Abstract
Akt kinase is activated by transforming growth factor-beta1 (TGF-β) in diabetic kidneys and plays important roles in fibrosis, hypertrophy and cell survival in glomerular mesangial cells (MC)1–11. However, the mechanisms of Akt activation by TGF-β are not fully understood. Here we show that TGF-β activates Akt in MC by inducing microRNA-216a (miR-216a) and miR-217, both of which target phosphatase and tensin homologue (PTEN). Both these miRs are located within the second intron of a non-coding RNA (RP23-298H6.1-001). The RP23 promoter was activated by TGF-β and also by miR-192 via E-box-regulated mechanisms as shown previously3. Akt activation by these miRs also led to MC survival and hypertrophy similar to TGF-β. These studies reveal a mechanism of Akt activation via PTEN downregulation by two miRs regulated by upstream miR-192 and TGF-β. Due to the diversity of PTEN function12, 13, this miR amplifying circuit may play key roles not only in kidney disorders, but also other diseases.
Diabetic Nephropathy (DN) is a major complication of diabetes. Key features of DN include glomerular mesangial expansion, hypertrophy and accumulation of extracellular matrix (ECM) proteins such as collagen in the kidney2, 3, 7, 10, 11. The phosphoinositide 3-kinase (PI3K)-Akt pathway is activated in animal models of DN4–7 and Akt1−/− mice are protected from hyperhexosemia-induced mesangial hypertrophy and ECM accumulation7. These results implicate Akt kinase as a key mediator of DN. Increased expression of TGF-β is observed in renal cells during DN progression1–3. PI3K-Akt activation by TGF-β4, 5, 8–10 has been related to increased ECM protein expression9, 10, hypertrophy5, cell survival and oxidant stress in MC4. However, the mechanism by which TGF-β activates Akt has not been fully elucidated.
MicroRNAs (miRs) are short non-coding RNAs that induce gene silencing mainly by blocking mRNA translation or promoting mRNA degradation14, 15. A number of miRs are highly expressed in the kidney16, 17. miR-192 was shown to be upregulated in TGF-β-treated mouse MC (MMC) and in diabetic mouse glomeruli, and to increase collagen type I α2 chain (Col1a2) expression by downregulating Zeb2 (also called SIP1 or Zfhx1b), an E-box repressor3. Another report showed that miR-377 regulates fibronectin expression in DN18. However, the functional roles and regulation of other renal miRs are unclear. Here we show that miR-216a is also upregulated by TGF-β in MMC in a dose- and time-dependent manner, similar to miR-1923 (Fig. 1a,b and Supplementary Fig. S1c,k). Col1a2 was induced in parallel. miR-216a levels were also increased in renal glomeruli isolated from type1 (streptozotocin [STZ] injected) and type 2 (db/db) diabetic mice relative to their respective controls (C57BL/6 and db/+) (Fig. 1c).
Figure 1. Expression and genomic organizations of miR-216a and miR-217.
a, Dose response of TGF-β (24hr) on miRs and related genes in MMC (qPCR data, *, p<0.05; #, p<0.01 vs control, n=3). See Fig. S1 for details. b, Time course of TGF-β effects in MMC (qPCRs, *, p<0.05; #, p<0.01, n=3). See Fig. S1 for details. c, Real time qPCRs showing significant increases in miR-216a and miR-217 levels in glomeruli of STZ-injected type 1, and db/db type 2 diabetic mice (n=4). d, Schematic representation of mouse non-coding RNA RP23-298H6.1-001 genomic region showing miR-216a and miR-217 locations in the second intron, with upstream CAGAs and E-box clusters. e, Genomic structure of the upstream region of the RP23 gene. CAGA repeats (blue triangles) and potential E-boxes (red diamonds) are found in the 2–5kb upstream region. f, Basal activity of RP23 promoter regions and response to TGF-β −4.8k and −3.5k constructs, but not −2.7k, responded to TGF-β (n=4). g, Response of deletion mutants of the −4.8–2.7k region to TGF-β,( n=4). −4.8–3.5k region includes three E-boxes and ten CAGAs while −3.5–2.7k region has four E-boxes and eight CAGAs. Significant increase in Luc activity of the 3.5–2.7k construct was observed but no change with −4.8–3.5k region (n=4). TGF-β had similar effects in the RP23-3.5-2.7k, 2xE-boxes and 1xE-box constructs. One E-box in the most proximal part of −3.5–2.7k was sufficient for TGF-β response. Mutation of this proximal E-box abrogated the TGF-β response. h, qPCRs showing that miR-192 mimic (10 nM) decreased the expression of Zeb2 and reciprocally increased RP23, miR-216a and miR-217 levels (n=3). i, qPCRs showing that, similar to miR-192, ON-TARGETplus SMART pool Zeb2 siRNA (10 nM) decreased the expression of Zeb2 but increased RP23, miR-216a and miR-217 levels (n=3). j, miR-192 mimic (10 nM) or SMART pool Zeb2 siRNA(10nM) increased Luc activity of RP23-3.5-2.7k-luc (n=4). NC, negative control. Bar graph data expressed as mean and s.e.m.
miR-216a lies in the second intron of a non-coding RNA (RP23-298H6.1-001, RP23) located in mouse chromosome 11 (miRBase, http://microrna.sanger.ac.uk)19 (Fig. 1d and Supplementary Fig. S2a). RP23 levels were also increased by TGF-β in MMC (Fig. 1a,b and Supplementary Fig. S1b,j). Interestingly, another miR-217 was present in the same intron, 6.6 kb-downstream of miR-216a (Fig. 1d). Indeed, miR-217 levels were increased in TGF-β-treated MMC (Fig. 1a, b and Supplementary Fig. S1d,l), and in the glomeruli of diabetic mice (Fig. 1c). Therefore, miR-216a and miR-217 were expressed along with RP23 and induced by diabetic conditions or TGF-β.
Next, the RP23 promoter region was examined. We focused on CAGA repeats (Smad binding elements) and E-boxes (CAXXTG), due to their function in TGF-β response3, 20. Multiple CAGAs and E-boxes were found in the RP23 upstream region, especially from −5 kb to −2 kb (Fig. 1e). Upstream fragments of the RP23 gene were cloned into pGL4-Luc reporter and transfected into MMC. The longest construct (RP23-4.8k-luc) had the lowest basal activity, but significant response to TGF-β (Fig. 1f). RP23-3.5k-luc had intermediate basal activity and also responded to TGF-β. The shortest construct (RP23-2.7kb-luc) had highest basal activity, but no TGF-β response. The 4.8-2.7kb region seemed to have negative elements for basal activity, and positive TGF-β-response elements. To identify the elements, partial fragments of this region were cloned into pGL3P-Luc. While RP23-3.5-2.7k-luc responded to TGF-β, neither RP23-4.8-3.5k-luc nor pGL3P alone did (Fig. 1g). Deletion of two or three upstream E-boxes from −3.5–2.7kb did not alter TGF-β effects (Fig. 1g). However, mutation of the most proximal E-box completely abrogated the TGF-β response, confirming that E-boxes within −3.5-2.7kb are essential for the TGF-β response in MMC.
E-boxes are negatively regulated by Zeb1 and Zeb23, 21, 22. miR-192 upregulates Col1a2 by targeting Zeb23. In time course experiments, TGF-β induced a significant increase in miR-192 by 30min, while induction of RP23, miR-216a and miR-217 required 1hr (Fig. 1b,Supplementary Fig. S1i-l), suggesting that miR-192 may upregulate RP23, miR216a and miR-217. Zeb1 and Zeb2 were also reduced at 1hr (Fig. 1b,Supplementary Fig S1m,n). Transfection of MMC with miR-192 mimic or Zeb2 siRNA significantly decreased Zeb2 levels and increased the expression of RP23, miR-216a and miR-217 (Fig. 1h,i) as well as RP23-3.5-2.7k-luc activity (Fig. 1j). By inhibiting Zeb2, miR-192 relieves the repression of RP23 (also miR-216a and miR-217) through its upstream E-boxes, similar to collagen induction by TGF-β3. Initial induction of miR-192 by TGF-β may subsequently increase RP23 and miR-216a/miR-217.
Transcription factor E3 (TFE3) is a positive regulator of E-boxes20, 23. Ectopic expression of Tfe3 significantly increased RP23-3.5-2.7k promoter activity (Supplementary Fig. S2b). Conversely, knockdown of Tfe3 by shRNAs23 significantly decreased reporter activity (Supplementary Fig. S2c). Chromatin immunoprecipitation (ChIP) assays revealed that Tfe3 occupancy at the RP23-3.5-2.7kb region was increased by TGF-β while Zeb1 occupancy was decreased (Supplementary Fig. S2d,e). E-box regulators may play key roles in the transcription of RP23 and miR-216a/217 by TGF-β, collaborating with Smads20 (Supplementary Fig. S2i).
Genomic structures of miR216a and miR-217 are conserved from human to mouse, rat and Zebrafish19, 24, 25. In the human genome, miR-216a and miR-217 also lie in the second intron of a non-coding RNA (DA732292, DA73) on chromosome 2. CAGA and E-box clusters were also found in the human DA73 promoter (7 kb upstream) (Supplementary Fig. S2f). The −7.3–6.5 kb upstream region of DA73 was cloned into pGL3P and transfected into MMC or human embryonic kidney (HEK-293) cells. Luc activity of this construct was significantly enhanced by TGF-β or miR-192 mimics (Supplementary Fig. S2g,h). These results suggest a conserved regulation of miR-216a and miR-217 from mouse to human.
Using miR target predictions (TargetScan, http://www.targetscan.org), we found that PTEN is a potential target of both miR-216a and miR-217 (Fig. 2a). PI3K generates phosphoinositide-3, 4, 5-triphosphate (PIP3) from PIP2, which then activates Akt12, 13. PTEN dephosphorylates PIP3 to inhibit Akt activation. PTEN mutations enhance Akt activity in certain cancer cells12. Akt activation and decreased PTEN are observed in MC treated with high glucose (HG) or TGF-β5. Therefore, PTEN might be a link between TGF-β and Akt activation.
Figure 2. Pten is a direct target of miR-216a and miR-217.
a, Alignment of human and mouse miR-216a or miR-217 with Pten 3′UTR sequences. Seed regions of these miRs are completely complementary to target sites in the Pten 3′UTR. Target sequences are highly conserved from human and mouse. Numbers next to the PTEN 3′UTR sequence are relative to 3′ UTR start sites. b–d, Immunostaining of Pten in mouse renal sections. Pten levels in glomeruli from db/db diabetic mice (c) were significantly lower than control db/+ (b) (n=3). Scale bar, 20 μm. e–g,j, Pten protein levels in MMC treated for 24 hr with TGF-β or HG (n=3) (e), for 48 hr with miR-216a mimic (10nM)(n=3)(f), miR-217 alone or with miR-216a (n=3) (g), miR-216a, miR-217 or miR-192 mimics (n=4) (j). h,i, Phospho-Akt levels in MMC treated for 24 hr with TGF-β (n=3) (h), for 48 hr with miR-216a, miR-217 or miR-192 mimics (10nM)(n=4) (i). k, Luc reporters containing full-length (6k) or shorter (1.4k) Pten 3′UTR, and mutants of miR-216a and miR-217 target sites (SVP, SV40 promoter). miR-216a and miR-217 mimics (10nM) significantly inhibited Pten 3′UTR Luc (full) activity relative to negative control (NC). The shorter 1.4k construct (WT) which includes the miR-216a and miR-217 target sites showed similar responses as full-length. miR-216a site mutant still responded to miR-217 but not to miR-216a, and vice versa. The double mutant did not respond to either of the miR mimics. Mean and s.e.m. (n=4). See also Fig. S4a for details of Pten 3′UTR constructs. l, Effects of TGF-β and miR-216a inhibitor on the Pten 3′UTR reporters. Plasmid containing luciferase fused to Pten 3′UTR (miR-216a target site) was used as reporter (Pten 3′UTR-S). Significant decrease in reporter activity was detected with Pten 3′UTR-S by TGF-β (n=4) and this was reversed by 10 nM miR-216a inhibitor (n=4). m, TGF-β significantly decreased Pten 3′UTR-S (miR-217 site) reporter activity (n=4), and this was reversed by 5 nM miR-217 inhibitor (n=4). See also Fig. S4. Bar graph data expressed as mean and s.e.m. Full or larger scan blots of e–j are shown in Supplementary Fig. S7.
Decreased levels of Pten were confirmed in glomeruli of diabetic db/db mice (Fig. 2b–d) and in MMC treated with TGF-β or HG (Fig. 2e). miR-216a or miR-217 mimic decreased Pten levels in MMC (30~40% of negative control)(Fig. 2f,g). Simultaneous transfection with these two mimics decreased Pten levels to a greater extent (~10% of negative control), suggesting an additive effect of these two miRs on Pten downregulation. Despite significant decrease in Pten protein levels in TGF-β-treated MMC, no decrease in Pten mRNA levels was observed (Fig. 1a,b and Supplementary Fig. S1h,p), suggesting post-transcriptional reduction of Pten by TGF-β in MMC.
TGF-β activated Akt (Ser-473 phosphorylation) in MMC (Fig. 2h), consistent with previous reports4, 5. A parallel increase in Akt activation in MMC transfected with miR-216a, miR-217 or both (Fig. 2i) further confirmed that miR-216a/217 target Pten in response to TGF-β. A biphasic Akt activation was observed in the time course results (Supplementary Fig. S3a,c). The rapid response (5 min)4 may presumably result from direct interaction of TGF-β receptor and PI3K8. The second, more robust Akt activation by 6 to 24hr (Supplementary Fig. S3a,c) occurs via induction of RP23 (miR-216a/217) and decrease in Pten (Fig. 2e and Supplementary Fig. S3a,b). Interestingly, miR-192 mimic decreased Pten levels and promoted Akt activation similar to miR-216a/miR-217 mimics (Fig. 2i,j), again suggesting that miR-192 acts upstream of miR-216a and miR-217.
To verify whether these miRs target the Pten 3′UTR, Luc reporters containing Pten 3′UTR (full-length and shorter form with miR-216a and miR-217 target sites) were constructed. miR-216a or miR-217 mimics significantly inhibited Pten 3′UTR Luc activity relative to negative controls, while Pten 3′UTR containing mutations at miR-216a and/or miR-217 sites lost this response (Fig. 2k and Supplementary Fig. S4a). Similar inhibition was observed with TGF-β, while miR-216a/217 inhibitors could override this TGF-β-induced decrease in Luc activity (Fig. 2l,m and Supplementary Fig. S4b–g). These results confirm that Pten 3′UTR is a target of miR-216a and miR-217.
Transgenic mice with the miR-17-92 cluster displayed lymphoproliferative diseases due to decreased Pten and Bim targeted by this cluster26. They also had enlarged kidney glomeruli, hypercellularity, mesangial expansion, and proteinuria (features similar to DN). These observations further support the physiological significance of our data that Pten downregulation by miRs is an important step in kidney dysfunction.
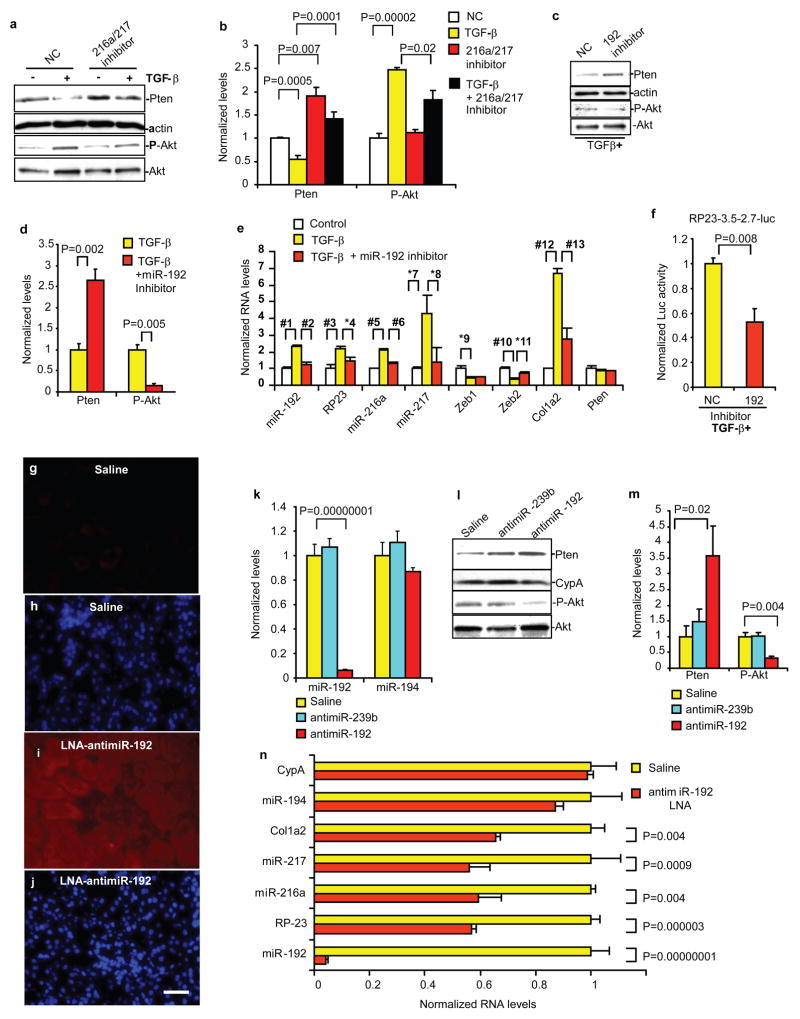
Inhibitors of miR-192, miR-216a and miR-217 were next tested to determine functional relevance. We first used MMC treated with TGF-β as these cells show features of DN (Akt activation and ECM accumulation). miR-216a and miR-217 inhibitors reversed the effects of TGF-β on Pten and P-Akt levels in MMC (Fig. 3a,b), as also the miR-192 inhibitor (Fig. 3c,d). Moreover, miR-192 inhibitor reversed the effects of TGF-β on miR-192, RP23, miR-216a, miR-217, Zeb2 and Col1a2 RNA expressions (Fig. 3e), and attenuated TGF-β-induced RP23 promoter activation (Fig. 3f). Therefore, the miR-192 inhibitor blocked key pathological effects of TGF-β in MMC related to DN (including Col1a2 and Akt activation).
Figure 3. Effects of miR inhibitors on downstream signaling in vitro and in vivo.
a,b, Effects of miR-216a and miR-217 inhibitors (10nM) on endogenous Pten, P-Akt levels (Western blots) in MMC treated with or without TGF-β. NC, negative control (n=3). c,d, Effects of miR-192 inhibitor (10nM) on TGF-β induced effects on Pten and P-Akt in MMC. MMC were transfected with inhibitors and then treated with TGF-β, followed by Western blotting (n=3). e, Effects of miR-192 inhibitor on endogenous RNA levels of miR-192, RP23, miR-216a, miR-217, Zeb1, Zeb2, Col1a2 and Pten in MMC treated with TGF-β (qPCRs, *, p<0.05; #, p<0.01, n=3). #1, p=0.005; #2, p=0.001; #3, p=0.001; *4, p=0.02; #5, p=0.006; #6, p=0.0007; *7, p=0.02; *8, p=0.02; *9, p=0.04; #10, p=0.01; *11, p=0.02; #12, p=0.002; #13, p=0.003. f, Effects of miR-192 inhibitor on TGF-β-induced RP23 promoter (RP23-3.5-2.7k-luc) activity in MMC (n=4). g–j, In vivo delivery of LNA-antimiR-192. LNA-antimiR-192 was subcutaneously injected (2.5 mg/kg) into normal C57BL/6 mice and renal cortical tissues harvested 6hr later. Representative data of in-situ hybridization to detect LNA-antimiR-192 in mouse renal cortical sections 6hr after injections. Very feeble fluorescence was detected in the kidney of control vehicle saline-injected mice (saline, g). Intense and widespread fluorescence in kidney glomerular and tubular compartments was observed in LNA-antimiR-192 injected mice (i). DAPI staining of kidney cortex of saline and LNA-antimiR-192 injected mice respectively (h,j). Scale bar, 20 μm. k, qPCRs of cortical tissues obtained from normal C57BL/6 mice injected subcutaneously with 2.5 mg/kg LNA-antimiR-192, LNA-antimiR-239b (targeting C. elegans miR-239b) or saline. Significant decrease in RNA levels of miR-192, but not miR-194, was observed with LNA-antimiR-192, while no change with saline or LNA-antimiR-239b. l,m, Concomitant significant increase in Pten protein levels and decrease in P-Akt levels (P-Akt to total Akt ratios) by LNA-antimiR-192 but not by LNA-antimiR-239b. n, Significant decrease in RNA levels of miR-192, RP23, miR-216a, miR-217, and Col1a2, but no change in miR-194 or CypA in LNA anti-miR-192 injected mice. (n=3). Bar graph data expressed as mean and s.e.m. Full or larger scan blots of a,c,l are shown in Supplementary Fig. S7.
We next examined the in vivo consequences of miR-192 inhibition using Locked nucleic acid (LNA)-modified oligonucleotides. LNA antimiRs are potent miR inhibitors even in primates27. Marked accumulation of LNA-modified miR-192 inhibitor (antimiR-192) was observed in mouse kidneys after subcutaneous injections relative to saline (Fig. 3g–j). LNA-antimiR-239b targeting C. elegans miR-239b (similar sequences not found in mammalian RNAs) was injected as a negative control. LNA-antimiR-192, but not LNA-antimiR-239b, significantly inhibited miR-192 levels in renal cortical tissues (Fig. 3k). miR-194 expression was unaffected, indicating specificity of LNA-antimiR-192. LNA-antimiR-192 also attenuated P-Akt while augmenting Pten levels relative to saline and LNA-antimiR-239b controls (Fig. 3l,m). Concomitant decreases in RP23, miR-216a, miR-217 and Col1a2 levels (but not miR-194 or CypA) were observed (Fig. 3n). These in vivo data further support miR-192 as a master regulator of other related miRs, and the therapeutic potential of antimiR-192 for DN. The possible cell-type differential effects of miRs should however be considered while designing such therapies.
Next, the biological significance and downstream consequences of Akt activation by this miR circuit were evaluated. Phosphorylation of Akt targets, FoxO3a, mTOR and GSK3β, associated with cell survival and hypertrophy4–6, 28, was induced by miR-192 and miR-216a/217 mimics as well as TGF-β (Fig. 4a–d). Interestingly, phosphorylation of these proteins by TGF-β was blocked or attenuated by inhibitors of these miRs (Fig. 4a–d), suggesting that this miR circuit affects key Akt targets. Since Akt-induced phosphorylation of FoxO3a transcription factor promotes nuclear to cytoplasmic translocation and inactivation, we next tested the effects of the miRs in MMC transfected with a vector expressing FoxO3a-GFP4. Serum depletion induced- nuclear localization of FoxO3a, while TGF-β treatment induced cytoplasmic translocation presumably due to Akt activation and FoxO3a phosphorylation (Fig. 4e), as previously reported4. miR-192 and miR-216a/217 mimics induced a similar cytoplasmic translocation of FoxO3a in serum-depleted MMC (Fig. 4e). Conversely, inhibitors of these miRs induced nuclear localization of FoxO3a in TGF-β-treated MMC (Fig. 4e). Fig. 4f showing quantification of the percentage of cells with cytoplasmic FoxO3a confirms that these miRs induced significant nuclear to cytoplasmic translocation of FoxO3a in serum-depleted MMC similar to TGF-β, while inhibitors of miRs attenuated TGF-β effects. Therefore, these miRs promote FoxO3a phosphorylation and cytoplasmic translocation by inhibiting Pten and activating Akt.
Figure 4. Effects of miR-192 and miR-216a/217 on Akt downstream targets.
a. Blots showing that miR-192 and miR-216/217 mimics (10nM) induced phosphorylation of Akt targets, FoxO3a, mTOR and GSK3β similar to the actions of TGF-β miR inhibitors (10nM) blocked or attenuated TGF-β effects. Actin and total FoxO3a used as loading controls. b–d. Bar graph quantifications show increased phosphorylation of FoxO3a (P-FoxO3a) (b), mTOR (P-mTOR) (c) and GSK3β (P-GSK3β) (d) in MMC treated with TGF-β, miR-192 and miR-216a/217 mimics and attenuation by the miR-192 or miR-216a/217 inhibitors (n=3). e, Cellular localization of FoxO3a in MMC. Left panels in each treatment show FoxO3a-GFP cellular localization. Right panels show Hoechst staining (nuclear)4. Serum depletion induced nuclear localization of FoxO3a and TGF-β reversed this localization to the cytoplasm. miR-192 and miR-216a/217 mimics induced cytoplasmic translocation of FoxO3a similar to TGF-β. Conversely, inhibitors of these miRs relocated FoxO3a into the nucleus in TGF-β– treated cells. Scale bar, 20 μm. f, Cells with cytoplasmic or nuclear localization were counted in three fields and the results depicted in the bar graph as percentage of cells with cytoplasmic fluorescence relative to total (n=3). FoxO3a was localized in the cytoplasm in 96% of cells under normal culture conditions (Serum +). However, after serum depletion (SD), 96% of cells had nuclear FoxO3a (4% in cytoplasm). FoxO3a translocated back to the cytoplasm upon treatment with miR-192 (76%) or miR- 216a/217 (90%) mimic, or TGF-β (95%). miR-192 or miR216a/217 inhibitor significantly attenuated cytoplasmic FoxO3a to 71% or 64% respectively in TGF-β– treated cells. Bar graph data expressed as mean and s.e.m. Full or larger scan blots of a are shown in Supplementary Fig. S7.
To test whether this miR circuit functionally inhibits FoxO3a transcriptional activity, MMC were transfected with a luciferase reporter containing Forkhead-response elements (pFRE-luc)4 along with the miRs. miR-192 and miR-216a/217 inhibited Luc activity of this reporter similar to TGF-β (Fig. 5a). Inhibitors of these miRs could override the reduction in Luc activity by TGF-β (Fig. 5b). By downregulating Pten and activating Akt, this miR circuit can inhibit FoxO3a function. The expression of the pro-apoptotic gene Bim, a FoxO3a target, was inhibited by miR-192 and miR-216a/217 as well as TGF-β while inhibitors of these miRs reversed the effects of TGF-β (Fig. 5c). Moreover, miR-192, miR-216a/217 and TGF-β prevented serum-depletion-induced MMC apoptosis, while the TGF-β effect was reversed by miR inhibitors (Fig. 5d). Therefore, similar to TGF-β, these miRs promote MMC survival by downregulating Bim via Akt activation (due to Pten reduction) and subsequent FoxO3a phosphorylation. The expression of the antioxidant gene manganese superoxide dismutase (MnSOD), another FoxO3a target4, was also inhibited by TGF-β, miR-192 or miR-216a/217 mimics, whereas miR inhibitors restored its expression (Supplementary Fig. S5a), suggesting that this miR circuit may also upregulate oxidant stress in MMC via FoxO3a phosphorylation.
Figure 5. Effects of miR-192 and miR-216a/217 on FoxO3a activity, MMC apoptosis and hypertrophy.
a, FoxO3a function was monitored by pFRE-luc. Luc activity was decreased by TGF-β. miR-192 and miR-216a/217 mimics (10nM) decreased reporter activity similar to TGF-β (n=4). b, Inhibitors (10nM) of miR-192 and miR-216a/217 increased basal activity of pFRE-luc and also restored the reporter activity inhibited by TGF-β (n=4). c, In serum-depleted cells, Bim expression levels (qPCRs) were decreased by miR-192 and miR-216a/217 mimics as well as TGF-β. Inhibitors of miR-192 and miR-216a/217 reversed the inhibitory effect of TGF-β on Bim expression (n=3). d, TGF-β, as well as miR-192 and miR-216/217 mimics, reduced serum depletion-induced apoptosis of MMC. Inhibitors of miR-192 and miR-216a/217 reversed the inhibitory effect of TGF-β (n=4). e, Cellular protein levels were calculated as ratio of total protein amount/total cell number. Protein levels (hypertrophy) were increased by TGF-β in serum-depleted cells. miR-192 and miR-216a/217 mimics induced hypertrophy to the same extent as TGF-β. Conversely, inhibitors of miR-192 and miR-216a/217 attenuated TGF-β-induced hypertrophy (n=3). Bar graph data expressed as mean and s.e.m. f, Schematic model of miR-dependent amplifying mechanisms in the pathogenesis of DN. TGF-β induced by diabetic conditions upregulates miR-192 which targets Zeb2. The ensuing decrease in Zeb2 upregulates the expression of RP23 which hosts miR-216a and miR-217, both of which target PTEN. The decrease in PTEN increases PIP3 and Akt activation, leading to the phosphorylation of Akt target proteins and increased MMC survival, oxidant stress, ECM expression and hypertrophy, major features of DN.
Finally, we evaluated MC hypertrophy, a major feature of DN and downstream consequence of Akt activation2, 5, 6, 11, 28. TGF-β significantly increased protein levels (Fig. 5e), confirming that it induces MMC hypertrophy. miR-192 or miR-216a/217 mimics similarly induced hypertrophy in serum-depleted MC, while inhibitors of these miRs attenuated TGF-β effects (Fig. 5e), suggesting that this miR circuit mediates TGF-β-induced MMC hypertrophy. Akt downstream proteins mTOR and GSK3β also regulate protein synthesis and hypertrophy6, 28. Our data with miR mimics and inhibitors on mTOR and GSK3β phosphorylation (Fig. 4a–d), further support a role for these miRs in TGF-β-induced MC hypertrophy. Since FoxO3a is implicated in the regulation of autophagy genes29, these miRs might also promote hypertrophy via reducing autophagy.
Notably, cyclin-dependent kinase inhibitor p21-mediated cell cycle arrest, but not apoptosis, of cancer cells by miR-192 via the p53 pathway was recently reported30, while miR-192 activated the promoter of anti-apoptotic Survivin gene31. Our results show that miR-192 also activates Akt and protects cells from apoptosis (Fig. 5d). Therefore, miR-192 likely induces only cell cycle arrest but not apoptosis. Since p21-mediated cell cycle arrest is related to MC hypertrophy11, miR-192 might also promote glomerular hypertrophy by activating p21 and p53. Further studies are needed to verify these aspects.
In summary, miR-216a and miR-217 are co-expressed with RP23 and downregulate the same target (PTEN), illustrating an efficient way to control target genes under certain disease states (Supplementary Fig. S6). Co-expression of miR-216a and miR-217 in the pancreas (high in acinar but low in islets)24, 32 and opposite expression of PTEN (high in islets but low in acinar)33 support such regulation. Our current results also demonstrate a new mechanism for Akt activation by TGF-β via PTEN downregulation by miR-216a/217, and controlled by upstream miR-192 (Fig. 5f). Akt activation by this miR circuit produced biological effects in MMC including ECM gene expression, cell survival and hypertrophy, all classic features of DN. Since E-box regulators, PTEN and Akt also play key roles in cancer development and insulin signaling12, 13, 23, miR-192, 216a and 217 may also regulate not only kidney disorders, but also cancer and metabolic diseases. Inhibitors of these miRs, their host non-coding RNAs or promoters, could therefore serve as novel therapeutic modalities for such diseases.
METHODS
Cell culture
Primary MMC were isolated and cultured as described3 and used in all experiments. Recombinant human TGF-β1 was from R&D Systems, Inc. (Minneapolis, MN). Cells in 12-well-plate were serum-depleted for 48 hr and then treated with TGF-β for indicated time periods.
Animal studies
All animal studies were approved by Institutional Animal Care and Use Committee. Induction of diabetes with STZ in C57BL/6 mice was carried out as described3. Mice were used seven weeks after the onset of diabetes. db/db mice and genetic control db/+ mice (10 weeks old) were from Jackson Laboratories (Bar Harbor, Maine). Glomeruli were sieved from mouse renal cortical tissues as described3.
Real time quantitative PCR (qPCR)
Real-time qPCRs were performed using SYBR Green PCR Master Mix and 7300 Realtime PCR System (Applied Biosystems, Foster City, CA USA). Real-time qPCRs were also used to detect miRs, as reported3. Sequences of miR-specific primers for cDNA synthesis and reverse primers for miR-216a were: 5′-CATGATCAGCTGGGCCAAGACACAGTTGCCAGCTG-3′ and 5′-TAATCTCAGCTGGCAA-3′. Primers for miR-217 were: 5′-CATGATCAGCTGGGCCAAGAATCCAGTCAGTT-3′ (cDNA synthesis) and 5′-TACTGCATCAGGAACT-3′ (reverse primer). miR expression levels were also confirmed by poly-adenylation of mature miR and cDNA synthesis primed by oligo-dT primer tagged with universal primer sequence (miScript System, Qiagen). cDNA was amplified using miR specific and universal primers. For miR-192, miR-194, miR-216a and miR-217 PCRs, the same reverse primers mentioned above and the primers provided by Qiagen were used as miR-specific primers. 5S RNA or 18S RNA served as internal control (Ambion). PCR primers for Col1a2, Zeb1, Zeb2 and MnSOD were described previously3, 4. Mouse Pten PCR primers were from SuperArray Bioscience Corporation (Frederick MD). RP23 PCR primers were: forward (exon 2) 5′-TACTGAACCATCCCCACATCCC -3′ and reverse (exon 3) 5′-TGGCCCAGCTGTAGTTCTGTGA -3′, reverse (exon 3 and 4 junction) 5′-CTTTCTCCAGCCTCAGAGTG -3′ reverse (exon 4) 5′-ATGCAGGCAGTAATCCTTGTG -3′. Bim PCR primers were: forward 5′-cCGGAGATACGGATTGCACAGG-3′ and reverse 5′-cgcttATACCAGACGGAAGATAAAGcG-3′. Cyclophilin A (CypA) primers were: forward 5′-ATGGTCAACCCCACCGTGT-3′ and reverse 5′-TTCTTGCTGTCTTTGGAACTTTGTC-3′.
Western blot analysis
These were performed as described previously3, 4. Antibodies against PTEN, P-Foxo3a, P-mTOR, P-GSK3β, Cyclophilin A (Cell Signaling, Beverly, MA) andβ-actin (Sigma-Aldorich, St. Louis, MO) were used.
Immunohistochmistry
Immunostaining of renal cortical sections was performed as reported4 using antibodies to PTEN and quantified with Image-Pro Plus 5.1 software (Media Cybernetics, Inc. Bethesda, MD).
Cloning of the promoter regions of RP23-298H6.1-001
The promoter regions of RP23 were cloned by PCR using primers, 5′-AAGGagatctTCACTTCATGAACAAACTGCCTT-3′ and 5′-CTTGaagcttCATCACTGGAATGTTATTAAGTCAAG-3′ for the proximal −0.5 kb fragment, 5′-CTActcgaGGCTACCCTGCATCCTTTTCTAG-3′ and 5′-CTTGaagcttCATCACTGGAATGTTATTAAGTCAAG-3′ for the proximal −2.7 kb fragment, 5′-CCAAGCTAGCCAGATCATAATAGTGAGGTTTGG-3′ and 5′-CTTGaagcttCATCACTGGAATGTTATTAAGTCAAG-3′ for the proximal −3.5 kb fragment, 5′-CCCAGCTAGCCAGATCAAAAGCAGCTTGGG-3′ and 5′-CTTGaagcttCATCACTGGAATGTTATTAAGTCAAG-3′ for the proximal −4.8 kb fragment, 5′-CCAAGCTAGCCAGATCATAATAGTGAGGTTTGG-3′ and 5′-CACTCGAGTCTGATTGGAGTTACCAGTG-3′ for the −3.5–2.7kb fragment, and 5′-CCCAGCTAGCCAGATCAAAAGCAGCTTGGG-3′ and 5′-GCACTCGAGTCTGTGTGCAGGCTAAGTGC-3′ for the −4.8–3.5 kb fragment. The entire −4.8 k, −3.5 k, −2.7 k and proximal regions were cloned into pGL4. −3.5–2.7 kb and −4.8–3.5 kb fragments were cloned into the pGL3-promoter (Promega). Reporter with 2x E-boxes was made by digestion of HpaI site. 1xE-box was generated by PCR using a primer, 5′-TGGGAGATCTTGGCACGTGCCTTCAATCCCA-3′ (E-box underlined) and −2.7 k reverse primer,and cloned into the pGL3 promoter. Mutant (Mut) E-box construct was generated by PCR using primers, 5′-TGGCTAGCGCCTTCAATCCCA-3′ (mutant E-box is underlined) and −2.7 k reverse primer, and cloned into the pGL3P.
Plasmids and 3′-UTR reporters
Full-length 3′UTR of mouse Pten was amplified using primers, 5′-CATGCTCGAGCTTATCAAGAGGGATAAAATACCATG-3′ and 5′-CATGCGGCCGCCACAGGAAGGTATTTCTAGTCTTATG-3′. The PCR fragment was digested with Xho I and Not I and cloned into psiCheck2 (Promega). The shorter 1.4k fragment of Pten 3′UTR (which includes the miR-216a and miR-217 sites) was amplified using primers, 5′-CATGCTCGAGCTTATCAAGAGGGATAAAATACCATG-3′ and 5′-GTTGCGGCCGCTTTACTGCAACTCTAATTTCTTTTTTAAC-3′. miR-216a site mutant in the 1.4k construct was made by Quick Change site directed mutagenesis (Stratagene) using primers, 5′-TTCACAGGAAATTTCAACTCCGAATTCAACAGTAA-3′ and 5′-TTACTGTTGAATTCGGAGTTGAAATTTCCTGTGAA-3′. Bases substituted are underlined. miR-217 site mutant was amplified using primers, 5′-CATGCTCGAGCTTATCAAGAGGGATAAAATACCATG-3′ and 5′-GTTGCGGCCGCTTGAGATCTACTCTAATTTCTTTTTTAAC-3′. Bases substituted are underlined. Sequences of all the inserts were confirmed by sequence analysis. For Pten 3′UTR minimum target site reporters, NotI site-tagged potential target sequences in the 3′UTR of mouse Pten (for miR-216a site, sense, 5′-GGCCTTCACAGGAAATTTCAACTTGAGATTC-3′; antisense, 5′-GGCCGAATCTCAAGTTGAAATTTCCTGTGAA-3′, for miR-217 site, sense, 5′-GGCCAAAAGAAATTAGAGTTGCAGTAA-3′; antisense, 5′-ggccTTACTGCAACTCTAATTTCTTTT-3′) were cloned into NotI site of psiCheck2 (Promega). Plasmid containing a sense Pten-3′UTR sequence was used as reporter (psiCheck2mPten-3′UTR-S) while the plasmid containing the 3′UTR sequence in the anti-sense orientation was used as negative control (psiCheck2mPten-3′UTR-AS). The plasmid expressing FoxO3aGFP and the luciferase reporter plasmid with pFRE-luc were described previously4.
Luciferase assays
MMC were transfected with Luc reporter plasmids and/or small RNAs using XtremeGENE (Roche, Indianapolis, IN USA) in 24-well-plates and treated with TGF-β as reported3. After 24hr, Luc activity was measured as described3. For miR and shRNA experiments, cells were treated for 48hr. MC were also transfected with plasmids and/or small RNAs using Nucleofector (Amaxa Biosystems, Gaithersburg, MD). Basic Nucleofector Kit for Primary Smooth Muscle Cells (Amaxa) was used. Five programs (A33, D33, P13, P24 and U25) were tested to examine the transfection efficiency of MC with pmaxGFP (Amaxa). Program U25 resulted in highest efficiency (~60%) and was used in subsequent experiments (Supplementary Figure S5b).
miR mimic and inhibitor Oligonucleotides (Oligos)
miR mimics, negative control #1 (NC) mimics, miR inhibitors, and negative control #1 inhibitors were obtained from Dharmacon (Lafayette, CO). Sequences of mimics and inhibitors are based on Sanger miRBase (http://microrna.sanger.ac.uk). LNA-modified antimiRs were obtained from Exiqon (Woburn, MA) and IDT (Integrated DNA Technologies, Inc. Coralville, IA). LNA-antimiR-192: 21mer, 5′-gGctGtcAatTcaTagGtcAg-3′ (uppercase: LNA; lowercase: DNA, full phosphorothioate) or control LNA-antimiR-239b; 21mer, 5′-cAgtActTttGtgTagTacAa-3′ were subcutaneously injected (2.5 mg/kg) into normal C57BL/6 mice and renal cortical tissues were harvested 6hr later.
in situ hybridization to detect LNA-antimiR-192 accumulation in vivo
in situ hybridization was performed as described27. Briefly, frozen kidney cortex sections from LNA-antimiR-192-injected and saline control mice (n=3/group) were fixed in paraformaldehyde (3%, 10 min, room temperature) followed by rinsing in PBS. Slides were pre-hybridized (48°C, 30 min) in hybridization solution (50% formamide, 5× SSC, 500 μg/ml yeast tRNA, 1× Denhardt’s solution), and then hybridized (48°C, 5 min) with Texas Red-labeled LNA-modified oligo probe complementary to LNA-antimiR-192 (21mer, 5′-TexasRed-ctgAccTatGaaTtgAcaGcc-3′ (Exiqon) followed by post-hybridization wash (3 × 10 min, 55°C) in 0.1× SSC. Slides were then stained with DAPI, mounted and analyzed with fluorescence microscope.
Zeb2 knockdown siRNA
To knockdown Zeb2 in MMC, ON-TARGETplus SMART pool siRNA (Dharmacon) was transfected using Amaxa Nucleofector. Target sequences are GAGCAGACAGGCUUACUUA, GUAAAUGGCCGAAUGAGAA, GCGACACGGCCAUUAUUUA and GCAGGUAACCGCAAGUUCA. ON-TARGETplus Non-targeting Pool (Dharmacon) was used as control.
Cell number and cellular protein levels
To calculate cellular protein content per cell, MMC were trypsinized and counted using a Coulter Counter with 100μm aperture (Beckman Coulter, Fullerton, CA). Cells were lysed, and total protein content determined using Bio-Rad Protein Assays (Bio-Rad, Hercules, CA).
Apoptosis assay
Apoptosis was evaluated with the ApopTag Fluorecein kit from Chemicon/Millipore. Slides were counterstained with DAPI (Sigma), washed and embedded in Vectashield (Vector Labs). Quantitative evaluations were performed with laser scanning cytometry34. Briefly, stained slides were scanned through 40x objective at 0.5μm step using iCys laser scanning cytometer (Compucyte, Cambridge, MA. U.S.A.) and iCys 3.3.2 software. Nuclei were contoured using the 405nm laser based on the DAPI fluorescence. TUNEL positive cells were recorded using the 488nm laser.
Data Analyses
Results are expressed as means and standard error (s.e.m.) of multiple experiments. Paired Student′s t-tests (two-tailed) were used to compare two groups. Statistical significance was detected at the 0.05 level.
Supplementary Material
Acknowledgments
We are grateful to on-line data bases, miRBase (http://microrna.sanger.ac.uk), TargetScan (http://www.targetscan.org), microRNA.org (http://www.microrna.org/microrna), NCBI (http://www.ncbi.nlm.nih.gov), Blat (http://genome.ucsc.edu/cgi-bin/hgBlat) and Ensembl (http://www.ensembl.org) for target prediction and analysis of genomic structure of human and mouse miRs.
This work was supported by grants from the National Institutes of Health (NIH-NIDDK) and the Juvenile Diabetes Research Foundation (to RN). We are grateful to Dr. Mickey C.-T. Hu for generous gift of plasmids (pFRE-luc and FoxO3aGFP), to Ms. Lin Wang and Ms. Jerlisa Arizala for technical assistance and to Ms. Sylvia DaCosta for editing the manuscript.
Footnotes
Author contributions
M.K. and R.N. designed research; M.K., S.P., M.W., H.Y., L.L., I.N., A.G., and I.T. performed research; M.K., M.W., Y.N,, H.S., and J.J.R. contributed new reagents/analytic tools; M.K., S.P. and I.T analyzed data; and M.K. and R.N. wrote the paper.
References
- 1.Yamamoto T, Nakamura T, Noble NA, Ruoslahti E, Border WA. Expression of transforming growth factor beta is elevated in human and experimental diabetic nephropathy. Proc Natl Acad Sci U S A. 1993;90:1814–1818. doi: 10.1073/pnas.90.5.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma K, Ziyadeh FN. Hyperglycemia and diabetic kidney disease. The case for transforming growth factor-beta as a key mediator. Diabetes. 1995;44:1139–1146. doi: 10.2337/diab.44.10.1139. [DOI] [PubMed] [Google Scholar]
- 3.Kato M, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato M, et al. Role of the Akt/FoxO3a pathway in TGF-beta1-mediated mesangial cell dysfunction: a novel mechanism related to diabetic kidney disease. J Am Soc Nephrol. 2006;17:3325–3335. doi: 10.1681/ASN.2006070754. [DOI] [PubMed] [Google Scholar]
- 5.Mahimainathan L, Das F, Venkatesan B, Choudhury GG. Mesangial cell hypertrophy by high glucose is mediated by downregulation of the tumor suppressor PTEN. Diabetes. 2006;55:2115–2125. doi: 10.2337/db05-1326. [DOI] [PubMed] [Google Scholar]
- 6.Nagai K, et al. Gas6 induces Akt/mTOR-mediated mesangial hypertrophy in diabetic nephropathy. Kidney Int. 2005;68:552–561. doi: 10.1111/j.1523-1755.2005.00433.x. [DOI] [PubMed] [Google Scholar]
- 7.Xin X, Chen S, Khan ZA, Chakrabarti S. Akt activation and augmented fibronectin production in hyperhexosemia. Am J Physiol Endocrinol Metab. 2007;293:E1036–1044. doi: 10.1152/ajpendo.00271.2007. [DOI] [PubMed] [Google Scholar]
- 8.Yi JY, Shin I, Arteaga CL. Type I transforming growth factor beta receptor binds to and activates phosphatidylinositol 3-kinase. J Biol Chem. 2005;280:10870–10876. doi: 10.1074/jbc.M413223200. [DOI] [PubMed] [Google Scholar]
- 9.Runyan CE, Schnaper HW, Poncelet AC. The phosphatidylinositol 3-kinase/Akt pathway enhances Smad3-stimulated mesangial cell collagen I expression in response to transforming growth factor-beta1. J Biol Chem. 2004;279:2632–2639. doi: 10.1074/jbc.M310412200. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh Choudhury G, Abboud HE. Tyrosine phosphorylation-dependent PI 3 kinase/Akt signal transduction regulates TGFbeta-induced fibronectin expression in mesangial cells. Cell Signal. 2004;16:31–41. doi: 10.1016/s0898-6568(03)00094-9. [DOI] [PubMed] [Google Scholar]
- 11.Wolf G, Ziyadeh FN. Molecular mechanisms of diabetic renal hypertrophy. Kidney Int. 1999;56:393–405. doi: 10.1046/j.1523-1755.1999.00590.x. [DOI] [PubMed] [Google Scholar]
- 12.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 13.Lazar DF, Saltiel AR. Lipid phosphatases as drug discovery targets for type 2 diabetes. Nat Rev Drug Discov. 2006;5:333–342. doi: 10.1038/nrd2007. [DOI] [PubMed] [Google Scholar]
- 14.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, et al. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 2004;32:e188. doi: 10.1093/nar/gnh186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res. 2008;18:404–411. doi: 10.1101/gr.6587008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. Faseb J. 2008;22:4126–4135. doi: 10.1096/fj.08-112326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua X, Liu X, Ansari DO, Lodish HF. Synergistic cooperation of TFE3 and smad proteins in TGF-beta-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev. 1998;12:3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekido R, et al. The delta-crystallin enhancer-binding protein delta EF1 is a repressor of E2-box-mediated gene activation. Mol Cell Biol. 1994;14:5692–5700. doi: 10.1128/mcb.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comijn J, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa Y, et al. TFE3 transcriptionally activates hepatic IRS-2, participates in insulin signaling and ameliorates diabetes. Nat Med. 2006;12:107–113. doi: 10.1038/nm1334. [DOI] [PubMed] [Google Scholar]
- 24.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 25.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 26.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 28.Glass DJ. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol. 2003;5:87–90. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- 29.Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braun CJ, et al. p53-Responsive MicroRNAs 192 and 215 Are Capable of Inducing Cell Cycle Arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gou D, Zhang H, Baviskar PS, Liu L. Primer extension-based method for the generation of a siRNA/miRNA expression vector. Physiol Genomics. 2007;31:554–562. doi: 10.1152/physiolgenomics.00005.2007. [DOI] [PubMed] [Google Scholar]
- 32.Bravo-Egana V, et al. Quantitative differential expression analysis reveals miR-7 as major islet microRNA. Biochem Biophys Res Commun. 2008;366:922–926. doi: 10.1016/j.bbrc.2007.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perren A, et al. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am J Pathol. 2000;157:1097–1103. doi: 10.1016/S0002-9440(10)64624-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omori K, et al. Improvement of human islet cryopreservation by a p38 MAPKinhibitor. Am J Transplant. 2007;7:1224–1232. doi: 10.1111/j.1600-6143.2007.01741.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.