Abstract
The protein-DNA and protein-protein interactions of Epstein-Barr virus nuclear antigen 1 (EBNA1) are known to play an important role in the many functions of this viral protein. Large quantities of pure EBNA1 protein would be useful in biochemical assays to elucidate such interactions. In particular, the crystal structure of the full-length protein would be important to show possible regions of interaction and/or post-translational modification. Recently, we described a novel approach to overexpress and purify EBNA1 from Escherichia coli; however, it is not ideal for large-scale production of EBNA1. We were able to optimize this protocol by 1) adding a polyethyleneimine precipitation step prior to Ni-NTA chromatography to reduce complexity of the sample and remove nucleic acid, 2) optimizing the Ni-NTA gradient to further separate EBNA1 from impurities, and 3) concluding with a MonoS cation-exchange chromatography step to further purify and concentrate EBNA1. We were able to recover 10-mg quantities of pure EBNA1 protein.
INTRODUCTION
Epstein-Barr virus (EBV) is the causative agent of infectious mononucleosis and a risk factor for developing a variety of malignancies, including Burkitt’s lymphoma, nasopharyngeal carcinoma, and post-transplant immunoproliferative disease [1, 2]. EBV nuclear antigen 1 (EBNA1) is a multifunctional protein that has been implicated in host cell immortalization and is necessary for the replication and maintenance of the EBV genome [3]. The N-terminal portion of EBNA1 primarily consists of a Glycine-Glycine-Alanine (GGA) repeat region that allows EBNA1 to escape immunosurveillance by preventing proteosomal degradation and presentation on the cell surface [4]. Other domains within the N-terminus have been implicated in the transcription, replication, and maintenance functions of EBNA1 [3, 5–7]. The C-terminal portion of EBNA1 encodes a nuclear localization sequence (NLS) followed by the DNA binding and dimerization domain. A C-terminal acidic tail has not been described functionally.
Recently, EBNA1 was overexpressed and purified from Escherichia coli [1, 8]. We sought to improve this purification strategy for large-scale production of EBNA1. The initial polyethyleneimine (PEI) precipitation reduced the complexity of the starting material and removed nucleic acid. This unusual fractionation resulted in the accumulation of the basic EBNA1 protein in the PEI pellet with nucleic acid and acidic protein. This advantageous separation strategy can be applied to other basic, DNA-binding proteins. The Ni-NTA chromatography was optimized with a shallower gradient and a cation-exchange chromatography step was added for further purification and concentration. Using this procedure, we have purified 10-mg quantities of EBNA1 in E. coli. It is a significant improvement over the previously described E. coli purification strategy [8].
This purification procedure will improve the study of the biochemistry of EBNA1 by providing a reliable protocol to obtain a large quantity of pure protein. For example, this substantial concentration of EBNA1 would be useful for crystallization trials. Full-length EBNA1 has not been crystallized, though the structure of the C-terminal portion was determined [9, 10]. Regions of the N-terminal half of the protein, however, are necessary for EBNA1’s functions [3, 11]. The full-length crystal structure would provide important information on additional EBNA1-DNA interactions as well as possible sites for protein-protein interactions and post-translational modifications that could affect EBNA1’s functions.
MATERIALS AND METHODS
Reagents and plasmids
All reagents were obtained from Sigma Chemical (St. Louis, MO) unless otherwise noted. TE buffer contains 50 mM Tris HCl and 0.1 mM EDTA, pH 7.9. The EBNA1 expression plasmid has been described previously [8]. A functionally wildtype derivative of EBNA1 lacking 224 amino acids from the GGA repeat region between amino acids 90 and 325 [12] was ligated into bacterial expression vector pET22b (Novagen, Madison, WI).
Electrophoresis and Western blotting
Proteins were separated by electrophoresis using 4–12% Bis/Tris NuPAGE polyacrylamide gels (Invitrogen, Carlsbad, CA). Western blots were prepared by probing a 0.45 μm nitrocellulose membrane with anti-EBNA1 mAb 1EB14 alone or in combination with anti-EBNA1 mAb 3EB7. After washing with PBST (phosphate-buffered saline + 0.1% Tween 20), a secondary antibody (goat anti-mouse IgG, Chemicon International, Temecula, CA) conjugated to alkaline phosphatase was added and the 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium (BCIP/NBT) reagent was used as substrate. Prestained molecular weight markers (Multimark, Invitrogen) were included on all gels.
Protein determinations
Protein concentration was determined by a modified Bradford assay [13] using Coomassie blue reagent and bovine serum albumin (Pierce, Rockford, IL) as a protein standard.
Cell preparation
BL21(DE3)Rosetta2 cells (Novagen) carrying pET22b-EBNA1 expression vector were cultured in LB broth containing 100 μg ampicillin/ml and 35 μg chloramphenicol/ml. Cultures were grown at 37°C and induced at an OD (600 nm) of 0.6 with 1 mM isopropylthiogalactoside (IPTG) for 4 h. Cells were harvested and stored at −20°C. E. coli cell pellets, approximately 10 g wet weight, were resuspended in 60 ml of TE + 0.1 M NaCl. Cells were lysed by sonication (3 rounds of 30 s bursts on ice) and cell debris was removed by centrifugation (27,000g for 20 min at 4°C).
PEI precipitation
Polyethyleneimine (PEI) was added as a neutralized 10% (v/v) stock solution to the soluble fraction of the E. coli whole cell extract to yield a final concentration of 0.15%. The solution was incubated on ice for 15 min. The PEI pellet was collected by centrifugation (9,000 g for 5 min at 4°C). The pellet should contain nucleic acid as well as DNA-bound and negatively-charged proteins. The PEI pellet was washed with TE + 0.3 M NaCl and collected by centrifugation (9,000 g for 5 min at 4°C). EBNA1 was eluted from the PEI pellet with TE + 0.8 M NaCl. Supernatant was collected after centrifugation (9,000 g for 5 min at 4°C). Ammonium sulfate (AS) was added to the 0.8 M NaCl eluate until 60% saturation (7.6 g AS was added to 20 ml eluate). The precipitation was carried out at 4°C for 18 h. The pellet was collected by centrifugation (27,000 g for 20 min at 4°C).
Ni-NTA chromatography
The AS pellet was resuspended in Buffer C (50 mM Tris-HCl, 100 mM NaCl, 0.1% Tween-20 (Pierce), 5% glycerol (Fisher Scientific, Waltham MA), 5 mM imidazole (Fisher), pH 7.9) + 0.5 M GuHCl. The solution was clarified by centrifugation (27,000 g for 10 min at 4°C). A 2-ml Ni-NTA Superflow column (Qiagen, Valencia, CA) was equilibrated with Buffer A (50 mM Tris-HCl, 700 mM NaCl, 0.1% Tween 20, 5% glycerol, 5 mM imidazole, pH 7.9). The sample was loaded onto the column and washed with 10 column volumes (CV) of 99.5% Buffer A and 0.5% Buffer B (50 mM Tris-HCl, 700 mM NaCl, 0.1% Tween 20, 5% glycerol, 800 mM imidazole). Bound proteins were eluted over 15 CV with a 0.5 – 30% Buffer B gradient and one-ml fractions were collected. The column was then washed with 5 CV of 100% Buffer B. The flow rate was 1 ml/min. Fractions were analyzed by gel electrophoresis and Bradford assay.
MonoS cation-exchange chromatography
A 1-ml MonoS HR 5/5 cation-exchange column (GE Healthcare, Piscataway, NJ) was equilibrated with TE + 0.25 M NaCl. Eluate from the Ni-NTA purification was diluted to approximately 0.23 M NaCl with TE buffer. The sample was applied to the column and washed with 5 CV of TE + 0.5 M NaCl. EBNA1 was eluted over 45 CV with a 0.5 – 1 M NaCl gradient in TE buffer and the column was washed with 2 CV of TE + 1 M NaCl. The flow rate was 1 ml/min. Fractions were analyzed by gel electrophoresis.
Spin concentration
An Amicon ultracentrifugal filter (4 ml volume, 5 kDa molecular weight cut off (MWCO), Millipore, Billerica, MA) was used to concentrate the EBNA1 protein from the pooled MonoS column peak. The sample was centrifuged at 9,000 g until the protein concentration reached approximately 8 mg/ml (~16-fold concentration).
Size-exclusion chromatography (SEC)
SEC using a 120-ml HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare) was initially used to further purify and generate a homogeneous EBNA1 sample for crystal trials. The Ni-NTA eluate was first spin concentrated (10 kDa MWCO, 15 ml volume, Millipore) to 5% of the CV following manufacturer instructions. The Superdex 200 column was equilibrated in TE + 0.5 M NaCl, the sample was added, and an isocratic elution in the same buffer was performed at 1 ml/min.
A 23-ml Superose 6 10/300 GL column (GE Healthcare) was used to analyze EBNA1 in the presence or absence of dithiothreitol (DTT). The Ni-NTA eluate was first spin concentrated as above to 5% of the CV. The column was equilibrated in either TE + 0.5 M NaCl or the same buffer + 0.1 mM DTT. The sample was added and an isocratic elution using the same buffer was performed at 0.5 ml/min.
RESULTS
PEI Precipitation
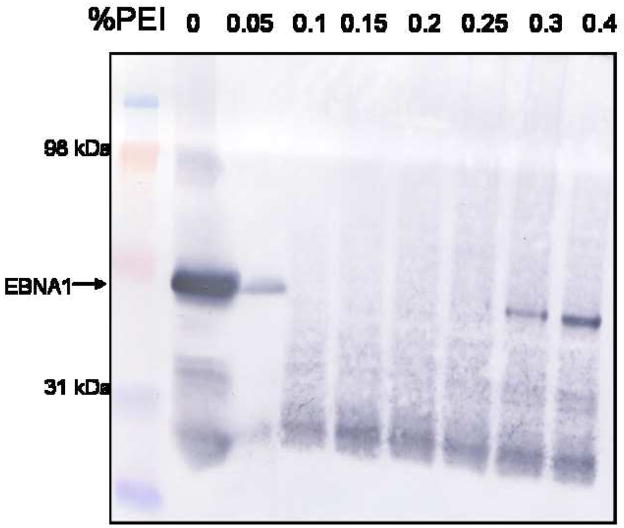
Due to the substantial volume of the starting material, it is important to remove a majority of unwanted protein and nucleic acid before applying the sample to a chromatography column. PEI is a positively-charged polymeric molecule that binds very tightly to nucleic acid and negatively-charged proteins. EBNA1 is positively-charged, with an isoelectric point of 9.5, and binds DNA tightly. To determine the concentration of PEI required to precipitate EBNA1, a precipitation trial was performed [14, 15]. Aliquots of whole cell extract from E. coli overexpressing EBNA1 were mixed with increasing concentrations of PEI. The pellet was recovered by centrifugation and the supernatant was tested by Western blot for the presence of EBNA1 (Fig. 1A). The starting material, 0% PEI, shows a strong signal for EBNA1. That signal drops significantly upon the addition of only 0.05% PEI and remains absent through 0.2% PEI. This result shows that EBNA1 has precipitated in the PEI pellet at these concentrations. However, at the higher PEI concentrations (0.3% and 0.4%), EBNA1 is present in the supernatant. This result suggests that at low concentrations of PEI, EBNA1 is bound to DNA and precipitates with DNA when PEI is added. Upon the addition of high levels of PEI, the PEI competes with EBNA1 for binding to DNA and releases EBNA1 into the supernatant. The mAb used in the Western blot binds to the N-terminal portion of the EBNA1 protein. In all samples, an EBNA1 degradation band is seen. This fragment may lack a functional DNA-binding domain and therefore may not be included in the PEI pellet. Based on these experiments, 0.15% PEI was selected as the ideal concentration to precipitate EBNA1.
Figure 1. PEI precipitation of EBNA1.
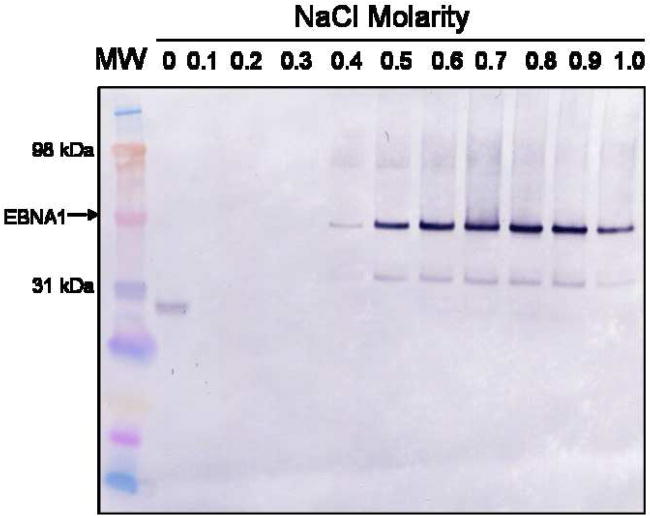
A) PEI trial to determine the percentage of PEI necessary to precipitate EBNA1. Portions of whole cell extract were mixed with varying concentrations of PEI, the pellets were collected, and the supernatants were tested for the presence of EBNA1 by Western blot analysis with anti-EBNA1 mAb 1EB14. mAb 1EB14 binds in the N-terminal portion of EBNA1 suggesting that the EBNA1 fragment seen in each PEI sample lacks a functional DNA binding domain and is therefore not precipitated. B) Salt trials to determine the optimal concentration of NaCl to both wash the PEI pellet and elute EBNA1 from the pellet. 0.15% PEI pellets were washed with varying concentrations of NaCl, recovered by centrifugation, and the supernatants were tested for the presence of EBNA1 as described in A. MW is molecular weight markers.
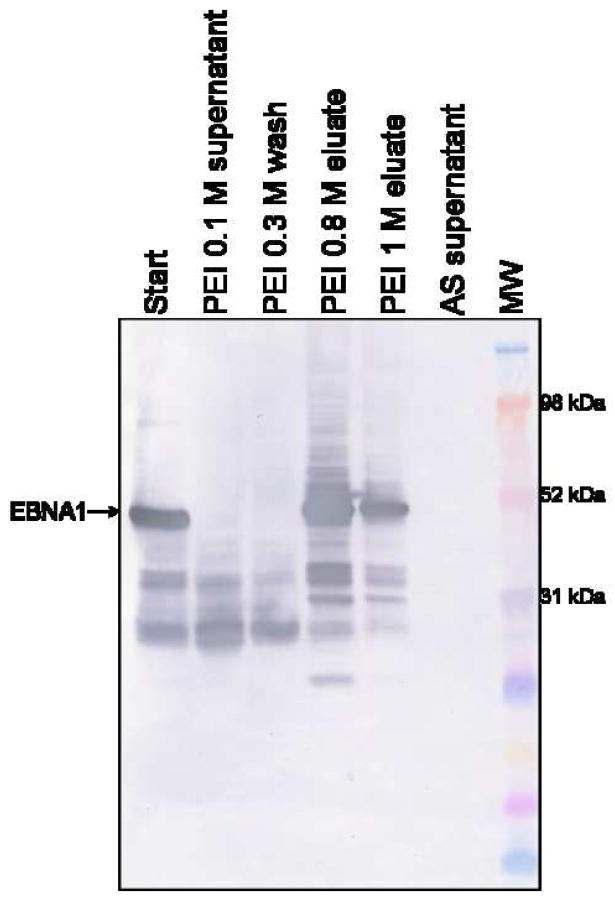
Next, it was necessary to determine the optimal salt concentration to both wash the pellet and elute EBNA1 from the pellet. The wash buffer should be the highest salt concentration that maintains EBNA1 in the PEI pellet, but releases loosely bound protein. The ideal elution buffer is the lowest salt concentration necessary to release EBNA1 from the pellet while leaving nucleic acid and acidic protein behind. A PEI pellet was washed with either TE buffer or TE buffer + various concentrations of NaCl. The supernatant was analyzed by Western blot to determine if EBNA1 had been released from the PEI pellet (Fig. 1B). There was no EBNA1 signal in the supernatant from 0 – 0.3 M NaCl. The strongest supernatant signal was reached by 0.8 M NaCl. Based on these results, 0.3 M NaCl was chosen as the salt concentration for the wash buffer and 0.8 M NaCl was chosen as the salt concentration for the EBNA1 elution buffer.
After the EBNA1 sample was isolated from the PEI pellet, it was necessary to remove any residual PEI to prevent later aggregation when the salt concentration was decreased. This removal was achieved by performing a 60% saturated ammonium sulfate precipitation, thereby pelleting all protein and removing residual PEI.
Ni-NTA Chromatography
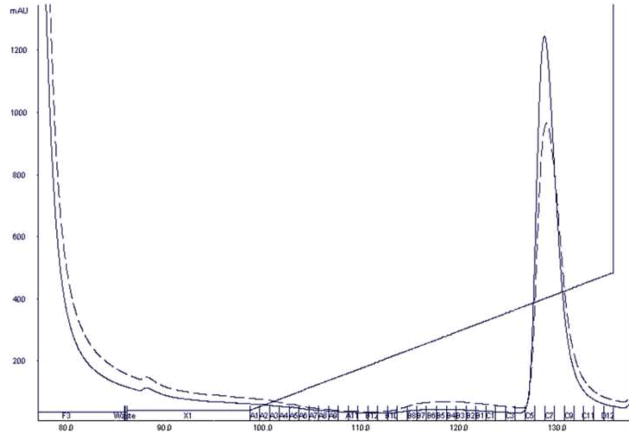
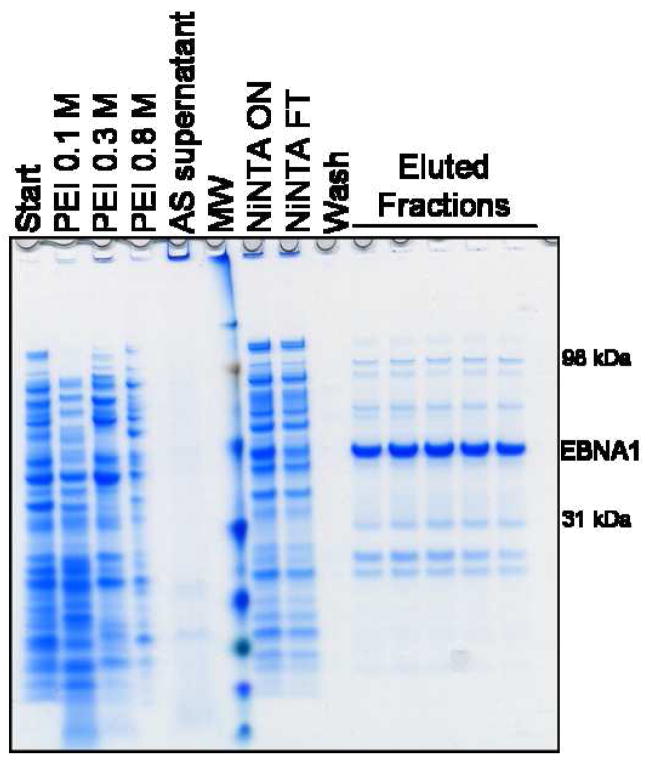
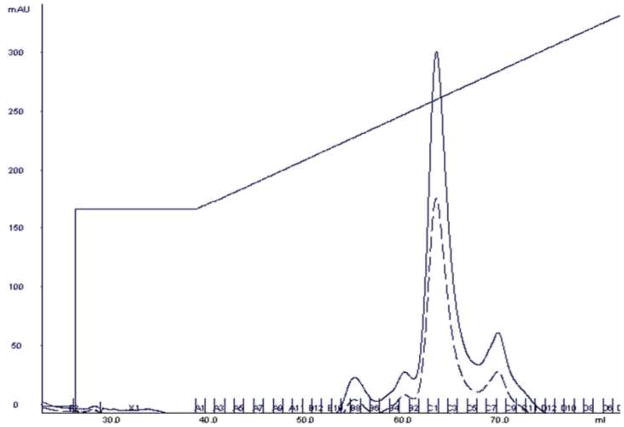
After the sample was enriched for EBNA1 through PEI precipitation, it was applied to a Ni-NTA Superflow immobilized metal affinity chromatography column. The sample was bound to the column, washed with low imidazole buffer, and then eluted with an imidazole gradient. The chromatogram can be seen in Fig. 2. EBNA1 eluted off the Ni-NTA column at approximately 215 mM imidazole. Following Ni-NTA chromatography, EBNA1 was the most abundant protein in the sample, but additional purification was needed (Fig. 3A).
Figure 2. Chromatogram showing the Ni-NTA purification of the protein eluted from the PEI pellet.
The absorbance at 280 nm is shown as a solid black line. The absorbance at 260 nm is shown as a dashed black line. The concentration gradient of Buffer B is shown as the slanted black line. EBNA1 eluted at approximately 215 mM imidazole. Fractions C5–C10 were pooled.
Figure 3. Analysis of PEI and Ni-NTA purification steps.
A) SDS-PAGE showing the PEI fractionation, ammonium sulfate (AS) precipitation, and Ni-NTA chromatography steps. FT is flowthrough. Gel is stained with Coomassie. B) Western blot probed with mAbs 1EB14 and 3EB7 shows that EBNA1 is not lost in the 0.1 M and 0.3 M NaCl wash steps. Some EBNA1 remains trapped in the PEI pellet and is released in a second high salt wash (TE + 1 M NaCl), and no EBNA1 is lost during the AS precipitation. PEI washes are listed with the NaCl concentration used. MW is molecular weight markers.
There were significant bands in the PEI wash samples that migrate similarly to EBNA1 in gel electrophoresis. Western blot analysis (probed with mAbs reactive to two different epitopes) showed that none of these bands were EBNA1 and the target protein was retained in the PEI pellet (Fig. 3B). Some EBNA1 degradation products are lost during the wash steps.
Size-Exclusion Chromatography
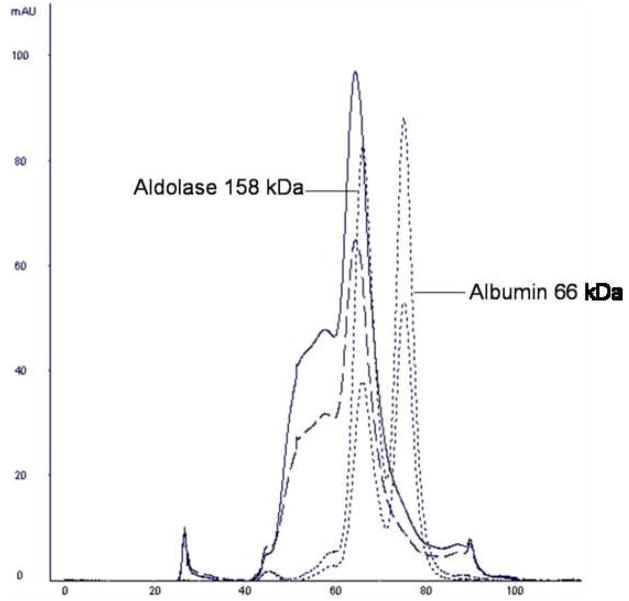
Initially, the final step in EBNA1 purification was size-exclusion chromatography (SEC) in order to separate impurities away from EBNA1 and to provide a homogeneous sample. However, SEC did not result in sufficient separation of EBNA1 from the impurities (Fig. 4). The Ni-NTA eluate was applied to a Superdex 200 column and purified in TE + 0.5 M NaCl buffer. Although the sample was further enriched for EBNA1, multiple bands could be seen representing both larger and smaller protein impurities.
Figure 4. Superdex 200 size-exclusion chromatography of EBNA1.
A) The chromatogram resulting from further purification of the Ni-NTA eluate over a Superdex 200 column. Absorbance at 280 nm is shown as a solid black line. Absorbance at 260 nm is shown as a dashed black line. Molecular weight standards are as marked. B) SDS-PAGE (Coomassie stained) showing EBNA1 purification by SEC. MW is molecular weight markers.
MonoS Chromatography
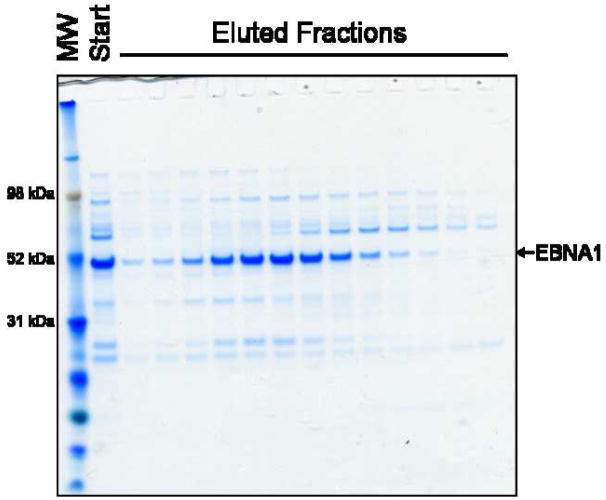
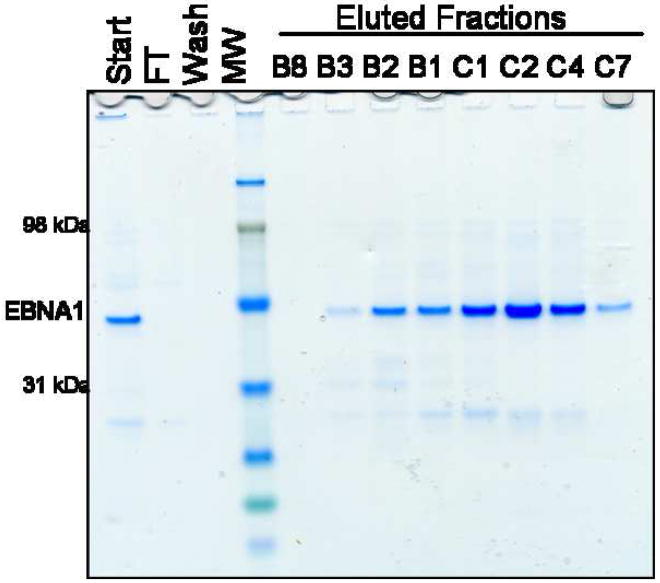
To take advantage of the unusually high isoelectric point of EBNA1, MonoS cation-exchange chromatography was employed. The Ni-NTA eluate was first diluted to approximately 0.23 M NaCl and applied to a MonoS column. The column was washed at 0.5 M NaCl and EBNA1 was eluted over a shallow salt gradient from 0.5 – 1 M NaCl. EBNA1 binds this column extremely well, eluting off at approximately 0.75 M NaCl. The MonoS purification resulted in higher purity of EBNA1 compared to SEC (Fig. 5).
Figure 5. MonoS cation-exchange chromatography of EBNA1.
A) The chromatogram resulting from further purification of the Ni-NTA eluate over a MonoS column. Absorbance at 280 nm is shown as a solid black line. Absorbance at 260 nm is shown as a dashed black line. The salt gradient utilized is shown as the slanted black line. B) SDS-PAGE (Coomassie stained) showing EBNA1 purification by MonoS chromatography. FT is flowthrough. MW is molecular weight markers.
Investigating the Primary Impurity
There is one small protein impurity (~27 kDa) consistently found eluting with EBNA1 during the MonoS purification (Fig. 5B). Western blot analysis of the eluate, using a variety of different monoclonal antibodies reactive to EBNA1 or the C-terminal His-tag, resulted in a protein of approximately 27 kDa reacting with those antibodies that bind epitopes between amino acids 219–417 (data not shown).
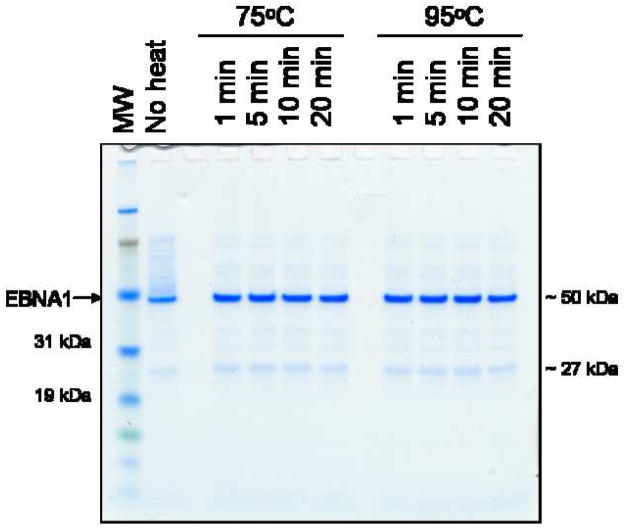
During the preparation of samples for gel electrophoresis, heat-induced cleavage can occur at Asp-Pro bonds. EBNA1 contains one Asp-Pro bond at amino acids 209–210. Cleavage of this bond could result in two fragments (209 aa and 208 aa) that would run at the approximate molecular weight of the impurity. Samples prepared at varying temperatures and incubation times showed no difference in EBNA1 fragmentation (Fig. 6).
Figure 6. Analysis of EBNA1 heat-induced Asp-Pro bond cleavage.
An EBNA1 sample (fraction B1 for the MonoS purification) was heated at either 75°C or 95°C for 1, 5, 10, or 20 minutes. The amount of sample breakdown, as shown by the accumulation of the lower band, was compared among the samples. MW is molecular weight markers.
This impurity may be a protein that is binding through hydrophobic interactions. Most of the purification steps are done in high salt buffer, which would increase the strength of a hydrophobic interaction. However, such an interaction would not be expected to survive the PEI precipitation and later low salt wash steps.
This impurity may also be a heterodimer between full-length EBNA1 and an EBNA1 fragment. We performed SEC to determine whether the size of EBNA1 differed in the presence of 1 mM DTT. EBNA1 eluted off the Superose 6 SEC column at the same place whether or not under reducing conditions and SDS-PAGE analysis showed the lower band in the peak fraction of both purifications (data not shown). These data suggest that a disulfide bond does not maintain an EBNA1 heterodimer; however, it is possible that a higher concentration of DTT is required to separate the dimer. We also analyzed the MonoS eluate by non-reducing gel electrophoresis and found no difference in the migration of the proteins compared to separation under reducing conditions (data not shown).
DISCUSSION
The previous purification strategy for E. coli-expressed EBNA1 [8], though an improvement upon prior protocols, still had weaknesses especially for large-scale production of EBNA1. The amount of DNA in the sample is problematic as well as the complexity of the sample to be added directly to the Ni-NTA chromatography column. To improve both of these problems, we incorporated a PEI precipitation step. EBNA1 has a significant nonspecific interaction with DNA, which was shown by the inclusion of EBNA1 with the DNA and acidic proteins in the initial PEI pellet at low salt. Wash buffers containing a minimum of 0.4 M NaCl were required to start eluting EBNA1 from the pellet. This unusual accumulation of a basic protein in the PEI pellet was an advantage for reducing complexity of the sample. Not only was DNA removed, but basic proteins were not precipitated initially and washed away, weakly bound proteins were eluted in the low salt wash, and very acidic proteins were maintained in the DNA/PEI pellet after elution of EBNA1. This PEI precipitation can be applied to other basic, DNA-binding proteins to provide a rapid, initial purification step that reduces the complexity of the sample significantly. Though the sample is still a mixture of protein, the amount of starting material on the Ni-NTA column is significantly reduced and EBNA1 is enriched for.
The Ni-NTA chromatography step is an improved version of the original protocol discussed in [8]. In particular, the elution gradient is shallower and optimized for EBNA1. Though size-exclusion chromatography was not successful in separating EBNA1 away from impurities, we were able to take advantage of another unusual property of EBNA1 during our final purification step. EBNA1 has a high isoelectric point of 9.5 and thus binds very tightly to a cation-exchange column. EBNA1 bound to a MonoS column while impurities either did not bind the column or were removed by a high salt wash. Pure EBNA1 was eluted at approximately 0.75 M NaCl.
While characterizing the protein fragment eluting with EBNA1, we were able to show that EBNA1 is not susceptible to heat-induced Asp-Pro bond cleavage under the conditions we used. A heterodimer is not maintained through hydrophobic interactions; however, a dimer linked by disulfide bonds cannot be excluded. Though this impurity is found at very low abundance in the final EBNA1 sample, this protein purification strategy is a significant improvement upon previous approaches and may be useful to optimize crystallization conditions for EBNA1 crystal trials. We started with approximately 10 g wet weight of E. coli cells overexpressing EBNA1 and were able to recover about 10 mg of pure EBNA1 protein at greater than 50% yield.
Acknowledgments
We would like to thank Dr. Bill Sugden for helpful discussion and Dr. Scott Lindner for discussion and assistance with protein modeling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 2.Crawford DH. Biology and disease associations of Epstein-Barr virus. Philos Trans R Soc Lond B Biol Sci. 2001;356:461–73. doi: 10.1098/rstb.2000.0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leight ER, Sugden B. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev Med Virol. 2000;10:83–100. doi: 10.1002/(sici)1099-1654(200003/04)10:2<83::aid-rmv262>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 4.Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc Natl Acad Sci U S A. 1997;94:12616–21. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–9. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984;81:3806–10. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gahn TA, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–6. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duellman SJ, Burgess RR. Overproduction in Escherichia coli and purification of Epstein-Barr virus EBNA-1. Protein Expr Purif. 2006;47:434–40. doi: 10.1016/j.pep.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 9.Bochkarev A, Barwell JA, Pfuetzner RA, Bochkareva E, Frappier L, Edwards AM. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 1996;84:791–800. doi: 10.1016/s0092-8674(00)81056-9. [DOI] [PubMed] [Google Scholar]
- 10.Bochkarev A, Barwell JA, Pfuetzner RA, Furey W, Jr, Edwards AM, Frappier L. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell. 1995;83:39–46. doi: 10.1016/0092-8674(95)90232-5. [DOI] [PubMed] [Google Scholar]
- 11.Yates JC. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. Cancer Cells. 1988;6:197–205. [Google Scholar]
- 12.Aiyar A, Sugden B. Fusions between Epstein-Barr viral nuclear antigen-1 of Epstein-Barr virus and the large T-antigen of simian virus 40 replicate their cognate origins. J Biol Chem. 1998;273:33073–81. doi: 10.1074/jbc.273.49.33073. [DOI] [PubMed] [Google Scholar]
- 13.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 14.Marshak DR, Kadonaga JT, Burgess RR, Knuth MW, Brennan WA, Lin SH. Strategies for protein purification and characterization: A laboratory manual. Woodbury, NY: Cold Spring Harbor Press; 1996. [Google Scholar]
- 15.Burgess RR, Jendrisak JJ. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975;14:4634–8. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]