Abstract
AppA is a blue-light and redox responding regulator of photosynthesis gene expression in Rb. sphaeroides. Detailed time-resolved fluorescence spectroscopy and sub-picosecond transient absorption spectroscopy study of the BLUF domain is presented for wild type AppA (AppAwt) and a photo-inactive Y21F mutant of AppA. The main findings discussed here are: 1) Time-resolved laser excitation studies on dark-adapted protein shows that AppAwt and Y21F mutant protein exhibits a fluorescence decay with a lifetime of 0.6 ns. Dark-adapted AppAwt, but not Y21F, also exhibits slower fluorescence decay with a lifetime of 1.7 ns. Analysis of AppAwt that was light excited to a stable light-adapted form prior to data collection shows mono-exponential fluorescence decay with a lifetime of 1.0 ns. This component disappeared after 1 minute of data collection after which the original “dark-adapted” values were recovered demonstrating the presence of a ∼ 1 minute lifetime intermediate during return of AppA from light to dark-adapted form. 2) Transient absorption spectral analysis reveals a very fast rising of transient absorption (<1 ps) for AppAwt. This fast component is missing in the Y21F mutant, which lacks Tyr21, giving rise to a slower transient absorption at 4-6 ps. In the AppAwt transient spectra, most ground state recovers within ∼30 ps, comparing to ∼90-130 ps in the mutant Y21F. We propose that a temporary electron transfer occurs from Tyr21 to the N5 of flavin in AppAwt and is a triggering event for subsequent hydrogen bond rearrangements. Dynamics of the AppA photocycle is discussed in view of the currently solved crystallographic structure of AppA.
Keywords: Flavin photoreceptor, blue light absorption, photocycle, time-resolved spectroscopy
A crystal structure of the AppA flavin-binding domain (also called Blue-Light sensing domain Using FAD) from Rb. sphaeroides was recently solved at 2.3 Å resolution (1) (PDB code 1YRX). The structure indicates that AppA represents a novel class of photoreceptors with a ferrodoxin-like fold that has an exceptionally long-lived photocycle. AppA is both a light and redox responding regulator of photosynthesis gene transcription in Rb. sphaeroides where it can be found in two different functional forms. In anaerobic low-light growth conditions, AppA is in a “dark adapted” form where it is able to bind and inactivate the repressor PpsR, thus allowing the RNA polymerase to maximally transcribe photosynthesis genes. In aerobic high light conditions, or under strong blue light illumination, FAD in AppA becomes photoexcited into a signaling state (called here the “light adapted” form) that is incapable of interacting with the photosynthesis repressor PpsR. Under these conditions there is maximal repression of photosynthesis gene expression (2).
Genome sequence analysis indicates that the BLUF domain is present in a variety of proteins in a number of different species. Typically, BLUF is present as an amino terminal “receiver” domain that is linked to a variety of different “output” domains. For example, the amino terminal third of AppA constitutes the light absorbing BLUF domain with the remainder C-terminal region containing the Cys-rich redox responding output domain. The E. coli protein YcgF contains an amino terminal BLUF domain that is covalently linked to an EAL output domain that is predicted to have cyclic-di-GMP phosphodiesterase activity (3). In Euglena gracilis, the BLUF domain is connected to an adenylate cyclase with blue light excitation controlling cyclase activity (4) that subsequently mediates photoavoidance. In Synechocystis sp., the product of the slr1694 gene, contains a BLUF domain that likely communicates with other proteins to participate in blue-light phototactic avoidance (5).
A major spectroscopic characteristic of the AppA photocycle is a reversible 10 nm red-shift of the visible flavin spectrum that occurs upon light excitation. The light excited spectral shift decays back to the dark state with a remarkably long half-life of ∼10 minutes. The mechanism of AppA light excitation was studied extensively over last years (6-9) with initial NMR studies (6) indicating that there is a rearrangement of H-bonds between flavin and surrounding amino acids that is responsible for the red-shift in the spectrum. FTIR studies on AppA, and the Slr1694 protein (8, 10), also indicate that light excitation resulted in formation of a new H-bond at the C4=O position of the flavin isocyclic ring. Ultrafast spectroscopy studies of dark adapted AppA (9) have revealed transient formation of several intermediates with life-times in the ps range that occur upon excitation of the flavin in the S1 state that decay into the red-shifted product within 10 ns. Triplet formation was also observed as a minor side-reaction during laser excitation. Deprotonation of the flavin at N3 was also suggested to occur upon illumination (7). Recently, Fukushima et al. (11) observed a 5 nm red-shifted intermediate in the BLUF protein Tl10078 from Thermosynechococcus elongatus at temperatures below 50 K that converted into a 10-nm shifted species above 50 K. Appearance of this intermediate is thought to involve minor structural changes as molecular motion is limited at these low temperatures.
In this paper, we report detailed spectroscopic studies on the BLUF domain of AppAwt and on a photo-inactive AppA mutant Y21F. The Y21F mutant does not exhibit the 10-nm red-shift and therefore it has been suggested that it is “locked” in the dark form. A possible mechanism for the initial steps in the photocycle dynamics is discussed in light of the recently solved crystal structure of AppA in its dark form (1). Electron transfer from Tyr21 to the flavin, and charge recombination on a ps timescale, is suggested as the event triggering the H-bond rearrangement.
EXPERIMENTAL PROCEDURES
Protein purification
The AppA wild-type BLUF fragment is photoactive with a photocycle indistinguishable from that of full length AppAwt. However the AppA BLUF fragment is significantly more amendable to purification and is more soluble, thus all spectral studies were undertaken with the BLUF fragment (amino acids 1-126) of AppA.
Wild type or Y21F AppA constructs containing entire BLUF FAD-binding domain were isolated by expressing an intein-chitin-binding domain-AppA fusion (plasmids pTY/AppA1-126 wt or pTY/Y21F1-126) in E.coli BL21(DE3) cells. Cells were grown at 37°C until OD600=0.6, then transferred to 16°C and induced for 16 hours by addition of 1 mM isopropyl-β-D-thiogalactopyranoside. AppA protein was purified on Chitin Beads (New England Biolabs) using buffers recommended by manufacturer followed by Superose 12 chromatography using ACTA FPLC in 20 mM Tris-HCl and 100 mM NaCl as described previously (1).
Time-resolved fluorescence
Fluorescence decays of samples that were either dark-adapted or light-exposed immediately before experiments were measured using the time-correlated single photon counting (TCSPC) method and a two-photon excitation scheme using a Ti:sapphire laser at 800 nm, 76 MHz repetition rate, and 150 fs pulse duration. The absorption of two photons has the advantages that there is no out-of-focus photobleaching, the excitation beam is not attenuated by out-of-focus absorption, and the light path to the detector is better defined with the result of cleaner transients than for one photon absorption. Moreover, the low number of fluorescent photons per pulse is adequate for the TCSPC technique. The fluorescence from the quartz sample cell was focused through a 540 nm bandpass (80 nm bandwidth) filter onto a fast photomultiplier that was connected to the TCSPC electronics (PicoQuant, TimeHarp 200). The time resolution of the TCSPC electronics is better than 40 ps. The acquired data were processed using PicoQuant software for fluorescence decay based on iterative reconvolution of the instrument response with nonlinear least-squares error minimization using the Levenberg-Marquardt or Simplex algorithms. Standard exponential (up to four exponential terms) and rate constant distribution models can be fitted to the observed data. The instrumental response function, the decay backgrounds, as well as time shift are included as fit parameters.
Transient absorption measurements
Transient absorption experiments used for pumping the S1 and S2 states of flavin the second harmonic of a fraction of the output of a Ti:sapphire regenerative amplifier (Coherent) at 250 kHz repetition rate, seeded by a mode-locked oscillator at 76 MHz. The remainder of the 800 nm beam was used to pump an optical parametric amplifier whose tunable wavelength output (490-700 nm) was used as the probe pulse. The density of energy per pump and probe pulses incident onto the sample was ∼ 1.7 μJ/mm2and ∼ 10 nJ/mm2, respectively. For measurements of relaxation decay, the probe beam was split into reference and a signal beams with the reference pulse arriving at a fixed time before the pump. The actual probe pulse arrived at a delayed time after the pump that was continuously adjusted using a variable length delay line (500 ps max.). A mechanical chopper was used to modulate the pump beam at 800 Hz. The differential signal between the reference and the probe beams at the modulation frequency was measured using a lock-in amplifier. Data were fitted using a multi-exponential decay model and common nonlinear regression methods provided by the Origin software:
RESULTS
Fluorescence decay analysis
Previous flash photo-activation experiments on AppAwt demonstrated that the red shifted absorption changes accompanying the transition from “dark” adapted” to “light adapted” forms of AppA involves a biphasic process with a fast phase occurring in less than 1 μs and a slow phase occurring at approximately 5 ms (6). To better resolve fast events, we undertook fluorescence lifetime experiments and ultrafast transient absorption spectroscopy on both AppAwt and Y21F mutant proteins. The Y21F mutant contains a substitution of phenylalanine for tyrosine at position 21 in AppA. Tyr21 has been implicated in being one of several amino acid side chains that form an intricate hydrogen bond network between the apoprotein and the flavin that is involved in promoting the photocycle (Figure 1) (1, 6). The Y21F mutant was previously shown to bind FAD but was defective in forming the long-lived red-shifted product of the photocycle (6) presumably due to disruption of a hydrogen-bond to Gln63 (1). This mutant also lacks the fluorescence quenching that is observed with wild type AppA upon light-excitation (data not shown).
Figure 1.
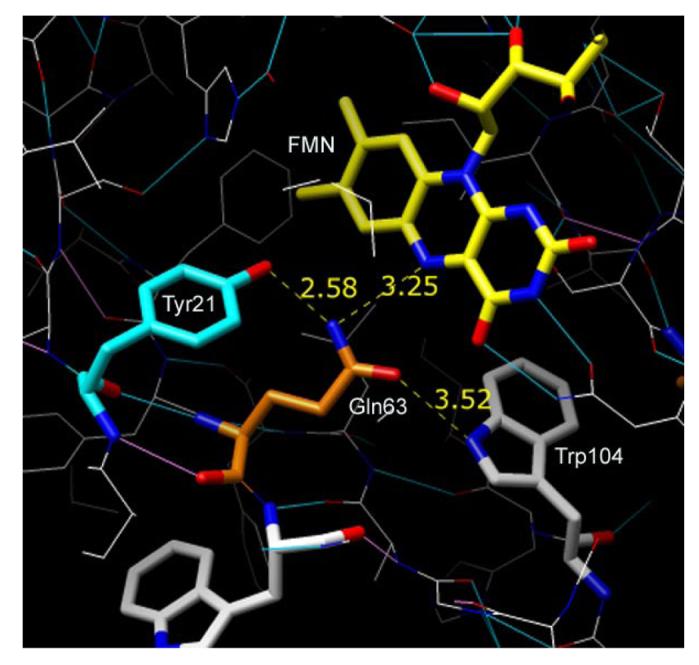
The FAD-binding pocket in the AppA crystal in the dark state. FMN and amino acids important in photocycle of AppA are indicated. In blue: nitrogen atoms, in red: oxygen atoms. H-bonds between amino acids and isoalloxazine ring (dashed yellow line) and their distances are indicated.
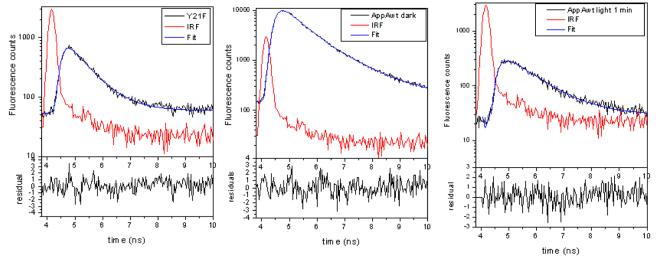
Even though previous steady-state studies have established that the Y21F mutant fails to form the final product, they did not reveal whether or not this mutant is able to undertake rapid photochemistry that may occur early in the photocycle. The results in Figure 2 show typical time-resolved fluorescence of the AppAwt and of the Y21F mutant in solution. Two-photon excitation with 800 nm light has been used to eliminate saturation of absorption in condensed phase and allowed us to measure the fluorescence life-times of AppAwt and Y21F mutant in samples that were either dark-adapted or exposed to strong white light (light-adapted) immediately before measurements. A comparison of the fluorescence decays in AppA wt and the Y21F mutant is provided in Table 1.
Figure 2.
Fluorescence decays and multi-exponential fits of the dark adapted Y21F mutant and AppA wt. Right image: AppAwt was illuminated with strong blue light for 2 minutes and data collection was done for 1 minute immediately after illumination. Red line represents instrument response function. Results of multiexponential deconvolution can be found at Table 1. The values of χ2 are following: 1.29 for Y21F (left), 1.32 for AppAwt dark form (middle), and 0.99 for AppAwt light form (right).
Table 1.
Fluorescence life-times of AppAwt and Y21F mutant. Negative amplitudes indicate a rising component.
| A1 | τ1(ns) | A2 | τ2 (ns) | A3 | τ3 (ns) | |
|---|---|---|---|---|---|---|
| Mutant Y21F | − 49 % | 0.124±0.022 | 51 % | 0.587±0.017 | ||
|
WT (dark adapted) |
− 50 % | 0.253 ±0.009 | 44 % | 0.551±0.023 | 6 % | 1.65 ±0.05 |
|
WT (light- exposed) 3min.collection 1min.collection |
50 % −61 % |
0.274 ±0.022 0.212 ±0.018 |
45 % 39 % |
0.56 ±0.06 1.00 ±0.034 |
5 % |
1.82 ±0.11 |
As indicated in Table 1, most of dark-adapted AppAwt and Y21F mutant protein shows a fluorescence decay with a lifetime of 0.55-0.59 ns. However, a small part, about 6%, of dark-adapted AppAwt shows fluorescence decay with a lifetime of 1.65 ns. Interestingly, after light-excitation, the AppAwt fluorescence showed mono-exponential decay with a lifetime of 1.0 ns. This component is missing in data collected for a time longer than 3 minutes, when lots of molecules of light-excited AppA returned back to the dark state. This result suggests that an intermediate is formed during the decay from light-excited to dark forms that lasts for approximately 1 minute. The values of fluorescence decay that we measured are similar to values published by Gauden et al. (9) who reported 25 ps, 150 ps, 670 ps and 3.8 ns species for AppAwt. Although we do not observe the first two short-lived species due the limit of resolution of our fluorescence lifetime setup, the reported 670 ps major component agrees well with our 0.5-0.6 ns major decay. However, our longest lifetime is somewhat shorter than the one reported (1.7 ns vs. 3.8 ns). Analysis of the Y21F mutant indicates the presence of only one major fluorescence decay component of 0.55 ns, which occurs when the mutant protein is either kept in the dark or illuminated just prior to fluorescence measurement. It is also interesting to note that the rise of the fluorescence signal is somewhat faster in the mutant (∼0.1 ns) than in AppAwt (∼0.2 ns) protein.
Transient absorption analysis of light excited AppA and Y21F mutant
Pump-probe transient absorption measurements were performed on both AppAwt and Y21F mutant protein to obtain information on events that occur at early steps of the AppA photocycle. Trial experiments indicated that more than 8 minutes of laser exposure resulted in the beginning of irreversible bleaching of the major flavin absorption band at 450 nm with an increase of a new band at 300 nm (data not shown), which indicates flavin reduction. A long-lived component (>1ns) also started to appear upon extended laser irradiation in addition to the short-lived ps components. We concluded that long exposure to the laser beam resulted in flavin being liberated from its binding pocket and subsequently reduced. Thus, each pump-probe transient absorption spectroscopy analysis utilized freshly prepared samples of AppAwt and Y21F mutant protein that was laser irradiated for less than 8 minutes. A UV-VIS absorption measurement was also performed at the end of each experiment to check that the native state of the protein was unaffected. We also observed that laser-irradiated AppAwt is converted to the “light-adapted” form during measurement as evidenced by UV-VIS spectral analysis of the sample following laser excitation (data not shown). Note that re-circulation of a sample in a cell would also not yield measurement of the “dark-adapted” state as the interval between consecutive laser pulses is too short (4 μs) to allow a flow to displace enough sample from the laser focal point between laser pulses. For example, with a flow of the sample at 20 cm/s, the sample would move only 0.8 μm between laser pulses. Since the laser beam is 0.5 mm in diameter, 625 laser pulses would occur before a sample is completely changed. Therefore, the laser intensity was kept at its lowest possible level, with no re-circulation, thus providing transient absorption measurements of AppAwt only in its light-adapted state.
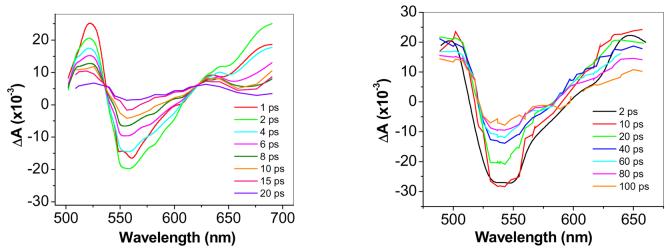
Figure 3 shows transient absorption spectra for both AppAwt and Y21F mutant protein taken at various times after laser excitation. AppAwt has three positive transient absorption peaks and one negative peak in the measured region between 480 nm and 695 nm. One positive absorption band is at 520 nm and a second one with near identical transients is beyond 675 nm. A third peak at 630 nm decays more slowly than the rest of transient absorption peaks. The entire AppA spectra is red-shifted ∼15 nm relative to that observed with the Y21F mutant that is locked in the dark-adapted state, which is in agreement with our expectations that transient absorption spectra of AppAwt is measuring transitions to its light-adapted state. Analysis of the Y21F mutant also shows an absence of the 630 nm peak as well as blue shifted peaks at 505 and 645 nm.
Figure 3.
Transient absorption spectra of AppAwt (a) and Y21F (b) mutant taken at different time delays. Sample concentrations were ∼ 3mg/ml.
No significant shifts in the position of absorption peaks were observed during the lifetime of the transient absorption signal. The negative transient absorption can be attributed to stimulated emission from S1 excited state. Positive peaks are known to occur in the case of excited flavin in the oxidized state and are due to absorption from the S1 excited state to the S2 and S3 manifold (12). However, similar large positive peaks of transient absorption are also present in the case of flavins in the semiquinone state and even in reduced state (13, 14). Therefore, it is difficult to assess, for example, whether an electron transfer has occurred by just looking at the transient spectra.
Due to similarities between the transient spectra, it is necessary to have a closer look at the individual decays in order to be able to differentiate between possible mechanisms of excitation decay. Figure 4 shows typical transient absorption decays for AppA wt and AppA Y21F mutant, taken at the transient absorption maxima, the minimum and around 630 nm where the slow decaying component in AppAwt was seen. The transients were fitted with a multiple exponential decay model with coefficients given in Table 2. One difference that stands out between AppAwt and Y21F upon inspection of Figure 4 and Table 2 is the different rise times of the positive absorption transients in these different proteins. For AppAwt, the rise completes in ∼ 400-500 fs at most wavelengths, which represents roughly twice the laser pulse duration. For Y21F, a longer, 4-6 ps, characteristic time is necessary to fit the positive edge of the transient peak absorption. In general, the AppAwt decay is fast, with τ1 = 4-7 ps and τ2 = 23-32 ps. Around 630 nm, a relatively long-lived species was detected decaying with τ = 196 ps. It is interesting to note that the stimulated emission of AppAwt (negative signal) decays very fast as a single exponential with a lifetime of τ = 4.8 ps.
Figure 4.
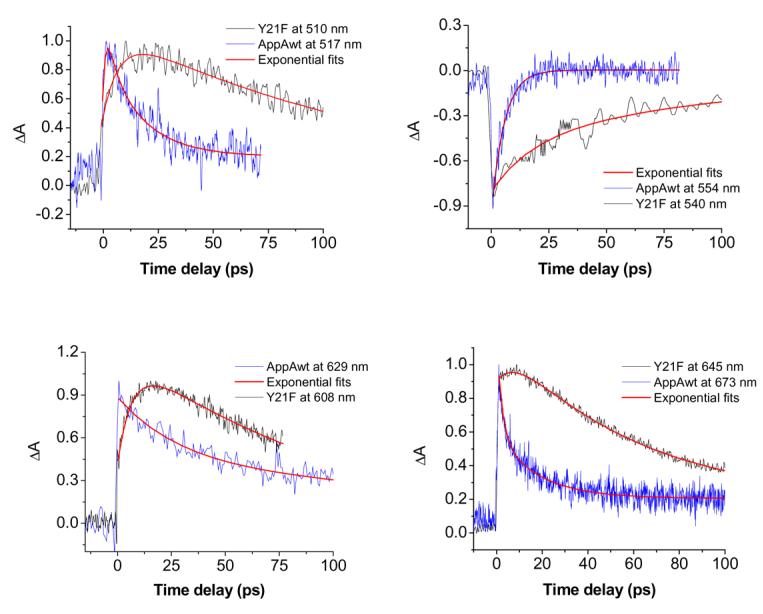
Representative curves of transient absorption of AppAwt (blue) and Y21F mutant (black) at different wavelengths (as indicated). Multi-exponential fits were made using Origin software and are in red.
Table 2.
Results of multi-exponential fits of transient absorption of AppAwt and Y21F mutant. Negative amplitude represents raising exponentials.
| λ (nm) |
A1 % |
T1(ps) |
A2 % |
T2 (ps) |
A3 % |
T3 (ps) |
A4 % |
T4 (ps) |
|---|---|---|---|---|---|---|---|---|
| Mut 510 | −32 | 6.6 ± 0.5 | 60 | 134.7 ± 2.4 | ||||
| 540 | 15 | 0.83 ± 0.11 | −29 | 19.1 ± 1.8 | −35 | 130.8 ± 3.8 | ||
| 608 | − 58 | 6.9 ± 0.2 | 87 | 97.4 ± 1.6 | ||||
| 645 | −11 | 4.0 ± 0.3 | 53 | 90.9 ± 0.4 | ||||
| WT 517 | −30 | 1.28 ± 0.7 | 35 | 5.2 ± 3.1 | 27 | 28.3 ± 9.0 | ||
| 554 | 20 | 0.53 ± 0.08 | −80 | 4.8 ± 0.25 | ||||
| 629 | 17 | 0.44 ± 0.12 | 32 | 23.5 ± 2.8 | 57 | 196 ± 7.7 | ||
| 673 | −25 | 0.38 ± 0.03 | 20 | 7.4 ± 0.3 | 7 | 175 ± 12.3 | ||
| 692 | −66 | 0.35 ± 0.03 | 25 | 3.9 ± 0.2 | 11 | 32.0 ± 1.0 |
As mentioned, the Y21F mutant can be fitted with a single rising exponential with τ = 4-6 ps at all wavelengths where positive signal is observed. Most of transient absorption of the mutant decays more slowly than AppAwt (within 90-135 ps) with one contributing fast component of 19 ps at the stimulated emission decay (540 nm). The 4-6 ps component is missing in the negative peak coming from the stimulated emission with fast formation of stimulated emission peak at 0.8 ps seen instead. This indicates that the S1 excited state of the flavin in the mutant is formed fast after light-excitation with laser and that other slower processes follow that contribute to the positive peaks of transient absorption.
DISCUSSION
To better understand the mechanism of the AppA photocycle, we have examined the fluorescence life-times and the transient absorption in both AppAwt and Y21F mutant. In general, very fast decays of fluorescence and transient absorption on ps timescale were seen for both AppAwt and mutant. Examining the mechanisms responsible for the observed decays indicates several possibilities. One possible mechanism could be intersystem crossing resulting in triplet formation. However, intersystem crossing can be discounted in this case since it usually occurs at a ns timeframe and is typically accompanied by a large absorption band beyond 650 nm.
The decay lifetime of a triplet excited state also typically occurs on a μs timescale, which was not observed. Moreover, it is generally accepted that enhanced intersystem crossing is not the principal mechanism of fluorescence quenching for most flavoproteins (15) with some exceptions, like the plant LOV domain photocycle, which does undergo triplet formation (16, 17).
Resonance energy transfer as a mechanism of fast decay can also be discarded since it is energetically unfavorable in flavoproteins as there is no overlap between flavin emission and excitation wavelengths of aromatic amino acids. Collision quenching could also play a role in the fluorescence quenching, however, it typically occurs on a much slower time scale of > ns (18).
The most likely mechanism for the observed fast decays of fluorescence and transient absorption is electron transfer, followed by proton transfer to the flavin. Many flavoproteins fall in this category with a few transient absorption studies indicating that electron transfer occurs on a ps timescale (13, 18), followed by charge recombination. The quenching of fluorescence from the S1 excited state could also occur through H-bond rearrangement since it is known that the rigidity of the chromophore plays an important role in determining the rate of radiationless transitions (15). H-bonding is also known to modify the redox potential of the flavin (19), having an indirect influence on electron transfer rates.
As the transient absorption spectra of AppAwt in the dark state have just been published (9), our data on transient absorption of light excited AppAwt are complementary and indeed allow direct comparison between the spectral properties of AppAwt in light and dark states. Six absorption transient species were reported for AppAwt in the dark state. Species at 250 fs and 1.2 ps were attributed to the flavin S1 excited state formation and species at 90 ps, 590 ps and 2.7 ns (+ one non-decaying comp.) as parallel decaying components (9). As mentioned earlier, we observed a component with >1 ns lifetime only in samples that were irradiated for more than 8 min. As this component is missing on shorter laser exposed measurements with fresh protein, we attribute this long-lived species to the formation of free flavin as a byproduct of laser excitation. Note that a small contribution from a ns component is observable in time-resolved fluorescence of fresh samples. The fact that we do not see it in transient absorption for fresh samples while we start to see it for irradiated samples is probably due to insufficient sensitivity.
When comparing dark and light states of AppAwt there are clearly different spectral species observed during decay with similar, fast, rising lifetimes found. The transient absorption spectrum of AppAwt in the light state is comparable with the dark state AppA on the measured timescale. However, several very fast spectral species are observed in the transient absorption decay of AppA in the light state (4-7 ps and 23-32 ps, and a ∼200 ps species around 630 nm) compared to the dark state (90 ps, 590 ps , 2.7 ps and a non-decaying component). The much faster decay of transient spectral species correlates well with the strong fluorescence quenching of AppAwt in the light state. In both light and dark states, the transient absorption rises very fast, within ∼1 ps. Interestingly, the transient absorption rises more slowly, within 4-6 ps, in the Y21F mutant indicating that the Tyr21 hydroxyl is important for the first picoseconds of the photocycle. Most of the transient absorption of mutant decays within 90-134 ps, which is close to the 90 ps component of AppA observed in the dark state (9). Therefore, this mutant is clearly functionally closer to the AppAwt in the dark state that has a similar spectrum, comparable fluorescence efficiency and similar transient absorption and fluorescence decay lifetimes.
Involvement of electron transfer in the AppA photocycle
It is well known that oxidized flavin is a strong electron acceptor when excited by light and that the redox properties of nearby aromatic amino acids such as Tyr or Trp provide a favorable driving force for electron transfer reactions. Indeed, Tyr and Trp are known to donate an electron to a proximal isoalloxazine ring in several flavoproteins (13, 18). This type of electron transfer process is usually very fast on the order of fs or a few ps depending on charge transfer distance and orientation of the aromatic residues relative to the isoalloxazine ring. For a few flavoproteins, the transient absorption signal is dominated by positive electron transfer absorption at wavelengths longer than 545 nm (18). The ps range of transient absorption observed for AppAwt in the dark and light states indicates that electron transfer may indeed be the first event of this photocycle. In such case, all the dynamics observed would be a superposition of FAD* decay, FAD− formation (rise) and FAD− decay (charge recombination), if neglecting Tyr+/Trp+ absorption.
Inspection of the flavin binding pocket of AppA (Figure 1), as defined by recent X-ray crystallographic analysis (1), indicates that Tyr21 and Trp104 are potential electron donors. Both residues are 4.5-5.5 Å from the isoalloxazine ring and also form hydrogen bond interactions with Gln63. In the dark form, the amine group of Gln63 is proposed to form an H-bond with both N5 of the flavin ring and the hydroxyl of Tyr21. At the same time the carbonyl of Gln63 is thought to form an H-bond to Trp104. Upon light excitation, Tyr21 and Trp104 are proposed to undergo hydrogen bond rearrangements with Gln63 undergoing a ∼180° rotation that allows the carbonyl of Gln63 to form a hydrogen bond to Tyr21 and the amine group to form a new hydrogen bond to the flavin at C(4)=0. Presumably, light excitation of the flavin isoalloxazine ring is the driving force that favors initiation of these electron transfer events. Fast electron transfer, and subsequent efficient charge recombination, can happen with the condition that Tyr is properly oriented. In the dark state of AppA, the Tyr21 ring is approximately perpendicular to the isoalloxazine ring and at convenient ∼ 4.5 Å distance (Fig 1), thus electron transfer could potentially occur between Tyr21 and N5 of flavin upon light excitation. This would be followed by charge recombination and H-bond re-arrangement as supported by NMR and FTIR studies on AppA (6, 8). Indeed, our results indicate that Tyr21 participates in an event leading to the formation of an intermediate within ∼1ps as this fast species is missing in the Y21F mutant (rise time: ∼ 4-6 ps).
Although our data on AppAwt in the light state is not a direct measurement of the biological process of light excitation from dark to light state, it still provides considerable insight to the mechanism of the photocycle. Considering the confined environment of Tyr21 in the hydrophobic flavin-binding pocket of AppA, Tyr21 is not likely to move significantly during light excitation, so electron transfer is likely to occur in both light and dark states each with different kinetics of charge recombination. Electron transfer and especially ground state recovery are slower for the dark form of AppA than is observed with the light form which indicates that the distance between Tyr21 and flavin is likely to be slightly larger (or their orientation less favorable) in the dark state, than in the light state. Analysis of AppA in the light state is also biologically relevant at high light conditions where fast fluorescence quenching can function as efficient protection against strong light.
For comparison, glucose oxidase has a similar orientation of Tyr/Trp and flavin as is observed in AppA and a reported photoinduced electron transfer rate from Tyr/Trp to FAD of 1.8 ps (18). In this case, the observed ground state recovery decay was 35 ps. For riboflavine (vitamin B-2)-binding protein, which has the Tyr parallel to the isoalloxazine ring, the characteristic transient absorption of the electron transfer-state took less than 0.7 ps to develop, while charge recombination and total ground state recovery took less than 9 ps.
Several lines of evidence suggest that there is formation of a flavin radical during excitation. The very fast 4.6 ps decay of stimulated emission at 553 nm (Table 2) in AppAwt light state is indicative of an electron transfer process followed by proton transfer to create a temporary neutral flavin semiquinone, which is non-fluorescent and doesn't show stimulated emission (13). Similar fast decaying species is seen in the positive peaks of transient absorption of AppAwt (between 4-7 ps). Neutral flavin semiquinone possesses red-shifted ground state absorption with maxima around 580 and 630 nm (20). If fast proton transfer from Tyr21 to N5 of flavin follows electron transfer, the 630 nm transient absorption peak with τ ∼ 200 ps identified in AppAwt could thus originate from temporary semiquinone ground state absorption.
The formation of excited radicals FAD•− and Tyr•+ was not observed in our room temperature studies which is not surprising given that these species are ultra-fast and difficult to spectrally discriminate at these time scales (21). A partial charge transfer could also lead to spin inversion within intermediate radical pair (Fl γ− ….Q γ+) that results in deactivation rather than formation of free radicals (15):
However, low-temperature studies of Tl10078 from T. elongatus (11) did detect a 5-nm red shifted intermediate that can be trapped only below 50 K. We speculate that this intermediate may be a stabilized anion flavin radical as electron transfer could still occur at low temperatures that would impede large structural changes in the protein. The ground state absorption spectrum of this putative anion flavin radical shows a large peak at about 480 nm with wide of absorption reaching beyond the red absorption edge of the oxidized flavin spectrum (22).
Fast electron and putative proton transfer cannot occur in the Y21F mutant because the hydroxyl group is missing. Instead, a slower rising component of ∼5 ps is seen that still decays on a picosecond timescale. The slower rising intermediate in the Y21F mutant cannot be rationalized through excited state absorption of oxidized flavin because transitions from S1 to the upper states occur rapidly in flavin regardless of whether or not a Tyr is present. It is known that the S2 excited state in flavoproteins will decay to S1 within 100 fs (12), which is faster than what we observe for AppAwt and very far from the observed mutant dynamics. In absence of fast electron transfer from Tyr21, it is possible that a slower electron transfer event may be occurring from a nearby aromatic amino acid such as Trp104 that is ∼5.5 Å from the flavin ring (1). As further reactions cannot proceed in the Y21F mutant, and the H-bonding network cannot be formed like that in the wild-type protein, the charges recombine within 90-130 ps, which is similar to the recombination life times of AppA in the dark.
It is interesting to compare transient absorption results with time-resolved fluorescence as there are several correlations. For example, transient absorption of the Y21F mutant rises within ∼4-7 ps and then decays within 90-130 ps (Table 2). Interestingly, time-resolved fluorescence of the Y21F mutant also rises within ∼120 ps and then decays within 0.6 ns (Table 1). It is therefore reasonable to conclude that the observed transient absorption decay product at τ= 90-130 ps may be forming the ∼120 ps fluorescent species that has the 0.6 ns lifetime. In the case of AppAwt, there are three spectral species observed by transient absorption at ∼5, 28 and 200 ps (Table 2) with the ∼200 ps spectral species potentially be the fluorescent species that forms at ∼200 ps with a lifetime of 1 ns. Furthermore, since the fast transient absorption species at ∼5, ∼28 ps in AppAwt are non-fluorescent, they could also potentially be responsible for formation of the 200 ps species.
The results of this, and other studies, support a model that places Tyr21 and Gln63 as critical residues in the BLUF photocycle. In the “dark” form, the amine group of Gln63 is proposed to form an H-bond with both N5 of the flavin as well as with the hydroxyl of Tyr21(Figure 1) (1). Upon light excitation, it has been proposed that Gln63 rotates such that the carbonyl of Gln63 now forms a hydrogen bond to Tyr 21 and the amine group forms a H-bond with C4=O of the flavin. Indeed, recent FTIR studies indicates that H-bond formation on C4=O does occur after light excitation (8). In this context, electron transfer from Tyr21 to flavin would place a negative charge on N5 of flavin that likely attracts a proton from Tyr21 to disturb Gln63 H-bonding. In this case, the H-bond between the Tyr21 hydroxyl and the amine of Gln63 would be broken due to the presence of a tyrosine radical. These early events would then destabilize the H-bonding network and provide energy for Gln63 re-orientation to occur, under which a new H-bond between C=O of Gln63 and Tyr21 would be formed. Further studies on several other AppA mutants are planned to confirm the involvement of specific amino acids in this photocycle.
Acknowledgements
Thanks to Dr. Shinji Masuda for cloning AppA. We are grateful to Dr. Spencer Anderson for his input and comments on this manuscript.
ABBREVIATIONS
- BLUF
Blue-Light sensing Using FAD
- TCSPC
time-correlated single photon counting
- DDMAB
N-dodecyl-N,N-(dimethylammonio)butyrate
Footnotes
This work was supported by a National Institutes of Health Grant GM53940 to CEB.
REFERENCES
- 1.Anderson S, Dragnea V, Masuda S, Ybe J, Moffat K, Bauer C. Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry. 2005;44:7998–8005. doi: 10.1021/bi0502691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110:613–623. doi: 10.1016/s0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 3.Rajagopal S, Key JM, Purcell EB, Boerema DJ, Moffat K. Purification and initial characterization of a putative blue light-regulated phosphodiesterase from Escherichia coli. Photochemistry and Photobiology. 2004;80:542–547. doi: 10.1562/2004-06-16-RA-203. [DOI] [PubMed] [Google Scholar]
- 4.Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Sugai M, Takahashi T, Hori T, Watanabe M. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
- 5.Masuda S, Ono T. Biochemical characterization of the major adenylyl cyclase, Cya1, in the cyanobacterium Synechocystis sp PCC 6803. Febs Letters. 2004;577:255–258. doi: 10.1016/j.febslet.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 6.Kraft BJ, Masuda S, Kikuchi J, Dragnea V, Tollin G, Zaleski JM, Bauer CE. Spectroscopic and mutational analysis of the blue-light photoreceptor AppA: A novel photocycle involving flavin stacking with an aromatic amino acid. Biochemistry. 2003;42:6726–6734. doi: 10.1021/bi030055o. [DOI] [PubMed] [Google Scholar]
- 7.Laan W, van der Horst MA, van Stokkum IH, Hellingwerf KJ. Initial characterization of the primary photochemistry of AppA, a blue-light-using flavin adenine dinucleotide-domain containing transcriptional antirepressor protein from Rhodobacter sphaeroides: A key role for reversible intramolecular proton transfer from the flavin adenine dinucleotide chromophore to a conserved tyrosine? Photochemistry and Photobiology. 2003;78:290–297. doi: 10.1562/0031-8655(2003)078<0290:icotpp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 8.Masuda S, Hasegawa K, Ono T. Light-induced structural changes of apoprotein and chromophore in the sensor of blue light using FAD (BLUF) domain of AppA for a signaling state. Biochemistry. 2005;44:1215–1224. doi: 10.1021/bi047876t. [DOI] [PubMed] [Google Scholar]
- 9.Gauden M, Yeremenko S, Laan W, van Stokkum IHM, Ihalainen JA, van Grondelle R, Hellingwerf KJ, Kennis JTM. Photocycle of the flavin-binding photoreceptor AppA, a bacterial transcriptional antirepressor of photosynthesis genes. Biochemistry. 2005;44:3653–3662. doi: 10.1021/bi047359a. [DOI] [PubMed] [Google Scholar]
- 10.Masuda S, Hasegawa K, Ishii A, Ono T. Light-induced structural changes in a putative blue-light receptor with a novel FAD binding fold sensor of blue-light using FAD (BLUF); Slr1694 of Synechocystis sp PCC6803. Biochemistry. 2004;43:5304–5313. doi: 10.1021/bi049836v. [DOI] [PubMed] [Google Scholar]
- 11.Fukushima Y, Okajima K, Shibata Y, Ikeuchi M, Itoh S. Primary intermediate in the photocycle of a blue-light sensory blue-light FAD-protein, Tll0078, of Thermosynechococcus elongatus BP-1. Biochemistry. 2005;44:5149–5158. doi: 10.1021/bi048044y. [DOI] [PubMed] [Google Scholar]
- 12.Stanley RJ, MacFarlane AW. Ultrafast excited state dynamics of oxidized flavins: Direct observations of quenching by purines. Journal of Physical Chemistry A. 2000;104:6899–6906. [Google Scholar]
- 13.Pan J, Byrdin M, Aubert C, Eker APM, Brettel K, Vos MH. Excited-state properties of flavin radicals in flavoproteins: Femtosecond spectroscopy of DNA photolyase, glucose oxidase, and flavodoxin. Journal of Physical Chemistry B. 2004;108:10160–10167. [Google Scholar]
- 14.Enescu M, Lindqvist L, Soep B. Excited-state dynamics of fully reduced flavins and flavoenzymes studied at subpicosecond time resolution. Photochemistry and Photobiology. 1998;68:150–156. doi: 10.1562/0031-8655(1998)068<0150:esdofr>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Heelis PF. The Photophysical and Photochemical Properties of Flavins (Isoalloxazines) Chemical Society Reviews. 1982;11:15–39. [Google Scholar]
- 16.Kottke T, Heberle J, Hehn D, Dick B, Hegemann P. Phot-LOV1: Photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii. Biophysical Journal. 2003;84:1192–1201. doi: 10.1016/S0006-3495(03)74933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennis JTM, Crosson S, Gauden M, van Stokkum IHM, Moffat K, van Grondelle R. Primary reactions of the LOV2 domain of phototropin, a plant blue-light photoreceptor. Biochemistry. 2003;42:3385–3392. doi: 10.1021/bi034022k. [DOI] [PubMed] [Google Scholar]
- 18.Zhong DP, Zewail AH. Femtosecond dynamics of flavoproteins: Charge separation and recombination in riboflavine (vitamin B-2)-binding protein and in glucose oxidase enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11867–11872. doi: 10.1073/pnas.211440398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yagi K, Ohishi N, Nishimoto K, Choi JD, Song PS. Effect of Hydrogen-Bonding on Electronic-Spectra and Reactivity of Flavins. Biochemistry. 1980;19:1553–1557. doi: 10.1021/bi00549a003. [DOI] [PubMed] [Google Scholar]
- 20.Jorns MS, Baldwin ET, Sancar GB, Sancar A. Action Mechanism of Escherichia-Coli DNA Photolyase .2. Role of the Chromophores in Catalysis. Journal of Biological Chemistry. 1987;262:486–491. [PubMed] [Google Scholar]
- 21.Mataga N, Chosrowjan H, Shibata Y, Tanaka F. Ultrafast fluorescence quenching dynamics of flavin chromophores in protein nanospace. Journal of Physical Chemistry B. 1998;102:7081–7084. [Google Scholar]
- 22.Niemz A, Imbriglio J, Rotello VM. Model systems for flavoenzyme activity: One- and two-electron reduction of flavins in aprotic hydrophobic environments. Journal of the American Chemical Society. 1997;119:887–892. [Google Scholar]