Abstract
Small molecules that reconstitute the binding mode(s) of a protein and in doing so elicit a programmed functional response offer considerable advantages in the control of complex biological processes. The development challenges of such molecules are significant, however. Many protein-protein interactions require multiple points of contact over relatively large surface areas. More significantly, several binding modes can be superimposed upon a single sequence within a protein, and a true small molecule replacement must be pre-programmed for such multi-modal binding. This is the case for the transcriptional activation domain or TAD of transcriptional activators as these mofifs utilize a poorly characterized multi-partner binding profile in order to stimulate gene expression. Here we describe a unique class of small molecules that exhibit both function and a binding profile analogous to natural transcriptional activation domains. Of particular note, the small molecules are the first reported to bind to the KIX domain within the CREB binding protein (CBP) at a site that is utilized by natural activators. Further, a comparison of functional and non-functional small molecules indicates that an interaction with CBP is a key contributor to transcriptional activity. Taken together, the evidence suggests that the small molecule TADs mimic both the function and mechanism of their natural counterparts and thus present a framework for the broader development of small molecule transcriptional switches.
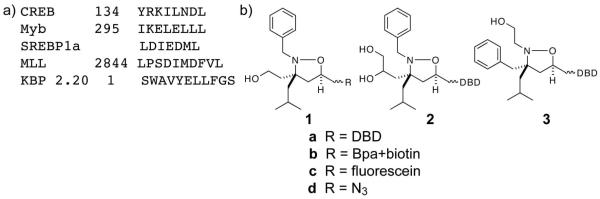
Transcriptional activators are essential for high fidelity transcription, responsible for seeking out particular genes and up-regulating them to precise levels in a signal-responsive fashion.(1, 2) Indeed, the altered transcription patterns observed in disease states can often be attributed to malfunctioning and/or mis-regulated transcriptional activators.(3-6) Alterations in the function of the tumor suppressor p53, for example, are found in >50% of all human cancers;(7, 8) similarly, constitutively active NF-κB, an activator that regulates genes responsible for apoptosis, inflammatory response, and proliferation, is observed in inflammatory disorders and most cancers.(9, 10) There is thus tremendous interest in the development of activator artificial transcription factors (activator ATFs), nonnatural molecules programmed to perform the same function as endogenous activators, as both mechanistic tools and as transcription-targeted therapeutic agents.(2, 11-14) The architecture of activator ATFs is analogous to that of their natural counterparts in that they minimally consist of a DNA binding domain (DBD) that confers gene-targeting specificity and a transcriptional activation domain (TAD) that controls the extent of gene activation. Of the two domains, it has proven more challenging to identify small molecule TAD replacements with functional properties comparable to the natural system despite their likely advantageous stability, delivery and/or immunogenic properties.(2)
The challenges associated with small molecule TAD discovery are due in large part to the scarcity of molecular-level details regarding natural TAD function. The largest and most well studied class of activators is the amphipathic class, characterized by interspersed polar and hydrophobic amino acid residues in the TAD (Figure 1a).(1, 2) As part of transcription initiation TADs facilitate assembly of the transcriptional machinery (RNA polymerase II and associated transcription factors) through direct binding interactions. Several lines of evidence suggest that TADs associate with three or more binding partners (coactivators) as part of this process, including components of the chromatin-remodeling machinery, the proteasome, and the Mediator complex.(15-19, 20, 21-26) However, the identity of coactivator targets in vivo remains a topic of significant debate. Thus, binding screens to identify novel TADs are difficult to implement, with only limited success with nonpeptide based molecules.(27-30) Further complicating small molecule TAD discovery is that there are few structures of natural TAD•coactivator complexes upon which molecular scaffolds could be based.(31-38) Indeed, although the prevailing model is that natural TADs interact with coactivators as amphipathic helices, there is evidence for other structural motifs.(39-41)
Figure 1. Natural and designer transcriptional activation domains (TADs).
a) Key sequences from amphipathic TADs that interact with the coactivator CBP. b) Isoxazolidine TADs (iTADs) that were designed to generically mimic their natural counterparts and up-regulate transcription when localized to a specific promoter (when R = DBD).(42-45)
We recently reported the first small molecule that reconstitutes the function of a transcriptional activation domain, isoxazolidine TAD (iTAD) 1 (Figure 1b).(42) (43) This molecule and related iTADs (2, 3) were designed to emulate amphipathic TADs, with hydrophobic and polar functional groups displayed on a conformationally constrained scaffold similar to a helix.(44, 45) However, an open question was if these small molecules were genuine TAD mimics, able to replicate the complex binding pattern(s) of their endogenous counterparts in addition to up-regulating transcription. Here a detailed study of the interaction with one binding partner, the kinase-inducible (KIX) domain of the histone acetyltransferase Creb binding protein (CBP)(15, 16, 46), reveals that the binding footprint of the iTADs and the binding-induced changes in CBP are remarkably similar to that of the endogenous TADs that target this site, including that of MLL (mixed lineage leukemia factor).(32, 47, 48) Molecular mutagenesis of the isoxazolidine scaffold further supports this model, as incorporation of functional groups into the iTAD scaffold that in the context of MLL promote or prohibit interaction with CBP similarly impact the small molecules. More broadly, we further demonstrate that iTADs exhibit a multi-partner binding profile analogous to natural TADs, interacting with several coactivator binding partners. Thus, the binding pattern and function of a natural transcriptional activation domain can be reconstituted with a small molecule despite a considerable difference in size and structural complexity.
RESULTS
iTAD 1 interacts with CBP
Although prevailing evidence suggests that transcriptional activators interact with several coactivators in the transcriptional machinery as part of pre-initiation complex assembly, the identity of those coactivators remains a topic of debate.(2) There is, however, a `short list' of transcriptional machinery proteins that through biochemical and genetic strategies have been often identified as likely targets of amphipathic transcriptional activators. Among these are CBP and the closely related p300,(15, 16, 49) several components of the Mediator complex (Med23, Med15, for example),(50-52) the SAGA chromatin-modifying complex (TRRAP(Tra1)), Taf12, and Sug2.(22-26, 53, 54) To identify potential coactivator targets we used 1 in `squelching' or competitive inhibition experiments against well-characterized activators to preliminarily test if the iTAD had a similar binding profile. Of particular note, 1 produced dose-dependent inhibition of the activator KBP 2.20, thought to function at least in part through interaction with CBP (Supp. Figure S1).(55) KBP 2.20 was originally identified in a `bottom-up' experiment as a ligand for the KIX domain of CBP.(55) Taken together, these results suggested CBP and more specifically the KIX domain of CBP as at least one cellular interaction partner of 1.
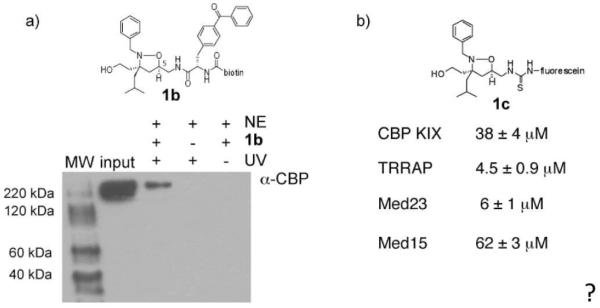
As a further assessment of this, a variant of 1 (1b) was prepared that contains a photoactivatable crosslinking group, p-benzoylphenylalanine, at the C5 position as well as a biotin tag (Figure 2, compound 1b). Upon irradiation with 365 nm light, the benzophenone moiety is converted to a diradical species that undergoes C-H insertion reactions with nearby amino acid residues.(56) Isoxazolidine 1b was combined with HeLa nuclear extracts and following irradiation, the mixture was affinity purified on avidin beads. Western blot analysis of the resulting mixture revealed seven clear binding partners (Supporting Figure S2) and one of these was confirmed as CBP (Figure 2a).
Figure 2. iTAD 1 interacts with CBP.
a) Cross-linking experiments with isoxazolidine 1b and HeLa nuclear extracts. HeLa nuclear extracts were incubated with 1b for 12 hours followed by 30 min irradiation of the mixture with 365 nm light. Following immunoprecipitation with streptavidin, Western blot analysis demonstrates that one interaction partner of 1b is CBP. In the absence of UV irradiation (final lane), no CBP is observed, consistent with a direct interaction between 1 and CBP. Lane 2 (`input' contains nuclear extracts alone. Nuclear extracts are indicated by `NE' and UV indicates irradiation with 365 nm light. b) Fluorescence polarization binding experiments with a fluorescein-labeled variant of 1 (1c) and CBP(KIX domain), TRRAP(Tra1)(3092-3524), Med23(Sur2)(352-625) or Med15(Gal11)(1-357) were used to obtain the indicated dissociation constants. Each experiment was performed in triplicate (R2>0.98) with the error indicated. See Supporting Information for additional details.
Focus on CBP: the KIX domain
CBP integrates signals from numerous transcriptional activators using several distinct domains.(15, 16) Given the competitive inhibition of KBP 2.20, it appeared most likely that 1 interacts with the so-called KIX domain of CBP, an approximately 90 residue module that consists of three α-helices and two 310 helices.(57) Originally identified in CBP/p300 this domain has now been found in several eukaryotic coactivators in mammals, plants and fungi and it is hypothesized to be a conserved TAD binding motif.(21, 58, 59) The CBP KIX domain interacts with >12 different TADs(16) and contains at least two distinct binding sites for TADs.(32, 57, 60) A larger, shallower binding site formed at the interface of the α1 and α3 helices interacts with the TADs of CREB and Myb whereas a deeper binding site on the other face of the protein formed by the sidechains of the C-terminus of α1, L12, the N-terminal half of α2 and the C-terminus of α3 is used by Jun, MLL, Tax, and Tat TADs.(31, 32, 35, 57, 60-62) It is difficult to predict which site a TAD will utilize. Thus, in addition to querying whether the iTAD interacts with the KIX domain, there was an additional question of binding site specificity. To assess the first question, a plasmid encoding hexahistidine-tagged murine KIX domain [CBP(586-672)] with a polar linker was constructed (His6KIX), and the protein was overexpressed and purified using established protocols.(35, 57) A fluorescence polarization binding experiment with a fluorescein-tagged variant of 1 and the isolated KIX domain yielded a KD of 38 μM ± 4 μM (Supp. Figure S1). This is consistent with dissociation constants for endogenous KIX ligands (KDs ranging from 300 nM to 40 μM).(35, 47, 57, 60, 63, 64)
Binding site identification
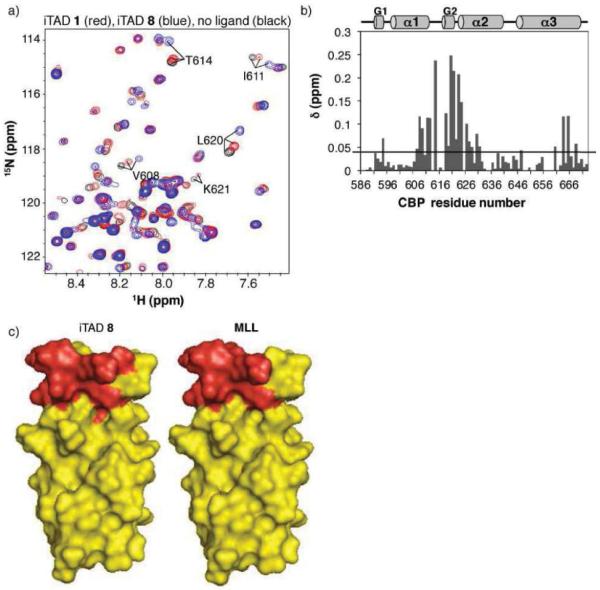
To identify the binding site(s) of the iTAD, the KIX domain was uniformly labeled with 15N and an 1H, 15N-HSQC spectrum was recorded for the protein in the absence and presence of excess isoxazolidine 1 (1d, R = N3). The spectrum of this fragment of CBP without a tag has been fully assigned,(35, 57) and this facilitated spectral assignment for this His6-tagged variant. In a titration experiment in which the concentration of 1 was gradually increased, the ligand induced gradual chemical shift perturbations, consistent with a fast exchange process. This is analogous to endogenous ligands such as Jun and Tax that also interact with the KIX domain in the fast exchange regime.(60, 61) Figure 3 shows the spectrum (red) of the protein in the presence of 5-fold excess isoxazolidine 1 overlayed on the spectrum of free protein (black). Chemical shift perturbation mapping was used to identify the iTAD binding site; this has proven to be a reliable method for characterizing the binding sites of natural TADs with the KIX domain.(31, 47, 60, 61) For the interaction with isoxazolidine 1, the average chemical shift change upon binding is 0.016 ppm and the largest shift is 0.066 ppm (for R623), comparable to changes observed with endogenous KIX ligands. For example, the average chemical shift observed for KIX residues upon binding to Jun is 0.02 ppm with the largest shifts just over 0.10 ppm.(60) The plot of chemical shifts in Figure 3b reveals that the significant perturbations occur for residues clustered in the structure indicative of specific binding. Residues experiencing significant chemical shifts map onto the C-terminus of α1, L12, G2, and the N-terminus of α2, corresponding to the MLL/Tax/Jun/Tat binding site, the smaller of the two binding sites of the CBP KIX domain (Figure 3c). Overall, the binding profile of iTAD 1 with KIX is remarkably similar to that of endogenous ligands that utilize the same site.
Figure 3. iTAD 1 binds the site on the CBP KIX domain bound by endogenous TADs MLL, Jun, Tat, and Tax.
a) The 1H, 15N-HSQC spectrum of 15N-His6KIX bound by iTAD 1 (1d, R = N3), red, is overlayed on the spectrum of free 15N-His6KIX. NMR samples were prepared with 400 μM protein in 90% H2O/10% D2O 10 mM phosphate buffer with 150 mM NaCl and 1% CD3OD; the spectrum in the presence of iTAD 1 contained a 5-fold excess of the ligand. b) The amide chemical shifts upon addition of ligand were quantitated (Δδ=[Δδ(1H)2 + 0.1Δδ(15N)2]1/2) and plotted against residue number. The average chemical shift is 0.016 ppm and the largest chemical shift is 0.066 for R623. c) The residues that experience the largest chemical shift perturbation upon binding iTAD 1, MLL and Jun are highlighted in red on the space filling diagrams of the CBP KIX domain.(47, 60) Residues experiencing chemical shifts greater than 2 standard deviations above the average are V608, A618, L620, K621, and R623. Residues experiencing shifts one standard deviation above the average are I611, T614, R624, and E665. Pymol figures were generated from 1kdx.(35)
Molecular mutagenesis impacts CBP binding and function
As outlined above, the KIX domain interacts with >12 amphipathic(16) TADs despite differences in sequence (Figure 1). At least in the case of the larger of the two binding sites significant changes in the identity and spacing of the hydrophobic side chains do not preclude binding to this domain.(65) Thus it might be predicted that iTADs with different side chains and/or side chain arrangements would maintain KIX binding ability. To test this, we examined the interaction of five additional isoxazolidines with CBP (Figure 4).
Figure 4. Additional isoxazolidines evaluated for their ability to interact with the KIX domain of CBP.
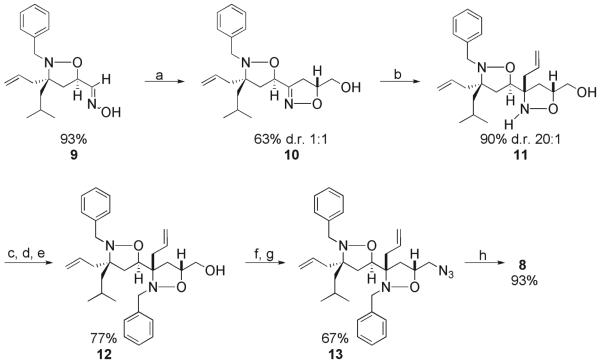
Isoxazolidine 4 has the same functional groups as 1 but in different orientation. Hydrophobic isoxazolidine 5 maintains the two key hydrophobic functional groups present in 1 (benzyl, isobutyl) but lacks the C3 hydroxyl moiety. In the case of isoxazolidine 6, the C3 substituents are identical to 1 but the N2 substituent is now a larger, biphenyl sidechain. In addition to these monomeric isoxazolidines, two dimeric versions, 7 and 8, were prepared for this study. Both of these molecules contain additional hydrophobic surface area through the incorporation of a second ring, surface area that would presumably enhance the interaction with coactivator binding surfaces. Monoisoxazolidines 4-6 were prepared in accordance with previously reported methods and characterization can be found in the Supporting Information.(44, 66-68) Bis-isoxazolidines 7 and 8 were prepared via an iterative synthetic strategy in which successive 1,3-dipolar cycloadditions were employed to install the two rings. In the case of 8, a cycloaddition with monoisoxazolidine 9 and allyl alcohol produced 10 as a 1:1 mixture of diastereomers in 63% yield (Figure 5). The diastereomer shown was isolated by HPLC purification and treated with allyl magnesium chloride to introduce a substituent at C3 of the second ring. This reaction proceeds with high selectivity (20:1). Alkylation of the nitrogen and oxidative functionalization of the allyl side chains then produced isoxazolidine 8. Similar to 1, isoxazolidine 8 functions as a TAD when localized to a promoter (Supporting Figure S3). Additional synthetic and characterization details can be found in the Supporting Information.
Figure 5. Synthesis of bis-isoxazolidine 8.
Reaction conditions: a) allyl alcohol, NaOCl, CH2Cl2, 0 °C to rt, 5h b) BF3•OEt2, allylmagnesium chloride, tol, -78 °C, 4h c) TBSOTf, Et3N, DMAP, THF, 0 °C, 2h d) BnBr, iPr2NEt, DMF, μwave e) TBAF, THF, 0 °C, 3h f) MsCl, Et3N, CH2Cl2, 0 °C, 30 min g) NaN3, DMSO, 100 °C, 12h h) OsO4, NMO, tBuOH/THF/H2O, rt, 5h.
Of these molecules, two (5 and 6) showed no interaction at concentrations up to 600 μM, 2-fold excess compared to KIX in these experiments (Supp. Figure S4). There is no evidence that these results are due to compromised solubility or aggregation. In the case of 5, for example, a 1H NMR spectrum of the molecule alone in the NMR buffer showed a sharp, well-defined spectrum analogous to that obtained in organic solvents (Supp. Figure S4). However, upon combination with 15N-labeled His6KIX, no specific interaction was observed. In addition, in a fluorescence polarization experiment with a fluorescein-labeled variant of 5 and the KIX domain no binding was detected. Analogous behavior was observed with 6. Evaluation of 7 was hindered by limited solubility in aqueous buffers; although some shifting of key KIX residues was observed at low concentrations of the small molecule, precipitation precluded examination at higher concentrations. In contrast, 4 and 8 exhibited binding behavior similar to 1. Indeed, when chemical shift perturbation analysis was carried out, the pattern for isoxazolidine 4 was nearly identical to that of 1 in terms of direction and magnitude (Supp. Figure S4). Bis-isoxazolidine 8 also specifically bound to the MLL binding site, although the detailed pattern of shifts was slightly altered compared to 1, consistent with 8 presenting a larger hydrophobic surface area (Figure 6). In addition to the residues that change upon titration of iTAD 1, perturbations observed with small molecule 8 extend along α2 and further changes are observed in the C-terminus of α3, analogous to MLL.(32, 47)
Figure 6. iTAD 8 binds the site on the CBP KIX domain bound by endogenous TADs MLL, Jun, Tat, and Tax.
iTAD 8 binds the site on the CBP KIX domain bound by endogenous TADs MLL, Tat, Tax, Jun, and others. a) A 1H, 15N-HSQC of His6KIXwas collected in the presence of excess iTAD 8 and the amide chemical shifts were quantitated (Δδ=[Δδ(1H)2 + 0.1 Δδ(15N)2]1/2) and plotted against residue number.(69) b) The average chemical shift is 0.04 ppm and the largest chemical shift is 0.25 ppm (L620). The black bar indicated the average chemical shift. c) The residues that experience the largest chemical shift upon binding iTAD 8 and MLL(47) are highlighted in red on the space filling diagrams of the CBP KIX domain. Pymol figures were generated from 1kdx.(35)
Interactions with other coactivators
Natural TADs typically interact with multiple coactivator proteins and the current model is that contact with at least three distinct transcriptional machinery proteins is a critical contributor to function.(22, 23) We thus assessed the interaction of 1 with other coactivators identified as common binding partners of endogenous amphipathic TADs. In each case fluorescence polarization binding experiments were carried out with the domain of the protein that had been previously identified as the TAD-interaction module.(52, 53, 70) iTAD 1 interacts with TRRAP(Tra1) (KD 4.5 μM) and Med23(Sur2) (KD 6 μM) with low micromolar KDs, similar to the interaction with CBP (Figure 2b and Supp. Information Figure S4). TRRAP(Tra1) is a component of the chromatin remodeling SAGA complex whereas Med23(Sur2) is a component of the Mediator complex. In addition, iTAD 1 binds to the amino terminus of Med15(Gal11) with a KD of 62 μM. In contrast, isoxazolidine 5 exhibits a different binding profile. As outlined above, 5 does not bind to the KIX domain of CBP; it does, however, interact with the Mediator protein Med23(Sur2) with a KD of 400 nM and with Med15 with a KD of 61 μM.* Thus, one distinguishing feature of iTAD 1 (and iTAD 8) is its ability to interact with the chromatin-modifying machinery in general and the KIX domain of the HAT CBP specifically.
DISCUSSION
CBP, along with the closely related coactivator p300, is ubiquitously expressed and is an important node for many signaling networks.(15, 16, 46) Deletion of CBP/p300 is embryonic lethal and loss of a single CBP allele leads to severe developmental defects.(71-73) At the heart of its importance is its ability to interact with a wide range of transcriptional activators and by doing so stimulate transcription initiation.(46) The KIX domain is one of five activator binding motifs within CBP, and it alone interacts with >12 activators through at least two binding sites.(16, 57, 60) Given this, it is perhaps not surprising that both the cross-linking and competitive inhibition experiments suggested that a small molecule transcriptional activation domain, iTAD 1, also interacts with CBP and more specifically the KIX domain (Figure 2 and Supp. Figure S1).
The HSQC experiments of iTADs 1, 4, and 8 (Figures 3, 6, and Supp. Figure S4) in complex with 15N-labeled KIX module revealed that the molecules interact with a single site within the protein, the site that is also utilized by the TADs of Jun, MLL, Tax, and Tat, among others.(47, 60-62) The iTADs discussed here are the first small molecules reported to bind this site in the KIX domain, although small molecules that bind a distinct site not known to be a target of endogenous TADs have been reported.(69) Of the natural TADs that target this site, only in the case of MLL has a solution structure in complex with the KIX domain been solved, enabling a more detailed comparison with the small molecules.(32) Within the MLL TAD only 10 amino acids (residues 2848-2857) comprise the structured region of the amphipathic helix that interacts with the KIX domain. One polar (T2857) and four hydrophobic (I2849, F2852, V2853, L2854) residues make extensive contacts with a predominantly hydrophobic binding groove (Figure 7), contributing to a KD of approximately 3 μM.(47) iTAD 1 has a smaller surface area for interaction consisting of one polar (hydroxyl at C3) and two hydrophobic (benzyl at N2 and isobutyl at C3) functional groups. The substituent at C5 (the point of attachment for DNA binding functionality in the context of activation, Figure 1b) does not contribute significantly to binding; molecules containing larger and more polar substituents (dimethylacetal, for example) show nearly identical binding characteristics. The difference in available binding surface is reflected in the reduced affinity (KD 38 μM). The three iTAD 1 functional groups are remarkably similar to those of the MLL residues that interact with the subsite (F2852, L2854 and T2857) and they also assume similar relative positions. The MLL binding site of KIX undergoes significant structural rearrangement upon binding endogenous TAD sequences;(32) this flexible nature may enable the binding site to tailor itself to the recognition of ligands of different size (iTAD 1) and functional group orientation (iTAD 4).(32) Indeed, the positional isomer of 1, iTAD 4, exhibits essentially identical binding and functional characteristics.(44) Further supporting this model, significant line broadening is observed for several resonances involved in binding to the iTADs (K621, R624, N627, for example), a characteristic of protein flexibility.
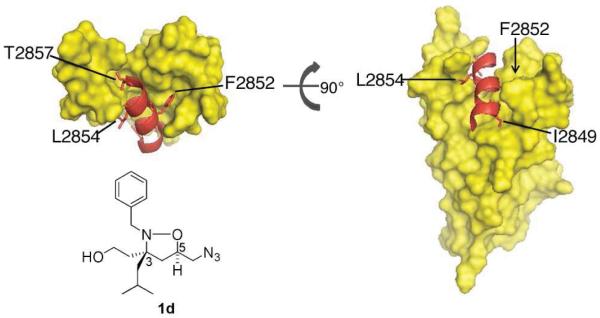
Figure 7. MLL•KIX solution structure.
The solution structure of MLL•KIX shows that one polar (T2857) and four hydrophobic (I2849, F2852, V2853, and L2854) residues make extensive contacts with KIX.(32) MLL residues F2852, L2854, and T2857 are predicted to be mimicked by iTAD 1 functional groups benzyl, isobutyl, and hydroxyl, respectively. MLL amino acid I2849 is predicted to be mimicked by a ring B substituent of 8 with ring A binding in a similar orientation to iTAD 1.
iTADs 6 and 8 have significantly larger hydrophobic surface area than 1 and 4 (Figures 1 and 4). Compound 6 has a more extended hydrophobic substituent at N2 whereas bis-isoxazolidine 8 has five substituents in addition to the azide group in the second ring. Compound 6 was not observed to bind to the KIX domain (Supp. Figure S4). In contrast bis-isoxazolidine 8 targets the KIX domain at the same site as iTADs 1 and 4 (Figure 6). Consistent with the larger size, the KIX residues that shift upon binding to 8 extend into α2 and α3. The additional perturbations along α2 are located in the same region that I2849 of MLL contacts (Figure 7). A binding model consistent with the observed chemical shift changes in α2 of the KIX domain would be ring A of iTAD 8 binding in the same orientation as iTAD 1 with one of the groups on the second ring making contacts similar to I2849 of MLL. However, additional experiments will be required to define the details of the interaction.
The ability of isoxazolidines 1 and 4-8 to interact with the KIX domain parallels the transcriptional activity of the molecules. (42-44) Compounds 1, 4, and 8 all bind the KIX domain and elicit high levels of activation when localized to DNA. Isoxazolidines 5 and 6 do not function as transcriptional activation domains and also do not detectably interact with KIX. This binding profile is consistent with a model in which the iTAD 1•KIX complex is analogous to the MLL•KIX complex and both of the hydrophobic functional groups as well as the pendant hydroxyl are required for interaction. In the case of 6, an examination of the natural TADs that interact with this KIX site reveals only a single large hydrophobic amino acid (W at residue 11 of Tat) that would be similar in size to the biphenyl of 6. Further, a F2852Y mutation in the MLL TAD abrogates binding to KIX and concomitantly reduces MLL-mediated transcription by 60%.(48) Thus, at least in the case of this TAD-binding site, large hydrophobic groups do not appear to be well tolerated. Isoxazolidine 6, however, does interact with other coactivators; it binds to Med23(Sur2), for example, with a KD of 700 nM. While iTAD 1 interacts with several coactivators, the correlation of KIX binding and ability to activate transcription for all analogs tested suggests that analogous to many natural activators CBP recruitment is an essential component of iTAD function.
Taken together, the results presented here suggest that the amphipathic iTADs are pre-programmed to exhibit a multi-partner coactivator binding profile that is analogous to natural transcriptional activation domains. The comparison with the transcriptional activation domain of MLL indicates some degree of structural mimicry, with the amphipathic mono- and bis-isoxazoldine scaffold presenting the amphipathic functional groups for interaction with the permissive binding surfaces present in coactivators. Perhaps more significant, however, is that at least in the case of the interaction with CBP, iTAD 8 reproduces nearly exactly the binding mode of the MLL TAD despite a considerable size differential. This suggests several future applications of isoxazolidines as transcriptional regulators. There is significant evidence that the two KIX sites are bound cooperatively in vitro and emerging evidence that a number of metazoan promoters utilize two KIX-binding activators to recruit CBP.(32, 48, 60, 74-76) For example, the MLL•KIX complex interacts with the TAD of Myb approximately 2-fold more tightly than the KIX domain alone.(32) Thus, the iTADs may synergize with activators such as CREB that target the second, larger binding site of KIX, enhancing their activity. Further, these data suggest that isoxazolidines, both activating and non-activating, should be excellent starting points for the design of inhibitors for activator-coactivator interactions, long a challenging endeavor. This will likely require, however, molecules that bind more tightly than iTAD 1. The observation that small structural changes alter the binding profile of the small molecules also implies that some degree of specificity for a given activator or activator class may indeed be achievable within this framework.
METHODS
Isoxazolidines 3, 5, and 6 were prepared as previously reported.(43, 45) Med23(352-625) and Med15(1-357) were bacterially expressed and purified as previously described.(52, 77) Fluorescence polarization binding experiments were conducted as previously reported.(78)
Photocrosslinking
HeLa nuclear extracts (25 mL, 13.5 mg/mL, Promega) were incubated with 30 μM compound 1b with gentle mixing in Buffer A (10 mM PBS, pH 7.4, 10% (v/v) glycerol) plus 1% (v/v) DMSO at 4 °C for 12 h. Samples were irradiated with a handheld UV lamp (365 nm) at 4 °C for 30 min. The irradiated solution was enriched for crosslinked products using Neutravidin beads (50 μL, Pierce) in Buffer A (plus 1% (v/v) BSA, 0.1% (v/v) Nonidet NP-40) by incubating for 1 h at 4 °C. The beads were then washed 3x with Wash Buffer (10 mM PBS, pH 7.4, 10% (v/v) glycerol, 0.1% (v/v) Nonidet NP-40), resuspended in elution buffer (10 mM PBS, pH 7.4, 10% (v/v) glycerol, 25% (v/v) 1x Nu-PAGE loading dye (Invitrogen)), and heated at 95 °C for 10 min. The eluted samples were subsequently analyzed by Western blots using standard conditions. The mouse monoclonal anti-CBP (SC-369) and horseradish peroxidase-labeled goat anti-mouse antibodies were obtained from Santa Cruz Biotechnology.
His6KIX plasmid preparation and protein expression
A plasmid encoding His6KIX, was generated by amplifying the DNA sequence encoding the KIX domain residues 586-672 of mouse CBP from pGEX KT KIX 10-672 and insertion into pRSET-B (Invitrogen) at restriction enzyme sites HindIII and BglII. For protein expression, the plasmid was transformed into Rosetta2(DE3) pLysS E. coli (Novagen) and grown in M9 minimal media containing 15N-labelled NH4Cl (for NMR studies) or LB media. After an OD600 of 0.6 was reached (37 °C, 250 rpm), the cultures were cooled to 25 °C for 30 min, and expression was induced with 0.1 mM IPTG for 12 h (250 rpm). The His-tagged protein was affinity purified using Ni-NTA beads (Qiagen) and buffer exchanged to CH3CN/H2O using PD-10 columns (GE Healthcare) before being lyophilized. The lyophilized protein was utilized immediately for NMR experiments.
1H, 15N-HSQC experiments
The uniformly 15N-labelled His6KIX protein was prepared as a 300-400 μM solution in 90% H2O/10% D2O 10 mM phosphate buffer with 150 mM NaCl at pH 7.2. Samples containing small molecule ligands were prepared by adding 2.0-5.0 equivalents of the small molecule as a solution in CD3OD to achieve a final CD3OD concentration of 1%. Samples recorded in the absence of small molecule ligands also contained 1% CD3OD. 1H, 15N-heteronuclear single quantum coherence experiments were recorded at 27 °C on an Avance Bruker 600 MHz NMR spectrometer equipped with a triple-resonance 5 mm cryogenic probe. Data was processed using NMRPIPE and analyzed using Sparky.
Tra1 plasmid preparation and protein expression
A plasmid encoding Tra1(3092-3524) fused to the maltose binding protein was generated by amplifying the Tra1 DNA sequence encoding amino acid residues 3092-3524 from S. cerevisiae genomic DNA and insertion into pMal-c2g (New England Biolabs) using standard molecular biology techniques. For protein expression, the plasmid was transformed into Rosetta2(DE3) pLysS E. coli (Novagen) and grown in Select APS Super Broth (Difco). After an OD600 of 0.3 was reached (37 °C, 300 rpm), the cultures were cooled to 16 °C for 1 h (100 rpm), and expression was induced with 0.1 mM IPTG for 12 h (250 rpm). The MBP-tagged protein was isolated using amylose resin (New England Biolabs). The protein solution was stored in Storage buffer (10 mM PBS, pH 7.0, 1 mM DTT, 10% glycerol (v/v), and 0.01% NP-40) at -80 °C until needed.
Supplementary Material
Acknowledgement
A.K.M. thanks the NIH (GM65330), the NSF (CAREER), and Novartis (Novartis Young Investigator award) for support of this work. S.J.B. and A.R.M. were supported by the UM CBI training program (GM08597) and S.J.B. by an ACS Division of Medicinal Chemistry Graduate Fellowship sponsored by Wyeth. C.A.B. was supported by the UM Pharmaceutical Sciences Training Program (GM007767). We thank Dr. J.P. Desaulnier for the preparation of compound 6 and for Med23 and R. J. Casey for the preparation of 1d. We thank Prof. Mark Montminy for the generous gift of pGEX KT KIX 10-672. We are grateful to E. Dethoff and M. Doucleff for technical assistance and helpful discussions.
Footnotes
It was not possible to obtain a KD for the interaction with TRRAP under these conditions.
REFERENCES
- 1.Ptashne M, Gann A. Genes & Signals. Cold Spring Harbor Laboratory; New York: 2001. [Google Scholar]
- 2.Mapp AK, Ansari AZ. A TAD further: exogenous control of gene activation. ACS Chem Biol. 2007;2:62–75. doi: 10.1021/cb600463w. [DOI] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Aksien LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji JF, Dudoit S, Ng IOL, van de Rijn M, Botstein D, Brown PO. Gene expression patterns in human liver cancers. Mol. Biol. Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darnell JE. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 6.Pandolfi PP. Transcription therapy for cancer. Oncogene. 2001;20:3116–3127. doi: 10.1038/sj.onc.1204299. [DOI] [PubMed] [Google Scholar]
- 7.Chene P. Inhibiting the p53-MDM2 interaction: An important target for cancer therapy. Nat Rev Cancer. 2003;3:102–109. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 8.Bargonetti J, Manfredi JJ. Multiple roles of the tumor suppressor p53. Curr Opin Oncol. 2002;14:86–91. doi: 10.1097/00001622-200201000-00015. [DOI] [PubMed] [Google Scholar]
- 9.Karin M, Cao Y, Greten FR, Li ZW. NF-κB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 10.Karin M, Lin A. NF-κB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 11.Denison C, Kodadek T. Small-molecule-based strategies for controlling gene expression. Chemistry & Biology. 1998;5:R129–R145. doi: 10.1016/s1074-5521(98)90167-3. [DOI] [PubMed] [Google Scholar]
- 12.Ansari AZ. Regulating gene expression: The design of synthetic transcriptional regulators. Curr Org Chem. 2001;5:903–921. [Google Scholar]
- 13.Ansari AZ, Mapp AK. Modular design of artificial transcription factors. Curr. Opin. Chem. Biol. 2002;6:765–772. doi: 10.1016/s1367-5931(02)00377-0. [DOI] [PubMed] [Google Scholar]
- 14.Arndt HD. Small molecule modulators of transcription. Angew Chem Int Ed Engl. 2006;45:4552–4560. doi: 10.1002/anie.200600285. [DOI] [PubMed] [Google Scholar]
- 15.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 16.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 17.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN'beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 18.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 19.Marr MT, 2nd, Isogai Y, Wright KJ, Tjian R. Coactivator cross'talk specifies transcriptional output. Genes Dev. 2006;20:1458–1469. doi: 10.1101/gad.1418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, Macol C, Iyer L, Tjian R, van den Heuvel S, Hart AC, Wagner G, Naar AM. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 22.Fishburn J, Mohibullah N, Hahn S. Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell. 2005;18:369–378. doi: 10.1016/j.molcel.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 23.Reeves WM, Hahn S. Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol. 2005;25:9092–9102. doi: 10.1128/MCB.25.20.9092-9102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang C, Gonzalez F, Rothermel B, Sun L, Johnston SA, Kodadek T. The Gal4 activation domain binds Sug2 protein, a proteasome component, in vivo and in vitro. J Biol Chem. 2001;276:30956–30963. doi: 10.1074/jbc.M102254200. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- 26.Ard PG, Chatterjee C, Kunjibettu S, Adside LR, Gralinski LE, McMahon SB. Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol Cell Biol. 2002;22:5650–5661. doi: 10.1128/MCB.22.16.5650-5661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alluri P, Liu B, Yu P, Xiao X, Kodadek T. Isolation and characterization of coactivator'binding peptoids from a combinatorial library. Mol Biosyst. 2006;2:568–579. doi: 10.1039/b608924k. [DOI] [PubMed] [Google Scholar]
- 28.Xiao X, Yu P, Lim HS, Sikder D, Kodadek T. Design and synthesis of a cell' permeable synthetic transcription factor mimic. J Comb Chem. 2007;9:592–600. doi: 10.1021/cc070023a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B, Alluri PG, Yu P, Kodadek T. A potent transactivation domain mimic with activity in living cells. J Am Chem Soc. 2005;127:8254–8255. doi: 10.1021/ja0515295. [DOI] [PubMed] [Google Scholar]
- 30.Alluri PG, Reddy MM, Bachhawat'Sikder K, Olivos HJ, Kodadek T. Isolation of protein ligands from large peptoid libraries. J Am Chem Soc. 2003;125:13995–14004. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 31.Zor T, De Guzman RN, Dyson HJ, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of c'Myb. J Mol Biol. 2004;337:521–534. doi: 10.1016/j.jmb.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 32.De Guzman RN, Goto NK, Dyson HJ, Wright PE. Structural basis for cooperative transcription factor binding to the CBP coactivator. J Mol Biol. 2006;355:1005–1013. doi: 10.1016/j.jmb.2005.09.059. [DOI] [PubMed] [Google Scholar]
- 33.Parker D, Jhala US, Radhakrishnan I, Yaffe MB, Reyes C, Shulman AI, Cantley LC, Wright PE, Montminy M. Analysis of an activator : coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol Cell. 1998;2:353–359. doi: 10.1016/s1097-2765(00)80279-8. [DOI] [PubMed] [Google Scholar]
- 34.Jonker HR, Wechselberger RW, Boelens R, Folkers GE, Kaptein R. Structural properties of the promiscuous VP16 activation domain. Biochemistry. 2005;44:827–839. doi: 10.1021/bi0482912. [DOI] [PubMed] [Google Scholar]
- 35.Radhakrishnan I, Pérez'Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Structural Analyses of CREB'CBP Transcriptional Activator'Coactivator Complexes by NMR Spectroscopy: Implication for Mapping the Boundaries of Structural Domains. J Mol Biol. 1999;287:859–865. doi: 10.1006/jmbi.1999.2658. [DOI] [PubMed] [Google Scholar]
- 36.Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 37.Uesugi M, Verdine GL. The alpha'helical FXX Phi Phi motif in p53: TAF interaction and discrimination by MDM2. Proc Natl Acad Sci U S A. 1999;96:14801–14806. doi: 10.1073/pnas.96.26.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langlois C, Mas C, Di Lello P, Jenkins LM, Legault P, Omichinski JG. NMR structure of the complex between the Tfb1 subunit of TFIIH and the activation domain of VP16: structural similarities between VP16 and p53. J Am Chem Soc. 2008;130:10596–10604. doi: 10.1021/ja800975h. [DOI] [PubMed] [Google Scholar]
- 39.Van Hoy M, Leuther KK, Kodadek T, Johnston SA. The acidic activation domains of the GCN4 and GAL4 proteins are not alpha helical but form beta sheets. Cell. 1993;72:587–594. doi: 10.1016/0092-8674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- 40.Leuther KK, Salmeron JM, Johnston SA. Genetic evidence that an activation domain of GAL4 does not require acidity and may form a beta sheet. Cell. 1993;72:575–585. doi: 10.1016/0092-8674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- 41.Ferreira ME, Hermann S, Prochasson P, Workman JL, Berndt KD, Wright AP. Mechanism of transcription factor recruitment by acidic activators. J Biol Chem. 2005;280:21779–21784. doi: 10.1074/jbc.M502627200. [DOI] [PubMed] [Google Scholar]
- 42.Minter AR, Brennan BB, Mapp AK. A small molecule transcriptional activation domain. J Am Chem Soc. 2004;126:10504–10505. doi: 10.1021/ja0473889. [DOI] [PubMed] [Google Scholar]
- 43.Rowe SP, Casey RJ, Brennan BB, Buhrlage SJ, Mapp AK. Transcriptional up-regulation in cells mediated by a small molecule. J Am Chem Soc. 2007;129:10654–10655. doi: 10.1021/ja0736865. [DOI] [PubMed] [Google Scholar]
- 44.Buhrlage SJ, Brennan BB, Minter AR, Mapp AK. Stereochemical Promiscuity in Artificial Transcriptional Activators. J Am Chem Soc. 2005;127:12456–12457. doi: 10.1021/ja0536567. [DOI] [PubMed] [Google Scholar]
- 45.Casey RJ, Desaulniers JP, Hojfeldt JW, Mapp AK. Expanding the repertoire of small molecule transcriptional activation domains. Bioorg Med Chem. 2008 doi: 10.1016/j.bmc.2008.02.045. [DOI] [PubMed] [Google Scholar]
- 46.Kasper LH, Fukuyama T, Biesen MA, Boussouar F, Tong C, de Pauw A, Murray PJ, van Deursen JM, Brindle PK. Conditional Knockout Mice Reveal Distinct Functions for the Global Transcriptional Coactivators CBP and p300 in T-Cell Development. Mol Cell Biol. 2006;26:789–809. doi: 10.1128/MCB.26.3.789-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goto NK, Zor T, Martinez'Yamout M, Dyson HJ, Wright PE. Cooperativity in Transcription Factor Binding to the Coactivator CREB-Binding Protein (CBP) J Biol Chem. 2002;227:43168–43174. doi: 10.1074/jbc.M207660200. [DOI] [PubMed] [Google Scholar]
- 48.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind Cooperatively to the Nuclear Coactivator CREB-Binding Protein. Mol Cell Biol. 2001;21:2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 50.Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 51.Malik S, Baek HJ, Wu W, Roeder RG. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol Cell Biol. 2005;25:2117–2129. doi: 10.1128/MCB.25.6.2117-2129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asada S, Choi Y, Yamada M, Wang SC, Hung MC, Qin J, Uesugi M. External control of Her2 expression and cancer cell growth by targeting a Ras-linked coactivator. Proc Natl Acad Sci U S A. 2002;99:12747–12752. doi: 10.1073/pnas.202162199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 54.Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frangioni JV, LaRiccia LM, Cantley LC, Montminy MR. Minimal activators that bind to the KIX domain of p300/CBP identified by phage display screening. Nature Biotechnology. 2000;18:1080–1085. doi: 10.1038/80280. [DOI] [PubMed] [Google Scholar]
- 56.Dorman G, Prestwich GD. Benzophenone photophores in biochemistry. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 57.Radhakrishnan I, Pérez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution Structure of the KIX Domain of CBP Bound to the Transactivation Domain of CREB: A Model for Activator:Coactivator Interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 58.Thakur JK, Arthanari H, Yang F, Pan SJ, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E, Struhl K, Moye-Rowley WS, Cormack BP, Wagner G, Näär AM. A Nuclear Receptor-Like Pathway Regulating Multidrug Resistance in Fungi. Nature. 2008;452:604. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- 59.Novatchkova M, Eisenhaber F. Linking transcriptional mediators via the GACKIX domain super family. Current biology : CB. 2004;14:R54–55. doi: 10.1016/j.cub.2003.12.042. [DOI] [PubMed] [Google Scholar]
- 60.Campbell KM, Lumb KJ. Structurally Distinct Modes of Recognition of the KIX Domain of CBP by Jun and CREB. Biochemistry. 2002;41:13956–13964. doi: 10.1021/bi026222m. [DOI] [PubMed] [Google Scholar]
- 61.Vendel AC, Lumb KJ. Molecular recognition of the human coactivator CBP by the HIV-1 transcriptional activator Tat. Biochemistry. 2003;42:910–916. doi: 10.1021/bi0270034. [DOI] [PubMed] [Google Scholar]
- 62.Vendel AC, Lumb KJ. NMR Mapping of the HIV-1 Tat Interaction Surface of the KIX Domain of the Human Coactivator CBP. Biochemistry. 2004;43:904–908. doi: 10.1021/bi035612l. [DOI] [PubMed] [Google Scholar]
- 63.Vendel AC, McBryant SJ, Lumb KJ. KIX-Mediated Assembly of the CBP-CREB-HTLV-1 Tax Coactivator-Activator Complex. Biochemistry. 2003;42:12481–12487. doi: 10.1021/bi0353023. [DOI] [PubMed] [Google Scholar]
- 64.Zor T, Mayr BM, Dyson HJ, Montminy MR, Wright PE. Roles of Phosphorylation and Helix Propensity in the Binding of the KIX domain of CREB-binding Protein by Constitutive (c-Myb) and Inducible (CREB) Activators. J Biol Chem. 2002;277:42241–42248. doi: 10.1074/jbc.M207361200. [DOI] [PubMed] [Google Scholar]
- 65.Rowe SP, Mapp AK. Assessing the permissiveness of transcriptional activator binding sites. Biopolymers. 2008;89:578–581. doi: 10.1002/bip.20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minter AR, Fuller AA, Mapp AK. A concise approach to structurally diverse beta-amino acids. J Am Chem Soc. 2003;125:6846–6847. doi: 10.1021/ja0298747. [DOI] [PubMed] [Google Scholar]
- 67.Kanemasa S, Nishiuchi M, Kamimura A, Hori K. 1st Successful Metal Coordination Control in 1,3-Dipolar Cycloadditions - High'Rate Acceleration and Regiocontrol and Stereocontrol of Nitrile Oxide Cycloadditions to the Magnesium Alkoxides of Allylic and Homoallylic Alcohols. J Am Chem Soc. 1994;116:2324–2339. [Google Scholar]
- 68.Bode JW, Fraefel N, Muri D, Carreira EM. A general solution to the modular synthesis of polyketide building blocks by Kanemasa hydroxy-directed nitrile oxide cycloadditions. Angew Chem Int Ed. 2001;40:2082–2085. doi: 10.1002/1521-3773(20010601)40:11<2082::AID-ANIE2082>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 69.Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, Zor T, Gardner KH, Montminy M. Identification of small-molecule antagonists that inhibit an activator: coactivator interaction. Proc Natl Acad Sci U S A. 2004;101:17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeong CJ, Yang SH, Xie Y, Zhang L, Johnston SA, Kodadek T. Evidence that Gal11 protein is a target of the Gal4 activation domain in the mediator. Biochemistry. 2001;40:9421–9427. doi: 10.1021/bi010011k. [DOI] [PubMed] [Google Scholar]
- 71.Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RCM, Masuno M, Tommerup N, Van ommen GB, Goodman RH, Peters DJM, Breuning MH. Rubenstein'Taybi Syndrome Caused by Mutations in the Transcriptional Co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 72.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal Skeletal Patterning in Embryos Lacking a Single Cbp Allele: A Partial Similarity with Rubenstein-Taybi Syndrome. Proc Natl Acad Sci U S A. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanaka Y, Naruse I, Hongo T, Xu M, Nakahata T, Maekawa T, Ishii S. Extensive Brain Hemmorage and Embryonic Lethality in a Mouse Null Mutant of CREb-binding Protein. Mech. Dev. 2000;95:133–145. doi: 10.1016/s0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 74.Geiger TR, Sharma N, Kim YM, Nyborg JK. The Human T-Cell Leukemia Virus Type 1 Tax Protein Confers CBP/p300 Recruitment and Transcriptional Activation Properties to Phosphorylated CREB. Mol Cell Biol. 2008;28:1383–1392. doi: 10.1128/MCB.01657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramírez JA, Nyborg JK. Molecular Characterization of HTLV-1 Tax Interaction with the KIX Domain of CBP/p300. J Mol Biol. 2007;372:958–969. doi: 10.1016/j.jmb.2007.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghee M, Baker H, Miller JC, Ziff EB. AP-1, CREB and CBP Transcription Factors Differentially Regulate the Tyrosine Hydroxylase Gene. Mol Brain Res. 1998:101–114. doi: 10.1016/s0169-328x(97)00370-7. [DOI] [PubMed] [Google Scholar]
- 77.DelProposto J, Majmudar CY, Smith JL, Brown WC. Mocr: a novel fusion tag for enhancing solubility that is compatible with structural biology applications. Protein Expr Purif. 2009;63:40–49. doi: 10.1016/j.pep.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Z, Belanger G, Brennan BB, Lum JK, Minter AR, Rowe SP, Plachetka A, Majmudar CY, Mapp AK. Targeting the transcriptional machinery with unique artificial transcriptional activators. J Am Chem Soc. 2003;125:12390–12391. doi: 10.1021/ja036685v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.