Abstract
Myc proteins are known to have an important function in stem cell maintenance. As Myc has been shown earlier to regulate microRNAs (miRNAs) involved in proliferation, we sought to determine whether c-Myc also affects embryonic stem (ES) cell maintenance and differentiation through miRNAs. Using a quantitative primer-extension PCR assay we identified miRNAs, including, miR-141, miR-200, and miR-429 whose expression is regulated by c-Myc in ES cells, but not in the differentiated and tumourigenic derivatives of ES cells. Chromatin immunoprecipitation analyses indicate that in ES cells c-Myc binds proximal to genomic regions encoding the induced miRNAs. We used expression profiling and seed homology to identify genes specifically downregulated both by these miRNAs and by c-Myc. We further show that the introduction of c-Myc-induced miRNAs into murine ES cells significantly attenuates the downregulation of pluripotency markers on induction of differentiation after withdrawal of the ES cell maintenance factor LIF. In contrast, knockdown of the endogenous miRNAs accelerate differentiation. Our data show that in ES cells c-Myc acts, in part, through a subset of miRNAs to attenuate differentiation.
Keywords: differentiation, embryonic stem cell, microRNA, myc
Introduction
MicroRNAs (miRNAs), 21–23 nucleotide non-protein coding RNAs, act as powerful regulators of gene expression at the post-transcriptional level by targeting specific mRNA degradation or by suppression or activation of translation (Carrington and Ambros, 2003; Dykxhoorn et al, 2003; Grewal and Moazed, 2003; Pickford and Cogoni, 2003; Vasudevan et al, 2007). Studies from many laboratories have showed that miRNAs influence a wide range of biological processes including development, lifespan, metabolism, and cancer (Kato and Slack, 2008; Stefani and Slack, 2008). Regulation of miRNA genesis occurs at multiple levels in response to differentiation and mitogenic signalling (Thomson et al, 2006; Viswanathan et al, 2008). Some miRNAs are characteristic of specific differentiated cell types, whereas others are specifically expressed in stem and progenitor cells during early development (Lim et al, 2003; Chen et al, 2004; Poy et al, 2004; Chang and Mendell, 2007; Hwang and Mendell, 2007; Ibarra et al, 2007). Recent evidence has shown that miRNA mutations or deregulations correlate with different human cancers, and has also shown that miRNAs can function as tumour suppressors or oncogenes (Calin and Croce, 2006; Esquela-Kerscher and Slack, 2006; Medina and Slack, 2008; Tavazoie et al, 2008). The most well-characterized miRNA cluster involved in tumourigenesis, miR-17-92, was found to be induced by Myc oncoprotein expression (He et al, 2005; Mendell, 2005; O'Donnell et al, 2005).
The Myc protein family comprises basic helix-loop-helix-zipper (bHLHZ) transcription factors (c-, N-, and L-Myc) that can each form obligate heterodimers with the small bHLHZ protein Max. Myc–Max heterodimers bind the E-box sequence CACGTG in many different cell types and activate transcription of a large number of genes, many of which are associated with cell growth. In addition, Myc has been implicated in transcriptional repression of many genes that normally limit cell-cycle progression (Adhikary and Eilers, 2005; Cole and Nikiforov, 2006; Kleine-Kohlbrecher et al, 2006). Myc's broad effects on both normal and abnormal cell behaviour have been generally assumed to relate to its regulation of RNA polymerase II transcription of protein coding target genes as well as RNA polymerase I and RNA polymerase III transcription of RNAs involved in translation and growth (Gomez-Roman et al, 2003; Arabi et al, 2005; Grandori et al, 2005). However, the demonstration that Myc also controls expression of a subset of miRNAs has added another class of critical Myc targets. Induced expression of c-Myc in the P493-6 B cell line showed upregulation of six miRNAs within the miR-17 cluster on chromosome 13 through direct binding of Myc to the miR-17 locus (He et al, 2005; O'Donnell et al, 2005). Recently, Myc was shown to be involved in repression of numerous miRNAs in tumour cell lines, including the let-7 tumour suppressor, which regulates expression of c-myc itself (Sampson et al, 2007; Chang et al, 2008) and miR-23a/b resulting in increased glutamine catabolism (Gao et al, 2009). Thus, Myc proteins seem to be intimately involved in the regulation of a broad range of miRNAs, many of which are likely to have key roles in cell proliferation and cancer (for reviews see Lotterman et al, 2008; Medina and Slack, 2008).
Another biological setting in which Myc and miRNA regulation may converge is in stem cell self-renewal and pluripotency. Genetic studies in mice indicate that c-Myc is involved in the growth, proliferation, and differentiation of epidermal, neural and lung stem/progenitor cells, and haematopoietic stem cells (Arnold and Watt, 2001; Knoepfler et al, 2002; Okubo et al, 2005; Dubois et al, 2008). In embryonic stem (ES) cells c-Myc has been shown to be required for the maintenance of self-renewal whereas Myc downregulation on withdrawal of leukaemia inhibitory factor (LIF) is critical for differentiation (Cartwright et al, 2005). Furthermore, myc family genes, together with Oct4, Klf4, and Sox2, act to reprogramme differentiated cells into induced pluripotent stem (iPS) cells with ES cell properties, suggesting that c-Myc is a driver of pluripotency (Takahashi et al, 2007; Wernig et al, 2007). Although iPS conversion can occur without introduction of Myc, the overall yield of converted cells is dramatically decreased, as is the rate of conversion (Nakagawa et al, 2008). miRNAs have also been implicated in the ES cell function. Targeted deletion of the Dicer pre-miRNA processing ribonuclease abrogated ES cell differentiation whereas self-renewal was less drastically reduced (Kanellopoulou et al, 2005; Murchison et al, 2005). Indeed, a characteristic miRNA profile has been defined in murine and human ES cell lines, which overlaps with miRNAs implicated earlier in proliferation and tumourigenesis (Suh et al, 2004; Calabrese et al, 2007). Moreover, two studies have shown that pluripotency-related transcription factors regulate many miRNAs and are themselves subject to miRNA regulation (Marson et al, 2008; Tay et al, 2008). Furthermore, recent reports show that ES cell-specific miRNAs belonging to the miR-290 family promote the G1-S transition and self-renewal in ES cells as well as induced pluripotency during reprogramming of fibroblasts (Wang et al, 2008; Judson et al, 2009). These findings prompted us to determine whether c-Myc might contribute to ES cell pluripotency through regulation of miRNAs.
Results
Identification of c-Myc-induced miRNAs in ES cells
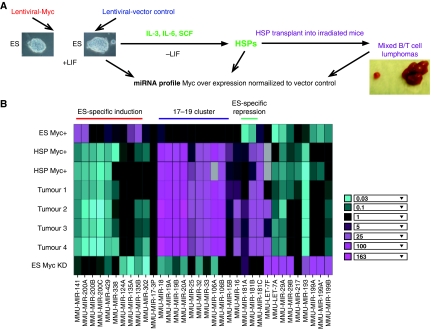
To identify ES-specific c-Myc-induced miRNAs, we analysed three cell populations that permit us to compare ES cells to their differentiated and tumourigenic derivatives: (i) murine AK7 ES cells (ii) induced haematopoietic stem/progenitor cells (HSPs) derived from these ES cells on differentiation, and (iii) tumours from mice transplanted with ES cell-derived HSPs (Figure 1A). To determine the effects of increasing c-Myc levels, we used a c-myc-expressing lentiviral vector in ES cells (hereafter referred to as c-Myc+ cells) and identified c-Myc-induced miRNAs after normalization to a lentiviral vector control. We observed an 8–14-fold increase in c-myc RNA and protein levels as determined by qRT–PCR and immunoblotting, respectively (Supplementary Figure S1). We also generated differentiated cells from control and c-Myc+ ES cells. Treatment of murine ES cells with the cytokines IL-3, IL-6, and SCF resulted in induction of HSPs (Kushida et al, 2001; Burt et al, 2004). Finally, mixed T- and B-cell tumours were collected from irradiated mice transplanted with cytokine-induced c-Myc+ HSPs and compared with vector control transplanted HSPs in the miRNA profiling experiment (Supplementary Figure S2).
Figure 1.
miRNAs regulated by c-Myc in murine ES cells. (A) Scheme showing the experimental design for identification of miRNAs regulated by c-Myc in ES cells. c-Myc level was increased by lentiviral-delivered overexpression of c-Myc in ES cells (see Supplementary Figure S1). These ES cells, their differentiated haematopoietic stem/progenitor cell derivatives (HSP Myc+) and tumours after injection of HSP c-Myc+ cells into irradiated mice, were subjected to miRNA profiling. (B) Heatmap of miRNAs displaying significant regulation by c-Myc in ES cells and ES cell-derived differentiated cells or tumours. HSP: haematopoietic stem/progenitor cells derived by differentiation of ES cells; Tumours1–4: mixed T- and B-cell tumours derived by transplantation of Myc+ HSPs into irradiated mice. Note that results are shown for two HSP, and four tumour cell isolates independently derived from the same ES cell line. Bottom row shows miRNAs levels after c-Myc knockdown in WT ES cells. Inset: Fold change in miRNA copy number represents the ratio of miRNA level in ES cell in which c-Myc has been introduced (Myc+), or knocked down by c-myc shRNA (ES Myc KD), relative to lentiviral vector (see Supplementary Figure S1 for quantitation). Grey boxes represent no detectable signal.
miRNA expression levels were measured using a quantitative primer-extension PCR assay as described earlier (Raymond et al, 2005; He et al, 2007). Primers representing 192 miRNAs were used in this study. To determine the effects of increasing c-Myc levels we calculated the fold change in miRNA copy number as the error-weighted average of the ratio in Myc+ cells relative to the lentiviral vector control (see Materials and methods). Figure 1B shows a heat map of the most significantly changed miRNAs in c-Myc+ versus vector controls in the ES cell-derived populations as described above. The figure displays results for two HSP, and four tumour cell isolates independently derived from the same ES cell line. Consistent with earlier findings (Mendell, 2005; O'Donnell et al, 2005), a subset of the oncogenic miRNA cluster, miR17-92, was highly induced in c-Myc+ differentiated cells, and in tumours from mice transplanted with c-Myc+ HSPs derived from our Myc+ ES cells (Figure 1B). However, the levels of these miRNAs were not substantially altered in c-Myc+ ES cells. We also identified four miRNAs, let-7, miR-29, miR-181, and miR-199 that are decreased in c-Myc+ ES cells relative to the vector control. Let-7 and miR-29 are downregulated by c-Myc in B-cell lymphomas (Chang et al, 2008). In addition, we find that miR-181A and miR-181B are sharply induced in Myc+ differentiated and tumour cells (Figure 1B). We also identified several miRNAs, miR-124, miR-135A, miR-135B, miR-141, miR-200, miR-302, miR-338, and miR-429 that seem to be upregulated specifically by c-Myc in ES cells (induced ∼four-fold in c-Myc+ cells; Figure 1B; see Supplementary Table S1 for complete list; Supplementary Figure S3A for northern blots). These miRNAs are induced by c-Myc in ES cells, but are either not increased or are strongly reduced in differentiated haematopoietic cells (Figure 1B; Supplementary Figure S3B) or in the tumours from transplanted mice. To further examine the regulation of these miRNAs by c-Myc we knocked down c-Myc levels two to four-fold in wild-type ES cells using lentiviral shRNA (Supplementary Figure S1B) and determined miRNA levels by quantitative primer-extension PCR as described. Our results show that after Myc knockdown the levels of miR-124, miR-135A, miR-135B, miR-141, miR-200, miR-302, miR-338, and miR-429 are sharply reduced (Figure 1B, compare top and bottom rows). The c-Myc knockdown experiment also confirms that expression of the miR17-92 cluster is diminished in shMyc-treated ES cells compared to the Myc+ differentiated and tumour cells, whereas miRNAs repressed in Myc+ ES cells are strongly activated after c-Myc knockdown (Figure 1B). Together, our data confirm that Myc activates or represses multiple miRNAs and identifies a subset of those whose specific expression in ES cells is Myc dependent.
Earlier studies had reported that among the group of c-Myc-regulated miRNAs in ES cells described above, miR-135 and miR-124 regulate neurogenesis during central nervous system (CNS) development (Cao et al, 2007; Visvanathan et al, 2007). miR-302 is related to the miR-290 family several of whom have been recently shown to promote ES cell proliferation by targeting cell-cycle regulators (Wang et al, 2008). Interestingly, miR-291-3p, -294, and -295 seem to be directly regulated by Myc during reprogramming of fibroblasts to induce pluripotency (Judson et al, 2009). However, the targets and functions of several c-Myc-induced miRNAs, such as miR-141, miR-200, miR-338, and miR-429 in ES cells, have not been established. These miRNAs are endogenously expressed in ES cells in a Myc-dependent manner and their levels are augmented on c-Myc overexpression (Figure 1B; Supplementary Figure S3A and B). We therefore chose to focus on identifying targets and functions of these miRNAs.
Analysis of c-Myc binding to selected miRNA loci
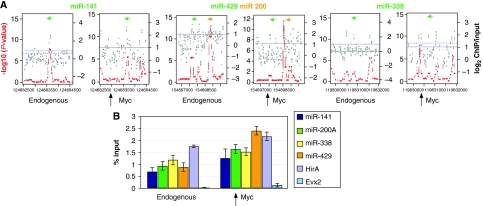
To address whether c-Myc might regulate these miRNAs by direct binding to their genomic loci, we mapped the genomic-binding sites of c-Myc in ES cells using DNA derived from anti-c-Myc chromatin immunoprecipitation (ChIP) to probe a custom-designed DNA microarray (chip) tiling 3 Kb regions of miR-124, miR-135, miR-141, miR-200, miR-302, miR-338, and miR-429. Three independent sets of ChIPs with c-Myc antibody versus input DNA were hybridized to the custom array. The enrichment ratio for ChIP/input DNA (Figure 2A, grey dots, plotted on a log2 scale in which dotted purple line indicates log2=1) and the statistics program Algorithm for Capturing Microarray Enrichment (ACME) (Scacheri et al, 2006b) were used to identify chromosomal regions with statistically significant c-Myc binding (Figure 2A, red lines, see Material and methods section for details). Figure 2 shows binding by endogenous c-Myc proximal (within 100–200 bp) to coding regions for miR141, miR200/miR429, miR338. When we applied the ChIP–chip analysis to c-Myc+ ES cells we detected significant binding and/or additional binding sites at the miR-141, miR-200, miR-338 loci (Figure 2A). We also observed little or no endogenous c-Myc binding to either miR124 or miR135a whereas increased Myc levels generates binding in the vicinity of these two miR transcripts (Supplementary Figure S4). Interestingly, neither endogenous nor overexpressed c-Myc binds to the miR302 locus (Supplementary Figure S4), suggesting c-Myc indirectly induces miR302. We further validated the binding of Myc to genomic loci proximal to miR-141, miR-200, miR-338, and miR429 by qChIP–PCR using primers to amplify the bound regions defined by our array analysis (Figure 2B). Myc binding to miRNA loci occurred to the same extent as endogenous binding to the Myc target gene HirA (Chen et al, 2008; Kim et al, 2008). Evx2 served as a negative control. In contrast to ES cells, we were unable to detect c-Myc binding to the same miRNA loci in HSPs (data not shown), consistent with our data showing that these miRNAs are not induced by Myc in these differentiated cells (Figure 1B; Supplementary Figure S3B).
Figure 2.
Association of c-Myc with genomic regions proximal to miRNA genes. (A) c-Myc genomic binding was assessed by ChIP–chip analysis. ACME statistical analysis (–log10 P-value scale on left side of each panel), plotted as red dots, indicates statistically significant enrichment. The ACME data are derived from the fold enrichment ratios (log2 scale on the right side of each panel) representing the ChIP enriched versus total input genomic DNA for all probes within the indicated genomic regions and plotted as grey dots. x axis represents the genomic region surrounding the miRNA loci with tiling oligonucleotide probes on a custom-designed tiling array. Positions of the mature miRNA transcripts are indicated by green or orange bars. Arrows indicate direction of transcription. (B) c-Myc binding at microRNA loci. qChIP–PCR was carried out as described in Materials and methods using AK7 mES cells. Shown is endogenous c-Myc binding to the known c-Myc target gene HirA as a positive control. The Evx2 locus was used as a negative control. Analysis of Myc binding to the indicated miRNA genomic loci was carried out using ChIP from wild-type ES cells (endogenous) as well as ES cells overexpressing c-Myc (overexpression). Equal amounts of anti-c-Myc ChIP DNA and total input DNA were used for quantitative PCR using SYBR Green detection with an ABI7900HT system. The sequences of all primers for qChIP–PCR were based on the Myc-binding sites (100–200 bp from microRNA coding regions) identified from ChIP–chip analysis and are available on request. Bar heights represent the Myc-bound DNA as a percentage of the total input DNA as determined from four independent sets of experiments.
Analysis of the targets of ES-specific c-Myc-induced miRNAs
RNAs targeted by miRNAs are generally degraded and can be identified through expression array analysis (Lim et al, 2005; Visvanathan et al, 2007; Liu et al, 2008; Ziegelbauer et al, 2009). Therefore, to assess the extent to which miR-141, miR-200a, miR-429, miR-338-3p, and miR-302a control gene expression in ES cells, we used expression array profiling to identify transcripts regulated by each miRNA. An RNA duplex was designed for each miRNA in which the guide (active) strand matches the mature form of the natural miRNA (see Supplementary Table S2 for sequences). This approach has been used extensively to identify and confirm miRNA targets (Lim et al, 2005; Visvanathan et al, 2007). Duplexes were transfected into ES cells, and RNA was collected 12 or 24 h later. The miRNA-regulated genes were identified using oligonucleotide microarrays representing 21 000 genes. Transcripts regulated by the miRNA were identified as those whose expression was significantly decreased with a P-value ⩽0.01 relative to mock-transfected cells (see Materials and methods). Two independent experiments were performed, one including both 12 and 24 h time points after transfection, and one analysing a 24 h time point. The 12 h sample was included as an early time point to show trends in kinetic regulation of transcripts and aid in identification of direct targets. Figure 3 shows a heat map for transcripts regulated by miR-141, miR-200, miR-302, miR-338, and miR-429. We observed good agreement among the transcripts regulated in the individual 24 h samples by miR-141 and miR-200. The duplexes corresponding to miR-141 and miR-200 had overlapping but non-identical seed regions (see Supplementary Table S2 for sequences). Consistent with this, the signatures for these miRNAs were partially overlapping, that is some transcripts were regulated in common by both miRNAs, whereas some transcripts were regulated only by the individual miRNAs. miR-429, with a seed region unrelated to miR-141 or miR-200, regulated an independent set of transcripts. The transcripts showing significant downregulation for each miRNA were enriched in 3′ UTR sequences complementary to the seed region of the transfected miRNA (Figure 3). The signatures for each miRNA target set showed enrichment for all three hexamers corresponding to the seed region of that miRNA. Hexamer enrichment ranged from an E-value (P-value with Bonferroni correction) of 10−2–10−13. Thus, the microarray analysis identified transcripts likely to be direct targets of the miRNAs. Similar data were obtained for miR-338 and miR-302 (see Supplementary Table S3).
Figure 3.
Expression profiling identifies putative targets of c-Myc-induced miRNAs. Heatmap of transcript levels after transfection of ES cells with miR-141, miR-200, miR-338, miR-302, or miR-429. ES cell RNA was hybridized to Agilent microarrays containing oligonucleotides corresponding to approximately 21 000 genes. Transcript regulation was calculated as the error-weighted mean log10 ratio for each transcript across the fluor-reversed pair. Inset shows colour scale indicating change in expression level. The white box(es) highlights transcripts downregulated (P-value <0.01 relative to mock-transfected cells) in common across time points and replicates (24 h time point) for the same miRNA. The seed region hexamers enriched in the signature are indicated for each miRNA (gold lettering). Hexamers are listed from top to bottom for each miRNA as hexamer 1 (positions 1–6), hexamer 2 (positions 2–7), and hexamer 3 (positions 3–8). Samples marked ‘a' and ‘b' represent independent experiments. The dendrogram at the top of the panel clusters transcripts with similar expression patterns.
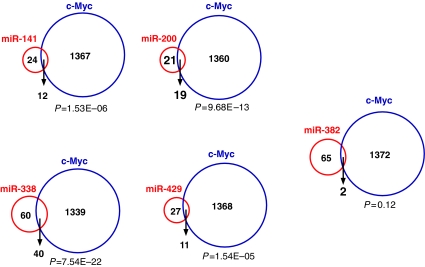
We reasoned that if the miRNAs identified here are part of a c-Myc-regulated pathway in ES cells, then the transcripts downregulated by the miRNAs should overlap with transcripts downregulated by c-Myc overexpression. We therefore looked for genes whose expression is downregulated by both miRNA transfection and in response to c-Myc overexpression (Figure 4). Myc-regulated genes in ES cells were identified using the 21 000 gene oligonucleotide microarray as described above. Approximately one-third of the targets for each miRNA overlapped with c-Myc-regulated transcripts (Figure 4). As a control we examined miR-382, an miRNA that has not been found to be regulated by c-Myc, and determined that only 2 of 65 miR-382 repressed targets overlap with genes downregulated by c-Myc, a number lacking statistical significance (P=0.12) (Figure 4).
Figure 4.
The intersection of putative targets of c-Myc-induced miRNAs and targets regulated by c-Myc. Venn diagrams illustrating the overlap among genes downregulated by c-Myc and by each miRNA tested. The numbers in the circles represent the number of genes significantly regulated (P<0.01) by c-Myc and by the miRNAs indicated. The hypergeometric probabilities for the overlaps are given below each Venn diagram. miR-382 is a control miRNA not induced by c-Myc in ES cells.
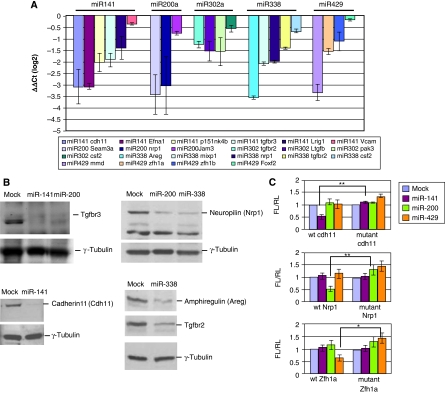
Among the genes significantly repressed by both c-Myc and the c-Myc-induced miRNAs, a subset has been implicated in growth arrest and differentiation of a number of cell types (Supplementary Table S3; see Discussion). To validate the expression profiling results, we employed qRT–PCR to assess expression of 20 potential targets 24 h after transfection of duplex miRNAs into ES cells (Figure 5A). We confirmed by northern blotting that the levels of introduced miRNAs are comparable to those following induction by c-Myc (see Supplementary Figure S5). Our results show that introduction of these miRNAs into ES cells is associated with decreased transcript levels for the predicted miRNA targets. Furthermore, using immunoblotting we found that the protein levels of several target genes, such as amphiregulin (Areg), Neuropilin 1 (Nrp1), cadherin 11 (Cdh11), and TGFβ receptor 3 (Tgfbr3), were sharply decreased at 48 h after miRNA transfection (Figure 5B).
Figure 5.
Target gene specificity of Myc-induced miRNAs. (A) The transcript levels of the indicated microRNA targets were validated by real-time PCR. RNA extracted 24 h after transfection of miRNA duplexes into ES cells was used to determine expression of target genes by real-time PCR. Each bar represents the average ΔΔCt on a log2 scale of triplicate sets of experiments after normalization to internal controls (ΔCt) and mock control (ΔΔCt). Error bars show standard error of the mean. (B) Total protein extracted from ES cells 48 h post transfection of miRNA duplexes was analysed by immunoblotting with commercially available antibodies. Mock-transfected cells were used as controls and γ-tubulin as loading control. (C) microRNA target specificity determined by dual luciferase reporter assay. Sequences complementary to the seed region of miR-141, miR-200, and miR-429 were present in the 3′ UTRs of Cdh11, Nrp1, and zfh1, respectively. To disrupt base pairing with miRNAs we mutated the 3′ UTR complementary seed sequences from ‘AACACT' to ‘ACAACT' for the 3′ UTR of Cdh11, from ‘AACAC' to ‘AGCGC' for Nrp1, and from ‘TAATAC' to ‘TCGTAC' for Zfh1a. The wild type or mutant 3′ UTRs derived from the indicated target genes were linked to luciferase and transfected into HeLa cells together with the indicated miRNAs. The fold change represents the ratio of firefly luciferase (FL)/Renilla luciferase (RL). Error bars represent standard error from four independent experiments. **P-value=0.002–0.008; *P-value=0.03.
To determine whether miRNA target downregulation is mediated through recognition of the miRNA seed sequences, we cloned 3′ UTRs from Cdh11, Nrp1, and the zinc-finger homeobox gene (Zfh1a), which are target genes of miR-141, miR-200, and miR-429, respectively (Figure 5C). These 3′ UTRs contain seed homology sequences specific for each miRNA. We next made point mutations by site-directed mutagenesis within the 3′ UTR seed homology elements to test the miRNA target site specificity. We generated reporter constructs containing the wild type and mutant target 3′ UTR sequences linked to luciferase. The wild type or mutant reporters were co-transfected with individual miRNA duplexes into HeLa cells. Data were normalized to a co-transfected Renilla luciferase (RL) control. Figure 5C shows that miR-141, miR-200, and miR-429 specifically inhibit activity from their cognate wildt-ype reporter genes whereas mutagenesis of the miRNA recognition sites abolishes the inhibition by these miRNAs.
As a further test of specificity we determined the effect of each miRNA duplex on endogenous Cdh11, Nrp1, and Zfh1a RNA levels by qRT–PCR (Supplementary Figure S6). We compared miRNA duplexes with wild-type and scrambled seed regions (Supplementary Figure S6A). In each case, downregulation of the specific target gene for a given miRNA is dependent on the miRNA having an intact seed region homologous to the target gene 3′ UTR. We also used LNA-anti-miRNAs (see below) along with scrambled controls to examine target gene expression. As shown in Supplementary Figure S6B, upregulation of RNA expression of the endogenous target gene is dependent on a wild-type seed region. We further extended these experiments by examining target gene protein levels. After introduction of c-Myc into ES cells we observe decreased levels of Nrp1, Cdh11, and Areg proteins, as determined by immunoblotting (Supplementary Figure S6C). However, treatment of the c-Myc-overexpressing cells with LNA-anti-miRNAs, directed against the endogenous Myc-induced miRNAs that specifically target these gene products, results in upregulation of their protein levels (Supplementary Figure S6C). Taken together, our transient reporter assays and determination of endogenous target gene levels support the idea that regulation of the miRNA target genes defined by our expression profiling experiments is specific and that Myc-mediated silencing of at least a subset of these targets is dependent on Myc-induced miRNAs.
c-Myc-induced miRNAs influence ES cell pluripotency
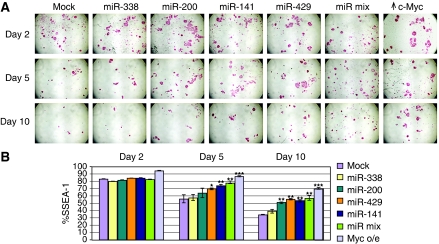
Our finding that c-Myc-regulated miRNAs miR-141, miR-200, miR-338, and miR-429 are expressed in ES cells and that a subset of their target genes is associated with lineage differentiation raises the question of whether these miRNAs affect the balance of ES self-renewal and differentiation. Addition of LIF to the culture medium maintains ES cells in a self-renewing pluripotent state whereas withdrawal of LIF is a widely used method to induce ES cell differentiation (Solter and Knowles, 1978; Houbaviy et al, 2003; Cartwright et al, 2005; Chen et al, 2007). LIF removal permits spontaneous, non-biased differentiation into multiple cell types including cardiac myocytes, striated skeletal muscle, neuronal cells, and haematopoietic progenitors. In ES cells, LIF has been shown to stimulate c-myc expression whereas c-Myc abundance sharply decreases on LIF removal (Cartwright et al, 2005). To evaluate whether enforced expression of miR-141, miR-200, miR-338, and miR-429 is involved in ES cell identity we therefore tested the ability of ES cells transduced with these miRNAs to differentiate subsequent to LIF withdrawal. ES cell pluripotency was evaluated over time after LIF removal (i) visually by staining for alkaline phosphatase (AP) activity (Cartwright et al, 2005) (Figure 6A) as well as by (ii) quantitative changes in the population of SSEA-1-positive stem cells (Solter and Knowles, 1978) (Figure 6B). The flow cytometric measurements of SSEA represent 10 000 gated cells for each time point. Figure 6A and B show decreases in both AP and SSEA-1-positive staining in wild-type mES cells between 2 and 10 days after LIF withdrawal whereas parallel cultures maintained in LIF have no evident change in these markers (see Figure 8B). The reduction in AP and SSEA-1 staining is partially abrogated by the introduction of wild-type c-Myc (Figure 6A, far right panel and Figure 6B). Transduction of miRNAs miR-141, miR-200, and miR-429 each results in levels of AP staining over 5 and 10 days that are well above those observed for mock-treated ES cells but less than that detected in c-Myc+ ES cells (Figure 6A). In parallel experiments, we tested miRNA duplexes with scrambled seed regions and found these to be considerably less effective than the wild-type miRNA duplexes in attenuating loss of AP staining (Supplementary Figure S7A). Moreover, the percentage of SSEA-positive cells was significantly higher on transduction of these individual miRNAs when compared with mock-transfected cells, although, again, the effect was not as pronounced as with c-myc overexpression (Figure 6B). We also found that combining miRNAs-141, -200, -338, and -429 (miR-mix) effectively attenuated loss of AP staining on LIF withdrawal (Figure 6A and B). miR-338 and miR-302 were relatively ineffective in maintaining AP as well as SSEA staining. It is important to note that the changes in AP and SSEA are not because of alterations in cell proliferation or cell numbers. After LIF removal, self-renewal ceases and the individual colonies that lose AP staining retain approximately the same number of cells (see Supplementary Figure S7B). Furthermore, we have determined directly that the Myc-induced miRNAs do not affect ES cell population doubling time (Supplementary Figure S7C) or proliferation as assessed by MTT and BrdU-labelling assays (Wang et al, 2007) (data not shown).
Figure 6.
c-Myc-induced miRNAs affect the balance between ES cell self-renewal and differentiation. ES cells were either mock transfected or transfected with the indicated miRNAs and/or c-myc. Twenty-four hours after transfection, LIF was withdrawn from the medium. miR-mix indicates transfection of a mixture of miR-141, -200, -338, -429. (A) Staining for alkaline phosphatase activity at the indicated times after LIF elimination. A single well is shown for each time point. (B) Flow cytometric determination of SSEA-1 staining at the indicated times after LIF elimination for ES cells into which miRNAs or constructs were introduced as indicated at right. The %SSEA indicates the fraction stained relative to mES cells in the presence of LIF. 10 000 gated cells were used for each dataset. *P-value<0.05, **P-value<0.01, ***P-value <0.001.
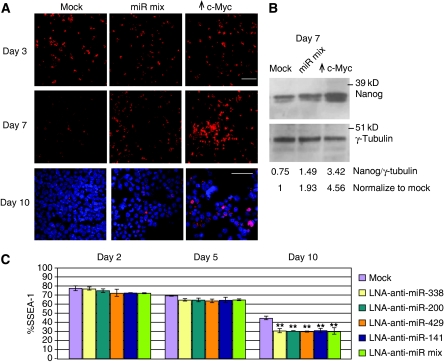
We also examined expression of the pluripotency factor Nanog by immunostaining after the introduction of the miR-mix or c-Myc at 3 and 7 days after LIF removal (Figure 7A). The miR-mix ES cultures show higher levels of Nanog than mock-treated controls although the Nanog levels are highest in the c-Myc-overexpressing cultures at day7 after LIF withdrawal. To provide a more quantitative analysis, we used immunoblotting with anti-Nanog and found that miR-mix and c-Myc-overexpressing cells had approximately two-fold and five-fold increased Nanog levels, respectively, compared with mock controls (Figure 7B). Although Nanog expression decreased makedly at day 10 after LIF withdrawal, the level of Nanog is still highest in c-Myc-overexpressing cells (Figure 7A, DAPI staining showed cells are present in the image). Therefore, our assessment of ES cell markers, AP, SSEA, and Nanog, indicates that the c-Myc-induced miRNAs act to attenuate ES cell differentiation on LIF removal.
Figure 7.
c-Myc-induced miRNAs affect the expression of pluripotency marker and differentiation. (A) Staining for the pluripotency marker Nanog at days 3 and 7 after LIF elimination. Each image was taken from a single well. Staining of Nanog and DAPI at day 10 after LIF removal shows cells within the wells are heterogeneous with respect to Nanog expression. Bar indicates 50 μm. (B) Nuclear extracts from ES cells at 7 day after LIF removal were electrophoresed and subjected to immunoblotting with anti-Nanog. γ-tubulin served as a loading control. Numbers represent ratios of Nanog:tubulin. (C) Flow cytometric determination of SSEA-1 staining at day 2 (left set), day 5 (middle set), and day 10 (right set) after LIF removal from ES cells into which LNA-anti-miRNAs were introduced as indicated at right. The %SSEA indicates the fraction stained relative to mES cells in the presence of LIF. **P-value<0.01.
To further assess the specificity of the observed miRNA biological activity we partially inhibited endogenous miRNA activity in ES cells using locked-nucleic acid anti-miRNAs (LNA-anti-miRNA; see Materials and methods). Representative LNA-anti-miRNA knockdowns of the c-Myc-induced miRNAs are shown in the northern blots in Supplementary Figure S8. We had earlier shown the seed specificity of these anti-miRNAs on several miRNA endogenous target genes (see Supplementary Figure S6) as well as their ability to reverse Myc-induced target repression (Supplementary Figure S6C). Introduction of LNA-anti-miRNAs against individual or mixed c-Myc-induced miRNAs resulted in a modest but significant acceleration of differentiation as measured by diminished SSEA and AP activity relative to mock-treated cells, most strongly at day 10 after LIF removal (Figure 7C; data not shown). We detected markers for both neuronal (TUJ1) and muscle (MF20) differentiation (data not shown) indicating that differentiation occurs along multiple lineages. Our data show that the overexpression of the c-Myc-induced miRNAs has the opposite effect as the LNA-anti-miRNAs on SSEA levels after LIF removal relative to the mock controls in each experiment (compare Figures 6B and 7C).
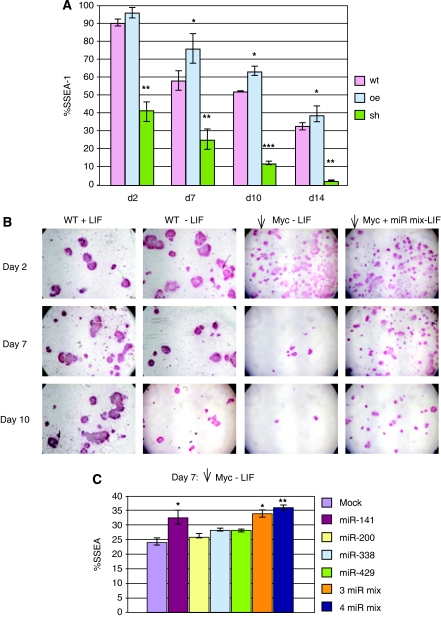
An earlier study had shown that an oncogenically activating point mutation in c-myc led to ES cell self-renewal in the absence of LIF and, further, that a putative dominant-negative mutant of c-Myc inhibited self-renewal and promoted differentiation (Cartwright et al, 2005). Here, we have used an shRNA against c-myc to directly decrease c-Myc levels and observed that reduction in c-Myc abundance enhances loss of both SSEA and AP markers as well as ES cell colony morphology after LIF withdrawal (Figure 8A and B; Supplementary Figure S10) in general agreement with the earlier study. We therefore assessed whether the c-Myc-induced miRNAs affect the rate of differentiation after c-myc knockdown. We introduced a mixture of miR-141, miR-200, miR-338, and miR-429 into ES cells that were treated with shRNA against c-myc. We then removed LIF and assessed loss of pluripotency markers. We find that the miRNAs reversed the loss of AP activity observed in c-myc knockdown ES cells over a 10-day period after LIF withdrawal (Figure 8B). Determination of SSEA staining at an intermediate time point (day 7 after LIF removal) also shows that an miRNA mixture, or miR-141 alone, significantly retards loss of the SSEA marker compared with mock-treated c-myc knockdown ES cells (Figure 8C). Nanog levels were also increased by the miR-mix as well as by individual miRNAs (Supplementary Figure S9). Our results indicate that co-transfection of Myc-induced miRNAs attenuates ES cells differentiation under conditions of LIF removal and c-Myc depletion.
Figure 8.
Myc-induced miRNAs attenuate differentiation of c-Myc-depleted ES cells on LIF withdrawal. (A) The percentage of cells staining positive for SSEA-1 at the indicated time points was quantitated by flow cytometry after removal of LIF from mES cells. Before LIF withdrawal mES cells were infected with empty lentiviral vector alone (red bars); with lentiviral vector expressing c-myc (blue bars); or lentiviral vector expressing shRNA against c-myc (green bars). Data are expressed as the percentage of SSEA-1-positive mES cells in the absence of LIF relative to mES cells in the presence of LIF. *P-value<0.05, **P-value<0.01, ***P-value <0.001. (B) mES cells were infected with empty lentiviral vector (WT) or lentiviral vector expressing shRNA against c-myc. After selection, the c-Myc knockdown ES cells (see A above) were either electroporated with a mixture of miRNAs (miR-141, -200, -338, -429) or electroporated without addition of miRNAs. Twenty-four hours after transfection, LIF was withdrawn from the medium. Left panel shows WT ES cells maintained in LIF for the time course of the experiment. A single well is shown for each time point. (C) SSEA-1 flow cytometry 7 days after LIF withdrawal from c-myc knockdown ES cells transfected as indicated in the inset. *P-value<0.05, **P-value<0.01, ***P-value <0.001.
Discussion
In this study we provide evidence that, in mES cells, introduction of c-Myc induces miRNAs whose targets include genes involved in differentiation. We focused on a subset of these miRNAs that are upregulated by c-Myc in ES cells but not in the differentiated and tumourigenic derivatives of these ES cells, even though they continue to express ectopic Myc. Although the levels of these miRNAs are increased by Myc overexpression, they are nonetheless expressed endogenously in ES cells, and knockdown of endogenous c-Myc reduces their expression.
We decided to concentrate on miR-141, miR-429, miR-200, miR-338, and miR-302, a subset of the miRNAs identified in our screen, as these had neither been characterized earlier nor been linked to Myc function. We established that both endogenous and overexpressed c-Myc protein binds to genomic DNA in the vicinity of the coding sequences of all these miRNAs with the exception of miR-302. However, we have not been able to detect potential E-box-related Myc–Max-binding sites associated with the Myc-bound regions, suggesting that a non-canonical mode of Myc binding occurs at these sites. E-box-independent Myc association with DNA has been reported earlier (Eilers and Eisenman, 2008) and further experiments will be required to delineate the nature of Myc binding to these miRNA encoding regions. As the promoter and regulatory regions of these miRNAs have not yet been analysed, we cannot definitively determine whether such binding by Myc directly results in transcriptional activation of these miRNA genes. Nonetheless, the induction of these miRNA genes by Myc, coupled with our finding of dosage-dependent Myc binding near their coding regions provides circumstantial evidence that they are directly targeted by Myc. Moreover, we show, by means of expression profiling using oligonucleotide microarrays, that gene sets downregulated by our identified miRNAs (and that possess their cognate seed-binding regions) display highly significant overlap with genes downregulated by c-Myc in mES cells. The fact that this overlap is only partial may be because of the inability of introduced c-Myc to maintain high levels of these miRNAs attained after duplex transfection. Furthermore, we show that the specific downregulation depends on the seed sequences of the miRNAs and on the miRNA response elements complementary to these seed sequences that are located within the 3′ UTRs of target genes. Finally, we show that specific LNA-anti-miRNAs result in seed-dependent upregulation of the cognate miRNA target genes. Taken together, these data suggest that c-Myc downregulates the genes identified in the microarray analysis through the c-Myc-induced mES cell-specific miRNAs we have defined here.
To determine whether the Myc–miRNA pathway is functional in mES cells we assessed the effects of introducing Myc-regulated miRNAs on ES cell differentiation. Importantly, we show that overexpressed wild-type c-Myc itself delays but does not completely block differentiation. Expression of miR-141 and miR-429 and a mixture of miRNAs (comprising miR-141, -200, -338, -429) were most effective in retarding loss of AP activity, SSEA staining, and Nanog expression in mES cells after LIF removal. In contrast, introduction of the corresponding LNA-anti-miRNAs facilitated differentiation. Moreover, knockdown of endogenous c-Myc strongly accelerates differentiation, an effect that is significantly reversed by introduction of the miRNA mixture. These data indicate that c-Myc levels control the rate of ES cell differentiation and that c-Myc-regulated miRNAs can function in this process. We note that in our experiments the myc-induced miRNAs attenuated, but did not entirely block, the loss of ES cell pluripotency markers. We surmise that these miRNAs represent only a part of the process that results in ES cell differentiation and act cooperatively or redundantly with other factors. Indeed, we have found that many of our miRNA target genes have been reported earlier to be bound by Polycomb group proteins (C-HL and RNE, unpublished). In addition, because of the limited nature of our miRNA screen and the use of RNA expression profiling rather than proteomics (which has not been effectively used with ES cells) we have most likely not identified all Myc-regulated miRNAs in ES cells.
One mechanism through which Myc-regulated miRNAs can influence ES cell pluripotency and attenuate differentiation is through the inhibition of differentiation-specific genes. Among the c-Myc/miRNA targets we found that a subset has been implicated in differentiation. These include the Tgfbr3 (miR-141 and miR-200) involved in multiple developmental pathways, and Cdh11 (miR-141). Cdh11 is an adhesion molecule that has an important role in developmental processes such as morphogenesis and induction of neurite outgrowth (Taniguchi et al, 2006; Boscher and Mege, 2008). Another Myc and miRNA downregulated target is Nrp1 (miR-200 and miR-338) a co-receptor for class 3 semaphorins with key roles in axonal guidance. Nrp1 is essential for neuronal and cardiovascular development and controls axon guidance through neural crest cells (Mizui and Kikutani, 2008; Pellet-Many et al, 2008; Schwarz et al, 2008). Interestingly, one of the Nrp1 interacting semaphorins (Sema3a) is also targeted by c-Myc through miR-200. Another overlapping target is Foxf2, a mesenchymal factor that controls epithelial proliferation and survival during murine gut development (Ormestad et al, 2004, 2006). Foxf2 is also expressed in the neural crest, CNS, and limb mesenchyme (Wang et al, 2003). Importantly, in independent experiments we have confirmed that these and other Myc/miRNA targets are downregulated at the protein and/or RNA level. Therefore, Myc-induced miRNAs in ES cells may contribute to inhibition of broad aspects of cell-type differentiation during development.
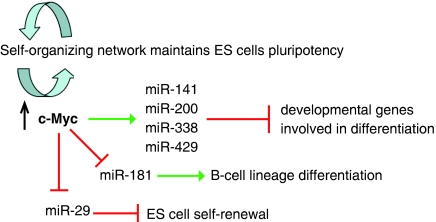
It has been suggested that ES cells have established a self-organizing network to maintain self-renewal and pluripotency (Bernstein et al, 2006; Boyer et al, 2006; Lee et al, 2006; Tay et al, 2008). Accumulating evidence indicates that Myc family proteins may contribute to this network as they are involved in the development of pluripotency and the control of ES cell self-renewal and differentiation. A great deal of earlier work has shown that differentiation-specific genes in ES cells are repressed but maintained in a poised state for transcriptional activation through the binding of Polycomb complexes (Azuara et al, 2006; Bernstein et al, 2006; Boyer et al, 2006; Bracken et al, 2006; Lee et al, 2006). The data presented in this paper show that at least a part of Myc's role in pluripotency lies in the inhibition of differentiation-specific genes in ES cells through modulation of the expression of a group of miRNAs (Figure 9). These miRNAs may reinforce repression of Polycomb-bound genes as well as act on an entirely different set of differentiation targets. In conclusion, we have characterized subsets of miRNAs that are regulated by c-Myc and are important in ES cell identity. Further parsing of these miRNAs and their targets may provide a means to drive ES cell differentiation along specific lineages and to facilitate conversion of somatic cells into iPS cells.
Figure 9.
c-Myc is involved in ES cell pluripotency through regulation of miRNAs. The figure summarizes a putative pathway through which c-Myc regulates groups of miRNAs in ES cells that in turn control genes involved in differentiation and self-renewal.
Materials and methods
Cell culture
ES cell lines R1 and AK7 were cultured in DMEM supplied with 10%FBS, L-glutamine, non-essential amino acid, sodium pyruvate, LIF, and β-mercaptoethanol. ES cells were thawed on plates with mitomycin C-inactivated mouse embryonic fibroblasts and then grown and expanded on 0.1% gelatin-coated plates for subsequent experiments.
Manipulating c-Myc level in ES cells by lentiviral-delivered overexpression
c-Myc cDNA was amplified from an ES-derived cDNA library and cloned into lentiviral vector pLenti6/V5-DEST (Invitrogen) for overexpression. Lentiviral production followed an earlier published protocol (Lois et al, 2002). Three days subsequent to viral infection of ES cells blasticidin was used to select for cells with integrated lentiviruses. Positive clones were picked and expanded for RNA extraction and for miRNA and gene expression profiling experiments. In parallel experiments, c-Myc+ ES cells were induced along haematopoietic lineages using an earlier established protocol (Burt et al, 2004) and then transplanted into irradiated (800 cGy) Balb/cJ mice and (1000 cGy) C57/BL6 by intra-bone marrow injection (Kushida et al, 2001). To identify ES-specific c-Myc-induced miRNAs, RNA obtained from ES cells, induced haematopoietic cells, and tumours from transplanted mice were used for miRNA profiling. This work was approved by FHCRC IACUC protocol 1195.
Quantitation of miRNA levels
Quantitative PCR: miRNA levels were determined using a quantitative primer-extension PCR assay (Raymond et al, 2005). Ct values were converted to copy numbers by comparison to standard curves generated using single-stranded mature miRNAs and are expressed as copies/10 pg (approximately equivalent to copies/cell). The fold change in miRNA copy number was then calculated as the error-weighted average of the ratio of c-Myc-overexpressing cells relative to their corresponding empty lentiviral vector controls. miRNAs showing copy numbers less than 1 or below the lower level of detection for the miRNA assay were not used for further analysis. For miRNA northern blotting, 20 mg total RNA was applied on 12% polyacrylamide (19:1)/8 M urea/1 × TBE gel. Subsequent to transfer, the membrane was probed with γ-P32-ATP end labelled oligonucleotide corresponding to the miRNA. A U6 probe was used as an internal loading control. Band intensity was measured using a Typhoon phosphoimager and ImageQuant program.
miRNA duplex and LNA-anti-miRNA transfection in ES cells
miR-141, miR-200, miR338, or miR-429 duplexes (Sigma) were transfected into ES cells at a final concentration of 100 nM by DharmaFECT1 (Dharmacon) transfection reagent following the manufacturer's instructions. DharmaFECT1 transfection reagent alone was used as a mock control. Total RNA was isolated at 12 and 24 h post transfection. Transfection of mixed miRNAs duplex and LNA-anti-miRNAs (50 nM, obtained from Exiqon) into ES cells was carried out by electroporation with Gene Pulser (Bio-Rad) at a constant voltage of 240 mV, 500 mF. The sequences of miRNA duplexes are listed in Supplementary Table S2. As control, the scrambled miRNA duplexes were designed with two nucleotides substitutions at the seed sequences of guide strand: AACACU to ACAACU (miR-141), UAACAC to UAGCGC (miR200a), and UAAUAC to UCGUAC (miR-429). The LNA-anti-miRNAs were purchased from Exiqon and the sequences of wild-type LNA-anti-miRNAs were listed in Supplementary Table S2. A similar design using two nucleotides substitutions at the seed sequences was achieved for scrambled LNA-anti-miRNAs: AGTGTT to AGTACT (LNA-anti-miR141), AGTGTT to AGGGGT (LNA-anti-miR200a), and GTATTA to GTAGCA (LNA-anti-miR429).
Microarray analysis
Total RNA was extracted by TRIzol reagent (Invitrogen), purified by Qiagen RNeasy kit, and processed for hybridization to custom Agilent microarrays containing oligonucleotides corresponding to approximately 21 000 genes. Hybridizations were performed with fluorescent label reversal to eliminate dye bias. Data shown are signature genes that display a difference in expression level (P<0.01) relative to mock-transfected cells. Data were analysed using Rosetta Resolver software. Transcript regulation was calculated as the error-weighted mean log10 ratio for each transcript across the fluor-reversed pair. To remove a common signature and show miRNA-specific signatures, the expression signatures for each treatment have been ratioed to the average of all treatments in the corresponding experiment. Microarray data are available at NCBI's Gene Expression Omnibus, Series GSE13127). miRNA target candidates were defined as those transcripts showing significant downregulation in miRNA-treated cells versus mock-treated cells (log10 ratio <0 and P-value <0.01). miRNA target candidates identified by microarray were examined for sequence complementarity to the seed region of the miRNA. Each individual hexamer within the seed region octamer (bases 1–8) was examined. miRNA target candidates were validated by RT–PCR. Briefly, cDNA was synthesized by SuperScript II kit (Invitrogen) according to the manufacturer's instructions. The transcript expression level was measured by real-time PCR with SYBR Green detection on ABI7900HT system (Applied Biosystems). S16 was used as internal control for normalization (ΔCt). The expression level (ΔΔCt) was measured by subtraction of ΔCt (derived from overexpressing cells) from the ΔCt (derived from the vector control). Primers for real-time PCR are available on request. Transcripts downregulated in MYC-overexpressing cells were determined as above, relative to empty lentiviral vector-infected cells.
Western blotting
Total protein was extracted in RIPA buffer 48 h post transfection of individual miR-141, miR-200, or miR-338 duplexes was examined whether the protein level of miRNA targets also decreased. All antibodies for western blot were obtained from Santa Cruz Biotechnology Inc and used according to the manufacturer's instructions. Subsequent to selection of ES cell colonies overexpressing c-Myc as described above, LNA-anti-miRNAs from Exiqon were introduced into these cells, incubated for 4 days, and total protein after extraction in RIPA buffer was subjected to western blot analysis.
Manipulating c-Myc level in ES cells by lentiviral-delivered shRNA knockdown
shRNA sequences against c-Myc was designed by using Dharmacon siDESIGN and then hairpin siRNA oligonucleotides were annealed and cloned into modified FUGW lentiviral vector with an H1 promoter (Lien et al, 2008)). Lentiviral production followed an earlier published protocol (Lois et al, 2002), then drug selection for positive integration in ES cells 3 days after infection. Positive clones were picked and expanded for differentiation experiments.
miRNA target specificity by dual luciferase reporter assay
The 3′ UTR of Cdh11, Nrp1, and Zfh1a were amplified by PCR from ES cells cDNA and cloned into pGL3-control vector (Promega) at the 3′ end of the firefly luciferase (FL) gene and then transformed into dam+ Escherichia coli strain. Point mutations within each putative miRNA target site were made by PCR-based site-directed mutagenesis. HeLa cells were plated at 104 cells per well in 24-well plates on the day before transfection. Subsequently, 100 nM of miRNA duplexes, 500 ng of FL expression construct carrying either wild-type or mutated 3′ UTR, and 125 ng of RL expression vector (pRL-TK, Promega) were co-transfected into cells by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Forty hours later, a luciferase assay was performed using the ‘Dual Luciferase Reporter Assay System' (Promega). The experiments were carried out in four independent replicates.
FACS analysis
Individual miRNA-duplex-transfected ES cells were cultured on 0.1% gelatin-coated plates. At days 2, 5, 7, or 10 after LIF withdrawal the cells were collected using trypsin and washed with PBS before SSEA-1 antibody incubation (Chemicon, mouse monoclonal 1:200 dilution) for 2 h. Cells were subsequently washed with PBS twice and incubated with PE-conjugated secondary antibody. Finally, cells were washed twice and resuspended in PBS before FACS analysis. The population (%) of positive SSEA-1-stained cells was compared with unstained cells, PE-isotype control, undifferentiated ES cells, and cells stained with secondary antibody alone. The two-sample t-test was applied to determine whether the average difference between miRNA transfection and mock control was significant.
Analysis of AP activity
Individual miRNA or combined miRNA-transfected ES cells were cultured on 0.1% gelatin-coated chamber slides and cells at days 2, 5, 7, or 10 after LIF withdrawal were fixed with 4% paraformaldehyde for 2 min before AP activity assay (Chemicon, Alkaline phosphatase detection kit).
Nanog immunocytochemistry
ES cells cultured on 0.1% gelatin-coated chamber slides were mock transfected or transfected with miRNA duplexes. At day 5 after LIF withdrawal, cells were fixed with 4% paraformaldehyde for 10 min before blocking with 10% goat serum in 1 × PBS/0.5%Triton X-100. Subsequently, Nanog antibody (Abcam:ab21603-100) 1:500 dilution in 1 × PBS/0.5%Triton X-100 was used for staining overnight. The AlexaFlur 594-conjugated secondary was used followed by three washes with 1 × PBS/0.5%Triton X-100. Images were taken using the Zeiss 510 confocal microscope.
ChIP and DNA microarrays (ChIP–chip)
Cells were crosslinked with 1.1% formaldehyde and then blocked with 0.125 M glycine before lysis and sonication to produce 0.2–1 kb DNA fragments. Protein-A-agarose beads (50% slurry) were blocked with 2 μg/ml sonicated salmon sperm DNA and 0.5 mg/ml BSA and washed twice in lysis/sonication buffer (50 mM Tris, 10 mM EDTA, 1%SDS, pH=8) before use. Chromatin was pre-cleared with control IgG before immunoprecipitation overnight with c-Myc rabbit polyclonal antibody (Santa Cruz, #SC764, Lot:F1307). ChIP and amplification (ligation-mediated PCR) for ChIP–chip analysis followed NimbleGen protocol. Custom-designed DNA microarrays including miR124, miR135a, miR135b, miR-141, miR-200, miR302, miR338, and miR-429 were tiled through 3 kb upstream and downstream of miRNA transcripts with isothermal probes. For endogenous Myc binding, triplicate sets of ChIP and input DNA were applied on this custom array and genomic sites enriched for c-Myc binding were calculated by ChIP versus input (log2 ratio provided by NimbleGen data output). We used both log2 ratios from ChIP/input normalization and P-values from computer program ACME (window=500 nucleotides, threshold=0.95) (Scacheri et al, 2006a, 2006b) to identify promoter regions significantly enriched for c-Myc binding (P<0/0001). c-Myc overexpression in ES cells by lentiviral delivery was performed as described earlier and triplicate sets of ChIP and input DNA were hybridized on this custom array. The ChIP–chip enrichment ratio was calculated and analysed as described above for endogenous Myc binding. To validate the ChIP–chip data, 1 ng input and ChIP DNA from independent sets of ChIP were applied to quantitative real-time PCR (qRT–PCR) by using SYBR green PCR mix (Applied Biosystems) on ABI7900HT detection system. The enrichment was calculated as % input as described (Frank et al, 2001). We evaluated enrichment from duplicates of two independent ChIP assays obtained from both the R1 and AK7 ES cell lines. The sequences of all primers for qChIP–PCR were based on the Myc binding (100–200 bp to miRNA coding regions) identified from ChIP–chip analysis (sequences are available on request).
Supplementary Material
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table Captions
Supplementary Information
Review Process File
Acknowledgments
We are grateful to Philippe Soriano, Valera Vasioukhin, and Andras Nagy for discussions and reagents. We thank Stephen Tapscott and Patrick Carroll for critical readings of the manuscript and Dan Kuppers for advice. We thank the Rosetta Gene Expression Laboratory for microarray hybridizations and microRNA quantitation assays. This work was supported by Grant RO1CA20525 from the National Cancer Institute/NIH to RNE.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adhikary S, Eilers M (2005) Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 [DOI] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, Larsson LG, Wright AP (2005) c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 7: 303–310 [DOI] [PubMed] [Google Scholar]
- Arnold I, Watt FM (2001) c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol 11: 558–568 [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boscher C, Mege RM (2008) Cadherin-11 interacts with the FGF receptor and induces neurite outgrowth through associated downstream signalling. Cell Signal 20: 1061–1072 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt RK, Verda L, Kim DA, Oyama Y, Luo K, Link C (2004) Embryonic stem cells as an alternate marrow donor source: engraftment without graft-versus-host disease. J Exp Med 199: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA (2007) RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci USA 104: 18097–18102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866 [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH (2007) A functional study of miR-124 in the developing neural tube. Genes Dev 21: 531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301: 336–338 [DOI] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S (2005) LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 132: 885–896 [DOI] [PubMed] [Google Scholar]
- Chang TC, Mendell JT (2007) microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet 8: 215–239 [DOI] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT (2008) Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon D, Lee CT, Blake J, Sun Y, Strauss WM (2007) Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm Genome 18: 316–327 [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Cole MD, Nikiforov MA (2006) Transcriptional activation by the Myc oncoprotein. Curr Top Microbiol Immunol 302: 33–50 [DOI] [PubMed] [Google Scholar]
- Dubois NC, Adolphe C, Ehninger A, Wang RA, Robertson EJ, Trumpp A (2008) Placental rescue reveals a sole requirement for c-Myc in embryonic erythroblast survival and hematopoietic stem cell function. Development 135: 2455–2465 [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM, Novina CD, Sharp PA (2003) Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol 4: 457–467 [DOI] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN (2008) Myc's broad reach. Genes Dev 22: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269 [DOI] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B (2001) Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 15: 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV (2009) c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman N, Grandori C, Eisenman RN, White RJ (2003) Direct activation of RNA polymerase III transcription by c-Myc. Nature 421: 290–294 [DOI] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ (2005) c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Moazed D (2003) Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ (2007) A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA (2003) Embryonic stem cell-specific MicroRNAs. Dev Cell 5: 351–358 [DOI] [PubMed] [Google Scholar]
- Hwang HW, Mendell JT (2007) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 96 (Suppl): R40–R44 [PubMed] [Google Scholar]
- Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ (2007) A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev 21: 3238–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R (2009) Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27: 459–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Slack FJ (2008) microRNAs: small molecules with big roles—C. elegans to human cancer. Biol Cell 100: 71–81 [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Adhikary S, Eilers M (2006) Mechanisms of transcriptional repression by Myc. Curr Top Microbiol Immunol 302: 51–62 [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN (2002) N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev 16: 2699–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushida T, Inaba M, Hisha H, Ichioka N, Esumi T, Ogawa R, Iida H, Ikehara S (2001) Intra-bone marrow injection of allogeneic bone marrow cells: a powerful new strategy for treatment of intractable autoimmune diseases in MRL/lpr mice. Blood 97: 3292–3299 [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K et al. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien WH, Gelfand VI, Vasioukhin V (2008) Alpha-E-catenin binds to dynamitin and regulates dynactin-mediated intracellular traffic. J Cell Biol 183: 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP (2003) Vertebrate microRNA genes. Science 299: 1540. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN (2008) microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev 22: 3242–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D (2002) Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295: 868–872 [DOI] [PubMed] [Google Scholar]
- Lotterman CD, Kent OA, Mendell JT (2008) Functional integration of microRNAs into oncogenic and tumor suppressor pathways. Cell Cycle 7: 2493–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Slack FJ (2008) microRNAs and cancer: an overview. Cell Cycle 7: 2485–2492 [DOI] [PubMed] [Google Scholar]
- Mendell JT (2005) MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle 4: 1179–1184 [DOI] [PubMed] [Google Scholar]
- Mizui M, Kikutani H (2008) Neuropilin-1: the glue between regulatory T cells and dendritic cells? Immunity 28: 302–303 [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ (2005) Characterization of dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA 102: 12135–12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S (2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 26: 101–106 [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Okubo T, Knoepfler PS, Eisenman RN, Hogan BL (2005) Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development 132: 1363–1374 [DOI] [PubMed] [Google Scholar]
- Ormestad M, Astorga J, Carlsson P (2004) Differences in the embryonic expression patterns of mouse Foxf1 and -2 match their distinct mutant phenotypes. Dev Dyn 229: 328–333 [DOI] [PubMed] [Google Scholar]
- Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P (2006) Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development 133: 833–843 [DOI] [PubMed] [Google Scholar]
- Pellet-Many C, Frankel P, Jia H, Zachary I (2008) Neuropilins: structure, function and role in disease. Biochem J 411: 211–226 [DOI] [PubMed] [Google Scholar]
- Pickford AS, Cogoni C (2003) RNA-mediated gene silencing. Cell Mol Life Sci 60: 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M (2004) A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230 [DOI] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM (2005) Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 11: 1737–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ (2007) MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 67: 9762–9770 [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Crawford GE, Davis S (2006a) Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol 411: 270–282 [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Davis S, Odom DT, Crawford GE, Perkins S, Halawi MJ, Agarwal SK, Marx SJ, Spiegel AM, Meltzer PS, Collins FS (2006b) Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Genet 2: e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz Q, Vieira JM, Howard B, Eickholt BJ, Ruhrberg C (2008) Neuropilin 1 and 2 control cranial gangliogenesis and axon guidance through neural crest cells. Development 135: 1605–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solter D, Knowles BB (1978) Monoclonal antibody defining a stage-specific mouse embryonic antigen (SSEA-1). Proc Natl Acad Sci USA 75: 5565–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ (2008) Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9: 219–230 [DOI] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, Kim VN, Kim KS (2004) Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270: 488–498 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Kawauchi D, Nishida K, Murakami F (2006) Classic cadherins regulate tangential migration of precerebellar neurons in the caudal hindbrain. Development 133: 1923–1931 [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I (2008) MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM (2006) Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20: 2202–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318: 1931–1934 [DOI] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK (2007) The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 21: 744–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI (2008) Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tamakoshi T, Uezato T, Shu F, Kanzaki-Kato N, Fu Y, Koseki H, Yoshida N, Sugiyama T, Miura N (2003) Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Dev Biol 259: 83–94 [DOI] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R (2008) Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40: 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R (2007) DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R (2007) In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448: 318–324 [DOI] [PubMed] [Google Scholar]
- Ziegelbauer JM, Sullivan CS, Ganem D (2009) Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet 41: 130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table Captions
Supplementary Information
Review Process File