Abstract
Eukaryotic transcription occurs within a chromatin environment, whose organization plays an important regulatory role and is partly encoded in cis by the DNA sequence itself1-6. Here, we examine whether evolutionary changes in gene expression are linked to changes in the DNA-encoded nucleosome organization of promoters. We find that in aerobic yeast species, where cellular respiration genes are active under typical growth conditions, the promoter sequences of these genes encode a relatively open (nucleosome-depleted) chromatin organization. This nucleosome-depleted organization requires only DNA sequence information, is independent of any co-factors and of transcription, and is a general property of growth-related genes. In contrast, in anaerobic yeast species, where cellular respiration genes are inactive under typical growth conditions, respiration gene promoters encode relatively closed (nucleosome-occupied) chromatin organizations. Thus, our results suggest a previously unidentified genetic mechanism underlying phenotypic diversity, consisting of DNA sequence changes that directly alter the DNA-encoded nucleosome organization of promoters.
Introduction
Changes in transcriptional regulation are important for generating phenotypic diversity among species, but the mechanisms underlying these regulatory changes are not well understood. Consistent with the centrality of transcription factors to transcriptional control, some phenotypic changes have been associated with changes in the binding-site content of promoters7 or with changes in the targets bound by transcription factors8. However, modulation of other processes key to transcriptional regulation may also lead to phenotypic diversity. Recent studies that measured nucleosome occupancy genome-wide have revealed strong associations between chromatin organization and gene expression4,9-14, and other studies have shown that the organization of nucleosomes is partly encoded in the genome through the sequence preferences of nucleosomes1-5. However, the relationship between evolutionary changes in DNA-encoded nucleosome organization and expression divergence has not been examined.
Here, we study the relationship between gene expression and the DNA-encoded nucleosome organization of promoters across two yeast species, the budding yeast Saccharomyces cerevisiae and the human pathogen Candida albicans, for which large compendia of gene expression data are available. These species exhibit several phenotypic differences. Most notably, in high glucose, C. albicans grows by respiration and correspondingly activates transcription of genes required for the TCA cycle and oxidative phosphorylation, while S. cerevisiae grows primarily by fermentation and correspondingly reduces transcription of respiration genes. We henceforth term the respiratory growth “aerobic” and the fermentative growth “anaerobic”. Our approach consists of three steps. First, we quantify the extent to which the gene expression patterns of biologically meaningful gene sets are conserved across the two species. Next, we examine the DNA-encoded nucleosome organization of these gene sets using both a computational model and in vitro reconstitutions of nucleosome on purified DNA from each species. Finally, we test whether orthologous gene sets with divergent expression patterns between the two species exhibit corresponding changes in their DNA-encoded nucleosome organization.
Identifying large-scale expression changes between S. cerevisiae and C. albicans
We downloaded two large collections of microarray-based gene expression data from ~1000 and ~200 different cellular states and environmental conditions in S. cerevisiae and C. albicans, respectively, compiled in ref.7. To compare the expression patterns of orthologous genes, we downloaded a yeast orthology map15 and quantified the degree to which the co-expression relationships of a gene in one species are similar to the co-expression relationships of its orthologous counterpart in the other species. Such an approach has been successfully used to compare expression patterns across distant species16,17. To obtain insights at the level of biological processes, we used biologically meaningful gene sets from Gene Ontology18 as the basic units of analysis19, and restricted ourselves only to the 796 gene sets (of the total 2152 gene sets) in which the average normalized correlation between all pairs of its member genes was above 0.5 in both expression compendia (Methods). We anchored our analysis around the cytosolic ribosomal protein (CRP) genes, since these genes exhibit coherent expression patterns across many conditions20 and their expression shows strong associations with cellular growth21. For each of the above 796 gene sets, we then computed, separately in each species, the average normalized correlation between the expression of every gene within the set and the expression of every one of the CRP genes (Methods). Thus, in this measure, gene sets that are active under typical growth conditions will have a high (positive) normalized correlation to the CRP genes, whereas gene sets that are inactive under typical growth conditions will have a low (negative) normalized correlation to the CRP genes.
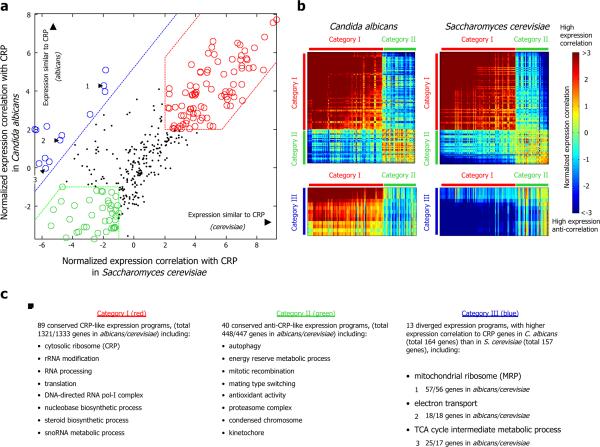
Comparing these expression correlations for every gene set between the two species, we find three main categories of gene sets (Fig. 1, Table S1). The first (“category I”) consists of 89 gene sets (totaling 1333 and 1321 genes in S. cerevisiae and C. albicans, respectively) whose expression in both species is highly correlated to that of the CRP genes. Many gene sets in this category are indeed related to cellular growth, including the CRPs themselves (by construction), amino acid biosynthesis pathways, and RNA processing genes. The second category (“category II”) consists of 40 gene sets (447 and 448 genes in S. cerevisiae and C. albicans, respectively) whose expression in both species exhibits a strong anti-correlation with the expression of the CRP genes. This category includes many gene sets that are activated only in specific cellular states, and gene sets that are induced in response to environmental stress conditions20, such as proteasome-, autophagy- and mating-related genes. The third category (“category III”) consists of 13 gene sets (157 and 164 genes in S. cerevisiae and C. albicans, respectively) whose transcriptional program diverged between the two species, such that the correlation between the expression of their member genes and the expression of the CRP genes is much higher in C. albicans than in S. cerevisiae. This category includes gene sets related to cellular respiration and mitochondrial functions, such as the TCA cycle, oxidative phosphoryation and mitochondrial ribosomal genes. The expression divergence of a subset of these genes was reported previously7, and reflects the difference in respiratory versus fermentative growth preferences between the species.
Figure 1. S. cerevisiae and C. albicans exhibit large-scale changes in the transcriptional programs of cellular respiration and mitochondrial function genes.
(a) For gene sets from Gene Ontology, shown is the average normalized Pearson correlation (see Methods) between the expression of their member genes and the expression of the cytosolic ribosomal proteins (CRP), computed separately for the expression compendia of C. albicans (y-axis) and S. cerevisiae (x-axis). Only gene sets that exhibit coherent co-expression in both species are shown, where we define coherently co-expressed gene sets as those in which the average normalized correlation between its member genes is above 0.5. From these gene set expression correlation measures, we define three categories of gene sets (category I, red, high correlation with CRP genes in both species; category II, green, anti-correlation with CRP genes in both species; and category III, blue, higher correlation with CRP genes in C. albicans than in S. cerevisiae. Three gene sets from category III are numbered for reference in other figures. (b) Shown are the normalized expression correlations, computed as in (a), between every pair of gene sets from all three categories defined in (a). Note the expression divergence of gene sets from category III (blue), which exhibit strong expression correlation with category I gene sets from C. albicans, but strong anti-correlation with category I gene sets from S. cerevisiae. (c) A subset of the list of gene sets in each of the categories defined in (a), along with the number of gene sets and number of genes in each category. More details are given for the three numbered categories from (a). See Table S1 or the full list.
Large-scale expression changes between S. cerevisiae and C. albicans are accompanied by changes in DNA-encoded nucleosome organization
To study the relationship between transcriptional programs and chromatin organization, we examined DNA-encoded nucleosome organization over the promoter regions of the gene sets in each of the three categories, both experimentally and using a computational model of the nucleosome sequence preferences22. These sequence preferences are represented by a probability distribution over nucleosome-length sequences, estimated from a large set of fully sequenced in vivo nucleosomes from S. cerevisiae. The model uses this distribution to compute the probability that each basepair in the genome is covered by a nucleosome, in an assumed equilibrium between all competing nucleosome configurations. Using a cross-validation scheme, this sequence-based model was shown to be highly predictive of the experimentally measured nucleosome organization, suggesting that nucleosome organization is partly encoded in cis by the DNA, and that we can reliably use this model to examine the DNA-encoded nucleosome organization (see ref.22 for an overview and evaluation of this model).
We used this model to compute the occupancy over the nucleosome-depleted region of every promoter in each of the two species. We focused on the 200 basepairs upstream of the translation start site, since in vivo measurements of nucleosome occupancy showed that promoters exhibit a stereotyped depleted region of length ~100-150bp within the 200bp upstream of the translation start site4,9-11,14. We defined the occupancy over this promoter nucleosome-depleted region (henceforth termed “PNDR”) as the lowest average nucleosome occupancy across any 100 basepair region in the 200 basepairs upstream of the translation start site. Other parameter choices that we tested for the region (in the range of 100-150bp for the width of the least-occupied region and 200-400bp for the overall length of the upstream region) resulted in equivalent results. Thus, when the PNDR score of each gene is computed by the model, it represents a predicted measure of the degree to which the gene's promoter encodes an open (nucleosome-depleted) or closed (nucleosome-occupied) nucleosome organization.
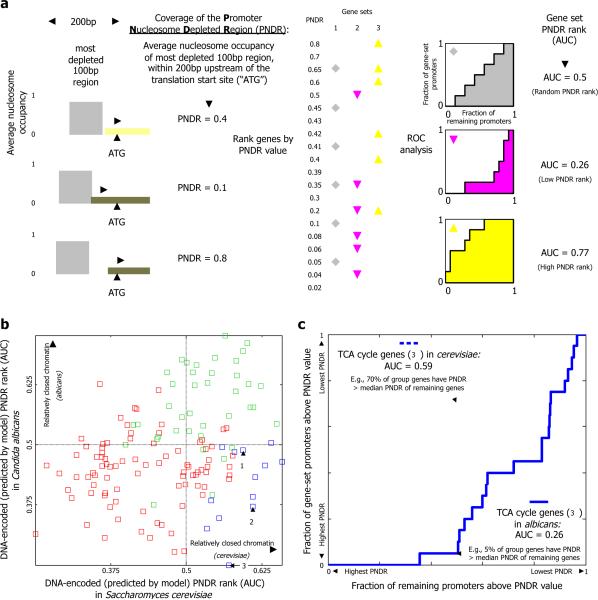
To test whether the DNA sequences of promoters from a given gene set encode a relatively open or closed nucleosome organization, we compared, separately for each species, the PNDR scores of the gene set's promoters to the PNDR scores of all other promoters. Specifically, we ranked all promoters by their PNDR score and measured the relative ranking of the gene set's promoters using a normalized Mann-Whitney rank statistic, which is equal to the area under the curve23 (AUC) when plotting the fraction of the gene set's promoters above a given PNDR score versus the fraction of all other promoters above that PNDR score, for all possible PNDR values (Fig. 2a). In this measure, a gene set with a relatively closed promoter organization, in which every promoter has a PNDR score above that of every other promoter, will receive an AUC score of 1. A gene set in which every promoter has a PNDR score below that of every other promoter will receive an AUC score of 0 (relatively open nucleosome organization), and a gene set composed of randomly selected promoters set will receive, on average, an AUC score of 0.5.
Figure 2. The expression divergence of cellular respiration genes is accompanied by changes in the DNA-encoded nucleosome organization of their promoters.
(a) A toy example illustrating the rank statistic used to assess whether the DNA-encoded nucleosome organization of promoters of a given gene set encode a relatively open or relatively closed organization. For each gene, we use the model of the nucleosome sequence preferences22 to compute the DNA-encoded nucleosome coverage over the nucleosome depleted region of its promoter (left, termed PNDR), and rank all genes by these PNDR scores (middle table, values were arbitrarily chosen for illustration). The rank statistic of each gene set is then obtained by computing the area under the curve (AUC) in a graph that plots the fraction of promoters from the gene set (y-axis) that are above a certain PNDR score versus the fraction of all other promoters above that PNDR score, for all possible PNDR values (three plots on right-end). Plots for three sample gene sets are shown: a gene set with a random AUC rank of 0.5 (top, gray); a gene set whose genes have relatively open nucleosome organizations and thus relatively low PNDR scores (middle, pink); and a gene set whose genes have relatively closed nucleosome organizations and thus relatively high PNDR scores (bottom, yellow). (b) For every gene set from the three categories defined in Fig. 1a, shown are its PNDR rank statistic, computed as explained in (a), in both C. albicans (y-axis) and S. cerevisiae. Gene sets from each category are colored as in Fig. 1a. The three numbered gene sets from category III in Fig. 1a are numbered here as well. (c) Example of an AUC plot for one of the gene sets from (b) (the TCA cycle gene set), in both S. cerevisiae (dashed line), and C. albicans (full line). The promoters of the gene set in this example have relatively high PNDR scores in S. cerevisiae and thus encode relatively closed nucleosome organizations, whereas in C. albicans, they have relatively low PNDR scores and thus encode relatively open nucleosome organizations. For example, only ~5% of the gene sets' promoters have higher PNDR scores than the PNDR score which is exceeded by ~50% of the promoters in C. albicans.
For each gene set from the three categories above, defined solely by their expression profiles, we then compare the predicted PNDR AUC value in S. cerevisiae to the AUC in C. albicans (Fig. 2b,c). Notably, in both species, the growth-related gene sets of category I, whose expression profile in both species is highly correlated to that of the CRP genes, have AUC scores significantly lower than all other gene sets (p<10-13 and p<10-9 in student t-test in S. cerevisiae and C. albicans, respectively), indicating that their promoters encode relatively open nucleosome architectures. Conversely, the condition-specific gene sets of category II, in which expression is anti-correlated to that of the CRP genes in both species, have AUC scores significantly higher than all other gene sets (p<10-5 and p<10-18), indicating that their promoters encode relatively closed nucleosome architectures. These results suggest that both S. cerevisiae and C. albicans preserve a system-level relationship between transcriptional programs and DNA-encoded nucleosome organizations, whereby promoters of growth-related genes encode relatively open nucleosome organizations, while promoters of condition-specific genes encode relatively closed nucleosome organizations.
In contrast to the largely conserved nucleosome organization of gene sets from the first two categories, the aerobic cellular respiration gene sets of category III exhibit many changes between the two species in the DNA-encoded nucleosome organization over their promoters. In C. albicans, aerobic respiration gene sets (category III) have AUC values significantly lower than all other gene sets (p<0.005 in student t-test) and thus their promoters encode relatively open nucleosome organizations, while in S. cerevisiae, these aerobic respiration gene sets have AUC values significantly above all other gene sets (p<10-6) and thus their promoters encode relatively closed chromatin architectures. Notably, these changes in the DNA-encoded nucleosome organization are coupled to the expression divergence that the category III gene sets exhibit between the two species, in a manner that may facilitate the transcriptional program of each species. Category III gene sets, which have higher expression correlation to growth-related genes in C. albicans than in S. cerevisiae, encode a relatively open nucleosome organization in C. albicans, in accordance with the trend observed for growth-related gene sets (category I), and a relatively closed nucleosome organization in S. cerevisiae, in accordance with the trend observed for gene sets whose expression is anti-correlated to growth-related genes (category II).
These results demonstrate that a global relationship between transcriptional programs and the DNA-encoded nucleosome organizations is remarkably conserved across two yeast species, even in the presence of expression divergence. Our results thus suggest a conserved design principle of transcriptional regulation in yeast, whereby the default repression of condition-specific genes (like the aerobic respiration genes of S. cerevisiae) is facilitated by the relatively closed nucleosome organization encoded over their promoters. In conditions where activation of these genes is required, this repression is actively alleviated, presumably by the combined action of transcription factors and chromatin remodeling complexes. In contrast, for growth-related genes most commonly used by the organism (like the aerobic respiration genes of C. albicans), the repression by nucleosomes is by default alleviated through the encoding of relatively open nucleosome organizations over their promoters. We note that although this global trend is strong in our analysis, it clearly does not apply to every growth-related or condition-specific gene set or individual gene within them, since some of these gene sets exhibit moderate AUC values.
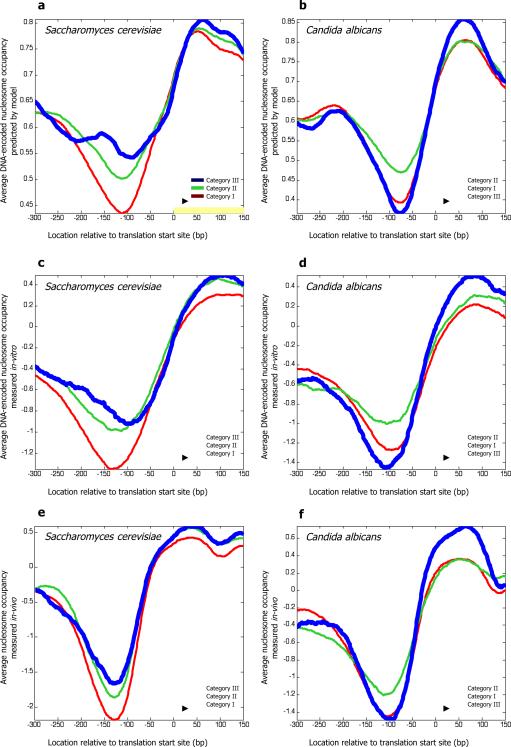
The same behavior is also evident when we create a single gene set for each of the three categories, consisting of all the genes from the gene sets of that category. In both species, when we plot the average nucleosome occupancy predicted by the model across the promoters, we find stronger predicted nucleosome depletion (lower PNDR score) in category I promoters relative to category II promoters (p<10-6 and p<10-9 in student t-test in S. cerevisiae and C. albicans, respectively). However, the cellular respiration promoters from category III differ in their average nucleosome occupancy between the two species, such that in S. cerevisiae, they encode the most closed nucleosome organization of all three categories (Fig. 3a), but in C. albicans, they encode the most open nucleosome organization of all three categories (Fig. 3b). Indeed, the category III promoters have a significantly more closed nucleosome organization in S. cerevisiae than in C. albicans, since the average difference in the predicted PNDR score between the two species is significantly higher than the difference obtained when randomly choosing the same number of promoters (p<10-4, 10,000 permutation tests).
Figure 3. The DNA-encoded nucleosome organization of cellular respiration promoters has diverged between S. cerevisiae and C. albicans.
(a) For each of the three categories defined in Fig. 1a, we created a single gene set per category that consists of all genes from all gene sets of that category. Shown is the average nucleosome occupancy in S. cerevisiae, predicted by our sequence-based model of nucleosome sequence preferences, across this unionized gene set per category. Average occupancy profiles are shown relative to the translation start site of the corresponding genes (we used translation start sites since transcription start sites are not well-annotated genome-wide for C. albicans). (b) Same as (a), but for C. albicans. (c) Same as (a), but using the in vitro map of nucleosome occupancy that we measured in S. cerevisiae. (d) Same as (c), but for C. albicans. (e) Same as (a), but using the in vivo map of nucleosome positions that we measured in S. cerevisiae. (f) Same as (e), but for C. albicans.
Genome-wide maps of nucleosomes assembled on purified genomic DNA from each species validate model-predicted changes in DNA-encoded nucleosome organization
As a direct experimental validation of the model predictions, we purified chicken erythrocyte histone octamers and assembled them on purified genomic DNA from both S. cerevisiae and C. albicans by salt gradient dialysis24. We then isolated mononucleosomes by standard micrococcal nuclease digestion, and used parallel sequencing to determine nucleosome positions. We performed two completely independent experiments in each species, and mapped more than 10 million reconstituted nucleosomes in each species. Thus, the resulting data provide a genome-wide map in each species, in which nucleosome positions are governed only by the intrinsic sequence preferences of nucleosomes6. For each map, we calculated the average nucleosome occupancy at every basepair as the log-ratio of the number of reads that cover that basepair and the median number of reads per basepair across the genome. The independent replicates of each species were in excellent agreement, so we averaged the replicates within each species to create two in vitro nucleosome occupancy maps, one in S. cerevisiae and one in C. albicans.
As a first validation, we compared the PNDR scores predicted by the model for each promoter, on which our above AUC analyses are based, to the PNDR scores computed from the in vitro maps. We find that these scores are in good agreement, with an overall correlation of 0.76 and 0.72 between the model PNDR scores and the PNDR scores computed from the in vitro maps in S. cerevisiae and C. albicans, respectively. Moreover, we find a correlation of 0.70 between the model-predicted and data-measured divergence in PNDR scores per promoter between S. cerevisiae and C. albicans (Fig. S1). We next examined the average nucleosome occupancy measured by these in vitro maps across the promoters of each of our three categories, and found that they are highly similar to those predicted by the model (Fig. 3c,d). As predicted by the model, the in vitro maps display stronger nucleosome depletion in category I promoters relative to category II promoters (p<10-4 and p<10-6 in student t-test in S. cerevisiae and C. albicans, respectively). The occupancy profiles of aerobic respiration promoters (category III) in the in vitro maps also agree with the model predictions, showing the most closed and open nucleosome organization of all three categories in S. cerevisiae and in C. albicans, respectively (Fig. 3c,d), and exhibiting a significantly more closed nucleosome organization in S. cerevisiae than in C. albicans (p<10-4 in 10,000 permutation tests). A model constructed from the S. cerevisiae in vitro data6 yielded equivalent predictions to those of the in vivo based model that we use here (Fig. S2). Thus, in accord with the model predictions, these in vitro nucleosome occupancy maps demonstrate that evolutionary changes in the DNA sequence of aerobic respiration gene promoters contributed to the divergence of nucleosome organization at these promoters in S. cerevisiae and C. albicans.
The nucleosome organization in vivo is similar to DNA-encoded organization
Next, we tested whether the in vivo nucleosome organization of promoters from each of the above three categories is similar to their DNA-encoded organizations, as predicted by the model and measured by the in vitro maps. To this end, we isolated mononucleosomes from both S. cerevisiae and C. albicans each cultured in their own “normal” growth conditions (Methods), and used parallel sequencing to obtain genome-wide maps of in vivo nucleosome positions. The maps consist of more than 10 million individual nucleosome reads in each species. We performed two completely independent experiments in each species, calculated the average nucleosome occupancy at every basepair, and averaged the highly similar replicates within each species to create two in vivo nucleosome occupancy maps, one in S. cerevisiae and one in C. albicans.
We subjected these in vivo maps to the same tests that we performed for the in vitro maps and found that indeed, the nucleosome organization of promoters in vivo is highly similar to the DNA-encoded nucleosome organization predicted by the model and measured by the in vitro maps6. As expected, the agreement between the model predictions and the in vitro maps is higher than the agreement between the model predictions and the in vivo maps. The PNDR scores predicted by the model for each promoter and those computed from the in vivo maps in each species are nonetheless in good agreement, with a correlation of 0.62 and 0.63 in S. cerevisiae and C. albicans, respectively. Similarly, there is good agreement (correlation=0.60) between the model-predicted and data-measured difference in PNDR scores per promoter between the two species (Fig. S3). The S. cerevisiae model-predicted PNDR scores are also in agreement with other published nucleosome occupancy maps (Table S2). In accord with the model predictions and the in vitro maps, the in vivo maps also reveal stronger nucleosome depletion in category I promoters relative to category II promoters in both species (Fig. 3e,f; p<10-3 and p<10-4 in student t-test in S. cerevisiae and C. albicans, respectively). Similarly, the in vivo maps indicate that aerobic respiration promoters (category III) have the most closed and open nucleosome organization of all three categories in S. cerevisiae and in C. albicans, respectively (Fig. 3e,f), and that they have a significantly more closed nucleosome organization in S. cerevisiae than in C. albicans (p<0.01 in 10,000 permutation tests).
The divergence of aerobic and anaerobic yeast species coincides with a large-scale change in DNA-encoded nucleosome organization
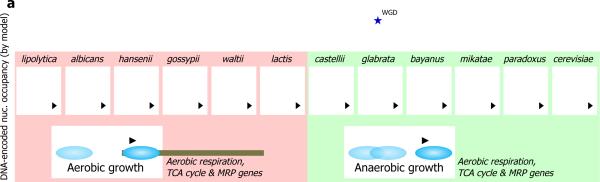
To obtain a broader evolutionary perspective, we used our model to examine the DNA-encoded nucleosome organization of promoters in ten additional yeast species. Notably, we found that the relation between the DNA-encoded nucleosome organization of promoters from category I and category II is conserved across all of the yeast species that we examined. In all species, promoters of the growth-related genes from category I encode relatively open chromatin organizations, while promoters of condition-specific genes from category II encode relatively closed chromatin organizations. In contrast, the nucleosome organization of respiration gene promoters from category III has diverged in evolution, exactly at the point in which the yeast species that we examined exhibit phenotypic divergence between aerobic and anaerobic growth. Specifically, the promoters of these genes are predicted to encode relatively open nucleosome organizations in all of the aerobic yeast species, and relatively closed nucleosome organizations in all of the anaerobic yeast species (Fig. 4). Thus, our results demonstrate that a major phenotypic change across yeast species, namely the emergence of anaerobic yeast species, was accompanied by evolution of DNA-encoded nucleosome organization in a large number of aerobic respiration gene promoters. Notably, this phenotypic divergence coincides with a whole-genome duplication event in the evolutionary history of yeast, such that the six anaerobic yeast species descend from a post-genome duplication ancestor. Several studies have pointed out other unique evolutionary changes that coincide with this whole-genome duplication event15,25.
Figure 4. The emergence of anaerobic yeast species coincides with an evolutionary change in the DNA-encoded nucleosome organization of cellular respiration promoters.
Shown is the average nucleosome occupancy, predicted by our model of nucleosome sequence preferences, across all genes from each of the three categories of gene sets defined in Fig. 1a, for each of 12 different yeast species whose genomic sequence is available. Average nucleosome occupancy profiles are shown relative to the translation start site of the corresponding genes in each species, color-coded as in Fig. 3. Yeast species are organized according to their phylogenetic tree, aerobic and anaerobic yeast species are indicated by pink (left) and green (right) boxes, respectively, and the point in evolution where the apparent whole-genome duplication event has occurred is indicated (blue asterisk). Note that in all of the aerobic yeast species, promoters of category III genes encode a relatively more open nucleosome organization compared to promoters of category II genes, while in all of the anaerobic yeast species, the situation is reversed, and promoters of category III genes encode a relatively more closed nucleosome organization compared to promoters of category II genes.
The relationship between evolutionary changes in DNA-encoded nucleosome organization and evolutionary changes in expression
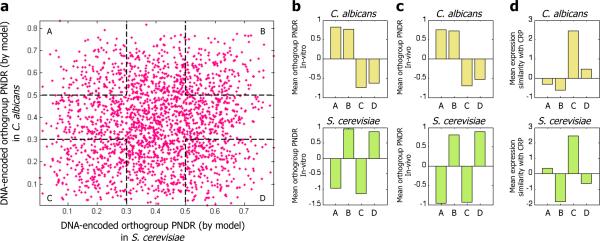
Finally, we used the model to obtain a genome-wide view of changes in the DNA-encoded nucleosome organization across all of the genes that are conserved between S. cerevisiae and C. albicans (Fig. 5a). From this view, we extracted the four extreme groups of genes based on whether they have relatively open or closed organizations in S. cerevisiae and C. albicans, as determined by their PNDR scores. In all cases, high or low PNDR scores predicted by the model indeed have significantly high or low PNDR scores in both the in vitro and in vivo nucleosome occupancy maps (Fig. 5b,c). Examining the expression profiles of every group in each of the two species, we find a notable global trend, in that the two groups (“B” and “C”) whose DNA-encoded nucleosome organization is conserved between the two species also exhibit conservation of their transcriptional programs, whereas the two groups (“A” and “D”) whose DNA-encoded organization diverged between the two species also exhibit divergence in their transcriptional programs (Fig. 5d). For example, group “A” genes, which have a relatively closed (high PNDR scores) and open (low PNDR scores) chromatin organization in C. albicans and S. cerevisiae, respectively, exhibit negative expression correlation to the CRP genes in C. albicans and positive expression correlation to the CRP genes in S. cerevisiae.
Figure 5. A global relationship between evolutionary changes in the DNA-encoded nucleosome organization and evolutionary changes in expression.
(a) Shown is the PNDR score predicted by our model of nucleosome sequence preferences, for each ortholog from C. albicans (y-axis) and S. cerevisiae (x-axis). For orthologs that contain more than one gene in one of the species, the average PNDR score is shown. We used this plot to define four groups (“A”, “B”, “C”, and “D”), for all combinations of high (above 0.5) and low (below 0.3) PNDR scores in the two species. (b) For each of the four groups defined in (a), shown are the PNDR scores computed from the in vitro nucleosome occupancy map in C. albicans (top) and S. cerevisiae (bottom). PNDR scores are shown as the difference between the actual PNDR score of the gene group and the average PNDR score of all genes in each respective species. Note that for all groups, high and low PNDR scores predicted by the model in each species have correspondingly high and low PNDR scores in the in vitro maps, respectively. (c) Same as (b), when computing the PNDR scores using the in vivo nucleosome occupancy maps in C. albicans (top) and S. cerevisiae (bottom). Here too, for all groups, the model predictions are upheld in the in vivo nucleosome maps in each species. (d) For each of the four groups from (a), shown is the average normalized correlation between the expression of the genes from the group and the expression of the CRP genes in C. albicans (top) and S. cerevisiae (bottom).
These results reinforce the trend that we observed for our three categories of gene sets, in that many individual genes whose DNA-encoded nucleosome organization has diverged between the two species also exhibit divergence in their transcriptional programs. Moreover, in all cases, the direction of change in the encoded nucleosome organization is opposite to the direction of change in expression, such that changes that result in relatively more open and more closed nucleosome organizations are accompanied by higher and lower expression correlation to the CRP genes, respectively. Notably, our gene-set level analysis (Fig. 1) did not identify gene sets that exhibit the expression divergence of group “A” genes, further highlighting the utility of this analysis at the level of individual genes. While these are the global trends observed in each group, many individual genes within each group behave differently.
Conclusions
Our results suggest that yeast species exhibit a simple relationship between transcriptional programs and nucleosome organizations encoded in promoter sequences. Promoters of genes that are required for the typical mode of growth tend to encode relatively open nucleosome organizations, while promoters of genes that are not part of the typical growth pathways of the organism (e.g., condition-specific and stress-response genes) tend to encode relatively closed nucleosome organizations. Notably, this relationship continues to hold even after the divergence of yeast into species that grow aerobically through pathways that involve cellular respiration and mitochondrial genes, and species that grow anaerobically through pathways that do not involve these genes26. We propose that this large-scale change in the expression of respiration genes is achieved, at least in part, through DNA sequence changes that alter the DNA-encoded nucleosome organization in their promoters. We provide strong support for this proposed mechanism by showing that these changes in nucleosome organization are also seen in a reconstitution of nucleosomes on purified DNA from S. cerevisiae and C. albicans. Our results thus show one case in which a system-level reprogramming of the yeast transcriptional network is associated with, and presumably achieved, in part, through evolution of intrinsic nucleosome organization encoded in the DNA sequence of promoters. This evolutionary mechanism for genetic change may also account for other types of phenotypic diversity observed across eukaryotic species.
Methods
Parallel sequencing of in vivo nucleosome maps in S. cerevisiae and C. albicans
Mononucleosomes were extracted from log-phase yeast cells using standard methods. Saccharomyces cerevisiae and Candida albicans mononucleosomes were prepared separately, and two independent replicates were taken from each species. The DNA was extracted, and protected fragments of length ~147bp were cloned and sequenced on an Illumina GA II instrument.
Creation of in vitro nucleosome maps in S. cerevisiae and C. albicans
S. cerevisiae genomic DNA was purified from strain YLC8 [MAT(a) ura3(Δ) leu2(Δ) his3(Δ) met15(Δ)] using standard methods. C. albicans DNA was purified from strain SC5314 using standard methods. For both species, additional steps were taken to remove contaminating RNA. After recovery by ethanol precipitation, DNA was resuspended in TE buffer (TE is 10 mM Tris pH 8.0, 1 mM EDTA), and the DNA concentration was determined by agarose gel electrophoresis using ethidium stain, followed by comparison to mass standards using quantitative fluorometry. The sample was then subjected to RNase A digestion at 50 °C overnight, using 100 μ of RNase A for every 10 μ of DNA, followed by ethanol precipitation of the DNA. After resuspension of the pellet in TE, the genomic DNA was sheared twice each through a 25 gauge needle and then a 27½ gauge needle. The entire mixture was then electrophoresed on a 20 × 20 cm, 1% agarose, 1× TAE gel at 100 V for 6-8 hours. The genomic DNA band was cut out, and the agarose slab containing the DNA was then reelectrophoresed inside a dialysis bag, with occasional UV-light monitoring, to elute the DNA.
Histone octamer (HO) was purified from chicken erythrocytes using salt extraction and hydroxyapatite column chromatography, as previously described27. Genomic DNA was reconstituted into nucleosomes under selective pressure for nucleosome-favoring sequences by salt gradient dialysis24. For S. cerevisiae, the reconstitution reaction used 40 μ HO + 100 μ DNA in a 200 μg volume. The resulting nucleosomes were biochemically isolated by micrococcal nuclease (MNase) digestion, in two independent experiments, using 6×10-3 or (separately) 6×10-4 units MNase (Sigma Chemical Company, St. Louis) per 10 μ competitively-reconstituted DNA, in 10 mM Tris pH 8.0, 1 mM CaCl2, for 5 min at 37 °C. For C. albicans, reconstitution reactions used 37 μ HO + 93 μ DNA in a 200 μl volume. Nucleosomes from two independent reconstitutions were digested (separately) with 6×10-3 units MNase per 10 μ competitively-reconstituted DNA, for 5 min at 37 °C. After digestion, DNA was extracted, and protected fragments of length ~147 bp were isolated by polyacrylamide gel electrophoresis, extracted from the gel. Samples were independently subjected to Illumina sequencing. A detailed comparison between the in vitro and in vivo nucleosome maps of S. cerevisiae will be presented elsewhere.
Mapping and post-processing of parallel sequencing reads
To map the reads resulting from the above sequencing experiments in each species we used NCBI BLAST28 requiring 32 matches and allowing at most 1 gap. To estimate the mean DNA fragment length in each experiment, we superimposed the nucleosome reads of one strand and examined the distribution of nucleosome reads of the opposite strand. As expected, this distribution shows a strong peak at ~140-170 bp for all experiments, with slight variations between experiments. We used the maximum of the peak as an estimation of the mean DNA fragment length and extended all nucleosome reads to this length. We defined repetitive regions as regions that were matched by a read that mapped to more than one place in the genome. We excluded repetitive regions and their 150bp vicinity from our analyses. To obtain genomic nucleosome occupancy tracks we summed for each position all reads covering it. We excluded basepairs covered by more than 10 times the median genomic basepair coverage (typically less than 1% of all basepairs). Finally, we normalized each track by the mean basepair coverage.
Datasets
The genome sequence and gene and chromosome annotations of the yeast species examined in this study were obtained from a recent compilation15. The member genes of Gene Ontology gene sets from S. cerevisiae were downloaded from the Gene Ontology repository18. The member genes of the same gene sets in other yeast species were defined as the orthologs of the original gene set from S. cerevisiae, using a recent orthology map across 17 yeast species15. For all pairwise comparisons between S. cerevisiae and C. albicans, we restricted our analysis only to genes that have orthologs in the other species, resulting in 2835 genes in S. cerevisiae and 2823 genes in C. albicans, representing 2225 orthogroups. Note that the number of genes in each species differs, since the orthology map includes many-to-many relationships (i.e., some orthogroups include more than one gene from one or both species). For the analysis across multiple yeast species (Fig. 4) the genes in each species were restricted only to those that have orthologs in all of the other species. Expression compendia of ~1000 and ~200 gene expression measurements in S. cerevisiae and C. albicans, respectively, were downloaded from a previous compilation7.
Computing expression correlations between gene sets
As our input gene sets, we used all of the gene sets from Gene Ontology18 that have at least 10 orthologous genes in each species. We then computed a transcription-program similarity measure between each pair of gene sets, separately in each species, as the average (over non-identical gene pairs) normalized Pearson correlation between expression profiles. The normalized Pearson correlation is the Pearson correlation after subtracting the mean and dividing by the standard deviation of the Pearson correlation between every pair of genes in that species. This standardization corrects for potential biases in the Pearson correlation that may arise due to size differences between the expression compendia of each species. We used these normalized correlations to further restrict the input gene sets to use only those for which the average normalized correlation between all pairs of genes from the gene set was above 0.5.
Model predictions
The model used for predicting nucleosome organization based on the genomic sequence has a single parameter that represents the apparent nucleosome concentration. We set this nucleosome concentration parameter to 1 in S. cerevisiae, and for each other species we set it such that the average genome-wide predicted nucleosome occupancy in that species is equal to that predicted for S. cerevisiae using a concentration of 1. To reduce potential biases that may arise from these concentration parameters, we restricted our analyses to examining the relative, rather than the absolute, relations between the nucleosome organization of different gene sets.
Acknowledgements
We acknowledge with gratitude the gift of strains, protocols and advice from Judith Berman, thank Hemant Kelkar for help with the Illumina sequencing data, and the members of our respective laboratories for discussions and comments on the manuscript. This work was supported by grants from the NIH to JDL, from the NIH to JW, and from the European Research Council (ERC) and NIH to ES. NK is a Clore scholar. ES is the incumbent of the Soretta and Henry Shapiro career development chair.
Footnotes
URLs. For our data, results and model predictions, see http://genie.weizmann.ac.il/pubs/nucleosomes09.
References
- 1.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ioshikhes IP, Albert I, Zanton SJ, Pugh BF. Nucleosome positions predicted through comparative genomics. Nat Genet. 2006;38:1210–5. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- 3.Peckham HE, et al. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–7. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–44. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 5.Yuan GC, Liu JS. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput Biol. 2008;4:e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2008 doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ihmels J, et al. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–40. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- 8.Borneman AR, et al. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–9. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- 9.Yuan GC, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–30. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 10.Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–6. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 11.Shivaswamy S, et al. Dynamic Remodeling of Individual Nucleosomes Across a Eukaryotic Genome in Response to Transcriptional Perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–5. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 13.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol. 2007;25:244–8. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 14.Mavrich TN, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008 doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 16.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–55. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 17.Bergmann S, Ihmels J, Barkai N. Similarities and differences in genome-wide expression data of six organisms. PLoS Biol. 2004;2:E9. doi: 10.1371/journal.pbio.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–8. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 20.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–57. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mager WH, Planta RJ. Coordinate expression of ribosomal protein genes in yeast as a function of cellular growth rate. Mol Cell Biochem. 1991;104:181–7. doi: 10.1007/BF00229818. [DOI] [PubMed] [Google Scholar]
- 22.Field Y, et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 24.Thastrom A, Bingham LM, Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J Mol Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 25.Man O, Pilpel Y. Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat Genet. 2007;39:415–21. doi: 10.1038/ng1967. [DOI] [PubMed] [Google Scholar]
- 26.Piskur J, Langkjaer RB. Yeast genome sequencing: the power of comparative genomics. Mol Microbiol. 2004;53:381–9. doi: 10.1111/j.1365-2958.2004.04182.x. [DOI] [PubMed] [Google Scholar]
- 27.Feng HP, Scherl DS, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering: test of a model for nucleosome transcription. Biochemistry. 1993;32:7824–31. doi: 10.1021/bi00081a030. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]