Abstract
Signal transducer and activator of transcription (STAT) proteins comprise a seven-member family of latent cytoplasmic transcription factors that are activated through tyrosine phosphorylation by a variety of cytokines and growth factors. Aberrant activation of STATs accompanies malignant cellular transformation with resultant leukemogenesis. Constitutive activation of STATs has been demonstrated in various leukemias. A better understanding of the mechanisms of dysregulation of the STAT pathway and understanding of the cause and effect relationship in leukemogenesis may serve as a basis for designing novel therapeutic strategies directed against STATs. Mechanisms of STAT activation, the potential role of STAT signaling in leukemogenesis, and recent advances in drug discovery targeting the STAT pathway are the focus of this review.
INTRODUCTION
Signal transducer and activator of transcription (STAT) proteins are a family of cytoplasmic transcription factors involved in cytokine, hormone, and growth factor signal transduction to mediate a variety of biologic processes including cellular growth, differentiation, and apoptosis (Fig 1).1 Seven members of the STAT family have been identified: STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6. The exact chromosomal localizations of the STAT genes in humans were identified during the sequencing of the human genome.2 Several domains are conserved in all STAT family members (Table 1; Fig 2).3,4
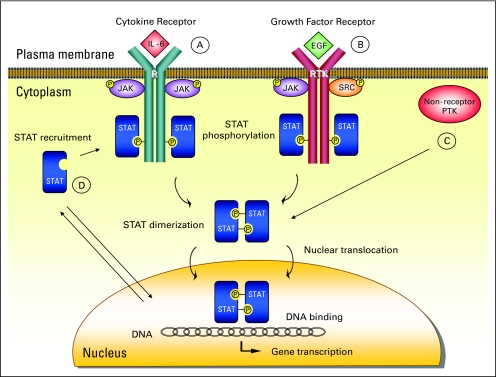
Fig 1.
Signal transducer and activator of transcription (STAT) proteins are activated by receptor and nonreceptor tyrosine kinases through several mechanisms. (A) The receptor-associated Janus family tyrosine kinases (JAKs) are activated on cytokine-receptor binding through cross-phosphorylation in the classical pathway. Activated JAKs phosphorylate tyrosine residues on the receptor (R), which become docking elements for cytoplasmic STAT proteins. STATs are subsequently phosphorylated on a single tyrosine residue in the carboxy (COOH) -terminal portion and form homo- or heterodimers through reciprocal interaction between the phosphotyrosine of one STAT and the Schmidt-Ruppin A-2 viral oncogene homolog (avian) (SRC) -homology-2 (SH2) domain of another. Dimerized STATs shift into the nucleus via importins to induce target gene transcription by binding to specific regulatory elements. (B) Receptors with intrinsic tyrosine kinase activities (RTK), including platelet-derived growth factor receptor (PDGFR), epidermal growth factor receptor (EGFR), and FMS (formerly McDonough feline sarcoma viral oncogene homolog) -related tyrosine kinase 3 (FLT3), may directly activate STATs without involvement of JAKs. (C) STATs can be phosphorylated by constitutively active nonreceptor protein tyrosine kinases (PTKs), such as SRC and BCR-ABL. (D) Unphosphorylated STATs can independently enter the nucleus to mediate gene transcription possibly by acting as a transcriptional coregulator to bind DNA. IL-6, interleukin-6; P, phosphorus.
Table 1.
STATs Structure
| Domain | Function |
|---|---|
| Oligomerization domain | Mediates oligomerization of STAT dimers to form tetramers and interactions with other proteins |
| DNA binding domain | Mediates distinct signals for specific ligands to define the DNA-binding specificity |
| SH2 domain | Mediates specific interactions between STAT-receptor, STAT-JAK, and STAT-STAT |
| COOH-terminal domain | Regulates the transcriptional activity of STATs and provides functional specificity |
| Tyrosine residue | Phosphorylation site in the COOH-terminal domain approximately 700 residues from the NH2 terminus that regulates the DNA-binding activity |
| Serine residue | A second phosphorylation site in the COOH-terminal domain except STAT2 and STAT6 |
Abbreviation: STAT, signal transducer and activator of transcription; JAK, Janus family tyrosine kinases; COOH, carboxy; NH2, N-terminal.
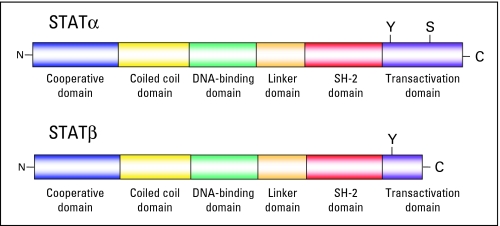
Fig 2.
Structure and functional domains of signal transducer and activator of transcription (STAT) molecules. Shown on the top is the full-length STATα. Below is the COOH-terminal (C) transactivation domain truncation resulting in STATβ isoforms. The following two phosphorylation sites in the COOH domain exist: a tyrosine phosphorylation site (Y) that controls dimerization yielding the DNA-binding activity of the STATs, and a serine phosphorylation residue (S) that further modulates the transcriptional activity of STATs.
STAT ISOFORMS
STAT isoforms lacking parts of the carboxy (COOH) –terminal domain (STATβ) have a competitive dominant negative (DN) effect counteracting the full-length isoform, STATα.5–7 The transcriptional activities of the different isoforms are distinct, suggesting that the balance of these isoforms regulates gene activation, leading to distinct biologic responses (Table 2). Truncated STATβ that lacks the tyrosine residues at the 699 to 705 position can still be recruited to tyrosine phosphorylated receptor proteins via the remaining SH2 domain, but STAT signaling terminates.
Table 2.
STAT3 Isoforms
| Isoform | Description | Molecular Weight (kDa) |
|---|---|---|
| STAT3α | Full length | 92 |
| STAT3β | Truncated COOH-terminal transactivation domain; functionally distinct, either dominant negative or altered binding | 83 |
| STAT3γ | Missing tyrosine residue; able to bind to the remaining SH-2 domain, but functionally inactive | 72 |
| STAT3δ | Unknown | 64 |
Abbreviation: STAT, signal transducer and activator of transcription; COOH, carboxy; SH-2, Schmidt-Ruppin A-2 viral oncogene homolog–homology 2.
STATβ isoforms are generated by alternative mRNA splicing5 or proteolytic processing.6,7 The characterization of this proteolytic activity revealed a serine endopeptidase capable of cleaving both STAT3 and STAT5, but not STAT6.7 A recent provocative study claimed cathepsin G as STAT5 protease and argued that COOH-terminally truncated STAT5 was in fact an artifact generated during in vitro sample preparation with no in vivo significance.8 Further studies are needed to clarify this controversy.
REGULATION OF STAT SIGNALING
Transcriptional activity of the STAT proteins is tightly regulated by endogenous inhibitory molecules and post-translational modification mechanisms for appropriate physiologic cellular functions including ubiquitination, ISGylation, sumoylation, methylation, and acetylation.9–11 Increasing evidence suggests that loss of function or methylation silencing of these negative regulators is likely involved in chronic constitutive activation of STATs.
The suppressor of cytokine signaling (SOCS) family of proteins (SOCS1 to SOCS7 and cytokine-inducible SH2-containing protein [CIS]) downregulates STAT signaling as a classic negative feedback loop.9,12 COOH-terminal domain SOCS box is responsible for the recruitment of the ubiquitin-transferase complex. SOCS1 directly binds to tyrosine phosphorylated Janus family tyrosine kinases (JAKs) to inhibit catalytic activity.12 In contrast, the SH2 domains of SOCS2 and SOCS3 proteins bind to phosphotyrosine residues of the activated cytokine receptors. Additionally, SOCS proteins induce ubiquitin-mediated proteasome-dependent degradation of the STATs. Finally, CIS inhibits STAT activation by competing with STATs for phosphotyrosine binding sites on the cytoplasmic portion of the cytokine receptors.
Protein tyrosine phosphatases (PTPs) neutralize the effects of kinases to dephosphorylate active JAKs/STATs in both the cytoplasm and the nucleus.9,13,14 Members of PTPs include SH2-containing phosphatase (SHP) -1, SHP-2, CD45, T-cell PTP (TCPTP), PTP1B, and serine/threonine phosphatase PP2A. Discovery of how these PTPs confer specificity in dephosphorylation of various STAT family members will be a huge step forward in understanding STAT-mediated leukemogenesis.
The protein inhibitors of activated STATs (PIAS) family of proteins (PIAS1, PIAS3, PIASx, and PIASy) is a negative regulator of STAT-mediated gene transcription.15,16 PIAS1 and PIASy interact with STAT1, PIAS3 interacts with STAT3 and STAT5, and PIASx interacts with STAT4.16 PIAS proteins inhibit STAT-DNA binding activity and recruit other transcriptional corepressors such as histone deacetylases (HDACs). Furthermore, they have small ubiquitin-related modifier (SUMO) E3 ligase activity.15 Consequently, transcriptional activity of STATs is inhibited by SUMO conjugation.
The ubiquitin-proteasome degradation pathway represents another negative feedback mechanism.17 Ubiquitylation involves sequential engagement of ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). The SOCS box of SOCS proteins associates with elongins B and C in addition to cullin-2 to form ubiquitin E3 ligase complex. Consequently, the JAK and STAT substrate proteins are selectively targeted to the 26S proteasome for degradation. STAT-interacting LIM protein (SLIM) is a nuclear ubiquitin E3 ligase that contains a PDZ (acronym for the following three proteins: postsynaptic density protein, Drosophila disc large tumor suppressor, and zonula occludens-1 protein) domain and a LIM (representing LIN11, Isl1, and MEC-3 proteins) domain.18 SLIM induces ubiquitination and proteasomal degradation of STAT1 and STAT4. The significance of SLIM in human diseases warrants further investigation.
SUMO and interferon-stimulated gene 15 (ISG15) proteins are members of the ubiquitin-like proteins family.19,20 Sumoylation and ISGylation pathways are analogous to ubiquitin conjugation with differences in the enzymes involved. However, unlike ubiquitination and sumoylation, protein ISGylation provides a positive feedback for JAK-STAT signaling.19 The exact roles and relevance of sumoylation and ISGylation in the regulation of STAT signal transduction remain to be fully elucidated.
Cytoplasmic and nuclear STAT-interacting proteins, such as STAT3-interacting protein (StIP1 = ELP2, elongation protein), minichromosome maintenance 5 protein (MCM5), N-c-myelocytomatosis viral oncogene homolog (MYC) –interacting protein (Nmi), and p300/CBP (cyclic AMP–responsive element-binding protein [CREB] –binding protein), have also been described to regulate transcriptional activation.21,22
STAT NUCLEOCYTOPLASMIC SHUTTLING
After ligand stimulation, cytoplasmic latent STATs rapidly accumulate in the nucleus by crossing through nuclear pore complexes.23 Most of the nuclear translocation is phosphorylation dependent, whereas some STAT proteins shuttle between the nucleus and cytoplasm independent of phosphorylation.24–28 Activated STATs shuttle more rapidly than nonactivated ones.24,28 Direct interaction of unphosphorylated STATs with the nuclear pore proteins (nucleoporins) Nup153 and Nup214 allows carrier-independent nuclear translocation.25 Nuclear translocation of activated STATs is mediated by the karyopherin-β family of transport proteins called importins or exportins depending on their moving direction.23 Specific sequence motifs on the surface of the STATs, known as nuclear localization signals and nuclear export signals, allow STAT-importin and STAT-exportin interactions. Specific adaptor molecules, the importin-α family, are involved in STAT–importin-β interaction. Distinct importin-α subtypes determine trafficking of different STATs.26 STAT activation with nuclear accumulation terminates within minutes.29 STATs dephosphorylated by nuclear phosphatases, such as TC45, are actively exported back to the cytoplasm by binding to chromosome region maintenance 1 (CRM1; also called exportin-1) protein. Interestingly, nuclear export of truncated STATβ isoforms was shown to be reduced with resultant prolonged nuclear retention, mainly as a result of the accumulation of unphosphorylated proteins.27,30 Nuclear export differences determine the cytokine sensitivity of STAT isoforms. Inhibition of nuclear export breaks up the STAT reactivation cycle and results in reduced STAT phosphorylation and gene induction with consequent apoptosis.
STATS IN CANCER: EMPHASIS ON LEUKEMIC TRANSFORMATION
Dysregulation of the STAT signaling pathway plays a role in oncogenesis and leukemogenesis.31,32 As a proof of concept, inhibition of aberrant STAT activity has been repeatedly shown to result in arrest of tumor development and apoptosis. Constitutive STAT activation is associated with malignant transformation induced by various oncoproteins, such as sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) (v-SRC), breakpoint cluster region (BCR) -abelson (ABL), and epidermal growth factor receptor (EGFR) tyrosine kinases.33–41 The candidate target genes regulated by the STAT pathways, such as cyclin D1/D2, MYC, B-cell chronic lymphocytic leukemia (CLL)/lymphoma (Bcl-x), and myeloid cell leukemia sequence-1 (Mcl-1), seem to contribute to oncogenesis through the control of cell cycle progression and/or the prevention of apoptosis.35–39,41–43 In addition, recombinantly altering the transactivation domain of STAT3 was shown to induce constitutive activation, leading to malignant transformation in the absence of tyrosine phosphorylation.42 Importantly, activated STAT3 was reported to mediate p53 inhibition by binding to the p53 promoter.44 This finding is significant because of the recent demonstration that activated STAT3 regulates murine thymoma viral oncogene homolog (AKT) gene expression,45 a crucial downstream target of phosphatidylinositol 3-kinase (PI3K), which is constitutively active in acute myeloid leukemia (AML) and regulates survival and chemotherapy resistance via nuclear factor-κB, mitogen-activated protein kinase (MAPK), and the p53 pathways (Fig 3).46 Similarly, a constitutively active STAT5 mutant was shown to form a complex with PI3K and the scaffolding adapter Gab2, resulting in AKT activation in myeloid leukemias.47 Moreover, a direct cross talk between the STAT and the MAPK pathways has been demonstrated as part of their involvement in oncogenic transformation.48,49 Finally, a particularly intriguing role for constitutive STAT3 activity in the upregulation of vascular endothelial growth factor expression and tumor angiogenesis was demonstrated.45,50 In summary, STAT3 seems to be the central transcription factor for many signaling pathways.
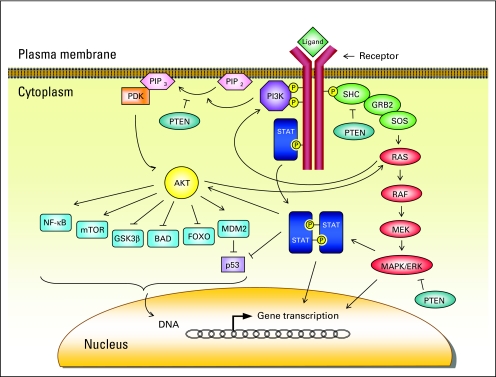
Fig 3.
Phosphatidylinositol 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) signaling pathways. Ligand binding to a receptor tyrosine kinase induces PI3K phosphorylation, which in turn converts phosphatidylinositol (4,5)-bisphosphate (PIP2) to phosphatidylinositol (3,4,5)-trisphosphate (PIP3). Phosphatidylinositol-dependent kinase (PDK) is then recruited and activated, which subsequently phosphorylates murine thymoma viral oncogene homolog (AKT). Activated AKT controls fundamental cellular processes such as cell cycle regulation, angiogenesis, survival, and apoptosis via its target molecules. However, the MAPK pathway is initiated by interaction of SRC homology 2 domain containing transforming protein (SHC) with growth factor receptor–bound protein 2 (GRB2) and son of sevenless (SOS), resulting in activation of RAS and downstream MAPK cascade. BAD, Bcl-2 antagonist of cell death; ERK, extracellular signal-regulated kinase; FOXO, forkhead transcription factor; GSK3β, glycogen synthase kinase-3β; MDM2, murine double minute; MEK, mitogen-activated ERK kinase; mTOR, mammalian target of rapamycin; NF-κB, nuclear factor-κB; PTEN, phosphatase and tensin homolog deleted on chromosome 10; STAT, signal transducer and activator of transcription.
CONSTITUTIVE STAT ACTIVITY IN LEUKEMIAS
STAT activation in leukemic cell lines51,52 and blasts from AML and acute lymphoblastic leukemia (ALL) patients has been demonstrated (Table 3).49,51,53–59 STAT3 activity is most prevalent in AML, whereas STAT5 activity is more common in ALL. Autocrine/paracrine stimulation of the JAK-STAT pathway by hematopoietic cytokines, such as interleukin (IL) -6, might cause constitutive STAT activity.60 However, the role of IL-6–induced STAT3 activation in leukemogenesis remains controversial because of the antiproliferative effects of IL-6 in AML.61 The clinical significance of constitutive STAT3 activity was demonstrated in AML patients. Disease-free survival (DFS) was significantly shorter in patients with constitutive STAT3 activity compared with patients without STAT3 activity.59 In a subgroup analysis, patients with both constitutive STAT3 activity and the truncated STAT3β isoform had the shortest DFS and shorter overall survival compared with all other patients. This was the first demonstration of the clinical prognostic significance for STAT proteins in any malignancy.
Table 3.
Constitutive STAT Activity in AML Blasts
| Study | STAT1 |
STAT3 |
STAT5 |
ERK |
FLT3 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of Patients/Total No. | % | No. of Patients/Total No. | % | No. of Patients/Total No. | % | No. of Patients/Total No. | % | No. of Patients/Total No. | % | |
| Hayakawa et al49 | — | — | — | — | 10/10 | 100 | 10/10 | 100 | 10/10* | 100* |
| Aronica et al53 | 10/20 | 50 | — | — | — | — | — | — | — | — |
| Gouilleux-Gruart et al54† | 1/5 | 20 | 5/5 | 100 | 2/5 | 40 | — | — | — | — |
| Weber-Nordt et al55‡ | 10/14 | 71 | 10/14 | 71 | 1/14 | 7 | — | — | — | — |
| Hayakawa et al56 | — | — | 17/23 | 74 | 40/50 | 80 | 50/50 | 100§ | — | — |
| Xia et al57 | — | — | 10/36 | 28 | 8/36 | 22 | — | — | — | — |
| Birkenkamp et al58 | — | — | — | — | 18/26 | 69 | — | — | 12/17 | 71 |
| Benekli et al59 | — | — | 28/63 | 44 | 11/50 | 22 | — | — | — | — |
Abbreviations: STAT, signal transducer and activator of transcription; AML, acute myeloid leukemia; ERK, extracellular signal-regulated kinase.
All samples had previously known tandem duplication of FLT3 gene.
In addition to AML, constitutive STAT1 and STAT5 activities were demonstrated in peripheral blood samples of one and three of three acute lymphoblastic leukemia (ALL) patients, respectively.
In addition to AML, constitutive STAT1 activity was found in one (4%) of 24 patients with ALL, and STAT5 activity was found in 15 (63%) of 24 patients.
All samples had previously known constitutive mitogen-activated protein kinase activity.
Constitutive serine, but not tyrosine, phosphorylation of STAT1 and STAT3 in CLL cells was demonstrated using specific antibodies against the phosphorylated Ser-727 residue.62 However, the significance of this finding in CLL pathobiology remains undetermined.
STAT ACTIVATION BY LEUKEMOGENIC FUSION PROTEINS AND TYROSINE KINASES
Aberrant STAT activation may be associated with leukemic transformation by various oncoproteins. Leukemic fusion proteins with protein tyrosine kinase (PTK) activity have been shown to activate STATs without the need for receptor activation.
BCR-ABL
The BCR-ABL chimeric protein is a constitutively activated tyrosine kinase that causes growth factor–independent proliferation and transformation of hematopoietic cells in chronic myeloid leukemia (CML), ALL, and rarely AML.33,34 The Philadelphia chromosome (Ph) is generated by reciprocal translocation of chromosomes 9 and 22, t(9;22)(q34;q11). As a result, two different fusion proteins, p190BCR-ABL (190 kDa) and p210BCR-ABL (210 kDa), are produced depending on the breakpoint site on the BCR gene. p210BCR-ABL is the characteristic feature of CML, whereas both p190 and p210 are involved in ALL and AML. Constitutive STAT5 and/or STAT1 activity was demonstrated in BCR-ABL–positive cell lines, peripheral-blood samples from CML patients, and hematopoietic cell lines transfected in vitro with BCR-ABL, leading to malignant transformation.33–39 STAT activation was stronger in cells transformed by the p190BCR-ABL isoform, in contrast to the p210BCR-ABL isoform,33,34 suggesting a decisive role for the magnitude of STAT phosphorylation on the biologic effects of BCR-ABL. Finally, DN STAT5 isoforms were shown to inhibit p210BCR-ABL-dependent STAT5 phosphorylation, with subsequent inhibition of cell growth confirming the central role for STAT proteins in BCR-ABL signaling.36
Possible downstream targets of BCR-ABL and STAT5 activity involve genes regulating apoptosis. BCR-ABL increases the expression of the antiapoptotic Bcl-xL protein via STAT5 phosphorylation in IL-3–dependent cell lines.36–39 Interestingly, the BCR-ABL-tyrosine kinase inhibitor imatinib mesylate was shown to induce apoptosis by suppressing STAT5 binding to the Bcl-x promoter and downregulating Bcl-xL expression in BCR-ABL–expressing cell lines and CD34+ cells from CML patients.38,39
Interestingly, a study by Sexl et al63 suggested that there may not be a definitive requirement for STAT5 and that redundant pathways (yet undiscovered) independent of STAT5 may be involved in BCR-ABL–mediated transformation.
TEL-JAK2
In some patients with T-cell ALL, pre–B-cell ALL, and atypical CML, t(9;12)(p24;p13) results in the fusion of the 3′ functional JH1 kinase domain of JAK2 to the 5′ pointed domain of translocated erythroblastosis (ETS) leukemia (TEL), a member of the ETS transcription factor family.64,65 The TEL-JAK2 fusion protein induces STAT1, STAT3, and STAT5 activation with subsequent cytokine-independent proliferation in the IL-3-dependent Ba/F3 pre–B-cell line.64,65 In addition, TEL-JAK2 transgenic mice develop T-cell leukemia with constitutive STAT5 and STAT1 activity.66 Finally, activation of STAT5 was demonstrated to cause myelo- and lymphoproliferative diseases by TEL-JAK2 in a murine bone marrow transplantation model.67 These data indicate a cardinal role for STAT5 activity in TEL-JAK2–induced growth factor–independent hematopoietic transformation.
TEL-PDGFβR
TEL-platelet-derived growth factor β receptor (PDGFβR) tyrosine kinase fusion protein results from t(5;12)(q33;p13) in chronic myelomonocytic leukemia.68,69 Constitutive STAT168 and STAT565 activities were observed in Ba/F3 cell lines transfected with TEL-PDGFβR. Transformation by TEL-PDGFβR causes hyperphosphorylation of STAT5 on tyrosine residues.69 However, full transformation required engagement of a combination of signaling intermediates, PI3K and phospholipase C-γ, as well as activation of STAT5, suggesting that constitutive activation of STAT5 by itself may not be sufficient for transformation in this model.
FLT3
FMS (formerly McDonough feline sarcoma viral oncogene homolog) -related tyrosine kinase 3 (FLT3) is a member of the class III receptor tyrosine kinase family expressed on hematopoietic progenitor cells.70 Its ligand promotes clonal expansion of stem cells. Activating FLT3 mutations, either involving internal tandem duplications (ITDs) or point mutations in the activating loop (tyrosine kinase domain), are observed in approximately 30% of AML patients and are associated with poorer prognosis. Mutated FLT3 by either mechanism is constitutively activated, leading to dual constitutive STAT5 and MAPK activation with factor-independent proliferation.49,71,72 Furthermore, STAT5 target genes such as CIS, Pim-2, and cyclin-dependent kinase (CDK) inhibitor p21 were highly induced by FLT3-ITD.72,73 Moreover, constitutive STAT5 activity was shown to be associated with spontaneous phosphorylation of mutated FLT3 in primary blasts from AML patients.49,58,72 Recently, activation of STAT5 by FLT3-ITD has been shown to be direct and independent of JAK and nonreceptor PTKs, such as SRC.74 Interestingly, Pallis et al75 suggested that phosphorylated STAT5 was a general feature of AML, not specifically associated with mutated FLT3, even though the FLT3 ligand was repeatedly shown to activate the PI3K/AKT and/or MAPK pathways rather than the STAT5 pathway to transmit proliferative signals in cells expressing the wild-type FLT3 (FLT3-WT).49,72 These data collectively indicate diversity and differential activation of pathways involved in FLT3 signaling.
Non-PTK Fusion Proteins
STAT proteins are also directly incorporated in non-PTK leukemic fusion proteins. Acute promyelocytic leukemia (APL) is characterized by reciprocal translocations between the retinoic acid receptor α (RARα) gene and five different partner genes including STAT5b.76 An interstitial deletion within chromosome 17 gives rise to the STAT5b-RARα fusion protein, which blocks myeloid differentiation through its interaction with a corepressor complex containing HDAC activity.77–79 Additionally, APL fusion proteins, including STAT5b-RARα, were shown to enhance STAT3 transcriptional activity through a mechanism involving interaction of HDAC and nuclear coactivators.76,77 The APL fusion proteins act to shift the balance between the corepressor and the coactivator associated with STAT3. All-trans-retinoic acid (ATRA) reverses this balance to switch off STAT3 activation.76 However, STAT1/STAT2 activation may be one of the mechanisms of ATRA-induced granulocytic differentiation. ATRA was shown to induce expression of the interferon (IFN)- stimulated transcription factors STAT1, STAT2, and IFN regulatory factor-1 (IRF-1) during myeloid cell differentiation.80,81 These results indicate involvement of an aberrant regulation of the STAT signal transduction pathway in APL.
ROLE OF STAT ISOFORMS IN LEUKEMOGENESIS
STATβ isoforms have been observed in several AML-derived cell lines and myeloblasts from AML patients.5,7,57,59,82 The demonstration of constitutive STAT3 activation with resultant neoplastic transformation in genetically engineered COOH-terminal STAT3 mutants was the first suggestion that the COOH-terminal transactivation domain might play a causative role in oncogenesis.42 In this context, STAT3β isoforms with distinct transcriptional activities and intracellular dynamics were proposed to play a role in leukemogenesis. Recent demonstration of reduced nuclear export and prolonged intranuclear accumulation of STAT1β and STAT3β mapping to their COOH-terminal end is particularly intriguing.27,30 It is likely that truncated STATβ forms have a more versatile multifaceted role in hematopoietic transformation, rather than a simple opposition to STAT function. This would lend further explanation(s) to our findings that constitutive STAT3β activity in leukemic cells identified a group of patients with shorter DFS and overall survival.59 Furthermore, truncated STAT proteins were demonstrated to be prevalent at relapse of AML, suggesting that STATβ isoform expression, rather than the level of constitutive activity, may be involved in disease progression.82 Aberrant constitutive activation of the STATβ proteins likely has important implications in the pathophysiology of AML, albeit with many unanswered questions.
TREATMENT STRATEGIES TARGETING STAT PROTEINS
Tampering with STAT signaling emerges as an attractive target to inhibit oncogenesis and leukemogenesis.83 Treatment strategies including inactivation of upstream STAT activators as well as direct targeting of STAT molecules and downstream effector end proteins are applicable to both hematologic and nonhematologic malignancies (Fig 4; Table 4). Because neoplastic cells are dependent on constitutive STAT activation (oncogene addiction phenomenon), targeting STATs causes preferential cancer cell killing with minimal effects on normal cells.
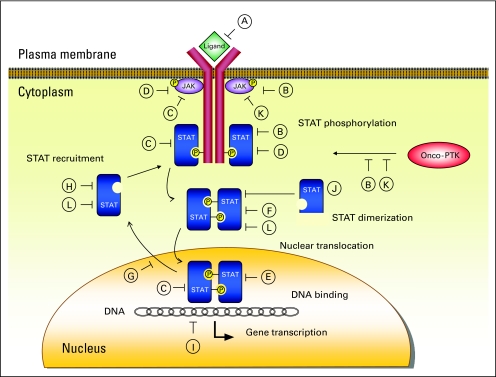
Fig 4.
Signal transducer and activator of transcription (STAT) targeting strategies. (A) Cytokine receptor antagonists or receptor-directed monoclonal antibodies. Blocking activation of growth factor/cytokine receptors with monoclonal antibodies might be beneficial.40,84 (B) Tyrosine kinase inhibitors. Inhibition of upstream tyrosine kinases, such as Janus family tyrosine kinase (JAK), breakpoint cluster region-abelson tyrosine kinase, FMS-related tyrosine kinase 3, and sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian), with resultant downregulation of STATs has proven to be the most effective STAT-targeting strategy.38,39,43,58,71,72,83,85–93 (C) Phosphotyrosine phosphatases,9,13,14 (D) suppressors of cytokine signaling proteins,9,12 and (E) protein inhibitors of activated STATs9,16 are negative regulators of STAT activity. Activating these molecules represents another promising therapeutic approach.10,95 (F) Peptidomimetic inhibitors of STAT dimerization directly and selectively target STATs.96,97 (G) Targeting STAT nucleocytoplasmic shuttling.28 (H) Antisense oligodeoxynucleotides and small interfering RNA molecules cause selective STAT mRNA inhibition.98–100 (I) Disruption of STAT-DNA binding by G-quartets and decoy oligonucleotides.98,101,102 (J) Dominant-negative STAT_isoforms inhibit STAT signaling pathway and tumor growth.7,57,59,82,83 (K) Arsenic trioxide indirectly decreases STAT activation by direct protein tyrosine kinase (PTK) inhibition.94,103–107 (L) Novel platinum (IV) compounds directly bind to STAT molecule and block DNA-binding activity.108,109 P, phosphorus; Onco-PTK, oncogenic PTK.
Table 4.
STAT Targeting Strategies
| Molecule | Strategy | Mechanism | Examples | References |
|---|---|---|---|---|
| Receptor antagonists, monoclonal antibodies | Blockade of cytokine/growth factor binding to the receptor | Indirect | Anti-CD20, anti-EGFR antibody | 40,84 |
| Tyrosine kinase (JAK, BCR-ABL, TEL-ABL, TEL-PDGFβR, FLT3, SRC) inhibitors | Inhibition of upstream tyrosine phosphorylation | Indirect | AG490, cucurbitacin, imatinib mesylate, AG1296, CEP701, GTP14564, PKC412, SU5614, indirubin | 38,39,43,58,71,72,83,85–93 |
| PTP activators | Activating negative regulators of STAT | Indirect | FTY720, forskolin | 9,13,14,95 |
| SOCS protein demethylators | Activating negative regulators of STAT | Indirect | Demethylating agents | 9,10,12 |
| PIAS demethylators | Activating negative regulators of STAT | Indirect | Demethylating agents | 9,10,16 |
| Small-molecule peptidomimetics | Disruption of STAT dimerization | Direct | PY*LKTK, PY*L, AY*L, ISS 610 | 96,97 |
| STAT nuclear export (exportin-1) inhibitors | Blockade of STAT nucleocytoplasmic shuttling | Indirect | Ratjadone A | 28 |
| Antisense oligodeoxynucleotides, siRNA molecules | Selective STAT mRNA inhibition | Direct | ISIS 345794, siRNAs | 98–100 |
| G-quartets, decoy oligonucleotides | Disruption of STAT-DNA binding and transcription | Direct | G-quartets, decoy oligonucleotides | 98,101,102 |
| STATβ isoforms | Dominant-negative STAT inhibition | Direct | STATβ isoforms | 7,57,59,82,83 |
| Arsenic trioxide | PTK inhibition | Indirect | Arsenic trioxide | 94,103–107 |
| Chemotherapy | Direct irreversible interaction with STAT to block DNA-binding activity | Direct | Novel platinum (IV) compounds | 108,109 |
Abbreviations: STAT, signal transducer and activator of transcription; EGFR, epidermal growth factor receptor; JAK, Janus family tyrosine kinase; BCR-ABL, breakpoint cluster region-abelson; TEL-ABL, translocated erythroblastosis (ets) leukemia-abelson; TEL-PDGFβR, translocated erythroblastosis (ets) leukemia–platelet-derived growth factor β receptor; FLT3, FMS-related tyrosine kinase 3; SRC, sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian); PTP, phosphotyrosine phosphatase; SOCS, suppressors of cytokine signaling; PIAS, protein inhibitors of activated STATs; siRNA, small interfering RNA; PTK, protein tyrosine kinase.
Receptor Inhibition
Blocking autocrine and paracrine activation loops of cytokine receptors with receptor antagonists or monoclonal antibodies targeting receptors might prove beneficial in the treatment of leukemias. As a proof of principle, inhibition of constitutive STAT3 activity has been demonstrated by the anti-CD20 (a transmembrane B-cell antigen) chimeric antibody rituximab in non-Hodgkin's lymphomas.84 Similarly, EGFR-directed antibodies were suggested to downregulate STAT3 signaling.40 However, this strategy remains to be elucidated in leukemias.
Tyrosine Kinase Inhibition
Tyrosine kinase inhibitors have become the focus of intensive investigation in disrupting STAT activation. Selective blockage of JAK2 activity by a tryphostin family tyrosine kinase inhibitor, AG490, was shown to impede in vitro and in vivo growth of leukemic cells.85 Furthermore, AG490 was shown to promote apoptosis by suppressing STAT3-mediated antiapoptotic Bcl-xL expression in U266 myeloma cells83 and Mcl-1 expression in large granular lymphocyte leukemia.43 Cucurbitacin I (JSI-124) is another selective JAK kinase inhibitor identified in the National Cancer Institute Diversity Set.86 This compound was reported to suppress STAT3 activation in various cancer cell lines, resulting in inhibition of STAT3-mediated gene transcription. Recently, another cucurbitacin derivative, cucurbitacin Q, was shown to suppress the growth of STAT3-transformed tumors in nude mice xenograft models.87
The BCR-ABL tyrosine kinase inhibitor imatinib mesylate inhibits the growth of cells expressing BCR-ABL, TEL-ABL, and TEL-PDGFβR fusion proteins,88 which are all known to transmit signals through the STAT5 pathway. As direct supporting evidence, blockade of BCR-ABL kinase activity by imatinib mesylate was demonstrated to induce apoptosis of BCR-ABL–positive cell lines and CD34+ cells from CML patients by suppressing the STAT5-dependent expression of Bcl-xL.38,39 Currently, imatinib mesylate is approved by the US Food and Drug Administration as first-line treatment for CML and BCR-ABL–positive ALL.
FLT3 tyrosine kinase inhibitors are under investigation for the treatment of FLT3-positive AML. Small-molecule FLT3 inhibitors, such as AG1296, CEP701, GTP14564, PKC412, and SU5614, have been shown to inhibit STAT5 activation, leading to growth arrest and apoptosis through downregulation of the STAT5 target genes (Bcl-xL and p21) in cells expressing FLT3 mutations.58,71,72,89 Cells harboring FLT3–tyrosine kinase domain are more sensitive in vitro to these inhibitors compared with FLT3-ITD cells.72 Moreover, cotreatment with PKC412/GTP14564 and 17-allylamino-demethoxygeldanamycin, an inhibitor of the heat shock protein 90 (Hsp90), was synergistically effective with associated downregulation of phosphorylated STAT5.89 Preliminary results of the phase I and II studies suggested that PKC412 had hematologic activity in patients with relapsed/refractory AML and myelodysplastic syndrome expressing FLT mutations; however, the responses were brief.90–92
Indirubin is a Chinese herbal medicine used for the treatment of CML that serves as a CDK inhibitor resulting in cell cycle arrest.93 Derivatives of indirubin were shown to directly block SRC kinase activity and constitutive STAT3 signaling in human breast and prostate cancer cells, resulting in apoptosis through downregulation of the antiapoptotic proteins Mcl-1 and survivin.93 Another CDK inhibitor, roscovitine, was shown to cause apoptosis of human T-cell leukemia virus-1–transformed MT-2 T cells by inhibiting STAT5 phosphorylation.94 It was suggested that roscovitine inhibited the interaction of STAT5 with PDGFRα receptor rather than JAK-dependent STAT5 activation. The exact mechanism of action of these CDK inhibitors in the blockade of STAT activation deserves further study.
Activating STAT Inhibitors
Pharmacologic modulation of STAT signaling by STAT-interacting proteins has been suggested as another promising therapeutic approach, although practical accomplishment is still elusive.9 PTPs negatively regulate JAKs and STATs by dephosphorylation of kinases.9,13,14 Specific customized molecules that induce phosphatase activities to dephosphorylate activated STATs may have potential as therapeutic agents. As a proof of principle, PP2A activating agents such as 2-amino-2-[2-(4-octylphenyl)ethyl]-1,3-propanediol hydrochloride (FTY720) and forskolin were shown to downregulate BCR-ABL and STAT5 and subsequently suppress leukemogenesis in both in vitro and in vivo CML and Ph-positive ALL models.95
The SOCS and PIAS families of proteins are negative regulators of STAT signaling.9,12,16 Their clinical significance has been put forward by the demonstration of sustained STAT activity as a result of epigenetic silencing by hypermethylation.10 Literature reports are conflicting regarding SOCS1 methylation in AML. Although two groups110,111 observed SOCS1 methylation resulting in transcriptional silencing, two other groups112,113 failed to detect any SOCS1 methylation in AML. Interestingly, hypermethylation of SHP-1 and PIASy were also reported variably in samples from AML patients.112–114 The reason for these inconsistencies might be the area of the gene promoter studied. Regardless, further work on this subject is warranted, especially in view of the rapidly developing field of treatment with demethylating agents in AML.
Peptidomimetic Inhibitors of STAT Dimerization
Disruption of the STAT3 dimerization by the SH2 domain–binding phosphotyrosyl peptide PY*LKTK and its tripeptide derivatives PY*L and AY*L was demonstrated to block STAT3-mediated DNA binding activity, gene regulation, and cell transformation in vitro and in vivo.96 On the basis of these tripeptide derivatives, more potent specific peptidomimetics (ISS610) were generated and shown to inhibit cell growth.97 Because STAT proteins are directly and selectively targeted, fewer nonspecific adverse effects are theoretically expected than with other strategies that block upstream STAT signaling.
Inhibition of STAT Nucleocytoplasmic Shuttling
Targeting STAT nuclear traffic is another promising strategy of developing cancer drugs.28 This is especially important for STAT3 because of its prominent nuclear presence independent of its phosphorylation. Specific structures determining STAT3 nuclear translocation should be targeted to avoid excessive toxicity as a result of complete block of nucleocytoplasmic shuttling.
mRNA Inhibition
Targeting STATs using single-stranded antisense oligonucleotides (ODNs) with a sequence complementary to the mRNA target has been proven to be effective in reducing intracellular STAT levels with resultant loss of function in in vitro and preclinical models.98 However, modulation of STAT3 expression in every cell is expected in the setting of systemic application of ODN. Preclinical studies of a potent second-generation antisense ODN, ISIS 345794, which selectively inhibits STAT3, are currently ongoing, and phase I trials are expected to begin in the near future.99 Similar to antisense ODNs, RNA interference phenomenon using STAT3-specific small interfering RNAs for mRNA inhibition was demonstrated to block STAT3-mediated prostate cancer cell growth in vitro.100
Targeting STAT3 DNA Binding
Using a nonantisense mechanism, guanosine-rich ODNs that form intramolecular G-quartet structures were developed to inhibit STAT3-DNA binding with a decrease in Bcl-2, Bcl-xL, and Mcl-1 and resultant tumor cell apoptosis in prostate and breast tumor xenografts.101
Decoy ODNs
Intracellular delivery of short double-stranded DNA pieces (decoy ODNs) carrying the consensus STAT-binding sequences has been shown to prevent STAT3 binding to the STAT3 response element within the c-FOS (FBJ murine osteosarcoma viral oncogene homolog) promoter and to interfere with head and neck squamous carcinoma growth in vitro.98 Furthermore, the use of a STAT1 decoy has been reported to inhibit bryostatin 1–induced differentiation of CLL cells with decreased immunoglobulin M production and CD22 expression.102 Modulation of endogenous gene transcription by introducing STAT decoy ODNs emerges as a novel approach to disrupt STAT activation, but it demands a firm understanding of the causative role of STATs in specific leukemia cases.
Targeting STAT3β
DN STAT isoforms have been shown to inhibit STAT signaling pathways with resultant loss of function in in vitro and in vivo tumor models.83 However, constitutively active truncated STAT3β isoforms have been suggested to be involved in leukemic transformation7,57,59,82 based on the reports that the COOH-terminal transactivation domain of STATs plays a causative role in oncogenesis.42 A novel serine-dependent proteolytic activity is responsible for the truncation of the STAT3 COOH-terminal domain in human AML blasts.7 Characterization and cloning of the proteolytic activity with subsequent design of custom-made targeted therapies might hold promise for AML treatment.
Arsenic trioxide
Arsenic trioxide (ATO), which is used as a second-line treatment for relapsed/refractory APL, induces apoptosis of non-APL AML cells in vitro.115 We have demonstrated that ATO indirectly decreases activation of STAT1, STAT3, and STAT5 proteins by direct inhibition of various PTKs in a dose-dependent manner.103 This effect was specific to the STAT pathway without any effect on the MAPK pathway. Furthermore, we have recently shown that ATO synergizes with Hsp90 inhibitors to downregulate constitutive STAT3 activity in an AML cell line model.104 Interestingly, Hayashi et al105 showed that ATO abrogates IL-6–induced phosphorylation of STAT3 in MM.1S myeloma cells via inhibition of JAK1 and JAK2 activity without affecting MAPK and PI3K pathways. Likewise, Cheng et al106 demonstrated inhibition of STAT3 activity in HepG2 hepatoma cells as a result of direct JAK1 and JAK2 suppression by sodium arsenite, independent of the MAPK pathway. STAT5 activity was also shown to be inhibited by ATO through undisclosed mechanisms, resulting in apoptosis of leukemic MT-2 cells.94 These results are encouraging, but a phase II study of ATO alone failed to show any survival advantage in relapsed/refractory elderly AML patients.107 Phase I and III clinical trials to study the in vivo effects of ATO on the STAT pathway in AML patients are ongoing.
Chemotherapy and Biologic Therapy
Pharmacologic modulation of STAT activity after chemotherapy and biologic therapy has been well-documented.108,109,116–118 The novel alkylator compounds of the platinum (IV) family, CPA-1, CPA-7, platinum tetrachloride, and IS3 295, were shown to disrupt STAT3 activity, block neoplastic proliferation, and induce apoptosis in solid tumor cell lines and animal models.108,109 Moreover, blockade of STAT3 by IS3 295 also suppressed the expression of STAT3-induced genes Bcl-x and cyclin D1.109 It seems that this compound interacts directly with the DNA-binding domain of STAT3, both the inactive monomer and the activated dimer, and irreversibly blocks the binding of activated STAT3 to its consensus DNA response element. Similarly, the antimetabolite fludarabine was shown to cause specific depletion of STAT1 mRNA and protein in CLL cells.117 Additionally, STAT proteins were suggested to be involved in ATRA-induced growth inhibition and myeloid differentiation of APL cells.81 In myeloid leukemia cell lines, ATRA was shown to activate STAT1, STAT2, p48, and IRF-1 expression as essential molecules in IFN-α signal transduction.80,81 Moreover, chronic systemic administration of IFN-α has been reported to cause loss of constitutively active STAT1 and STAT3 DNA-binding abilities in precursor melanoma lesions, with associated STAT3 dephosphorylation.118 The cross talk between retinoic acid and IFN signaling suggests a potentially useful synergistic combination in the treatment of leukemias.
CONCLUSION AND FUTURE DIRECTIONS
Dysregulated STAT signaling is associated with precipitous cellular proliferation, disturbed differentiation, and arrested apoptosis, which are the hallmarks of leukemogenesis. Constitutive activation of STAT3 and the presence of COOH-terminally truncated STAT3β isoform have been demonstrated to be correlated with poor clinical outcome in AML. However, it is still unclear whether constitutive STAT activity itself is the cause or the result of a transforming process. Although blocking the STAT pathway has been shown to be sufficient to inhibit malignant transformation, the STAT pathway is probably not the only transcription pathway involved in leukemogenesis.
Identification of STAT-regulated genes by gene expression profiling may provide important insights into the role of STATs in the development of leukemias. Understanding the molecular and biologic mechanisms of how aberrant STAT signaling is involved in cellular transformation is of paramount importance for the development of tailored therapeutic approaches to interrupt STAT signaling. Important developments in drug discovery targeting STATs have been made in recent years, and clinical interest is rapidly growing to construct more selective and efficacious agents.
Footnotes
Supported in part by Grants No. CA16056, CA85580, and CA99238 from the National Cancer Institute, Bethesda, MD, and The Heidi Leukemia Research Fund, Buffalo, NY.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Mustafa Benekli, Meir Wetzler
Collection and assembly of data: Mustafa Benekli
Data analysis and interpretation: Heinz Baumann, Meir Wetzler
Manuscript writing: Mustafa Benekli
Final approval of manuscript: Mustafa Benekli, Heinz Baumann, Meir Wetzler
REFERENCES
- 1.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.National Center for Biotechnology Information. NCBI genome resource guides. http://www.ncbi.nlm.nih.gov/genome/guide/
- 3.Chen X, Vinkemeier U, Zhao Y, et al. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 4.Becker S, Bernd G, Muller CW. Three-dimensional structure of the Stat3 beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 5.Chakraborty A, White SM, Schaefer TS, et al. Granulocyte colony-stimulating factor activation of STAT3 alpha and STAT3 beta in immature normal and leukemic human myeloid cells. Blood. 1996;88:2442–2449. [PubMed] [Google Scholar]
- 6.Azam M, Lee C, Strehlow I, et al. Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity. 1997;6:691–701. doi: 10.1016/s1074-7613(00)80445-8. [DOI] [PubMed] [Google Scholar]
- 7.Xia Z, Salzler RR, Kunz DP, et al. A novel serine-dependent proteolytic activity is responsible for truncated signal transducer and activator of transcription proteins in acute myeloid leukemia blasts. Cancer Res. 2001;61:1747–1753. [PubMed] [Google Scholar]
- 8.Schuster B, Hendry L, Byers H, et al. Purification and identification of the STAT5 protease in myeloid cells. Biochem J. 2007;404:81–87. doi: 10.1042/BJ20061877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shuai K, Liu B. Regulation of JAK-STAT signaling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 10.Ghoshal Gupta S, Baumann H, Wetzler M. Epigenetic regulation of signal transducer and activator of transcription 3 in acute myeloid leukemia. Leuk Res. 2008;32:1005–1014. doi: 10.1016/j.leukres.2007.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim CP, Cao X. Structure, function, and regulation of STAT proteins. Mol Biosyst. 2006;2:536–550. doi: 10.1039/b606246f. [DOI] [PubMed] [Google Scholar]
- 12.Krebs DL, Hilton DJ. SOCS proteins: Negative regulators of cytokine signaling. Stem Cells. 2001;19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
- 13.Ram PA, Waxman DJ. Interaction of growth hormone-activated STATs with SH2-containing phosphotyrosine phosphatase SHP-1 and nuclear JAK2 tyrosine kinase. J Biol Chem. 1997;272:17694–17702. doi: 10.1074/jbc.272.28.17694. [DOI] [PubMed] [Google Scholar]
- 14.Woetmann A, Nielsen M, Christensen ST, et al. Inhibition of protein phosphatase 2A induces serine/threonine phosphorylation, subcellular redistribution, and functional inhibition of STAT3. Proc Natl Acad Sci U S A. 1999;96:10620–10625. doi: 10.1073/pnas.96.19.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson PK. A new RING for SUMO: Wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 2001;15:3053–3058. doi: 10.1101/gad.955501. [DOI] [PubMed] [Google Scholar]
- 16.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 17.Kim TK, Maniatis T. Regulation of interferon-gamma-activated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka T, Soriano MA, Grusby MJ. SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity. 2005;22:729–736. doi: 10.1016/j.immuni.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Malakhova OA, Yan M, Malakhov MP, et al. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 2003;17:455–460. doi: 10.1101/gad.1056303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ungureanu D, Vanhatupa S, Grönholm J, et al. SUMO-1 conjugation selectively modulates STAT1-mediated gene responses. Blood. 2005;106:224–226. doi: 10.1182/blood-2004-11-4514. [DOI] [PubMed] [Google Scholar]
- 21.Collum RG, Brutsaert S, Lee G, et al. A Stat3-interacting protein (StIP1) regulates cytokine signal transduction. Proc Natl Acad Sci U S A. 2000;97:10120–10125. doi: 10.1073/pnas.170192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Decker T, Kovarik P. Transcription factor activity of STAT proteins: Structural requirements and regulation by phosphorylation and interacting proteins. Cell Mol Life Sci. 1999;55:1535–1546. doi: 10.1007/s000180050393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reich NC, Liu L. Tracking STAT nuclear traffic. Nat Rev Immunol. 2006;6:602–612. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 24.Pranada AL, Metz S, Herrmann A, et al. Real time analysis of STAT3 nucleocytoplasmic shuttling. J Biol Chem. 2004;279:15114–15123. doi: 10.1074/jbc.M312530200. [DOI] [PubMed] [Google Scholar]
- 25.Marg A, Shan Y, Meyer T, et al. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J Cell Biol. 2004;165:823–833. doi: 10.1083/jcb.200403057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc Natl Acad Sci U S A. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lödige I, Marg A, Wiesner B, et al. Nuclear export determines the cytokine sensitivity of STAT transcription factors. J Biol Chem. 2005;280:43087–43099. doi: 10.1074/jbc.M509180200. [DOI] [PubMed] [Google Scholar]
- 28.Herrmann A, Vogt M, Monnigmann M, et al. Nucleocytoplasmic shuttling of persistently activated STAT. J Cell Sci. 2007;120:3249–3261. doi: 10.1242/jcs.03482. [DOI] [PubMed] [Google Scholar]
- 29.ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu Y, et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Qiu J, Dong S, et al. Stat3 isoforms, alpha and beta, demonstrate distinct intracellular dynamics with prolonged nuclear retention of Stat3beta mapping to its unique C-terminal end. J Biol Chem. 2007;282:34958–34967. doi: 10.1074/jbc.M704548200. [DOI] [PubMed] [Google Scholar]
- 31.Benekli M, Baer MR, Baumann H, et al. Signal transducer and activator of transcription proteins in leukemias. Blood. 2003;101:2940–2954. doi: 10.1182/blood-2002-04-1204. [DOI] [PubMed] [Google Scholar]
- 32.Yu H, Jove R. The STATS of cancer: New molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 33.Ilaria RL, Jr, van Etten RA. P210 and P190(Bcr-Abl) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem. 1996;271:31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- 34.Frank DA, Varticovski L. Bcr-Abl leads to the constitutive activation of Stat proteins, and shares an epitope with tyrosine phosphorylated Stats. Leukemia. 1996;10:1724–1730. [PubMed] [Google Scholar]
- 35.Bromberg JF, Horvath CM, Besser D, et al. Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gesbert F, Griffin JD. Bcr-Abl activates transcription of the Bcl-x gene through STAT5. Blood. 2000;96:2269–2276. [PubMed] [Google Scholar]
- 37.de Groot RP, Raaijmakers JA, Lammers JW, et al. STAT5-dependent CyclinD1 and Bcl-XL expression in Bcr-Abl-transformed cells. Mol Cell Biol Res Commun. 2000;3:299–305. doi: 10.1006/mcbr.2000.0231. [DOI] [PubMed] [Google Scholar]
- 38.Horita M, Andreu EJ, Benito A, et al. Blockade of the Bcr-Abl kinase activity induces apoptosis of chronic myelogenous leukemia cells by suppressing signal transducer and activator of transcription 5-dependent expression of Bcl-XL. J Exp Med. 2000;191:977–984. doi: 10.1084/jem.191.6.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donato NJ, Wu JY, Zhang L, et al. Down-regulation of interleukin-3/granulocyte-macrophage colony-stimulating factor receptor beta-chain in Bcr-Abl (+) human leukemic cells: Association with loss of cytokine-mediated Stat-5 activation and protection from apoptosis after Bcr-Abl inhibition. Blood. 2001;97:2846–2853. doi: 10.1182/blood.v97.9.2846. [DOI] [PubMed] [Google Scholar]
- 40.Hynes NE, Lane HA. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 41.Ozawa Y, Williams AH, Estes ML, et al. Src family kinases promote AML cell survival through activation of signal transducers and activators of transcription (STAT) Leuk Res. 2008;32:893–903. doi: 10.1016/j.leukres.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 42.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 43.Epling-Burnette PK, Liu JH, Catlett-Falcone R, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest. 2001;107:351–362. doi: 10.1172/JCI9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu G, Wright KL, Ma Y, et al. Role of Stat3 in regulating p53 expression and function. Mol Cell Biol. 2005;25:7432–7440. doi: 10.1128/MCB.25.17.7432-7440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Q, Briggs J, Park S, et al. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24:5552–5560. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- 46.Grandage VL, Gale RE, Linch DC, et al. PI3-kinase/Akt is constitutively active in primary acute myeloid leukaemia cells and regulates survival and chemoresistance via NF-kappaB, Mapkinase and p53 pathways. Leukemia. 2005;19:586–594. doi: 10.1038/sj.leu.2403653. [DOI] [PubMed] [Google Scholar]
- 47.Harir N, Pecquet C, Kerenyi M, et al. Constitutive activation of Stat5 promotes its cytoplasmic localization and association with PI3-kinase in myeloid leukemias. Blood. 2007;109:1678–1686. doi: 10.1182/blood-2006-01-029918. [DOI] [PubMed] [Google Scholar]
- 48.Goh KC, Haque SJ, Williams BR. P38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation induced by interferons. EMBO J. 1999;18:5601–5608. doi: 10.1093/emboj/18.20.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayakawa F, Towatari M, Kiyoi H, et al. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19:624–631. doi: 10.1038/sj.onc.1203354. [DOI] [PubMed] [Google Scholar]
- 50.Niu G, Wright KL, Huang M, et al. Constitutive STAT3 activity upregulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 51.Biethahn S, Alves F, Wilde S, et al. Expression of granulocyte colony-stimulating factor- and granulocyte-macrophage colony-stimulating factor-associated signal transduction proteins of the JAK/STAT pathway in normal granulopoiesis and in blast cells of acute myelogenous leukemia. Exp Hematol. 1999;27:885–894. doi: 10.1016/s0301-472x(99)00017-x. [DOI] [PubMed] [Google Scholar]
- 52.Spiekermann K, Biethahn S, Wilde S, et al. Constitutive activation of STAT transcription factors in acute myelogenous leukemia. Eur J Haematol. 2001;67:63–71. [PubMed] [Google Scholar]
- 53.Aronica MG, Brizzi MF, Dentelli P, et al. P91 STAT1 activation in interleukin-3-stimulated primary acute myeloid leukemia cells. Oncogene. 1996;13:1017–1026. [PubMed] [Google Scholar]
- 54.Gouilleux-Gruart V, Gouilleux F, Desaint C, et al. STAT-related transcription factors are constitutively activated in peripheral blood cells from acute leukemia patients. Blood. 1996;87:1692–1697. [PubMed] [Google Scholar]
- 55.Weber-Nordt RM, Egen C, Wehinger J, et al. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 1996;88:809–816. [PubMed] [Google Scholar]
- 56.Hayakawa F, Towatari M, Iida H, et al. Differential constitutive activation between STAT-related proteins and MAP kinase in primary acute myelogenous leukaemia. Br J Haematol. 1998;101:521–528. doi: 10.1046/j.1365-2141.1998.00720.x. [DOI] [PubMed] [Google Scholar]
- 57.Xia Z, Baer MR, Block AM, et al. Expression of signal transducers and activators of transcription proteins in acute myeloid leukemia blasts. Cancer Res. 1998;58:3173–3180. [PubMed] [Google Scholar]
- 58.Birkenkamp KU, Geugien M, Lemmink HH, et al. Regulation of constitutive STAT5 phosphorylation in acute myeloid leukemia blasts. Leukemia. 2001;15:1923–1931. doi: 10.1038/sj.leu.2402317. [DOI] [PubMed] [Google Scholar]
- 59.Benekli M, Xia Z, Donohue KA, et al. Constitutive activity of signal transducer and activator of transcription 3 protein in acute myeloid leukemia blasts is associated with short disease-free survival. Blood. 2002;99:252–257. doi: 10.1182/blood.v99.1.252. [DOI] [PubMed] [Google Scholar]
- 60.Schuringa JJ, Wierenga AT, Kruijer W, et al. Constitutive Stat3, Tyr705, and Ser727 phosphorylation in acute myeloid leukemia cells caused by the autocrine secretion of interleukin-6. Blood. 2000;95:3765–3770. [PubMed] [Google Scholar]
- 61.Koistinen P, Saily M, Poromaa N, et al. Complex effects of interleukin 6 on clonogenic blast cell growth in acute myeloblastic leukemia. Acta Haematol. 1997;98:14–21. doi: 10.1159/000203547. [DOI] [PubMed] [Google Scholar]
- 62.Frank DA, Mahajan S, Ritz J. B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J Clin Invest. 1997;100:3140–3148. doi: 10.1172/JCI119869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sexl V, Piekorz R, Moriggl R, et al. Stat5a/b contribute to interleukin 7-induced B-cell precursor expansion, but abl- and bcr/abl-induced transformation are independent of Stat5. Blood. 2000;96:2277–2283. [PubMed] [Google Scholar]
- 64.Lacronique V, Boureux A, Valle VD, et al. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 65.Lacronique V, Boureux A, Monni R, et al. Transforming properties of chimeric TEL-JAK proteins in Ba/F3 cells. Blood. 2000;95:2076–2083. [PubMed] [Google Scholar]
- 66.Carron C, Cormier F, Janin A, et al. TEL-JAK2 transgenic mice develop T-cell leukemia. Blood. 2000;95:3891–3899. [PubMed] [Google Scholar]
- 67.Schwaller J, Parganas E, Wang D, et al. STAT5 is essential for the myelo- and lymphoproliferative disease induced by TEL-JAK2. Mol Cell. 2000;6:693–704. doi: 10.1016/s1097-2765(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 68.Palmer AM, Mahajan S, Frank D, et al. The TEL-PDGFβR transforming protein activates STAT1. Blood. 1997;90:178a. abstr. [Google Scholar]
- 69.Sternberg DW, Tomasson MH, Carroll M, et al. The TEL-PDGFR fusion in chronic myelomonocytic leukemia signals through STAT5-dependent and STAT5-independent pathways. Blood. 2001;98:3390–3397. doi: 10.1182/blood.v98.12.3390. [DOI] [PubMed] [Google Scholar]
- 70.Choudhary C, Muller-Tidow C, Berdel WE, et al. Signal transduction of oncogenic Flt3. Int J Hematol. 2005;82:93–99. doi: 10.1532/IJH97.05090. [DOI] [PubMed] [Google Scholar]
- 71.Spiekermann K, Dirschinger RJ, Schwab R, et al. The protein tyrosine kinase inhibitor SU5614 inhibits FLT3 and induces growth arrest and apoptosis in AML-derived cell lines expressing a constitutively activated FLT3. Blood. 2003;101:1494–1504. doi: 10.1182/blood-2002-04-1045. [DOI] [PubMed] [Google Scholar]
- 72.Choudhary C, Schwable J, Brandts C, et al. AML-associated Flt3 kinase domain mutations show signal transduction differences compared with Flt3 ITD mutations. Blood. 2005;106:265–273. doi: 10.1182/blood-2004-07-2942. [DOI] [PubMed] [Google Scholar]
- 73.Takahashi S, Harigae H, Kaku M, et al. Flt3 mutation activates p21WAF1/CIP1 gene expression through the action of STAT5. Biochem Biophys Res Commun. 2004;316:85–92. doi: 10.1016/j.bbrc.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 74.Choudhary C, Brandts C, Schwable J, et al. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–374. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- 75.Pallis M, Seedhouse C, Grundy M, et al. Flow cytometric measurement of phosphorylated STAT5 in AML: Lack of specific association with FLT3 internal tandem duplications. Leuk Res. 2003;27:803–805. doi: 10.1016/s0145-2126(03)00012-2. [DOI] [PubMed] [Google Scholar]
- 76.Dong S, Tweardy DJ. Cross-talk between retinoic acid and STAT3 signaling pathways in acute promyelocytic leukemia. Leukemia Lymphoma. 2003;44:2023–2029. doi: 10.1080/1042819031000116670. [DOI] [PubMed] [Google Scholar]
- 77.Arnould C, Philippe C, Bourdon V, et al. The signal transducer and activator of transcription STAT5b gene is a new partner of retinoic acid receptor alpha in acute promyelocytic-like leukaemia. Hum Mol Genet. 1999;8:1741–1749. doi: 10.1093/hmg/8.9.1741. [DOI] [PubMed] [Google Scholar]
- 78.Dong S, Tweardy DJ. Interactions of STAT5b-RARα, a novel acute promyelocytic leukemia fusion protein, with retinoic acid receptor and STAT3 signaling pathways. Blood. 2002;99:2637–2646. doi: 10.1182/blood.v99.8.2637. [DOI] [PubMed] [Google Scholar]
- 79.Maurer AB, Wichmann C, Gross A, et al. The Stat5-RARα fusion protein represses transcription and differentiation through interaction with a corepressor complex. Blood. 2002;99:2647–2652. doi: 10.1182/blood.v99.8.2647. [DOI] [PubMed] [Google Scholar]
- 80.Matikainen S, Ronni T, Lehtonen A, et al. Retinoic acid induces signal transducer and activator of transcription (STAT) 1, STAT2, and p48 expression in myeloid leukemia cells and enhances their responsiveness to interferons. Cell Growth Differ. 1997;8:687–698. [PubMed] [Google Scholar]
- 81.Dimberg A, Nilsson K, Oberg F. Phosphorylation-deficient Stat1 inhibits retinoic acid-induced differentiation and cell cycle arrest in U-937 monoblasts. Blood. 2000;96:2870–2878. [PubMed] [Google Scholar]
- 82.Xia Z, Sait SN, Baer MR, et al. Truncated STAT proteins are prevalent at relapse of acute myeloid leukemia. Leuk Res. 2001;25:473–482. doi: 10.1016/s0145-2126(00)00158-2. [DOI] [PubMed] [Google Scholar]
- 83.Jing N, Tweardy DJ. Targeting Stat3 in cancer therapy. Anticancer Drugs. 2005;16:601–607. doi: 10.1097/00001813-200507000-00002. [DOI] [PubMed] [Google Scholar]
- 84.Alas S, Bonavida B. Rituximab inactivates signal transducer and activation of transcription 3 (STAT3) activity in B-non-Hodgkin's lymphoma through inhibition of the interleukin 10 autocrine/paracrine loop and results in down-regulation of Bcl-2 and sensitization to cytotoxic drugs. Cancer Res. 2001;61:5137–5144. [PubMed] [Google Scholar]
- 85.Meydan N, Grunberger T, Dadi H, et al. Inhibition of acute lymphoblastic leukaemia by a Jak-2 inhibitor. Nature. 1996;379:645–648. doi: 10.1038/379645a0. [DOI] [PubMed] [Google Scholar]
- 86.Blaskovich MA, Sun J, Cantor A, et al. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63:1270–1279. [PubMed] [Google Scholar]
- 87.Sun J, Blaskovich MA, Jove R, et al. Cucurbitacin Q: A selective STAT3 activation inhibitor with potent antitumor activity. Oncogene. 2005;24:3236–3245. doi: 10.1038/sj.onc.1208470. [DOI] [PubMed] [Google Scholar]
- 88.Carroll M, Ohno-Jones S, Tamura S, et al. CGP 57148, a tyrosine kinase inhibitor, inhibits the growth of cells expressing BCR-ABL, TEL-ABL and TEL-PDGFR fusion proteins. Blood. 1997;90:4947–4952. [PubMed] [Google Scholar]
- 89.Yao Q, Nishiuchi R, Kitamura T, et al. Human leukemias with mutated FLT3 kinase are synergistically sensitive to FLT3 and Hsp90 inhibitors: The key role of the STAT5 signal transduction pathway. Leukemia. 2005;19:1605–1612. doi: 10.1038/sj.leu.2403881. [DOI] [PubMed] [Google Scholar]
- 90.Propper DJ, McDonald AC, Man A, et al. Phase I and pharmacokinetic study of PKC412, an inhibitor of protein kinase C. J Clin Oncol. 2001;19:1485–1492. doi: 10.1200/JCO.2001.19.5.1485. [DOI] [PubMed] [Google Scholar]
- 91.Estey EH, Fisher T, Giles F, et al. A Randomized phase II trial of the tyrosine kinase inhibitor PKC412 in patients (pts) with acute myeloid leukemia (AML)/high-risk myelodysplastic syndromes (MDS) characterized by wild-type (WT) or mutated FLT3. Blood. 2003;102(suppl):2270. abstr. [Google Scholar]
- 92.Stone RM, DeAngelo DJ, Klimek V, et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood. 2005;105:54–60. doi: 10.1182/blood-2004-03-0891. [DOI] [PubMed] [Google Scholar]
- 93.Nam S, Buettner R, Turkson J, et al. Indirubin derivatives inhibit Stat3 signaling and induce apoptosis in human cancer cells. Proc Natl Acad Sci U S A. 2005;102:5998–6003. doi: 10.1073/pnas.0409467102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mohapatra S, Chu B, Wei S, et al. Roscovitine inhibits STAT5 activity and induces apoptosis in the human leukemia virus type 1-transformed cell line MT-2. Cancer Res. 2003;63:8523–8530. [PubMed] [Google Scholar]
- 95.Perrotti D, Neviani P. Protein phosphatase 2A (PP2A), a drugable tumor suppressor in Ph1(+) leukemias. Cancer Metastasis Rev. 2008;27:159–168. doi: 10.1007/s10555-008-9119-x. [DOI] [PubMed] [Google Scholar]
- 96.Turkson J, Ryan D, Kim JS, et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276:45443–45455. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- 97.Turkson J, Kim JS, Zhang S, et al. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther. 2004;3:261–269. [PubMed] [Google Scholar]
- 98.Coppelli FM, Grandis JR. Oligonucleotides as anticancer agents: From the benchside to the clinic and beyond. Curr Pharm Des. 2005;11:2825–2840. doi: 10.2174/1381612054546752. [DOI] [PubMed] [Google Scholar]
- 99.Gleave ME, Monia BP. Antisense therapy for cancer. Nat Rev Cancer. 2005;5:468–479. doi: 10.1038/nrc1631. [DOI] [PubMed] [Google Scholar]
- 100.Lee SO, Lou W, Qureshi KM, et al. RNA interference targeting Stat3 inhibits growth and induces apoptosis of human prostate cancer cells. Prostate. 2004;60:303–309. doi: 10.1002/pros.20072. [DOI] [PubMed] [Google Scholar]
- 101.Jing N, Sha W, Li Y, et al. Rational drug design of G-quartet DNA as anti-cancer agents. Curr Pharm Des. 2005;11:2841–2854. doi: 10.2174/1381612054546761. [DOI] [PubMed] [Google Scholar]
- 102.Battle TE, Frank DA. STAT1 mediates differentiation of chronic lymphocytic leukemia cells in response to Bryostatin 1. Blood. 2003;102:3016–3024. doi: 10.1182/blood-2002-09-2972. [DOI] [PubMed] [Google Scholar]
- 103.Wetzler M, Brady MT, Tracy E, et al. Arsenic trioxide affects signal transducer and activator of transcription proteins through alteration of protein tyrosine kinase phosphorylation. Clin Cancer Res. 2006;12:6817–6825. doi: 10.1158/1078-0432.CCR-06-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wetzler M, Earp JC, Brady MT, et al. Synergism between arsenic trioxide and heat shock protein 90 inhibitors on signal transducer and activator of transcription protein 3 activity: Pharmacodynamic drug-drug interaction modeling. Clin Cancer Res. 2007;13:2261–2270. doi: 10.1158/1078-0432.CCR-06-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hayashi T, Hideshima T, Akiyama M, et al. Arsenic trioxide inhibits growth of human multiple myeloma cells in the bone marrow microenvironment. Mol Cancer Ther. 2002;1:851–860. [PubMed] [Google Scholar]
- 106.Cheng HY, Li P, David M, et al. Arsenic inhibition of the JAK-STAT pathway. Oncogene. 2004;23:3603–3612. doi: 10.1038/sj.onc.1207466. [DOI] [PubMed] [Google Scholar]
- 107.Parmar S, Rundhaugen LM, Boehlke L, et al. Phase II trial of arsenic trioxide in relapsed and refractory acute myeloid leukemia, secondary leukemia and/or newly diagnosed patients at least 65 years old. Leuk Res. 2004;28:909–919. doi: 10.1016/j.leukres.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 108.Turkson J, Zhang S, Palmer J, et al. Inhibition of constitutive signal transducer and activator of transcription 3 activation by novel platinum complexes with potent antitumor activity. Mol Cancer Ther. 2004;3:1533–1542. [PubMed] [Google Scholar]
- 109.Turkson J, Zhang S, Mora LB, et al. A novel platinum compound inhibits constitutive Stat3 signaling and induces cell cycle arrest and apoptosis of malignant cells. J Biol Chem. 2005;280:32979–32988. doi: 10.1074/jbc.M502694200. [DOI] [PubMed] [Google Scholar]
- 110.Chen CY, Tsay W, Tang JL, et al. SOCS1 methylation in patients with newly diagnosed acute myeloid leukemia. Genes Chromosomes Cancer. 2003;37:300–305. doi: 10.1002/gcc.10222. [DOI] [PubMed] [Google Scholar]
- 111.Watanabe D, Ezoe S, Fujimoto M, et al. Suppressor of cytokine signalling-1 gene silencing in acute myeloid leukaemia and human haematopoietic cell lines. Br J Haematol. 2004;126:726–735. doi: 10.1111/j.1365-2141.2004.05107.x. [DOI] [PubMed] [Google Scholar]
- 112.Chim CS, Wong AS, Kwong YL. Epigenetic dysregulation of the Jak/STAT pathway by frequent aberrant methylation of SHP1 but not SOCS1 in acute leukaemias. Ann Hematol. 2004;83:527–532. doi: 10.1007/s00277-004-0843-1. [DOI] [PubMed] [Google Scholar]
- 113.Johan MF, Bowen DT, Frew ME, et al. Aberrant methylation of the negative regulators RASSFIA, SHP-1 and SOCS-1 in myelodysplastic syndromes and acute myeloid leukaemia. Br J Haematol. 2005;129:60–65. doi: 10.1111/j.1365-2141.2005.05412.x. [DOI] [PubMed] [Google Scholar]
- 114.Ueda M, Ota J, Yamashita Y, et al. DNA microarray analysis of stage progression mechanism in myelodysplastic syndrome. Br J Haematol. 2003;123:288–296. doi: 10.1046/j.1365-2141.2003.04601.x. [DOI] [PubMed] [Google Scholar]
- 115.Lehmann S, Bengtzen S, Paul A, et al. Effects of arsenic trioxide (As2O3) on leukemic cells from patients with non-M3 acute myelogenous leukemia: Studies of cytotoxicity, apoptosis and the pattern of resistance. Eur J Haematol. 2001;66:357–364. doi: 10.1034/j.1600-0609.2001.066006357.x. [DOI] [PubMed] [Google Scholar]
- 116.Chelbi-Alix MK, Pelicano L. Retinoic acid and interferon signaling cross-talk in normal and RA-resistant APL cells. Leukemia. 1999;13:1167–1174. doi: 10.1038/sj.leu.2401469. [DOI] [PubMed] [Google Scholar]
- 117.Frank DA, Mahajan S, Ritz J. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nat Med. 1999;5:444–447. doi: 10.1038/7445. [DOI] [PubMed] [Google Scholar]
- 118.Kirkwood JM, Farkas DL, Chakraborty A, et al. Systemic interferon-alpha (IFN-alpha) treatment leads to Stat3 inactivation in melanoma precursor lesions. Mol Med. 1999;5:11–20. [PMC free article] [PubMed] [Google Scholar]