Abstract
Purpose
The EML4-ALK fusion oncogene represents a novel molecular target in a small subset of non–small-cell lung cancers (NSCLC). To aid in identification and treatment of these patients, we examined the clinical characteristics and treatment outcomes of patients who had NSCLC with and without EML4-ALK.
Patients and Methods
Patients with NSCLC were selected for genetic screening on the basis of two or more of the following characteristics: female sex, Asian ethnicity, never/light smoking history, and adenocarcinoma histology. EML4-ALK was identified by using fluorescent in situ hybridization for ALK rearrangements and was confirmed by immunohistochemistry for ALK expression. EGFR and KRAS mutations were determined by DNA sequencing.
Results
Of 141 tumors screened, 19 (13%) were EML4-ALK mutant, 31 (22%) were EGFR mutant, and 91 (65%) were wild type (WT/WT) for both ALK and EGFR. Compared with the EGFR mutant and WT/WT cohorts, patients with EML4-ALK mutant tumors were significantly younger (P < .001 and P = .005) and were more likely to be men (P = .036 and P = .039). Patients with EML4-ALK–positive tumors, like patients who harbored EGFR mutations, also were more likely to be never/light smokers compared with patients in the WT/WT cohort (P < .001). Eighteen of the 19 EML4-ALK tumors were adenocarcinomas, predominantly the signet ring cell subtype. Among patients with metastatic disease, EML4-ALK positivity was associated with resistance to EGFR tyrosine kinase inhibitors (TKIs). Patients in the EML4-ALK cohort and the WT/WT cohort showed similar response rates to platinum-based combination chemotherapy and no difference in overall survival.
Conclusion
EML4-ALK defines a molecular subset of NSCLC with distinct clinical characteristics. Patients who harbor this mutation do not benefit from EGFR TKIs and should be directed to trials of ALK-targeted agents.
INTRODUCTION
Lung cancer is the leading cause of cancer deaths in the world, as greater than 1 million deaths from lung cancer occur each year.1 Although cytotoxic chemotherapy remains the mainstay of treatment for the majority of patients with advanced non–small-cell lung cancer (NSCLC),2,3 tyrosine kinase–based therapeutics have assumed an increasingly important role, particularly in genetically defined subsets of patients. For example, activating mutations in the receptor tyrosine kinase epidermal growth factor receptor (EGFR) define a small subset of patients with NSCLC who have sensitivity to EGFR tyrosine kinase inhibitors (TKIs), such as gefitinib or erlotinib.4,5 In mutation-positive patients with previously untreated, advanced disease, gefitinib recently has been shown to be superior to cytotoxic chemotherapy.6 The remarkable success of EGFR TKIs highlights the importance of identifying genotype-specific subsets of patients to guide the appropriate selection of targeted therapies.
The EML4-ALK fusion oncogene represents one of the newest molecular targets in NSCLC. First described in 2007,7,8 the fusion results from a small inversion within chromosome 2p, which leads to expression of a chimeric tyrosine kinase, in which the N-terminal half of echinoderm microtubule-associated protein-like 4 (EML4) is fused to the intracellular kinase domain of anaplastic lymphoma kinase (ALK).9 EML4-ALK possesses potent oncogenic activity both in vitro and in vivo.7,10 This activity can be effectively blocked by small-molecule inhibitors that target ALK,10,11 which supports a role for EML4-ALK as a key driver of lung tumorigenesis.
Several studies have examined the frequency of EML4-ALK in patients with NSCLC. In the original report of EML4-ALK, five of 75 lung tumors demonstrated expression of the fusion transcript, which corresponded to a frequency of 6.7%.7 In subsequent studies that primarily involved Asian patients with early-stage, resectable disease, EML4-ALK has been detected in a lower percentage of patients, which ranged from 1% to 4.9%.8,11–17 These findings suggest that, in unselected NSCLC populations, the EML4-ALK rearrangement is a relatively rare event. In part because of the small number of positive instances identified per study, the key pathologic, epidemiologic, and demographic features associated with EML4-ALK have not been definitively established. Furthermore, whether patients with this chromosomal rearrangement share similar outcomes to other genetically defined subsets of NSCLC, particularly in the metastatic setting, also is unknown.
Here, we present the largest series to date of EML4-ALK–positive patients with NSCLC. We describe the clinical and pathologic characteristics of patients with EML4-ALK, and we also examine treatment response and survival in patients who have metastatic disease with and without EML4-ALK.
PATIENTS AND METHODS
Study Population
The majority of patients were seen at the Massachusetts General Hospital Cancer Center. Three patients were observed at the Beth Israel Deaconess Medical Center, and one patient was observed at the Peter MacCallum Cancer Centre. Patients were selected for genetic screening on the basis of two or more of the following clinical characteristics: female sex, Asian ethnicity, never/light smoking history (defined in Table 1), and adenocarcinoma histology. All study patients had biopsy-proven NSCLC, and the majority of patients had metastatic disease. Patients with insufficient tissue for genetic testing, or for whom EML4-ALK fluorescent in situ hybridization (FISH) was inconclusive, were excluded. This study was approved by the institutional review boards at each of the participating centers.
Table 1.
Clinical Characteristics of Genetically Screened Patients With Non–Small-Cell Lung Cancer
| Characteristic | Patients (N = 141) |
|
|---|---|---|
| No. | % | |
| Age, years | ||
| Median | 63 | |
| Range | 29-90 | |
| Sex | ||
| Male | 48 | 34 |
| Female | 93 | 66 |
| Smoking history† | ||
| Never smoker | 59 | 42 |
| Light smoker | 26 | 18 |
| Smoker | 56 | 40 |
| Ethnicity | ||
| Asian | 9 | 6 |
| Non-Asian | 132 | 94 |
| Pathology | ||
| Adeno | 89 | 63 |
| BAC* | 41 | 29 |
| Adenosquamous | 4 | 3 |
| Squamous | 2 | 1 |
| Large cell/NOS | 5 | 4 |
| Stage‡ | ||
| IA | 14 | 10 |
| IB | 11 | 8 |
| IIA | 1 | 1 |
| IIB | 0 | 0 |
| IIIA | 5 | 4 |
| IIIB | 4 | 3 |
| IV | 96 | 68 |
| Multifocal BAC | 10 | 7 |
Abbreviations: adeno, adenocarcinoma; BAC, bronchioloalveolar carcinoma; NOS, not otherwise specified.
Adenocarcinoma with any element of BAC was listed as BAC.
Never smokers have smoked < 100 cigarettes in their lifetime; light smokers have smoked ≤ 10 pack years; and smokers have smoked > 10 pack years.
Clinical stage represents stage at time of mutation testing. Stage was determined according to current American Joint Commission on Cancer guidelines; however, patients with malignant pleural effusions were classified as stage IV.
Data Collection
For all patients, medical records were reviewed to extract data on clinicopathologic characteristics. For patients with stage IV disease at the time of genetic screening, we examined treatment regimens, response rates, and outcomes. Patients with multifocal bronchioloalveolar carcinoma (BAC) were excluded from this analysis. In the majority of patients, interval computed tomography scans were available for review by one thoracic radiologist. Responses were classified by using standard RECIST (Response Evaluation Criteria in Solid Tumors).18 Time to progression (TTP) was measured from the first day of treatment until radiologic or clinical progression. Overall survival (OS) was measured from the date of diagnosis of metastatic NSCLC until the date of death. Patients without a known date of death were censored at the time of last follow-up.
Tumor Pathology and Mutation Analysis
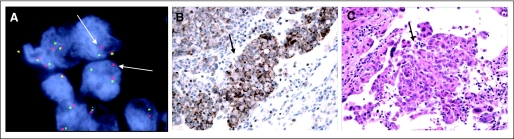
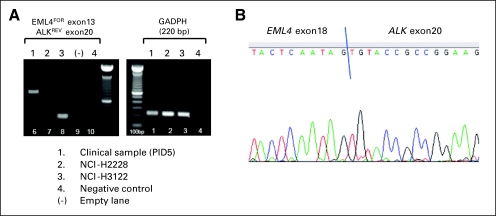
Tumor histology was classified by using WHO criteria.19 To identify ALK rearrangements, FISH was performed on formalin-fixed, paraffin-embedded tumors by using a break-apart probe to ALK (Vysis LSI ALK Dual Color, Break Apart Rearrangement Probe; Abbott Molecular, Abbott Park, IL; Fig 1A). All FISH-positive occurrences (defined as > 15% of tumor cells with split signals) were confirmed by immunohistochemistry (IHC) by using a mouse monoclonal antibody against ALK (clone ALK1; DAKO USA, Carpinteria, CA; Fig 1B). A subset of FISH-positive occurrences also was confirmed by reverse transcriptase polymerase chain reaction (Fig A1). EGFR and KRAS mutations were determined by direct DNA sequencing.
Fig 1.
Diagnostic features of EML4-ALK–positive non–small-cell lung cancer (NSCLC). (A) Fuorescent in situ hybridization (FISH) reveals a split of red and green probes that flank the ALK translocation site in an EML4-ALK–positive tumor (arrows). (B) ALK immunohistochemistry reveals cytoplasmic ALK staining. (C) Hematoxylin and eosin staining of the same tumor. Arrows in (B) and (C) indicate signet ring cells, which are commonly found in EML4-ALK–positive tumors.
Statistical Analysis
For clinical characteristics, treatment types, and response rates, Fisher's exact test was used to assess the association of genotype with dichotomous factors, whereas the Wilcoxon rank sum test was applied to continuous data. The Kaplan-Meier method was used to estimate TTP and OS, and the difference between genotypes was compared by using the log-rank test. General data analysis was conducted with SAS 9.1 (SAS Institute, Cary, NC), whereas StatXact 6.1 (Cytel Software, Cambridge, MA) was used to compute exact P values. All P values were based on a two-sided hypothesis.
RESULTS
Between November 2007 and October 2008, we screened 141 patients with NSCLC for ALK rearrangements, hereafter referred to as EML4-ALK. The criteria used to select patients for genetic screening were based on clinical features commonly associated with EGFR mutation.20,21 As a result, the cohort of screened patients was enriched for women, for never/light smokers, and for patients with adenocarcinomas or adenocarcinomas with bronchioloalveolar features (ie, BAC; Table 1). This enrichment strategy was chosen to target the population of never/light smokers and to enable identification of patients who harbored either EML4-ALK or EGFR mutation within the same study group.
Clinicopathologic Characteristics of EML4-ALK–Positive Patients
Of the 141 tumors screened, 19 (13%) harbored the EML4-ALK rearrangement, 31 (22%) harbored an activating EGFR mutation, and 91 (65%) were wild type for both ALK and EGFR (designated WT/WT). Of note, this WT/WT cohort included at least six patients with activating KRAS mutations (Table A1). Compared with patients who had EGFR mutant and WT/WT, EML4-ALK–positive patients were significantly younger; the median age was 52 years in EML4-ALK–positive patients compared with 66 years in patients with EGFR mutation and 64 years in patients with WT/WT status (Table 2; P < .001 and P = .005, respectively). EML4-ALK–positive patients also were more likely than either EGFR or WT/WT patients to be men (Table 2; P = .036 and P = .039, respectively). Although the EML4-ALK fusion oncogene was first discovered in a patient with a history of smoking,7 the EML4-ALK–positive patients in this series, like the EGFR patients, were significantly more likely to be never/light smokers compared with the WT/WT patients (P < .001).
Table 2.
Clinical Characteristics of Genotype-Specific Subsets of Patients With Non–Small-Cell Lung Cancer
| Characteristic | Genotype |
P |
||||||
|---|---|---|---|---|---|---|---|---|
|
ALK (n = 19) |
EGFR (n = 31) |
WT/WT (n = 91) |
||||||
| No. | % | No. | % | No. | % | ALK v EGFR | ALK v WT/WT | |
| Age, years | ||||||||
| Median | 52 | 66 | 64 | < .001 | .005 | |||
| Range | 29-76 | 36-90 | 29-87 | |||||
| Sex | ||||||||
| Male | 11 | 58 | 8 | 26 | 29 | 32 | .036 | .039 |
| Female | 8 | 42 | 23 | 74 | 62 | 68 | ||
| Smoking history | ||||||||
| Never smoker | 14 | 74 | 21 | 68 | 24 | 26 | .366 | < .001 |
| Light smoker | 5 | 26 | 6 | 19 | 15 | 16 | ||
| Smoker | 0 | 0 | 4 | 13 | 52 | 57 | ||
| Ethnicity | ||||||||
| Asian | 0 | 0 | 2 | 6 | 7 | 8 | .519 | .602 |
| Non-Asian | 19 | 100 | 29 | 94 | 84 | 92 | ||
| Pathology | ||||||||
| Adeno | 16 | 84 | 24 | 77 | 49 | 54 | .380* | .686* |
| BAC† | 2 | 11 | 7 | 23 | 32 | 35 | ||
| Adenosquamous | 1 | 5 | 0 | 0 | 3 | 3 | ||
| Squamous | 0 | 0 | 0 | 0 | 2 | 2 | ||
| Large cell/NOS | 0 | 0 | 0 | 0 | 5 | 6 | ||
| Stage | ||||||||
| IA | 2 | 11 | 2 | 6 | 10 | 11 | ||
| IB | 0 | 0 | 1 | 3 | 10 | 11 | ||
| IIA | 0 | 0 | 0 | 0 | 1 | 1 | ||
| IIB | 0 | 0 | 0 | 0 | 0 | 0 | ||
| IIIA | 0 | 0 | 2 | 6 | 3 | 3 | ||
| IIIB | 0 | 0 | 0 | 0 | 4 | 4 | ||
| IV | 17 | 89 | 26 | 84 | 53 | 58 | .695‡ | .051‡ |
| Multifocal BAC | 0 | 0 | 0 | 0 | 10 | 11 | ||
Abbreviations: WT, wild type; adeno, adenocarcinoma; BAC, bronchioloalveolar carcinoma; NOS, not otherwise specified.
Adeno and BAC v all others.
Adeno with any element of BAC is listed as BAC.
Stages I to III v IV.
Because requests for genetic screening originated primarily from medical oncology clinics, the majority of patients had metastatic disease at the time of screening (Table 1). Within the EML4-ALK cohort, 17 (89%) of 19 had stage IV disease. Similarly, 26 (84%) of 31 patients with EGFR mutations had stage IV disease (Table 2; P = .695). Only 53 (58%) of 91 WT/WT patients had metastatic disease, which suggests a trend toward higher clinical stage among EML4-ALK–positive patients compared with WT/WT patients (P = .051). The majority of screened patients also had adenocarcinoma, including adenocarcinoma with BAC features (Table 1). Of the 19 EML4-ALK–positive tumors, 18 were adenocarcinomas, and one was a mixed adenosquamous carcinoma. Interestingly, compared with either EGFR mutant or WT/WT tumors, EML4-ALK–positive tumors were significantly more likely to have abundant signet ring cells (defined as ≥ 10% of tumor cells; Fig 1C). In addition, among the evaluable adenocarcinomas that harbored EML4-ALK, 61% showed solid growth as the predominant pattern, whereas acinar growth and BAC patterns were seen in only 31% and 8%, respectively.
Molecular Genotyping of Patients
Consistent with previous studies, which showed that EML4-ALK and EGFR mutation are mutually exclusive,7,15,17 we identified no EGFR mutations in the EML4-ALK cohort and no instances of ALK rearrangement in the EGFR cohort (Table 3). Similarly, among the patients screened for KRAS mutation, we found six positive patients in the WT/WT cohort, but none in either the EML4-ALK or EGFR mutant cohorts (Table 3; P = .022). These findings demonstrate that the molecular subsets of NSCLC defined by EML4-ALK, EGFR, or KRAS mutations are distinct and nonoverlapping.
Table 3.
Mutation Analysis of Screened Patients With Non–Small-Cell Lung Cancer
| Analysis | Genotype |
||
|---|---|---|---|
| ALK | EGFR | WT/WT | |
| ALK rearrangement | |||
| Positive | 19 | 0 | 0 |
| Total | 19 | 31 | 91 |
| EGFR mutation | |||
| Positive | 0 | 31 | 0 |
| Total | 19 | 31 | 74 |
| KRAS mutation* | |||
| Positive | 0 | 0 | 6 |
| Total | 11 | 10 | 23 |
Abbreviation: WT, wild type.
KRAS mutation testing was not performed on all patients because of limited amounts of tissue.
Treatment Response and Clinical Outcome of Patients With and Without EML-ALK
We determined best clinical response after treatment with an EGFR TKI or a platinum-based chemotherapy regimen in patients with metastatic disease. Among 10 patients with EML4-ALK and with evaluable disease, none had a documented clinical response to erlotinib (Table 4). Four patients (40%) had stable disease (SD), and six patients (60%) had progressive disease (PD) on erlotinib. In the WT/WT cohort, three (13%) of 23 treated patients had a partial response (PR), seven (30%) had SD, and 11 (48%) had PD on gefitinib or erlotinib. The small difference in response rates between EML4-ALK and WT/WT patients treated with an EGFR TKI was not statistically significant (P = .536). By contrast, 16 (70%) of 23 patients with EGFR mutations had a documented clinical response to an EGFR TKI. The higher response rate of EGFR mutation–positive patients compared with EML4-ALK–positive or WT/WT patients was highly statistically significant (P < .001).
Table 4.
Summary of Treatments and Responses by Genotype in Metastatic Non–Small-Cell Lung Cancer
| Variable | Genotype |
P |
||||||
|---|---|---|---|---|---|---|---|---|
|
ALK (n = 15) |
EGFR (n = 25) |
WT/WT (n = 49) |
||||||
| No. | % | No. | % | No. | % | ALK v EGFR | ALK v WT/WT | |
| No. of treatment regimens | ||||||||
| Median | 3 | 1 | 2 | .083 | .178 | |||
| Range | 1-4 | 1-6 | 0-9 | |||||
| Type of treatment | ||||||||
| Chemotherapy* | 12 | 80 | 9 | 36 | 37 | 76 | .010 | 1.000 |
| EGFR TKI | 10 | 67 | 24 | 96 | 23 | 47 | .021 | .254 |
| Best response to chemotherapy* | ||||||||
| No. of patients evaluated | 12 | 8† | 34‡ | |||||
| CR | 0 | 0 | 0 | 0 | 0 | 0 | ||
| PR | 3 | 25 | 4 | 50 | 12 | 35 | ||
| SD | 7 | 58 | 3 | 38 | 19 | 56 | ||
| PD | 2 | 17 | 0 | 0 | 2 | 6 | ||
| Unevaluable§ | 0 | 0 | 1 | 13 | 1 | 3 | ||
| Best response to TKI | ||||||||
| No. of patients evaluated | 10 | 23† | 23 | |||||
| CR | 0 | 0 | 1 | 4 | 0 | 0 | ||
| PR | 0 | 0 | 15 | 65 | 3 | 13 | ||
| SD | 4 | 40 | 6 | 26 | 7 | 30 | ||
| PD | 6 | 60 | 0 | 0 | 11 | 48 | ||
| Unassessable§ | 0 | 0 | 1 | 4 | 2 | 9 | ||
| Response rate, % | ||||||||
| Chemotherapy* | 25 | 50 | 35 | .356 | .723 | |||
| TKI | 0 | 70 | 13 | < .001 | .536 | |||
NOTE. Patients with no documentation of treatment history were excluded from this analysis.
Abbreviations: WT, wild type; EGFR TKI, epidermal growth factor receptor tyrosine kinase inhibitor; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease.
Chemotherapy refers specifically to a platinum-based combination chemotherapy regimen.
Excludes chemotherapy plus TKI (n = 1).
Excludes chemotherapy plus radiation therapy (n = 3).
No assessment because of early death, short follow-up, or lack of documentation.
To evaluate response to platinum-based chemotherapy, we examined all metastatic patients who had received carboplatin or cisplatin in combination with one or more therapeutic agents. These agents included standard chemotherapies, such as taxanes, as well as targeted agents, such as bevacizumab. Patients who had previously received a platinum combination as adjuvant therapy were excluded from this analysis. Within the EML4-ALK cohort, three (25%) of 12 evaluable patients had a PR, seven (58%) had SD, and two (17%) had PD on platinum-based chemotherapy (Table 4). A similar response was seen among 34 treated, WT/WT patients: 12 (35%) had PRs, 19 (56%) had SD, and two (6%) had PD (P = .723). Compared with patients who harbored EGFR mutations, EML4-ALK–positive patients showed a lower response rate to platinum-based chemotherapy, but this difference was not statistically significant (P = .356).
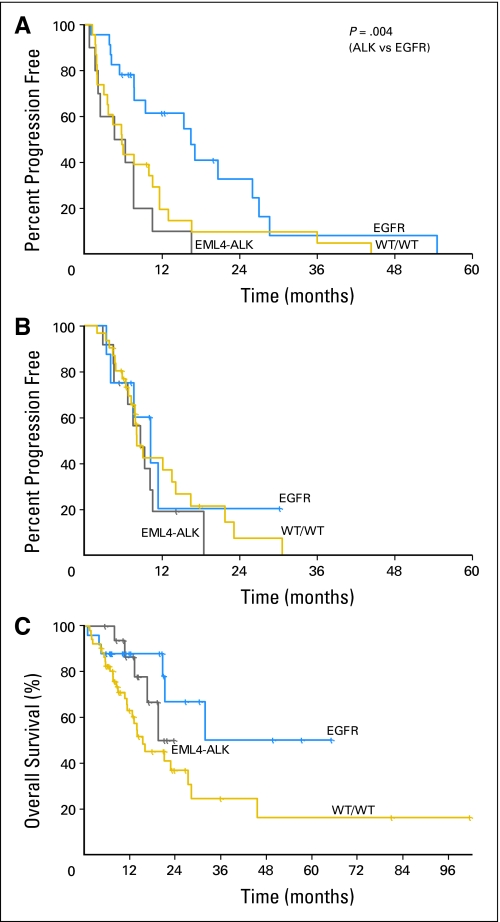
At the time of review, median follow-up of patients with metastatic NSCLC was 13 months among the 55 patients (57%) still alive; at that time, 39 patients (41%) had died, and two patients (2%) had been lost to follow-up. We analyzed both TTP and OS of patients according to genotype. For EML4-ALK–positive patients treated with an EGFR TKI, the median TTP was only 5 months, compared with 6 months for WT/WT patients (P = .337) and 16 months for patients with EGFR mutation (P = .004; Fig 2A). The median TTP for patients who received platinum-based chemotherapy was in the same range of 8 to 10 months across all three genotypes (Fig 2B). The median OS of EML4-ALK patients was 20 months, compared with 32 months for patients with EGFR mutation and 16 months for WT/WT patients, although these differences were not statistically significant (P = .468 and P = .152; Fig 2C).
Fig 2.
Time to progression (TTP) and overall survival (OS) of EML4-ALK–positive patients compared with patients who have EGFR mutant and wild-type (WT)/WT tumors. (A) TTP on epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor monotherapy. (B) TTP on any first-line, platinum-based, combination regimen. (C) Kaplan-Meier survival plots of OS.
DISCUSSION
The EML4-ALK translocation defines a new molecular subset of NSCLC with distinct clinical and pathologic features. Previous studies have reported a low frequency of EML4-ALK that has ranged from 1.5% to 6.7% in unselected populations.7,11,13–17 These studies have involved predominantly Asian patients with surgically resected disease. In this study, we show that, in a select subpopulation of predominantly white patients, the majority of whom had metastatic NSCLC, the frequency of EML4-ALK is significantly higher than that reported for unselected patients. Among the 141 patients screened, we identified 19 with EML4-ALK and 31 with EGFR mutation, which corresponded to frequencies of 13% and 22%, respectively. Within the group of never/light smokers in this study, the frequencies of EML4-ALK and EGFR were 22% and 32%, respectively; among never/light smokers without EGFR mutation, the frequency of EML4-ALK was 33%. These findings suggest that, in patients with NSCLC who have clinical characteristics associated with EGFR mutation but who have negative EGFR testing, as many as one in three patients may harbor EML4-ALK.
Previous reports that describe the frequency of EML4-ALK in NSCLC have been inconsistent in terms of clinical features that define this molecular subset. For example, in the first report of EML4-ALK in NSCLC, ALK rearrangement was detected in five patients, two of whom were noted to have a smoking history.7 In subsequent studies, EML4-ALK has been variably detected in both smokers and nonsmokers,8,11,15,16 which suggests a lack of association between smoking history and presence of EML4-ALK. Here, 19 of 85 patients classified as never/light cigarette smokers were positive for EML4-ALK, whereas all 56 patients with a smoking history (> 10 pack years) were negative. This result suggests that EML4-ALK is, in fact, strongly associated with never/light smoking history. This association was likely obscured in other studies because of small sample sizes and, possibly, differences in ethnic background.
Although EML4-ALK patients share several clinical features with patients who have EGFR mutant, including never/light smoking history and adenocarcinoma histology, this study demonstrates that EML4-ALK is associated with at least three distinct features. First, compared with EGFR or WT/WT patients, EML4-ALK patients are more likely to be men. As female sex was used as one of the clinical selection criteria for genetic screening, our study tested almost twice as many women as men. However, we found that a significantly greater percentage of men than women were positive for EML4-ALK (23% v 9%). The sex difference observed in this study cannot be explained by differences in smoking history, as 60% of men and 60% of women were never/light smokers. Second, compared with EGFR or WT/WT patients, EML4-ALK patients are significantly younger. The difference in median age between EML4-ALK patients and either EGFR or WT/WT patients exceeded 10 years. Of note, the median age of our EGFR cohort was similar to that reported in other studies.22–24 Among the 19 patients with EML4-ALK, four were younger than 40 years old. One recent study of EML4-ALK in Asian patients with NSCLC noted a nonstatistically significant trend toward younger median age.17 Interestingly, several other cancers known to harbor ALK rearrangements, such as anaplastic large cell lymphomas, neuroblastomas, and inflammatory myofibroblastic tumors, are also associated with younger age and are, in fact, most common in children and young adults. Third, EML4-ALK–positive tumors appear histologically distinct from EGFR mutant and WT/WT tumors. The diagnostic and clinical implications of this finding will be discussed in a separate report (Rodig et al, manuscript submitted for publication), but this observation suggests that EML4-ALK may represent a unique pathologic subtype of nonsmoking-related NSCLC.
In the clinic, the distinction between EML4-ALK and EGFR mutant tumors has important therapeutic implications. Whereas EGFR mutation confers sensitivity to EGFR TKIs, EML4-ALK is strongly associated with resistance. Among the 19 patients in this study with any response to erlotinib or gefitinib, 16 (84%) harbored an activating EGFR mutation, whereas none harbored EML4-ALK. Conversely, among the 34 patients refractory to EGFR TKIs, 10 (29%) were positive for EML4-ALK. These findings are consistent with preclinical studies, which showed that the EML4-ALK–containing NSCLC cell line H3122 is resistant to erlotinib.11 These findings are also reminiscent of the resistance to EGFR TKIs conferred by activating mutations in KRAS.25 However, whereas KRAS mutations are more commonly found in smokers,26 both EML4-ALK and EGFR mutations are found in a similar population of never/light smokers. As a result, in the absence of genetic testing, EML4-ALK patients are likely to be treated like patients with EGFR mutation. Indeed, in this study, five of 15 EML4-ALK patients with metastatic NSCLC received erlotinib in the first-line setting. These results illustrate the importance of pretreatment genetic testing to guide clinical treatment recommendations, especially with regard to EGFR TKIs.
Overall, the clinical response of EML4-ALK patients more closely resembles that of WT/WT patients rather than patients with EGFR mutation. Both EML4-ALK and WT/WT patients are unlikely to respond to EGFR TKIs and have lower rates of response to platinum-based chemotherapy than patients with EGFR mutation. This difference does not appear to be related to imbalances among the cohorts in terms of type of platinum or inclusion of bevacizumab. The higher response rate associated with EGFR mutation is consistent with previous studies, including the recently presented IPASS study (IRESSA Pan Asia Study), in which clinically selected patients with metastatic NSCLC were randomly assigned in the first-line setting to either carboplatin/paclitaxel or gefitinib. Among patients treated with chemotherapy, the objective response rates were 47.3% in EGFR mutation–positive patients and 23.5% in EGFR mutation–negative patients.6 This study suggests that EML4-ALK, in contrast to EGFR mutation, is not associated with enhanced chemosensitivity.
Previous studies have not examined the outcome of patients with NSCLC who harbor EML4-ALK. Here, we evaluated outcome by determining TTP and OS among patients with metastatic disease. This analysis was limited by the retrospective design of the study, by the relatively short duration of follow-up, and by the small number of events in the mutant cohorts. Nevertheless, EML4-ALK patients had a longer median survival compared with WT/WT patients, though the power was too low to detect a significant difference. Patients who harbored EGFR mutations showed a statistically significant improvement in survival compared with WT/WT patients (P = .018), which is consistent with previous reports that demonstrated a more favorable outcome among patients with EGFR mutation.27 This survival analysis is additionally complicated by baseline differences in demographic features, particularly age and smoking history, between EML4-ALK–positive and –negative patients, as well as by differences in the number and types of therapies received. In addition, to date, seven of 17 EML4-ALK patients with metastatic disease have participated in a phase 1 study of PF-02341066, a dual MET/ALK TKI.28 The clinical activity of this novel agent has not yet been reported, but its use in a significant proportion of EML4-ALK patients may have influenced the outcome of this cohort.
In conclusion, EML4-ALK defines a new molecular subset of NSCLC with distinct clinical and pathologic features. The patients most likely to harbor EML4-ALK are young, never/light smokers with adenocarcinoma. As some of these features are also associated with EGFR mutation, it is essential to screen such patients by mutation testing and not to rely solely on the presence of clinical predictors. We recommend screening first for EGFR mutation, because EGFR mutations are more common than EML4-ALK rearrangements and because, importantly, EGFR TKIs are now used as first-line agents in advanced, mutation-positive disease. In the absence of an EGFR mutation, patients then should be screened for EML4-ALK. Preclinical studies have shown that EML4-ALK confers sensitivity to ALK inhibitors,10,11,29 and studies suggest that patients with this chromosomal translocation may derive clinical benefit from specific ALK inhibition. This hypothesis currently is being tested in the clinic and if confirmed will validate ALK as a therapeutic target in NSCLC.
Appendix
The Appendix is included in the full-text version of this article, available online at www.jco.org. It is not included in the PDF version (via Adobe® Reader®).
Fig A1.
Example of reverse transcriptase polymerase chain reaction (RT-PCR) detection of EML4-ALK in a primary tumor sample and cell lines (RT-PCR for GAPDH expression was also performed as a control for RNA extraction and cDNA synthesis). In this patient (PID5), (A) RT-PCR and (B) DNA sequencing demonstrate the presence of EML4-ALK variant 5 (Wong DW, et al: Cancer 115:1723-1733, 2009), in which exon18 of EML4 is fused to exon20 of ALK (lane 1). NCI-H2228 and NCI-H3122 have previously been shown to harbor EML4-ALK variants 3 and 1, respectively (Koivunen JP, et al: Clin Cancer Res 14:4275-4283, 2008). Variant 1, which results from a fusion of EML4 exon13 with exon20 of ALK, is also detected in this assay (lane 3), whereas variant 3, in which EML4 exon6 is fused to exon20 of ALK, is not (lane 2).
Table A1.
Clinical Characteristics of Patients With WT/WT Genotype According to KRAS Mutation Status
| Characteristic |
KRAS Status |
P | |||||
|---|---|---|---|---|---|---|---|
| MUT (n = 6) |
WT (n = 17) |
ND (n = 68) |
|||||
| No. | % | No. | % | No. | % | ||
| Age, years | .046 | ||||||
| Median | 72 | 51 | 65 | ||||
| Range | 39-83 | 29-83 | 41-87 | ||||
| Sex | .660 | ||||||
| Male | 2 | 33 | 8 | 47 | 19 | 28 | |
| Female | 4 | 67 | 9 | 53 | 49 | 72 | |
| Smoking history | .843 | ||||||
| Never smoker | 1 | 17 | 6 | 35 | 17 | 25 | |
| Light smoker | 2 | 33 | 5 | 29 | 8 | 12 | |
| Smoker | 3 | 50 | 6 | 35 | 43 | 63 | |
| Ethnicity | 1.000 | ||||||
| Asian | 0 | 0 | 2 | 12 | 5 | 7 | |
| Non-Asian | 6 | 100 | 15 | 88 | 63 | 93 | |
| Pathology | 1.000* | ||||||
| Adeno | 4 | 67 | 12 | 71 | 33 | 49 | |
| BAC | 1 | 17 | 2 | 12 | 29 | 43 | |
| Adenosquamous | 0 | 0 | 1 | 6 | 2 | 3 | |
| Squamous | 0 | 0 | 1 | 6 | 1 | 1 | |
| Large cell/NOS | 1 | 17 | 1 | 6 | 3 | 4 | |
| Stage | 1.000† | ||||||
| IA | 0 | 0 | 1 | 6 | 9 | 13 | |
| IB | 0 | 0 | 0 | 0 | 10 | 15 | |
| IIA | 0 | 0 | 0 | 0 | 1 | 1 | |
| IIB | 0 | 0 | 0 | 0 | 0 | 0 | |
| IIIA | 0 | 0 | 1 | 6 | 2 | 3 | |
| IIIB | 1 | 17 | 1 | 6 | 2 | 3 | |
| IV | 4 | 67 | 14 | 82 | 35 | 51 | |
| Multifocal BAC | 1 | 17 | 0 | 0 | 9 | 13 | |
Abbreviations: MUT, mutation; WT, wild type; ND, not determined; adeno, adenocarcinoma; BAC, bronchioloalveolar carcinoma; NOS, not otherwise specified.
Adeno and BAC v all others.
Stages I to III v IV.
Footnotes
Supported in part by the National Institutes of Health Grant No. CA090578, by American Association for Cancer Research Grant No. 07-40-12-COST (D.B.C), and by internal funds from the Massachusetts General Hospital Cancer Center and MGH Pathology Department.
Presented in part at the 45th Annual Meeting of the American Society of Clinical Oncology, May 29-June 2, 2009, Orlando, FL.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Alice T. Shaw, Pfizer (C); A. John Iafrate, Pfizer (U); Daniel B. Costa, Pfizer (C); Thomas J. Lynch, Genentech (C), AstraZeneca (C), OSI Pharmaceuticals (C), Boehringer-Ingelheim (C), sanofi-aventis (C), Millennium Pharmaceuticals (C), EMD Serono (C) Stock Ownership: None Honoraria: Alice T. Shaw, Wyeth, Merck, GlaxoSmithKline Research Funding: None Expert Testimony: None Other Remuneration: Thomas J. Lynch, Genzyme
AUTHOR CONTRIBUTIONS
Conception and design: Alice T. Shaw, A. John Iafrate
Financial support: Thomas J. Lynch, A. John Iafrate
Administrative support: Thomas J. Lynch, A. John Iafrate
Provision of study materials or patients: Alice T. Shaw, Daniel B. Costa, Rebecca S. Heist, Benjamin Solomon, Thomas J. Lynch
Collection and assembly of data: Alice T. Shaw, Mari Mino-Kenudson, Subba R. Digumarthy, Daniel B. Costa, Hannah Stubbs, Sonal Admane, Ultan McDermott, Susumu Kobayashi, Scott J. Rodig, Lucian R. Chirieac, A. John Iafrate
Data analysis and interpretation: Alice T. Shaw, Beow Y. Yeap, Mari Mino-Kenudson, Subba R. Digumarthy, Rebecca S. Heist, Eugene J. Mark, A. John Iafrate
Manuscript writing: Alice T. Shaw, Beow Y. Yeap, A. John Iafrate
Final approval of manuscript: Alice T. Shaw, Beow Y. Yeap, Mari Mino-Kenudson, Subba R. Digumarthy, Daniel B. Costa, Rebecca S. Heist, Benjamin Solomon, Hannah Stubbs, Sonal Admane, Jeffrey Settleman, Eugene J. Mark, Scott J. Rodig, Lucian R. Chirieac, Eunice L. Kwak, Thomas J. Lynch, A. John Iafrate
REFERENCES
- 1.Shibuya K, Mathers CD, Boschi-Pinto C, et al. Global and regional estimates of cancer mortality and incidence by site: II. Results for the global burden of disease 2000. BMC Cancer. 2002;2:37. doi: 10.1186/1471-2407-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non–small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Sandler A, Gray R, Perry MC, et al. Paclitaxel-carboplatin alone or with bevacizumab for non–small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 6.Mok T, Wu Y-L, Thongprasert S, et al. Phase III, randomised, open-label, first-line study of gefitinib vs carboplatin/paclitaxel in clinically selected patients with advanced non–small-cell lung cancer (IPASS). Presented at the 33rd European Society for Medical Oncology Congress; September 12-16, 2008; Stockholm, Sweden. [Google Scholar]
- 7.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non–small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 8.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 9.Chiarle R, Voena C, Ambrogio C, et al. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8:11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- 10.Soda M, Takada S, Takeuchi K, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci U S A. 2008;105:19893–19897. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukuyoshi Y, Inoue H, Kita Y, et al. EML4-ALK fusion transcript is not found in gastrointestinal and breast cancers. Br J Cancer. 2008;98:1536–1539. doi: 10.1038/sj.bjc.6604341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeuchi K, Choi YL, Soda M, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res. 2008;14:6618–6624. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 14.Perner S, Wagner PL, Demichelis F, et al. EML4-ALK fusion lung cancer: A rare acquired event. Neoplasia. 2008;10:298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol. 2008;3:13–17. doi: 10.1097/JTO.0b013e31815e8b60. [DOI] [PubMed] [Google Scholar]
- 16.Shinmura K, Kageyama S, Tao H, et al. EML4-ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non–small-cell lung carcinomas. Lung Cancer. 2008;61:163–169. doi: 10.1016/j.lungcan.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Wong DW, Leung EL, So KK, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 18.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors: European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 19.Beasley MB, Brambilla E, Travis WD. The 2004 World Health Organization classification of lung tumors. Semin Roentgenol. 2005;40:90–97. doi: 10.1053/j.ro.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer: Molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 21.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 22.Inoue A, Suzuki T, Fukuhara T, et al. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non–small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24:3340–3346. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- 23.Asahina H, Yamazaki K, Kinoshita I, et al. A phase II trial of gefitinib as first-line therapy for advanced non–small-cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer. 2006;95:998–1004. doi: 10.1038/sj.bjc.6603393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non–small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 25.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 27.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non–small-cell lung cancer treated with chemotherapy alone and in combinationwith erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 28.Christensen JG, Zou HY, Arango ME, et al. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther. 2007;6:3314–3322. doi: 10.1158/1535-7163.MCT-07-0365. [DOI] [PubMed] [Google Scholar]
- 29.McDermott U, Iafrate AJ, Gray NS, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68:3389–3395. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]