Abstract
The subunit number and stoichiometry of membrane-bound proteins are difficult to determine without disrupting their membrane environment. Here we describe a single-molecule technique for counting subunits of proteins in live cell membranes by observing bleaching steps of GFP fused to a protein of interest. After testing the method with proteins of known stoichiometry expressed in Xenopus laevis oocytes, we resolved the composition of NMDA receptors composed of NR1 and NR3 subunits.
Many membrane proteins form multimers before they achieve a functional state and are transported to the plasma membrane of the cell. The stoichiometry of a complex is precisely regulated and is fundamental to its functional properties. Subunit stoichiometry is usually assessed via bulk biochemical and macroscopic functional analyses. For example, the multimeric state of K+ channels in the squid giant axon was originally predicted from the shape of the current trace during opening of the channels in voltage clamp1. A kinetic analysis of inactivation2 and an analysis of currents in mixed subunit channels3 indicated that the channels are probably composed of four similar or identical subunits, which was later confirmed by crystallography4. Sometimes macroscopic functional recordings do not distinguish stoichiometry precisely. For example, CNG channels for years had been assumed to be composed of a 2:2 stoichiometry of CNGA1 and CNGB1 subunits5, but has been shown to actually be composed of a 3:1 CNGA1 to CNGB1 stoichiometry by biochemical analysis6 and using fluorescence energy resonance transfer between fluorescent proteins fused to CNGA1 and CNGB1 (ref. 7).
The well-studied glutamate-gated NMDA receptors are tetramers containing two NR1 and two NR2 subunits. This stoichiometry had been deduced from macroscopic functional analysis, in which coexpression of wild-type and mutant subunits resulted in a triphasic response to agonists that could be explained by mixture of receptors containing zero, one or two mutant NR1 or NR2 subunits8, and from single-channel recordings that showed distinct behaviors consistent with the possible mixed stoichiometries9, and confirmed by crystallography10. Less is known about NMDA receptors containing the more recently discovered NR3 subunit, which is thought to be involved in synaptic development11. Initially it was thought that NR3 coassembles with NR1 and NR2 to form glutamate-gated receptors with unique properties11. More recently, and quite surprisingly, the NR3 subunit ligand binding domain had been found to bind glycine and not glutamate, and NR1:NR3 receptors have been shown to be activated by glycine12. The subunit composition of NR1:NR3 receptors has not yet been determined and, indeed, some doubt has remained because they have been found to not form functional receptors in some cell types.
Our aim was to develop a method of determining subunit composition of membrane proteins that focuses exclusively on the fraction that is on the cell surface and, in doing so, to resolve the subunit composition of the NR1:NR3 NMDA receptor. The method can be used to directly determine the number of subunits in membrane proteins by counting discrete steps of photobleaching of single fluorescent molecules. We first tested the method on three channels of known composition, which contained one, two or four fluorophores. We then used the method to determine the composition of the NR1:NR3 receptor.
We genetically engineered fusions of the protein of interest with GFP and expressed them in Xenopus laevis oocytes (Supplementary Methods online). The use of the genetically encoded GFP tag ensures that every tagged subunit carries exactly one chromophore and this avoids nonspecific labeling that could occur with organic dyes. The photobleaching of a single GFP is a discrete process; thus the fluorescence intensity of a protein complex with one or several GFP molecules drops in a stepwise fashion, and the number of steps reveals the number of GFP-tagged subunits in the complex.
To avoid the problem of autofluorescence from the cell cytoplasm, we used total internal reflection fluorescence microscopy, which restricts illumination to the interface between the coverslip and the cell resting on it13 (Supplementary Methods). We imaged areas of the membrane where the density of fluorescent spots was between 20 and 200 in a 15 × 15 μm area. At such densities we could obtain enough spots for statistical analysis, while keeping the probability low that two channels would lie within a diffraction-limited spot. We extracted the emission intensities of the fluorescent spots from the acquired movies (Fig. 1a and Supplementary Video 1 online) according to the procedure described in Supplementary Methods.
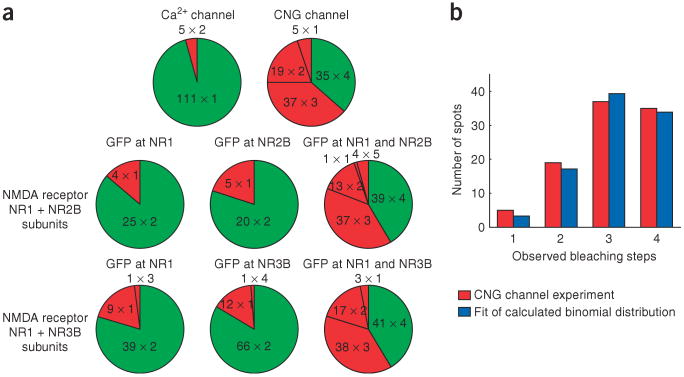
Figure 1.

Bleaching steps of single fluorescent protein complexes reveal the number of GFP-labeled subunits. (a) A single image of the acquired sequence shows the selected fluorescent spots in the cell membrane (blue circles). Scale bar, 2 μm. (b) The different labeling schemes include 1, 2 or 4 subunits of a membrane protein complex fused to GFP. (c) Time courses of fluorescence emission for the labeling schemes in b (two examples each from spots bleaching with the expected number of bleaching steps). Green arrows indicate the bleaching steps. The y-axis is scaled in photons per second for comparison purposes (for calibration see Supplementary Methods).
First we tested the α1E Ca2+ channel, which forms a functional channel on its own, without other Ca2+ channel subunits. This channel is tetrameric, but all of the subunits are connected into one polypeptide (Fig. 1b). The vast majority of α1E-GFP fluorescent spots (96%) displayed one bleaching step (Figs. 1c and 2a). The dominance of spots with one bleaching step is consistent with the α1E channel being composed of one polypeptide. A small number of spots (4%) bleached in two steps, and these probably arise from the rare colocalization of two channels within a diffraction-limited area.
Figure 2.
Distribution of bleaching steps for channels with different numbers of labeled subunits. (a) Numbers A × B in the sectors indicate that A spots with B bleaching steps were observed. Green sector, number of bleaching steps expected from biochemical and functional studies. Red sectors, number of bleaching steps different from what is expected. (b) The observed numbers of spots having 1, 2, 3 and 4 bleaching steps in the experiment with the tetrameric CNG channel (red) match closely a calculated binomial distribution (blue), assuming a probability of 77.5% that the GFP is fluorescent.
As an example of a channel assembled from four identical subunits, we examined the homotetrameric CNG channel. We used the X-fA4 chimera of the bovine CNGA1 and catfish CNGA2 and CNGA4, which had been shown to form functional channels14. Although many of the spots bleached in 4 steps, as expected (Fig. 1b), an equally large number bleached in 3 steps, and some showed 1 or 2 steps (Fig. 2a). Given the distribution of dwell times at each fluorescence level, we calculated that 10% of tetramers would be seen as trimers because two bleaching steps would occur too closely in time to be seen as separate steps, leading to an underestimate of the number of GFPs in each spot (see Supplementary Methods). Still, even after correction for these missed events there still remained a sizable number of spots with fewer than 4 bleaching steps. This suggests that not all of the GFPs are fluorescent at the start of data acquisition, perhaps due to misfolding or incomplete maturation of the GFP. With this in mind, we calculated the expected distribution of tetrameric channels containing 1, 2, 3 or 4 functional GFP tags and fitted it to the observed distribution of bleaching steps, allowing the probability that the GFP tag is fluorescent to be a free parameter. The resulting binomial distribution with a probability of 77.5% of the GFP to be fluorescent (79.5% after correction for missed events) matches very well the observed statistics (Fig. 2b). This supports the idea that ∼20% of the GFPs are in a nonfluorescent state, reminiscent of what has been observed for DsRed15. The confidence level that the observed distribution represents a tetramer is high. For example, we calculated a probability of 10−5 that this distribution is due to five subunits instead of four.
Next we coexpressed the NR1 and NR2B subunits of the NMDA receptor. These are known to assemble into tetrameric channels with a 2:2 stoichiometry8. In our first experiment, GFP was fused to the C terminus of the NR1 subunit, and the NR2B subunit was not labeled (Fig. 1b). We observed mainly two-step bleach events (Fig. 1c), and a minority of single step bleaching spots (Fig. 2a). The preponderance of two-step bleaching spots is consistent with there being two NR1 subunits in each channel. In the second experiment, we fused GFP to the NR2B subunit, and the NR1 subunit was not labeled. Again, the majority of the spots exhibited two bleaching steps (Fig. 2a). Finally, we labeled both NR1 and NR2B with GFP. We observed a distribution of 1–4 bleaching steps (Fig. 2a) similar to what we observed for the homotetrameric CNG channel, which, once again, was consistent with only ∼80% of the GFPs being fluorescent.
For the NMDA receptor containing the NR1 and NR3B subunits we used the same strategy as for the NR1:NR2B receptor. We expressed three combinations of subunits: (i) NR1-GFP and NR3B, (ii) NR1 and NR3B-GFP, or (iii) NR1-GFP and NR3B-GFP. We mainly observed spots with two bleaching steps and some with one bleaching step in the cases when only one subunit was tagged with GFP (Fig. 2a and Supplementary Fig. 1 online). When both subunits were labeled, the distribution of 1–4 subunits again was consistent with a tetrameric channel with about 80% of the GFP being fluorescent (Fig. 2a). The distributions are nearly identical to the ones obtained for the NR1:NR2B receptor. This shows that the subunit composition of the NR1:NR3B receptor is 2:2. This result indicates that although the NR3 ligand-binding pocket has more in common with NR1, its protein-protein interactions are similar those of NR2.
The single-molecule fluorescence method presented here provides a tool for quickly determining subunit stoichiometry of membrane proteins in live cells by counting bleaching steps of GFP tags. The optical measurements require a simple total internal reflection fluorescence microscope equipped with a sensitive camera, and the evaluation does not require specialized software, which makes the method broadly usable. A small number of single-molecule measurements is sufficient to provide clear reflections of the number and mixture of subunits. The method can be applied to fixed-stoichiometry channels and receptors, as we have done here, and should work equally well for auxiliary subunits, such those of voltage-gated channels, and for G-protein coupled receptors, which are known to dimerize in some cases, and where dimeric partnerships could be key to the combinatorial code of chemical sensing. The method may also prove useful for detecting function in signaling complexes, which recruit cellular partners when activated.
One limitation of the method is that for complexes with more than 5 subunits, the distributions of bleaching steps for n and n + 1 subunits look similar, and detection of discrete steps becomes more difficult. In this case, estimates for the subunit number can be made based on the size of a single bleaching step and the total starting fluorescence, as done recently in the flagellar motor16. Our results indicate that the GFP variant that we used (monomeric eGFP) has a probability of ∼80% of being fluorescent in distinct fusion proteins. It will be necessary to make similar determinations if one is interested in using other fluorescent proteins.
Supplementary Material
Acknowledgments
We thank E.C. Young for the X-fA4 channel DNA. This work was supported by postdoctoral fellowships from the Deutsche Forschungsgesellschaft and the American Heart Association to M.H.U., and by a grant from the US National Institutes of Health.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
Competing Interests Statement: The authors declare no competing financial interests.
References
- 1.Hodgkin AL, Huxley AF. J Physiol (Lond) 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacKinnon R, Aldrich RW, Lee AW. Science. 1993;262:757–759. doi: 10.1126/science.7694359. [DOI] [PubMed] [Google Scholar]
- 3.Liman ER, Tytgat J, Hess P. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 4.Doyle DA, et al. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 5.He Y, Ruiz ML, Karpen JW. Proc Natl Acad Sci USA. 2000;97:895–900. doi: 10.1073/pnas.97.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong H, Molday LL, Molday RS, Yau KW. Nature. 2002;420:193–198. doi: 10.1038/nature01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng J, Trudeau MC, Zagotta WN. Neuron. 2002;36:891–896. doi: 10.1016/s0896-6273(02)01099-1. [DOI] [PubMed] [Google Scholar]
- 8.Laube B, Kuhse J, Betz HJ. Neurosci. 1998;18:2954–2961. doi: 10.1523/JNEUROSCI.18-08-02954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Premkumar LS, Auerbach A. J Gen Physiol. 1997;110:485–502. doi: 10.1085/jgp.110.5.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa H, Singh SK, Mancusso R, Gouaux E. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 11.Das S, et al. Nature. 1998;393:377–381. doi: 10.1038/30748. [DOI] [PubMed] [Google Scholar]
- 12.Chatterton JE, et al. Nature. 2002;415:793–798. doi: 10.1038/nature715. [DOI] [PubMed] [Google Scholar]
- 13.Sonnleitner A, Mannuzzu LM, Terakawa S, Isacoff EY. Proc Natl Acad Sci USA. 2002;99:12759–12764. doi: 10.1073/pnas.192261499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young EC, Krougliak N. J Biol Chem. 2004;279:3553–3562. doi: 10.1074/jbc.M310545200. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Parajo MF, Koopman M, van Dijk EMHP, Subramaniam V, van Hulst NF. Proc Natl Acad Sci USA. 2001;98:14392–14397. doi: 10.1073/pnas.251525598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leake MC, et al. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.