Abstract
Nuclear receptors (NRs) are a class of hormone-gated transcription factors found in metazoans that regulate global changes in gene expression when bound to their cognate ligands. Despite species diversification, NRs act similarly across taxa to play fundamental roles in detecting intrinsic and environmental signals, and subsequently in coordinating transcriptional cascades that direct reproduction, development, metabolism, and homeostasis. These endocrine receptors function in vivo in part as molecular switches and timers that regulate transcriptional cascades. Here we discuss in detail how several C. elegans NRs integrate intrinsic and extrinsic signals to regulate the dauer diapause and longevity, molting, and heterochronic circuits of development, and draw parallels to similar in vivo endocrine regulated processes in other animals.
Introduction
Nuclear receptors (NRs) comprise an ancient family of hormone-gated transcription factors, which regulate metazoan gene expression in response to lipophilic ligands [1]. Ligand-gated transcription provides a direct and powerful means to couple environmental and nutrient signals to coordination of metabolism, development, reproduction, and homeostasis. Given the central role of NRs in animal biology, it is perhaps not surprising that their dysfunction accounts for many major human diseases, including diabetes, obesity, cancer, and cardiovascular disease [2, 3]. Because they can be pharmacologically manipulated by agonists or antagonists, NRs represent an important avenue for disease intervention.
A common molecular architecture, including a conserved N-terminal DNA binding domain (DBD) and a more variable C-terminal ligand binding domain (LBD) underlies NR signaling capabilities [1]. DNA binding is achieved by the association of two C4-Zn fingers with specific DNA response elements in the promoters of target genes. Transcriptional activation is achieved when, upon ligand binding, an NR undergoes a conformational change in which a C-terminal activation helix, AF-2, folds back onto the LBD core, thereby locking the NR in an active conformation [4–6].
NRs, having no ligand or for which no ligand(s) has been identified, are dubbed orphans [7]. Efforts to identify cognate ligands for these orphans, such as oxysterols and bile acids for the liver X receptor (LXR) [8, 9] and phospholipids and sphingolipids for steroidogenic factor 1 (SF-1) [8, 10, 11], have helped illuminate their physiological roles and mechanism. Other NRs can be constitutively actived by virtue of hydrophobic amino acid side chains occupying the ligand binding pocket [12] or can undergo ligand-independent regulation via intrinsic activation domains [13]. Moreover, covalent modifications of NRs, such as phosphorylation, acetylation, sumoylation, and ubiquitylation often modulate their transcriptional activity [14]. Finally, depending on type and context, NRs can act as monomers, heterodimers, and/or homodimers, as well as work in heterologous transcriptional complexes, giving rise to great combinatorial diversity and mechanistic complexity [1].
An important instructive component of NR signaling arises from their association with coregulators, adapator molecules that couple the NR to activating or repressive transcriptional machinery. NR-ligand binding typically results in the recruitment of coactivators, which consequently stimulates transcription, whereas unliganded NRs often dock corepressors, which repress transcription [15]. In fact, numerous coregulator complexes have been identified, and an emerging theme is that they may serve to coordinate diverse transcription factors that work together [15]. Mechanistically, transcriptional activation potentials of ligand-bound NRs are thought to be modulated by competition between coactivators and corepressors, which allows activation-response curves to range from continuous gradients to sharp thresholds [15, 16].
A comparison of the origins and functions of NRs has revealed important insights into their structural and functional diversification. Speciation, contributed in part by gene duplication events, has resulted in the amplification and subsequent divergence of NRs among metazoans. The NR superfamily is thought to have undergone two waves of expansion during metazoan evolution, giving rise to several paralogs [17]. The genome of C. elegans is predicted to contain a remarkable 284 NRs, whereas humans have 48, mice 49, and Drosophila 18 [18–21]. Roughly 15 worm NRs are homologous across taxa, whereas the remainder is thought to have evolved from an explosive expansion of the HNF4 lineage [22]. Although the reasons for this expansion are unknown, it is plausible that these novel receptors are deployed in processes such as chemical defense or immunity that require recognition of diverse molecules.
Despite the wealth of information on the mammalian NRs, studies in a simple model organism such as C. elegans offer distinct advantages. The worm’s streamlined anatomy (959 cells) and completely described development provide an unparalleled view of gene regulation at the single cell level. Most importantly, genetic analysis permits a facile examination of NR mechanisms, physiology, and signal transduction in an in vivo setting.
In general, the C. elegans receptors, like their mammalian counterparts, can be classified as contributing to either reproduction and development or to nutrient cycling and metabolism [23]. Significantly, in mammals, the expression patterns, rhythmic cycling, and physiological pathways affected by NRs do not strictly correlate with their known phylogenetic relationship within the NR subfamilies [23]. Notably, even within a species, structurally-related paralogs have undergone substantial functional diversification [17]. Thus, function and mechanism of action could be more informative than structural orthology in dissecting NR biology.
Indeed, such functional studies in C. elegans have already led to important insights into the role of NRs in sex determination, developmental timing, molting, aging, cell fate determination, neural differentiation, and metabolism, with implications for higher animals [24]. From another perspective, NRs can act as molecular switches, timers, homeostatic devices, or gradient regulators (rheostats). These devices are utilized in a variety of biological processes. Here, we focus in detail on several C. elegans NRs that function as switches or timing devices in the context of dauer formation, molting, and heterochrony, while also drawing parallels to NRs of other organisms.
Dauer Formation
As transcription factors that can activate or repress, NRs have the ability to act as switches that decide between alternate fates. We discuss here how the DAF-12 NR works as a switch to regulate the choice between reproductive growth and dauer arrest.
C. elegans larval development and reproductive maturation are responsive to nutrient cues and governed by an NR-dependent switch. Development and reproduction are energetically costly processes that require proper assessment of environmental and nutritional inputs for maximal fitness. C. elegans develops through four larval stages (L1 to L4) to become a reproductive adult. In an environment of limited food, high temperature, or overcrowding, the animal diverts its development into an alternative third larval stage called the dauer diapause (L3d) (Figure 1). Dauer larvae are developmentally arrested, sexually immature, stress resistant, long lived, and geared for survival. Upon return to favorable conditions, these larvae exit the dauer state to resume development and reproduction [25]. Importantly, a molecular-genetic dissection of dauer formation has provided key insights into metazoan longevity, growth control, and cancer, as well as fat metabolism and diabetes. More specifically, it has revealed how environmental cues are transformed into a hormone-regulated developmental switch.
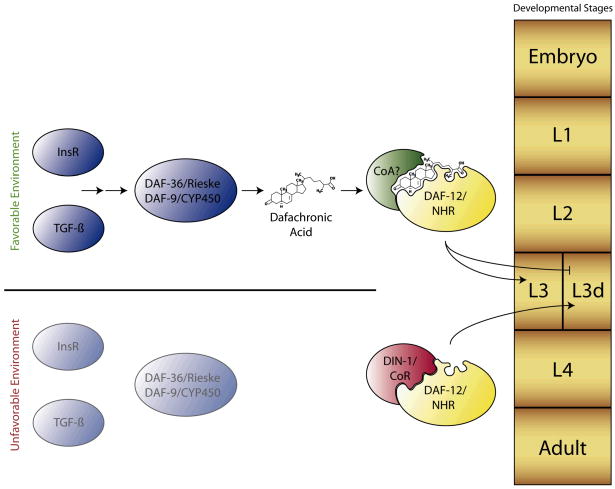
Figure 1.
The choice between two life history fates, reproductive (L3) or dauer (L3d), results from graded environmental cues impacting production of a ligand for DAF-12, a key nuclear receptor that controls the switch between these two fates. Depending upon its ligand-bound state, DAF-12 governs this switch through the DIN-1 corepressor (CoR) and unidentified coactivator(s) (CoA). In favorable conditions, signals from the environment are translated via the Insulin/IGF-1 (InsR) and TGF-β pathways to promote transformation of cholesterol by the DAF-36/Rieske-like oxygenase and DAF-9/CYP450 (and others) to produce dafachronic acid, the ligand for DAF-12. DAF-12 drives transcription of genes involved in reproductive development and fat metabolism. In unfavorable environments (e.g., low food, overcrowding, other stresses), low Insulin/IGF-1 and TGF-β signaling translates to little or no dafachronic acid production, resulting in unliganded DAF-12 binding to its corepressor DIN-1, and promoting the dauer specific lineage characterized by stress resistance, increased fat storage, and longevity.
DAF-12 is perhaps the best understood C. elegans NR and plays a key role in the choice between the dauer diapause and reproductive development. It also influences fat metabolism, developmental timing, and adult longevity. Though most closely related to the vertebrate vitamin D (VDR) and LXR, and the Drosophila hormone receptor HR96 [26], DAF-12 may work analogously to mammalian estrogen receptor (ER) by coupling nutrient cues to maturation. Epistasis analysis indicates that DAF-12 works downstream of several signaling cascades including the insulin/IGF-1 signaling (IIS) and TGF-β pathways [25, 27–31]. Cellular and molecular genetic studies suggest a model whereby in favorable environments, cues integrated by sensory neurons result in the graded production of TGF-β and insulin-like peptides (Figure 1), with their respective signal transduction pathways converging in endocrine tissues on biosynthetic enzymes involved in the production of the DAF-12 ligands, the dafachronic acids (DA) (see below) [27, 28, 32, 33]. Notably, when bound to its ligand, DAF-12, along with presumptive coactivators, promotes reproductive growth (Figure 1). In unfavorable conditions, during which levels of TGF-β and insulin-like peptides decrease, DA production is thought to be repressed. Unliganded DAF-12, together with its corepressor DIN-1, a homolog of the mammalian SHARP corepressor, promotes dauer diapause and longevity [34]. Thus, DAF-12 and associated co-regulator complexes work as hormone-regulated molecular switches that specify two different life history modes, reproductive development or the dauer diapause. Importantly, hormone deficient mutants such as daf-9 are long lived as adults and this longevity is dependent upon daf-12(+) [28, 29]. Similarly, animals that are germline deficient are also long lived, and this too depends upon daf-12(+) [35]. These studies provide pioneering evidence for NR control of somatic endurance and longevity, thereby suggesting potential research avenues for the vertebrate receptors.
Endogenous ligands for DAF-12 were recently identified, the first for any of the 284 C. elegans NRs, which has been instrumental in elucidating the mechanisms underlying the dauer switch [33]. Δ4- and Δ7-DA are cholestenoic acid derivatives that, at nanomolar concentrations, bind and activate DAF-12, thereby promoting downstream transcription [33]. Evidence that the DAF-9/cytochrome-P450 (CYP450) enzyme acted directly upstream of DAF-12 in a cell non-autonomous fashion suggested that it was likely to help synthesize a steroidal ligand for DAF-12 [27–29, 36]. Accordingly, lipid extracts from wild-type worms contained both DAs, whereas extracts from daf-9 mutants contained neither. Moreover, exogenous Δ4-DA could rescue all daf-9/CYP450 phenotypes, including those of dauer formation, gonadal maturation, and aging [32, 33]. Δ4-DA could also rescue the dauer-constitutive phenotypes of animals carrying mutations in daf-2/Insulin receptor, daf-7/TGF-β, or sterol trafficking Niemann-Pick type C1 homlogs, ncr-1 and ncr-2, but not daf-12, supporting the predicted epistatic relationship of these genes [33].
The discovery of DA has also led to insights into the nature and regulation of the hormone biosynthetic pathway. Biochemical studies revealed that daf-9/CYP450 catalyzes the last step in DA synthesis, the successive oxidation of the terminal side chain to the acid [33], in a manner similar to that of CYP27A1 (a regulator of mammalian bile acid synthesis) [37, 38]. This raises the possibility that similar metabolites are found in mammals. In addition, a Rieske-like oxygenase, DAF-36, was proposed to carry out the first step, converting cholesterol to 7-dehydrocholesterol, thus outlining a pathway for DA biosynthesis [39]. Interestingly, a similar activity is seen for the Drosophila homolog neverland in ecdysteroid production [40]. The expression patterns of DAF-9 (XXX cells, epidermis, spermatheca) and DAF-36 (intestine) are non-overlapping, revealing that biosynthesis is distributed and subject to inputs from various tissues. Notably, the XXX cells are a pair of neuroendocrine cells in which several dauer signaling molecules are found. These include sdf-9, a protein tyrosine phosphatase homolog that may be important for insulin signaling, ncr-1 and ncr-2 Niemann-Pick homologs involved in sterol trafficking [41], akt-1 kinase, and several enhancers of akt-1 [42] Somewhat surprisingly, epidermal daf-9 is dynamically regulated by daf-12, mostly in response to cholesterol availability, as well as to inputs from TGF-β and IIS [27, 36]. Moreover, excess ligand is inferred to inhibit daf-9 expression through DAF-12. Although the mechanisms underlying these various observations are currently unknown, determining the extent to which the worm homologs show similar regulatory circuitry to their mammalian counterparts will be important. For example, 25S-cholestenoic acid (akin to DA) is known to serve as a ligand for both LXR and DAF-12 [8, 43], thus hinting at a potentially broader overlap. In addition, these interactions may also help us to understand how insulin, TGF-β, and steroid hormone receptor signaling converge to control reproduction in higher animals.
The identification of DA has clarified our understanding of coregulator activity, an essential component of the switching mechanism. In particular, molecular genetic data suggest that DIN-1 and DAF-12 interact to form a corepressor complex in the absence of ligand production (i.e., in daf-9 mutants). In support of this mechanism, Δ4-DA was shown to dissociate the DAF-12/DIN-1 complex, abrogating repression in human cell culture [33, 34]. Studies of DIN-1 also provide compelling in vivo evidence for a key biological role of the unliganded receptor and its corepressor in specifying alternate metabolic states, dauer diapause, and longevity [33, 34]. The mammalian homolog SHARP works as a corepressor with several transcription factors, including unliganded retinoic acid receptor (RAR) and peroxisome proliferator activated receptor δ (PPARδ) [44, 45], illustrating that this corepressor-NR interaction is evolutionarily ancient.
Altogether, these studies reveal that steroid-like control of reproduction from biology to mechanism is evolutionarily conserved, and point in particular to the importance of bile acid-like steroids as signaling molecules, which is an emerging theme in vertebrates [46].
Molting
Timing circuits play essential roles in regulating daily, monthly, seasonal, and maturational life history events, and often rely on positive and negative feedback loops or transcriptional cascades. NRs are uniquely poised to drive such circuits because they can integrate intrinsic and environmental inputs to coordinate programs throughout the body. Moreover, ligand gating provides precise temporal control and is well suited to homeostatic feedback. Here, we discuss how the invertebrate NRs govern various timing devices, including the molt cycle and heterochronic timers.
The molt cycle, the synthesis of the new exoskeleton and shedding of the old, is a developmental clock that utilizes several coordinating NRs. During each larval stage (L1-L4), C. elegans undergoes a cycle of cuticle synthesis and shedding before emerging as adults. Both the nematode C. elegans, as well as the arthropod Drosophila, are considered Ecdysozoans, a clade of animals that undergo molting [47]. This process is best understood in Drosophila, in which pulses of 20-hydroxyecdysone stimulate a transcriptional cascade that drives molting via the ecdysone receptor (EcR) and its heterodimeric partner ultraspiracle (USP) [48]. Several downstream NRs are activated, including the Drosophila hormone receptor DHR3, Fushi tarazu-F1 (FTZ-F1), ecdysone-inducible proteins (E75/78), DHR38, and DHR78, which are activated in strict temporal sequence to drive molting [49–51]. These NRs are largely conserved across taxa. Humans homologs include RAR-related orphan receptor (ROR), SF-1, REV-ERB, Nur-related protein 1 (Nurr1), and testicular orphan receptor 2 and 4 (TR2/TR4); and all these mammalian NRs, except SF-1, display circadian expression levels [52]. C. elegans has homologs to all except EcR and USP; these include NHR-23, NHR-25, SEX-1 and NHR-85, NHR-6, and NHR-41. Only a few of these C. elegans receptors are currently known to affect molting: NHR-23, NHR-25, NHR-41 and NHR-67/TLL (Tailless) [19]. Here, we discuss these few in more detail as well as possible ligand(s) that drive ecdysis.
NHR-23 and NHR-25 are the most extensively-characterized NRs that govern molting. Expression of nhr-23 and nhr-25 (also nhr-41) mRNA oscillates with each molt cycle, with their highest expression during intermolts (Figure 2) [19].
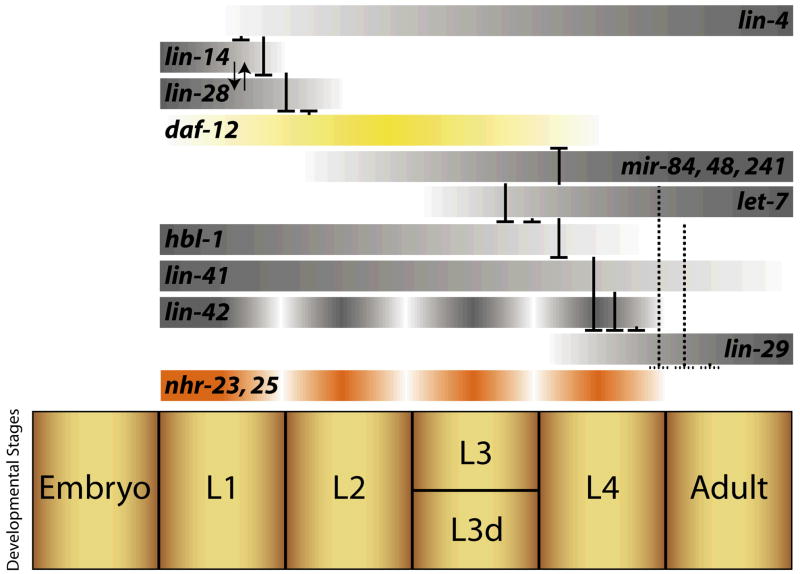
Figure 2.
Heterochronic genes control stage-specific programs of development and regulate expression of the DAF-12, NHR-23, and NHR-25 NRs. Graded expression levels of several selected heterochronic genes are shown, with higher expression levels indicated by increasing darkness. Together, these genes form an elegant network of gene regulation (repression is indicated by lines with blocked ends) that helps drive development. Dashed lines represent interactions in which it is unclear whether regulation is direct.
Reduction of nhr-23 by RNAi knockdown at any intermolt results in defective subsequent molts, and, in addition, disrupts collagen synthesis, tail development, and placement of epidermal seam cells [53]. NHR-23 is highly similar to Drosophila DHR3, an NR required for stage transitions and cuticle synthesis, as well as for proper expression of EcR and FTZ-F1 [54]. NHR-23 is orthologous to vertebrate RORα, which has roles in circadian rhythms, development of Purkinje cells, bone maintenance, immunity, and cholesterol and lipid metabolism [55]. In particular, RORα works reciprocally with the REV-ERB NR as a core component of the circadian clock, with REV-ERB repressing and RORα activating Bmal1 expression, respectively [56]. Similarly, Drosophila DHR3 and E75 (nhr-85 and sex-1 homolog) reciprocally regulate one another during the molt cycle [57, 58]. However, as of yet there is no solid evidence for sex-1 and nhr-85 showing a similar reciprocal relationship with nhr-23; sex-1 is involved in sex determination and nhr-85 functions in ovulation [19, 59].
NHR-25 is required for embryonic and post-embryonic functions and is homologous to Drosophila FTZ-F1, mammalian SF-1, and liver receptor homolog 1 (LRH-1). Embryonic loss of nhr-25 results in failure of ventral closure and defects in epidermal elongation, causing arrest at the two-fold stage [60]. Like nhr-23, loss of nhr-25 during larval stages results in severe defects in molting. In addition, nhr-25 mutants display defects in epidermal cell fusion, abnormal seam cell morphology and number, altered vulval cell differentiation, gonadal defects and germline overproliferation [60, 61]. In various developmental contexts, nhr-25 works alongside a variety of other transcription factors, including NOB-1 and LIN-39 Hox proteins, which regulate embryogenesis and vulval cell differentiation, respectively [60], as well as with β-catenin in the establishment of distal-proximal fates in the somatic gonad precursor cells [62]. Comparably, Drosophila, FTZ-F1 has embryonic roles in segmentation and larval metamorphosis, and often works together with the homeodomain protein FTZ [63–66]. The mammalian SF-1 orchestrates the development of the gonad, adrenal glands, hypothalamus, and pituitary, as well as male differentiation and steroidogenesis [67, 68]. Interestingly, it also works with β-catenin and Wnt signaling in a variety of contexts [67]. Lastly, LRH-1 has roles in cholesterol transport, bile acid homeostasis, and ovarian function [68].
In Drosophila, 20-hydroxyecdysone drives the molt process; however, the identity of such a ligand in C. elegans remains elusive. The ligands for the vertebrate NHR-25 homologs SF-1 and LRH surprisingly are various phospholipids and sphingolipids [8, 10, 11]. Given the high degree of conservation, the worm receptor, too, could have related ligands. Although nematodes lack ecdysteroids, evidence suggests that the ligand(s) driving C. elegans ecdysis could alternatively be a sterol, as cholesterol deprivation results in molting defects, and molecules implicated in cholesterol disposition, such as the LDL-like receptor protein, lrp-1, are required for molting [69]. Additionally mutations in let-767, encoding a 17-β-hydroxysteroid dehydrogenase homologue, results in molting defects, suggesting a possible role in production of a steroid hormone that drives ecdysis [70].
Genome-wide RNAi screens have identified scores of genes affecting the molting process, but surprisingly, none of them obviously give clear clues to the identity of a molting hormone [53, 71]. Aside from hormonal regulation, ecdysis may also be regulated by microRNAs. In particular, the let-7 microRNA and its paralog mir-84 regulate exit from molt cycling, and are thought to work through inhibitory feedback on nhr-23 and nhr-25, though it is unclear whether this regulation is direct (Figure 2) [72].
Finally, the DAF-12 NR may have a role in integrating developmental timing programs with the molt cycle. Disrupting nicotinic acetylcholine receptors (nAChR) delays L2 cell division programs, but not the molt cycle, a process suppressed by mutations in daf-9 and daf-12 [73]. This suggests that nAChRs could act via ligand (DA)-bound DAF-12 to repress L2 cell divisions and differentiation [73]. Still, a detailed mechanism illustrating how DAF-12 could integrate the molting and heterochronic circuits awaits.
Heterochrony
During development, cells acquire temporal identity, often with distinct stage-specific programs for embryo, juvenile, and adult. Heterochrony is a change in the relative timing of such stage specific events. NRs play a critical role in developmental timing in diverse taxa. Examples include the thyroid and ecdysone receptors (TR and EcR), which mediate metamorphosis in amphibians and insects, respectively [74, 75]. Additionally, androgen and estrogen receptors (AR and ER) promote reproductive maturation in mammals [76]. The vertebrate vitamin D receptor (VDR) has a role in the cycling of the hair follicles [77]. Post-developmentally, the ER and progesterone receptors (PR) orchestrate the menstrual cycle [78].
In the worm, DAF-12 promotes reproductive maturation in the context of the heterochronic pathway (Figure 2). Heterochronic loci control stage-specific programs from L1 to adult. Mutations in these genes result in temporal transformations during post-embryonic development, resulting in the misexpression of larval programs in adults (retarded) or conversely, adult programs in larva (precocious), usually in a stage- and tissue-specific manner [79, 80]. Many of the heterochronic genes are highly conserved, and hence, study of this elegant circuit has led to novel insights into metazoan developmental timing, and regulation of cell proliferation, migration, and stem cell division patterns [81].
Among these highly conserved genes are the first discovered microRNAs, lin-4 [82, 83] and let-7 [84], 22-nucleotide RNAs that inhibit expression by base pairing with the 3′ UTR of their target mRNAs. Interestingly, transitions in stage programs are often triggered by up-regulation of distinct sets of microRNAs, which down-regulate their targets. For example, lin-4 down-regulates the nuclear factor lin-14 and the cold shock domain protein lin-28 during L1/L2 transitions [83, 85]. The microRNAs mir-84, 48, and 241 down-regulate hbl-1/hunchback during L2/L3 transitions [86] and let-7 microRNA down-regulates lin-41/RBCC ring finger protein and hbl-1, as well as daf-12 during L4 to adult transitions [86–88] (Figure 2). Components involved in the processing and assembly of microRNA complexes also have heterochronic phenotypes [89–91]
NR DAF-12 primarily promotes L2/L3 transitions in gonadal and extragonadal tissues, as daf-12 mutants repeat L2 programs during the L3 stage [92]. In general, daf-12 LBD mutants have more severe heterochronic phenotypes than DBD mutants [26], possibly because they lock the receptor in a repressive state and interfere with other heterochronic activities [34]. One potential target of daf-12 regulation is lin-28, which is genetically epistatic to daf-12 [92] and whose protein accumulates at late stages in daf-12 LBD mutants [93, 94]. However, lin-28 mRNA levels remain unchanged during development, suggesting that regulation must be post-transcriptional and likely indirect. Presumably, other loci involved in the L2/L3 transition could be daf-12 targets.
Remarkably, homologs of several circadian clock genes, including lin-42/period, tim-1/timeless, and kin-20/double-time, also function in the C. elegans heterochronic circuit, but lack aspects of the feedback regulation and diurnal fluctuations found in classical circadian circuits [95, 96]. Instead, lin-42 mRNA levels fluctuate with the molt cycle (Figure 2) and functions to prevent early expression of adult fates. While RNAi knockdown of tim-1 and kin-1 resembles the precocious cell fusion patterns seen in lin-42 mutants [96], it is unclear whether they directly regulate lin-42. Interestingly, lin-42 and daf-12 act antagonistically with lin-42 mutants producing a precocious dorsal turn of the gonad at the L2 molt, whereas daf-12 mutant animals fail to make this turn appropriately at the L3 molt [92, 97, 98]. Epistasis analysis further suggests that daf-12 functions in parallel or downstream of lin-42. Speculatively, this interaction may mimic the relationship of PERIOD and the NRs REV-ERB and RORα in the circadian clock.
DAF-12 acts at the nexus of the dauer and heterochronic pathways, telling of its importance in regulating developmental programs. By integrating inputs from the dauer pathways, DAF-12 conveys information about stress, nutrients, and the environment into the heterochronic circuit, thereby either advancing or arresting development through a hormone-dependent mechanism (Figure 1). In a similar manner, the ER may govern puberty in response to environmental input [76]. Conceivably, global transitions such as mammalian puberty or tissue-specific clocks such as the hair follicle cycle may be embedded in the rich circuitry of similar heterochronic timers, which drive forward regulatory hierarchies and specify stage appropriate events.
Summary
The basic biology of metazoans dictates tight regulatory control of reproduction, development, metabolism, and homeostasis. NRs are well-poised to govern these processes because they can couple environmental and physiologic cues to coordinate organism-wide events. Although diversified through evolution, NRs across taxa regulate similar fundamental biological processes. Here, we have discussed in detail how a few of the best understood NRs from C. elegans function as switches and timers to control diapause, molting, and developmental timing. A comparison of these conserved circuits and mechanisms promises to illuminate in vivo transcriptional networks that underlie analogous processes in other animals.
A key challenge for the future will be to further dissect the molecular basis of how NRs act as switching and timing devices, as well as homeostatic controllers and rheostats in various biological contexts. To this end, it will important to identify and understand all the major components that control the regulatory circuitry. A systems approach, which takes into account not only the dimensions of ligand, receptor, and target gene, but also ratios of coactivator and corepressor complexes, tissue-specific licensing factors, and signaling modulators, will be needed to fully describe such circuits. In this light, in vivo quantitative readouts in a genetically tractable organism such as C. elegans should prove invaluable. A second critical challenge will be to understand the transcriptional cascades and target genes that underlie the physiology controlled by NRs. Such studies should reveal important insights into conserved NR mechanisms across taxa.
Acknowledgments
The authors wish to acknowledge grant support from the NURSA, NIA, NIH, and AFAR for A.A. and NIA for D.B.M.
References
- 1.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehrke M, Lazar MA. The many faces of PPARgamma. Cell. 2005;123:993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 3.Smith AG, Muscat GE. Orphan nuclear receptors: therapeutic opportunities in skeletal muscle. Am J Physiol Cell Physiol. 2006;291:C203–217. doi: 10.1152/ajpcell.00476.2005. [DOI] [PubMed] [Google Scholar]
- 4.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 5.Bledsoe RK, et al. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- 6.Cronet P, et al. Structure of the PPARα and -γ ligand binding domain in complex with AZ 242; ligand selectivity and agonist activation in the PPAR family. Structure. 2001;9:699–706. doi: 10.1016/s0969-2126(01)00634-7. [DOI] [PubMed] [Google Scholar]
- 7.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 8.Song C, Liao S. Cholestenoic acid is a naturally occurring ligand for liver X receptor alpha. Endocrinology. 2000;141:4180–4184. doi: 10.1210/endo.141.11.7772. [DOI] [PubMed] [Google Scholar]
- 9.Janowski BA, et al. An oxysterol signalling pathway mediated by the nuclear receptor LXRα. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 10.Krylova IN, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 11.Urs AN, et al. Steroidogenic factor-1 is a sphingolipid binding protein. Mol Cell Endocrinol. 2007:265–266. 174–178. doi: 10.1016/j.mce.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suino K, et al. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 13.Smith CL, et al. Modulation of the ligand-independent activation of the human estrogen receptor by hormone and antihormone. Proc Natl Acad Sci U S A. 1993;90:6120–6124. doi: 10.1073/pnas.90.13.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu M, et al. Acetylation of nuclear receptors in cellular growth and apoptosis. Biochem Pharmacol. 2004;68:1199–1208. doi: 10.1016/j.bcp.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 15.Lonard DM, et al. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 16.Rossi FM, et al. Transcriptional control: rheostat converted to on/off switch. Mol Cell. 2000;6:723–728. doi: 10.1016/s1097-2765(00)00070-8. [DOI] [PubMed] [Google Scholar]
- 17.Escriva H, et al. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11–26. doi: 10.1042/bse0400011. [DOI] [PubMed] [Google Scholar]
- 18.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 19.Gissendanner CR, et al. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev Biol. 2004;266:399–416. doi: 10.1016/j.ydbio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Otte K, et al. Identification of farnesoid X receptor beta as a novel mammalian nuclear receptor sensing lanosterol. Mol Cell Biol. 2003;23:864–872. doi: 10.1128/MCB.23.3.864-872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson-Rechavi M, et al. How many nuclear hormone receptors are there in the human genome? Trends Genet. 2001;17:554–556. doi: 10.1016/s0168-9525(01)02417-9. [DOI] [PubMed] [Google Scholar]
- 22.Robinson-Rechavi M, et al. Explosive lineage-specific expansion of the orphan nuclear receptor HNF4 in nematodes. J Mol Evol. 2005;60:577–586. doi: 10.1007/s00239-004-0175-8. [DOI] [PubMed] [Google Scholar]
- 23.Bookout AL, et al. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antebi A. Nuclear hormone receptors in C. elegans. In: Community TCeR., editor. WormBook. 2006. WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu PJ. Dauer. In: Community TCeR., editor. WormBook. 2007. WormBook. [Google Scholar]
- 26.Antebi A, et al. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- 27.Gerisch B, Antebi A. Hormonal signals produced by DAF-9/cytochrome P450 regulate C. elegans dauer diapause in response to environmental cues. Development. 2004;131:1765–1776. doi: 10.1242/dev.01068. [DOI] [PubMed] [Google Scholar]
- 28.Gerisch B, et al. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- 29.Jia K, et al. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development. 2002;129:221–231. doi: 10.1242/dev.129.1.221. [DOI] [PubMed] [Google Scholar]
- 30.Thomas JH, et al. Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics. 1993;134:1105–1117. doi: 10.1093/genetics/134.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riddle DL, et al. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- 32.Gerisch B, et al. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci USA. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Motola DL, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 34.Ludewig AH, et al. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev. 2004;18:2120–2133. doi: 10.1101/gad.312604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 36.Mak HY, Ruvkun G. Intercellular signaling of reproductive development by the C. elegans DAF-9 cytochrome P450. Development. 2004;131:1777–1786. doi: 10.1242/dev.01069. [DOI] [PubMed] [Google Scholar]
- 37.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 38.Goodwin B, et al. Identification of bile acid precursors as endogenous ligands for the nuclear xenobiotic pregnane X receptor. Proc Natl Acad Sci USA. 2003;100:223–228. doi: 10.1073/pnas.0237082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rottiers V, et al. Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev Cell. 2006;10:473–482. doi: 10.1016/j.devcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Yoshiyama T, et al. Neverland is an evolutionally conserved Rieske-domain protein that is essential for ecdysone synthesis and insect growth. Development. 2006;133:2565–2574. doi: 10.1242/dev.02428. [DOI] [PubMed] [Google Scholar]
- 41.Ohkura K, et al. SDF-9, a protein tyrosine phosphatase-like molecule, regulates the L3/dauer developmental decision through hormonal signaling in C. elegans. Development. 2003;130:3237–3248. doi: 10.1242/dev.00540. [DOI] [PubMed] [Google Scholar]
- 42.Hu PJ, et al. Two membrane-associated tyrosine phosphatase homologs potentiate C. elegans AKT-1/PKB signaling. PLoS Genet. 2006;2:e99. doi: 10.1371/journal.pgen.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Held JM, et al. DAF-12-dependent rescue of dauer formation in Caenorhabditis elegans by (25S)-cholestenoic acid. Aging Cell. 2006;5:283–291. doi: 10.1111/j.1474-9726.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- 44.Shi Y, et al. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi Y, et al. The peroxisome proliferator-activated receptor d, an integrator of transcriptional repression and nuclear receptor signaling. Proc Natl Acad Sci USA. 2002;99:2613–2618. doi: 10.1073/pnas.052707099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houten SM, et al. Endocrine functions of bile acids. Embo J. 2006;25:1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aguinaldo AM, et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 48.Buszczak M, Segraves WA. Drosophila metamorphosis: the only way is USP? Curr Biol. 1998;8:R879–882. doi: 10.1016/s0960-9822(07)00550-7. [DOI] [PubMed] [Google Scholar]
- 49.Ashburner M. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis. Dev Biol. 1974;39:141–157. doi: 10.1016/s0012-1606(74)80016-3. [DOI] [PubMed] [Google Scholar]
- 50.Huet F, et al. Sequential gene activation by ecdysone in Drosophila melanogaster: the hierarchical equivalence of early and early late genes. Development. 1995;121:1195–1204. doi: 10.1242/dev.121.4.1195. [DOI] [PubMed] [Google Scholar]
- 51.Sullivan AA, Thummel CS. Temporal profiles of nuclear receptor gene expression reveal coordinate transcriptional responses during Drosophila development. Mol Endocrinol. 2003;17:2125–2137. doi: 10.1210/me.2002-0430. [DOI] [PubMed] [Google Scholar]
- 52.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 53.Kostrouchova M, et al. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:7360–7365. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lam G, et al. DHR3 is required for the prepupal-pupal transition and differentiation of adult structures during Drosophila metamorphosis. Dev Biol. 1999;212:204–216. doi: 10.1006/dbio.1999.9343. [DOI] [PubMed] [Google Scholar]
- 55.Jetten AM, et al. The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes. Prog Nucleic Acid Res Mol Biol. 2001;69:205–247. doi: 10.1016/s0079-6603(01)69048-2. [DOI] [PubMed] [Google Scholar]
- 56.Guillaumond F, et al. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 57.Hiruma K, Riddiford LM. Differential control of MHR3 promoter activity by isoforms of the ecdysone receptor and inhibitory effects of E75A and MHR3. Dev Biol. 2004;272:510–521. doi: 10.1016/j.ydbio.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 58.Lam GT, et al. Coordination of larval and prepupal gene expression by the DHR3 orphan receptor during Drosophila metamorphosis. Development. 1997;124:1757–1769. doi: 10.1242/dev.124.9.1757. [DOI] [PubMed] [Google Scholar]
- 59.Carmi I, et al. The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature. 1998;396:168–173. doi: 10.1038/24164. [DOI] [PubMed] [Google Scholar]
- 60.Chen Z, et al. The Caenorhabditis elegans nuclear receptor gene nhr-25 regulates epidermal cell development. Mol Cell Biol. 2004;24:7345–7358. doi: 10.1128/MCB.24.17.7345-7358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev Biol. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- 62.Asahina M, et al. Crosstalk between a nuclear receptor and β-catenin signaling decides cell fates in the C. elegans somatic gonad. Dev Cell. 2006;11:203–211. doi: 10.1016/j.devcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 63.Broadus J, et al. The Drosophila beta FTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol Cell. 1999;3:143–149. doi: 10.1016/s1097-2765(00)80305-6. [DOI] [PubMed] [Google Scholar]
- 64.Lavorgna G, et al. Potential role for a FTZ-F1 steroid receptor superfamily member in the control of Drosophila metamorphosis. Proc Natl Acad Sci USA. 1993;90:3004–3008. doi: 10.1073/pnas.90.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ueda H, et al. A sequence-specific DNA-binding protein that activates fushi tarazu segmentation gene expression. Genes Dev. 1990;4:624–635. doi: 10.1101/gad.4.4.624. [DOI] [PubMed] [Google Scholar]
- 66.Yu Y, et al. The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature. 1997;385:552–555. doi: 10.1038/385552a0. [DOI] [PubMed] [Google Scholar]
- 67.Gummow BM, et al. Convergence of Wnt signaling and steroidogenic factor-1 (SF-1) on transcription of the rat inhibin α gene. J Biol Chem. 2003;278:26572–26579. doi: 10.1074/jbc.M212677200. [DOI] [PubMed] [Google Scholar]
- 68.Fayard E, et al. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 69.Yochem J, et al. A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development. 1999;126:597–606. doi: 10.1242/dev.126.3.597. [DOI] [PubMed] [Google Scholar]
- 70.Kuervers LM, et al. The sterol modifying enzyme LET-767 is essential for growth, reproduction and development in Caenorhabditis elegans. Mol Genet Genomics. 2003;270:121–131. doi: 10.1007/s00438-003-0900-9. [DOI] [PubMed] [Google Scholar]
- 71.Frand AR, et al. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hayes GD, et al. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133:4631–4641. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- 73.Ruaud AF, Bessereau JL. Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development. 2006;133:2211–2222. doi: 10.1242/dev.02392. [DOI] [PubMed] [Google Scholar]
- 74.Buszczak M, Segraves WA. Insect metamorphosis: out with the old, in with the new. Curr Biol. 2000;10:R830–833. doi: 10.1016/s0960-9822(00)00792-2. [DOI] [PubMed] [Google Scholar]
- 75.Sato Y, et al. A role of unliganded thyroid hormone receptor in postembryonic development in Xenopus laevis. Mech Dev. 2007;124:476–488. doi: 10.1016/j.mod.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mauras N, et al. Sex steroids, growth hormone, insulin-like growth factor-1: neuroendocrine and metabolic regulation in puberty. Horm Res. 1996;45:74–80. doi: 10.1159/000184763. [DOI] [PubMed] [Google Scholar]
- 77.Skorija K, et al. Ligand-independent actions of the vitamin D receptor maintain hair follicle homeostasis. Mol Endocrinol. 2005;19:855–862. doi: 10.1210/me.2004-0415. [DOI] [PubMed] [Google Scholar]
- 78.Tabibzadeh S. The signals and molecular pathways involved in human menstruation, a unique process of tissue destruction and remodelling. Mol Hum Reprod. 1996;2:77–92. doi: 10.1093/molehr/2.2.77. [DOI] [PubMed] [Google Scholar]
- 79.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 80.Rougvie AE. Intrinsic and extrinsic regulators of developmental timing: from miRNAs to nutritional cues. Development. 2005;132:3787–3798. doi: 10.1242/dev.01972. [DOI] [PubMed] [Google Scholar]
- 81.Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 82.Lee RC, et al. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 83.Wightman B, et al. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 84.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 85.Moss EG, et al. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 86.Abbott AL, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grosshans H, et al. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 88.Slack FJ, et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 89.Zhang L, et al. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ding L, et al. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 91.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 92.Antebi A, et al. daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development. 1998;125:1191–1205. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- 93.Seggerson K, et al. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 94.Morita K, Han M. Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 2006;25:5794–5804. doi: 10.1038/sj.emboj.7601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jeon M, et al. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–1146. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- 96.Banerjee D, et al. Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev Cell. 2005;8:287–295. doi: 10.1016/j.devcel.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 97.Fielenbach N, et al. DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev Cell. 2007;12:443–455. doi: 10.1016/j.devcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 98.Tennessen JM, et al. Novel heterochronic functions of the Caenorhabditis elegans period-related protein LIN-42. Dev Biol. 2006;289:30–43. doi: 10.1016/j.ydbio.2005.09.044. [DOI] [PubMed] [Google Scholar]