Abstract
The low-density lipoprotein (LDL) receptor-related protein (originally called LRP, but now referred to as LRP1) is a large endocytic receptor that is widely expressed in several tissues. LRP1 is a member of the LDL receptor family that plays diverse roles in various biological processes including lipoprotein metabolism, degradation of proteases, activation of lysosomal enzymes and cellular entry of bacterial toxins and viruses. Deletion of the LRP1 gene leads to lethality in mice, revealing a critical, but as of yet, undefined role in development. Tissue-specific gene deletion studies reveal an important contribution of LRP1 in the vasculature, central nervous system, in macrophages and in adipocytes. Three important properties of LRP1 dictate its diverse role in physiology: first, its ability to recognize more than thirty distinct ligands; second, its ability to bind a large number of cytoplasmic adaptor proteins via determinants located on its cytoplasmic domain in a phosphorylation-specific manner; and third, its ability to associate with and modulate the activity of other transmembrane receptors such as integrins and receptor tyrosine kinases.
I. Introduction
Protease activity in the blood is carefully regulated by a variety of proteinase inhibitors which are usually found circulating at high concentrations in the plasma. Early work led to the idea that once a proteinase forms a complex with its inhibitor, it is cleared from the circulation via a receptor system. Proof of this concept was demonstrated by showing that the proteinase inhibitor, α2-macroglobulin (α2M), was rapidly cleared by a liver receptor after forming a complex with a protease (73). These and other studies provided evidence for the existence of a receptor responsible for removal of α2M-proteinase complexes, and, using affinity chromatography approaches, two groups isolated the receptor responsible for the clearance of these complexes (3,169).
While this work was ongoing, a large hepatic receptor with remarkable resemblance to the LDL-receptor was identified (92). This receptor, originally termed the LDL receptor-related protein (LRP), but now termed LRP1 or occasionally CD91, was shown to bind to apolipoprotein E (12), raising the possibility that LRP1 might function in lipoprotein metabolism as a chylomicron remnant receptor. Sequencing studies soon revealed that the α2M receptor was identical to LRP1 (124,264), revealing that LRP1 is capable of interacting with more than one ligand. In fact, since its original discovery, LRP1 has been shown to bind to a number of ligands with high affinity (Table I) thereby impacting a variety of biological processes (93). Deletion of the LRP1 gene in mice leads to lethality demonstrating an essential, but still undefined, role during development (90). While originally believed to function exclusively as an endocytic receptor, LRP1 is now thought to also function in signaling pathways, and data reveal its ability to interact with other cellular receptors such as the PDGFR-β and integrins to protect the vasculature by modulating the response of smooth muscle cells to growth factors (23,24,142,184), to regulate cell migration by modulating integrin function (33,48,201,253), and to modulate the integrity of the blood brain barrier (213,305). This review outlines the structural organization of LRP1, its trafficking properties and the role of chaperones in this process, and this receptor’s diverse roles in biology.
Table I.
Ligands known to bind to LRP1
| Proteins involved in lipoprotein metabolism | ||
| apolipoprotein E-enriched lipoproteins (chylomicron and VLDL remnants) | ||
| β-VLDL | hepatic lipase | |
| lipoprotein lipase | sphingolipid activator protein | |
| Proteases and protease/inhibitor complexes | ||
| α2M* & α2M* protease complexes | ||
| pregnancy zone protein-protease complexes | ||
| aprotinin | pro-uPA, uPA | |
| uPA/PAI-1 complexes | tPA | |
| tPA/PAI-1 complexes | thrombin/PAI-1 complexes | |
| thrombin/anti-thrombin III | thrombin/heparin cofactor II | |
| thrombin/protease nexin-1 | neuroserpin | |
| neuroserpin/tPA complexes | elastase/α1-anti-trypsin | |
| C1s/C1q inhibitor | protease/protein C inhibitor | |
| MMP-9 | MMP-13 | |
| TSP-2/MMP-2 complexes | TFPI | |
| factor VIIa/TFPI | fVIII/fVIIIa | |
| factor Ixa | fXIa/protease nexin-1 | |
| β-amyloid precursor protein (KPI containing isoforms) | ||
| Matrix proteins | ||
| thrombospondin-1 | thrombospondin-2 | |
| fibronectin | ||
| Intracellular proteins | ||
| RAP | HIV Tat protein | |
| Calreticulin | ||
| Growth factors | ||
| PDGF | midkine | |
| connective tissue growth factor (CTGF/CCN2) | ||
| transforming growth factor-β | ||
| Others | ||
| circumsporozoite protein | gentamicin | |
| lactoferrin | polymycin B | |
| ricin A | Pseudomonas exotoxin A | |
| saposin | complement C3 | |
| rhinovirus | collectins (via calreticulin) | |
| β peptide (monomer) | ||
II. Structural organization of LRP1
LRP1 is a member of the LDL receptor family, which contains several structurally homologous receptors that are composed of modular structures. This receptor family includes seven family members that are closely related and include the LDL receptor, very low density lipoprotein (VLDL) receptor, apoE receptor 2, multiple epidermal growth factor-like domains 7 (MEGF7), glycoprotein 330 (gp330/megalin/LRP2), LRP1 and LRP1B (Figure 1). In addition, the family also includes additional members that are more distantly related, such as LRP5, LRP6 and SorLa/LRP11. Like other members of the LDL receptor family, the modular structures within LRP1 include cysteine-rich complement-type repeats, EGF repeats, β-propeller domains, a transmembrane domain and a cytoplasmic domain.
Figure 1. Modular domain organization of LDL receptor family members.
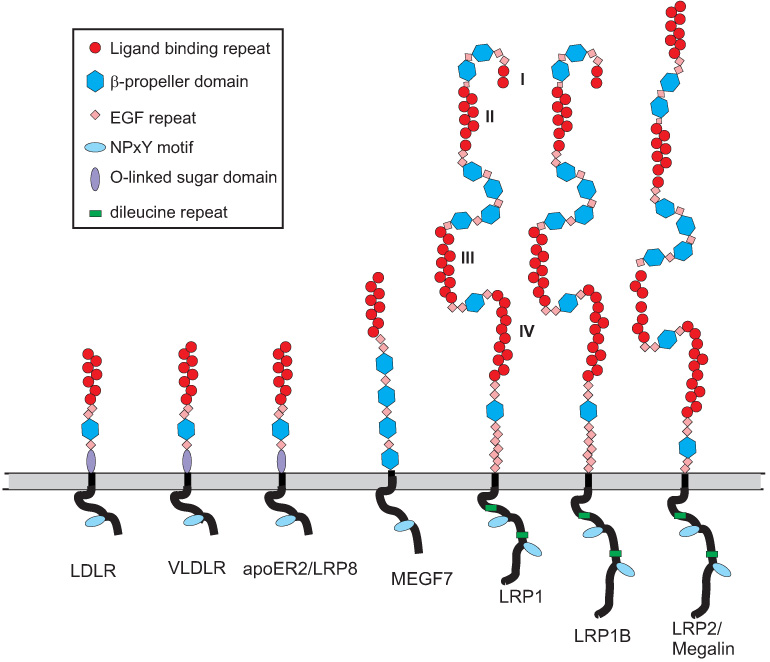
In LRP1, the four clusters of complement-type repeats are numbered I – IV.
A. Cysteine-rich complement-type repeats
All members of this receptor family contain clusters of two or more cysteine-rich complement-type repeats (CR) that are also commonly referred to as ligand-binding repeats, since most of the ligands bind to these repeats. The first insight into the folding properties of CR came from the NMR spectroscopy study of Daly et al. (50). This group solved the structure of the first repeat of the LDL receptor (CR1) revealing that this module consists of a β-hairpin structure followed by a series of β-turns. Subsequently, a crystal structure of CR5 from the LDL receptor was solved (65) revealing that the module forms a cage surrounding a calcium ion that stabilizes the structure. Since these early studies, a number of additional structures of CR from various members of the LDL receptor family have been reported, and include CR3 (60), CR7 (248), CR5–CR6 (104) and CR8 (97) from LRP1 (Figure 2A), and CR2 (49), CR1–CR2 (125), and CR6 (188) from the LDL receptor. In LRP1, the CR are localized into regions as clusters and are termed cluster I–IV, each containing variable numbers of CR. Binding experiments indicate that most LRP1 ligands bind to clusters II and IV (183,292).
Figure 2. Structure of modules from LDL receptor family members.
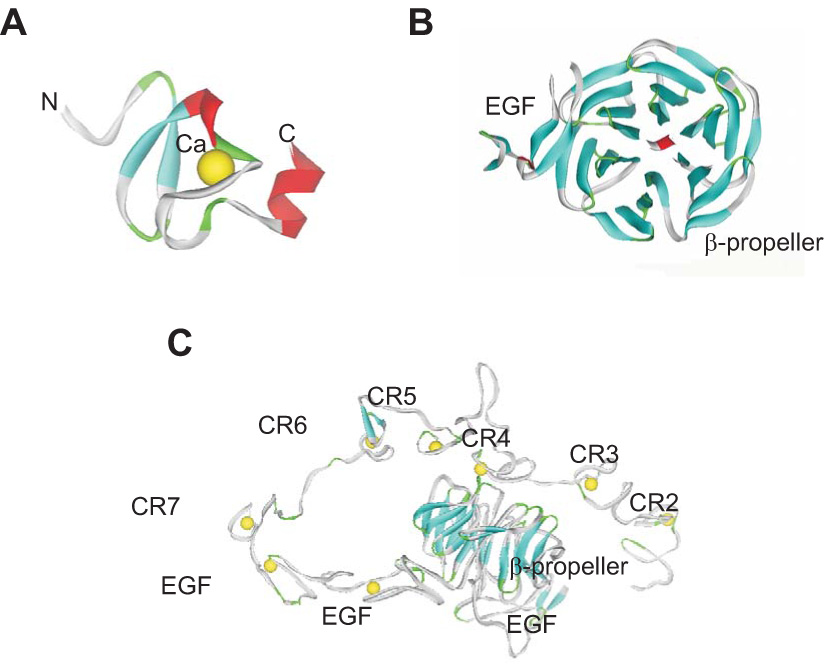
A. X-ray structure of CR7 from LRP1 (248) showing the basic folding of these modules with the structural calcium residue. B. X-ray structure of EGF and β-propeller (YWTD) domain from the LDL receptor (105) showing the six-bladed β-propeller domain. C. X-ray structure of the LDL receptor ectodomain solved at pH 5.2 (236) showing the interaction of CR4 and CR5 with the β-propeller domain at this reduced pH.
B. EGF and β-propeller (YWTD) domains
In addition to the CR, all LDL receptor family members contain one or more regions that are homologous to the EGF precursor which consists of two cysteine-rich EGF repeats, a YWTD repeat predicted from modeling to be folded as a β-propeller domain (254), followed by a third EGF-like repeat. Crystallization of the β-propeller domain along with the C-terminal EGF repeat from the LDL receptor (105) confirmed that the YWTD repeat forms a six-bladed β-propeller that packs tightly against the C-terminal EGF module (Figure 2B). The function of this region was discovered when investigators deleted it from the LDL (51) and VLDL receptor (162) and found that the mutant receptors failed to release their ligands in the low pH environment of the endosomal compartments. The structural basis for the involvement of the β-propeller domains in ligand uncoupling was finally understood when a crystal structure of the LDL receptor extracellular domain at pH 5.3 was solved (236). In this structure, shown in Figure 2C, CR2 through CR7 fold back over the two EGF-repeats and the β-propeller domain. At this low pH, CR4 and CR5 were found to associate with the β-propeller domain via their calcium-binding loop. This observation led to the proposal that the β-propeller domain functions as an alternate ligand for CR4 and CR5, which bind in a calcium-dependent manner promoting ligand release.
C. Transmembrane and cytoplasmic domains
Each member of the LDL receptor family contains a single-pass transmembrane domain and a cytoplasmic domain of varying length. In the case of LRP1, the cytoplasmic domain consists of 100 amino acid residues and includes two dileucine (LL) motifs and two NPxY motifs. The terminal NPxY motif is tyrosine phosphorylated by the PDGF receptor-β(23,142), by connective tissue growth factor (CTGF, also known as CCN2) (302) and by v-Src (6). The LRP1 cytoplasmic domain interacts with numerous adaptor molecules (Table II) (80,274) including Shc, disabled and Fe65 which are involved in directing cellular traffic or in cell signaling events. Additionally, LRP1 has been shown to undergo regulated intramembrane proteolysis (154), and, in vitro, its intracellular domain (LRP1-ICD) has been implicated in transcriptional modulation (113).
Table II.
Adaptor proteins known to bind to the cytoplasmic domain of LRP1
| Disabeled-1 (Dab1) | Src activation, neuronal migration |
| Shc | signal transduction by protein-tyrosine kinases |
| PKCα | proliferation, apoptosis, differentiation and motility |
| FE65 | actin, APP processing |
| PSD95 | coupling to NMDA receptors |
| SEMCAP-1 | axon guidance |
| JIP1, JIP2 | MAPK pathway |
| GULP | phagocytosis |
| Talin-like protein | coupling to actin cytoskeleton |
| OMP25 | mitochondrial transport |
| CAPON | NO synthase |
| PIP4,5 kinase like protein | Inositol signaling |
| ICAP1 | Integrin-mediated signaling |
| Cbl | E3 ligase, receptor tyrosine kinase downregulation |
III. Cellular trafficking of LRP1 and the role of the receptor associated protein
Because LRP1 recognizes such a wide variety of different ligands, mechanisms exist to prevent newly translated LRP1 from prematurely associating with ligands in the endoplasmic reticulum (ER) which leads to aggregation and degradation instead of proper targeting to the plasma membrane. A chaperone, termed the receptor associated protein (RAP), binds tightly to LRP1 and other members of the LDL receptor family at neutral pH values and antagonizes ligand binding while these receptors are in the ER enabling them to be successfully delivered to the plasma membrane.
A. Discovery of RAP
RAP was discovered as a protein that co-purified with LRP1 by ligand affinity chromatography (3,264). While it was originally thought to represent the carboxy-terminal region of the rat Heyman nephritis antigen (LRP2/megalin) (209), subsequent work revealed that RAP is a distinct ER-resident protein (263) that binds tightly to multiple sites on LRP1 and prevents ligands from binding to this receptor (91,289).
B. Structure of RAP
Attempts to crystallize the entire RAP molecule have not been successful, and our knowledge of the structure is derived from solved structures of individual domains. A three-domain structure of RAP was originally proposed by Bu and colleagues (30) based on the prediction of an internal triplication in the primary structure of RAP. Experimental evidence supporting this proposal was later obtained by Ellgaard et al. (62) and Lazic et al. (129) who prepared recombinant fragments representing domains 1, 2 and 3, and showed that the functional integrity of these domains is preserved when isolated. The structure of domain 1 (D1) (187,298), domain 2 (D2) (130) and domain 3 (D3) (131) of RAP were solved using NMR spectroscopy (Figure 3A). RAP D3 was also solved by X-ray crystallography (68) as a complex with CR4 and CR5 of the LDL receptor. These studies suggest that each RAP domain can be represented by a three-bundle helix connected by flexible loops. In D1, three distinct α-helices are present and consist of residues 23–35 (α1), 39–65 (α2) and 72–88 (α3). D2 is also comprised of three α-helices consisting of the residues 117–127 (α4), 132–161 (α5), and 184–210 (α6) (the α-helices are numbered in the context of the full length RAP). The N-terminal peptide segment of D2 comprising residues 101–116 is flexible and disordered. The linker between α4 and α5 is well defined, whereas the linker between α5 and α6 is 23 amino acids long and is disordered and susceptible to protease digestion (219). The D3 structure reveals that this RAP domain is also composed of a three-helical bundle containing a short helix followed by two longer helices. These helices consist of residues 222–230 (α7), 238–274 (α8), and 281–315 (α9). D1, D2 and D3 show a remarkably similar topology. Each domain is stabilized by hydrophobic interactions within the core of each structure, with the relative arrangement of the three helices in each domain is mainly determined by a number of hydrophobic contacts. It is noteworthy that the linkers connecting the two long helices in D1 and D3 are short and relatively well structured.
Figure 3. A. NMR structure of RAP domains 1 (D1) (298), D2 (130), and D3 (131) showing the three helical bundle organization of each domain.
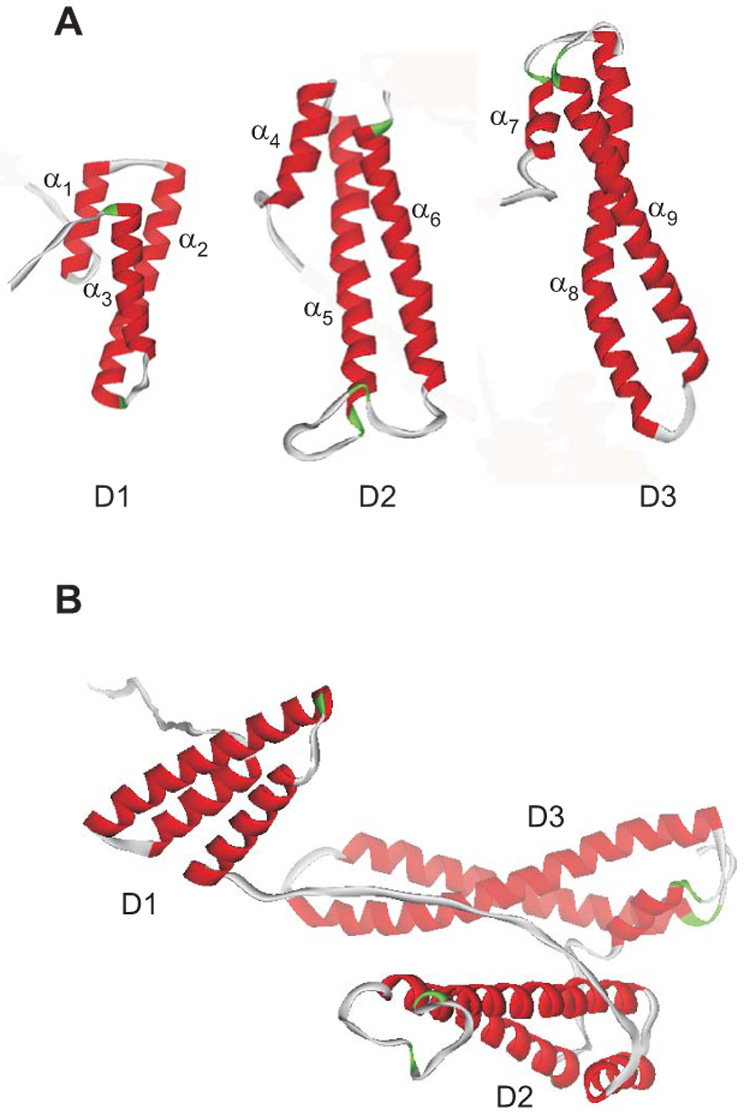
The helices are numbered as α1 – α9. B. Since the three domains of RAP are independent and do not interact, but are connected by long flexible loops, the protein is expected to adopt a variety of conformations in solution, one of which is shown.
To determine if the structures of individual domains of RAP are representative of those in full length RAP, the chemical shifts of the backbone amide groups of the individual domain constructs were compared with those of intact RAP using a combination of 2D-[15N,1H]-TROSY and 3D-TROSY-HNCA spectra (130). The chemical shift differences between the individual domains and those in full-length RAP are less than 0.05 ppm, except for the linker regions where domain constructs break off, suggesting that the structures of the individual domains are preserved and that there is no detectable chemical shift perturbation due to the presence of other domains. Thus, the individual domain structures are faithful representations of their structure in the full-length protein.
An idea of the overall structure of RAP was obtained by employing experimental small-angle neutron scattering (SANS) data and a novel simulated annealing protocol to characterize the overall structure of RAP (130). Since RAP consists of three independent domains joined by two flexible linkers, the protein is expected to have an ensemble of conformers in solution, one of which is shown in Figure 3B. RAP adopts a unique structural architecture consisting of three independent three-helix bundles that are connected by long and flexible linkers. The flexible linkers and the quasi-repetitive structural architecture may allow RAP to adopt various possible conformations when interacting with the LDL receptors, which are also made of repetitive substructure units.
C. Chaperone function of RAP
To understand the in vivo function of RAP, gene targeting was utilized to generate mice in which the RAP gene was deleted (291). The studies revealed that in RAP-deficient mice the amount of mature, processed LRP1 is substantially reduced in both the brain and liver. Loss of liver LRP1 function was confirmed by measuring delayed clearance of α2M-protease complexes from the circulation.
RAP is not secreted by cells (263) and is retained in the ER due to a tetrapeptide sequence (HNEL) at its carboxy-terminus (30). Pulse-chase and cross-linking experiments reveal that RAP associates transiently with newly synthesized LRP1. This association prevents ligands within the ER from associating with LRP1 and inducing receptor aggregation. In RAP-deficient fibroblasts, over-expression of certain ligands, such as apoE, resulted in the formation of complexes with LRP1 that were not effectively delivered to the cell surface; co-expression of RAP was shown to rescue surface LRP1 levels (293).
D. A histidine switch in RAP modulates LRP1 binding
A major function of RAP is to escort LRP1 from the ER to the Golgi apparatus, where RAP then dissociates from LRP1 as a result of a lowered pH encountered later in the secretory pathway (30,293). This function of RAP has been ascribed to the RAP D3 domain (194). When the surface charge distribution of RAP D3 at pH 7.2 (corresponding to the ER) was compared to the distribution at a more acidic pH, i.e. pH < 6.5, corresponding to the Golgi, the positively charged regions on the surface of D3 increased dramatically under the acidic conditions, mainly due to protonation of solvent-exposed histidine sidechains (131). These results suggested that histidine residues, especially highly conserved histidines, may function as a switch in response to the environmental change encountered when the RAP/LRP1 complex shuttles from the ER to the Golgi, leading to dissociation of RAP from LRP1. To test the involvement of histidine residues in modulating the pH-dependent binding of RAP to LRP1, mutant molecules were prepared in which all the conserved histidine residues in D2 and D3 were replaced with alanine residues. Binding studies revealed that mutation of the histidines in RAP D3 significantly reduced the pH sensitivity of D3 binding to LRP1 and failed to promote the secretion of soluble fragments of LRP1 from cells (131). Thus structure-based mutagenesis studies confirm that the protonation of histidine residues as a consequence of the pH changes modulate the binding/release of RAP from LRP1.
IV. Model for ligand recognition by LRP1
One of the major structural questions that remains to be solved is how LRP1 is capable of recognizing such a wide variety of structurally-distinct ligands. Site-directed mutagenesis studies point to an important role of basic residues present on the ligand that contribute to LRP1 recognition, while the first solved structure of a receptor fragment/RAP complex gives some insight into the mode by which these residues may interact with CR present within LDL receptor family members.
A. Role of basic residues in the recognition of ligands by LRP1
Apo E is a ligand recognized by most LDL receptor family members. Earlier studies highlighted the importance of lysines found between residues 140 to 160 (126) and Arg-172 (172) within the apo E molecule that contributed to its interaction with the LDL receptor. In the case of lipoprotein lipase, its interaction with LRP1 has been localized to the carboxyl-terminal domain (191,290) and involves two regions within this domain that include residues 380–384 and residues 404–430 (186). Mutation of Lys-407 to alanine resulted in a 10-fold reduction in the affinity of the carboxy-terminal domain of lipoprotein lipase for LRP1 (290).
A number of serpin enzyme complexes have been identified that interact with LRP1, including complexes consisting of proteases with plasminogen activator inhibitor 1 (PAI-1). Mutagenesis studies have identified basic residues in PAI-1 that appear important for its interaction with LRP1 (229). Thus conversion of Lys-82 and Arg-120 to alanine reduced the ability of LRP1 to recognize complexes of PAI-1 complexed to uPA. Likewise, mutation of Arg-78 and Lys-124 to alanine also resulted in loss of binding of the complex to LRP1. Stefansson et al. (259) found that a PAI-1 molecule with Arg-76 mutated to glutamic acid resulted in a loss of binding to LRP1.
Critical lysine residues were located in another ligand for LRP1, α2M. This molecule only binds to LRP1 following a conformational change induced by complex formation with proteases. Site directed mutagenesis implicated two lysine residues, Lys-1370 and Lys-1374, in binding to LRP1, and mutation of these two residues significantly reduced the affinity of the α2M receptor-binding domain for LRP1 (185).
The D3 domain of RAP binds with high affinity to LRP1, and in order to gain insight into amino acids that are required for the binding of RAP to LRP1, Migliorini et al. (160) performed random mutagenesis of D3 of RAP, which identified two critical lysine residues, Lys-256 and Lys-270, within the α8 helix of the third domain of RAP (Figure 4A) that are necessary for binding of D3 to LRP1. Mutation of either lysine residue significantly reduced the affinity RAP D3 for LRP1.
Figure 4. Structure of the RAP D3 in complex with two CR from the LDL receptor (68).
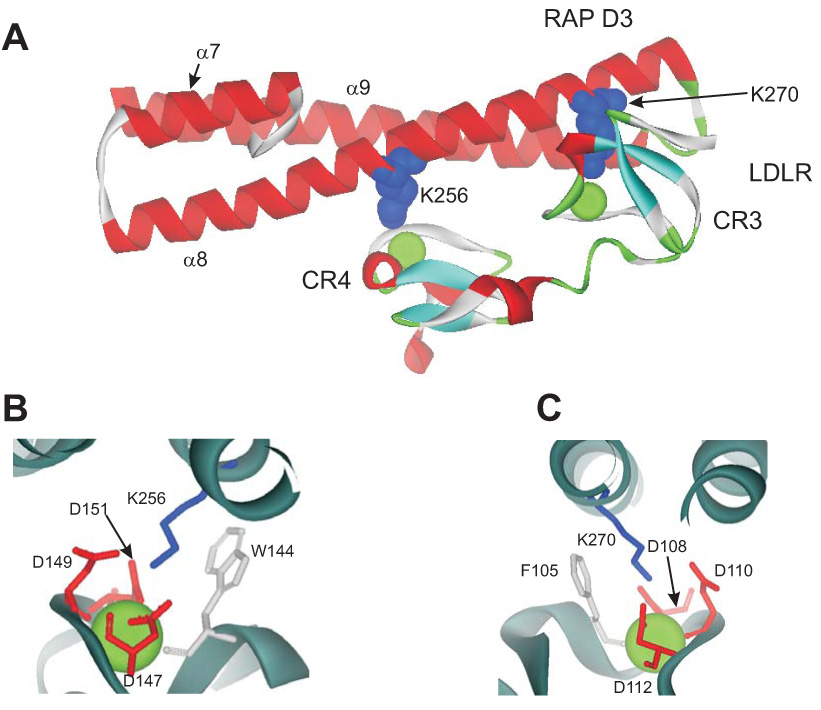
A. Lysines 256 and 270 are located in helix α8 of the D3 domain and provide the primary contacts with CR4 and CR3 of the LDLR, respectively. B and C. Detailed structure shows the acidic pocket surrounding K256 (B) and K270 (C). The structural calcium ion is shown. W144 and F105 are close to the aliphatic portion of the lysine residues in the pocket.
Not all LRP1 binding sites are composed of lysine residues, and, in the case of another serpin, protease nexin 1 (PN-1), a region has been identified corresponding to Pro-47 to Ile-58 of PN-1 that appears responsible for interacting with LRP1 (118). Thus, a synthetic peptide representing this region (PHDNIVISPHGI) was shown to competitively inhibit the LRP1-dependent endocytosis of thrombin:PN1 complexes. An antibody prepared against this synthetic peptide inhibited degradation of the PN1:thrombin complex by 70%, but it had no effect on binding of the complex to cell surfaces (117). Further, point mutations within the corresponding region of PN-1 (His48A and Asp49A) reduced the catabolism rate of mutated PN-1 to 15% of wild-type (117).
B. Structure of the RAP D3 receptor complex
Recently, Fisher et al. (68) solved the X-ray structure of a complex between a two-module region of the CR3 and CR4 of the LDL receptor and the third domain of RAP. In the complex, RAP D3 contains two docking sites for the LDL receptor CR involving Lys-256 and Lys-270 (Figure 4A). A relatively small interface between D3 and CR3–CR4 of the LDL receptor is dominated by electrostatic interactions between the two basic residues (Lys-256 and Lys-270) and the surface-exposed aspartate residues that participate in calcium coordination on CR3 and CR4 (Figure 4B,C). Each CR consists of four residues that provide a “docking” site for a lysine side chain protruding for helix α8 of the RAP D3 domain. Carboxylate oxygen atoms from three aspartates (Asp-147, Asp-149, and Asp-151 in CR4 and Asp-108, Asp-110 and Asp-112 in CR3) surround the ε-amino lysine group to form a salt bridge. In addition, two aromatic residues, (Phe-105 in CR3 and Trp-144 in CR4) pack up against the aliphatic portion of the lysine side chain. Importantly, all four residues participate in coordination of the calcium ion, and their position is therefore fixed in the structure.
C. Summary
As highlighted by Fisher et al. (68) the aspartic acid residues that form the acidic pocket responsible for docking the basic lysine residues in RAP are highly conserved amongst the CR of LDL receptor family members, and thus are representative of most CR repeats in these receptors. This suggests that lysine docking may represent a general mechanism for ligand recognition by LRP1 and other LDL receptor family members (68). If true, high affinity binding would require avidity effects resulting from the interaction of multiple lysine residues with multiple CR on the receptors. Interestingly, optimal high affinity (KD = 1.6 nM) binding of RAP to CR in cluster II of LRP1 requires three CR (CR5–CR7) (279); no binding of RAP to two repeats was detected in these experiments. Titration calorimetry experiments reveal that the binding of RAP or RAP D3 to two repeats, CR7–CR8 of LRP, is relatively weak (KD = 1 µM). These studies suggest that optimal binding of RAP D3 to LRP1 may require contact with at least three CR. Other ligands, such as activated forms of α2M also appear to optimally require three CR for their binding as well (59).
V. Hepatic function of LRP1 in the clearance of plasma proteins
LRP1 is abundantly expressed in the liver in hepatocytes and resident macrophages (Kupffer cells). Here, LRP1 recognizes a variety of distinct molecules in the circulation, including proteinase-inhibitor complexes, activated coagulation factors and chylomicron remnants, and mediates their endocytosis and intracellular degradation.
A. α2Macroglobulin
α2Macroglobulin is a highly conserved proteinase inhibitor capable of inhibiting target proteinases from all four major classes. The target proteinase cleaves α2M at a “bait” region, which triggers a conformational change in the molecule that entraps the proteinase in a cage-like structure and exposes a receptor binding site that is recognized by LRP1 on hepatocytes (251). Abundant evidence exists indicating that LRP1 is the key hepatic receptor responsible for clearing α2M-proteinase complexes. First, LRP1 was purified from tissue extracts by ligand affinity chromatography using the activated form of α2M coupled to Sepharose (3,169). Second, cells genetically deficient in LRP1 lack the ability to mediate the internalization and degradation of 125I-labeled α2M-proteinase complexes (69,214). Third, RAP was demonstrated to inhibit the clearance of 125I-labeled α2M-protease complexes from the circulation when co-injected with α2M (122) or when overexpressed in the liver (294). Finally, decreased hepatic levels of LRP1 using RAP-deficient mice resulted in delayed clearance of 125I-labeled α2M-proteinase complexes from the circulation (291). Together, these data provide compelling evidence that LRP1 participates in vivo in the clearance of α2M-proteinase complexes.
B. Serpin enzyme complexes
The serine proteinase inhibitors (serpins) are a large family of proteins, some of which are found circulating in the plasma where they function as inhibitors of serine proteases (247). These proteinase inhibitors form a complex with target proteinases that is initiated when the protease cleaves an exposed loop present in the inhibitor which in turn triggers a conformation change in the serpin. This results in the formation of a covalent complex with the target proteases. Serpin-enzyme complexes (SECs) are unstable, and will slowly break down releasing the active enzyme. Fortunately, SECs are recognized by a receptor system which is responsible for mediating their endocytosis and subsequent degradation. The existence of a hepatic receptor-based clearance mechanism was first suggested from the early work of Ohlsson and colleagues (196) who investigated the clearance of trypsin-inhibitor complexes from the circulation. Further studies revealed that the SEC receptor is specific for the serpin only after it has complexed with an enzyme, and does not effectively recognize the cleaved or native serpin (73,153).
Attempts to identify the receptor system responsible for the clearance of SECs led to the description of an SEC receptor that recognizes a pentapeptide sequence located at the carboxyl terminal fragment of α1-antitrypsin (107). This peptide appeared to bind to a cell-surface receptor and was reported to prevent the internalization and degradation of a number of SECs by HepG2 cell lines. However, mutation of this region in heparin cofactor II failed to diminished the binding, internalization, or degradation of thrombin:heparin cofactor II complexes by Hep G2 cells (147) revealing that other regions on the serpin are involved in receptor recognition.
Substantial evidence now indicates that LRP1 and other members of the LDL receptor family (LRP2/gp330/megalin and the VLDL receptor) function as prominent receptors in mediating the clearance of SECs (2,89,111,122,192,258). Given that LRP1 is the only one of these receptors that is abundant in the liver, this receptor is likely important in the hepatic removal of SECs from the plasma. LRP1 binds numerous SECs and, as expected for the SEC clearance receptor, does not recognize the native or cleaved serpin (122). The role of LRP1 in mediating the cellular uptake of SECs has been demonstrated by using cell lines genetically deficient in LRP1 and by in vivo clearance studies showing that RAP blocks removal of 125I-labeled SECs from the circulation (122).
C. Factor VIII
Factor VIII (fVIII) is a key plasma protein and a member of the coagulation cascade which is deficient in the well characterized bleeding disorder, hemophilia A. FVIII, which normally circulates in a complex with its carrier protein von Willebrand factor, is an inactive cofactor (66). Upon injury within the vasculature, this cofactor is activated to fVIIIa by limited proteolysis. This results in its dissociation from von Willebrand factor and subsequent assembly on the membrane surface with an enzymatically active form of factor IX (fIXa) to form a macromolecular Xase complex. This complex effectively activates factor X, the next proenzyme in the coagulation cascade. The fact that deficiencies in both fVIIIa and fIXa lead to bleeding disorders attests to the significant role that the macromolecular Xase complex plays in the blood coagulation cascade. While the functional and structural properties of fVIII are well described, only recently have the mechanisms by which this protein is metabolized become a key focus of investigation. Key hepatic receptors contributing to the clearance of fVIII are both members of the LDL receptor family, LRP1 and the LDL receptor.
Saenko et al. (238) and Lenting et al. (135) were the first to describe the potential of LRP1 to mediate the catabolism of fVIII. Both studies reported that LRP1 binds to fVIII with KD values between 60 – 116 nM (135,238). Further, both studies found that cells expressing LRP1, but not cells genetically deficient in LRP1, were able to mediate the uptake of fVIII in an LRP1-dependent manner. The in vivo significance of these observations was demonstrated by showing that RAP blocked the in vivo clearance of 125I-labeled fVIII (238) from the circulation. Importantly, von Willebrand factor was shown to inhibit the LRP1-mediated clearance of fVIII (135).
Genetic studies confirmed an important role for LRP1 in the metabolism of fVIII (25). This was demonstrated using cre/loxP-mediated recombination strategy to develop mice with LRP1 specifically deleted in the liver. This mutation resulted in an increase in the plasma levels of fVIII, from 1.9 U/ml in control mice to 3.4 U/ml in LRP1-deficient mice. Further, the clearance of fVIII was delayed in the hepatic LRP1-deficient mice. Together, these studies reveal that LRP1 functions in vivo and modulates fVIII plasma levels. More recent work has suggested that the LDL receptor, in addition to LRP1, also contributes to the clearance of fVIII from the plasma (26). Using hepatic LRP1 and LDL receptor double-deficient mice, Bovenschen et al. (26) demonstrated that mice with combined deficiency displayed a much greater increase of fVIII levels (~4–5-fold) than mice lacking LRP1 alone. In clearance studies, the mean residence time of fVIII was also dramatically prolonged (~5-fold) in mice with combined receptor deficiency (26). These findings, together with the fact that both LRP1 and the LDL receptor are predominantly expressed in the liver, reveal that LRP1 and the LDL receptor cooperate in regulating fVIII levels and clearance in vivo.
A puzzling question raised by these studies is how LRP1 and the LDL receptor function to effectively remove fVIII from the plasma. These receptors’ affinity for fVIII is relatively weak (KD values from 60 to 116 nM) which is well above the levels of fVIII circulating in the plasma. It could be that other cofactor molecules, such as cell surface heparan sulfate proteoglycans (HSPG), facilitate the uptake of fVIII by LRP1 and the LDL receptor. Additionally, it is now known that activation of fVIII generates a molecule with a significantly higher affinity for LRP1 (27). Thus, the removal of fVIII from the circulation may require prior activation and dissociation from its carrier protein, von Willebrand factor.
Interestingly, a familial study of factors influencing plasma fVIII levels revealed an association of fVIII levels with polymorphisms within the LRP1 gene (171). Specifically, the N allele of the LRP1/D2080N polymorphism was associated with slightly decreased plasma levels of fVIII (90.4 +/−8.7 vs 102.2 +/−3.5 IU/dl, p=0.02) (171).
D. Chylomicron remnants
Dietary lipids, cholesterol and fat-soluble vitamins are incorporated into large lipoproteins in the intestine known as chylomicrons (45). These triglyceride-rich lipoproteins are absorbed into the lymphatics and transferred to the general circulation via the thoracic duct (45). The enzyme lipoprotein lipase, expressed on endothelial cells especially in muscle and adipose tissue, selectively removes and hydrolyzes triglycerides, transferring free fatty acids to the tissue (260). The residual lipoprotein particles, called chylomicron remnants, are enriched in cholesteryl esters and contain apoE and apolipoprotein B48. After using exogenous fats, the liver can release excess lipids in the form of VLDL into the blood (11). VLDL is another substrate for lipoprotein lipase, and VLDL remnants can be taken up by the liver, in an apoE-mediated process or hydrolyzed to low density lipoproteins (LDL) (11).
Remnant lipoproteins are rapidly cleared from the plasma by the liver. This process requires apoE, which mediates binding of the lipoprotein particle to members of the LDL receptor family (LRP1 and LDL receptor) and to HSPG, which have been shown to play independent and cooperative roles in remnant lipoprotein clearance (148). The findings that the absence of normal LDL receptor activity leads to accumulation of LDL, but not remnant lipoproteins (116,235) led to the search for additional receptors that might be involved in remnant lipoprotein uptake, and the early discovery that LRP1 recognizes apoE (12) led to the notion that LRP1 might function as a remnant receptor. Evidence that LRP1 plays an in vivo role in remnant removal was provided by Willnow and colleagues (291). They bred mice genetically deficient in RAP, which have reduced hepatic LRP1 levels, to mice lacking the LDL receptor and demonstrated that the progeny mice have high levels of remnant-like lipoproteins in their circulation. Additionally, infection of mice lacking LDL receptors with an adenovirus that expressed RAP resulted in the accumulation of remnant lipoproteins in the plasma, supporting a role for a hepatic RAP-sensitive receptor, most likely LRP1, in the clearance of these particles (294). Rohlmann et al. (230) confirmed a role for LRP1 in remnant metabolism by using a viral Cre-mediated recombination technique to reduce LRP1 expression in the livers of mice on an LDLR-deficient background. Inactivation of LRP1 in the livers of these mice led to accumulation of cholesterol-rich remnant lipoproteins in their circulation, confirming a contribution of LRP1 in this process.
In addition to the two receptors, LRP1 and the LDL receptor, heparan sulfate proteoglycans are also known to participate in the removal of chylomicron remnants. Interestingly, inactivation of the biosynthetic gene GlcNAc N-deacetylase/Nsulfotransferase 1 (Ndst1) in hepatocytes, which results in a reduction of the sulfation of liver heparan sulfate, was found to have a dramatic effect on the accumulation of triglyceride-rich lipoprotein particles (145), revealing that hepatic HSPG directly contribute to the clearance of triglyceride-rich lipoproteins.
The current concept of remnant lipoprotein uptake recently was reviewed (148) and is summarized in Figure 5 (adapted from Mahley et al. (148) and MacArther et al. (145)). The first step in this process involves sequestering of the remnant lipoprotein particles in the Space of Disse via association with HSPG. Here the binding of remnant lipoproteins is primarily mediated by apoE. The purpose of this step is to assemble the participants in remnant clearance. In the second step, lipases (lipoprotein lipase and hepatic lipase) continue their lipolytic processing of the particles that began before their entry into the space of Disse, preparing them for the third step in the process: uptake into hepatocytes. It has been proposed that the LDL receptor, HSPG and the HSPG/LRP1 complex all serve as receptors or co-receptors mediating lipoprotein uptake. Interestingly, Wilsie and Orlando (295) discovered that LRP1 immunoprecipitates with HSPGs, although they reported that the LRP1/HSPG complex is unable to bind VLDL particles, which suggests a distinctly different model than the synergistic model proposed in Figure 5.
Figure 5. Proposed model for the involvement of LRP1 in remnant metabolism in the liver.
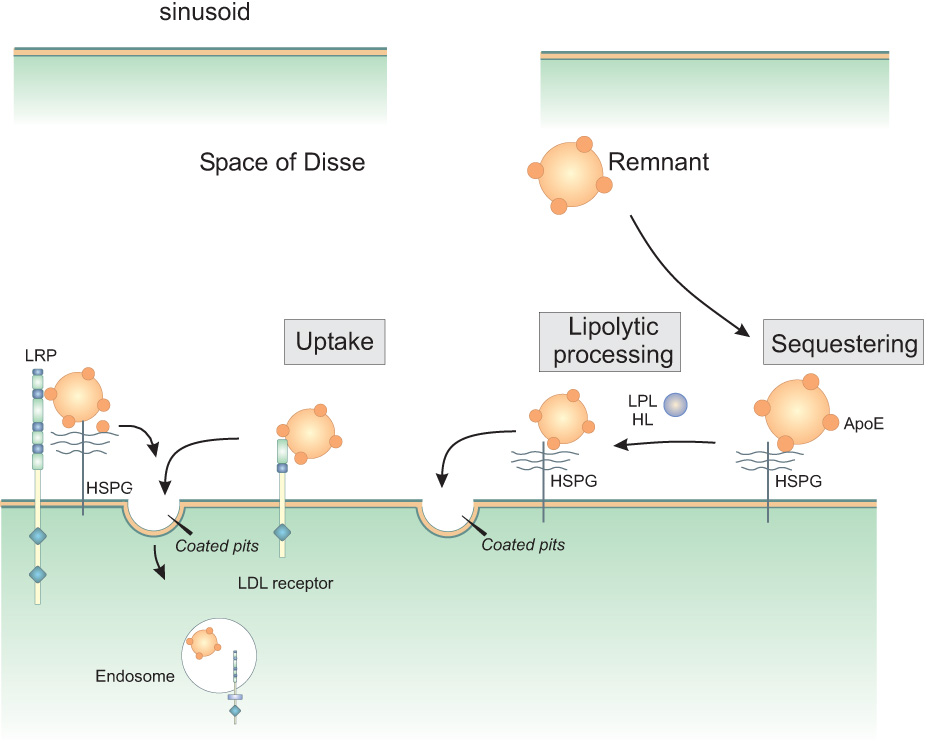
The model is adapted from (145,148). Remnant lipoprotein particles entering the space of Disse in the liver are first thought to be sequestered by association with heparan sulfate proteoglycans (HSPG). Here they are remodeled by the action of lipoprotein lipase (LPL) and hepatic lipase (HL). Internalization by the hepatocytes is mediated directly by HSPG, the LDL receptor, or HSPG/LRP1 complexes.
E. Summary
LRP1 in the liver plays an important role in facilitating the plasma removal of a number of molecules, including enzymes and co-factors involved in blood coagulation and fibrinolysis, enzyme-inhibitor complexes, and certain lipoprotein particles. Its function in the liver is important for normal homeostasis of these pathways. Deletion of hepatic LRP leads to increased plasma levels of certain molecules and accelerates the development of atherosclerosis (see below).
VI. Role of LRP1 in protecting the vasculature
Atherosclerosis is a leading cause of death and disability in industrialized nations, including the United States. It is the underlying medical problem in most patients with coronary artery disease, stroke, abdominal angina, and peripheral vascular disease. Elevated levels of serum lipoproteins, especially modified lipoproteins and triglyceride-rich lipoproteins, play a critical role in the development of this disease. Lipoproteins accumulate in the intima of large vessels where monocytes are then recruited in a step critical for initiating and sustaining lesion formation. There, monocytes differentiate into macrophages, scavenge sub-endothelial lipoproteins, transform into foam cells and accelerate plaque formation and lesion progression (143,233). Substantial evidence now exists from gene knockout studies in vascular smooth muscle cells (24), hepatocytes (64), and macrophages (95,203), that LRP1 functions to protect the vessel wall from injury.
A. Role of vascular smooth muscle cell LRP1 in modulating PDGF receptor function
Steps leading to the development of atherosclerosis are complex and are thought to result from an excessive response of the vascular endothelium and smooth muscle cells (SMC) in the artery wall to insult (232). SMC respond to the insult by undergoing proliferation and migration, mediated in part by platelet-derived growth factor (PDGF) released from endothelial cells. The mammalian family of PDGFs is comprised of four molecules (PDGF-A, PDGF-B, PDGF-C and PDGF-D), which differentially associate with two receptor tyrosine kinases: PDGFR-α and PDGFR-β. PDGF is a potent mitogen for fibroblasts and smooth muscle cells, and studies in mice in which either the PDGF-B or PDGFR-β gene has been deleted exhibit an almost complete lack of pericytes, mesenchymal-like cells which can differentiate into smooth muscle cells, fibroblasts or macrophages, in certain vascular beds (16,140).
Involvement of PDGF in the development of atherosclerosis has been demonstrated by employing balloon-catheterization injury of rat carotid arteries as a model. Balloon-catheterization results in damage to the endothelial cells and an increase in the level of activated PDGF receptors in the vessel wall (1,204). Furthermore, the intimal thickening that follows this treatment is inhibited by administration of neutralizing PDGF antibodies (67). In addition, infusion of PDGF-B into rats after carotid injury (103), or expression of recombinant PDGF-B in porcine arteries (181), caused a significant increase in vessel wall thickening. Within atherosclerotic lesions, PDGF stimulates smooth muscle cells to migrate from the media of the vessel to the intimal layer and to proliferate and produce matrix molecules at this site (215).
Recent in vivo and in vitro studies reveal that LRP1 is a physiological modulator of the platelet-derived growth factor (PDGF) signaling pathway. A tissue-specific deletion of the LRP1 gene in vascular SMC (smcLRP1−/−) on a background of LDL receptor deficiency led to SMC proliferation, aneurysm formation, and increased susceptibility to cholesterol-induced atherosclerosis (24). The smcLRP1−/− /LDL receptor −/− mice demonstrated significantly more atherosclerotic lesions and abnormal activation of PDGFR-β when compared to the smLRP1−/− mice. These effects could be inhibited by treatment of the mice with Gleevec, a known inhibitor of PDGF signaling. These studies indicate that LRP1 plays a role in protecting the integrity of the vascular wall and preventing atherosclerosis by suppressing PDGFR activation. At this time, the mechanism by which LRP1 modulates PDGFR function remains incompletely understood, but several possibilities exist. First, it was discovered that LRP1 directly binds PDGF-B (142) and thus LRP1 may function to reduce PDGF-BB levels, thereby reducing PDGFR-β activation. However, in vitro binding experiments reveal that the affinity of PDGF-B for LRP1 is somewhat weaker than its affinity for PDGFR-β, and thus a simple clearance mechanism is probably not the entire story (142).
A second possibility is that LRP1 may direct the trafficking of the PDGFR-β following its activation. Newton et al. (184) found that activated forms of the PDGFR-β co-immunoprecipitate with LRP1, and in cells, PDGFR-β associates with LRP1 within endosomes following addition of PDGF and mediates the tyrosine phosphorylation of the LRP1 cytoplasmic domain (23,142). Takayama et al. (270) also found that LRP1 binds to Cbl, a ubiquitin-protein ligase that associates with the PDGFR and other receptor tyrosine kinases (167,168), mediating their mono-ubiquitination which seems necessary for lysosomal-mediated degradation of the PDGFR complex. The potential of Cbl to associate with LRP1 provides a plausible mechanism that LRP1 might modulate PDGFR-β recycling/degradation. At this time, it is not known whether LRP1 phosphorylation is required for Cbl association. Using pulse-chase experiments, Takayama et al. (270) found that the steady-state turnover rate of PDGFR-β was accelerated in LRP1-deficient fibroblasts. While these effects are not consistent with the in vivo experiments indicating that LRP1 downregulates PDGFR signaling, the results do indicate that LRP1 can modulate PDGFR-β levels.
Finally, it is possible that LRP1 may modulate other signaling pathways that, in turn, could modulate PDGFR levels. In this regard, it is interesting to note that LRP1 has been identified as a receptor for TGF-β (see below), which itself is known induce expression of the PDGFR (86,101).
B. LRP1 as a TGF-β receptor
Transforming growth factor-β (TGF-β) regulates multiple biological processes, in a context-dependent and cell-specific manner, including proliferation, extracellular matrix biosynthesis, angiogenesis, immune response, apoptosis and differentiation (32). The biological activities of TGF-β are mediated by cellular receptors, and a variety of cell surface receptors have been identified by crosslinking 125I-labeled TGF-β to cells. One of these receptors, termed the TGF-βR-V was recently identified as LRP1 (96). LRP1 appears to be required for mediating the growth inhibitory response of TGF-β, in conjunction with Smad2/3 signaling through TGF-βR-I and II (96,275). In vivo, LRP1 appears to regulate TGF-β signaling pathways as well, as smooth muscle cell deletion of LRP1 also results in a Marfan-like syndrome with nuclear accumulation of phosphorylated Smad 2/3, disruption of elastic layers and increased expression of thrombospondin 1 and PDGFR-β in the vessel wall (22).
C. Hepatic LRP1 protects against development of atherosclerosis
To investigate the role of hepatic LRP1 in atherogenesis independent of its role in the removal of apoE-rich remnant lipoproteins, Espirito Santo et al. (64) crossed mice that are susceptible to inducible inactivation of hepatic LRP1 with mice deficient in both the LDL receptor and apoE (MX1Cre+LRP1flox/floxLDLR−/−APOE−/−). On an LDLR−/− APOE−/− background, hepatic LRP1 deficiency resulted in decreased plasma cholesterol and triglycerides. Interestingly, these mice showed a 2-fold higher atherosclerotic lesion area compared to control mice, revealing that hepatic LRP1 plays a protective role in the development of atherogenesis that is independent of plasma cholesterol levels. The mechanism by which LRP1 exerts its protective effect is not clear, but may be due to its ability to reduce plasma levels of proatherogenic ligands such as coagulation fVIII, whose levels are increased in the hepatic LRP1 knockout mice.
D. Macrophage LRP1 protects against the development of atherosclerosis
A crucial role of macrophages in the development of atherosclerosis has been demonstrated by studies in which mice with a defective macrophage colony-stimulating factor (M-CSF) gene were bred into an apoE-deficient background (249). M-CSF is a hematopoetic growth factor that stimulates survival, proliferation, differentiation, and multiple functions of cells derived from the mononuclear phagocytic lineage (70). The double-mutant mice had significantly smaller lesions in the aortic root region than their apoE-deficient control littermates, revealing that monocyte-derived macrophages play a key role in atherogenesis (225).
To investigate the in vivo role of LRP1 in macrophages and its contribution to the development of atherosclerosis, two groups (95,203) prepared mice with targeted deletion of LRP1 in macrophages. The first study (95) generated LRP1-deficient macrophages on a combined apoE/LDL receptor deficient background and found a 1.8-fold increase in atherosclerotic lesion area in the aortic root of 18-week old LRP-deficient mice. There were no changes in the lipoprotein profiles in these mice and the mechanism by which macrophage LRP1 is protective to the vessel wall remains unclear. The second study (203) generated the targeted deletion of LRP1 in macrophages and then performed a bone marrow transplantation into sub-lethally irradiated female LDL receptor −/− recipient mice. This resulted in a 40% increase in atherosclerosis as determined by measuring lesion area in the proximal aorta. The increased lesion area was not caused by altered serum lipoprotein levels but was speculated to result from a putative role for LRP1 in regulating inflammatory responses. In vitro studies using macrophages isolated from LRP1-deficient mice revealed increased production of TNFα by these macrophages upon LPS treatment although this was not confirmed in vivo.
In summary, the two separate studies performed in different mouse models (apoE/LDLR deficient mice versus LDLR deficient mice), confirmed an atheroprotective effect of macrophage LRP1. The mechanism by which macrophage LRP1 protects against the development and progression of atherosclerosis remains to be elucidated.
E. Potential role of LRP1 in facilitating LDL oxidation
Formation and uptake of oxidized LDL are thought to be critical to foam cell formation and the progression of atherosclerosis (143). Oxidative modification of LDL can occur by a variety of processes (261) including the action of lipoxygenases (LOs). One of these enzymes, 12/15-LO, which is capable of oxidizing esterified unsaturated fatty acids in LDL particles (35,252), is implicated in the development of atherosclerosis. Thus, disruption of the 12/15-LO gene in apoE-deficient mice (47) or in LDLR−/− mice (77) was found to retard the initiation and progression of atherosclerosis.
To determine the mechanism by which 12/15-LO oxidizes extracellular LDL, resident peritoneal macrophages from LDLR−/− mice were utilized. It was confirmed that the LDL receptor is not required for cell-mediated LDL oxidation (271). However, incubation of thioglycollate-elicited peritoneal macrophages with anti-LRP1 antibodies inhibited LDL oxidation by 56% (300), implicating LRP1 in this process. These studies were confirmed by using 12/15-LO-transfected J774A.1 cells and showing that anti-LRP1 antibodies, RAP, and antisense oligo-deoxyribonucleotides to knock down LRP1 reduced cell-mediated oxidation of LDL (300). Together, this work suggests that LRP1 mediates oxidation of LDL by 12/15-LO in macrophages. In further studies investigating the potential mechanism, LRP1 was found to promote the translocation of 12/15-LO from the cytosol to the plasma membrane (308), which is thought to be important for its activity.
F. Summary
Tissue selective gene deletion studies in vascular smooth muscle cells, hepatocytes and macrophages have all revealed a protective role for LRP1 in the development of atherosclerosis. The mechanism by which LRP1 expressed in these distinct cell types protects the vasculature is not fully understood. In the case of smooth muscle cells, LRP1 suppresses PDGF signaling pathways. In hepatocytes and macrophages, however, the pathway by which LRP1 alters the progression of atherosclerosis is not known. Finally, if LRP1 is definitively shown to play a role in the production or uptake of oxidized LDL by macrophages, this would add yet another facet to LRP1’s roles in macrophage function, especially in inflammatory states.
VII. Role of LRP1 in adipocytes
LRP1 is abundantly expressed in adipocytes (55), and some insight into its function in this tissue was derived from generating mice with an adipocyte-specific inactivation of the LRP1 gene (94). The adipocyte LRP1 knockout mice (adLRP1−/−) displayed delayed postprandial lipid clearance, smaller fat stores and lipid-depleted brown adipocytes which resulted in reduced body weight. This work highlights the importance of adipocyte LRP1 in postprandial triglyceride metabolism, where LRP1 in collaboration with lipoprotein lipase mediates both the lipolytic and endocytic processes responsible for triglyceride catabolism (37,38,157,189). In addition to the delay in postprandial triglyceride clearance, the adLRP1−/− mice also had an overall decrease in fat mass, and were resistant to diet-induced obesity. While the molecular mechanism by which deletion of the LRP gene in adipocytes leads to resistance of diet-induced obesity is not known, it was speculated that this may be due to a lack of LRP1-mediated lipid delivery to white adipocytes which in turn results in increased muscular activity in order for the mice to maintenance their core body temperature. Although many questions remain, the study reveals a prominent role of adipocyte LRP1 in modulating energy metabolism and sensitivity to diet-induced obesity.
VIII. Modulation of blood brain barrier function by LRP1
A. The neurovascular unit
The endothelial lining of vessels functions as a permeability barrier. In the brain, endothelial cells are one component of a functional unit that forms a barrier, termed the blood brain barrier (BBB), which protects the brain from the entrance of potentially harmful substances present in the blood and maintains the homeostatic environment of the central nervous system (CNS) (234). This functional unit, often called the neurovascular unit, is composed of endothelial cells with extensive tight junctions, astrocytes, neurons and a contractile apparatus of either SMCs or pericytes. A number of grafting and cell culture experiments have suggested that the barrier property of CNS endothelial cells also requires the cooperation of astrocytes (28,102,262) which appear to secrete factors that initiates signaling pathways necessary for BBB development (133). Although BBB permeability is carefully regulated, in pathological situations such as stroke, dysregulation of the BBB leads to vascular leakage resulting in severe edema (4,74).
B. LRP1 expression in the brain
In the adult human brain, LRP1 immunoreactivity is abundant on neuronal cell bodies and proximal processes (31,224,273,296). In situ hybridization assays revealed that within the cerebellum, LRP1 expression is observed in neurons diffusely scattered throughout the granular cell layer and is more intensely noted in the large Purkinje cells, but is not found in the molecular cell layer. In the dentate gyrus region of the hippocampus, LRP1 is expressed in neurons of both the granule and pyramidal cell layers (31). LRP1 immunostaining has been identified in astrocytic foot processes (213,296) and discontinually along capillary membranes (273). Electron microscopy confirmed that along the capillaries, LRP1 is expressed in the pericytes but not the endothelial cells. This early study was confirmed by immunohistochemical analysis of mouse brain sections (139), where prominent neuronal staining of LRP1 was detected, but no endothelial cell staining was observed. In contrast to these reports, Shibata et al. (245) reported expression of LRP1 in brain microvessels by immunocytochemical approaches. Curiously, this study did not observe the prominent neuronal staining of LRP1 that has been reported by others. The investigation also reported that LRP1 expression was decreased in the brains of older animals. The reason for the differences between this study and prior work is not apparent at this time. In cells, LRP1 message is extremely low in human umbilical cord vein endothelial cells (243), while LRP1 antigen has been detected at low levels in human cerebral microvascular endothelial cells by immunoblot analysis (283). Curiously, LRP1 is abundantly expressed in bovine aortic endothelial cells (201). Thus, to summarize all of the data, LRP1 is abundantly expressed in vascular smooth muscle cells, pericytes, astrocytes and neurons, but is not as abundant in the endothelium.
C. Role of LRP1 in maintaining the integrity of the blood brain barrier
A contribution of LRP1 to maintaining blood brain barrier function was discovered by Yepes et al. (305) who found that tPA regulates permeability at the blood-brain barrier (BBB) via a process that appears to be dependent upon LRP1. This was discovered when tPA injected into the cerebrospinal fluid led to increased vascular permeability even in the absence of ischemia (305). Further, blockade of LRP1 by co-injection of RAP abolished this effect (305). This study identifies an important role for LRP1 in controlling the permeability of the BBB in response to tPA. Interestingly, treatment with either RAP or anti-LRP1 IgG results in a faster recovery of motor activity and protection of the integrity of the neurovascular unit following middle cerebral artery occlusion (213). These findings are of significant clinical importance, as thrombolytic therapy for ischemic stroke with tPA may be accompanied by significant intracerebral bleeding. The mechanism by which tPA and LRP1 modulate BBB integrity is not yet known but may involve the potential of tPA and LRP1 to modulate or initiate signaling pathways. Interestingly, ischemic insult appears to induce shedding of LRP1's ectodomain from perivascular astrocytes into the basement membrane (213), which appears to be associated with a detachment of astrocytic end-feet processes and the formation of areas of perivascular edema. The shedding of LRP1's ectodomain is significantly decreased in tPA−/− mice. Further, shedding of LRP1’s ectodomain occurs in cultured astrocytes under conditions of oxygen and glucose deprivation, is increased when tPA is added, and is inhibited by the receptor-associated protein (RAP).
IX. Role of LRP1 in neurons
A. Alzheimer’s disease: Amyloid precursor protein and the amyloid hypothesis
Alzheimer’s disease is the most common age-related neurodegenerative disorder. Pathological findings include neuronal loss, neurofibrillary tangle formation and the extracellular deposition of insoluble protein fibrils called plaques (244). Neurofibrillary tangles are bundles of protein filaments found in the cytoplasm of neurons, while plaques are composed of a small, hydrophobic peptide termed β-amyloid (Aβ), which is derived from a ubiquitous type-I transmembrane protein, β-amyloid precursor protein (APP) (272). Aβ generation is thought to be central to the development of the disease (244).
Generation of Aβ from APP occurs in both secretory and endocytic compartments by regulated intramembrane proteolysis (RIP) (29), a sequential, two-step cleavage of transmembrane proteins with the second cleavage occurring within the transmembrane domain. In the case of APP, RIP is initiated by the β-site APP-cleaving enzyme BACE (280), an aspartyl proteinase that cleaves APP’s ectodomain and liberates the N-terminus of Aβ. Aβ generation is completed by intramembrane cleavage of APP, which requires presenilin-1(PS-1), an unusual aspartic proteinase with eight transmembrane domains (138). This cleavage can occur at slightly different positions, resulting in two principal forms of Aβ: Aβ40 and Aβ42, peptides with 40 and 42 amino acid residues, respectively. Once formed, the Aβ is released outside the cell. While Aβ40 constitutes about 90% of the total Aβ generated, the slightly longer Aβ42 has a higher tendency to form fibrils. Since all known genetic risk factors for AD impact Aβ metabolism, it is believed that the accumulation of Aβ fibrils into amyloid plaques plays a key role in the onset and/or progression of the disease.
B. Interaction of LRP1 with APP and the effect on Aβ production
Kounnas et al. (123) were the first to demonstrate that LRP1 can bind and mediate the cellular catabolism of the longer forms of APP (APP751, APP770) which contain Kunitz-type protease inhibitor (KPI) domains. Knauer et al. (119) subsequently found that transmembrane isoforms of APP containing KPI domains form complexes with a proteinase ligand, EGF-binding protein, and are internalized by a RAP-sensitive receptor, most likely LRP1. Together, these studies suggested a common LRP1-mediated internalization pathway for both soluble and transmembrane forms of APP containing KPI domains. Following these findings, Ulery et al. (276) tested the hypothesis that LRP1 can alter the trafficking of APP thereby modulating the production of the Aβ peptide. This study found that restoring LRP1 function in LRP1-deficient CHO cells increased the amyloidogenic pathway of APP processing, reducing the amount of soluble forms of APP generated by α-secretase cleavage (sAPPα) detected in the media, and increasing the production of the Aβ peptide.
Subsequent work (210) confirmed this initial study and further found that, not only does LRP1 affect Aβ production and the amount of sAPP released from the cell, but that it also effects APP internalization, turnover of full-length APP and the stability of APP C-terminal fragments. These LRP1-dependent changes occurred in all APP isoforms. Using deletion constructs, the critical region in LRP1 that modulates APP processing was mapped to the LRP1 cytoplasmic domain at the second NPXY motif, and appears to be dependent upon Fe65 (211), an adaptor protein that binds to the cytoplasmic domains of LRP1 and APP, linking them together. Together, all of these studies suggest that LRP1 functionally modulates APP steps critical for Aβ production and APP processing. Exactly how association of LRP1 with APP leads to enhanced Aβ production is not clear at present. One possibility is that the association of APP with LRP1 leads to increased trafficking of APP through the endosomal compartments where BACE and PS1 are known to reside, leading to enhanced proteolysis of APP (Figure 6).
Figure 6. Proposed model of LRP1 involvement in the trafficking of APP and Aβ production.
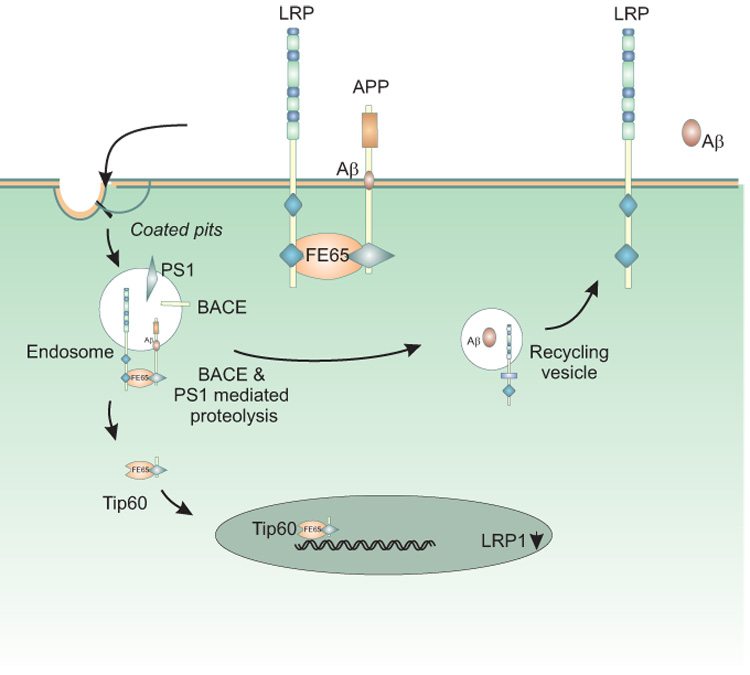
Fe65 bridges LRP1 and APP via cytoplasmic domain interactions, resulting in enhanced delivery of APP into endosomal compartments where BACE and PS1 are known to reside. Here, regulated intramembrane proteolysis of APP occurs, generating the Aβ peptide and releasing its intracellular domain. The APP intracellular domain forms a multimeric complex with Tip60 and Fe65, diffuses to the nucleus and modulates gene expression, including suppression of LRP1 gene transcription. The Aβ peptide is released into the media in recycling vesicles.
Konoshita et al. (114) confirmed that LRP1 interacts with APP in cells via both ectodomain and cytoplasmic domain interactions using fluorescence resonance energy transfer (FRET) measurements. The study identified interactions that were sensitive to RAP and assumed to be mediated by ectodomain interactions, as well as interactions that were insensitive to RAP and therefore assumed to represent cytoplasmic domain interactions. By using C-terminally tagged LRP1 and APP, the potential of the C termini of both APP695 and APP770 to interact with the C terminus of LRP1 was confirmed. These interactions were not sensitive to RAP treatment. FRET studies also confirmed a close proximity between the amino Fe65 phosphotyrosine binding (PTB) domain and LRP1 cytoplasmic domain and between the carboxyl Fe65 PTB domain and the APP cytoplasmic domain. These findings demonstrate that LRP1 and APP interact in cells.
To determine if LRP1 influences Aβ production in vivo, Zerbinatti et al. (306) generated a transgenic mouse overexpressing a functional LRP1 mini-receptor in neurons and crossed this mouse with the PDAPP mice, a well-known mouse model of amyloid deposition in which mice express a mutated version of human amyloid precursor protein under the control of the platelet-derived growth factor promoter (152). Overexpression of a functional LRP1 minireceptor in the brain of PDAPP mice results in age-dependent increase of soluble brain Aβ, with no changes in Aβ plaque burden. Importantly, soluble brain Aβ was found to be primarily in the form of monomers/dimers and to be highly correlated with deficits in spatial learning and memory. These results provide in vivo evidence that LRP1 may contribute to memory deficits typical of Alzheimer's disease by modulating the pool of small soluble forms of Aβ.
C. Regulation of LRP1 expression by APP
Cao and Sudhof (34) were the first to discover that the intracellular tail of APP, which is released following γ-secretase cleavage, formed a multimeric complex with the nuclear adaptor protein Fe65 and the histone acetyltransferase Tip60. This complex was found to stimulate transcription via heterologous Gal4 or LexA-DNA binding domains, and led these investigators to propose that the APP cytoplasmic tail may function to regulate gene expression. Interestingly, Liu et al (141) discovered that LRP1 expression is increased in mouse embryonic fibroblasts from APP knockout mice. They further showed that expression of the APP intracellular domain together with Fe65 and Tip60 interacts with the LRP1 promoter and suppresses its transcription. These studies uncovered an unexpected role for APP in suppressing LRP1 expression. Given that LRP1 recognizes numerous molecules, this observation may have a drastic impact on our understanding of neuronal physiology.
D. Role of LRP1 in clearance of A β from the brain
While the Aβ peptide is produced as a normal consequence of APP metabolism (85), Aβ fibrils do not accumulate in large quantities in healthy individuals, indicating the existence of clearance mechanisms. To date, three known pathways have been characterized that reduce the levels of Aβ: extracellular proteolysis, transport across the blood brain barrier, and receptor-mediated endocytosis. A number of proteinases are known to cleave the Aβ peptide (for review, see (150)) and include insulin degrading enzyme (216) and a neutral endopeptidase similar or identical to neprilysin (98). The significance of proteolytic pathways have been demonstrated by infusion of neutral endopeptidase inhibitors in the rat brain resulting in abnormal deposition of endogenous Aβ (98).
Aβ transport across the blood brain barrier is less well understood and the relative importance of this pathway to the overall removal of the Aβ peptide in vivo has not yet been demonstrated. However, injection of 125I-Aβ40 into the brain resulted in a rapid removal mainly by transport across the blood brain barrier (52). This process was significantly reduced by RAP, antibodies against LRP1, and α2M, implicating LRP1 in the removal of the Aβ peptide. These studies will have to be confirmed with tissue-selective LRP1 knockout studies to establish the contribution of LRP1 to this process in vivo.
The final mechanism that results in reduction of Aβ levels involves direct uptake by endocytic receptors. The class A and class B scavenger receptors can bind to and internalize fibrillar forms of Aβ (205). On the other hand, Aβ can form complexes with LRP1 ligands such as apoE (301), lactoferrin (217), and activated α2M (109,217), which can then be internalized via LRP1. More recent work (52) reveals that Aβ40 directly binds to LRP1 clusters II and IV with relatively high affinity, while Aβ42 binds with slightly weaker affinity. Interestingly, it appears that the affinity of Aβ for LRP1 decreases with increasing β-sheet content, suggesting that LRP1 binds with higher affinity to monomeric forms of Aβ and, therefore, has the potential to mediate the cellular uptake of Aβ.
Studies (110) have reported that a C766T polymorphism in exon 3 of LRP1 is under-represented in AD and associated with later age of disease onset; however, this is controversial and has been reproduced (120) and refuted by additional studies (36,134,240). Interestingly, Kang et al. (109) also suggested that lower levels of LRP1 in AD correlated with the CC genotype within the LRP1 exon 3 polymorphism locus and reported a reduction in the levels of LRP1 in the brains of patients with Alzheimer’s disease. Based on the suspected ability of LRP1 to mediate the transport of the Aβ peptide out of the brain, the study proposed that decreased levels of LRP1 may reduce Aβ clearance, thereby contributing to increased Aβ levels and enhanced disease. In contrast, Causevic et al. (36) found no correlation between LRP1 levels and Alzheimer’s disease. Thus, any connection between levels of LRP1 and Alzheimer’s disease requires further clarification.
E. Tissue selective LRP1 deletion in neurons supports a neurotransmitter role for LRP1
While LRP1 is abundantly expressed in neurons, its exact function here is unknown. To investigate the role of LRP1 in neurons, tissue selective deletion of LRP1 was accomplished (155). Mice lacking LRP1 in differentiated neurons develop severe behavioral and motor abnormalities, including hyperactivity, tremor, and dystonia. In these mice, no histological abnormalities were noted, indicating that gross developmental processes were not impaired. The hypothesis that LRP1 may participate in neurotransmitter-dependent postsynaptic responses resulted from the finding that LRP1 is in close proximity to the N-methyl-D-aspartate (NMDA) receptor in dendritic synapses in neurons and co-precipitates with NMDA receptor subunits and the postsynaptic density protein PSD-95 from neuronal cell lysates. If true, LRP1, like other ApoE receptors, may modulate synaptic transmission in the brain.
F. Summary
LRP1 is abundantly expressed in neurons where its function is yet to be established. Studies have raised the possibility that LRP1 may function in synaptic transmission in the brain, perhaps in cooperation with tPA. Additional work is required to determine if this is the case. In the brain, LRP1 can also associate with APP and modulate the trafficking of this molecule resulting in increased production of the Aβ peptide. On the other hand, LRP1 may also participate in the removal of the Aβ peptide by directly binding to it and mediating its cellular uptake and degradation. Thus LRP1 appears positioned to modulate the levels of this peptide and, in so doing, possibly to regulate the progression of AD.
X. Role of LRP1 in cell migration and integrin function
The interaction of cells with the extracellular matrix is important for cellular physiology, as these interactions regulate cell survival, proliferation, migration, and differentiation. The bi-directional communication between the extracellular matrix (ECM) and the actin cytoskeleton is regulated by integrins, a large family of cell surface receptors that regulate cell adhesion and migration. Cell migration is important in many physiological and pathological processes including wound healing, bone remodeling, development, angiogenesis, and invasion of cancer cells. During the process of cell migration, cells form and remodel their focal adhesions, both through reorganization of the cytoskeleton and through modulation of integrin signaling (72,170,228). Inside-out and outside-in signaling events activate integrins, which lead to conformational changes in the integrin dimer and increased affinity for its ECM ligands. Activated integrins are recruited to the leading edge of the migrating cell (115) where they also recruit proteases to enhance degradation of the ECM (174). While not yet fully understood, LRP1 is able to modulate integrin action by directly associating with integrins or by cooperation with other molecules, such as thrombospondin.
A. Interaction with calreticulin and thrombospondin 1: role of LRP1 in focal adhesion disassembly
The adhesive remodeling that is requisite for cell migration requires disassembly or restructuring of the integrin-linked focal adhesion scaffold. Focal adhesion disassembly can be triggered by a number of proteolytic and stimulatory signals. Members of a class of ECM proteins termed “matricellular” proteins can all perform such a trigger function. This class of proteins includes SPARC, tenascin-C, and thrombospondins-1 and -2, all of which function in focal adhesion disassembly. Thrombospondin-1 (TSP1), a large 420 kDa, homotrimeric, extracellular matrix protein (21), is released from platelet α-granules following platelet aggregation. TSP1 is also expressed by most cell types in culture, including epithelial cells, fibroblasts, endothelial cells, smooth muscle cells, and immune cells (99,100,128,173,222,278). Expression of TSP1 is highly-regulated: it is induced by growth factors, serum, hypoxia, and oxidative stress (61,149,208). Consequently, TSP1 expression is increased where there is tissue remodeling, which occurs in response to injury and fibrosis, during wound healing, and in development (193,223,226,227,278,287). Structurally, TSP1 is comprised of different domains that interact with a variety of cellular receptors. The N-terminal domain (NTD) of TSP1 binds LRP1 (164), calreticulin (CRT), heparan sulfate proteoglycans and integrins (156,158,257). This TSP domain can be cleaved from the remaining C-terminal portion of TSP1 by a wide array of serine proteases and has functions distinct from those of the intact TSP1 molecule (58,127,132,218).
TSP1 in its soluble form has anti-adhesive properties and causes reorganization of actin stress fibers and focal adhesion disassembly (83,177). Focal adhesions are signaling scaffolds composed of both structural and signaling proteins that link the extracellular environment to the cytoskeleton (41,46). Signaling through focal adhesions regulates cell shape, motility, survival, and differentiation (241,242,297). The stimulation of focal adhesion disassembly by TSP1 is thought to enable cell migration, potentially by triggering changes in cytoskeletal organization that are optimal for cell motility. Both intact TSP1 and the NTD can stimulate focal adhesion disassembly (176). This activity is localized to a 19 amino acid sequence in the NTD of TSP1 (amino acids 17–35) that binds to cell surface calreticulin (CRT) (78,176). A peptide mimetic of this sequence (hep I) has been used to probe TSP1 actions specific to this sequence (176).
CRT isolated from bovine aortic endothelial cells was identified as a TSP1 (hep I) binding protein, and it was shown that expression of CRT on the cell surface is necessary for TSP1 to signal focal adhesion disassembly and cell motility in endothelial cells and fibroblasts (79,198). The TSP1 binding sequence in CRT has been localized to an 18 amino acid sequence, aa 19–36, in the NTD of CRT (79). CRT, also identified as a C1q receptor, is best known as an endoplasmic reticulum (ER) chaperone protein that serves as an important regulator of both intracellular Ca2+ stores and antigen presentation (165,250,281). However, CRT is also localized on the cell surface of many cell types, where its expression is upregulated by cellular stress (78,81,106,212,286,299). CRT binds to integrins, LRP1, and collagens (201,207,231). Mice lacking the CRT gene die during embryogenesis due to defects in myocardial development (159). Initial studies, which showed that cell-surface CRT mediated TSP1 signaling, were perplexing since CRT neither contains a transmembrane domain nor is GPI-anchored. This suggested that CRT may form a complex with a binding partner in order to signal in response to TSP1 binding.
Studies from Orr et al. (201) identified LRP1 as the co-receptor which mediates TSP1 signaling of focal adhesion disassembly and stimulation of cell motility through binding to CRT (Figure 7A). This work revealed that an antibody to LRP1 or RAP blocks focal adhesion disassembly by TSP1 and hep I (201). Further, mouse embryonic fibroblasts (MEFs) deficient in LRP1 are unable to undergo focal adhesion disassembly in response to either TSP1 or hep I, although they retain the ability to undergo focal adhesion disassembly in response to tenascin A-D, a matricellular protein that induces focal adhesion disassembly through annexin II (44,178,198,201). Basal CRT-LRP1 interactions are not sufficient to trigger this signaling. Binding of TSP1 (hep I) is necessary to stimulate both increased association of CRT with LRP1 and downstream signaling events in endothelial cell membranes (198,201). The binding site(s) between CRT and LRP1 have not yet been identified. It is likely that the extracellular domain of LRP1 is important for CRT signaling since RAP can inhibit cellular responses to TSP1 (201). Furthermore, cells expressing LRP1 mini-receptor constructs that lack most of the extracellular domain of LRP1, fail to respond to TSP1/hep I (Van Duyn, Murphy-Ullrich, and Strickland, unpublished results). Focal adhesion disassembly by hep I and TSP1 also requires the surface expression of Thy-1, a GPI-linked protein: the role of Thy-1 is unclear, and there is no evidence that Thy-1 interacts directly with either CRT or LRP1 (5).
Figure 7. Role of LRP1 in mediating TSP1 signaling and metabolism.
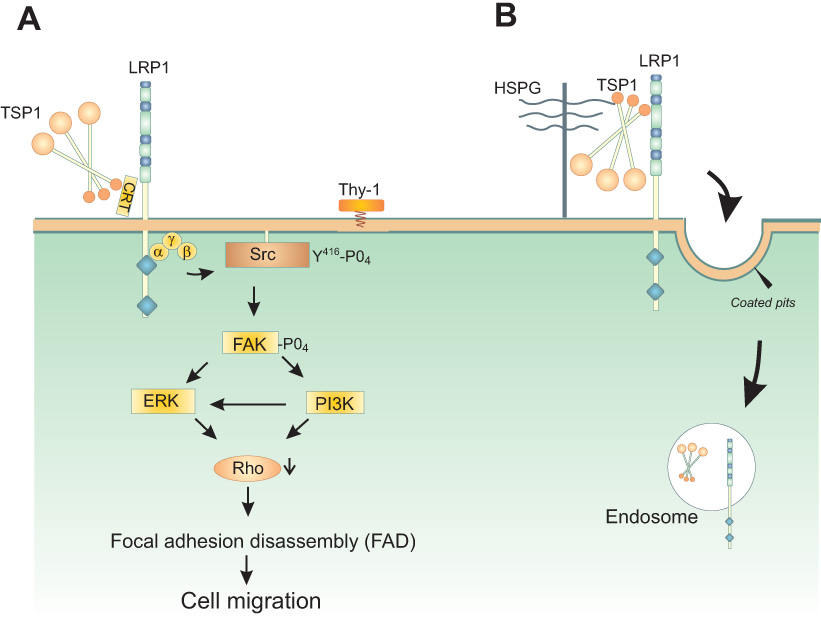
A. The hep I sequence (aa 17–35) of the N-terminal domain of TSP1 binds cell surface CRT (aa 19–36). When bound to TSP1 or the hep I peptide, CRT binding to LRP1 is enhanced and signaling through the CRT-LRP1 co-receptor complex is initiated. TSP1 binding to the CRT-LRP1 complex induces association of the Gαi2 protein subunit with LRP1. Phosphorylation of Src and FAK occurs downstream and leads to ERK and PI3K activation. This signaling cascade triggers inactivation of RhoA, resulting in focal adhesion disassembly (FAD) and stimulation of cell migration. In addition, TSP1 signaling through the hep I sequence requires the participation of Thy-1, a GPI-linked protein, to affect Src activation, although it does not appear that Thy-1 directly binds to either LRP1 or CRT(5) B. LRP1 mediates endocytosis of TSP1 through binding of the N-terminal domain of TSP1, a process which requires heparan sulfate proteoglycans (HSPG) for internalization (163,164,179,180).
Stimulation of the CRT-LRP1 co-complex by the hep I sequence of TSP1 induces the transient association of the Gαi2 protein subunit with LRP1. This pertussis toxin (PTX)-sensitive event triggers phosphorylation of FAK and Src, activation of ERK and PI3K, and culminates in RhoA inactivation. Cells lacking either CRT or LRP1 fail to activate FAK, PI3K, or ERK in response to TSP1 (5,198–201). PTX-sensitive G proteins also are involved in focal adhesion disassembly by fibroblast-derived motility factor and uPA (53,267). Although heterotrimeric G protein signaling is typically associated with seven-transmembrane spanning receptors, LRP1 has been linked to PTX-sensitive G proteins in other systems. For example, apolipoprotein E4 induces apoptosis of neuronal cells in a process thought to involve LRP1 and PTX-sensitive G proteins (88). In addition, lactoferrin signaling through LRP1 in macrophages induces a PTX-sensitive increase in inositol 1,4,5-trisphosphate (IP3) and intracellular calcium (166).
TSP1 signaling through the CRT-LRP1 co-complex is distinct from LRP1’s role in the endocytosis of TSP1 (Figure 7B). Thus, LRP1 can bind, internalize, and degrade TSP1 through LRP1 binding to the NTD of TSP1 (163,164). The sequence of TSP1 recognized by LRP1, and necessary for its endocytosis, is contained within amino acids 1–90 of the NTD (282). However, endocytosis of TSP1 by LRP1 does not involve the NTD sequence mimicked by hep I, since the hep I peptide does not bind LRP1 (201). Importantly, TSP1 also acts as a bridging molecule to facilitate clearance of other proteins by LRP1. TSP1 interacts with both MMP2 and MMP9 (10). LRP1 endocytoses pro-MMP2 that is bound to TSP1, and clearance of pro-MMP2 can be blocked by an anti-TSP1 antibody (304). Interestingly, endocytosis of the pro-MMP2-TIMP2 complex by LRP1 is TSP1 independent (63). TSP2 interactions with LRP1 are important for internalization of MMP2 and studies in TSP2 knockout animals suggest that a deficiency in MMP2 clearance in the absence of TSP2 results in defective connective tissue organization (303,304). Recently, it also has been shown that TSP1 mediates the clearance of vascular endothelial growth factor (VEGF) through LRP1 (82). These results suggest that LRP1 plays an important role in tissue and vascular remodeling through endocytosis of factors involved in matrix degradation and angiogenesis.
Studies suggest that engagement of LRP1’s signaling function impedes its ability to act as a scavenger receptor. Thus, Ranganathan et al. (221) discovered that the association of various adaptor proteins with the LRP cytoplasmic domain is modulated by its phosphorylation state and that serine and threonine phosphorylation reduces the association of LRP with adaptor molecules of the endocytic machinery. In contrast, serine and threonine phosphorylation was necessary for the interaction of LRP with Shc, an adaptor protein that participates in signaling events. Furthermore, serine and threonine phosphorylation increased the interaction of LRP with other adaptor proteins such as Dab-1 and CED-6/GULP. These results indicate that phosphorylation of LRP modulates the endocytic and signaling function of LRP by modifying its association with adaptor proteins. Grey et al. found that lactoferrin, a mitogen for osteoblasts, requires LRP1 for signaling (84). However, blocking endocytosis by placing cells in a hypertonic solution, lowering the temperature to 4°C, or using a pharmacological inhibitor of endocytosis did not affect lactoferrin signaling through LRP1. This suggests that the signaling and endocytic functions of LRP1 are independent of each other (84). Gotthardt et al. (80) found the adaptor protein DAB1, which regulates tyrosine kinase signaling and microtubule function in neurons, binds to the cytoplasmic tail of both LRP1 and the LDL receptor. In the presence of DAB1, LDL receptor degradation is reduced, suggesting that adaptor binding competes with the endocytic machinery. This suggests that engagement of the LDL receptor in signaling reactions precludes its ability to mediate endocytosis (80). Since TSP1, through binding to CRT, can engage LRP1 in G protein-mediated signaling and direct interactions of TSP1/2 with LRP1 stimulate endocytosis, it will be interesting to determine whether LRP1 signaling and scavenger activities are differentially regulated by TSPs.
B. Role of LRP1 in cell migration
Cell migration is regulated by proteases such as matrix metalloproteinases and the serine proteinases, urokinase plasminogen activator (uPA) and tissue-type plasminogen activator (tPA). uPA and tPA activate plasminogen to plasmin, which can digest the ECM and activate MMPs (161,285). Because LRP1 is involved in the processing of multiple enzymes which regulate matrix turnover and cell adhesion, it is not surprising that LRP1 was found to modulate cell migration. This was first demonstrated by Okada et al. (197) who used a Transwell filter migration assay with fibronectin-coated filters and found that anti-LRP antibodies or RAP inhibited cell migration. A similar inhibitory effect of RAP on smooth muscle cell migration and invasion was observed by Wijnberg et al. (288).
The exact mechanism by which LRP1 influences cell migration is not known, but it appears that multiple and distinct mechanisms exist. First, accumulating data suggest that certain LRP1 ligands can stimulate cell migration through engagement of LRP1 signaling. Okada et al. (197) observed that addition of either uPA or tPA stimulated vascular smooth muscle cell migration. Other mesenchymal cell types also appear to be stimulated by ligand binding to LRP1. Mouse embryonic fibroblasts (MEFs) were stimulated to migrate on a vitronectin and fibronectin matrix by TSP1 signaling through the CRT-LRP1 receptor co-complex (198). The migration was blocked with RAP, indicating that ligand binding (CRT) to LRP1 is required for this process. Further, LRP1- and CRT-null MEFs failed to migrate in response to TSP1 and hep I, confirming the involvement of LRP1 and CRT in this process (198). Degryse et al. (54) similarly reported that PAI-1-stimulated rat smooth muscle cell migration was blocked by an anti-LRP1 antibody or RAP. Interestingly, LRP1-deficient cells also exhibited defective migration in response to serum, and time lapse microscopy of these cells suggests that LRP1-deficient cells have impaired lamellipodia formation (198).
In contrast to these reports citing LRP1-ligand induced stimulation of cell migration, Weaver et al. (284) found that LRP1-deficient MEFs grown in serum-containing media migrated faster than wild-type MEFs when subjected to an in vitro scratch assay, revealing that LRP1 expression delayed cell migration. In these experiments, MEFs were grown in serum-containing medium until 95% confluent on bacterial plates coated with either serum, vitronectin, fibronectin, Matrigel, or type I collagen. Cell layers were scratched, and cell migration was assessed under serum-free conditions in media supplemented with 20 ng/ml of PDGF-BB. LRP1-deficient cells migrated faster in response to PDGF-BB than did wild type MEFs when plated on serum, vitronectin, and fibronectin coated plates. However, there was no difference in migration rates between LRP1-expressing and LRP1-deficient MEFs in cultures plated on either Matrigel or type I collagen, indicating the importance of the matrix environment. The increased migration noted in LRP1-deficient fibroblasts may relate to increased surface expression of the urokinase receptor (uPAR) noted in these cells (284).
The urokinase receptor (uPAR) is a three domain molecule attached to the plasma membrane by a GPI anchor (9,19,256) that binds tightly to uPA. uPAR plays an important role in a cell-based proteolytic system and is also known to stimulate signaling pathways (202). uPA activity is regulated by a serpin, PAI-1, and, upon complex formation with this inhibitor, a cryptic site is exposed that is recognized by LRP1 (192). The consequence of this interaction is LRP1-dependent endocytosis of cell-associated uPAR complexed to uPA-PAI-1 (190), which leads to reduced steady-state levels of uPAR (284). Under high serum conditions, LRP1-deficient fibroblasts have elevated levels of Rac, the small GTPase that is critical for cell spreading and lamellipodia formation (144). Thus, elevated pericellular proteolysis and/or persistence of Rac activating factors in the LRP1-deficient MEFs, due to the absence of LRP1 scavenger activity, could account for the increased motility in these cells. This also suggests that LRP1-mediated clearance of pericellular proteases and other growth factors can attenuate cell motility (284).
Another mechanism by which LRP1 can regulate cell migration is through the ability of certain LRP1 ligands to block the effect of stimulatory ligands on cell migration. For example, apoE inhibits SMC migration induced by PDGF-B (268). This occurs when apoE binds to LRP1 and initiates a signaling pathway that results in increased intracellular cAMP levels and protein kinase A activity, counteracting the stimulatory effects of PDGF (309).
LRP1 also can directly modulate cell migration by mediating the internalization of integrins under certain conditions. Czekay et al. (48) found that, paradoxically, addition of exogenous plasminogen activator inhibitor (PAI-1) and uPA to HT-1080 fibrosarcoma cells resulted in their detachment. This occurred through formation of a complex of PAI-1 with uPA bound to uPAR associated with integrins. Formation of the covalent PAI-1:uPA:uPAR complex led to internalization of the attached integrin by an LRP1-mediated process, which was detected by accumulation of αvβ3 and αvβ5 integrins in early endosomal fractions. This accumulation was completely blocked by anti-LRP1 IgG or RAP, showing that LRP1 is required for the uPA/PAI-1 induced internalization of integrins.
LRP1 also has been reported to directly associate with β2-integrins on leukocytes (33,253). In monocytes, immunofluorescence studies showed that LRP1 co-localizes with β2 integrins (33,253). In vitro binding experiments showed that recombinant cluster IV of LRP1 binds directly to purified immobilized αMβ2. The association of LRP1 with β2 integrins is thought to regulate integrin recycling during cell migration and may depend upon other LRP1 ligands, such as tPA (33).
C. Role of LRP1 in integrin processing
Unexpectedly, it was observed that loss of LRP1 expression correlated with reduced cell-surface expression of β1 integrin (239). Further studies showed that LRP1 plays a role in the post-translational processing and delivery of mature integrins to the cell surface. Mature β1 integrin is glycosylated in the endoplasmic reticulum (ER) and Golgi, whereas the immature form is not fully glycosylated. Alterations in integrin glycosylation have variably been shown to alter three aspects of their maturation and function: transport through the Golgi, pairing with the α-subunit, and their ligand binding affinity (13). Although there was less mature β1 integrin on the cell surface of cells lacking LRP1, the total amount of β1 integrin protein in the cell was unchanged, and there was increased β1 integrin in the ER, suggesting that LRP1 plays a role in β1 integrin trafficking from the ER to the cell surface. The effects of LRP1 on β1 integrin maturation were TGF-β and ECM substrate-independent, but dependent on culture confluency (239). Apparently, neither LRP1 ligand binding nor endocytosis is involved in integrin maturation, since RAP did not affect maturation. It is unclear whether LRP1 associates with chaperones or adaptor proteins in the ER during the transit of β1 integrins or whether LRP1 affects integrin glycosylation directly. Direct interactions between LRP1 and integrins do not appear to be involved, since the authors were unable to co-immunoprecipitate LRP1 with β1 integrin. It was suggested that chaperones or adaptor proteins such as calreticulin, hsp90, Fe65 or ICAP-1 might act as a bridge between LRP1 and β1 integrin.
D. LRP1 and tumor invasion
Because of LRP1’s complex role in regulating pericellular proteases and cell migration, a role for LRP1 in tumor cell invasion has been investigated. Various groups have investigated whether LRP1 expression on tumor cells correlates with invasiveness. Kancha et al. (108) examined a panel of breast carcinoma cells with different degrees of invasiveness including non-tumorigenic MCF10A breast cells, pre-neoplastic MCF10AT cells, non-invasive subclones of MCF10AT cells, and invasive subclones of MCF10AT cells. They evaluated LRP1 expression by Northern blot analysis and through the use of a binding assay with radioactively-labeled activated α2M to assess functional LRP1 at the cell surface. These studies showed that MCF10ATs and their invasive subclones had decreased mRNA levels and expression of LRP1 surface protein compared to the less invasive MCF10A and non-invasive subclones (108). In addition, invasion through Matrigel by a follicular thyroid carcinoma cell line was found to inversely correlate with LRP1 expression, and inhibition of LRP1 or increasing uPA levels increased invasiveness (246). These studies suggest that LRP1 is associated with a less invasive phenotype, perhaps by mediating endocytosis of proteases, and that decreased LRP1 levels correlate with increased invasiveness. Consistent with this idea is the observation that primary breast tumors did not exhibit levels of LRP1 detectable by immunohistochemical approaches, although LRP1 was found on stromal fibroblasts (40).
In contrast, other investigators provide evidence of a role for LRP1 in promoting breast cancer cell invasiveness (39,136). Flow cytometric analyses of different breast cancer cell lines showed varying LRP1 surface expression levels. Cell lines MDA-MB-231, T47D, BT-20, and HS-578T, which are more highly invasive in in vitro Matrigel invasion assays, had higher LRP1 expression levels compared to the less invasive cell lines HMEC, MCF-7, and MDA-MB-361, which exhibited lower LRP1 expression (136,137). Furthermore, LRP1 was localized to the leading edge of breast cancer cells, suggesting that LRP1 might regulate cell-matrix interactions and/or cytoskeletal organization to enhance the protrusive activity needed for cell migration (39).
E. Summary
These results highlight the difficulty in determining the function of a protein as complex as LRP1. The inherent differences between tissues and established cell lines and the variability of in vitro culture conditions all potentially influence the results. The dual nature of LRP1—both scavenger and signaling receptor— and the variable functions of its numerous ligands adds to the complexity of deciphering the role of LRP1 in biological processes. However, evidence to date supports a role for LRP1 in regulating the pericellular microenvironment through clearance of adhesion and matrix-altering proteases. In this capacity, LRP1 would likely act to stabilize the matrix and cell adhesion, thus reducing migration and invasiveness. However, depending on the ligand, engagement of LRP1 as a signaling receptor can also directly trigger cellular de-adhesion and cytoskeletal reorganization to support increased cell motility. Clearly, the nature of the matrix and the extracellular milieu of LRP1 ligands will determine its function in a cell-, tissue-, and context-specific manner.
XI. Function of LRP1 in inflammation and phagocytosis
The removal of apoptotic cells, necrotic debris or infectious agents by the process of phagocytosis is essential for maintenance of homeostasis, organogenesis, resolution of inflammation and prevention of autoimmune responses (146). Phagocytosis requires receptor-mediated recognition of such targets followed by their delivery into phagosomes. In the case of apoptotic cells, recognition and phagocytosis are complex events modulated by several known, and probably many as of yet unidentified, interactions between phagocyte receptors and ligands on the surface of apoptotic cells (reviewed in (265)). Evidence is beginning to accumulate to suggest an important role for LRP1 in this process.
A. Potential of LRP1 to mediate phagocytosis
Several lines of evidence suggest a role for LRP1 in the process of phagocytosis. Pathways involved in apoptotic cell recognition and phagocytic removal are highly conserved across species from fly to human. Transmembrane receptors responsible for recognizing apoptotic cells and initiating downstream signaling events to mediate actin rearrangement and phagocytosis have been identified in C. elegans (112,307) and Drosophila (151,237). Work in C. elegans revealed that two pathways regulate this process. In one of these pathways, a transmembrane receptor (CED-1) recognizes an unidentified ligand on apoptotic cells and recruits the adaptor protein CED-6 which binds to an NPxY motif on the CED-1 cytoplasmic tail (112). This recruitment activates CED-10, a Rac GTPase that initiates actin reorganization necessary for phagocytosis. LRP1 has been suggested to be a possible functional mammalian homologue of CED-1 (112) largely based on its status as a single-pass transmembrane receptor, its NPxY-containing intracellular domain (ICD), and the ability of its cytoplasmic tail to interact with phosphorylated forms of the adaptor protein GULP, the mammalian homologue of CED-6 (220,266).
A critical role for the Drosophila CED-1 homologue, Draper, has also been identified. Draper is expressed in two types of phagocytes: glia and hemocytes/macrophages (71). Freeman et al. (71) demonstrated that deletion of the draper locus in embryos resulted in an increased number of apoptotic neurons in the central nervous system, suggesting the involvement of this molecule in glial phagocytosis of apoptotic neurons. Manaka et al. confirmed the role of Draper in glia and, importantly, found that Draper is also involved in the phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages (151). The importance of CED-1 and Draper in C. elegans and Drosophila, respectively, has been clearly established, and while it has not been shown as definitively, evidence is beginning to accumulate suggesting that LRP1 may play a similar role in mammalian cells, as discussed below.
By employing a chimeric receptor, the hypothesis that the LRP1 cytoplasmic domain contains the necessary structural information to mediate phagocytosis was confirmed (206). RAW 264.7 cells (a macrophage cell line) were transfected with a chimeric receptor consisting of the extracellular domain of CD2, a T-cell surface protein capable of binding non-opsonized sheep red blood cells (SRBC), fused with the transmembrane domain and cytoplasmic tail of LRP1. As a control, a construct containing the LDL receptor transmembrane and intracellular domains was also generated. Cells transfected with the hybrid receptor containing the LDL receptor ICD were able to bind, but could not internalize, SRBC. In contrast, the phagocytes transfected with the chimeric receptor containing the LRP1 transmembrane and ICD were capable of internalizing SRBC, indicating that sufficient information is present within the LRP1 cytoplasmic domain to mediate phagocytosis. This work highlighted the potential of LRP1 to participate in phagocytosis.
B. Putative role of LRP1 in calreticulin-mediated phagocytosis of apoptotic cells
Members of the defense collagen family, including C1q and the collectins (lung surfactant proteins A and D (SP-A and SP-D), mannose binding lectin (MBL) and ficolin), assist in the recognition of particles and enhance the phagocytic activity of professional phagocytes (reviewed in (20)). These molecules are characterized by their structural similarity and contain a globular head and conserved collagen-like tails connected via a hinge region. C1q, a component of the initiator of the classical pathway of complement activation, was shown to bind to apoptotic cells via its globular head region (121,182). In 2001, Ogden et al. provided evidence that LRP1 on phagocytes, in a complex with cell-surface CRT, mediates enhanced ingestion of apoptotic cells opsonized with C1q and MBL (195). CRT, previously shown along with TSP1 and LRP1 to mediate focal adhesion disassembly (see Section IX), is also suspected to participate in phagocytosis (175). It may be delivered to the plasma membrane upon ER-mediated phagocytosis, an engulfment process during which portions of the ER are thought to supply membrane for development of the phagosome (56,57). In any case, CRT does not have a transmembrane domain of its own and, as such, would require interaction with another receptor in order to signal the initiation of events necessary for phagocytosis, e.g. cytoskeletal rearrangement. Ogden and colleagues reported that anti LRP1 and anti-CRT antibodies inhibited the uptake of apoptotic Jurkat T cells, coated with C1q or MBL, by human monocyte-derived macrophages in vitro (195), suggesting that LRP1, complexed with CRT, on phagocytes is a crucial player in the recognition events required for triggering the enhancement of phagocytosis (195). Interestingly, Ogden and colleagues reported that it is the collagen-like tails of C1q which actually interact with CRT/LRP1 on the phagocyte, linking apoptotic cells to phagocytes, and mediating enhanced ingestion (195).
Vandivier and colleagues extended these observations and demonstrated that other defense collagens, in this case lung surfactant proteins A and D, enhance ingestion of apoptotic Jurkat cells by murine alveolar macrophages in an LRP1 and CRT-dependent fashion (277). Again, anti-LRP1 and anti-CRT antibodies were the primary means used to implicate these two proteins in enhancement of phagocytosis.
While the aforementioned study by Ogden and colleagues proposed that LRP1 and CRT form a complex on the phagocyte to bind apoptotic cells opsonized with C1q or MBL, in 2005, Gardai and colleagues proposed an alternative model in which CRT on the surface of apoptotic cells binds as a ligand to LRP1 on the phagocyte and triggers enhanced ingestion, independent of defense collagens (75). They reported reduced uptake of apoptotic fibroblasts, neutrophils and Jurkat cells by fibroblasts in the presence of anti-CRT antibodies. Further, they showed a defect in clearance of apoptotic, CRT-deficient, murine embryonic fibroblasts (MEFs) from the peritoneum of normal mice compared to apoptotic MEFs which expressed CRT normally. These results led them to suggest that CRT is a surface ligand found on apoptotic cells that is critical for their removal. Furthermore, anti-LRP1 antibodies and treatment with RAP resulted in a decrease in LRP1-mediated phagocytosis of apoptotic neutrophils by the J774 macrophage cell line.
In addition to triggering engulfment, receptors responsible for mediating phagocytosis also act to define the consequences of phagocytosis as either pro-inflammatory or anti-inflammatory, depending on the combination of the target bound and the receptor involved (265). Gardai and colleagues suggested that certain defense collagens may serve as surveillance molecules which are capable of signaling either pro-or anti-inflammatory responses, depending on conditions in vivo (76). They proposed a model whereby, under normal conditions, the globular head regions of the lung collectins SP-A and SP-D are free to bind signal inhibitory regulatory protein α (SIRP-α), activating the tyrosine phosphatase SHP-1 with downstream blockade of signaling through src-family kinases and p38 MAP kinase leading to suppression of pro-inflammatory mediator production. Alternatively, in the setting of an infected or damaged lung, SP-A or SP-D bind foreign organisms or cell debris with their globular head regions while their tails interact with CRT/LRP1 and enhance phagocytosis and, via the p38 MAP kinase pathway, signal upregulation of NFκB-mediated transcription of pro-inflammatory mediators. While a direct role for LRP1 was not demonstrated in this work (76), the concept that LRP1 may be involved in regulating both phagocytosis and the overall inflammatory responses to environmental challenges in mammalian organisms is of great interest and will surely be further investigated in the future.
C. Summary
While the molecular mechanism governing defense collagen-mediated recognition of apoptotic cells and the role of LRP1/CRT dependent or independent of these molecules remains to be elucidated, evidence suggesting an important role for LRP1 in the process of phagocytosis is accumulating. To this point, indirect means of evaluating LRP1’s role in these processes have been employed, i.e. blocking antibodies and ligand-binding inhibitors. Use of the newly generated tissue-specific LRP1-deficient mice should enable direct testing of the role of LRP1 in this exciting area.
XII. Role of LRP in regulating immune responses
The receptor-mediated uptake of foreign molecules by endocytic receptors present on dendritic cells is an effective means of presenting antigens to MHC class II molecules (269). Although additional work is required, an emerging role for LRP1 in this process is suggested by studies showing that LRP1-mediated uptake of molecules covalently bound to α2M significantly enhances antigen presentation. Likewise, it is thought that LRP1 facilitates antigen presentation of peptides associated with various heat shock proteins by mediating their endocytosis.
A. Receptor-mediated antigen delivery mediated via α2-macroglobulin
In addition to its ability to inhibit proteases, α2M can form covalent complexes with diverse proteases during a transient protease-activated state (42,43). The resulting complexes are then internalized after binding to LRP1. To determine if α2M enhances antigen delivery and presentation, Chu and Pizzo (43) used T hybridoma clones that respond only to hen egg lysozyme in a MHC-restricted manner. Macrophages that were incubated with lysozyme-α2M-elastase complexes required 200 to 250 times less antigen than those incubated with free lysozyme in order to achieve effective presentation to T cells. Further, adding equimolar amounts of α2M-elastase complexes effectively blocked the presentation of lysozyme-α2M-elastase complexes but had no effect on free lysozyme presentation. These results indicate that LRP1-competent forms of α2M can enhance antigen processing by delivering antigens into macrophages through this LRP1-mediated process.
B. Role of LRP1 in mediating the uptake of heat shock proteins
Heat shock proteins are conserved peptide binding molecules that control the folding of proteins by preventing their aggregation (87). In addition, heat shock proteins appear to be very effective in interacting with antigen presenting cells (APCs) and facilitating the delivery of peptides to the MHC complex (255). Since heat shock proteins are released from cells as a result of necrotic death (8), their ability to deliver peptides to the MHC complex provides a potential pathway through which antigens unique to cancer cells are cross-presented by the APCs to naïve T-cells within the lymph node (7).
To identify the cellular receptor(s) responsible for the endocytosis of heat shock proteins, Binder et al. (17) performed affinity chromatography experiments with immobilized gp96, and identified an 80 kDa polypeptide that was eluted from the affinity column. Protein sequencing of this fraction identified four peptides that originated from the amino-terminal portion of the LRP1 515 kDa heavy chain, and thus represent a proteolytic fragment of this chain. Curiously, an antibody prepared against the material eluted from the affinity column failed to recognize the 515 kDa LRP1 heavy chain from macrophage extracts, as one would expect if the 80 kDa polypeptide was a proteolytic cleavage product of the LRP1 515 kDa subunit. Based on this work and additional studies, it has been suggested that LRP1 functions as a receptor for gp96, hsp90, and hsp70 (7,17) and plays an essential role in presentation of peptides to MHC complexes (18). However, the contribution of LRP1 to this process appears controversial at this time, as Berwin et al. (14) found that excess forms of activated α2M or RAP failed to compete for the binding and uptake of gp96 in macrophages, revealing that receptors distinct from LRP1 are involved in gp96 internalization. Indeed, subsequent work identified scavenger receptor A as the primary receptor for mediating gp96 internalization in macrophages (15). Thus, the exact role that LRP1 plays in mediating the internalization of various heat shock proteins remains to be firmly established.
XIII. Conclusions
The complexity of LRP1’s role in biology arises from its ability to interact with a variety of ligands, each of which uniquely contributes to different aspects of physiology. Additionally, the cytoplasmic domain of LRP1 has the potential to engage a variety of adaptor molecules involved in endocytosis, phagocytosis and cell signaling. Together, these important properties place LRP1 in a unique position to impact normal and abnormal mammalian physiology in a variety of ways. Indeed, studies employing tissue-selective deletion of LRP1 in murine neurons, vascular smooth muscle cells, hepatocytes, adipocytes and macrophages have revealed additional unique and distinct functions for LRP1. We eagerly anticipate development of additional tissue-selective LRP1 knockout mice and look forward to the continued clarification of both normal and pathological processes that future investigations of LRP1 promise to bring.
ACKNOWLEDGMENTS
This work was supported by grants HL 050784 (DKS), HL054710 (DKS), HL072929 (DKS) and HL079644 (JMU). APL was supported by the Duke University Medical Scientist Training Program, grant T32GM07171, as well as grant T32HL007698 while LBV was supported by the UAB Medical Scientist Training Program, grant T32GM008361-16.
References
- 1.Abe Ji, Deguchi J, Matsumoto T, Takuwa N, Noda M, Ohno M, Makuuchi M, Kurokawa K, Takuwa Y. Stimulated Activation of Platelet-Derived Growth Factor Receptor In Vivo in Balloon-Injured Arteries : A Link Between Angiotensin II and Intimal Thickening. Circulation. 1997;96:1906–1913. doi: 10.1161/01.cir.96.6.1906. [DOI] [PubMed] [Google Scholar]
- 2.Argraves KM, Battey FD, MacCalman CD, McCrae KR, Gåfvels M, Kozarsky KF, Chappell DA, Strauss JF, Strickland DK. The very low density lipoprotein receptor mediates the cellular catabolism of lipoprotein lipase and urokinase-plasminogen activator inhibitor type I complexes. J Biol Chem. 1995;270:26550–26557. doi: 10.1074/jbc.270.44.26550. [DOI] [PubMed] [Google Scholar]
- 3.Ashcom JD, Tiller SE, Dickerson K, Cravens JL, Argraves WS, Strickland DK. The human α2-macroglobulin receptor: identification of a 420-kD cell surface glycoprotein specific for the activated conformation of α2-macroglobulin. J Cell Biol. 1990;110:1041–1048. doi: 10.1083/jcb.110.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker RN, Cancilla PA, Pollock PS, Frommes SP. The movement of exogenous protein in experimental cerebral edema. An electron microscopic study after freeze-injury. J Neuropathol Exp Neurol. 1971;30:668–679. doi: 10.1097/00005072-197110000-00010. [DOI] [PubMed] [Google Scholar]
- 5.Barker TH, Pallero MA, MacEwen MW, Tilden SG, Woods A, Murphy-Ullrich JE, Hagood JS. Thrombospondin-1-induced focal adhesion disassembly in fibroblasts requires Thy-1 surface expression, lipid raft integrity, and Src activation. J Biol Chem. 2004;279:23510–23516. doi: 10.1074/jbc.M402169200. [DOI] [PubMed] [Google Scholar]
- 6.Barnes H, Ackermann EJ, van Der GP. v-Src induces Shc binding to tyrosine 63 in the cytoplasmic domain of the LDL receptor-related protein 1. Oncogene. 2003;22:3589–3597. doi: 10.1038/sj.onc.1206504. [DOI] [PubMed] [Google Scholar]
- 7.Basu S, Binder RJ, Ramalingam T, Srivastava PK. CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity. 2001;14:303–313. doi: 10.1016/s1074-7613(01)00111-x. [DOI] [PubMed] [Google Scholar]
- 8.Basu S, Binder RJ, Suto R, Anderson KM, Srivastava PK. Necrotic but not apoptotic cell death releases heat shock proteins, which deliver a partial maturation signal to dendritic cells and activate the NF-kappa B pathway. Int Immunol. 2000;12:1539–1546. doi: 10.1093/intimm/12.11.1539. [DOI] [PubMed] [Google Scholar]
- 9.Behrendt N, Ronne E, Dano K. The structure and function of the urokinase receptor, a membrane protein governing plasminogen activation on the cell surface. Biol Chem Hoppe Seyler. 1995;376:269–279. [PubMed] [Google Scholar]
- 10.Bein K, Simons M. Thrombospondin type 1 repeats interact with matrix metalloproteinase 2. Regulation of metalloproteinase activity. J Biol Chem. 2000;275:32167–32173. doi: 10.1074/jbc.M003834200. [DOI] [PubMed] [Google Scholar]
- 11.Beisiegel U. Lipoprotein metabolism. Eur Heart J. 1998;19 Suppl A:A20–A23. [PubMed] [Google Scholar]
- 12.Beisiegel U, Weber W, Ihrke G, Herz J, Stanley KK. The LDL-receptor related protein, LRP, is an apolipoprotein E binding protein. Nature. 1989;341:162–164. doi: 10.1038/341162a0. [DOI] [PubMed] [Google Scholar]
- 13.Bellis SL. Variant glycosylation: an underappreciated regulatory mechanism for beta1 integrins. Biochim Biophys Acta. 2004;1663:52–60. doi: 10.1016/j.bbamem.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Berwin B, Hart JP, Pizzo SV, Nicchitta CV. Cutting edge: CD91-independent cross-presentation of GRP94(gp96)-associated peptides. J Immunol. 2002;168:4282–4286. doi: 10.4049/jimmunol.168.9.4282. [DOI] [PubMed] [Google Scholar]
- 15.Berwin B, Hart JP, Rice S, Gass C, Pizzo SV, Post SR, Nicchitta CV. Scavenger receptor-A mediates gp96/GRP94 and calreticulin internalization by antigen-presenting cells. EMBO J. 2003;22:6127–6136. doi: 10.1093/emboj/cdg572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betsholtz C, Lindblom P, Bjarnegard M, Enge M, Gerhardt H, Lindahl P. Role of platelet-derived growth factor in mesangium development and vasculopathies: lessons from platelet-derived growth factor and platelet-derived growth factor receptor mutations in mice. Curr Opin Nephrol Hypertens. 2004;13:45–52. doi: 10.1097/00041552-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 17.Binder RJ, Han DK, Srivastava PK. CD91: a receptor for heat shock protein gp96. Nat Immunol. 2000;1:151–155. doi: 10.1038/77835. [DOI] [PubMed] [Google Scholar]
- 18.Binder RJ, Srivastava PK. Essential role of CD91 in re-presentation of gp96-chaperoned peptides. Proc Natl Acad Sci U S A. 2004;101:6128–6133. doi: 10.1073/pnas.0308180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blasi F. Urokinase and Urokinase receptor: A paracrine/Autocrine System regulating Cell Migration and Invasiveness. BioEssays. 1993;15:105–111. doi: 10.1002/bies.950150206. [DOI] [PubMed] [Google Scholar]
- 20.Bohlson SS, Fraser DA, Tenner AJ. Complement proteins C1q and MBL are pattern recognition molecules that signal immediate and long-term protective immune functions. Mol Immunol. 2007;44:33–43. doi: 10.1016/j.molimm.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 21.Bornstein P. Thrombospondins as matricellular modulators of cell function. J Clin Invest. 2001;107:929–934. doi: 10.1172/JCI12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boucher P, Li WP, Matz RL, Takayama Y, Auwerx J, Anderson RG, Herz J. LRP1 Functions as an Atheroprotective Integrator of TGFbeta and PDFG Signals in the Vascular Wall: Implications for Marfan Syndrome. PLoS ONE. 2007;2:e448. doi: 10.1371/journal.pone.0000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boucher P, Liu P, Gotthardt M, Hiesberger T, Anderson RG, Herz J. Platelet-derived Growth Factor Mediates Tyrosine Phosphorylation of the Cytoplasmic Domain of the Low Density Lipoprotein Receptor-related Protein in Caveolae. J Biol Chem. 2002;277:15507–15513. doi: 10.1074/jbc.M200428200. [DOI] [PubMed] [Google Scholar]
- 24.Boucher P, Gotthardt M, Li WP, Anderson RGW, Herz J. LRP: Role in Vascular Wall Integrity and Protection from Atherosclerosis. Science. 2003;300:329. doi: 10.1126/science.1082095. [DOI] [PubMed] [Google Scholar]
- 25.Bovenschen N, Herz J, Grimbergen JM, Lenting PJ, Havekes LM, Mertens K, van Vlijmen BJ. Elevated plasma factor VIII in a mouse model of low-density lipoprotein receptor-related protein deficiency. Blood. 2003;101:3933–3939. doi: 10.1182/blood-2002-07-2081. [DOI] [PubMed] [Google Scholar]
- 26.Bovenschen N, Mertens K, Hu L, Havekes LM, van Vlijmen BJ. LDL receptor cooperates with LDL receptor-related protein in regulating plasma levels of coagulation factor VIII in vivo. Blood. 2005;106:906–912. doi: 10.1182/blood-2004-11-4230. [DOI] [PubMed] [Google Scholar]
- 27.Bovenschen N, van Stempvoort G, Voorberg J, Mertens K, Meijer AB. Proteolytic cleavage of factor VIII heavy chain is required to expose the binding-site for low-density lipoprotein receptor-related protein within the A2 domain. J Thromb Haemost. 2006;4:1487–1493. doi: 10.1111/j.1538-7836.2006.01965.x. [DOI] [PubMed] [Google Scholar]
- 28.Broadwell RD, Charlton HM, Ebert P, Hickey WF, Villegas JC, Wolf AL. Angiogenesis and the blood-brain barrier in solid and dissociated cell grafts within the CNS. Prog Brain Res. 1990;82:95–101. doi: 10.1016/s0079-6123(08)62595-9. [DOI] [PubMed] [Google Scholar]
- 29.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 30.Bu G, Geuze HJ, Strous GJ, Schwartz AL. 39 kDa receptor-associated protein is an ER resident protein and molecular chaperone for LDL receptor-related protein. EMBO J. 1995;14:2269–2280. doi: 10.1002/j.1460-2075.1995.tb07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bu G, Maksymovitch EA, Nerbonne JM, Schwartz AL. Expression and function of the low density lipoprotein receptor-related protein (LRP) in mammalian central neurons. J Biol Chem. 1994;269:18521–18528. [PubMed] [Google Scholar]
- 32.Byfield SD, Roberts AB. Lateral signaling enhances TGF-beta response complexity. Trends Cell Biol. 2004;14:107–111. doi: 10.1016/j.tcb.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Cao C, Lawrence DA, Li Y, Von Arnim CA, Herz J, Su EJ, Makarova A, Hyman BT, Strickland DK, Zhang L. Endocytic receptor LRP together with tPA and PAI-1 coordinates Mac-1-dependent macrophage migration. EMBO J. 2006;25:1860–1870. doi: 10.1038/sj.emboj.7601082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 35.Cathcart MK, McNally AK, Chisolm GM. Lipoxygenase-mediated transformation of human low density lipoprotein to an oxidized and cytotoxic complex. J Lipid Res. 1991;32:63–70. [PubMed] [Google Scholar]
- 36.Causevic M, Ramoz N, Haroutunian V, Davis KL, Buxbaum JD. Lack of association between the levels of the low-density lipoprotein receptor-related protein (LRP) and either Alzheimer dementia or LRP exon 3 genotype. J Neuropathol Exp Neurol. 2003;62:999–1005. doi: 10.1093/jnen/62.10.999. [DOI] [PubMed] [Google Scholar]
- 37.Chappell DA, Fry GL, Waknitz MA, Muhonen LE, Pladet MW, Iverius P-H, Strickland DK. Lipoprotein lipase induces catabolism of normal triglyceride- rich lipoproteins via the low density lipoprotein receptor- related protein/α2-macroglobulin receptor in vitro. A process facilitated by cell-surface proteoglycans. J Biol Chem. 1993;268:14168–14175. [PubMed] [Google Scholar]
- 38.Chappell DA, Inoue I, Fry GL, Pladet MW, Bowen SL, Iverius P-H, Lalouel J-M, Strickland DK. Cellular catabolism of normal very low density lipoproteins via the low density lipoprotein receptor-related protein/α2-macroglobulin receptor is induced by the C-terminal domain of lipoprotein lipase. J Biol Chem. 1994;269:18001–18006. [PubMed] [Google Scholar]
- 39.Chazaud B, Ricoux R, Christov C, Plonquet A, Gherardi RK, Barlovatz-Meimon G. Promigratory effect of plasminogen activator inhibitor-1 on invasive breast cancer cell populations. Am J Pathol. 2002;160:237–246. doi: 10.1016/S0002-9440(10)64367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christensen L, Simonsen ACW, Heegaard CW, Moestrup SK, Andersen JA, Andreasen PA. Immunohistochemical localization of urokinase-type plasminogen activator, type-1 plasminogen-activator inhibitor, urokinase receptor and α2-macroglobulin receptor in human breast carcinomas. Int J Cancer. 1996;66:441–452. doi: 10.1002/(SICI)1097-0215(19960516)66:4<441::AID-IJC6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 41.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu CT, Oury TD, Enghild JJ, Pizzo SV. Adjuvant-free in vivo targeting. Antigen delivery by alpha 2- macroglobulin enhances antibody formation. J Immunol. 1994;152:1538–1545. [PubMed] [Google Scholar]
- 43.Chu CT, Pizzo SV. Receptor-mediated antigen delivery into macrophages: Complexing antigen to α2-macroglobulin enhances presentation to T cells. J Immunol. 1993;150:48–58. [PubMed] [Google Scholar]
- 44.Chung CY, Erickson HP. Cell surface annexin II is a high affinity receptor for the alternatively spliced segment of tenascin-C. J Cell Biol. 1994;126:539–548. doi: 10.1083/jcb.126.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooper AD. Hepatic uptake of chylomicron remnants. J Lipid Res. 1997;38:2173–2192. [PubMed] [Google Scholar]
- 46.Craig SW, Johnson RP. Assembly of focal adhesions: progress, paradigms, and portents. Curr Opin Cell Biol. 1996;8:74–85. doi: 10.1016/s0955-0674(96)80051-2. [DOI] [PubMed] [Google Scholar]
- 47.Cyrus T, Witztum JL, Rader DJ, Tangirala R, Fazio S, Linton MF, Funk CD. Disruption of the 12/15-lipoxygenase gene diminishes atherosclerosis in apo E deficient mice. Journal of Clinical Investigation. 1999;103:1597–1604. doi: 10.1172/JCI5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Czekay RP, Aertgeerts K, Curriden SA, Loskutoff DJ. Plasminogen activator inhibitor-1 detaches cells from extracellular matrices by inactivating integrins. J Cell Biol. 2003;160:781–791. doi: 10.1083/jcb.200208117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daly NL, Djordjevic JT, Kroon PA, Smith R. Three-dimensional structure of the second cysteine-rich repeat from the human low-density lipoprotein receptor. Biochemistry. 1995;34:14474–14481. doi: 10.1021/bi00044a025. [DOI] [PubMed] [Google Scholar]
- 50.Daly NL, Scanlon MJ, Djordjevic JT, Kroon PA, Smith R. Three-dimensional structure of a cysteine-rich repeat from the low- density lipoprotein receptor. Proc Natl Acad Sci U S A. 1995;92:6334–6338. doi: 10.1073/pnas.92.14.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis CG, Goldstein JL, Sudhof TC, Anderson RGW, Russell DW, Brown MS. Acid-dependent ligand dissociation and recylcing of LDL receptor mediated by growth factor homology region. Nature. 1987;326:760–765. doi: 10.1038/326760a0. [DOI] [PubMed] [Google Scholar]
- 52.Deane R, Wu Z, Sagare A, Davis J, Du YS, Hamm K, Xu F, Parisi M, LaRue B, Hu HW, Spijkers P, Guo H, Song X, Lenting PJ, Van Nostrand WE, Zlokovic BV. LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron. 2004;43:333–344. doi: 10.1016/j.neuron.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 53.Degryse B, Resnati M, Rabbani SA, Villa A, Fazioli F, Blasi F. Src-dependence and pertussis-toxin sensitivity of urokinase receptor-dependent chemotaxis and cytoskeleton reorganization in rat smooth muscle cells. Blood. 1999;94:649–662. [PubMed] [Google Scholar]
- 54.Degryse B, Neels JG, Czekay RP, Aertgeerts K, Kamikubo Yi, Loskutoff DJ. The LDL-receptor-related protein is a motogenic receptor for PAI-1. J Biol Chem. 2004 doi: 10.1074/jbc.M313004200. M313004200. [DOI] [PubMed] [Google Scholar]
- 55.Descamps O, Bilheimer D, Herz J. Insulin stimulates receptor-mediated uptake of ApoE-enriched lipoproteins and activated α2-macroglobulin in adipocytes. J Biol Chem. 1993;268:974–981. [PubMed] [Google Scholar]
- 56.Desjardins M. ER-mediated phagocytosis: a new membrane for new functions. Nat Rev Immunol. 2003;3:280–291. doi: 10.1038/nri1053. [DOI] [PubMed] [Google Scholar]
- 57.Desjardins M, Griffiths G. Phagocytosis: latex leads the way. Current Opinion in Cell Biology. 2003;15:498–503. doi: 10.1016/s0955-0674(03)00083-8. [DOI] [PubMed] [Google Scholar]
- 58.Dixit VM, Grant GA, Santoro SA, Frazier WA. Isolation and characterization of a heparin-binding domain from the amino terminus of platelet thrombospondin. J Biol Chem. 1984;259:10100–10105. [PubMed] [Google Scholar]
- 59.Dolmer K, Gettins PG. Three complement-like repeats compose the complete alpha2-macroglobulin binding site in the second ligand binding cluster of the low density lipoprotein receptor-related protein. J Biol Chem. 2006;281:34189–34196. doi: 10.1074/jbc.M604389200. [DOI] [PubMed] [Google Scholar]
- 60.Dolmer K, Huang W, Gettins PG. NMR solution structure of complement-like repeat CR3 from the low density lipoprotein receptor-related protein. Evidence for specific binding to the receptor binding domain of human alpha(2)-macroglobulin. J Biol Chem. 2000;275:3264–3269. doi: 10.1074/jbc.275.5.3264. [DOI] [PubMed] [Google Scholar]
- 61.Donoviel DB, Amacher SL, Judge KW, Bornstein P. Thrombospondin gene expression is associated with mitogenesis in 3T3 cells: Induction by basic fibroblast growth factor. J Cell Physiol. 1990;145:16–23. doi: 10.1002/jcp.1041450104. [DOI] [PubMed] [Google Scholar]
- 62.Ellgaard L, Holtet TL, Nielsen PR, Etzerodt M, Gliemann J, Thogersen HC. Dissection of the domain architecture of the α2macroglobulin- receptor-associated protein. European Journal of Biochemistry. 1997;244:544–551. doi: 10.1111/j.1432-1033.1997.00544.x. [DOI] [PubMed] [Google Scholar]
- 63.Emonard H, Bellon G, Troeberg L, Berton A, Robinet A, Henriet P, Marbaix E, Kirkegaard K, Patthy L, Eeckhout Y, Nagase H, Hornebeck W, Courtoy PJ. Low density lipoprotein receptor-related protein mediates endocytic clearance of pro-MMP-2.TIMP-2 complex through a thrombospondin-independent mechanism. J Biol Chem. 2004;279:54944–54951. doi: 10.1074/jbc.M406792200. [DOI] [PubMed] [Google Scholar]
- 64.Espirito Santo SMS, Pires NMM, Boesten LSM, Gerritsen G, Bovenschen N, van Dijk KW, Jukema JW, Princen HMG, Bensadoun A, Li WP, Herz J, Havekes LM, van Vlijmen BJM. Hepatic low-density lipoprotein receptor-related protein deficiency in mice increases atherosclerosis independent of plasma cholesterol. Blood. 2004;103:3777–3782. doi: 10.1182/blood-2003-11-4051. [DOI] [PubMed] [Google Scholar]
- 65.Fass D, Blacklow S, Kim PS, Berger JM. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module [see comments] Nature. 1997;388:691–693. doi: 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- 66.Fay PJ, Jenkins PV. Mutating factor VIII: lessons from structure to function. Blood Rev. 2005;19:15–27. doi: 10.1016/j.blre.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 67.Ferns GA, Raines EW, Sprugel KH, Motani AS, Reidy MA, Ross R. Inhibition of neointimal smooth muscle accumulation after angioplasty by an antibody to PDGF. Science. 1991;253:1129–1132. doi: 10.1126/science.1653454. [DOI] [PubMed] [Google Scholar]
- 68.Fisher C, Beglova N, Blacklow SC. Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol Cell. 2006;22:277–283. doi: 10.1016/j.molcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 69.Fitzgerald DJ, Fryling CM, Zdanovsky A, Saelinger CB, Kounnas M, Winkles JA, Strickland D, Leppla S. Pseudomonas exotoxin-mediated selection yields cells with altered expression of low-density lipoprotein receptor-related protein. J Cell Biol. 1995;129:1533–1541. doi: 10.1083/jcb.129.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fixe P, Praloran V. M-CSF: haematopoietic growth factor or inflammatory cytokine? Cytokine. 1998;10:32–37. doi: 10.1006/cyto.1997.0249. [DOI] [PubMed] [Google Scholar]
- 71.Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- 72.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 73.Fuchs HE, Shifman MA, Pizzo SV. In vivo catabolism of α1-proteinase inhibitor-trypsin, antithrombin III-thrombin, and α2-macroglobulin-methylamine. Biochem Biophys Acta. 1982;716:151–157. doi: 10.1016/0304-4165(82)90263-x. [DOI] [PubMed] [Google Scholar]
- 74.Garcia JH, Lossinsky AS, Kauffman FC, Conger KA. Neuronal ischemic injury: light microscopy, ultrastructure and biochemistry. Acta Neuropathol (Berl) 1978;43:85–95. doi: 10.1007/BF00685002. [DOI] [PubMed] [Google Scholar]
- 75.Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE, Bratton DL, Oldenborg PA, Michalak M, Henson PM. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell. 2005;123:321–334. doi: 10.1016/j.cell.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 76.Gardai SJ, Xiao YQ, Dickinson M, Nick JA, Voelker DR, Greene KE, Henson PM. By binding SIRPalpha or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. 2003;115:13–23. doi: 10.1016/s0092-8674(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 77.George J, Afek A, Shaish A, Levkovitz H, Bloom N, Cyrus T, Zhao L, Funk CD, Sigal E, Harats D. 12/15-Lipoxygenase gene disruption attenuates atherogenesis in LDL receptor-deficient mice. Circulation. 2001;104:1646–1650. doi: 10.1161/hc3901.095772. [DOI] [PubMed] [Google Scholar]
- 78.Goicoechea S, Orr AW, Pallero MA, Eggleton P, Murphy-Ullrich JE. Thrombospondin mediates focal adhesion disassembly through interactions with cell surface calreticulin. J Biol Chem. 2000;275:36358–36368. doi: 10.1074/jbc.M005951200. [DOI] [PubMed] [Google Scholar]
- 79.Goicoechea S, Pallero MA, Eggleton P, Michalak M, Murphy-Ullrich JE. The anti-adhesive activity of thrombospondin is mediated by the N-terminal domain of cell surface calreticulin. J Biol Chem. 2002;277:37219–37228. doi: 10.1074/jbc.M202200200. [DOI] [PubMed] [Google Scholar]
- 80.Gotthardt M, Trommsdorff M, Nevitt MF, Shelton J, Richardson JA, Stockinger W, Nimpf J, Herz J. Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. J Biol Chem. 2000;275:25616–25624. doi: 10.1074/jbc.M000955200. [DOI] [PubMed] [Google Scholar]
- 81.Gray AJ, Park PW, Broekelmann TJ, Laurent GJ, Reeves JT, Stenmark KR, Mecham RP. The mitogenic effects of the B beta chain of fibrinogen are mediated through cell surface calreticulin. J Biol Chem. 1995;270:26602–26606. doi: 10.1074/jbc.270.44.26602. [DOI] [PubMed] [Google Scholar]
- 82.Greenaway J, Lawler J, Moorehead R, Bornstein P, LaMarre J, Petrik J. Thrombospondin-1 inhibits VEGF levels in the ovary directly by binding and internalization via the low density lipoprotein receptor-related protein-1 (LRP-1) J Cell Physiol. 2007;210:807–818. doi: 10.1002/jcp.20904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Greenwood JA, Murphy-Ullrich JE. Signaling of de-adhesion in cellular regulation and motility. Microsc Res Tech. 1998;43:420–432. doi: 10.1002/(SICI)1097-0029(19981201)43:5<420::AID-JEMT8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 84.Grey A, Banovic T, Zhu Q, Watson M, Callon K, Palmano K, Ross J, Naot D, Reid IR, Cornish J. The low-density lipoprotein receptor-related protein 1 is a mitogenic receptor for lactoferrin in osteoblastic cells. Mol Endocrinol. 2004;18:2268–2278. doi: 10.1210/me.2003-0456. [DOI] [PubMed] [Google Scholar]
- 85.Haass C, Schlossmacher MG, Hung AY, Vigo-Pelfrey C, Mellon A, Ostaszewski BL, Lieberburg I, Koo EH, Schenk D, Teplow DB. Amyloid beta-peptide is produced by cultured cells during normal metabolism. Nature. 1992;359:322–325. doi: 10.1038/359322a0. [DOI] [PubMed] [Google Scholar]
- 86.Haberstroh U, Zahner G, Disser M, Thaiss F, Wolf G, Stahl RA. TGF-beta stimulates rat mesangial cell proliferation in culture: role of PDGF beta-receptor expression. Am J Physiol Renal Physiol. 1993;264:F199–F205. doi: 10.1152/ajprenal.1993.264.2.F199. [DOI] [PubMed] [Google Scholar]
- 87.Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 88.Hashimoto Y, Jiang H, Niikura T, Ito Y, Hagiwara A, Umezawa K, Abe Y, Murayama Y, Nishimoto I. Neuronal Apoptosis by Apolipoprotein E4 through Low-Density Lipoprotein Receptor-Related Protein and Heterotrimeric GTPases. J Neurosci. 2000;20:8401–8409. doi: 10.1523/JNEUROSCI.20-22-08401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heegaard CW, Simonsen ACW, Oka K, Kjoller L, Christensen A, Madsen B, Ellgaard L, Chan L, Andreasen PA. Very low density lipoprotein receptor binds and mediates endocytosis of urokinase-type plasminogen activator-type- 1 plasminogen activator inhibitor complex. J Biol Chem. 1995;270:20855–20861. doi: 10.1074/jbc.270.35.20855. [DOI] [PubMed] [Google Scholar]
- 90.Herz J, Clouthier DE, Hammer RE. LDL receptor-related protein internalizes and degrades uPA- PAI-1 complexes and is essential for embryo implantation. Cell. 1992;71:411–421. doi: 10.1016/0092-8674(92)90511-a. [DOI] [PubMed] [Google Scholar]
- 91.Herz J, Goldstein JL, Strickland DK, Ho YK, Brown MS. 39-kDa protein modulates binding of ligands to low density lipoprotein receptor-related protein/α2-macroglobulin receptor. J Biol Chem. 1991;266:21232–21238. [PubMed] [Google Scholar]
- 92.Herz J, Hamann U, Rogne S, Myklebost O, Gausepohl H, Stanley KK. Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hofmann SM, Zhou L, Perez-Tilve D, Greer T, Grant E, Wancata L, Thomas A, Pfluger PT, Basford JE, Gilham D, Herz J, Tschop MH, Hui DY. Adipocyte LDL receptor-related protein-1 expression modulates postprandial lipid transport and glucose homeostasis in mice. J Clin Invest. 2007;117:3271–3282. doi: 10.1172/JCI31929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu L, Boesten LS, May P, Herz J, Bovenschen N, Huisman MV, Berbee JF, Havekes LM, van Vlijmen BJ, Tamsma JT. Macrophage low-density lipoprotein receptor-related protein deficiency enhances atherosclerosis in ApoE/LDLR double knockout mice. Arterioscler Thromb Vasc Biol. 2006;26:2710–2715. doi: 10.1161/01.ATV.0000249641.96896.e6. [DOI] [PubMed] [Google Scholar]
- 96.Huang SS, Ling TY, Tseng WF, Huang YH, Tang FM, Leal SM, Huang JS. Cellular growth inhibition by IGFBP-3 and TGF-{beta}1 requires LRP-1. The FASEB Journal. 2003;17:2068–2081. doi: 10.1096/fj.03-0256com. [DOI] [PubMed] [Google Scholar]
- 97.Huang W, Dolmer K, Gettins PG. NMR solution structure of complement-like repeat CR8 from the low density lipoprotein receptor-related protein. J Biol Chem. 1999;274:14130–14136. doi: 10.1074/jbc.274.20.14130. [DOI] [PubMed] [Google Scholar]
- 98.Iwata N, Tsubuki S, Takaki Y, Watanabe K, Sekiguchi M, Hosoki E, Kawashima-Morishima M, Lee HJ, Hama E, Sekine-Aizawa Y, Saido TC. Identification of the major Abeta1-42-degrading catabolic pathway in brain parenchyma: suppression leads to biochemical and pathological deposition. Nat Med. 2000;6:143–150. doi: 10.1038/72237. [DOI] [PubMed] [Google Scholar]
- 99.Jaffe EA, Ruggiero JT, Falcone DJ. Monocytes and macrophages synthesize and secrete thrombospondin. Blood. 1985;65:79–84. [PubMed] [Google Scholar]
- 100.Jaffe EA, Ruggiero JT, Leung LK, Doyle MJ, McKeown-Longo PJ, Mosher DF. Cultured human fibroblasts synthesize and secrete thrombospondin and incorporate it into extracellular matrix. Proc Natl Acad Sci U S A. 1983;80:998–1002. doi: 10.1073/pnas.80.4.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Janat MF, Liau G. Transforming growth factor beta 1 is a powerful modulator of platelet-derived growth factor action in vascular smooth muscle cells. J Cell Physiol. 1992;150:232–242. doi: 10.1002/jcp.1041500203. [DOI] [PubMed] [Google Scholar]
- 102.Janzer RC, Raff MC. Astrocytes induce blood-brain barrier properties in endothelial cells. Nature. 1987;325:253–257. doi: 10.1038/325253a0. [DOI] [PubMed] [Google Scholar]
- 103.Jawien A, Bowen-Pope DF, Lindner V, Schwartz SM, Clowes AW. Platelet-derived growth factor promotes smooth muscle migration and intimal thickening in a rat model of balloon angioplasty. J Clin Invest. 1992;89:507–511. doi: 10.1172/JCI115613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jensen GA, Andersen OM, Bonvin AM, Bjerrum-Bohr I, Etzerodt M, Thogersen HC, O'Shea C, Poulsen FM, Kragelund BB. Binding site structure of one LRP-RAP complex: implications for a common ligand-receptor binding motif. J Mol Biol. 2006;362:700–716. doi: 10.1016/j.jmb.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 105.Jeon H, Meng W, Takagi J, Eck MJ, Springer TA, Blacklow SC. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat Struct Biol. 2001;8:499–504. doi: 10.1038/88556. [DOI] [PubMed] [Google Scholar]
- 106.Jethmalani SM, Henle KJ, Kaushal GP. Heat shock-induced prompt glycosylation. Identification of P-SG67 as calreticulin. J Biol Chem. 1994;269:23603–23609. [PubMed] [Google Scholar]
- 107.Joslin G, Fallon RJ, Bullock J, Adams SP, Perlmutter DH. The SEC receptor recognizes a pentapeptide neodomain of alpha 1- antitrypsin-protease complexes. J Biol Chem. 1991;266:11282–11288. [PubMed] [Google Scholar]
- 108.Kancha RK, Stearns ME, Hussain MM. Decreased expression of the low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor in invasive cell clones derived from human prostate and breast tumor cells. Oncol Res. 1994;6:365–372. [PubMed] [Google Scholar]
- 109.Kang DE, Pietrzik CU, Baum L, Chevallier N, Merriam DE, Kounnas MZ, Wagner SL, Troncoso JC, Kawas CH, Katzman R, Koo EH. Modulation of amyloid beta-protein clearance and Alzheimer's disease susceptibility by the LDL receptor-related protein pathway. J Clin Invest. 2000;106:1159–1166. doi: 10.1172/JCI11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kang DE, Saitoh T, Chen X, Xia Y, Masliah E, Hansen LA, Thomas RG, Thal LJ, Katzman R. Genetic association of the low-density lipoprotein receptor- related protein gene (LRP), an apolipoprotein E receptor, with late-onset Alzheimer's disease. Neurology. 1997;49:56–61. doi: 10.1212/wnl.49.1.56. [DOI] [PubMed] [Google Scholar]
- 111.Kasza A, Petersen HH, Heegaard CW, Oka K, Christensen A, Dubin A, Chan L, Andreasen PA. Specificity of serine proteinase/serpin complex binding to very- low-density lipoprotein receptor and alpha2-macroglobulin receptor/low-density-lipoprotein-receptor-related protein. Eur J Biochem. 1997;248:270–281. doi: 10.1111/j.1432-1033.1997.00270.x. [DOI] [PubMed] [Google Scholar]
- 112.Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, Schnabel R, Hengartner MO. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 2005;434:93–99. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- 113.Kinoshita A, Shah T, Tangredi MM, Strickland DK, Hyman BT. The Intracellular Domain of the Low Density Lipoprotein Receptor-related Protein Modulates Transactivation Mediated by Amyloid Precursor Protein and Fe65. J Biol Chem. 2003;278:41182–41188. doi: 10.1074/jbc.M306403200. [DOI] [PubMed] [Google Scholar]
- 114.Kinoshita A, Whelan CM, Smith CJ, Mikhailenko I, Rebeck GW, Strickland DK, Hyman BT. Demonstration by Fluorescence Resonance Energy Transfer of Two Sites of Interaction between the Low-Density Lipoprotein Receptor-Related Protein and the Amyloid Precursor Protein: Role of the Intracellular Adapter Protein Fe65. J Neurosci. 2001;21:8354–8361. doi: 10.1523/JNEUROSCI.21-21-08354.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kiosses WB, Shattil SJ, Pampori N, Schwartz MA. Rac recruits high-affinity integrin alphavbeta3 to lamellipodia in endothelial cell migration. Nat Cell Biol. 2001;3:316–320. doi: 10.1038/35060120. [DOI] [PubMed] [Google Scholar]
- 116.Kita T, Goldstein JL, Brown MS, Watanabe Y, Hornick CA, Havel RJ. Hepatic uptake of chylomicron remnants in WHHL rabbits: a mechanism genetically distinct from the low density lipoprotein receptor. Proc Natl Acad Sci U S A. 1982;79:3623–3627. doi: 10.1073/pnas.79.11.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Knauer MF, Crisp RJ, Kridel SJ, Knauer DJ. Analysis of a structural determinant in thrombin-protease nexin 1 complexes that mediates clearance by the low density lipoprotein receptor-related protein. J Biol Chem. 1999;274:275–281. doi: 10.1074/jbc.274.1.275. [DOI] [PubMed] [Google Scholar]
- 118.Knauer MF, Hawley SB, Knauer DJ. Identification of a binding site in protease nexin I (PN1) required for the receptor mediated internalization of PN1-thrombin complexes. J Biol Chem. 1997;272:12261–12264. doi: 10.1074/jbc.272.19.12261. [DOI] [PubMed] [Google Scholar]
- 119.Knauer MF, Orlando RA, Glabe CG. Cell surface APP751 forms complexes with protease nexin 2 ligands and is internalized via the low density lipoprotein receptor-related protein (LRP) Brain Res. 1996;740:6–14. doi: 10.1016/s0006-8993(96)00711-1. [DOI] [PubMed] [Google Scholar]
- 120.Kolsch H, Ptok U, Mohamed I, Schmitz S, Rao ML, Maier W, Heun R. Association of the C766T polymorphism of the low-density lipoprotein receptor-related protein gene with Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet. 2003;121:128–130. doi: 10.1002/ajmg.b.20043. [DOI] [PubMed] [Google Scholar]
- 121.Korb LC, Ahearn JM. C1q binds directly and specifically to surface blebs of apoptotic human keratinocytes: complement deficiency and systemic lupus erythematosus revisited. J Immunol. 1997;158:4525–4528. [PubMed] [Google Scholar]
- 122.Kounnas MZ, Church FC, Argraves WS, Strickland DK. Cellular internalization and degradation of antithrombin III-thrombin, heparin cofactor II-thrombin, and α1-antitrypsin- trypsin complexes is mediated by the low density lipoprotein receptor-related protein. J Biol Chem. 1996;271:6523–6529. doi: 10.1074/jbc.271.11.6523. [DOI] [PubMed] [Google Scholar]
- 123.Kounnas MZ, Moir RD, Rebeck GW, Bush AI, Argraves WS, Tanzi RE, Hyman BT, Strickland DK. LDL receptor-related protein, a multifunctional apoE receptor, binds secreted β-amyloid precursor protein and mediates its degradation. Cell. 1995;82:331–340. doi: 10.1016/0092-8674(95)90320-8. [DOI] [PubMed] [Google Scholar]
- 124.Kristensen T, Moestrup SK, Gliemann J, Bendtsen L, Sand O, Sottrup-Jensen L. Evidence that the newly cloned LRP is the α2M receptor. FEBS Lett. 1990;276:151–155. doi: 10.1016/0014-5793(90)80530-v. [DOI] [PubMed] [Google Scholar]
- 125.Kurniawan ND, Atkins AR, Bieri S, Brown CJ, Brereton IM, Kroon PA, Smith R. NMR structure of a concatemer of the first and second ligand-binding modules of the human low-density lipoprotein receptor. Protein Sci. 2000;9:1282–1293. doi: 10.1110/ps.9.7.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lalazar A, Weisgraber KH, Rall SC, Jr, Giladi H, Innerarity TL, Levanon AZ, Boyles JK, Amit B, Gorecki M, Mahley RW. Site-specific mutagenesis of human apolipoprotein E. Receptor binding activity of variants with single amino acid substitutions. J Biol Chem. 1988;263:3542–3545. [PubMed] [Google Scholar]
- 127.Lawler JW, Slayter HS. The release of heparin binding peptides from platelet thrombospondin by proteolytic action of thrombin, plasmin and trypsin. Thromb Res. 1981;22:267–279. doi: 10.1016/0049-3848(81)90119-5. [DOI] [PubMed] [Google Scholar]
- 128.Lawler JW, Slayter HS, Coligan JE. Isolation and characterization of a high molecular weight glycoprotein from human blood platelets. J Biol Chem. 1978;253:8609–8616. [PubMed] [Google Scholar]
- 129.Lazic A, Dolmer K, Strickland DK, Gettins PG. Structural organization of the receptor associated protein. Biochemistry. 2003;42:14913–14920. doi: 10.1021/bi035779e. [DOI] [PubMed] [Google Scholar]
- 130.Lee D, Walsh JD, Migliorini M, Ping Y, Cai T, Schweiters CD, Krueger S, Strickland DK, Wang Y-X. The structure of receptor-associated protein (RAP) Protein Sci. 2007;16:1628–1640. doi: 10.1110/ps.072865407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee D, Walsh JD, Mikhailenko I, Yu P, Migliorini M, Wu Y, Krueger S, Curtis JE, Harris B, Lockett S, Blacklow SC, Strickland DK, Wang Y-X. RAP Uses a Histidine Switch to Regulate Its Interaction with LRP in the ER and Golgi. Molecular Cell. 2006;22:423–430. doi: 10.1016/j.molcel.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 132.Lee NV, Sato M, Annis DS, Loo JA, Wu L, Mosher DF, Iruela-Arispe ML. ADAMTS1 mediates the release of antiangiogenic polypeptides from TSP1 and 2. EMBO J. 2006;25:5270–5283. doi: 10.1038/sj.emboj.7601400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lee SW, Kim WJ, Choi YK, Song HS, Son MJ, Gelman IH, Kim YJ, Kim KW. SSeCKS regulates angiogenesis and tight junction formation in blood-brain barrier. Nat Med. 2003;9:900–906. doi: 10.1038/nm889. [DOI] [PubMed] [Google Scholar]
- 134.Lendon CL, Talbot CJ, Craddock NJ, Han SW, Wragg M, Morris JC, Goate AM. Genetic association studies between dementia of the Alzheimer's type and three receptors for apolipoprotein E in a Caucasian population. Neurosci Lett. 1997;222:187–190. doi: 10.1016/s0304-3940(97)13381-x. [DOI] [PubMed] [Google Scholar]
- 135.Lenting PJ, Neels JG, van Den Berg BM, Clijsters PP, Meijerman DW, Pannekoek H, van Mourik JA, Mertens K, Van Zonneveld AJ. The light chain of factor VIII comprises a binding site for low density lipoprotein receptor-related protein. J Biol Chem. 1999;274:23734–23739. doi: 10.1074/jbc.274.34.23734. [DOI] [PubMed] [Google Scholar]
- 136.Li Y, Wood N, Grimsley P, Yellowlees D, Donnelly PK. In vitro invasiveness of human breast cancer cells is promoted by low density lipoprotein receptor-related protein. Invasion Metastasis. 1998;18:240–251. doi: 10.1159/000024517. [DOI] [PubMed] [Google Scholar]
- 137.Li Y, Wood N, Parsons PG, Yellowlees D, Donnelly PK. Expression of alpha2-macroglobulin receptor/low density lipoprotein receptor-related protein on surfaces of tumour cells: a study using flow cytometry. Cancer Lett. 1997;111:199–205. doi: 10.1016/s0304-3835(96)04520-x. [DOI] [PubMed] [Google Scholar]
- 138.Li YM, Xu M, Lai MT, Huang Q, Castro JL, DiMuzio-Mower J, Harrison T, Lellis C, Nadin A, Neduvelil JG, Register RB, Sardana MK, Shearman MS, Smith AL, Shi XP, Yin KC, Shafer JA, Gardell SJ. Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405:689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- 139.Lillis AP, Mikhailenko I, Strickland DK. Beyond endocytosis: LRP function in cell migration, proliferation and vascular permeability. J Thromb Haemost. 2005;3:1884–1893. doi: 10.1111/j.1538-7836.2005.01371.x. [DOI] [PubMed] [Google Scholar]
- 140.Lindahl P, Johansson BR, Leveen P, Betsholtz C. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science. 1997;277:242–245. doi: 10.1126/science.277.5323.242. [DOI] [PubMed] [Google Scholar]
- 141.Liu Q, Zerbinatti CV, Zhang J, Hoe HS, Wang B, Cole SL, Herz J, Muglia L, Bu G. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron. 2007;56:66–78. doi: 10.1016/j.neuron.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Loukinova E, Ranganathan S, Kuznetsov S, Gorlatov N, Migliorini MM, Loukinov D, Ulery PG, Mikhailenko I, Lawrence DA, Strickland DK. PDGF-induced tyrosine phosphorylation of the LDL receptor-related protein (LRP): Evidence for integrated co-receptor function between LRP and the PDGF receptor. J Biol Chem. 2002;277:15499–15506. doi: 10.1074/jbc.M200427200. [DOI] [PubMed] [Google Scholar]
- 143.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ma Z, Thomas KS, Webb DJ, Moravec R, Salicioni AM, Mars WM, Gonias SL. Regulation of Rac1 activation by the low density lipoprotein receptor-related protein. J Cell Biol. 2002;159:1061–1070. doi: 10.1083/jcb.200207070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.MacArthur JM, Bishop JR, Stanford KI, Wang L, Bensadoun A, Witztum JL, Esko JD. Liver heparan sulfate proteoglycans mediate clearance of triglyceride-rich lipoproteins independently of LDL receptor family members. Journal of Clinical Investigation. 2007;117:153–164. doi: 10.1172/JCI29154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Maderna P, Godson C. Phagocytosis of apoptotic cells and the resolution of inflammation. Biochim Biophys Acta. 2003;1639:141–151. doi: 10.1016/j.bbadis.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 147.Maekawa H, Tollefsen DM. Role of the proposed serpin-enzyme complex receptor recognition site in binding and internalization of thrombin-heparin cofactor II complexes by hepatocytes. J Biol Chem. 1996;271:18604–18610. doi: 10.1074/jbc.271.31.18604. [DOI] [PubMed] [Google Scholar]
- 148.Mahley RW, Huang Y. Atherogenic remnant lipoproteins: role for proteoglycans in trapping, transferring, and internalizing. Journal of Clinical Investigation. 2007;117:94–98. doi: 10.1172/JCI30889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Majack RA, Mildbrandt J, Dixit VM. Induction of thrombospondin messenger RNA levels occurs as an immediate primary response to platelet-derived growth factor. J Biol Chem. 1987;262:8821–8825. [PubMed] [Google Scholar]
- 150.Makarova A, Williams SE, Strickland DK. Proteases and lipoprotein receptors in Alzheimer's disease. Cell Biochem Biophys. 2004;41:139–178. doi: 10.1385/CBB:41:1:139. [DOI] [PubMed] [Google Scholar]
- 151.Manaka J, Kuraishi T, Shiratsuchi A, Nakai Y, Higashida H, Henson P, Nakanishi Y. Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J Biol Chem. 2004;279:48466–48476. doi: 10.1074/jbc.M408597200. [DOI] [PubMed] [Google Scholar]
- 152.Masliah E, Sisk A, Mallory M, Mucke L, Schenk D, Games D. Comparison of neurodegenerative pathology in transgenic mice overexpressing V717F β-amyloid precursor protein and Alzheimer's disease. J Neurosci. 1996;16:5795–5811. doi: 10.1523/JNEUROSCI.16-18-05795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mast AE, Enghild JJ, Pizzo SV, Salvesen G. Analysis of the plasma elimination kinetics and conformational stabilities of native, proteinase-complexed, and reactive site cleaved serpins: Comparison of α1-proteinase inhibitor, α1-antichymotrypsin, antithrombin III, α2-antiplasmin, angiotensinogen, and ovalbumin. Biochemistry. 1991;30:1723–1730. doi: 10.1021/bi00220a039. [DOI] [PubMed] [Google Scholar]
- 154.May P, Reddy YK, Herz J. Proteolytic processing of LRP mediates regulated release of its intracellular domain. J Biol Chem. 2002;277:18736–18743. doi: 10.1074/jbc.M201979200. [DOI] [PubMed] [Google Scholar]
- 155.May P, Rohlmann A, Bock HH, Zurhove K, Marth JD, Schomburg ED, Noebels JL, Beffert U, Sweatt JD, Weeber EJ, Herz J. Neuronal LRP1 functionally associates with postsynaptic proteins and is required for normal motor function in mice. Mol Cell Biol. 2004;24:8872–8883. doi: 10.1128/MCB.24.20.8872-8883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McKeown-Longo PJ, Hanning R, Mosher DF. Binding and degradation of platelet thrombospondin by cultured fibroblasts. J Cell Biol. 1984;98:22–28. doi: 10.1083/jcb.98.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Merkel M, Kako Y, Radner H, Cho IS, Ramasamy R, Brunzell JD, Goldberg IJ, Breslow JL. Catalytically inactive lipoprotein lipase expression in muscle of transgenic mice increases very low density lipoprotein uptake: Direct evidence that lipoprotein lipase bridging occurs in vivo. Proc Natl Acad Sci USA. 1998;95:13841–13846. doi: 10.1073/pnas.95.23.13841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Merle B, Malaval L, Lawler J, Delmas P, Clezardin P. Decorin inhibits cell attachment to thrombospondin-1 by binding to a KKTR-dependent cell adhesive site present within the N-terminal domain of thrombospondin-1. J Cell Biochem. 1997;67:75–83. [PubMed] [Google Scholar]
- 159.Mesaeli N, Nakamura K, Zvaritch E, Dickie P, Dziak E, Krause KH, Opas M, MacLennan DH, Michalak M. Calreticulin is essential for cardiac development. J Cell Biol. 1999;144:857–868. doi: 10.1083/jcb.144.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Migliorini MM, Behre EH, Brew S, Ingham KC, Strickland DK. Allosteric Modulation of Ligand Binding to Low Density Lipoprotein Receptor-related Protein by the Receptor-associated Protein Requires Critical Lysine Residues within Its Carboxyl-terminal Domain. J Biol Chem. 2003;278:17986. doi: 10.1074/jbc.M212592200. [DOI] [PubMed] [Google Scholar]
- 161.Mignatti P, Rifkin DB. Biology and biochemistry of proteinases in tumor invasion. Physiol Rev. 1993;73:161–195. doi: 10.1152/physrev.1993.73.1.161. [DOI] [PubMed] [Google Scholar]
- 162.Mikhailenko I, Considine W, Argraves KM, Loukinov D, Hyman BT, Strickland DK. Functional domains of the very low density lipoprotein receptor: molecular analysis of ligand binding and acid-dependent ligand dissociation mechanisms. J Cell Sci. 1999;112:3269–3281. doi: 10.1242/jcs.112.19.3269. [DOI] [PubMed] [Google Scholar]
- 163.Mikhailenko I, Kounnas MZ, Strickland DK. Low density lipoprotein receptor-related protein/α2-macroglobulin receptor mediates the cellular internalization and degradation of thrombospondin. A process facilitated by cell-surface proteoglycans. J Biol Chem. 1995;270:9543–9549. doi: 10.1074/jbc.270.16.9543. [DOI] [PubMed] [Google Scholar]
- 164.Mikhailenko I, Krylov D, Argraves KM, Roberts DD, Liau G, Strickland DK. Cellular internalization and degradation of thrombospondin-1 is mediated by the amino-terminal heparin binding domain (HBD) - High affinity interaction of dimeric HBD with the low density lipoprotein receptor-related protein. J Biol Chem. 1997;272:6784–6791. doi: 10.1074/jbc.272.10.6784. [DOI] [PubMed] [Google Scholar]
- 165.Milner RE, Baksh S, Shemanko C, Carpenter MR, Smillie L, Vance JE, Opas M, Michalak M. Calreticulin, and not calsequestrin, is the major calcium binding protein of smooth muscle sarcoplasmic reticulum and liver endoplasmic reticulum. J Biol Chem. 1991;266:7155–7165. [PubMed] [Google Scholar]
- 166.Misra UK, Chu CTC, Gawdi G, Pizzo SV. The relationship between low density lipoprotein-related protein/α2-macroglobulin (α2M) receptors and the newly described α2M signaling receptor. J Biol Chem. 1994;269:18303–18306. [PubMed] [Google Scholar]
- 167.Miyake S, Lupher ML, Jr, Druker B, Band H. The tyrosine kinase regulator Cbl enhances the ubiquitination and degradation of the platelet-derived growth factor receptor alpha. Proc Natl Acad Sci U S A. 1998;95:7927–7932. doi: 10.1073/pnas.95.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Miyake S, Mullane-Robinson KP, Lill NL, Douillard P, Band H. Cbl-mediated negative regulation of platelet-derived growth factor receptor-dependent cell proliferation. A critical role for Cbl tyrosine kinase-binding domain. J Biol Chem. 1999;274:16619–16628. doi: 10.1074/jbc.274.23.16619. [DOI] [PubMed] [Google Scholar]
- 169.Moestrup SK, Gliemann J. Purification of the Rat Hepatic α2-Macroglobulin Receptor as an Approximately 440 kDa Single Chain Polypeptide. J Biol Chem. 1989;264:15574–15577. [PubMed] [Google Scholar]
- 170.Moissoglu K, Schwartz MA. Integrin signalling in directed cell migration. Biol Cell. 2006;98:547–555. doi: 10.1042/BC20060025. [DOI] [PubMed] [Google Scholar]
- 171.Morange PE, Tregouet DA, Frere C, Saut N, Pellegrina L, Alessi MC, Visvikis S, Tiret L, Juhan-Vague I. Biological and genetic factors influencing plasma factor VIII levels in a healthy family population: results from the Stanislas cohort. Br J Haematol. 2005;128:91–99. doi: 10.1111/j.1365-2141.2004.05275.x. [DOI] [PubMed] [Google Scholar]
- 172.Morrow JA, Arnold KS, Dong J, Balestra ME, Innerarity TL, Weisgraber KH. Effect of arginine 172 on the binding of apolipoprotein E to the low density lipoprotein receptor. J Biol Chem. 2000;275:2576–2580. doi: 10.1074/jbc.275.4.2576. [DOI] [PubMed] [Google Scholar]
- 173.Mosher DF, Doyle MJ, Jaffe EA. Synthesis and secretion of thrombospondin by cultured human endothelial cells. J Cell Biol. 1982;93:343–348. doi: 10.1083/jcb.93.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mueller SC, Ghersi G, Akiyama SK, Sang QX, Howard L, Pineiro-Sanchez M, Nakahara H, Yeh Y, Chen WT. A novel protease-docking function of integrin at invadopodia. J Biol Chem. 1999;274:24947–24952. doi: 10.1074/jbc.274.35.24947. [DOI] [PubMed] [Google Scholar]
- 175.Muller-Taubenberger A, Lupas AN, Li H, Ecke M, Simmeth E, Gerisch G. Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 2001;20:6772–6782. doi: 10.1093/emboj/20.23.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Murphy-Ullrich JE, Gurusiddappa S, Frazier WA, Hook M. Heparin-binding peptides from thrombospondins 1 and 2 contain focal adhesion-labilizing activity. J Biol Chem. 1993;268:26784–26789. [PubMed] [Google Scholar]
- 177.Murphy-Ullrich JE, Hook M. Thrombospondin modulates focal adhesions in endothelial cells. J Cell Biol. 1989;109:1309–1319. doi: 10.1083/jcb.109.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Murphy-Ullrich JE, Lightner VA, Aukhil I, Yan YZ, Erickson HP, Hook M. Focal adhesion integrity is downregulated by the alternatively spliced domain of human tenascin. J Cell Biol. 1991;115:1127–1136. doi: 10.1083/jcb.115.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Murphy-Ullrich JE, Mosher DF. Interactions of thrombospondin with endothelial cells: Receptor-mediated binding and degradation. J Cell Biol. 1987;105:1603–1611. doi: 10.1083/jcb.105.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Murphy-Ullrich JE, Westrick LG, Esko JD, Mosher DF. Altered metabolism of thrombospondin by chinese hamster ovary cells defective in glycosaminoglycan synthesis. J Biol Chem. 1988;263:6400–6406. [PubMed] [Google Scholar]
- 181.Nabel EG, Yang Z, Liptay S, San H, Gordon D, Haudenschild CC, Nabel GJ. Recombinant platelet-derived growth factor B gene expression in porcine arteries induce intimal hyperplasia in vivo. J Clin Invest. 1993;91:1822–1829. doi: 10.1172/JCI116394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Navratil JS, Watkins SC, Wisnieski JJ, Ahearn JM. The globular heads of C1q specifically recognize surface blebs of apoptotic vascular endothelial cells. J Immunol. 2001;166:3231–3239. doi: 10.4049/jimmunol.166.5.3231. [DOI] [PubMed] [Google Scholar]
- 183.Neels JG, van Den Berg BM, Lookene A, Olivecrona G, Pannekoek H, Van Zonneveld AJ. The second and fourth cluster of class A cysteine-rich repeats of the low density lipoprotein receptor-related protein share ligand-binding properties. J Biol Chem. 1999;274:31305–31311. doi: 10.1074/jbc.274.44.31305. [DOI] [PubMed] [Google Scholar]
- 184.Newton CS, Loukinova E, Mikhailenko I, Ranganathan S, Gao Y, Haudenschild C, Strickland DK. Platelet-derived growth factor receptor-beta (PDGFR-beta) activation promotes its association with the low density lipoprotein receptor-related protein (LRP). Evidence for co-receptor function. J Biol Chem. 2005;280:27872–27878. doi: 10.1074/jbc.M505410200. [DOI] [PubMed] [Google Scholar]
- 185.Nielsen KL, Holtet TL, Etzerodt M, Moestrup SK, Gliemann J, Sottrup-Jensen L, Thogersen HC. Identification of residues in α-macroglobulins important for binding to the α2-macroglobulin receptor low density lipoprotein receptor-related protein. J Biol Chem. 1996;271:12909–12912. doi: 10.1074/jbc.271.22.12909. [DOI] [PubMed] [Google Scholar]
- 186.Nielsen MS, Brejning J, García R, Zhang HF, Hayden MR, Vilaró S, Gliemann J. Segments in the C-terminal folding domain of lipoprotein lipase important for binding to the low density lipoprotein receptor- related protein and to heparan sulfate proteoglycans. J Biol Chem. 1997;272:5821–5827. doi: 10.1074/jbc.272.9.5821. [DOI] [PubMed] [Google Scholar]
- 187.Nielsen PR, Ellgaard L, Etzerodt M, Thogersen HC, Poulsen FM. The solution structure of the N-terminal domain of alpha2- macroglobulin receptor-associated protein. Proc Natl Acad Sci U S A. 1997;94:7521–7525. doi: 10.1073/pnas.94.14.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.North CL, Blacklow SC. Solution structure of the sixth LDL-A module of the LDL receptor. Biochemistry. 2000;39:2564–2571. doi: 10.1021/bi992087a. [DOI] [PubMed] [Google Scholar]
- 189.Nykjaer A, Bengtsson-Olivecrona G, Lookene A, Moestrup SK, Petersen CM, Weber W, Beisiegel U, Gliemann J. The alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein binds lipoprotein lipase and beta-migrating very low density lipoprotein associated with the lipase. J Biol Chem. 1993;268:15048–15055. [PubMed] [Google Scholar]
- 190.Nykjaer A, Conese M, Christensen EI, Olson D, Cremona O, Gliemann J, Blasi F. Recycling of the urokinase receptor upon internalization of the uPA:serpin complexes. EMBO J. 1997;16:2610–2620. doi: 10.1093/emboj/16.10.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nykjaer A, Nielsen M, Lookene A, Meyer N, Roigaard H, Etzerodt M, Beisiegel U, Olivecrona G, Gliemann J. A carboxyl-terminal fragment of lipoprotein lipase binds to the low density lipoprotein receptor-related protein and inhibits lipase-mediated uptake of lipoprotein in cells. J Biol Chem. 1994;269:31747–31755. [PubMed] [Google Scholar]
- 192.Nykjær A, Petersen CM, Moller B, Jensen PH, Moestrup SK, Holtet TL, Etzerodt M, Thogersen HC, Munch M, Andreasen PA, Gliemann J. Purified α2-macroglobulin receptor/LDL receptor-related protein binds urokinase•plasminogen activator inhibitor type-1 complex. Evidence that the α2-macroglobulin receptor mediates cellular degradation of urokinase receptor-bound complexes. J Biol Chem. 1992;267:14543–14546. [PubMed] [Google Scholar]
- 193.O'Shea KS, Liu LH, Kinnunen LH, Dixit VM. Role of the extracellular matrix protein thrombospondin in the early development of the mouse embryo. J Cell Biol. 1990;111:2713–2723. doi: 10.1083/jcb.111.6.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Obermoeller LM, Warshawsky I, Wardell MR, Bu G. Differential functions of triplicated repeats suggest two independent roles for the receptor-associated protein as a molecular chaperone. J Biol Chem. 1997;272:10761–10768. doi: 10.1074/jbc.272.16.10761. [DOI] [PubMed] [Google Scholar]
- 195.Ogden CA, deCathelineau A, Hoffmann PR, Bratton D, Ghebrehiwet B, Fadok VA, Henson PM. C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J Exp Med. 2001;194:781–795. doi: 10.1084/jem.194.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ohlsson K, Ganrot PO, Laurell CB. In vivo interaction beween trypsin and some plasma proteins in relation to tolerance to intravenous infusion of trypsin in dog. Acta Chir Scand. 1971;137:113–121. [PubMed] [Google Scholar]
- 197.Okada SS, Grobmyer SR, Barnathan ES. Contrasting effects of plasminogen activators, urokinase receptor, and LDL receptor-related protein on smooth muscle cell migration and invasion. Arterioscler Thromb Vasc Biol. 1996;16:1269–1276. doi: 10.1161/01.atv.16.10.1269. [DOI] [PubMed] [Google Scholar]
- 198.Orr AW, Elzie CA, Kucik DF, Murphy-Ullrich JE. Thrombospondin signaling through the calreticulin/LDL receptor-related protein co-complex stimulates random and directed cell migration. J Cell Sci. 2003;116:2917–2927. doi: 10.1242/jcs.00600. [DOI] [PubMed] [Google Scholar]
- 199.Orr AW, Pallero MA, Murphy-Ullrich JE. Thrombospondin stimulates focal adhesion disassembly through Gi- and phosphoinositide 3-kinase-dependent ERK activation. J Biol Chem. 2002;277:20453–20460. doi: 10.1074/jbc.M112091200. [DOI] [PubMed] [Google Scholar]
- 200.Orr AW, Pallero MA, Xiong WC, Murphy-Ullrich JE. Thrombospondin induces RhoA inactivation through FAK-dependent signaling to stimulate focal adhesion disassembly. J Biol Chem. 2004;279:48983–48992. doi: 10.1074/jbc.M404881200. [DOI] [PubMed] [Google Scholar]
- 201.Orr AW, Pedraza CE, Pallero MA, Elzie CA, Goicoechea S, Strickland DK, Murphy-Ullrich JE. Low density lipoprotein receptor-related protein is a calreticulin coreceptor that signals focal adhesion disassembly. J Cell Biol. 2003;161:1179. doi: 10.1083/jcb.200302069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ossowski L, Aguirre-Ghiso JA. Urokinase receptor and integrin partnership: coordination of signaling for cell adhesion, migration and growth. Curr Opin Cell Biol. 2000;12:613–620. doi: 10.1016/s0955-0674(00)00140-x. [DOI] [PubMed] [Google Scholar]
- 203.Overton CD, Yancey PG, Major AS, Linton MF, Fazio S. Deletion of macrophage LDL receptor-related protein increases atherogenesis in the mouse. Circ Res. 2007;100:670–677. doi: 10.1161/01.RES.0000260204.40510.aa. [DOI] [PubMed] [Google Scholar]
- 204.Panek RL, Dahring TK, Olszewski BJ, Keiser JA. PDGF Receptor Protein Tyrosine Kinase Expression in the Balloon-Injured Rat Carotid Artery. Arterioscler Thromb Vasc Biol. 1997;17:1283–1288. doi: 10.1161/01.atv.17.7.1283. [DOI] [PubMed] [Google Scholar]
- 205.Paresce DM, Ghosh RN, Maxfield FR. Microglial cells internalize aggregates of the Alzheimer's disease amyloid beta-protein via a scavenger receptor. Neuron. 1996;17:553–565. doi: 10.1016/s0896-6273(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 206.Patel M, Morrow J, Maxfield FR, Strickland DK, Greenberg S, Tabas I. The cytoplasmic domain of the low density lipoprotein (LDL) receptor-related protein, but not that of the LDL receptor, triggers phagocytosis. J Biol Chem. 2003;278:44799–44807. doi: 10.1074/jbc.M308982200. [DOI] [PubMed] [Google Scholar]
- 207.Peerschke EI, Ghebrehiwet B. Platelet C1q receptor interactions with collagen- and C1q-coated surfaces. J Immunol. 1990;145:2984–2988. [PubMed] [Google Scholar]
- 208.Phelan MW, Forman LW, Perrine SP, Faller DV. Hypoxia increases thrombospondin-1 transcript and protein in cultured endothelial cells. J Lab Clin Med. 1998;132:519–529. doi: 10.1016/s0022-2143(98)90131-7. [DOI] [PubMed] [Google Scholar]
- 209.Pietromonaco S, Kerjaschki D, Binder S, Ullrich R, Farquhar MG. Molecular cloning of a cDNA encoding a major pathogenic domain of the Heymann nephritis antigen gp330. Proc Natl Acad Sci U S A. 1990;87:1811–1815. doi: 10.1073/pnas.87.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pietrzik CU, Busse T, Merriam DE, Weggen S, Koo EH. The cytoplasmic domain of the LDL receptor-related protein regulates multiple steps in APP processing. EMBO J. 2002;21:5691–5700. doi: 10.1093/emboj/cdf568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pietrzik CU, Yoon IS, Jaeger S, Busse T, Weggen S, Koo EH. FE65 constitutes the functional link between the low-density lipoprotein receptor-related protein and the amyloid precursor protein. J Neurosci. 2004;24:4259–4265. doi: 10.1523/JNEUROSCI.5451-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Plakidou-Dymock S, McGivan JD. Calreticulin--a stress protein induced in the renal epithelial cell line NBL-1 by amino acid deprivation. Cell Calcium. 1994;16:1–8. doi: 10.1016/s0143-4160(05)80002-8. [DOI] [PubMed] [Google Scholar]
- 213.Polavarapu R, Gongora MC, Yi H, Ranganthan S, Lawrence DA, Strickland D, Yepes M. Tissue-type plasminogen activator-mediated shedding of astrocytic low-density lipoprotein receptor-related protein increases the permeability of the neurovascular unit. Blood. 2007;109:3270–3278. doi: 10.1182/blood-2006-08-043125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Poller W, Willnow TE, Hilpert J, Herz J. Differential recognition of α1-antitrypsin-elastase and α1-antichymotrypsin-cathepsin G complexes by the low density lipoprotein receptor-related protein. J Biol Chem. 1995;270:2841–2845. doi: 10.1074/jbc.270.6.2841. [DOI] [PubMed] [Google Scholar]
- 215.Pompili VJ, Gordon D, San H, Yang Z, Muller DWM, Nabel GJ, Nabel EG. Expression and Function of a Recombinant PDGF B Gene in Porcine Arteries. Arterioscler Thromb Vasc Biol. 1995;15:2254–2264. doi: 10.1161/01.atv.15.12.2254. [DOI] [PubMed] [Google Scholar]
- 216.Qiu WQ, Walsh DM, Ye Z, Vekrellis K, Zhang J, Podlisny MB, Rosner MR, Safavi A, Hersh LB, Selkoe DJ. Insulin-degrading enzyme regulates extracellular levels of amyloid beta- protein by degradation. J Biol Chem. 1998;273:32730–32738. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- 217.Qiu Z, Strickland DK, Hyman BT, Rebeck GW. Alpha2-macroglobulin enhances the clearance of endogenous soluble beta- amyloid peptide via low-density lipoprotein receptor-related protein in cortical neurons. J Neurochem. 1999;73:1393–1398. doi: 10.1046/j.1471-4159.1999.0731393.x. [DOI] [PubMed] [Google Scholar]
- 218.Rabhi-Sabile S, Pidard D, Lawler J, Renesto P, Chignard M, Legrand C. Proteolysis of thrombospondin during cathepsin-G-induced platelet aggregation: Functional role of the 165-kDa carboxy- terminal fragment. FEBS Lett. 1996;386:82–86. doi: 10.1016/0014-5793(96)00408-5. [DOI] [PubMed] [Google Scholar]
- 219.Rall SC, Jr, Ye P, Bu G, Wardell MR. The domain structure of human receptor-associated protein. Protease sensitivity and guanidine HCl denaturation. J Biol Chem. 1998;273:24152–24157. doi: 10.1074/jbc.273.37.24152. [DOI] [PubMed] [Google Scholar]
- 220.Ranganathan S, Liu CX, Migliorini MM, Von Arnim CA, Peltan ID, Mikhailenko I, Hyman BT, Strickland DK. Serine and threonine phosphorylation of the LDL receptor-related protein (LRP) by protein kinase Calpha regulates endocytosis and association with adaptor molecules. J Biol Chem. 2004 doi: 10.1074/jbc.M407592200. [DOI] [PubMed] [Google Scholar]
- 221.Ranganathan S, Liu CX, Migliorini MM, Von Arnim CA, Peltan ID, Mikhailenko I, Hyman BT, Strickland DK. Serine and threonine phosphorylation of the low density lipoprotein receptor-related protein by protein kinase Calpha regulates endocytosis and association with adaptor molecules. J Biol Chem. 2004;279:40536–40544. doi: 10.1074/jbc.M407592200. [DOI] [PubMed] [Google Scholar]
- 222.Raugi GJ, Mumby SM, Abbott-Brown D, Bornstein P. Thrombospondin: synthesis and secretion by cells in culture. J Cell Biol. 1982;95:351–354. doi: 10.1083/jcb.95.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Raugi GJ, Olerud JE, Gown AM. Thrombospondin in early human wound tissue. J Invest Dermatol. 1987;89:551–554. doi: 10.1111/1523-1747.ep12461198. [DOI] [PubMed] [Google Scholar]
- 224.Rebeck GW, Reiter JS, Strickland DK, Hyman BT. Apolipoprotein E in sporadic Alzheimer's disease: Allelic variation and receptor interactions. Neuron. 1993;11:575–580. doi: 10.1016/0896-6273(93)90070-8. [DOI] [PubMed] [Google Scholar]
- 225.Reddington M, Priller J, Treichel J, Haas C, Kreutzberg GW. Astrocytes and microglia as potential targets for calcitonin gene related peptide in the central nervous system. Can J Physiol Pharmacol. 1995;73:1047–1049. doi: 10.1139/y95-148. [DOI] [PubMed] [Google Scholar]
- 226.Reed MJ, Iruela-Arispe L, O'Brien ER, Truong T, LaBell T, Bornstein P, Sage EH. Expression of thrombospondins by endothelial cells - Injury is correlated with TSP-1. Am J Pathol. 1995;147:1068–1080. [PMC free article] [PubMed] [Google Scholar]
- 227.Reed MJ, Puolakkainen P, Lane TF, Dickerson D, Bornstein P, Sage EH. Differential expression of SPARC and thrombospondin 1 in wound repair: immunolocalization and in situ hybridization. J Histochem Cytochem. 1993;41:1467–1477. doi: 10.1177/41.10.8245406. [DOI] [PubMed] [Google Scholar]
- 228.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 229.Rodenburg KW, Kjoller L, Petersen HH, Andreasen PA. Binding of urokinase-type plasminogen activator-plasminogen activator inhibitor-1 complex to the endocytosis receptors alpha2- macroglobulin receptor/low-density lipoprotein receptor-related protein and very-low-density lipoprotein receptor involves basic residues in the inhibitor. Biochem J. 1998;329:55–63. doi: 10.1042/bj3290055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Rohlmann A, Gotthardt M, Hammer RE, Herz J. Inducible inactivation of hepatic LRP gene by cre-mediated recombination confirms role of LRP in clearance of chylomicron remnants. J Clin Invest. 1998;101:689–695. doi: 10.1172/JCI1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rojiani MV, Finlay BB, Gray V, Dedhar S. In vitro interaction of a polypeptide homologous to human Ro/SS-A antigen (calreticulin) with a highly conserved amino acid sequence in the cytoplasmic domain of integrin alpha subunits. Biochemistry. 1991;30:9859–9866. doi: 10.1021/bi00105a008. [DOI] [PubMed] [Google Scholar]
- 232.Ross R. The pathogeneis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 233.Ross R. Atherosclerosis--an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 234.Rubin LL, Staddon JM. The Cell Biology of the Blood-Brain Barrier. Annual Review of Neuroscience. 1999;22:11–28. doi: 10.1146/annurev.neuro.22.1.11. [DOI] [PubMed] [Google Scholar]
- 235.Rubinsztein DC, Cohen JC, Berger GM, Vander Westhuyzen DR, Coetzee GA, Gevers W. Chylomicron remnant clearance from the plasma is normal in familial hypercholesterolemic homozygotes with defined receptor defects. J Clin Invest. 1990;86:1306–1312. doi: 10.1172/JCI114839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 237.Rutherford C, Martin W, Salame M, Carrier M, Anggard E, Ferns G. Substantial inhibition of neo-intimal response to balloon injury in the rat carotid artery using a combination of antibodies to platelet-derived growth factor-BB and basic fibroblast growth factor. Atherosclerosis. 1997;130:45–51. doi: 10.1016/s0021-9150(96)06042-x. [DOI] [PubMed] [Google Scholar]
- 238.Saenko EL, Yakhyaev AV, Mikhailenko I, Strickland DK, Sarafanov AG. Role of the low density lipoprotein-related protein receptor in mediation of factor VIII catabolism. J Biol Chem. 1999;274:37685–37692. doi: 10.1074/jbc.274.53.37685. [DOI] [PubMed] [Google Scholar]
- 239.Salicioni AM, Gaultier A, Brownlee C, Cheezum MK, Gonias SL. Low Density Lipoprotein Receptor-related Protein-1 Promotes {beta}1 Integrin Maturation and Transport to the Cell Surface. J Biol Chem. 2004;279:10005–10012. doi: 10.1074/jbc.M306625200. [DOI] [PubMed] [Google Scholar]
- 240.Sanchez-Guerra M, Combarros O, Infante J, Llorca J, Berciano J, Fontalba A, Fernandez-Luna JL, Pena N, Fernandez-Viadero C. Case-control study and meta-analysis of low density lipoprotein receptor-related protein gene exon 3 polymorphism in Alzheimer's disease. Neurosci Lett. 2001;316:17–20. doi: 10.1016/s0304-3940(01)02342-4. [DOI] [PubMed] [Google Scholar]
- 241.Schlaepfer DD, Mitra SK, Ilic D. Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim Biophys Acta. 2004;1692:77–102. doi: 10.1016/j.bbamcr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 242.Schoenwaelder SM, Burridge K. Bidirectional signaling between the cytoskeleton and integrins. Curr Opin Cell Biol. 1999;11:274–286. doi: 10.1016/s0955-0674(99)80037-4. [DOI] [PubMed] [Google Scholar]
- 243.Schulz S, Birkenmeier G, Schagdarsurengin U, Wenzel K, Muller-Werdan U, Rehfeld D, Suss T, Kabisch A, Werdan K, Glaser C. Role of LDL receptor-related protein (LRP) in coronary atherosclerosis. Int J Cardiol. 2003;92:137–144. doi: 10.1016/s0167-5273(03)00090-1. [DOI] [PubMed] [Google Scholar]
- 244.Selkoe DJ. Translating cell biology into therapeutic advances in Alzheimer's disease. Nature. 1999;399:A23–A31. doi: 10.1038/399a023. [DOI] [PubMed] [Google Scholar]
- 245.Shibata M, Yamada S, Kumar SR, Calero M, Bading J, Frangione B, Holtzman DM, Miller CA, Strickland DK, Ghiso J, Zlokovic BV. Clearance of Alzheimer's amyloid-ss(1–40) peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier. J Clin Invest. 2000;106:1489–1499. doi: 10.1172/JCI10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sid B, Dedieu S, Delorme N, Sartelet H, Rath GM, Bellon G, Martiny L. Human thyroid carcinoma cell invasion is controlled by the low density lipoprotein receptor-related protein-mediated clearance of urokinase plasminogen activator. Int J Biochem Cell Biol. 2006;38:1729–1740. doi: 10.1016/j.biocel.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 247.Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PGW, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O'Donnell E, Salvesen GS, Travis J, Whisstock JC. The Serpins Are an Expanding Superfamily of Structurally Similar but Functionally Diverse Proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 248.Simonovic M, Dolmer K, Huang W, Strickland DK, Volz K, Gettins PG. Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor. Biochemistry. 2001;40:15127–15134. doi: 10.1021/bi015688m. [DOI] [PubMed] [Google Scholar]
- 249.Smith JD, Trogan E, Ginsberg M, Grigaux C, Tian J, Miyata M. Decreased atherosclerosis in mice deficient in both macrophage colony-stimulating factor (op) and apolipoprotein E. Proc Natl Acad Sci USA. 1995;92:8264–8268. doi: 10.1073/pnas.92.18.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Solheim JC, Harris MR, Kindle CS, Hansen TH. Prominence of beta 2-microglobulin, class I heavy chain conformation, and tapasin in the interactions of class I heavy chain with calreticulin and the transporter associated with antigen processing. J Immunol. 1997;158:2236–2241. [PubMed] [Google Scholar]
- 251.Sottrup-Jensen L. Alpha-macroglobulins: structure, shape, and mechanism of proteinase complex formation. J Biol Chem. 1989;264:11539–11542. [PubMed] [Google Scholar]
- 252.Sparrow CP, Parthasarathy S, Steinberg D. Enzymatic modification of low density lipoprotein by purified lipoxygenase plus phospholipase A2 mimics cell-mediated oxidative modification. J Lipid Res. 1988;29:745–753. [PubMed] [Google Scholar]
- 253.Spijkers PPEM, Costa Martins P, Westein E, Gahmberg CG, Zwaginga JJ, Lenting PJ. LDL-receptor-related protein regulates {beta}2-integrin-mediated leukocyte adhesion. Blood. 2005;105:170–177. doi: 10.1182/blood-2004-02-0498. [DOI] [PubMed] [Google Scholar]
- 254.Springer TA. An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J Mol Biol. 1998;283:837–862. doi: 10.1006/jmbi.1998.2115. [DOI] [PubMed] [Google Scholar]
- 255.Srivastava P. Interaction of heat shock proteins with peptides and antigen presenting cells: chaperoning of the innate and adaptive immune responses. Annu Rev Immunol. 2002;20:395–425. doi: 10.1146/annurev.immunol.20.100301.064801. [DOI] [PubMed] [Google Scholar]
- 256.Stahl A, Mueller BM. The urokinase-type plasminogen activator receptor, a GPI- linked protein, is localized in caveolae. J Cell Biol. 1995;129:335–344. doi: 10.1083/jcb.129.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Staniszewska I, Zaveri S, Del Valle L, Oliva I, Rothman VL, Croul SE, Roberts DD, Mosher DF, Tuszynski GP, Marcinkiewicz C. Interaction of alpha9beta1 integrin with thrombospondin-1 promotes angiogenesis. Circ Res. 2007;100:1308–1316. doi: 10.1161/01.RES.0000266662.98355.66. [DOI] [PubMed] [Google Scholar]
- 258.Stefansson S, Kounnas MZ, Henkin J, Mallampalli RK, Chappell DA, Strickland DK, Argraves WS. gp330 on type II pneumocytes mediates endocytosis leading to degradation of pro-urokinase, plasminogen activator inhibitor- 1 and urokinase-plasminogen activator inhibitor-1 complex. J Cell Sci. 1995;108:2361–2368. doi: 10.1242/jcs.108.6.2361. [DOI] [PubMed] [Google Scholar]
- 259.Stefansson S, Muhammad S, Cheng XF, Battey FD, Strickland DK, Lawrence DA. Plasminogen activator inhibitor-1 contains a cryptic high affinity binding site for the low density lipoprotein receptor-related protein. J Biol Chem. 1998;273:6358–6366. doi: 10.1074/jbc.273.11.6358. [DOI] [PubMed] [Google Scholar]
- 260.Stein Y, Stein O. Lipoprotein lipase and atherosclerosis. Atherosclerosis. 2003;170:1–9. doi: 10.1016/s0021-9150(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 261.Steinbrecher UP, Parthasarathy S, Leake DS, Witztum JL, Steinberg D. Modification of low density lipoprotein by endothelial cells involves lipid peroxidation and degradation of low density lipoprotein phospholipids. Proc Natl Acad Sci U S A. 1984;81:3883–3887. doi: 10.1073/pnas.81.12.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Stewart PA, Wiley MJ. Developing nervous tissue induces formation of blood-brain barrier characteristics in invading endothelial cells: a study using quail--chick transplantation chimeras. Dev Biol. 1981;84:183–192. doi: 10.1016/0012-1606(81)90382-1. [DOI] [PubMed] [Google Scholar]
- 263.Strickland DK, Ashcom JD, Williams S, Battey F, Behre E, McTigue K, Battey JF, Argraves WS. Primary structure of α2-macroglobulin receptor-associated protein. Human homologue of a Heymann nephritis antigen. J Biol Chem. 1991;266:13364–13369. [PubMed] [Google Scholar]
- 264.Strickland DK, Ashcom JD, Williams S, Burgess WH, Migliorini M, Argraves WS. Sequence identity between the α2-macroglobulin receptor and low density lipoprotein receptor-related protein suggests that this molecule is a multifunctional receptor. J Biol Chem. 1990;265:17401–17404. [PubMed] [Google Scholar]
- 265.Stuart LM, Ezekowitz RA. Phagocytosis: elegant complexity. Immunity. 2005;22:539–550. doi: 10.1016/j.immuni.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 266.Su HP, Nakada-Tsukui K, Tosello-Trampont AC, Li Y, Bu G, Henson PM, Ravichandran KS. Interaction of CED-6/GULP, an Adapter Protein Involved in Engulfment of Apoptotic Cells with CED-1 and CD91/Low Density Lipoprotein Receptor-related Protein (LRP) J Biol Chem. 2002;277:11772–11779. doi: 10.1074/jbc.M109336200. [DOI] [PubMed] [Google Scholar]
- 267.Sugiura T, Shirasuna K, Hayashido Y, Sakai T, Matsuya T. Effects of human fibroblasts on invasiveness of oral cancer cells in vitro: isolation of a chemotactic factor from human fibroblasts. Int J Cancer. 1996;68:774–781. doi: 10.1002/(SICI)1097-0215(19961211)68:6<774::AID-IJC15>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 268.Swertfeger DK, Bu G, Hui DY. Low density lipoprotein receptor-related protein mediates apolipoprotein E inhibition of smooth muscle cell migration. J Biol Chem. 2002;277:4141–4146. doi: 10.1074/jbc.M109124200. [DOI] [PubMed] [Google Scholar]
- 269.Tacken PJ, de Vries IJ, Torensma R, Figdor CG. Dendritic-cell immunotherapy: from ex vivo loading to in vivo targeting. Nat Rev Immunol. 2007;7:790–802. doi: 10.1038/nri2173. [DOI] [PubMed] [Google Scholar]
- 270.Takayama Y, May P, Anderson RG, Herz J. Low Density Lipoprotein Receptor-related Protein 1 (LRP1) Controls Endocytosis and c-CBL-mediated Ubiquitination of the Platelet-derived Growth Factor Receptor {beta} (PDGFR{beta}) J Biol Chem. 2005;280:18504–18510. doi: 10.1074/jbc.M410265200. [DOI] [PubMed] [Google Scholar]
- 271.Tangirala RK, Mol MJ, Steinberg D. Macrophage oxidative modification of low density lipoprotein occurs independently of its binding to the low density lipoprotein receptor. J Lipid Res. 1996;37:835–843. [PubMed] [Google Scholar]
- 272.Tanzi RE, Gusella JF, Watkins PC, Bruns GAP, St.George-Hyslop PH, Van Keuren ML, Paterson D, Pagan S, Kurnit DM, Neve RL. The amyloid β protein gene: cDNA cloning, mRNA distribution, and genetic linkage near the Alzheimer locus. Science. 1987;235:880–994. doi: 10.1126/science.2949367. [DOI] [PubMed] [Google Scholar]
- 273.Tooyama I, Kawamata T, Akiyama H, Kimura H, Moestrup SK, Gliemann J, Matsuo A, McGeer PL. Subcellular localization of the low density lipoprotein receptor-related protein (α2-macroglobulin receptor) in human brain. Brain Res. 1995;691:235–238. doi: 10.1016/0006-8993(95)00735-9. [DOI] [PubMed] [Google Scholar]
- 274.Trommsdorff M, Borg JP, Margolis B, Herz J. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J Biol Chem. 1998;273:33556–33560. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- 275.Tseng WF, Huang SS, Huang JS. LRP-1/TbetaR-V mediates TGF-beta1-induced growth inhibition in CHO cells. FEBS Lett. 2004;562:71–78. doi: 10.1016/S0014-5793(04)00185-1. [DOI] [PubMed] [Google Scholar]
- 276.Ulery PG, Beers J, Mikhailenko I, Tanzi RE, Rebeck GW, Hyman BT, Strickland DK. Modulation of beta-amyloid precursor protein processing by the low density lipoprotein receptor-related protein (LRP). Evidence that lrp contributes to the pathogenesis of Alzheimer's disease. J Biol Chem. 2000;275:7410–7415. doi: 10.1074/jbc.275.10.7410. [DOI] [PubMed] [Google Scholar]
- 277.Vandivier RW, Ogden CA, Fadok VA, Hoffmann PR, Brown KK, Botto M, Walport MJ, Fisher JH, Henson PM, Greene KE. Role of surfactant proteins A, D, and C1q in the clearance of apoptotic cells in vivo and in vitro: calreticulin and CD91 as a common collectin receptor complex. J Immunol. 2002;169:3978–3986. doi: 10.4049/jimmunol.169.7.3978. [DOI] [PubMed] [Google Scholar]
- 278.Varani J, Riser BL, Hughes LA, Carey TE, Fligiel SE, Dixit VM. Characterization of thrombospondin synthesis, secretion and cell surface expression by human tumor cells. Clin Exp Metastasis. 1989;7:265–276. doi: 10.1007/BF01753679. [DOI] [PubMed] [Google Scholar]
- 279.Vash B, Phung N, Zein S, DeCamp D. Three complement-type repeats of the low-density lipoprotein receptor-related protein define a common binding site for RAP, PAI-1, and lactoferrin. Blood. 1998;92:3277–3285. [PubMed] [Google Scholar]
- 280.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE [see comments] Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 281.Wada I, Imai S, Kai M, Sakane F, Kanoh H. Chaperone function of calreticulin when expressed in the endoplasmic reticulum as the membrane-anchored and soluble forms. J Biol Chem. 1995;270:20298–20304. doi: 10.1074/jbc.270.35.20298. [DOI] [PubMed] [Google Scholar]
- 282.Wang S, Herndon ME, Ranganathan S, Godyna S, Lawler J, Argraves WS, Liau G. Internalization but not binding of thrombospondin-1 to low density lipoprotein receptor-related protein-1 requires heparan sulfate proteoglycans. J Cell Biochem. 2004;91:766–776. doi: 10.1002/jcb.10781. [DOI] [PubMed] [Google Scholar]
- 283.Wang X, Lee SR, Arai K, Lee SR, Tsuji K, Rebeck GW, Lo EH. Lipoprotein receptor-mediated induction of matrix metalloproteinase by tissue plasminogen activator. Nat Med. 2003;9:1313–1317. doi: 10.1038/nm926. [DOI] [PubMed] [Google Scholar]
- 284.Weaver AM, Hussaini IM, Mazar A, Henkin J, Gonias SL. Embryonic fibroblasts that are genetically deficient in low density lipoprotein receptor-related protein demonstrate increased activity of the urokinase receptor system and accelerated migration on vitronectin. J Biol Chem. 1997;272:14372–14379. doi: 10.1074/jbc.272.22.14372. [DOI] [PubMed] [Google Scholar]
- 285.Werb Z, Mainardi CL, Vater CA, Harris ED., Jr Endogenous activiation of latent collagenase by rheumatoid synovial cells. Evidence for a role of plasminogen activator. N Engl J Med. 1977;296:1017–1023. doi: 10.1056/NEJM197705052961801. [DOI] [PubMed] [Google Scholar]
- 286.White TK, Zhu Q, Tanzer ML. Cell surface calreticulin is a putative mannoside lectin which triggers mouse melanoma cell spreading. J Biol Chem. 1995;270:15926–15929. doi: 10.1074/jbc.270.27.15926. [DOI] [PubMed] [Google Scholar]
- 287.Wight TN, Raugi GJ, Mumby SM, Bornstein P. Light microscopic immunolocation of thrombospondin in human tissues. J Histochem Cytochem. 1985;33:295–302. doi: 10.1177/33.4.3884704. [DOI] [PubMed] [Google Scholar]
- 288.Wijnberg MJ, Quax PH, Nieuwenbroek NM, Verheijen JH. The migration of human smooth muscle cells in vitro is mediated by plasminogen activation and can be inhibited by alpha2- macroglobulin receptor associated protein. Thromb Haemost. 1997;78:880–886. [PubMed] [Google Scholar]
- 289.Williams SE, Ashcom JD, Argraves WS, Strickland DK. A novel mechanism for controlling the activity of α2-macroglobulin receptor/low density lipoprotein receptor-related protein. Multiple regulatory sites for 39-kDa receptor-associated protein. J Biol Chem. 1992;267:9035–9040. [PubMed] [Google Scholar]
- 290.Williams SE, Inoue I, Tran H, Fry GL, Pladet MW, Iverius P-H, Lalouel J-M, Chappell DA, Strickland DK. The Carboxyl-terminal Domain of Lipoprotein Lipase Binds to the Low Density Lipoprotein Receptor-related Protein/α2-Macroglobulin receptor (LRP) and mediates Binding of Normal Very Low Density Lipoproteins to LRP. J Biol Chem. 1994;269:8653–8658. [PubMed] [Google Scholar]
- 291.Willnow TE, Armstrong SA, Hammer RE, Herz J. Functional expression of low density lipoprotein receptor-related protein is controlled by receptor-associated protein in vivo. Proc Natl Acad Sci U S A. 1995;92:4537–4541. doi: 10.1073/pnas.92.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Willnow TE, Orth K, Herz J. Molecular dissection of ligand binding sites on the low density lipoprotein receptor-related protein. J Biol Chem. 1994;269:15827–15832. [PubMed] [Google Scholar]
- 293.Willnow TE, Rohlmann A, Horton J, Otani H, Braun JR, Hammer RE, Herz J. RAP, a specialized chaperone, prevents ligand-induced ER retention and degradation of LDL receptor-related endocytic receptors. EMBO J. 1996;15:2632–2639. [PMC free article] [PubMed] [Google Scholar]
- 294.Willnow TE, Sheng Z, Ishibashi S, Herz J. Inhibition of hepatic chylomicron remnant uptake by gene transfer of a receptor antagonist. Science. 1994;264:1471–1474. doi: 10.1126/science.7515194. [DOI] [PubMed] [Google Scholar]
- 295.Wilsie LC, Orlando RA. The low density lipoprotein receptor-related protein complexes with cell surface heparan sulfate proteoglycans to regulate proteoglycan-mediated lipoprotein catabolism. J Biol Chem. 2003;278:15758–15764. doi: 10.1074/jbc.M208786200. [DOI] [PubMed] [Google Scholar]
- 296.Wolf BB, Lopes MBS, VandenBerg SR, Gonias SL. Characterization and immunohistochemical localization of α2-macroglobulin receptor (low-density lipoprotein receptor- related protein) in human brain. Am J Pathol. 1992;141:37–42. [PMC free article] [PubMed] [Google Scholar]
- 297.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochim Biophys Acta. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 298.Wu Y, Migliorini M, Walsh J, Yu P, Strickland DK, Wang YX. NMR structural studies of domain 1 of receptor-associated protein. J Biomol NMR. 2004;29:271–279. doi: 10.1023/B:JNMR.0000032507.96325.6d. [DOI] [PubMed] [Google Scholar]
- 299.Xiao G, Chung TF, Pyun HY, Fine RE, Johnson RJ. KDEL proteins are found on the surface of NG108–15 cells. Brain Res Mol Brain Res. 1999;72:121–128. doi: 10.1016/s0169-328x(99)00188-6. [DOI] [PubMed] [Google Scholar]
- 300.Xu W, Takahashi Y, Sakashita T, Iwasaki T, Hattori H, Yoshimoto T. Low density lipoprotein receptor-related protein is required for macrophage-mediated oxidation of low density lipoprotein by 12/15- lipoxygenase. J Biol Chem. 2001;39:36454–36459. doi: 10.1074/jbc.M105093200. [DOI] [PubMed] [Google Scholar]
- 301.Yang DS, Small DH, Seydel U, Smith JD, Hallmayer J, Gandy SE, Martins RN. Apolipoprotein E promotes the binding and uptake of beta-amyloid into Chinese hamster ovary cells in an isoform-specific manner. Neuroscience. 1999;90:1217–1226. doi: 10.1016/s0306-4522(98)00561-2. [DOI] [PubMed] [Google Scholar]
- 302.Yang M, Huang H, Li J, Li D, Wang H. Tyrosine phosphorylation of the LDL receptor-related protein (LRP) and activation of the ERK pathway are required for connective tissue growth factor to potentiate myofibroblast differentiation. FASEB J. 2004;18:1920–1921. doi: 10.1096/fj.04-2357fje. [DOI] [PubMed] [Google Scholar]
- 303.Yang Z, Kyriakides TR, Bornstein P. Matricellular proteins as modulators of cell-matrix interactions: adhesive defect in thrombospondin 2-null fibroblasts is a consequence of increased levels of matrix metalloproteinase-2. Mol Biol Cell. 2000;11:3353–3364. doi: 10.1091/mbc.11.10.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Yang Z, Strickland DK, Bornstein P. Extracellular matrix metalloproteinase 2 levels are regulated by the low density lipoprotein-related scavenger receptor and thrombospondin 2. J Biol Chem. 2001;276:8403–8408. doi: 10.1074/jbc.M008925200. [DOI] [PubMed] [Google Scholar]
- 305.Yepes M, Sandkvist M, Moore EG, Bugge TH, Strickland DK, Lawrence DA. Tissue-type plasminogen activator induces opening of the blood-brain barrier via the LDL receptor-related protein. Journal of Clinical Investigation. 2003;112:1533–1540. doi: 10.1172/JCI19212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Zerbinatti CV, Wozniak DF, Cirrito J, Cam JA, Osaka H, Bales KR, Zhuo M, Paul SM, Holtzman DM, Bu G. Increased soluble amyloid-{beta} peptide and memory deficits in amyloid model mice overexpressing the low-density lipoprotein receptor-related protein. PNAS. 2004 doi: 10.1073/pnas.0305803101. 0305803101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 308.Zhu H, Takahashi Y, Xu W, Kawajiri H, Murakami T, Yamamoto M, Iseki S, Iwasaki T, Hattori H, Yoshimoto T. Low density lipoprotein receptor-related protein-mediated membrane translocation of 12/15-lipoxygenase is required for oxidation of low density lipoprotein by macrophages. J Biol Chem. 2003;278:13350–13355. doi: 10.1074/jbc.M212104200. [DOI] [PubMed] [Google Scholar]
- 309.Zhu Y, Hui DY. Apolipoprotein E Binding to Low Density Lipoprotein Receptor-related Protein-1 Inhibits Cell Migration via Activation of cAMP-dependent Protein Kinase A. J Biol Chem. 2003;278:36257–36263. doi: 10.1074/jbc.M303171200. [DOI] [PubMed] [Google Scholar]


