Abstract
Mechanisms of action of non-mutagenic carcinogens such as asbestos remain poorly characterized. As pleural mesothelioma is known to have limited numbers of genetic mutations, we aimed to characterize the relationships among gene-locus specific methylation alterations, disease status, asbestos burden, and survival in this rapidly-fatal asbestos-associated tumor. Methylation of 1505 CpG loci associated with 803 cancer-related genes were studied in 158 pleural mesotheliomas and 18 normal pleura. After false-discovery rate correction, 969 CpG loci were independently associated with disease status (Q < 0.05). Classifying samples based upon CpG methylation profile with a mixture model approach, methylation classes discriminated tumor from normal pleura (permutation P < 0.0001). In a random forests classification the overall misclassification error rate was 3.4%, with <1% (n=1) of tumors misclassified as normal (P < 0.0001). Among tumors, methylation class membership was significantly associated with lung tissue asbestos body burden (P < 0.03), and significantly predicted survival (likelihood ratio P < 0.01). Consistent with prior work, asbestos burden was associated with an increased risk of death (HR = 1.4, 95% CI, 1.1 – 1.8). Our results have shown that methylation profiles powerfully differentiate diseased pleura from non-tumor pleura and that asbestos burden and methylation profiles are independent predictors of mesothelioma patient survival. We have added to the growing body of evidence that cellular epigenetic dysregulation is a critical mode of action for asbestos in the induction of pleural mesothelioma. Importantly, these findings hold great promise for using epigenetic profiling in the diagnosis and prognosis of human cancers.
Keywords: Methylation, asbestos, mesothelioma
Introduction
A central tenet of cancer biology states that cancer is clonal, with tumors arising as the result of expansion of increasingly dysregulated cells. This insight yielded a now well known paradigm that selective expansion of cells with a growth advantage occurs in an ordered fashion, driven primarily by genetic changes (1). This model has expanded to now include the thesis that cancers also evolve a “mutator phenotype” and become malignant as a result of somatic genetic events (2). While this is almost certainly true of some cancers, particularly those induced by well characterized mutagens (e.g. tobacco smoke and ionizing radiation), other known human carcinogens are not mutagenic (or are very poor mutagens) and may be less prone to induce cancers via this mechanism. Asbestos, which is known to induce mesothelioma, is an example of a non-mutagenic carcinogen. In the case of carcinogens such as asbestos, it may be that dysregulation of the somatic epigenome is equally, if not more crucial for cancer development.
Aberrant epigenetic events, including DNA hypermethylation-induced gene silencing, are well recognized as important contributors to carcinogenesis. Methylation associated gene silencing occurs when certain cytosines in specific clustered regions primarily located in gene promoters are hypermethylated. These regulatory CpG islands often occur in tumor suppressor genes and are thought to remain largely unmethylated in noncancerous cells. Approximately half of all human genes contain CpG islands and are, therefore, potentially subject to this type of aberrant silencing (3). Recent technologic advances allow for the simultaneous resolution of hundreds of specific, phenotypically defined cancer-related methylation events, providing a platform for the rapid epigenetic profiling of gene silencing in human tumors (4).
Malignant pleural mesothelioma is a rapidly fatal malignancy associated with asbestos exposure in approximately 80% of patients (5). In the United States, Great Britain, and Japan, over 5000 cases occur annually and median survival of patients with pleural mesothelioma is less than one year (6–8). The economic burden of treating this disease and the litigation associated with asbestos exposure is estimated to exceed $265 billion over the next four decades in the United States (9). Despite the decline in asbestos use among industrialized nations, the incidence of mesothelioma continues to rise, and it is not expected to peak until 2020, as disease latency can be as long as fifty years (10). Importantly, asbestos is currently mined and exported throughout the world, with heavy use evident in developing nations such as China, India, and Central America (11). Asbestos-containing products are still imported to the U.S., and many asbestos exposure hazards remain from earlier applications; one well publicized example being dust from the World Trade Center towers collapse in New York City (12). A more complete understanding the molecular-genetic consequences of asbestos exposure and the mechanism of action of these mineral fibers in inducing mesothelioma is critically needed to develop more effective approaches for identifying and treating this devastating disease.
The causal link between asbestos and pleural mesothelioma has been widely accepted since 1960 (13), and the carcinogenic mechanisms of asbestos have been investigated in earnest since that time; establishing that asbestos fibers are not point mutagens, but rather both clastogenic and cytotoxic in vitro (14, 15). Additionally, methylation-induced tumor suppressor gene silencing has been observed in recent studies of mesothelioma (16–20) leading to the hypothesis that asbestos fibers contribute to epigenetic silencing of tumor suppressor genes in this disease. Consistent with this, Tsou et al. observed a significant association between self-reported asbestos exposure and methylation at the MT1A, and MT2A gene loci in mesotheliomas (18). Concomitantly, work in our laboratory, using quantitative asbestos body counts as a measure of asbestos exposure burden, revealed an association between cell cycle control tumor suppressor gene methylation and increased asbestos burden in mesothelioma (20).
To comprehensively investigate aberrant tumor-specific, phenotypically relevant methylation events in pleural mesothelioma, we profiled 158 tumors and 18 non-tumorigenic parietal pleura samples for methylation at 1505 CpG dinucleotides associated with 803 cancer-related genes using the Illumina GoldenGate® methylation bead array. We have definitively delineated the relationship between a comprehensive, phenotypically important CpG methylation profile and disease status, in addition to defining the tumor methylation profiles associated with patient clinical course and asbestos exposure.
Methods
Study population
Tumor material was obtained following surgical resection at Brigham and Women’s Hospital through the support of the International Mesothelioma Program. Similarly, grossly non-tumorigenic parietal pleura samples were taken as residual tissue during extrapleural pneumonectomy from uninvolved anatomic sites. Patients were drawn in near equivalent numbers from a pilot study conducted in 2002 (n=70), and an incident case series beginning in 2005 (n=88). Among identified cases the participation rate was 85%. All patients provided informed consent under the approval of the appropriate Institutional Review Boards. All patients underwent surgical resection prior to other treatments. Clinical information, including histologic diagnosis was obtained from pathology reports. Each patient was assessed for history of exposure to asbestos as well as additional demographic and environmental data by obtaining their medical and occupational history with an in-person questionnaire or interview. Additionally, we quantified asbestos bodies in samples of lung tissue from multiple sites in the resected lung (21) as previously described (22). Each tumor was pathologically examined and the amount of tumor in every sample estimated by direct microscopic evaluation and recorded as the percent tumor for that specimen. Patients were followed for survival using the National death index and last known clinic visit.
Methylation analysis
Tumor and non-tumor pleural DNA was extracted from frozen tissue using the QIAamp DNA mini kit according to the manufacturer’s protocol (Qiagen, Valencia, CA). DNA was modified by sodium bisulfite to convert unmethylated cytosines to uracil using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) according to the manufacturer’s protocol. Illumina GoldenGate® methylation bead arrays were used to simultaneously interrogate 1505 CpG loci associated with 803 cancer-related genes. The Illumina array interrogates approximately two CpGs per gene and although sequencing methods would provide additional details, CpGs were cultivated from reports which have demonstrated the methylation-expression relationship in large part through sequencing experiments. Bead arrays have a similar sensitivity as quantitative methylation-specific PCR and were run at the UCSF Institute for Human Genetics, Genomics Core Facility according to the manufacturer’s protocol and as described by Bibikova et al (4).
Statistical analysis
BeadStudio Methylation software from the array manufacturer Illumina (SanDiego, CA) was used for dataset assembly. All array data points are represented by fluorescent signals from both M (methylated) and U (unmethylated) alleles, and methylation level is given by β= (max(M , 0))/(|U| + |M| + 100), the average methylation (β) value is derived from the ~30 replicate methylation measurements and a Cy3/Cy5 methylated/unmethylated ratio. At each locus for each sample the detection P-value was used to determine sample performance, three samples (2%) with >25% of loci having a detection P–value > 1e-5 were dropped from analysis. Similarly, CpG loci with a median detection P–value > 0.05 (n=8, 0.5%), were eliminated from analysis.
Subsequent analyses were carried out using the R software (23). For exploratory and visualization purposes, hierarchical clustering was performed using R function hclust with Manhattan metric and average linkage. Associations between sample type, or covariates such as age or gender and methylation at individual CpG loci were tested with a generalized linear model (GLM). The beta-distribution of average beta values was accounted for with a quasi-binomial logit link with an estimated scale parameter constraining the mean between 0 and 1, in a manner similar to that described by Hsuing et al. (24). CpG loci where an a priori hypothesis existed were tested independently. In contrast, array-wide scanning for CpG loci associations with sample type or covariate used false discovery rate correction and Q–values computed by the qvalue package in R (25).
For inference, data were clustered using a mixture model with a mixture of beta distributions, and the number of classes was determined by recursively splitting the data via 2-class models, with Bayesian information criterion (BIC) used at each potential split to decide whether the split was to be maintained or abandoned as described in (26). Permutation tests (running 10,000 permutations) were used to test for association with methylation class by generating a distribution of the test statistic for the null distribution for comparison to the observed distribution. For continuous variables, the permutation test was run with the Kruskal-Wallis test statistic. For categorical variables we used a Mutual information test statistic; (27) equivalent to a likelihood ratio test comparing the saturated model for a contingency table against a model that assumes independent margins. Significant associations from permutation tests were controlled for potential confounders where appropriate using logistic regression with methylation classes and potential confounders and a likelihood ratio test of the model with and without methylation classes. For survival analyses, Cox proportional hazards models were utilized, and likelihood ratio tests were used to examine the significance of inclusion of the methylation classes in the models.
The R Package was also used to build classifiers with the Random Forest (RF) approach. RF is a tree-based classification algorithm similar to Classification and Regression Tree (CART) (28) and was performed on CpG average beta values using RandomForest R package version 4.5–18 by Liaw and Wiener. RF builds each individual tree by taking a bootstrap sample (sampling with replacement) of the original data and on average about 1/3 of the original data are not sampled (out of bag or OOB). Those sampled are used as the training set to grow the trees, and the OOB data are used as the test set. At each node of the tree, a random sample of m out of the total M variables is chosen and the best split is found among the m variables. The default value for m in the Random Forest R package is √M. In this analysis we will test a range of m from half of √M to two times the √M and will use the m that gives the lowest prediction error. The OOB error rate is the percentage of time the RF prediction is incorrect.
Results
To characterize the epigenetic profile of mesothelioma and non-tumorigenic parietal pleura we used the Illumina GoldenGate® bead array that simultaneously interrogates 1505 CpG sites associated with 803 cancer-related genes to generate a methylation value based upon ~30 replicate measurements for each locus in each sample. In this study GoldenGate® arrays were used to assess methylation in 158 incident cases of mesothelioma and 18 non-tumorigenic parietal pleura specimens. Exposure, demographic and tumor characteristic data for these samples are presented in Table 1.
Table 1.
Subject gender, age, histology and exposure for mesothelioma patients and non-tumor pleural samples.
| Mesothelioma patients |
Pleura donors | |
|---|---|---|
| Gender, n (%) | ||
| Female | 38 (24) | 4 (22) |
| Male | 120 (76) | 14 (78) |
| Age | ||
| Range | 30 – 80 | 38 – 77 |
| Mean (sd) | 62 (9.8) | 58 (11.3) |
| Histology, n (%) |
||
| Epithelioid | 116 (73) | - |
| Mixed | 37 (23) | - |
| Sarcomatoid | 5 (3) | - |
| Asbestos exposure, n (%) | ||
| Yes | 112 (74) | 13 (72) |
| No | 39 (26) | 5 (28) |
| Log Asbestos Body | ||
| Available n (%) | 108 (68) | - |
| Range | 0 – 5.5 | - |
| Mean (sd) | 2.16 (1.18) | - |
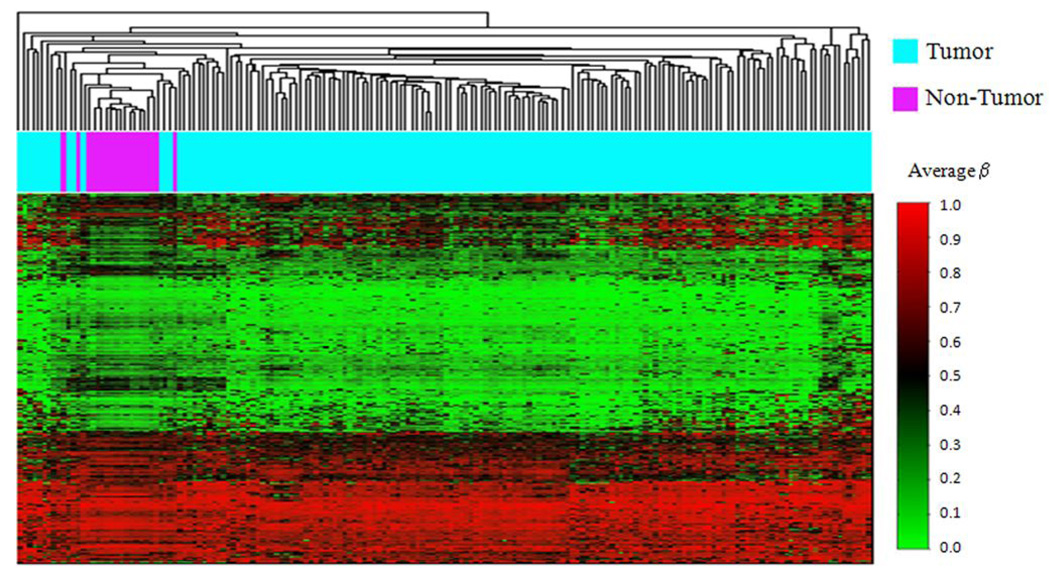
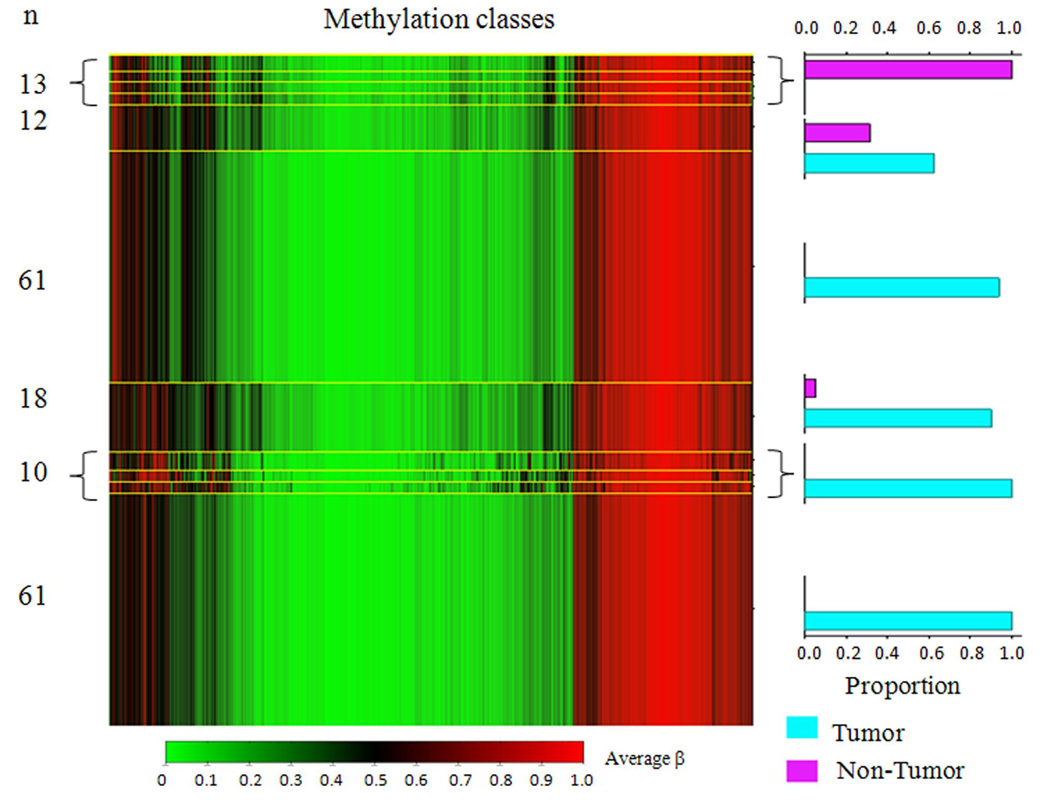
Array methylation data were first explored with unsupervised hierarchical clustering using Manhattan distance and average linkage for the 750 most variable autosomal CpG loci (Figure 1). Striking differences between the epigenetic profiles of mesothelioma and non-tumor pleura are observed, with almost perfect clustering of epigenetic profiles based on disease status. Next, in a univariate approach, we tested all CpG loci individually for an association between methylation and disease status, and 969 CpG loci had methylation levels that differed (Q < 0.05) comparing tumor and non-tumor pleura following FDR correction. Of these, 727 loci associated with 493 genes had enhanced methylation in non-tumor pleura, and 242 loci associated with 153 genes had more methylation in the tumors (Supplemental Table S1). Since so many loci were differentially methylated between tumor and non-tumor pleura, we next applied a modified model-based form of unsupervised clustering known as mixture modeling. This approach built classes of samples based on profiles of methylation with data from all autosomal loci using a mixture of beta distributions to recursively split the tumors into parsimoniously differentiated classes (29–31). All posterior class membership probabilities were numerically indistinct from 0 or 1. Applying a beta mixture model to methylation data from all autosomal loci in tumors and non-tumor pleura returned eleven methylation classes, their average methylation profiles, and their sample type distributions (Figure 2). Methylation class membership was a highly significant predictor of diseased versus non-diseased tissue (permutation P < 0.0001). Among the 11 classes in the model, 9 classes perfectly captured only tumor or only normal, and there were 2 methylation classes containing both tumor and normal samples. To follow up, a supervised random forest classification of non-tumor and tumor samples was performed. Only 1 tumor (< 1%) was misclassified as a non-tumor sample, and 5 non-tumor samples (28%) were misclassified as tumors. The overall misclassification error rate was 3.4%, significantly lower than the expected error rate under the null hypothesis (P < 0.0001).
Figure 1. Unsupervised clustering of average beta values in tumor and non-tumor pleura.
Using the R software package normal tissue sample average beta values were subjected to unsupervised hierarchical clustering based on Manhattan distance and average linkage. Each column represents a sample and each row represents a CpG locus (750 most variable autosomal loci). Above the heatmap blue indicates a tumor sample, and purple indicates a non-tumor pleural sample. In the heat map green = average beta of zero, or unmethylated, and red = average beta of one, or methylated
Figure 2. Beta mixture model of methylation profiles in mesothelioma and non-tumor pleura.
Methylation average β is green for unmethylated and red for methylated. Methylation profile classes are stacked in rows separated by yellow lines, and class height corresponds to the number of samples in each class. Class methylation at each locus is a mean of methylation for all samples within a class. Bar charts display the proportion of tumors and non-tumor pleura samples in each class. Methylation profile classes differentiate tumor from non-tumor pleura (P < 0.0001).
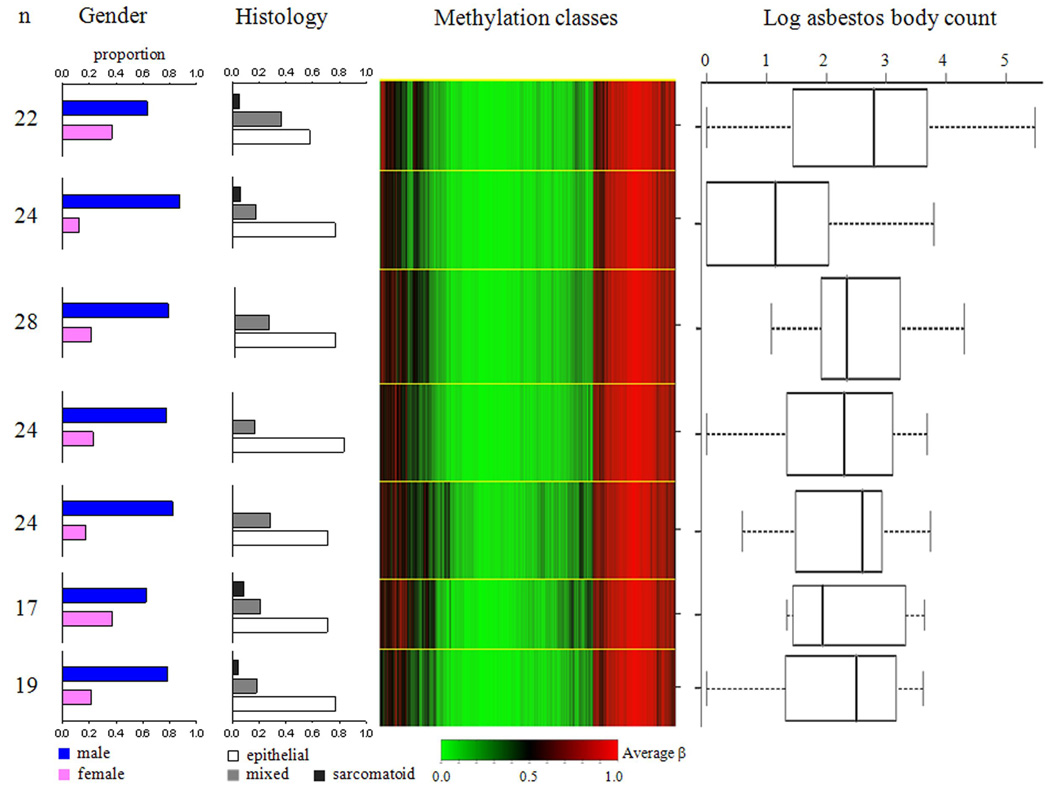
We next restricted our analyses to tumors, (n = 158) first applying our beta mixture model approach, and Figure 3 shows the seven methylation classes that resulted. This figure also displays the distributions of gender, histology, and asbestos body counts by methylation class. Methylation class membership was not a significant predictor of patient gender or tumor histology (data not shown). Again, methylation profile class membership was not associated with the amount of tumor in the sample. However, methylation class membership significantly predicted lung asbestos body count (permutation P < 0.04). Since men with pleural mesothelioma have higher asbestos body counts compared to women (P < 0.0001) (32) we controlled for gender, and methylation class membership remained a significant predictor of asbestos burden (likelihood ratio test P < 0.03). Based upon prior published work, specific CpG loci were tested for associations between methylation and asbestos body counts; consistent with our prior data,(20) tumor methylation average β values at CDKN2A (P < 0.02), CDKN2B (P < 0.02), and RASSF1 (P < 0.03) were significantly and positively associated with asbestos body counts. In addition, methylation of MT1A (previously reported as asbestos exposure-associated by Tsou et al. (18)) was significantly positively associated with asbestos burden; promoter associated CpG49 (P < 0.04), and exonic CpG13 (P < 0.02). When testing all autosomal loci for an association between methylation and asbestos burden using the MTA1 promoter CpG 49 Q–value (Q = 0.32) as a cutoff, there were 110 loci with an association between methylation status and asbestos burden (Supplemental Table S2). The vast majority of these 110 loci (94%) had a positive correlation between CpG methylation and asbestos body counts, indicating gene silencing was the dominant phenotype associated with asbestos associated epigenetic change.
Figure 3. Beta mixture model of methylation profiles in pleural mesothelioma.
Methylation average β is green for unmethylated and red for methylated. Methylation profile classes are stacked in rows separated by yellow lines, and class height corresponds to the number of samples in each class. Class methylation at each locus is a mean of methylation for all samples within a class. On the left, bar charts show proportions for gender and tumor histology among samples within each class. On the right, box plots of log asbestos body counts for each class. Controlling for gender, methylation class membership predicts asbestos burden (P < 0.03).
Lastly, we examined the relationships between methylation profiles and patient outcome using Cox proportional hazards models of survival controlling for age, gender, and tumor histology. Median survival time of this population was 12.5 months with 67 months of follow-up time. In a proportional hazards model including all cases (n = 158), women had half the risk of death of men (HR = 0.5, 95% CI, 0.3 – 0.96), and patients with mixed histology tumors were at greater risk of death compared to those with epithelial tumors (HR = 2.7, 95% CI, 1.7 – 4.4). Importantly, methylation class membership was also a significant predictor of patient outcome (P < 0.01). In particular, membership in methylation classes four and seven were both independently associated with a significant 3-fold increased risk of death compared to the class with the lowest median asbestos count (95% CIs, class four: 1.4 – 7.0, class seven: 1.3 – 7.4) (Table 2). Where data were available (n = 108), and after adjustment for methylation class membership, asbestos burden was associated with a significant 1.4-fold increased risk of death (95% CI, 1.1 – 1.8) (Table 2). In this model, membership in methylation class four remained associated with a significant, nearly 3-fold increased risk of death (HR = 2.8, 95% CI, 1.1 – 7.1). Again, in this model including asbestos exposure, likelihood ratio tests indicate that methylation classes were significant predictors of patient outcome (P < 0.005).
Table 2.
Gender, tumor histology, methylation profile class membership, and asbestos burden are predictors of pleural mesothelioma patient survival.
| All Cases | Cases with asbestos burden data | |||||||
|---|---|---|---|---|---|---|---|---|
| Co-Variate | n (%) Total n=158 |
HR | 95% CI | P -value | n (%) Total n=108 |
HR | 95% CI | P-value |
| Age, mean (sd) | 62 (9.8) | 1.02 | 1.0 – 1.05 | 0.09 | 61 (9.5) | 1.03 | 1.0 – 1.1 | 0.18 |
| Gender | ||||||||
| Male | 120 (76) | 1.0 | (reference) | - | 84 (78) | 1.0 | (reference) | - |
| Female | 38 (24) | 0.5 | 0.3 – 0.96 | < 0.04 | 24 (22) | 1.5 | 0.6 – 3.5 | 0.38 |
| Histology | ||||||||
| Epithelial | 109 (69) | 1.0 | (reference) | - | 74 (68) | 1.0 | (reference) | - |
| Mixed | 44 (28) | 2.7 | 1.7 – 4.4 | < 0.0001 | 31 (29) | 2.1 | 1.2 – 3.8 | < 0.02 |
| Sarcomatoid | 5 (3) | 2.8 | 0.95 – 8.2 | 0.06 | 3 (3) | 1.2 | 0.3 – 5.2 | 0.83 |
| Asbestos burden, mean (sd) | - | - | - | - | 2.2 (1.2) | 1.4 | 1.1 – 1.8 | < 0.04 |
| Methylation Class | ||||||||
| 2 | 24 (15) | 1.0 | (reference) | - | 17 (16) | 1.0 | (reference) | - |
| 1 | 22 (14) | 1.4 | 0.6 – 3.4 | 0.47 | 10 (9) | 0.5 | 0.1 – 2.2 | 0.37 |
| 3 | 28 (18) | 0.9 | 0.4 – 2.0 | 0.75 | 19 (18) | 0.4 | 0.1 – 1.2 | 0.11 |
| 4 | 24 (15) | 3.1 | 1.4 – 7.0 | < 0.01 | 24 (22) | 2.8 | 1.1 – 7.1 | < 0.03 |
| 5 | 24 (15) | 1.4 | 0.6 – 3.5 | 0.44 | 17 (16) | 0.9 | 0.3 – 2.8 | 0.89 |
| 6 | 17 (11) | 2.0 | 0.8 – 5.4 | 0.16 | 11 (10) | 1.2 | 0.3 – 4.8 | 0.79 |
| 7 | 19 (12) | 3.1 | 1.3 – 7.4 | < 0.01 | 10 (9) | 1.7 | 0.6 – 5.0 | 0.36 |
Controlled for all variables in table, model log liklihood P < 0.01
Classes 1 to 7 corresppond top to bottom from figure 3
Controlled for all variables in table, model log liklihood P < 0.005
Discussion
Exposure to asbestos is the single most important risk factor for pleural mesothelioma and prior research has established that somatic mutations (33) and alterations in gene expression (34) are a feature of this disease. Interestingly, relatively few pathologically important mutations arise in this cancer, and there is no characteristic somatic genetic change that can be attributed to the action of asbestos (33). Further, although there is consensus that gene expression (at the mRNA level) is significantly altered in mesothelioma, there is no gene expression signature representative of the action of asbestos in this disease, and there remains debate about the clinical utility of mRNA expression profiling (35–37). Our work sought to definitively characterize phenotypically important alterations in the epigenome of mesothelioma. We enumerated the epigenetic status of over 800 known cancer-related genes that stably control mRNA expression; comparing normal pleura with mesotheliomas. Our findings indicate that an extremely large number of loci are epigenetically altered in mesothelioma, asbestos exposure is associated with the degree of epigenetic alteration, and that profiles of gene silencing are associated with clinical outcome. This work demonstrates that the epigenome is a primary target of asbestos in the genesis of mesothelioma.
Precisely why epigenetic alterations are a prominent feature of pleural mesothelioma is not clear. However, it is well known that chronic inflammation is a primary tissue response to asbestos exposure (38). Epigenetic alterations have been associated with inflammation in colon cancer, and it has been suggested that inflammation-related epigenetic alterations are common in human cancers (39). Infiltrating macrophages, neutrophils, and eosinophils have been observed at sites of fiber deposition; these cells are known to generate reactive oxygen, and nitrogen, and to induce the release of peroxidases (38, 40). Treatment of human mesothelial cells with crocidolite asbestos has been shown to increase reactive oxygen species and oxidized DNA residues such as 8-oxoguanine (41). Further, it has been reported that 5-hydroxymethylcytosine can be generated by oxidation of 5-methylcytosine (42). Both 5-methylcytosine adjacent to 8- oxoguanine, and 5-hydroxymethylcytosine have been shown to inhibit binding of methyl-CpG binding protein 2, a critical epigenetic regulator that recruits cytosine methyltransferases and histone deacetylases (43). It is also known that 5-hydroxymethylcytosine is not recognized as 5- methylcytosine by the maintenance methyltransferase DNMT1, and hence, may lead to aberrant loss of methylation during cell replication (44). Additional base alterations occur via neutrophil and eosinophil peroxidase-derived HOCl and HOBr which can react with DNA to form 5- chlorocytosine and 5-bromocytosine respectively (45). These halogenated cytosines can be mistaken by DNMT1 as 5-methylcytosine during replication, thus providing a potential mechanism for asbestos-related, inflammation-induced aberrant hypermethylation (44). This is consistent with our data suggesting that asbestos burden (and the resultant inflammatory response) is associated with epigenetic alterations. Furthermore, the decades-long latency of MPM would allow ample time for cellular turnover and selection of cells with altered epigenetic programs that favor survival and deregulated proliferation
We also found that epigenetic profiles differentiate tumor from non-tumor pleura (P < 0.0001). Similarly, a genome-wide approach profiling of gene expression at the mRNA level in mesothelioma has demonstrated significant differential expression of over 600 genes between mesothelioma and non-tumor pleura, although the precise pattern of gene expression differs in the hands of different investigators (34, 37). Epigenetic gene inactivation is inherently more stable and less prone to variability than measures of mRNA, making our approach for differentiating tumors from non-tumor pleura likely more reproducible. Although our method differentiates tumors from non-tumor pleura extremely well, there remained two methylation profile classes with a minor mixture of tumor and non-tumor samples. It is clear that “contaminating” non-tumor tissue, or samples containing a lower percent tumor cannot explain this result, as we were able to reject this based upon direct observation of the amount of tumor in each specimen. Consequently, it may be that the relative epigenetic similarity between certain tumors and non-tumor pleura is explained by a difference in the prevalence of somatic genetic aberrations (such as gene deletions) among these tumors. In other words, tumors resembling nontumor pleura epigenetically may harbor significantly greater genetic alterations (relative to more epigenetically divergent tumors) that contribute to their malignant phenotype. An integrative genomics approach using common tumor sets for both epigenetic alteration profiling and genome-wide copy number alteration profiling will begin to address this question.
Hundreds of loci had different methylation levels between tumor and non-tumor pleura after correcting for multiple comparisons (Q < 0.05). Some of the pathways and processes whose genes are differentially methylated include epigenetic regulation, cell cycle control, and inflammation, among others. For instance, HIC1 was hypermethylated in tumors, which may prevent its transcriptional repression of the histone deacetylase SIRT1, and DNMT3B had significantly lower methylation in tumors, suggesting epigenetic dysregulation. Cell cycle control genes WEE1 and RASSF1 both had significantly higher methylation in tumors, suggesting uncontrolled proliferative potential, a hallmark of cancer (46). Another hypermethylated gene in tumors was ASB4; a SOCS box-containing protein that inhibits c-Jun N-terminal kinases, is thought to contribute to chronic inflammation (47). Among tumors, JAK2 had significantly decreased methylation, and concomitant silencing of SOCS genes may then result in increased, or constitutive STAT activation, which is a recognized inflammation associated mechanism of tumor initiation and promotion (48). In addition, HOXA9 was hypermethylated in tumors; HOXA9 is an inhibitor of NF-κB-dependent activation of leukocyte adhesion molecules ICAM-1, VCAM-1 and E-selectin which recruit leukocytes, exacerbating the inflammatory response (49).
At the same time, it is tenable to posit that, although many genes harbor altered epigenetic states, only a small number of genes with epigenetic alterations are chiefly responsible for the phenotypic differences between tumor and normal tissue, and between more deadly mesothelioma compared with less deadly forms. However, if relatively few “epigenetic gatekeepers” determine phenotype, there must be a very large number of genes that are inactivated consequent to the clonal evolution of the tumor; this may be analogous to ‘hitchhiker’ mutations that arise during tumor clonal expansion. Alternatively, a ‘hit’ at a locus essential for epigenetic regulation, the chronic inflammatory process, or altered tumor metabolism, for example, may induce a change in overall epigenetic regulation that results in the targeting and aberrant methylation of many genes in a wholesale fashion. It is not possible to directly distinguish the alternative explanations of our data.
Among the 158 mesotheliomas studied, seven distinct methylation profile classes were identified. Previous genome-wide mRNA expression analysis of mesotheliomas was able to define two distinct subclasses of tumors that loosely correlated with histology (34). Although our epigenetic profiling data revealed more tumor subclasses, the genome-wide approach using mRNA expression as an outcome encompassed over ten-times as many loci, and used fewer tumors, both of which potentially decreased the power to resolve tumor subclasses compared to our larger sample set restricted to cancer-associated loci. We found that methylation profiles were associated with asbestos body burden in an analysis controlling for gender (P <0.03), again suggesting asbestos-associated inflammation as a mechanism driving tumors into distinct epigenetic subclasses. There were also distinct methylation profile classes comprised of tumors with similar asbestos burden distributions, arguing that asbestos burden per se does not account for all of the observed epigenetic alterations in mesothelioma. However, in the context of a decades-long latency and a non-specific, asbestos-related chronic inflammation state, alterations of different epigenetic gatekeeper genes could occur at different stages in tumorigenesis and tumor progression. Furthermore, specific genes, or gene pathways, may be more likely to be selected for, or more susceptible to asbestos-related epigenetic alterations; consistent with this, associations between CpG methylation of specific genes and asbestos previously reported by our group and others were confirmed in this study (18, 20). Here, we extended these findings to include several apoptosis-related genes such as AATK, CASP2, CASP10, and PYCARD.
Finally, overall methylation profile class membership was a significant, independent predictor of patient survival (P < 0.01), suggesting that epigenetic dysregulation is strongly associated with disease progression. Although general epigenetic deregulation may be highly correlated with disease prognosis, we cannot exclude epigenetic events at specific loci as responsible for predicting survival. Continued investigation into the CpG loci whose differential methylation status is associated both with membership in specific classes and prognosis is needed in order to elucidate the biological mechanism(s) which critically influence patient survival. Additionally, consistent with our previous finding, we confirmed that an increased asbestos burden is a significant, independent predictor of reduced survival in pleural mesothelioma (32).
In summary, our data show that epigenetic alterations are extraordinarily common in mesothelioma and discriminate the malignant phenotype from normal pleura. Epigenetic changes are also significantly associated with asbestos burden and significantly predict clinical outcome. Hence, our data suggest that phenotypically important somatic epigenetic modification is a major mode of action of asbestos in mesothelioma. Further investigation into the underlying mechanism responsible may assist in diagnosis, assessment of prognosis and design of therapies for this rare, but rapidly fatal disease.
Supplementary Material
Acknowledgments
Financial Support: National Cancer Institute (R01 CA126939); National Institutes of Environmental Health Sciences (T32ES007155, P42ES05947); International Mesothelioma Program at Brigham and Women's Hospital (Research grant); Mesothelioma Applied Research Foundation (Research grant).
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature reviews. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 4.Bibikova M, Lin Z, Zhou L, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome research. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson BW, Lake RA. Advances in malignant mesothelioma. The New England journal of medicine. 2005;353:1591–1603. doi: 10.1056/NEJMra050152. [DOI] [PubMed] [Google Scholar]
- 6.Morinaga K, Kishimoto T, Sakatani M, Akira M, Yokoyama K, Sera Y. Asbestos-related lung cancer and mesothelioma in Japan. Ind Health. 2001;39:65–74. doi: 10.2486/indhealth.39.65. [DOI] [PubMed] [Google Scholar]
- 7.Price B, Ware A. Mesothelioma trends in the United States: an update based on Surveillance, Epidemiology, and End Results Program data for 1973 through 2003. Am J Epidemiol. 2004;159:107–112. doi: 10.1093/aje/kwh025. [DOI] [PubMed] [Google Scholar]
- 8.Roushdy-Hammady I, Siegel J, Emri S, Testa JR, Carbone M. Genetic-susceptibility factor and malignant mesothelioma in the Cappadocian region of Turkey. Lancet. 2001;357:444–445. doi: 10.1016/S0140-6736(00)04013-7. [DOI] [PubMed] [Google Scholar]
- 9.Bhagavatula R, Moody R, Russ J. Asbestos: a moving target. Best's Review. 2001;102:85–90. [Google Scholar]
- 10.Peto J, Decarli A, La Vecchia C, Levi F, Negri E. The European mesothelioma epidemic. British journal of cancer. 1999;79:666–672. doi: 10.1038/sj.bjc.6690105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joshi TK, Gupta RK. Asbestos in developing countries: magnitude of risk and its practical implications. International journal of occupational medicine and environmental health. 2004;17:179–185. [PubMed] [Google Scholar]
- 12.Landrigan PJ, Lioy PJ, Thurston G, et al. Health and environmental consequences of the world trade center disaster. Environmental health perspectives. 2004;112:731–739. doi: 10.1289/ehp.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner JC, Sleggs CA, Marchand P. Diffuse pleural mesothelioma and asbestos exposure in the North Western Cape Province. Br J Ind Med. 1960;17:260–271. doi: 10.1136/oem.17.4.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaurand MC. Mechanisms of fiber-induced genotoxicity. Environmental health perspectives. 1997;105 Suppl 5:1073–1084. doi: 10.1289/ehp.97105s51073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelsey KT, Yano E, Liber HL, Little JB. The in vitro genetic effects of fibrous erionite and crocidolite asbestos. British journal of cancer. 1986;54:107–114. doi: 10.1038/bjc.1986.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B, Lee AY, Dadfarmay S, et al. Secreted frizzled-related protein 4 is silenced by hypermethylation and induces apoptosis in beta-catenin-deficient human mesothelioma cells. Cancer research. 2005;65:743–748. [PubMed] [Google Scholar]
- 17.Hirao T, Bueno R, Chen CJ, Gordon GJ, Heilig E, Kelsey KT. Alterations of the p16(INK4) locus in human malignant mesothelial tumors. Carcinogenesis. 2002;23:1127–1130. doi: 10.1093/carcin/23.7.1127. [DOI] [PubMed] [Google Scholar]
- 18.Tsou JA, Galler JS, Wali A, et al. DNA methylation profile of 28 potential marker loci in malignant mesothelioma. Lung Cancer. 2007 doi: 10.1016/j.lungcan.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsou JA, Shen JY, Siegmund KD, et al. Distinct DNA methylation profiles in malignant mesothelioma, lung adenocarcinoma, and non-tumor lung. Lung Cancer. 2005;47:193–204. doi: 10.1016/j.lungcan.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Christensen BC, Godleski JJ, Marsit CJ, et al. Asbestos exposure predicts cell cycle control gene promoter methylation in pleural mesothelioma. Carcinogenesis. 2008;29:1555–1559. doi: 10.1093/carcin/bgn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Vuyst P, Karjalainen A, Dumortier P, et al. 1998 ERS Task Force Report Guidelines for mineral fibre analyses in biological samples: report of the ERS working group. Eur Respir J. 1998;11:1416–1426. doi: 10.1183/09031936.98.11061416. [DOI] [PubMed] [Google Scholar]
- 22.Churg A, Warnock ML. Correlation of quantitative asbestos body counts and occupation in urban patients. Archives of pathology & laboratory medicine. 1977;101:629–634. [PubMed] [Google Scholar]
- 23.R Development CT. A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2007. [Google Scholar]
- 24.Hsiung DT, Marsit CJ, Houseman EA, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 25.Storey J, Taylor J, Siegmund D. Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J Royal Stat Soc. 2004;(Series B):187–205. [Google Scholar]
- 26.Houseman EA, Christensen BC, Marsit CJ, et al. Model-based clustering of DNA methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics. 2008:9. doi: 10.1186/1471-2105-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao Y. Information-theoretic measures for knowledge discovery and data mining. In: Karmeshu, editor. Entropy Measures. Maximum Entropy Principle and Emerging Applications: Springer; 2003. pp. 115–136. [Google Scholar]
- 28.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 29.Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegmund KD, Connor CM, Campan M, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siegmund KD, Laird PW, Laird-Offringa IA. A comparison of cluster analysis methods using DNA methylation data. Bioinformatics (Oxford, England) 2004;20:1896–1904. doi: 10.1093/bioinformatics/bth176. [DOI] [PubMed] [Google Scholar]
- 32.Christensen BC, Godleski JJ, Roelofs CR, et al. Asbestos burden predicts survival in pleural mesothelioma. Environmental health perspectives. 2008;116:723–726. doi: 10.1289/ehp.11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugarbaker DJ, Richards WG, Gordon GJ, et al. Transcriptome sequencing of malignant pleural mesothelioma tumors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3521–3526. doi: 10.1073/pnas.0712399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon GJ, Rockwell GN, Jensen RV, et al. Identification of novel candidate oncogenes and tumor suppressors in malignant pleural mesothelioma using large-scale transcriptional profiling. The American journal of pathology. 2005;166:1827–1840. doi: 10.1016/S0002-9440(10)62492-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon GJ, Jensen RV, Hsiao LL, et al. Using gene expression ratios to predict outcome among patients with mesothelioma. Journal of the National Cancer Institute. 2003;95:598–605. doi: 10.1093/jnci/95.8.598. [DOI] [PubMed] [Google Scholar]
- 36.Gordon GJ, Rockwell GN, Godfrey PA, et al. Validation of genomics-based prognostic tests in malignant pleural mesothelioma. Clin Cancer Res. 2005;11:4406–4414. doi: 10.1158/1078-0432.CCR-04-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Rios F, Chuai S, Flores R, et al. Global gene expression profiling of pleural mesotheliomas: overexpression of aurora kinases and P16/CDKN2A deletion as prognostic factors and critical evaluation of microarray-based prognostic prediction. Cancer research. 2006;66:2970–2979. doi: 10.1158/0008-5472.CAN-05-3907. [DOI] [PubMed] [Google Scholar]
- 38.Moalli PA, MacDonald JL, Goodglick LA, Kane AB. Acute injury and regeneration of the mesothelium in response to asbestos fibers. The American journal of pathology. 1987;128:426–445. [PMC free article] [PubMed] [Google Scholar]
- 39.Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer research. 2001;61:3573–3577. [PubMed] [Google Scholar]
- 40.Quinlan TR, Marsh JP, Janssen YM, Borm PA, Mossman BT. Oxygen radicals and asbestos-mediated disease. Environmental health perspectives. 1994;102 Suppl 10:107–110. doi: 10.1289/ehp.94102s10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q, Marsh J, Ames B, Mossman B. Detection of 8-oxo-2'-deoxyguanosine, a marker of oxidative DNA damage, in culture medium from human mesothelial cells exposed to crocidolite asbestos. Carcinogenesis. 1996;17:2525–2527. doi: 10.1093/carcin/17.11.2525. [DOI] [PubMed] [Google Scholar]
- 42.Masuda T, Shinoara H, Kondo M. Reactions of hydroxyl radicals with nucleic acid bases and the related compounds in gamma-irradiated aqueous solution. Journal of radiation research. 1975;16:153–161. doi: 10.1269/jrr.16.153. [DOI] [PubMed] [Google Scholar]
- 43.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic acids research. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer research. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 45.Henderson JP, Byun J, Takeshita J, Heinecke JW. Phagocytes produce 5-chlorouracil and 5-bromouracil, two mutagenic products of myeloperoxidase, in human inflammatory tissue. The Journal of biological chemistry. 2003;278:23522–23528. doi: 10.1074/jbc.M303928200. [DOI] [PubMed] [Google Scholar]
- 46.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 47.Li JY, Chai BX, Zhang W, Liu YQ, Ammori JB, Mulholland MW. Ankyrin repeat and SOCS box containing protein 4 (Asb-4) interacts with GPS1 (CSN1) and inhibits c-Jun NH2-terminal kinase activity. Cellular signalling. 2007;19:1185–1192. doi: 10.1016/j.cellsig.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–565. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 49.Trivedi CM, Patel RC, Patel CV. Homeobox gene HOXA9 inhibits nuclear factor-kappa B dependent activation of endothelium. Atherosclerosis. 2007;195:e50–e60. doi: 10.1016/j.atherosclerosis.2007.04.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.