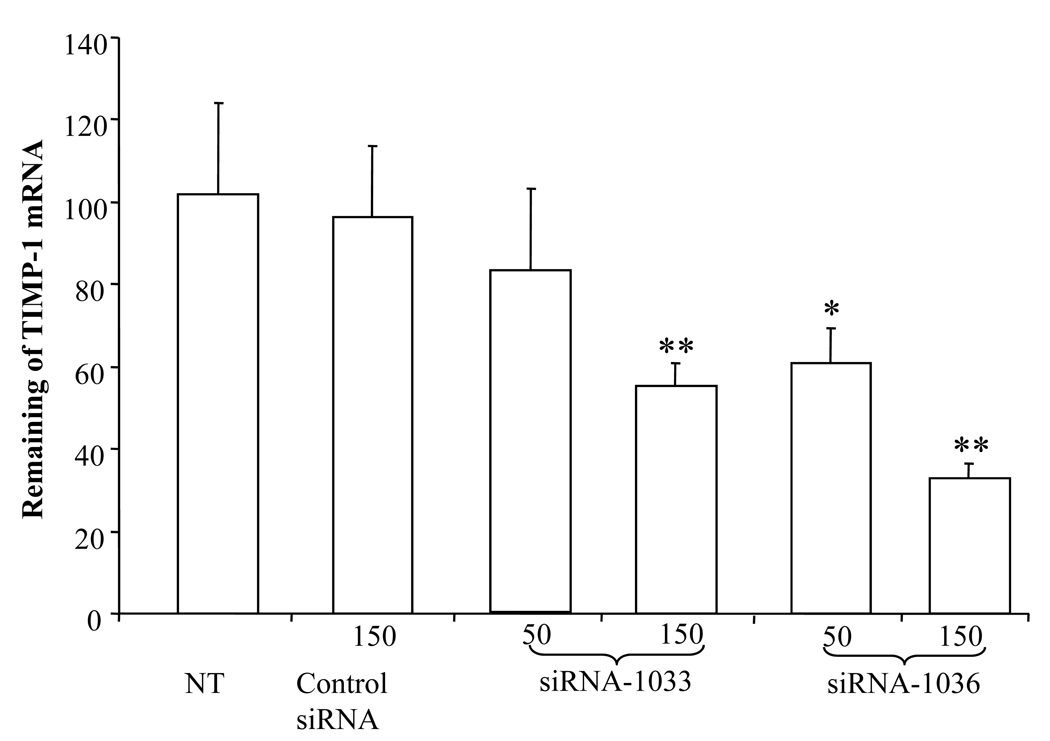
TGF-β1 is the key profibrogenic cytokine in response to chronic liver injuries. It not only enhances ECM synthesis, but also inhibits ECM degradation by down-regulating matrix-degrading enzymes and inducing tissue inhibitor of metalloproteinases-1 (TIMP-1).22 High level of TIMP-1 in the fibrotic accounts for the slow degradation of type I collagen. Therefore, we measured TIMP-1 mRNA expression of HSC-T6 cells after transfection with TGF-β1 siRNAs. As shown in Figure 6, both siRNA-1033 and siRNA-1036 inhibited TIMP-1 expression, while the control siRNA did not show any effect on TIMP-1 expression. These results suggested the similar inhibition profile of TIMP-1 and TGF-β1 after transfection of HSC-T6 cells with siRNA targeting TGF-β1.
Figure 6.
Effect of Transforming Growth Factor-β1 (TGF-β1) gene silencing on Tissue inhibitor of metalloproteinase 1 (TIMP-1) gene expression after transfection of HSC-T6 cells with Lipofectamine 2000/siRNA complexes. At 48h post transfection, cells were harvested, total RNA was extracted, and TIMP-1 gene expression was determined at mRNA level using real time RT-PCR. Results were represented as the mean ± SD (n=3). *: p < 0.05, **:p<0.01
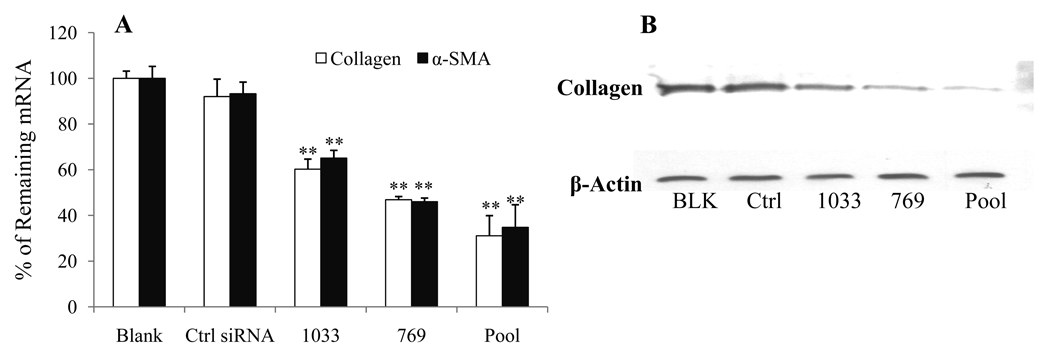
Effect of siRNA on Type I Collagen and α-SMA Expression
Inhibition of TGF-β1 should enhance the degradation of ECM, in which type I collagen is the main component. Therefore, we designed collagen α1(I) mRNA specific primers to determine the effect of TGF-β1 siRNA on collagen α1(I) mRNA expression. At 150 nM, siRNA-1033, siRNA-769 and the pool of these two siRNAs showed significant inhibition of collagen α1(I) mRNA (Figure 7A), while the control siRNA had little effect on collagen α1(I) mRNA expression. We also determined the effect of TGF-β1 gene silencing on type α(I) collagen protein expression by western blot analysis of lysates of HSC-T6 cells after transfection with siRNAs targeting TGF-β1 start sites of 769 and 1033 as well as the pool of these two siRNAs (Figure 7B). Consistent with the RT-PCR results, there was significant decrease in collagen of siRNA-769, siRNA-1033 and siRNA pool, while there was no decrease in collagen concentration for the control-siRNA treated sample.
Figure 7.
Effect of Transforming Growth Factor-β1 (TGF-β1) gene silencing on type α1(I) collagen and α-SMA expression after transfection of HSC-T6 cells Lipofectamine 2000/siRNA-1033, 769 and pool complexes. A) At 48h post transfection, cells were harvested, total RNA was extracted, and type α1(I) collagen and α-SMA expression was determined at mRNA levels using Real Time RT-PCR. Results were represented as the mean ± SD (n=3). *: P < 0.05, **:P<0.01 B) Western blot analysis for type α1(I) collagen expression after TGF-β1 gene silencing.
Construction of shRNA Expression Vector
Following screening of different siRNA sequences for TGF-β1 gene silencing, we selected two potent siRNA sequences, which contain unique restriction sites at the 5’ and 3’ ends for cloning and a TTTTTT stretch in the antisense to create the pol III terminal signal and TTCAAGAGA as a loop (Table 2). The constructed pshRNA-769 and pshRNA-1033 were confirmed by DNA sequencing using the forward primer: 5’- TAAGTTGGGTAACGCCAGGG-3’; and reverse primer: 5’- GCTATGACCATGATTACGCC-3'.
Effect of shRNA on TGF-β1 Silencing
TGF-β1 gene expression was determined by real time RT-PCR and Western blot after transfection of HSC-T6 cells with pshRNA-769 and pshRNA-1033 complexed with pyridinium lipid (C16:1, amide linker, trans)/DOPE (1:1 mol/mol) cationic liposomes.20 Both pshRNA-769 and pshRNA-1033 showed significantly higher TGF-β1 gene silencing effect compared to control shRNA (Figure. 8A). There was also similar decrease in the protein levels of TGF-β1 and type 1 collagen (Figure 8B), which is in good agreement with the results of synthetic siRNAs.
Figure 8.
Effect of Transforming Growth Factor-β1 (TGF-β1) gene silencing on type α1(I) collagen and TGF-β1 expression after transfection of HSC-T6 cells with pshRNA-1033 or pshRNA-769 after complex formation with pyridinium lipid (C16:1, amide linker, transisomer)/DOPE (1:1 mol/mol) cationic liposomes.20 At 42h post-transfection, cells were harvested, total RNA was extracted, TGF-β1 expression was determined at mRNA levels using Real Time RT-PCR (A). In addition, protein levels of TGF-β1 and type I collagen in the supernatant were determined using western blot (B). Results were represented as the mean ± SD (n=3). *: p < 0.05, **:p<0.01
Effect of TGF-β1 Gene Silencing on TNF-α and IL-1β
Since liver inflammation promotes liver fibrosis, we wanted to determine whether TGF-β1 gene silencing could decrease collagen gene expression by decreasing inflammation in the liver. Therefore, we measured the amount of TNF-α and IL-1β secreted in the culture media upon transfection with pshRNA-1033 after complex formation with pyridinium lipid (C16:1, amide linker, trans)/DOPE (1:1 mol/mol) cationic liposomes20 using ELISA kits. As can be seen in Figure 9A, the amount of TNF-α was decreased to 43.41±4.03% and 51.85±4.75%, respectively compared to the control group not treated with pshRNA-TGF-β1. In the same trend, the level of IL-1β also decreased to 49.12±5.78% and 59.49±9.67% respectively compared to the control shRNA (Figure 9B).
Figure 9.
Effect of Transforming Growth Factor-β1 (TGF-β1) gene silencing on Tumor necrosis factor-α (TNF-α) and Interleukin-1β (IL-1β) secretion. HSC-T6 cells were transfected with pshRNA-1033 and pshRNA-769 after complex formation with pyridinium lipid (C16:1, amide linker, transisomer)/DOPE (1:1 mol/mol) cationic liposomes. Following transfection, TNF-α and IL-1β concentrations were measured by Enzyme-Linked ImmunoSorbent Assay (ELISA). Results were represented as the mean ± SD (n=3). *: p < 0.05, **:p<0.01
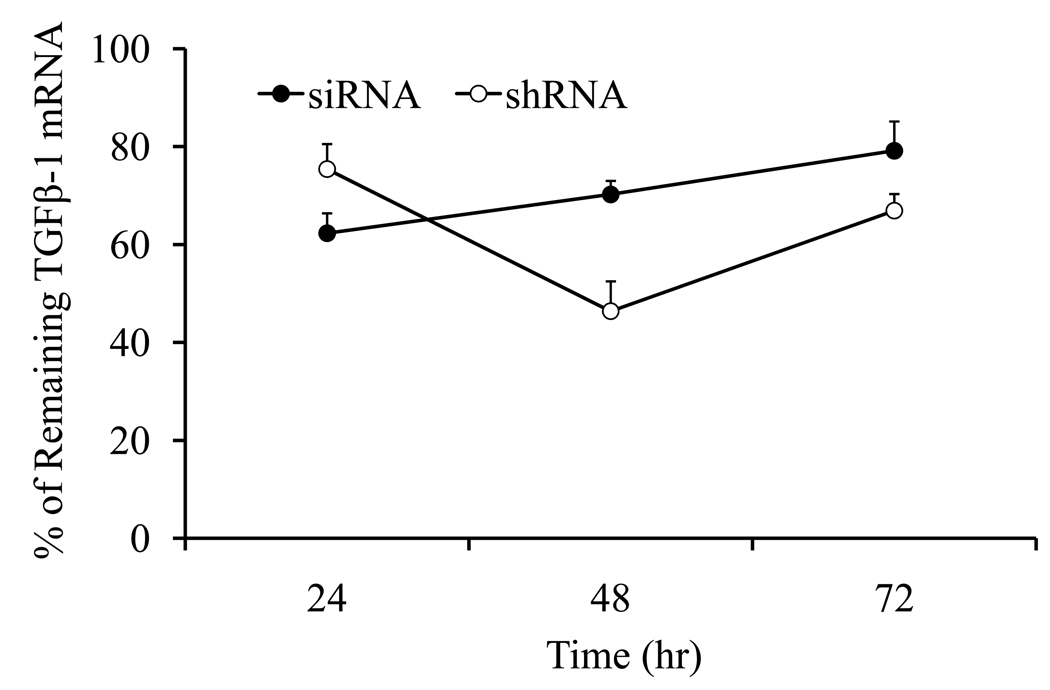
Persistence of TGF-β1 Gene Silencing
To compare siRNA and shRNA on TGF-β1 gene silencing, we transfected HSC-T6 cells with siRNA-1033 and pshRNA-TGF-β1-1033 after complex formation with our pyridinium lipid (C16:1, amide linker, trans)/DOPE (1:1 mol/mol) cationic liposomes at 3.5/1 (+/−) ratio. As shown in Figure 10, the level of TGF-β1 mRNA was significantly lower for the pSilencer-TGF-β1 group, which was lowest at 48h post-transfection compared to 24h post-transfection for the siRNA-TGF-β1 group. For siRNA treated group, the level of TGF-β1 gene silencing significantly decreased with time, being the lowest at 72h post-transfection. However, for the shRNA group, there was still significant silencing even after 72h post-transfection.
Figure 10.
Persistence of siRNA and shRNA gene silencing. HSC-T6 cells were transfected with siRNA-1033 and shRNA-769 after complex formation with pyridinium lipid (C16:1, amide linker, trans)/DOPE (1:1 mol/mol) cationic liposomes as described above. After 3 h of transfection, 200 µl FBS was added and incubated for another 24 h, 48h and 72h. At different time points, the cells are collected and total RNA was isolated for real time RT-PCR analysis.
DISCUSSION
Liver fibrosis results from chronic injuries to the liver by chronic hepatitis, alcohol abuse, and toxic agents. Among all fibrogenic cytokines, TGF-β is the key mediator of liver fibrosis and thus has gained great attention.1 TGF-β contains three isoforms including TGF-β1, TGF-β2 and TGF-β3. Among them TGF-β1 is the dominant stimulus to ECM production from HSCs and its expression increases during fibrogenesis.8 TGF-β1 can be secreted by Kupffer cells, biliary cells and infiltrated inflammatory cells, while the autocrine expression of TGF-β1 by HSCs is the most important.4 Compared to quiescent HSCs, fully activated HSCs are relatively unresponsive to TGF-β1, possibly due to the down-regulation of TGF-β receptor. TGF-β1 knockout mice have shown reduced collagen accumulation in response to liver injury compared to that of normal mice.23 Disrupting TGF-β1 synthesis or signaling pathways significantly decreased fibrosis in experimental animal model.6, 9 However, due to its multiple actions, TGF-β represents a potentially important link between neoplasia and fibrosis in the liver. Mutation of TGF-β receptor or signaling intermediates has been found in various epithelial cancers.11 Strategies aimed at disrupting TGF-β1 expression or signaling pathways are being extensively investigated for treating liver fibrosis and various animal studies have demonstrated antifibrotic effect.6, 9–14 Currently neutralizing antibodies, soluble TGF-β receptors, antisense oligonucleotide and siRNA were explored to block TGF-β1 signaling pathway.
Selection of a potent siRNA sequence targeting a specific gene is a crucial step in developing its therapeutic applications. In this study, we used BLOCK-iT™ RNAi Designer from Invitrogen to design ten synthetic siRNAs (Table 1) targeting at different coding regions of TGF-β1 mRNA. As shown in Figure 3, only 4 siRNAs showed significant silencing effect on TGF-β1 gene expression, while other 6 siRNAs did not show any effect. All these 4 siRNAs are located in the coding regions of TGF-β1 mRNA starting from 769 to 1036. Among these 4 siRNAs, siRNAs targeting 1033 and 1036 coding sites showed the highest TGF-β1 gene silencing effect. These may be due to the different local secondary structures of the TGF-β1 mRNA, and the 769∼1036 region is more accessible for siRNAs.
Two selected siRNAs, siRNA-1033 and siRNA-1036, showed dose dependent inhibition of TGF-β1 mRNA (Figure 4). The silencing effect of siRNA-1036 was similar to that of siRNA-1033 at low concentration, but more significant at high concentration (150 nM). At low concentrations, both siRNAs showed little inhibition and therefore it is difficult to distinguish which one is more effective. Considering there is only 3 base sequence shift between siRNA-1033 and siRNA-1036, the significant difference in silencing indicated that the silencing effect was very sequence specific.
Even though the dose increased to three fold, from 50 nM to 150 nM, the level of TGF-β1 gene silencing did not increase that much. The gene silencing ability of siRNAs had no linear relationship with their dose. However, at the same total concentration, application of more than one siRNA targeting at different sites of mRNA produced enhanced TGF-β1 gene silencing and significant decrease in type I collagen (Figure 3, Figure 5 and Figure 7), suggesting the use of more than one siRNA sequences as a pool may produce synergistic effect. These results are in good agreement with our recently published work where we demonstrated that a pool of siRNAs targeting different regions of HBsAg showed high gene silencing in HepG2.2.15 cells.24
The in vivo stability of synthetic siRNA and the high cost for in vivo studies hamper its therapeutic application. Therefore, we decided to construct a plasmid based shRNA targeting the most potent region of the TGF-β1 mRNA. The silencing effect of shrNAs targeted at 769 and 1033 showed relatively higher gene silencing, which lasted longer than synthetic siRNAs targeted at the same positions (Figure 3, Figure 8 and Figure 10). One reason might be the relatively higher stability of shRNA inside the cells than siRNA. Its consistent expression ability in the cells and ease to produce makes it a promising vector for therapeutic application. It is worth mentioning that HSC-T6 cells grow relatively fast and thus 72h after transfection was the maximum time we could maintain healthy HSCs with 100% confluence. Beyond 72h, the cells were too crowded and became unhealthy. Therefore, we could not determine gene silencing beyond 72h post-transfection.
The development of chronic liver diseases is mediated by sustained hepatic inflammation. Many studies have shown various cytokines, which are crucial indicators of inflammation, such as TGF-β1 and TNF-α, are important activators during the process of liver fibrosis.25–27 TNF-α has been reported closely related to liver fibrosis and cirrhosis.27 Therefore, we determined the level of TNF-α secreted in the culture media after transfection of HSC-T6 cells with TGF-β1 shRNA. TGF-β1 gene silencing significantly decreased TNF-α concentration, suggesting TGF-β1 gene silencing will decrease liver inflammation. IL-1β is another important cytokine, which is known to promote local inflammatory response and consequently promotes chronic liver fibrosis.28 Our results also showed that there is significant decrease in secreted IL-1β upon TGF-β1 gene silencing (Figure 9B). Both TNF-α and IL-1β levels correlated with TGF-β1 gene silencing levels.
In summary, we have successfully designed and validated TGF-β1-specific siRNAs and then converted two potent siRNA sequences into shRNAs, which effectively silenced TGF-β1 gene expression in HSC-T6 cells. TGF-β1 gene silencing significantly reduced the production of type I collagen, TIMP-1, and inflammatory cytokines. Our results suggested that silencing of TGF-β1 by siRNA and shRNA may be an efficient and more specific approach for therapy of liver fibrosis.
ACKNOWLEDGEMENTS
This work was supported by grants R01 EB003922 and R01 DK064633 from the NIH. We would also like to thank Xiangxu Jia for technical assistance, Lin Zhu for preparing pyridinium/DOPE liposomes, and Feng Li for useful discussion on shRNA vector construction.
REFERENCES
- 1.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115(2):209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahato RI, Cheng K, Guntaka RV. Modulation of gene expression by antisense and antigene oligodeoxynucleotides and small interfering RNA. Expert Opin. Drug Deliv. 2005;2(1):3–28. doi: 10.1517/17425247.2.1.3. [DOI] [PubMed] [Google Scholar]
- 3.Iredale JP. Cirrhosis: new research provides a basis for rational and targeted treatments. Bmj. 2003;327(7407):143–147. doi: 10.1136/bmj.327.7407.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gressner AM, Weiskirchen R, Breitkopf K, Dooley S. Roles of TGF-beta in hepatic fibrosis. Front Biosci. 2002;7:d793–d807. doi: 10.2741/A812. [DOI] [PubMed] [Google Scholar]
- 5.Bauer M, Schuppan D. TGFbeta1 in liver fibrosis: time to change paradigms? FEBS Lett. 2001;502(1–2):1–3. doi: 10.1016/s0014-5793(01)02655-2. [DOI] [PubMed] [Google Scholar]
- 6.Shek FW, Benyon RC. How can transforming growth factor beta be targeted usefully to combat liver fibrosis? Eur J Gastroenterol Hepatol. 2004;16(2):123–126. doi: 10.1097/00042737-200402000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Knittel T, Mehde M, Kobold D, Saile B, Dinter C, Ramadori G. Expression patterns of matrix metalloproteinases and their inhibitors in parenchymal and non-parenchymal cells of rat liver: regulation by TNF-alpha and TGF-beta1. J Hepatol. 1999;30(1):48–60. doi: 10.1016/s0168-8278(99)80007-5. [DOI] [PubMed] [Google Scholar]
- 8.Friedman SL. Liver fibrosis--from bench to bedside. J Hepatol. 2003;38 Suppl 1:S38–S53. doi: 10.1016/s0168-8278(02)00429-4. [DOI] [PubMed] [Google Scholar]
- 9.Qi Z, Atsuchi N, Ooshima A, Takeshita A, Ueno H. Blockade of type beta transforming growth factor signaling prevents liver fibrosis and dysfunction in the rat. Proc Natl Acad Sci U S A. 1999;96(5):2345–2349. doi: 10.1073/pnas.96.5.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Gouville AC, Boullay V, Krysa G, Pilot J, Brusq JM, Loriolle F, Gauthier JM, Papworth SA, Laroze A, Gellibert F, Huet S. Inhibition of TGF-beta signaling by an ALK5 inhibitor protects rats from dimethylnitrosamine-induced liver fibrosis. Br J Pharmacol. 2005;145(2):166–177. doi: 10.1038/sj.bjp.0706172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George J, Roulot D, Koteliansky VE, Bissell DM. In vivo inhibition of rat stellate cell activation by soluble transforming growth factor beta type II receptor: a potential new therapy for hepatic fibrosis. Proc Natl Acad Sci U S A. 1999;96(22):12719–12724. doi: 10.1073/pnas.96.22.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okuno M, Akita K, Moriwaki H, Kawada N, Ikeda K, Kaneda K, Suzuki Y, Kojima S. Prevention of rat hepatic fibrosis by the protease inhibitor, camostat mesilate, via reduced generation of active TGF-beta. Gastroenterology. 2001;120(7):1784–1800. doi: 10.1053/gast.2001.24832. [DOI] [PubMed] [Google Scholar]
- 13.Arias E, Anderson RN, Kung HC, Murphy SL, Kochanek KD. Deaths: final data for 2001. Natl Vital Stat Rep. 2003;52(3):1–115. [PubMed] [Google Scholar]
- 14.Kim KH, Kim HC, Hwang MY, Oh HK, Lee TS, Chang YC, Song HJ, Won NH, Park KK. The antifibrotic effect of TGF-beta1 siRNAs in murine model of liver cirrhosis. Biochem Biophys Res Commun. 2006;343(4):1072–1078. doi: 10.1016/j.bbrc.2006.03.087. [DOI] [PubMed] [Google Scholar]
- 15.Li GM, Shi Y, Li DG, Xie Q, Guo Q, Jin YX. [Effect of small interfering RNA targeting connective tissue growth factor on the synthesis and secretion of extracellular matrix in hepatic stellate cells] Zhonghua Gan Zang Bing Za Zhi. 2004;12(9):526–529. [PubMed] [Google Scholar]
- 16.Zhou X, Murphy FR, Gehdu N, Zhang J, Iredale JP, Benyon RC. Engagement of alphavbeta3 integrin regulates proliferation and apoptosis of hepatic stellate cells. J Biol Chem. 2004;279(23):23996–24006. doi: 10.1074/jbc.M311668200. [DOI] [PubMed] [Google Scholar]
- 17.Lindquist JN, Parsons CJ, Stefanovic B, Brenner DA. Regulation of alpha1(I) collagen messenger RNA decay by interactions with alphaCP at the 3'-untranslated region. J Biol Chem. 2004;279(22):23822–23829. doi: 10.1074/jbc.M314060200. [DOI] [PubMed] [Google Scholar]
- 18.Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, Chen J, Shankar P, Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9(3):347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 19.Kushibiki T, Nagata-Nakajima N, Sugai M, Shimizu A, Tabata Y. Delivery of plasmid DNA expressing small interference RNA for TGF-beta type II receptor by cationized gelatin to prevent interstitial renal fibrosis. J Control Release. 2005;105(3):318–331. doi: 10.1016/j.jconrel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 20.Zhu L, Lu Y, Miller DD, Mahato RI. Structural and formulation factors influencing pyridinium lipid-based gene transfer. Bioconjug Chem. 2008;19(12):2499–2512. doi: 10.1021/bc8004039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Hu H, Yin JQ. Therapeutic strategies against TGF-beta signaling pathway in hepatic fibrosis. Liver Int. 2006;26(1):8–22. doi: 10.1111/j.1478-3231.2005.01192.x. [DOI] [PubMed] [Google Scholar]
- 23.Hellerbrand C, Stefanovic B, Giordano F, Burchardt ER, Brenner DA. The role of TGFbeta1 in initiating hepatic stellate cell activation in vivo. J Hepatol. 1999;30(1):77–87. doi: 10.1016/s0168-8278(99)80010-5. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Mahato RI. siRNA pool targeting different sites of human hepatitis B surface antigen efficiently inhibits HBV infection. J Drug Target. 2008;16(2):140–148. doi: 10.1080/10611860701878750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malizia G, Brunt EM, Peters MG, Rizzo A, Broekelmann TJ, McDonald JA. Growth factor and procollagen type I gene expression in human liver disease. Gastroenterology. 1995;108(1):145–156. doi: 10.1016/0016-5085(95)90019-5. [DOI] [PubMed] [Google Scholar]
- 26.Jonsson JR, Barrie HD, O'Rourke P, Clouston AD, Powell EE. Obesity and steatosis influence serum and hepatic inflammatory markers in chronic hepatitis C. Hepatology. 2008;48(1):80–87. doi: 10.1002/hep.22311. [DOI] [PubMed] [Google Scholar]
- 27.Tipoe GL, Liong EC, Casey CA, Donohue TM, Jr, Eagon PK, So H, Leung TM, Fogt F, Nanji AA. A voluntary oral ethanol-feeding rat model associated with necroinflammatory liver injury. Alcohol Clin Exp Res. 2008;32(4):669–682. doi: 10.1111/j.1530-0277.2008.00623.x. [DOI] [PubMed] [Google Scholar]
- 28.Bortolami M, Kotsafti A, Cardin R, Farinati F. Fas / FasL system, IL-1beta expression and apoptosis in chronic HBV and HCV liver disease. J Viral Hepat. 2008;15(7):515–522. doi: 10.1111/j.1365-2893.2008.00974.x. [DOI] [PubMed] [Google Scholar]