Abstract
Distinct protein assemblies are nucleated at telomeric DNA to both guard the ends from damage and lengthen the DNA following replication. In yeast, Cdc13 recruits either Stn1/Ten1 to form a protective cap or the telomerase holoenzyme to extend the DNA. We have established an in vitro yeast telomere system in which Stn1/Ten1-unextendable or telomerase-extendable states can be observed. Both assemblies are Cdc13-dependent, as the Cdc13 C-terminal region supports Stn1/Ten1 interactions and the N-terminal region contains a telomerase activation function. Notably, the yeast Hsp90 chaperone Hsp82 mediates the switch between the telomere capping and extending structures by modulating the DNA binding activity of Cdc13. Taken together, our data demonstrate that the Hsp82 chaperone facilitates telomere DNA maintenance by promoting transitions between different operative complexes and by reducing the potential for binding events that would otherwise block the assembly of downstream structures.
Introduction
Telomeres are heterogeneous nucleoprotein assemblies localized at eukaryotic chromosome termini that serve both to protect the ends from damage and to extend the DNA length following replication. To accomplish these activities various proteins are nucleated at telomeric DNA to form distinct structures, including capping and extending complexes1,2. While the exact composition of each assembly is not entirely understood, genetic analysis in the budding yeast Saccharomyces cerevisiae has revealed many critical components, including EST2, STN1, TEN1 and CDC13. EST2 encodes the telomerase reverse transcriptase subunit that is responsible for extending telomeric DNA and the Stn1 and Ten1 proteins function to cap the chromosome ends. Cdc13 promotes both DNA extension and protection—in vivo studies support a model in which Cdc13 recruits and stabilizes both Est2-extending and Stn1/Ten1-capping structures3–7. Remarkably, the telomere protein system functions effectively in vivo despite a common binding specificity for single-stranded telomeric DNA by Cdc13, Stn1/Ten1 and telomerase4,8,9. Why the proteins do not interfere with each other, how transitions between the different telomere structures are mediated and how Cdc13 serves in these seemingly opposed telomere activities (i.e., capping vs. extending) has been unclear.
In addition to Cdc13, the yeast Hsp90 molecular chaperones Hsp82 and Hsc82 affect both capping and extending components. Hsp82 and Hsc82 are high-copy suppressors for the capping mutants stn1-1 and cdc13-1 yet in a wild type background elevated chaperone levels result in shortened telomeric DNA10. Neither the mechanism for the suppression nor the reduced telomere DNA length upon Hsp82-overexpression in wild type cells is understood. In contrast, the means by which Hsp82 modulates telomerase has been revealed. Human Hsp90 was the first telomerase cofactor discovered to directly influence DNA extension activity in vitro11; work on the yeast system showed that Hsp82 promotes both telomerase DNA binding and nucleotide affinity during DNA extension12. Hence, Hsp90 chaperones affect both capping and extending telomere components though the mechanistic contributions to the capping factors had yet to be revealed.
We sought to test directly whether the Hsp82 molecular chaperone might function with Cdc13 alone or in association with Stn1 and Ten1, by [a] establishing the functional effect of Cdc13 alone and in conjunction with Stn1/Ten1 in vitro, [b] demonstrating whether Hsp82 affects the different protein assemblies, and [c] determining how a switch occurs between different operative telomere-protein states.
Results
Cdc13 stimulates telomerase DNA extension activity
As a first step we determined the Cdc13 effect on telomerase DNA extension activity in vitro. We anticipated that Cdc13 would inhibit telomerase function by competing for the DNA substrate, as observed for the human Cdc13 ortholog Pot113. To discriminate Cdc13 DNA binding-dependent effects we used DNA substrates with either 23- or 7-nucleotide 3′-overhangs in the telomerase extension assays. Prior biochemical studies indicated that Cdc13 requires a minimum of 11 single-stranded nucleotides to bind DNA14. In accordance, we found that our Cdc13 protein did not bind to DNA with a 7-base 3′-overhang but did bind to a 22-nucleotide overhang substrate (Supplementary Fig. 1). Unexpectedly, Cdc13 activated telomerase DNA extension activity independent of 3′-overhang DNA length (Fig. 1a). The DNA products below the +1 position for the 23-base substrate are likely produced by a previously described telomerase-associated endonuclease activity15.
Figure 1.
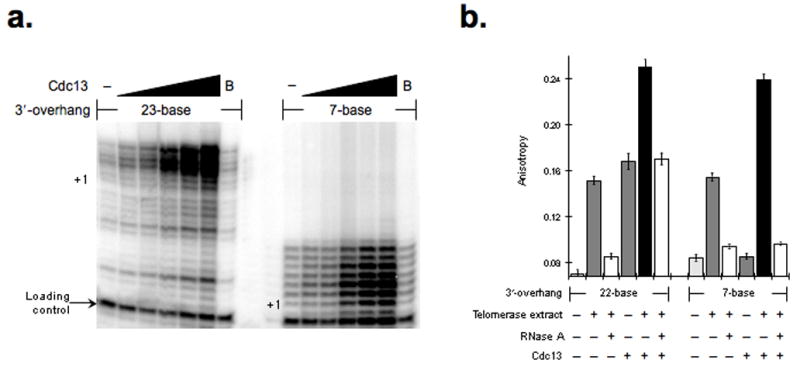
Cdc13 stimulates telomerase DNA extension activity independent of single-stranded 3′-overhang DNA length. (a) The Cdc13 effect on telomerase-mediated DNA extension was determined using DNA substrates with either 23- or 7-nucleotide 3′-overhangs and a Cdc13 protein titration (50, 100, 250, 500 and 1000 nM), as marked. To control for possible non-specific protein affects the impact of BSA (B) (1000 nM) addition was tested. All extension reactions were supplemented with a loading control primer (arrow) prior to precipitation and electrophoretic resolution and the +1 position for each DNA substrates is marked. (b) Cdc13 can tether to DNA-bound telomerase. Fluorescence anisotropy with a fluorescein-labeled telomeric oligonucleotide was used to detect DNA association. The binding activities of Cdc13 (250 nM) or telomerase alone on 22- and 7-base DNA substrates were examined and the ability of Cdc13 to join DNA-bound telomerase was determined by adding purified Cdc13 (250 nM) to telomerase-DNA complexes using either the 22- and 7-base DNA substrates, as indicated. To demonstrate telomerase-dependent DNA binding RNaseA treatment was included19. The quantified data represent average values (mean +/− s.d.) from 5 independent assays.
Cdc13 enhanced the extension of the 23-base substrate to a greater extent relative to the 7-nucleotide 3′-overhang DNA (~18- vs. ~9-fold) suggesting that DNA binding by Cdc13 positively contributed to telomerase activity. Nonetheless, the Cdc13 activation with a 7-base substrate suggested that Cdc13 could affect telomerase independent of direct DNA binding. Notably, we found that Cdc13 assembled with telomerase or potentially some other RNase-sensitive component of the extract on 7- or 22-base 3′-overhang substrates but that the 7-base DNA structure was dependent upon telomerase-DNA binding activity, which suggests that Cdc13 tethers to the DNA-bound enzyme (Fig. 1b). While stimulation by Cdc13 contrasts with the inhibitory effect of Pot1, it should be noted that Pot1 activates telomerase in conjunction with the telomeric protein Tpp116. Hence, Cdc13 apparently functions in a manner comparable to the combined effects of Pot1 and Tpp1.
The Cdc13 amino-terminus has a telomerase activation function
To better understand how Cdc13 regulates telomerase activity we produced and characterized a series of Cdc13 protein derivatives that included full-length (FL), Cdc13-2 point mutant (2), DNA binding domain (DBD), amino-terminus through the DBD (N) and DBD through the carboxyl-terminus (C) (Fig. 2); all derivatives displayed high-affinity DNA binding activity (data not shown). The FL, Cdc13-2 and the N-fragment activated telomerase DNA extension activity on either DNA substrate. Hence, in addition to its suggested telomere recruitment function6, the Cdc13 N-terminus has a telomerase activation surface. In contrast, the C or DBD derivatives inhibited extension of the 23-base substrate but did not affect basal extension of the 7-base 3′-overhang DNA (Fig. 2).
Figure 2.
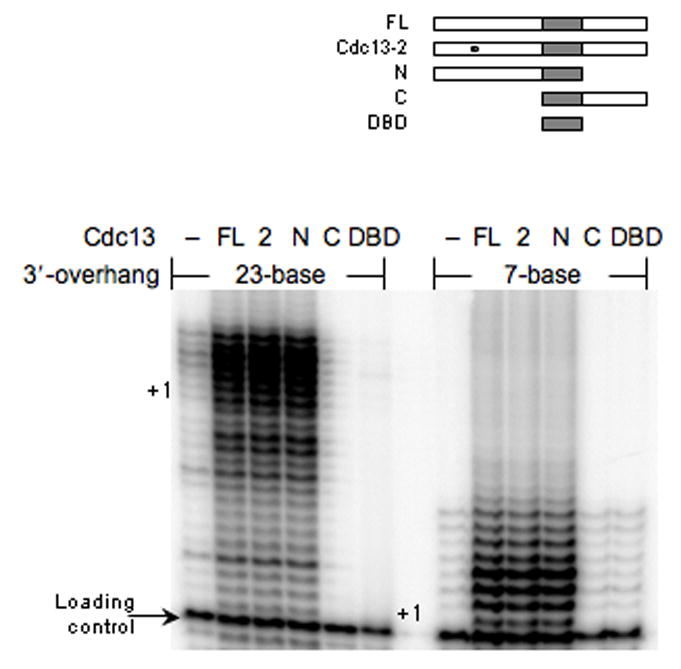
The amino-terminal domain of Cdc13 has a telomerase-activation function. The effect of full-length (FL) Cdc13, Cdc13-2 (2), amino-terminus (N), carboxyl-terminus (C) or DNA binding domain (DBD) proteins (250 nM) on telomerase DNA extension using 7- or 23-base 3′-overhang DNA substrates was determined. All extension reactions were supplemented with a loading control primer (arrow) prior to precipitation and electrophoretic resolution and the +1 position for each DNA substrates is marked.
The ability of Cdc13-2 to stimulate telomerase indicates that the up-regulation does not involve interactions with the holoenzyme component Est1, as the Cdc13-2 mutation disrupts Cdc13-Est1 interactions, and likely is distinct from the Cdc13 telomere recruitment function7. We believe that the observed telomerase activation potential likely occurs downstream from nucleation of telomerase to a telomere in vivo. The positive effect of the N-domain on telomerase extension activity might account, in part, for the increased telomere DNA length in cdc13-5 yeast6. Assuming telomerase remains telomere engaged following a single Cdc13-mediated recruitment event, then an additional Cdc13 function would be required to not only account for the hyper elongation of the DNA in the cdc13-5 background but to also counter the DNA attrition that would be anticipated by the loss of the carboxyl-terminal Stn1 interaction site18.
In contrast to activation, telomerase inhibition by the C and DBD fragments was dependent upon the length of the single-stranded DNA, which suggests the decline in activity likely results from competitive binding to the DNA substrate. If these suppositions are correct then proper telomere regulation in vivo would require a mechanism to prevent Cdc13 from competing with telomerase for DNA binding without interfering with Cdc13-regulatory capabilities.
Cdc13, Stn1 and Ten1 cooperate to form an unextendable telomere DNA complex
In addition to Cdc13, Stn1 and Ten1 serve in telomere capping in vivo1. To determine whether FL-Cdc13 can function with Stn1 and Ten1 we purified the proteins (Supplementary Fig. 2a) and tested the effects of the proteins on telomerase activity. In the presence of Cdc13 we observed a striking reduction in DNA extension with increasing Stn1/Ten1 levels using the 23-base 3′-overhang DNA but not the 7-base substrate (Fig. 3a and Supplementary Fig. 2b). In the absence of Cdc13, Stn1/Ten1 had no apparent effect on DNA extension (Fig. 3a). Importantly, the point mutant Cdc13-2, which does not interact with Stn1 in vivo6, fails to repress telomerase activity with Stn1/Ten1 in vitro (Fig. 3b) indicating that the Stn1/Ten1 interaction is specific for wild type Cdc13. Our in vitro data support recent in vivo work suggesting Cdc13 and Stn1 function to block telomerase action at a telomere17.
Figure 3.

Stn1 and Ten1 cooperate with Cdc13 to form an unextendable telomere protein-DNA complex in vitro. (a) The effect of an Stn1/Ten1 protein titration (50, 100, 250, 500 and 1000 nM) on telomerase DNA extension in the presence or absence of Cdc13 (250 nM) was determined using 23- or 7-base 3′-overhang DNAs. For comparison, the activity of unsupplemented and Cdc13 supplemented telomerase extract is shown. (b) Cdc13-2 does not form an unextendable DNA complex with Stn1/Ten1. Telomerase DNA extension reactions were performed using a 23-nucleotide 3′-overhang substrate and the reactions were supplemented with Cdc13 (FL) (250 nM) or Cdc13-2 (2) (250 nM) with or without Stn1/Ten1 (250 nM), as marked. All extension reactions were supplemented with a loading control primer (arrow) prior to precipitation and electrophoretic resolution and the +1 position for each DNA substrates is indicated. (c) The carboxyl-terminus of Cdc13 harbors an Stn1/Ten1 interaction surface. Fluorescence anisotropy with a fluorescein-labeled 15-base 3′-overhang telomeric oligonucleotide was used to detect DNA association; the 15-base DNA section provides a single Cdc13 binding site. The anisotropy was measured for each Cdc13 derivative (100 nM) alone and in the presence of Stn1/Ten1 (100 nM). The quantified data represent average values (mean +/− s.d.) from 5 independent assays.
The negative effect of the DBD and C-domain fragments on telomerase activity complicated our ability to assess whether these proteins can form an Stn1/Ten1-cap structure since DNA extension activity is suppressed in the absence of Supplementary Stn1/Ten1. To determine which Cdc13 domain assembles with Stn1/Ten1 on DNA we performed fluorescence anisotropy DNA binding experiments. We found that both FL and the C-domain associated with Stn1/Ten1 on DNA in vitro (Fig. 3c), which agrees with prior genetic data demonstrating that the C-terminus can interact with Stn1 to protect telomere ends in vivo18. In contrast, the N-terminus and Cdc13-2 fail to assemble with Stn1/Ten1 on DNA in vitro (Fig. 3c). Comparable results were observed using the electro-mobility shift assay (Supplementary Fig. 2c). Hence, Cdc13 has three distinct domains—an amino-terminal telomerase activation region, a central DNA binding domain and a carboxyl-terminal Stn1/Ten1 interaction site.
Hsp82 converts the telomere unextendable state to an extendable form
The ability of the DBD and C-domain to interfere with DNA extension indicates that Cdc13 DNA binding activity is sufficient to inhibit telomerase, yet full-length Cdc13 only activates, unless Stn1/Ten1 is also supplemented. To account for these results we believed that a telomerase cofactor was present in the reactions, preventing Cdc13 from interfering with telomerase but still permitting DNA-bound telomerase to associate with Cdc13. Given the established Hsp82 roles with capping and extending telomere components, we tested whether Hsp82 might affect Cdc13. As an initial test Hsp82 was titrated into DNA extension reactions containing a Cdc13/Stn1/Ten1-capped telomeric DNA substrate (Fig. 4a). In the absence of Supplementary Hsp82, Cdc13/Stn1/Ten1 suppressed the DNA extension activity below basal levels (Fig. 4a lanes 1 & 3). However, in an Hsp82-dependent manner the extension activity increased ~15-fold, which indicates that Hsp82 is sufficient to switch the Cdc13/Stn1/Ten1-capping structure into a Cdc13/telomerase-extending complex (Fig. 4a).
Figure 4.
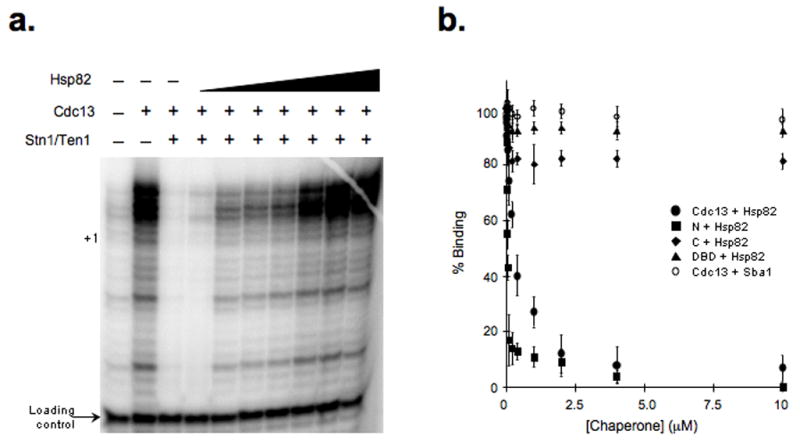
Hsp82 promotes conversion of the Cdc13-capping structure into a Cdc13-extending complex by dissociating Cdc13 from DNA. (a) The ability of Hsp82 to convert an unextendable Cdc13/Stn1/Ten1-capped DNA complex into a telomerase accessible DNA structure was determined using a 23-base 3′-overhang DNA substrate, telomerase extract and an Hsp82 protein titration (0.25, 0.5, 1.0, 2.5, 5.0, 10.0 and 20.0 μM). The cellular Hsp82 concentration is 17 μM34. (b) Hsp82 dissociates amino-terminal Cdc13 protein derivates from DNA. Cdc13 DNA binding was monitored by fluorescence anisotropy. The impact of Hsp82 on the binding activities of FL, C, N or DBD Cdc13 protein derivatives (100 nM) was determined along with the effect of Sba1 on FL, as indicated.
To investigate how Hsp82 affects the protein-DNA complexes, we determined the impact of Hsp82 on the DNA binding activities of the Cdc13 derivatives. We found that Hsp82 dissociated FL Cdc13 and the N-domain from DNA but the chaperone did not extensively affect DNA binding by the DBD or C-terminal fragment (Fig. 4b). Hence, Hsp82 modulates Cdc13 through interactions with the N-terminus. In addition to direct DNA binding, we found that Cdc13 and the DBD fragment could protect single-stranded DNA from nuclease digestion (Supplementary Fig. 3). However, in the presence of Hsp82 the full-length Cdc13 no longer protected the DNA. Based on the anisotropy data, Hsp82 displayed a higher affinity for the N-fragment (Kd ~35 nM) relative to FL (Kd ~325 nM) suggesting contacts between the amino- and carboxyl-domains of Cdc13 impact modulation by Hsp82. In contrast to Hsp82, Sba1 had no apparent effect on Cdc13 DNA binding despite the ability of Sba1 to dissociate telomerase from DNA (Fig. 4b)19.
Notably, we found that Hsp82 actively prevented Cdc13 DNA binding, as Cdc13 binding activity was restored upon Hsp82 inhibition (Fig. 5a). Furthermore, treatment of telomerase extract with an Hsp82 small-molecule inhibitor (radicicol) resulted in Cdc13-mediated suppression of telomerase activity using the 23-base substrate rather than activation (Fig. 5b). Thus, Hsp82 specifically converts the Cdc13-capping structure into a Cdc13/telomerase-extending complex by interfering with DNA binding by Cdc13 but not with its ability to associate with DNA-bound telomerase. In addition, we found that alternate stepwise addition of Hsp82 and Stn1/Ten1 revealed a competitive equilibrium effect on the DNA binding activity in vitro (Fig. 5c). Thus, Hsp82 can dissociate the Cdc13/Stn1/Ten1 bound complex yet the capacity of additional Stn1/Ten1 to counter the Hsp82 dissociation activity provides a mechanism to reestablish a Cdc13 DNA-bound state, which has important implications for Cdc13-mediated DNA structures.
Figure 5.
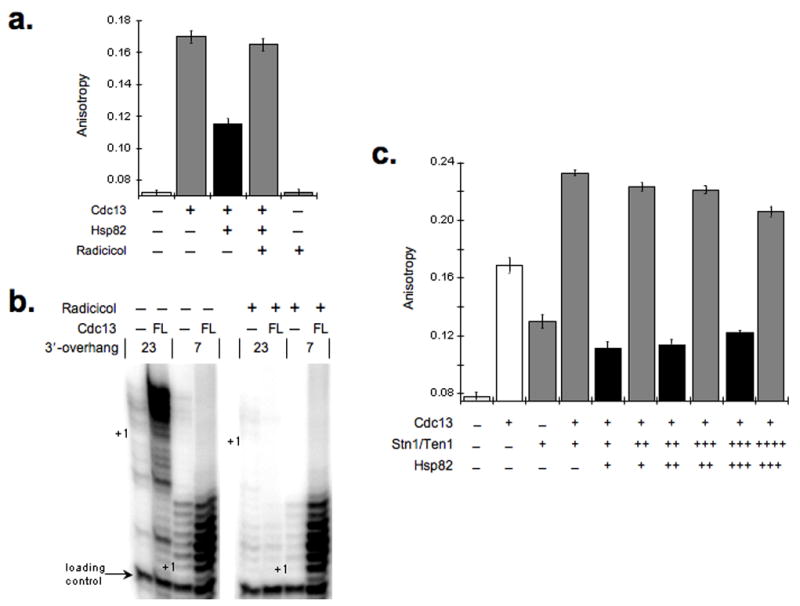
Hsp82-mediated Cdc13-DNA dissociation is an active process. (a) Addition of the Hsp82 small-molecule inhibitor Radicicol reestablishes Cdc13 DNA binding activity. Anisotropy values for a fluorescein-telomeric oligonucleotide upon the consecutive addition of Cdc13 (100 nM) then Hsp82 (1.0 μM) and finally Radicicol (20 μM) is shown. (b) Wild type Cdc13 impedes telomerase activity on a 23-base DNA substrate upon small molecule inhibition of Hsp82. Telomerase DNA extension reactions were performed using 23- or 7-nucleotide 3′-overhang substrates. The reactions were supplemented with Cdc13 (250 nM) alone or with Radicicol (20 μM). As expected, loss of Hsp82 function resulted in a declined telomerase DNA extension activity12. All extension reactions were supplemented with a loading control primer (arrow) prior to precipitation and electrophoretic resolution and the +1 position for each DNA substrates is marked. (c) Hsp82 and Stn1/Ten1 can drive a competitive equilibrium for Cdc13 DNA binding. Aliquots of Stn1/Ten1 (250 nM) and Hsp82 (1 μM) were added sequentially to a reaction mix of fluorescein-telomeric oligonucleotide and Cdc13 (250 nM). The anisotropy values were measured after each addition. The quantified data represent average values (mean +/− s.d.) from 5 independent assays.
Hsp82 modulates Cdc13 telomere occupancy and telomere activities in vivo
Given the impact of Hsp82 on Cdc13 in vitro, one might predict that Hsp82 overexpression would have a marked effect on telomere DNA length in vivo. Yet, our studies and prior work indicate that ectopic Hsp82 overexpression results in an ~50% reduction in telomere DNA length (Supplementary Fig. 4a)10. Though the change in DNA length does correlate with the relative increase in Hsp82 protein amounts (~2-fold; Supplementary Fig. 4a), the modest telomere effect might also occur if Hsp82 is affecting competing telomere functions. For example, an increase in cellular Hsp82 might lower Cdc13-mediated DNA end protection by limiting formation of capping complexes; however, a reduction in Cdc13 DNA-binding would promote telomerase-DNA interactions by elevating telomerase accessibility to the extreme 3′ DNA end.
To test this model we examined the Hsp82 effect on the cell cycle telomere occupancies by Cdc13 and telomerase and the Hsp82 impact on telomere capping and extending activities. We found that telomere association by Cdc13 declined upon Hsp82 overexpression throughout the cell cycle except during late S phase (Fig. 6a). Thus, we observed the occupancy pattern that would be predicted based upon our in vitro data. Cdc13 telomere interactions are reduced during cell cycle phases associated with capping events but are unaffected at late S phase when Cdc13 would activate telomerase by tethering to the DNA-bound enzyme. To determined if the declined Cdc13 telomere occupancy affects DNA end protection we checked whether the relative levels of single-stranded TG1–3 DNA fluctuated with Hsp82 expression using an in-gel native Southern blot assay20,21. As might be anticipated, we observed a TG1–3 signal increase in yeast overexpressing Hsp82 (Supplementary Fig. 4b), which suggests DNA end protection declines with increased Hsp82 protein in vivo.
Figure 6.
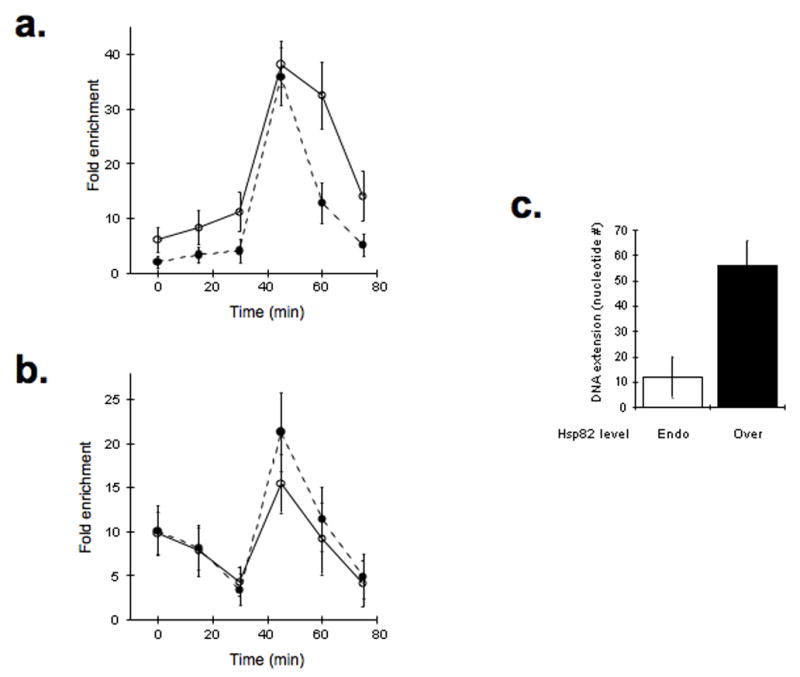
Hsp82 affects telomere events in vivo. Hsp82 affects Cdc13-telomere association in vivo. The cell cycle-dependent telomere occupancy by Cdc13 (a) and the telomerase reverse transcriptase subunit Est2 (b) were determined using the chromatin immunoprecipitation (ChIP) assay in either CDC13-9xMYC or EST2-9xMYC, respectively. Yeast transformants with endogenous (open symbols) or overexpressed (closed symbols) Hsp82 levels were synchronized with α-factor. (c) Telomeric DNA extension is HSP82-dependent. Elongation of inducibly truncated telomeric DNA was monitored in yeast with endogenous (Endo) or overexpressed (Over) Hsp82, as indicated. The quantified data represent average values (mean +/− s.d.) from 5 independent assays. The average number of nucleotides added to the shorten-telomeric DNA was determined using an established protocol22.
In contrast to Cdc13, we found that Est2 telomere interactions did not change appreciably with Hsp82 expression except for a modest but reproducible (albeit statistically insignificant) increase in late S phase (Fig. 6b). Minimally, the data show that additional Hsp82 does not interfere with the ability of Cdc13 to efficiently recruit telomerase to telomeres. We suspect that the mild rise in Est2-telomere association results from an increase in direct DNA binding by telomerase (i.e., Est2-DNA crosslinking efficiency has increased as more Est2 is in direct contact with the DNA). If elevated Hsp82 levels do facilitate telomerase access to the telomeric DNA then there might be an increase in telomere DNA extension rates since telomerase would now be directly engaged with the DNA.
To investigate telomere DNA lengthening activity, we exploited an established, engineered yeast strain that permits controlled, rapid reduction in telomere DNA length and assessment of telomerase DNA extension rates in vivo22. In brief, the left arm terminus of chromosome VII contains an internal telomere tract that is flanked by Flp1-recognition target sites, the tract is specifically removed upon transient Flp1 expression and DNA elongation can be monitored in subsequent cell divisions. To limit the impact of Hsp82 effects on capping events, Hsp82 overexpression was induced in conjunction with Flp1 while the yeast were under α-factor cell cycle arrest. This permitted a focus on the DNA extension levels achieved during the first cell cycle after the Flp1-mediated telomeric DNA reduction. In the presence of elevated Hsp82 protein levels, we found that the extension length was faster (~5-fold) relative to yeast with endogenous Hsp82 levels (Fig. 6c). While there are a number of possibilities for the increased DNA lengthening including direct effects on telomerase activity by Hsp8212, we suggest that the change also involves modulation of Cdc13 DNA binding by Hsp82. In brief, the telomerase-mediated DNA extension increases since Hsp82 inhibits assembly of the unextendable telomere structure (i.e., reduces steric interference by Cdc13) without disrupting positive Cdc13 functions (i.e., telomere recruitment and telomerase activation). Taken together, our data suggest that steady-state telomere DNA length is reduced upon Hsp82 overexpression due to declined capping activities. However, the magnitude of the shortening is limited since telomerase accessibility to the telomeric DNA has increased and therefore telomerase-mediated DNA extension is enhanced, which counters the impact of the declined telomere capping.
Discussion
The presented work provides a potential resolution to the apparent conundrum of how proteins with identical binding specificities coordinately function at a single site. Often DNA binding proteins form long-lived complexes with target DNAs in vitro that would interfere with the assembly of other structures and telomere-protein assemblies are no exception2. For example, a stable Cdc13-telomere association at the extreme 3′ DNA end would block telomerase DNA binding yet Cdc13 binding at the extreme termini likely is required to protect the DNA from degradation in vivo. Our data supports a model in which Hsp82 frees the DNA end of competing binding proteins without interfering with critical regulatory events.
In our model, telomerase recruitment to a telomere initiates through transient contacts between Cdc13 and the holoenzyme (Fig. 7). To permit proper telomerase DNA association Hsp82 displaces proteins bound at the extreme 3′ DNA end including Cdc13. Recent genetic studies highlight the potential importance of maintaining a dynamic telomere environment, as the peak of telomere association by Cdc13, Stn1 and Est1 all occur in S-phase17,23. Presumably, both capping (Stn1) and extending (Est1) proteins are telomere recruited each S-phase since not all telomeres are extended each cell cycle24. Hence, depending upon the DNA length of a particular telomere a molecular choice is made to either permit or preclude telomerase function (i.e., build an Est1-extending or Stn1-capping complex). If Hsp82 maintains Cdc13 in a dynamic DNA binding cycle, then any modifications (e.g., phosphorylation) meant to guide a telomere to a distinct operative phase would be immediately incorporated.
Figure 7.
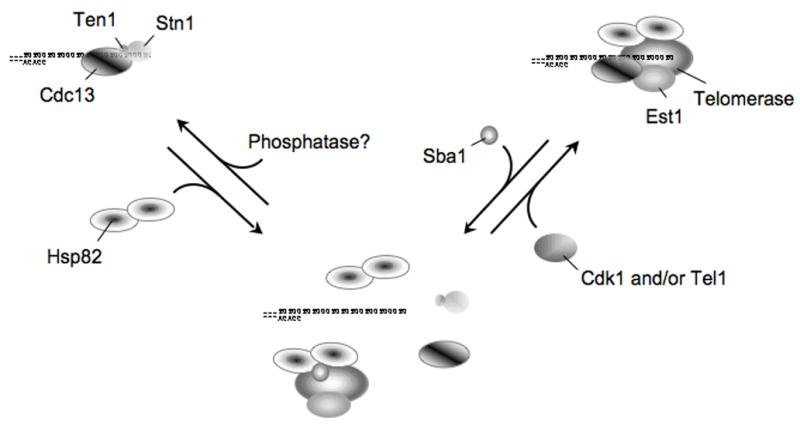
Hsp82 mediates telomere-protein dynamics. Cdc13 recruits select proteins to telomeric DNA including Stn1/Ten1 and telomerase/Est1. As the independent structures form stable and long-lived interactions with telomeric DNA, transitions between the distinct complexes requires molecular chaperone-mediated disassembly2,19. Specific post-translational modifications would then guide the assembly of the requisite structure, which minimally would vary with the phase of the cell cycle. For example, phosphorylation of Cdc13 by Cdk1 and/or Tel1 would promote telomerase nucleation and DNA extension events25,26.
Recent genetic studies suggest post-translational modifications mediated by the Tel1 (ATM homolog) and Cdk1 kinases are critical for proper telomere DNA maintenance25,26. For example, Tel1 is recruited to critically short telomeric DNA tracts, which are preferentially elongated, and Cdc13 has been shown to be a Tel1 target 25,27. In addition, Cdk1 phosphorylates Cdc13 and the modification appears to influence the preference between Stn1- and Est1-containing Cdc13 nucleated telomere complexes26. Hence, depending upon the Cdc13 phosphorylation-state, telomeric DNA would be in a Cdc13-Stn1/Ten1 unextendable state or a Cdc13-Est1/telomerase extendable form. Dynamic Cdc13 action, mediated by Hsp82, would enable the telomere system to rapidly transition between the different structures as needed.
In addition to protein dynamics, our studies reveal a Cdc13 telomerase activation function, which may account for the telomere lengthening phenotypes for several cdc13 mutations5,6. Of note, the Cdc13 telomerase stimulation activity that we observe is in stark contrast to a recent report showing telomerase inhibition by full-length Cdc1328. Currently, the reason for the different effect of full-length Cdc13 on telomerase activity is unclear. We did find that the Cdc13 activation function was lost following a brief (5 min) incubation at 42°C while DNA binding activity was unaffected (data not shown), which suggests that the conformation of the activation surface is labile. Perhaps by expressing full-length Cdc13 in a specialized bacterial strain (i.e., streptomycin-resistant strain), which was critical for obtaining soluble recombinant protein29, the activation surface was provided an opportunity to properly fold. Opposed to the data on full-length Cdc13, both studies did observe telomerase inhibition by the Cdc13 DNA binding domain fragment and the DBD displayed no apparent difference when expressed in either classic or streptomycin-resistant bacterial cells29.
Nevertheless, prior genetic data suggests a role for Cdc13 with telomerase that is downstream of telomere nucleation. For example, the cdc13-4 yeast have short but stable telomeric DNA despite an apparent capacity of Cdc13-4 to recruit telomerase to telomeres and protect the DNA from degradation30. While the effected residue (P235S) occurs within the amino-terminal domain, the mechanism for the telomere defect was not identified. However, the in vivo phenotype is consistent with a post-recruitment function for Cdc1330. Intriguingly, the Cdc13-109 and Cdc13-231 mutants lead to over-elongated telomeric DNA despite a decline in DNA binding activity5. If the sole positive function of Cdc13 was to recruit telomerase to a telomere, then a decline in DNA binding should result in shorter not longer telomeric DNA. Additionally, the hyper-elongation of telomeric DNA in the cdc13-5 yeast is consistent with a post-recruitment role for Cdc13. The Cdc13-5 protein (N-domain) consists of the amino-terminus and DNA binding domain6. In the absence of the Cdc13 carboxyl-terminal Stn1 interaction site the telomeric DNA should be vulnerable to nuclease attack. Yet, the telomeric DNA is not degraded in the cdc13-5 yeast but rather is over-elongated6. We suggest that, in addition to recruiting telomerase, Cdc13-5 activates telomerase to not only hyper-extend the DNA but also to counter any potential DNA degradation. Of note, the stimulatory function is downstream from Cdc13-mediated recruitment since the effect is abrogated when combined with the Cdc13 point mutation cdc13-2 (i.e., cdc13-2,5)6. Taken together, established genetic data support our contention that Cdc13 modulates the DNA-bound enzyme. We suggest that telomerase cofactors, including Cdc13, likely reengage the telomere protein assembly by tethering to the DNA-bound telomerase enzyme.
A chaperone-mediated protein dynamics model has been previously proposed for transcription pathways31. In brief, molecular chaperones promote a dynamic action for transcription factors that is necessary to permit rapid functional recruitment of multiple coactivating complexes to a gene promoter. An important distinction, however, is the impact of the Hsp90 chaperone: Hsp90 promotes DNA binding by transcription factors but facilitates dissociation of Cdc13-DNA complexes. Perhaps the dual role for Hsp82 at telomeres provides an explanation for the difference. By exploiting the telomerase-associated cofactor Hsp82 to both support telomerase function and to clear the telomeric DNA of competing proteins, an elegant means to ensure telomeric DNA extension within the short working period allotted telomerase at the end of S phase is provided.
Traditionally Hsp90 has been viewed as a cytoplasmic molecular chaperone required for the late folding stages of signaling molecules32. However, recent studies including high-throughput screens have identified a broad-range of potential nuclear client proteins2. Given the impact of Hsp90/Hsp82 both on telomeric and transcriptional targets, there appears to be a general cellular role for Hsp90 chaperones in controlling protein-DNA dynamics. In brief, multi-step pathways, including the telomere system, move forward through high affinity interactions between the low abundant proteins unique to that system (e.g., Cdc13, telomerase, Stn1) while proper structure composition (i.e., competitive interactions) along with efficient transitions between the different assemblies are mediated by transient, low affinity interactions with the highly abundant molecular chaperones.
Methods
Protein purification
We expressed the Cdc13 protein derivatives (pET28) and Stn1 (pETDuet1) protein as amino-terminal (His)6 fusions using T7 expression vectors in streptomycin resistant Escherichia coli cells CH184. Protein expression in the CH184 background alleviated solubility problems associated with Cdc13 and Stn1/Ten1. The Ten1 (pETDuet1) protein was coexpressed and copurified with the His-tagged Stn1. The Hsp82 was expressed as an amino-terminal (His)6 fusion using a T7 expression vector (pET28) in codon optimized Rosetta Escherichia coli cells (Novagen Inc.). For the telomeric proteins the cells were grown at 18°C, LB media lacking streptomycin was seeded to an O.D.595 = 0.2 with transformants from an overnight culture grown in LB supplemented with streptomycin (0.2 mg mL−1), the cells were grown to an O.D.595 = 0.8, protein expression was induced with IPTG and the cells were incubated for an additional 4 h. For Hsp82 expression the cells were grown comparably except streptomycin was not used in the overnight seed culture and cells were incubated at 24°C. After IPTG induction the cultures were clarified by centrifugation, resuspended in ice-cold 1x Talon binding buffer, and snap frozen; NP40 (1% v/v final) was added to the Cdc13 preparations. The proteins were purified using metal affinity chromatography (Talon resin) as per manufacture’s instructions (Clontech Inc.). The telomeric proteins were further resolved over MonoQ anion exchange resin and a Superdex-200 size exclusion column (GE Inc.). The Hsp82 was purified as previously described12. The identities of the purified proteins were confirmed by mass spectrometry and/or western blot analysis.
Telomerase DNA extension assays
Telomerase extracts were prepared using wild type (YPH499) Saccharomyces cerevisiae and the telomerase DNA extension reactions were performed as described 19. In brief, the DNA extension reactions used MonoQ telomerase extract in extension buffer (50 mM Tris pH 8.0, 1.0 mM MgCl2, 1.0 mM spermidine, 1.0 mM DTT, 0.5% (v/v) glycerol, 50 μM dTTP, 20 mCi [α-32P] dGTP) with 2.0 pmol telomeric DNA substrate immobilized on paramagnetic beads and supplemented with the indicated proteins. Following cleavage of the DNA substrates with EcoRI the released DNA fragments were magnetically separated from the beads, an end-labeled precipitation/loading control primer was added, the DNAs were subjected to ethanol precipitation, the products were reconstituted in formamide/NaOH loading buffer, subjected to denaturing gel electrophoresis and visualized using a Molecular Dynamics PhosphoImager.
Fluorescence anisotropy assay
The anisotropy reactions were performed in 25 μL volumes containing TMG-30 supplemented with a fluorescein-labeled telomere oligonucleotide (12.5 nM; fl- GTGGGTGTGTGTGTGGGGTGGTGTGTGTGG). To form the hybrid single-/double-stranded DNA substrates shorter complimentary oligonucleotides were annealed to the 5′ end of the telomere primer—7-base (CACCACCCCACACACACACCCAC), 15-base (CACACACACACCCAC) or 22-base (ACACCCAC) 3′-overhang. The reactions were incubated 5 min at 25°C before determining the anisotropy values using an Ultra Evolution plate reader (Tecan Inc.). RNase sensitivity of telomerase DNA binding was determined by preincubating aliquots of the MonoQ extracts with 10 mg of RNase A for 10 min at room temperature19. All anisotropy data represent average values (mean +/− SD) from 5 independent assays.
Chromatin immunoprecipitation assay
ChIP analysis was essentially performed as described19. For the cell cycle experiments yeast were arrested in late G1 with α-factor (10 μg mL−1), following a 2 h incubation the cultures were clarified by centrifugation and resuspended in rich media lacking α-factor to allow synchronous progression through the cell cycle at 30°C. Aliquots were removed to monitor cell cycle synchronization by FACS and budding assay or treated with formaldehyde (1% v/v final). To determine the relative telomere occupancy by Cdc13 or Est2 transformants (pRS425-GPD or pRS425-GPD-HSP82) of either the CDC13-MYC9x or EST2-MYC9x yeast23 were utilized. The fold-enrichments were determined by normalizing the ratio of the specific/nonspecific signal (e.g., α-Myc ChIP threshold cycle (Ct values)/normal mouse IgG ChIP Ct value) at the telomeric DNA targets to the ratio of the specific/nonspecific signal produced by amplifying a non-telomeric region of the genome (YJL052W).
Telomere DNA length analysis
Genomic DNA was prepared from yeast (YPH499) transformants (pRS425-GPD or pRS425-GPD-HSP82) after the indicated passages (~8–10 generations per passage). The DNA was digested overnight with XhoI, resolved on a 25 cm 1% (w/v) agarose gel for 20–24 h at 40 V and the DNA was transferred to Immobilon-Ny+ membrane (Millipore Inc.). The telomeric DNA was identified following hybridization in Ekono buffer (ISC BioExpress Inc.) supplemented with a radiolabeled telomeric oligonucleotide ((TGTGGGT)4). As a loading and migration control we used an established probe that recognizes a 1,621 bp fragment of chromosome IV33. The probe was generated by PCR amplification of genomic DNA in the presence of α-dGTP using specific oligonucleotides (GTACCTCGGTTTAGTTAAGCG and CTCATTCGAATCCATACGACC). Following appropriate washes using SSC/SDS buffers the telomeric and control DNA was visualized using a PhosphorImager (Molecular Dynamics Inc.).
Telomere DNA extension assay
The telomere elongation assay was performed as described22. Three representative data sets are shown in Supplementary Fig. 5.
Supplementary Material
Acknowledgments
We dedicate the presented work to our enduring memories of Oyetunji A. Toogun. We thank Jose Barral (University of Texas, Galveston) for the Ch184 E. coli strain, Victoria Lundblad (Salk Institute) for the Cdc13 constructs, Virginia Zakian (Princeton University) for the CDC13-Myc9x and EST2-Myc9x yeast strains and David Shore (Geneve University) for Stn1 and Ten1 vectors. D.C.D. designed and performed experiments, O.A.T. designed and performed experiments, F.J.E. performed experiments and B.C.F. designed and performed experiments and wrote the manuscript. We also appreciate the helpful comments on the manuscript by A. Belmont, W. Brieher, P. Newmark and D. Rivier. D.C.D. was supported by the CMB NIH training grant and B.C.F. was supported by the Public Service grant DK074270.
References
- 1.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 2.DeZwaan DC, Freeman BC. HSP90: the Rosetta stone for cellular protein dynamics? Cell Cycle. 2008;8:1006–1012. doi: 10.4161/cc.7.8.5723. [DOI] [PubMed] [Google Scholar]
- 3.Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci U S A. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 5.Grandin N, Damon C, Charbonneau M. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol Cell Biol. 2000;20:8397–8408. doi: 10.1128/mcb.20.22.8397-8408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes & Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 8.Lue NF. Sequence-specific and conformation-dependent binding of yeast telomerase RNA to single-stranded telomeric DNA. Nucleic Acids Res. 1999;12:2560–2567. doi: 10.1093/nar/27.12.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 10.Grandin N, Charbonneau M. Hsp90 levels affect telomere length in yeast. Mol Genet. 2001;265:126–134. doi: 10.1007/s004380000398. [DOI] [PubMed] [Google Scholar]
- 11.Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, Wright WE, White MA. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toogun OA, DeZwaan DC, Freeman BC. The hsp90 molecular chaperone modulates multiple telomerase activities. Mol Cell Biol. 2008;28:457–467. doi: 10.1128/MCB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelleher C, Kurth I, Lingner J. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol Cell Biol. 2005;25:808–818. doi: 10.1128/MCB.25.2.808-818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J Mol Biol. 2004;338:241–255. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 15.Niu H, Xia J, Lue NF. Characterization of the interaction between the nuclease and reverse transcriptase activity of the yeast telomerase complex. Mol Cell Biol. 2000;20:6806–6815. doi: 10.1128/mcb.20.18.6806-6815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 17.Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 2008;27:2328–2339. doi: 10.1038/emboj.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang MJ, Lin YC, Pang TL, Lee JM, Chou CC, Lin JJ. Telomere-binding and Stn1p-interacting activities are required for the essential function of Saccharomyces cerevisiae Cdc13p. Nucleic Acids Res. 2000;28:4733–4741. doi: 10.1093/nar/28.23.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toogun OA, Zieger W, Freeman BC. The p23 molecular chaperone promotes functional telomerase complexes through DNA dissociation. Proc Nat l Acad Sci USA. 2007;104:5765–5770. doi: 10.1073/pnas.0701442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dionne I, Wellinger RJ. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc Natl Acad Sci USA. 1996;93:13902–13907. doi: 10.1073/pnas.93.24.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertuch AA, Lundblad V. The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol Cell Biol. 2003;23:8202–8215. doi: 10.1128/MCB.23.22.8202-8215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcand S, Brevet V, Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taggart AK, Teng SC, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 24.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 25.Tseng SF, Lin JJ, Tseng SC. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 2007;34:6327–6336. doi: 10.1093/nar/gkl786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Makovet S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arneric M, Lingner J. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO J. 2007;8:1080–1085. doi: 10.1038/sj.embor.7401082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zappulla DC, Roberts JN, Goodrich KJ, Cech TR, Wuttke DS. Inhibition of yeast telomerase action by the telomeric ssDNA-binding protein, Cdc13. Nucleic Acids Res. 2009;37:354–367. doi: 10.1093/nar/gkn830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siller E, DeZwaan DC, Anderson JF, Freeman BC, Barral JM. Slow bacterial translation rates enhance eukaryotic protein folding efficiency. doi: 10.1016/j.jmb.2009.12.042. (submitted) [DOI] [PubMed] [Google Scholar]
- 30.Meier B, Driller L, Jaklin S, Feldmann HM. New function of CDC13 in positive telomere length regulation. Mol Cell Biol. 2001;21:4233–4245. doi: 10.1128/MCB.21.13.4233-4245.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeman BC, Yamamoto KR. Continuous recycling: a mechanism for modulatory signal transduction. Trends Biochem Sci. 2001;26:285–290. doi: 10.1016/s0968-0004(01)01834-5. [DOI] [PubMed] [Google Scholar]
- 32.Wegele H, Muller L, Buchner J. Hsp70 and Hsp90 a relay team for protein folding. Physiol Biochem Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- 33.Friedman KL, Cech TR. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 1999;13:2863–74. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picard D. Intracellular dynamics of the Hsp90 co-chaperone p23 is dictated by Hsp90. Exp Cell Res. 2006;312:198–204. doi: 10.1016/j.yexcr.2005.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


