Abstract
G protein-coupled receptor (GPCR) kinases (GRKs) were discovered by virtue of their ability to phosphorylate activated GPCRs. They constitute a branch of the AGC kinase superfamily, but their mechanism of activation is largely unknown. To initiate a study of GRK2 activation, we sought to identify sites on GRK2 remote from the active site that are involved in interactions with their substrate receptors. Using the atomic structure of GRK2 in complex with Gβγ as a guide, we predicted that residues on the surface of the kinase domain that face the cell membrane would interact with the intracellular loops and carboxyl (C) terminal tail of the GPCR. Our study focused on two regions: the kinase large lobe and an extension of the kinase domain known as the C-tail. Conserved residues in the GRK2 large lobe whose side-chains are solvent exposed and facing the membrane were targeted for mutagenesis. Residues in the C-tail of GRK2, although not ordered in the crystal structure, were also targeted because this region has been implicated in receptor binding and in the regulation of AGC kinase activity. Four substitutions out of twenty, all within or adjacent to the C-tail, resulted in significant deficiencies in the ability of the enzyme to phosphorylate activated GPCRs, rhodopsin and the β2-adrenergic receptor (β2AR). The mutant exhibiting the most dramatic impairment, V477D, also showed significant defects in phosphorylation of non-receptor substrates. Interestingly, Michaelis-Menten kinetics suggested that V477D had 12-fold lower kcat, but no changes in KM, suggesting a defect in acquisition or stabilization of the closed state of the kinase domain. V477D was also resistant to activation by agonist-treated β2AR. Therefore, Val477 and other residues in the C-tail are expected to play a role in the activation of GRK2 by GPCRs.
G protein-coupled receptors (GPCRs), distinguished by their seven transmembrane helices and their ability to couple to heterotrimeric G proteins (Gαβγ), are activated by hormones, neurotransmitters and sensory signals, and regulate many diverse processes. Receptor stimulation catalyzes the binding of GTP to the Gα subunit, allowing Gα and Gβγ to directly modulate effectors such as adenylyl cyclase, phospholipase Cβ, cGMP phosphodiesterase, ion channels and Rho family guanine-nucleotide exchange factors (1). Appropriate levels of GPCR desensitization are critical for cellular function. One mechanism of receptor desensitization is initiated by the activation-dependent phosphorylation of their third intracellular loops or C-terminal tails catalyzed by a small family of G protein-coupled receptor kinases (GRKs) (2). Phosphorylated GPCRs are recognized by arrestins, which uncouple the receptors from heterotrimeric G proteins and target them for internalization (3).
The seven mammalian GRKs are grouped into three subfamilies: the GRK1/7 subfamily, the GRK2/3 subfamily, and the GRK4/5/6 subfamily. Their central ~350 amino acid catalytic domain belongs to the protein kinase A/protein kinase G/protein kinase C (AGC) family of protein kinases (4). Like other AGC kinases, the GRK kinase domain contains an N-terminal (small) lobe and a C-terminal (large) lobe, followed by an extension to the conserved core called the C-terminal tail (C-tail). The C-tail begins in the large lobe of the kinase domain (the C-terminal lobe tether, or “CLT”), passes over the active site where it contributes to the nucleotide and phosphoacceptor binding sites (the active site tether or “AST”), and ends by donating additional structural elements to the small lobe (the N-terminal lobe tether, or “NLT”). The AST regions of AGC kinases often contain motifs that confer additional levels of regulation on kinase activity (5).
Three-dimensional structures of GRKs representing each GRK subfamily (GRK1, GRK2 and GRK6) have been determined by X-ray crystallography, which reveal that they all share a core structure consisting of a regulator of G protein signaling (RGS) homology (RH) domain intimately associated with the kinase domain, which is inserted between the ninth and tenth helices of the RH domain (Fig. 1) (6–8). In all of the reported structures, the kinase domains exist in a relatively open, presumably inactive conformation, with AST regions that are either entirely or partially disordered.
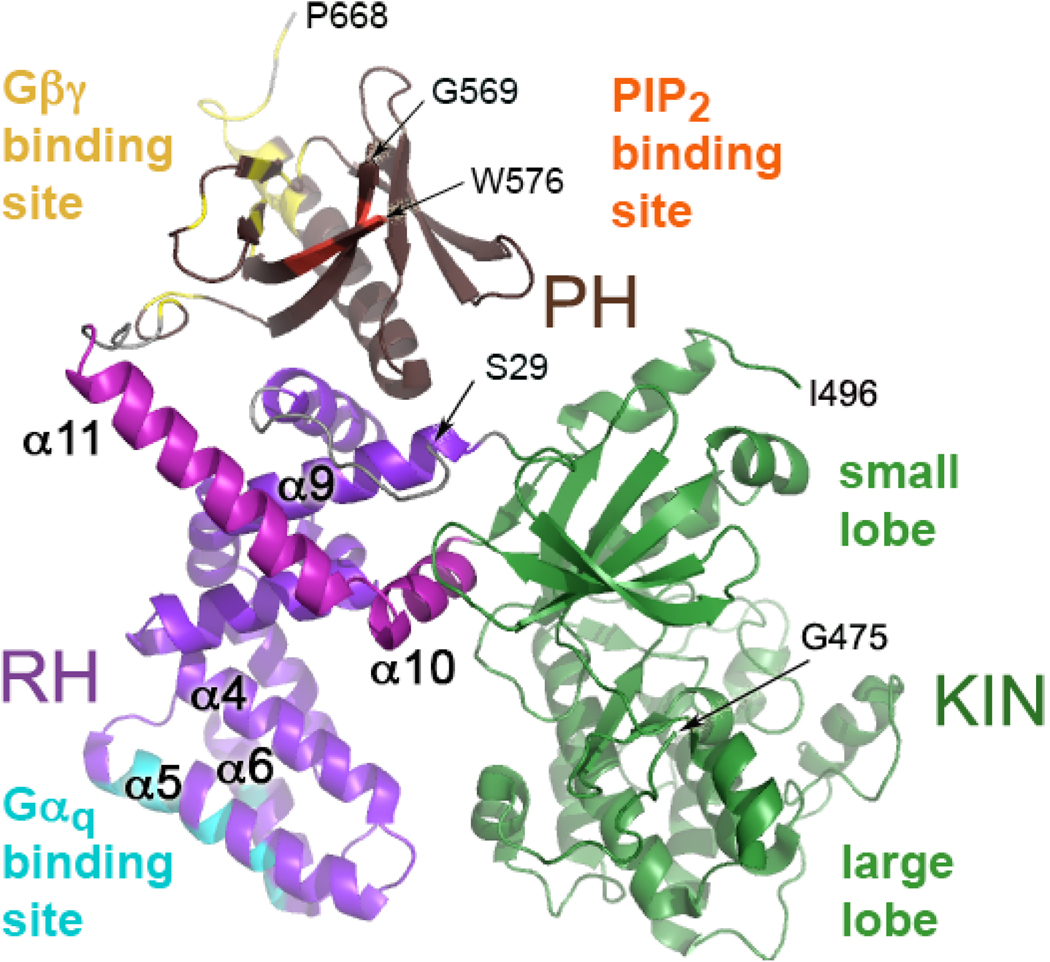
Figure 1. GRK2 Structure.
Full-length GRK2 is oriented to display its predicted membrane-proximal surface, regions of which are expected to interact with the cytoplasmic loops and tails of an activated GPCR (6). In GRK2, the RH domain contacts both the kinase domain (green) and the PH domain (brown). In the RH domain, α-helices 1–9 are purple while α-helices 10 and 11, which are unique to the RH domains of GRKs, are magenta. The regions that interact with Gαq are colored cyan, that interact with Gβγ are yellow, and that interact with phosphatidylinositol bisphosphate (PIP2) are orange. The N-terminal 28 amino acids, a large portion of the kinase extension (476–495), a loop of the PH domain, and the C-terminal 21 residues are not structured in GRK2 crystals. The termini of ordered regions are indicated by Ser29, Gly475, Ile496, Gly569, Trp576, and Pro668 (arrows).
Activation of AGC kinases typically involves changes in the orientation of the C helix, kinase domain closure such that the small lobe rotates relative to the large lobe, ordering of the AST region, and phosphorylation and reorientation of the activation loop (5, 9, 10). Several lines of evidence suggest that GRKs employ only a subset of these events to achieve their activated state. First, in the crystal structures of GRK1, GRK2 and GRK6, the C-helix is already in its “active” conformation, and active site residues engage the substrate Mg2+·ATP as expected in high-resolution structures of GRK1 (8). Second, GRKs lack two or all of the three phosphorylation motifs found in other AGC kinases, including the activation loop site, whose phosphorylation in PKA and PKB orders the loop and stabilizes residues that contribute to catalysis. In the GRK structures reported thus far, the activation loops are well-ordered, even in the absence of phosphorylation. Thus, it appears that GRKs simply need to reorient their small and large lobes to achieve a closed state that properly aligns the catalytic machinery. Unlike PKA, the binding of nucleotides is apparently insufficient to drive this conformational change (8).
While the detailed mechanism of GRK activation has yet to be delineated, activated GPCRs are clearly the most important regulators of kinase activity. GRKs exhibit low activity when peptides are used as substrates, but this activity can be stimulated in the presence of activated receptors (11–14). Such experiments support the existence of a non-catalytic, allosteric “docking site” on GRKs that also serves to discriminate between inactive and active receptors. The molecular basis for how GRK2 recognizes activated GPCRs is also poorly understood. The extreme N-terminal region of GRKs, which is disordered in all but one of the published GRK crystal structures, is predicted to form a helix that is important for receptor phosphorylation either via binding directly to receptors or to phospholipids (15, 16). The RH domain may also play a role in receptor interaction. The GRK2-D527A mutation in the α11 helix of the RH domain hinders quenching of ligand-induced inositol phosphate formation and ablates co-immunoprecipitation of GRK2 with the metabotropic glutamate receptor, mGluR1a (17).
The predicted model of the GRK2-Gβγ complex at the membrane (6) suggested that the cytoplasmic loops and tails of receptors could be accommodated in the space between the membrane plane and the large lobe of the kinase domain (Fig. 2 inset). This simple model suggested that two regions of the kinase domain could be in close proximity to the receptor. The first region consists of the αD helix and the αF-αG loop, which form the walls of the putative peptide-binding channel on the large lobe (Fig. 2). There is further rationale to believe that the large lobe might contain elements that contribute to receptor binding. In the crystal structure of PKA with the PKI inhibitor peptide, PKI not only binds to the phosphoacceptor site adjacent to the active site, but also forms extensive interactions with the αF-αG loop. These latter interactions are required for high affinity binding (18) (Fig. 2). By analogy, an extension of the receptor containing the phosphoacceptor Ser or Thr, such as a portion of the carboxyl tail or third intracellular loop, may bind to the peptide channel of the large lobe. The second region consists primarily of the AST, which is conserved among GRKs but less so with other AGC kinases. This region is disordered in structures of GRK2 and GRK6, and partially ordered in nucleotide-bound structures of GRK1. In the AGC kinase family, the C-tail is recognized as a site used for regulation of activity via covalent modification or protein-protein interactions (5) Indeed, this region is near the hinge of the kinase domain and could make direct contacts with both ATP and phosphoacceptor. A glutathione-S-transferase (GST) fusion protein, GST-GRK2(457–546), which includes the entire C-tail, was also reported to bind rhodopsin and inhibit GRK2-mediated desensitization, while a truncated GRK2 containing residues 1–457 could phosphorylate peptides, but not rhodopsin (20). Furthermore, an autophosphorylation-deficient mutant of GRK5, which eliminated two phosphoacceptor sites in the AST region, has a 15-fold lower catalytic efficiency resulting from a combined higher KM (2.8-fold) and a lower Vmax (5.3-fold) using rhodopsin as a substrate (21). A proline-rich region on the N-terminal side of the AST has also been reported to be important for GRK interaction with activated GPCRs (22).
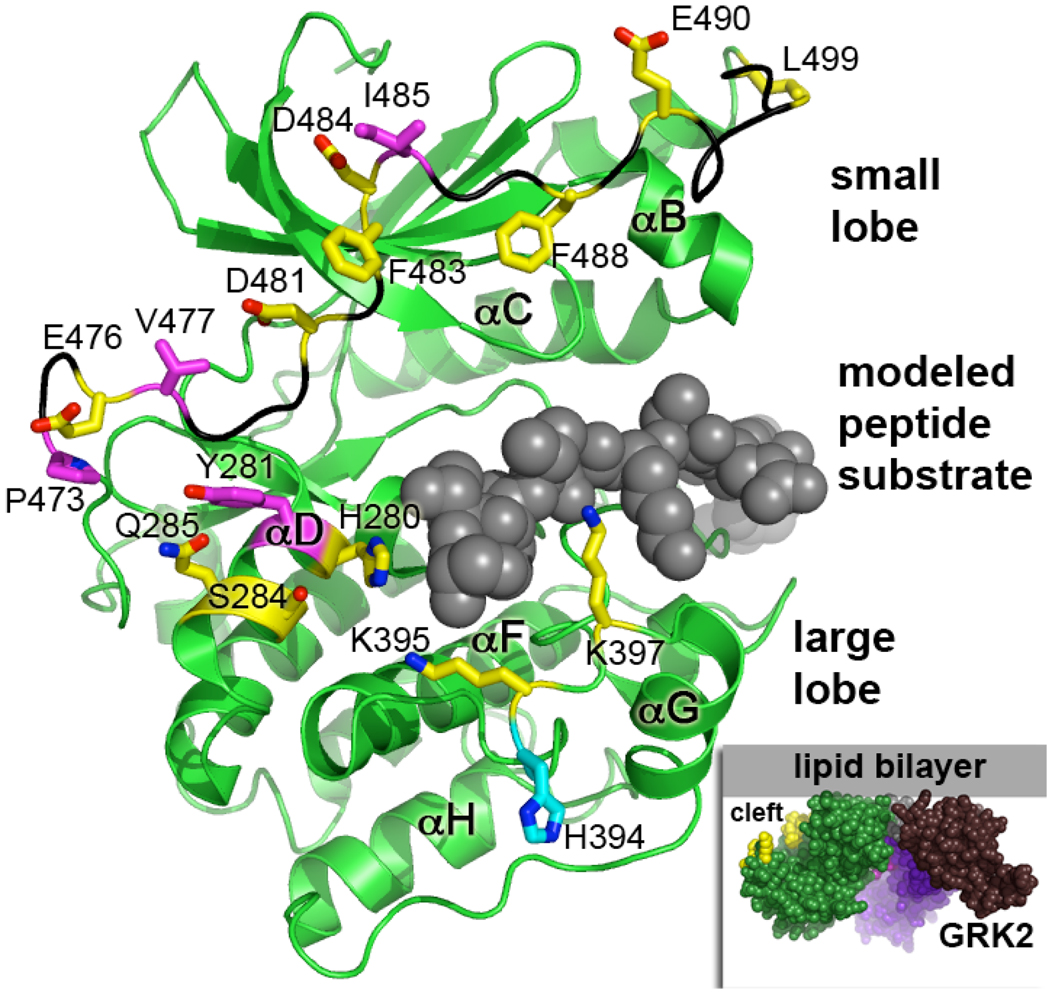
Figure 2. Kinase Domain Residues Targeted for Mutagenesis.
The GRK2 kinase domain was modeled in a closed conformation based on an activated structure of PKA (PDB 1L3R). Two regions of the kinase domain were targeted for mutagenesis: the large lobe and the C-tail. To indicate the expected position of the phosphoacceptor binding site, the PKA inhibitor PKI (space-filling atoms in gray) was mapped onto the GRK2 structure. Nearby residues that could interact with other regions of the receptor (His280, Tyr281, Gln285, Ser284, His394, Lys395, and Lys397) were substituted with alanine. The AST of GRK2 has also been implicated in receptor interaction (20) but residues Glu476-Leu499 have been unstructured in all crystals of GRK2 reported thus far. However, a nearly fully ordered AST loop (black backbone) was observed in GRK1 (8). This structure was mapped onto GRK2 to provide an estimate of the position of amino acids in the AST. Pro473, Glu476, Val477, Asp481, Phe483, Asp484, Ile485, Phe488, Glu490, Gly495, and Leu499 in the AST were selected for site-directed mutagenesis. Substituted residues that showed diminished capacity to phosphorylate receptor and at least some of the non-receptor substrates have carbon atoms colored magenta, whereas those that did not have significant effects on rhodopsin phosphorylation have carbons colored yellow. (Inset) A space-filling model of GRK2 at the plasma membrane using the Fig. 1 coloring scheme (RH domain is purple and magenta, and the PH domain is colored brown). We speculated that the cleft between the lipid bilayer and the kinase large lobe could serve to accommodate the intracellular loops and carboxyl tail of a GPCR. Mutated residues in the kinase domain are shown as yellow spheres.
We therefore tested whether specific residues of the kinase domain large lobe and C-tail were critical for docking with activated receptors. Twenty surface residues of the large lobe and the AST were selected for mutagenesis. Four mutants showed significant defects in their ability to phosphorylate two GPCRs: rhodopsin and the β2-adrenergic receptor (β2AR). However, these mutants also showed lower but appreciable defects in their ability to phosphorylate non-receptor substrates. Furthermore, Michaelis-Menten kinetics of the most defective mutant, V477D in the AST, was most consistent with a catalytic but not a binding defect. In summary, our studies indicate that several residues in the AST are involved in the mechanism by which receptors activate GRKs and that, except for Tyr281, residues in the large lobe do not thus far appear to significantly contribute to receptor docking or kinase domain closure.
EXPERIMENTAL PROCEDURES
Materials
African Green monkey kidney cells (COS-1) and Spodoptera frugiperda (Sf9) insect cells were from the American Tissue Culture Collection. Bovine GRK2 cDNA in a mammalian cell expression vector, pcDNA3-GRK2, was a gift from Dr. Jeffrey Benovic. β2 ARs with an amino terminal Flag tag and a carboxyl terminal hexahistidine tag in phospholipid vesicles (23, 24) and in dodecylmaltoside (DDM) were a generous gift of Dr. Robert J. Lefkowitz (Duke University). An E. coli plasmid for expression of human α-synuclein (pRK172) was provided by Dr. Michel Goedert (University of Cambridge). The PyMOL Molecular Graphics System was used to generate protein structure figures.
Mutagenesis of GRK2
Site-directed mutagenesis (QuikChange, Stratagene) with minor modifications (25) was used to make 20 mutants in the pcDNA3-GRK2 construct, generating single amino acid substitutions in the AST region and large lobe of the kinase domain. The mutants were: H280A, Y281A, S284A, Q285A, Q285K, H394A, K395A, K397A, P473A, P473E, E476K, V477D, D481A, F483D, D484A, I485A, F488D, E490K, G495A, and L499D. Each mutant was verified by DNA sequencing.
COS-1 Cell Transient Transfection and Quantification of GRK2 Expression
In three independent experiments, COS-1 cells (7.4 × 105) grown in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 µg/ml streptomycin at 37 °C with 5% CO2 were plated on 6 cm dishes and transfected with 4 µg plasmid DNA using 6 µl FuGENE-6 (Roche). After 48 h, cells were washed twice in STE (20 mM Tris, 150 mM NaCl, 1 mM EDTA, pH 8), and scraped in 0.4 ml/plate of 20 mM HEPES, pH 7.5, 250 mM NaCl, 10 mM EDTA, 0.02% Triton-X100, 0.5 mM PMSF, 20 µg/ml leupeptin, and 100 µg/ml benzamidine (Buffer A). Lysates were sonicated with a microtip probe sonicator 30 times in 1 sec pulses with 2 sec rest periods between pulses. Soluble fractions were prepared by centrifugation for 10 min at 40,000×g. The GRK2 concentration in the soluble fraction was determined by Western analysis of three separate transfections (26). Under the conditions used, endogenous GRK2 was not detected.
Purification of WT and Mutant GRK2
COS-1 Cells
GRK2 mutants with apparent defects in their ability to phosphorylate rhodopsin were purified for more detailed analysis of their catalytic activity. COS-1 cells (2.4 × 106) propagated as described above were plated on four 10 cm plates and transfected ~20 hours later with 10 µg of plasmid DNA using 25 µL FuGENE-6 reagent (Roche). After 48 hours, cells were washed twice with STE and scraped into 0.8 mL/plate of Buffer A. Lysates were sonicated in 1 sec bursts with 2 sec rest periods for 3 min and high-speed supernatant (HSS) fractions were obtained by centrifugation for 10 min at 40,000×g. HSS fractions were then diluted to ~50 mM NaCl with Buffer B (20 mM HEPES, 5 mM EDTA, 0.02% Triton X-100, 1 mM PMSF, 10 µg/mL leupeptin, 0.2 mg/mL benzamidine) and loaded at 0.5 mL/min onto tandem 1 mL High Q and High S columns (Bio-Rad) pre-equilibrated with Buffer B containing 50 mM NaCl. The High Q column was removed and the High S column was washed with 10 mL of Buffer B containing 50 mM NaCl and then eluted with a 20 mL 50–400 mM NaCl gradient at 0.5 mL/min. Fractions (0.5 mL) were analyzed for purity by Coomassie staining of 8% SDS polyacrylamide gels and a kinase assay, using rhodopsin as a substrate, was conducted as described below in a 20 µL reaction with 6 µL of High S column fractions. Fractions were subsequently pooled, diluted to ~50 mM NaCl with Buffer B, and loaded onto a 0.5 mL heparin-Sepharose CL-6B (Pharmacia Biotech/GE Healthcare) column at 0.5 mL/min. Following washing, the heparin-Sepharose column was eluted with a 20 mL 100–600 mM NaCl gradient in Buffer B. Fractions were analyzed for purity by 8% SDS-PAGE and Coomassie staining, and a 20 µL kinase reaction with 6 µL of heparin-Sepharose fractions was used to identify fractions containing GRK2. GRK2 fractions were pooled, diluted to achieve a salt concentration of ~100 mM NaCl in Buffer B and then protein was concentrated by centrifugation (Microcon or Centricon) at 5000 rpm (~2500×g) and stored at −20 °C in 25% glycerol.
Baculovirus Production and Purification of WT and Mutant GRK2 from Sf9 Insect Cells
The Bac-to-Bac Baculovirus Expression System (Invitrogen) was used to express GRK2 in Sf9 insect cells as previously described (27). A HindIII fragment containing the WT or mutant GRK2 cDNA was excised from the pcDNA3-GRK2 constructs and ligated into HindIII-cut pFastBac1 (pFB1). Pellets from ~200 mL of baculovirus-infected cells were resuspended in 50 ml of Buffer B and cell lysis was achieved using a polytron tissue disruptor. The lysate was clarified by centrifugation at 35,000×g for 20 min and a HSS fraction was generated by centrifugation at 138,000×g for 60 min in a SW28 rotor. The HSS fraction was diluted five-fold to achieve 50 mM NaCl and loaded at 2 mL/min on tandem 5 mL High Q and High S columns. After removal of the High Q column, the High S column was washed and then eluted with a 100 mL 50–400 mM NaCl gradient. Fractions containing GRK2 were pooled, diluted to 100 mM NaCl with Buffer B, and loaded on a 1 mL heparin-Sepharose CL-6B (Pharmacia Biotech/GE Healthcare) column at 1 mL/min. GRK2 was eluted with a 20 mL 100–600 mM NaCl gradient in Buffer B. GRK2-containing fractions were pooled, glycerol was added to 25%, and fractions were stored at −20 °C.
GRK2 Activity with Receptor and Non-Receptor Substrates
Rhodopsin
Rod outer segments were isolated from frozen dark-adapted bovine retinas (W. L. Lawson Co., Lincoln, NE) and washed with urea as previously described (28). WT and mutant GRK2 (~165 nM) purified from COS-1 cells were analyzed in 20 µL reactions with GRK2 Kinase Buffer (20 mM Tris-HCl, pH 7.5, 2 mM EDTA, 7.5 mM MgCl2, 200 µM ATP) containing ~0.2 dpm/fmol [γ-32P] ATP and 15 µM rhodopsin for 3 min at 30 °C in ambient room light. Reactions were quenched by the addition of 20 µL of SDS-PAGE sample buffer (Laemmli sample buffer containing 8% SDS, 30% glycerol, and 25 mM β-mercaptoethanol), incubated for 30 minutes at 60 °C, and then 20 µL was resolved by 10% SDS-PAGE. Gels were stained with Coomassie, dried, rhodopsin bands were excised, and the amount of 32P incorporation was measured by liquid scintillation counting. An empty region of the gel was used to obtain a background measurement. Values were expressed in terms of mean kinase activity ± standard error for three experiments carried out in duplicate. Statistical significance of mutant GRK2 activity was assessed by ANOVA comparison to WT GRK2. The phosphorylation of dark-adapted rhodopsin was negligible.
β2AR
WT and mutant GRK2 (60 nM) purified from COS-1 cells were analyzed in 20 µL reactions with GRK2 Kinase Buffer containing ~2.0 dpm/fmol [γ-32P] ATP and 50 nM β2AR reconstituted in phosphatidylcholine vesicles (23) for 40 min at 30 °C in the presence of either 100 µM alprenolol hydrocholoride (Sigma) or 100 µM (−)-isoproterenol (Sigma). Reactions were analyzed as described above for rhodopsin, except that autoradiography was required to identify β2AR bands. Values are expressed in terms of mean kinase activity ± standard error for three experiments carried out in duplicate.
Tubulin
Lyophilized bovine brain tubulin (Cytoskeleton, Denver, CO) was resuspended in 50 mM Tris pH 7.5. The ability of WT or mutant GRK2 to phosphorylate tubulin was assessed by incubation of ~165 nM COS-1 purified GRK2 in 20 µL reactions with GRK2 Kinase Buffer, [γ-32P] ATP (~1 dpm/fmol), and 500 nM tubulin for 30 min at 30 °C. Reactions were analyzed as described above for rhodopsin. Three experiments were performed in duplicate and mean values are expressed ± standard error.
α-Synuclein
Human α-synuclein was expressed in E. coli and isolated by boiling and source Q purification as described (29). The ability of Sf9 purified WT and mutant GRK2 (100 nM) to phosphorylate 10 µM α-synuclein was assessed by incubation for 30 min at 30 °C in a 15 µL reaction containing [γ-32P] ATP (~1 dpm/fmol) in a modified Kinase Buffer that lacks EDTA (20 mM Tris-HCl, pH 7.5, 4.0 mM MgCl2, 200 µM ATP). Synuclein was resolved on 8–16 % gradient acrylamide gels and then 32P transfer was determined as described above for rhodopsin. Values are expressed in terms of mean kinase activity ± standard error for three experiments carried out in duplicate.
RESA Peptide
The peptide R3E5SA3 (RESA) was synthesized and purified by the Protein Microanalysis Core at the University of Texas at Austin Institute for Cellular and Molecular Biology, and suspended at 30 mM in 50 mM Tris pH 7.4. WT and mutant GRK2 (~273 nM) purified from COS-1 cells were incubated in 20 µL reactions with Kinase Buffer, [γ-32P] ATP (~0.2 dpm/fmol), and 1 mM RESA for one hour at 30 °C. Reactions were quenched by the addition of 10 µL of 45% trichloroacetic acid and denatured protein was pelleted by centrifugation for 10 min at 12,000×g. Supernatants were spotted on P81 paper in duplicate, washed six times with 75 mM phosphoric acid, and the extent of 32P transfer to RESA was evaluated by liquid scintillation counting. Background counts from an unspotted piece of P81 paper were subtracted from all measurements. Values are expressed as means ± standard error for three experiments carried out in duplicate.
RASASA Peptide
The peptide R3ASA3SA2 (RASASA) was synthesized and purified by Keck Biotechnology Resource Laboratory (Yale University) and a 10 mM stock solution was prepared in 50 mM Tris, pH 7.4. Sf9-purified WT and mutant GRK2 (100 nM) were assayed for 50 min at 30 °C in 15 µL reactions containing 1 mM RASASA in GRK2 Kinase Buffer with [γ-32P] ATP at ~1 dpm/fmol. Reactions were quenched with 9 µL SDS-PAGE sample buffer, heated to 60 °C for 15 min, and 10 µL were analyzed on 20 % SDS polyacrylamide gels. The RASASA peptide was visualized by Coomassie stain and bands were excised from the destained gel to determine the amount of 32P transferred. A lane from a reaction that lacked enzyme was used to determine background. Values are expressed as means ± standard error for three experiments carried out in duplicate.
Michaelis-Menten Kinetic Analysis
The rhodopsin concentration in rod outer segments was varied between 2.5–20 µM in 15 µL reactions containing 100 nM GRK2, GRK2-Y281A, GRK2-P473E, and GRK2-I485A or 400 nM GRK2-V477D (baculovirus expressed and purified), 100 mM HEPES pH 7.5, 1 mM EDTA, 10 mM MgCl2, 1 mM ATP, and [γ-32P] ATP (~0.2 dpm/fmol). Reactions were carried out for 2 min and processed as described above. Preliminary experiments verified that rhodopsin phosphorylation was linear during this time frame and <5% of the substrate was phosphorylated. Experiments were performed three times in duplicate. To determine KM and kcat for WT and mutant GRK2, the initial rate data were fit to the Michaelis-Menten rate equation using GraphPad Prism 4.0 (GraphPad Software, Inc., San Diego, CA).
Activation of GRK2 by β2AR in Phosphatidylinositol (PI)/DDM Mixed Micelles
PI/DDM mixed micelles were prepared using minor modifications of procedure described by Onorato et al. (30). Bovine liver PI in chloroform (Avanti Polar Lipids, Alabaster, AL) was dried with a stream of nitrogen gas and resuspended at 2 mg/mL by incubation 30 °C, 30 min in 0.8 mM dodecylmaltoside (Anatrace, Maumee, OH), 20 mM Tris-HCl, pH 7.5, 2 mM EDTA, 7.5 mM MgCl2. β2AR in DDM was inserted into PI/DDM mixed micelles by incubation on ice for 1 hr yielding a stock concentration of 320 nM β2AR in 2 mg/mL PI and 0.91 mM DDM. The ability of β2AR to stimulate GRK2 activity (100–200 nM WT or 200 nM V477D) was assayed by comparing RASASA phosphorylation (as described above) in the absence of receptor or in the presence of isoproterenol (100 µM)-treated β2AR that had been diluted 4-fold from the 320 nM stock into the kinase assay. The effects of alprenolol-treated β2AR or mixed micelles lacking β2AR on RASASA phosphorylation were also assessed. Values are expressed as means ± standard error for five experiments. Two-way ANOVA with a Bonferroni’s post-test was used to assess statistical significance.
RESULTS
In an initial screen to identify receptor phosphorylation defects mediated by residues in the large lobe and C-tail of GRK2 (Fig. 2), wild type (WT) and mutant GRK2 were over-expressed in COS-1 cells, and extracts were prepared to assess the level of GRK2 in the HSS fraction. All mutants were expressed at normal levels with the exception of H394A, which was not detectable in the soluble fraction (Fig. 3A). The remaining WT and GRK2 mutant proteins from COS-1 lysates were then assayed for their ability to phosphorylate a model GPCR, the visual pigment rhodopsin. Two mutants, P473E and V477D, showed statistically significant defects in their ability to phosphorylate light-activated rhodopsin. Three other mutants, Y281A, D481A, and I485A, showed subtle but not statistically significant impairment (Fig. 3B). All five mutants were subsequently purified to homogeneity from lysates and reassessed for their ability to phosphorylate rhodopsin (Fig 4A). The V477D mutant was the most severely impaired, phosphorylating rhodopsin at only 2.3% of the WT level. Y281A, P473E, and I485A, showed lesser (30–60% of WT) but statistically significant defects (P < 0.01). In contrast, purified D481A did not exhibit any phosphorylation defect (data not shown) and was not studied further. All four mutants reside in the AST or in the αD helix of the large lobe, which contains residues that interact with the AST. Notably, mutation of residues equivalent to Y281, V477 and I485 in GRK1 were also found to be defective in receptor phosphorylation in a mutagenesis study (31).
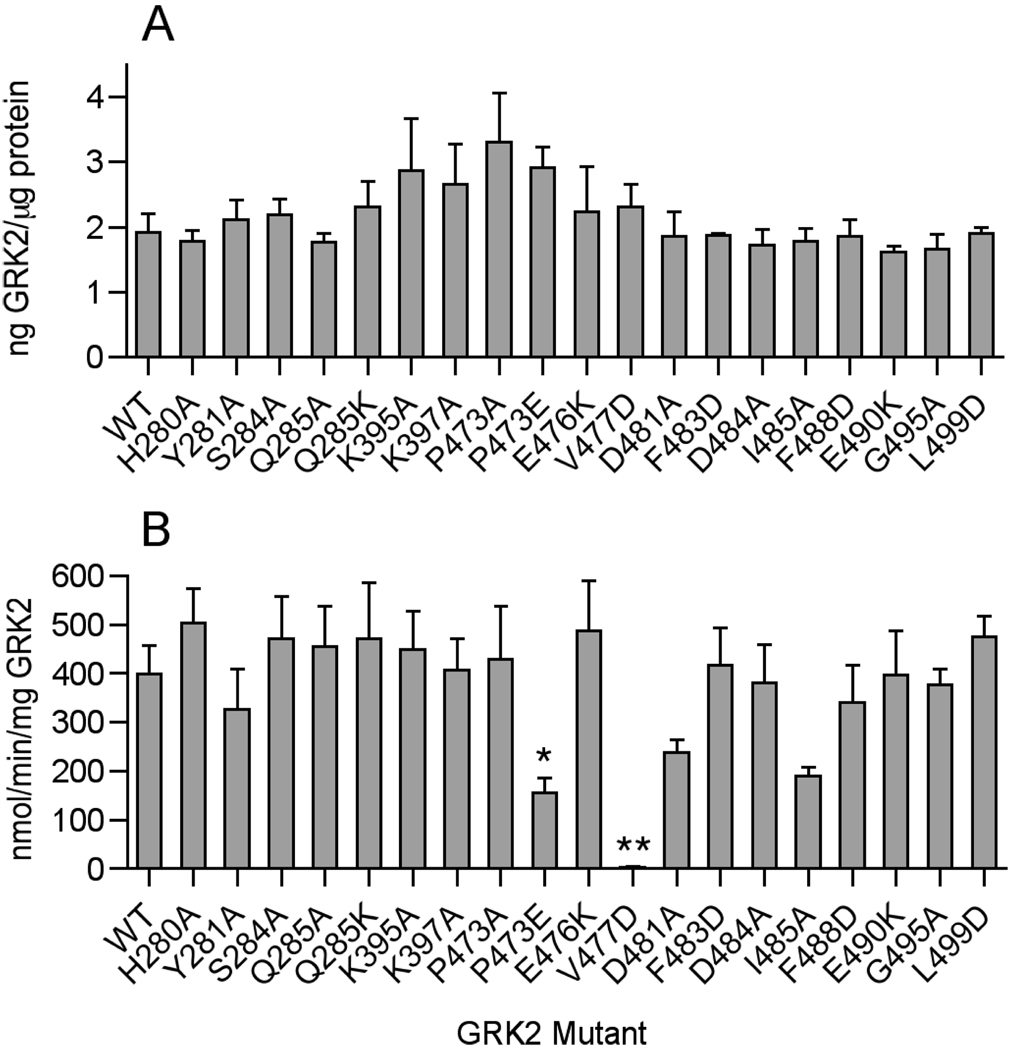
Figure 3. Expression and Phosphorylation of Light-activated Rhodopsin by GRK2 Kinase-Domain Mutants.
A. WT and GRK2 mutant cDNAs were transiently transfected into COS-1 cells, and the amount of GRK2 in the HSS fraction of the cells was quantified by immunoblotting and densitometry. B. Kinase activity of WT and mutant GRK2 (~10 nM) in cell lysates was assessed using 15 µM light-activated rhodopsin and 200 µM γ-32P-ATP (0.2 dpm/fmol) as substrates. Coomassie-stained rhodopsin bands separated by SDS-PAGE were excised and the phosphate transferred was quantified by liquid scintillation counting. Error bars indicate SEM. One-way ANOVA was used to compare statistical significance of differences relative to WT. *, p<0.05; **, p<0.01.
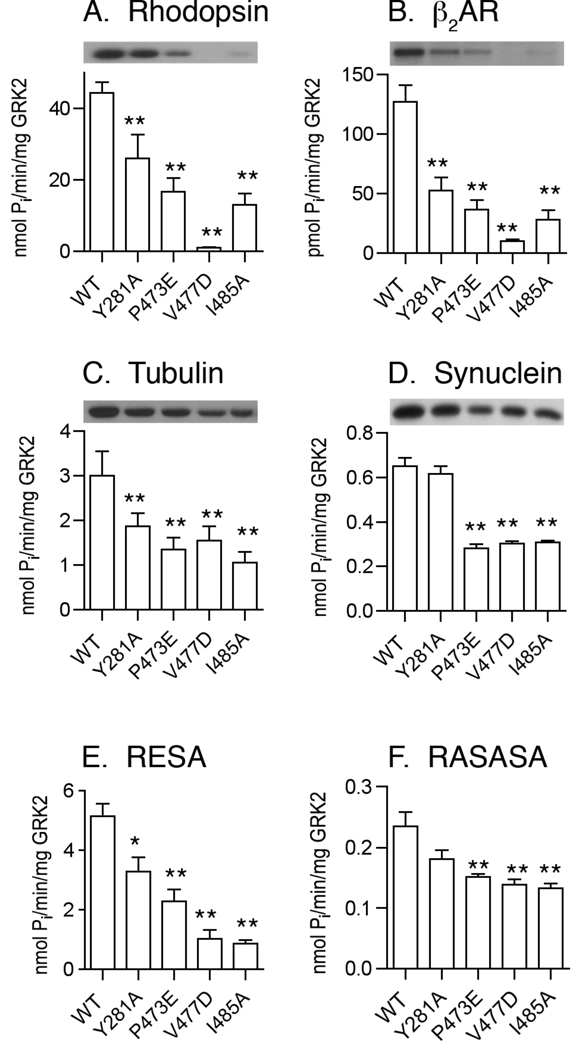
Figure 4. Phosphorylation of GPCR and Non-receptor Substrates by GRK2 Kinase-Domain Mutants.
Four mutants (Y281A, P473E, V477D, and I485A) were over-expressed in COS-1 or Sf9 insect cells and purified. Kinase assays were carried out with rhodopsin (A), β2AR in phosphatidylcholine vesicles (B), tubulin (C), α-synuclein (D), RESA peptide (E), and RASASA peptide (F) as described in Experimental Procedures. Error bars indicate SEM. One-way ANOVA was used to compare statistical significance of differences relative to WT. *, p<0.05; **, p<0.01.
The four remaining mutants were also impaired in the ability to phosphorylate another GPCR, isoproterenol-activated β2AR reconstituted into phospholipids vesicles (Fig. 4B). The rank order of the β2AR phosphorylation defect was similar to the rhodopsin phosphorylation defect with V477D being most defective (7.8% of WT), followed by I485A (22% of WT), P473E (29% of WT), and Y281A (41% of WT).
We then tested the ability of WT and mutant GRK2 enzymes to phosphorylate soluble, non-receptor substrates. Non-receptor substrates have been used to distinguish receptor-dependent effects. Early studies aiming to characterize the substrate specificity of GRK2 focused on the phosphorylation of free peptides, either representing portions of β2AR (32) or synthetic peptides (33). Peptides harboring acidic residues on the amino terminal side of the phosphoacceptor were the best substrates with Michaelis constants of ~1 mM. Some non-receptor proteins such as tubulin and α-synuclein also act as substrates for GRK2 and are more convenient to assay than peptides. For both tubulin and α-synuclein, the phosphoacceptor sites are embedded within acidic regions and the Michaelis constant for tubulin is lower than that of free peptides and rhodopsin at ~0.5 µM (34, 35). One assumption is that these non-receptor substrates bind directly to the phosphoacceptor site of the kinase domain, and can therefore serve to assess intrinsic kinase activity.
The primary difference observed between tubulin and GPCR phosphorylation was that the defect of the V477D mutant is not nearly as severe. V477D phosphorylated tubulin at 47% of WT, on par with the 32–64% defect observed with the other mutants (Fig. 4C). Similarly, the P473E, V477D and I485A mutants phosphorylated α-synuclein at ~46% of the WT level, while Y281A showed no defect in α-synuclein phosphorylation (Fig. 4D). All mutants were impaired in their ability to phosphorylate the highly acidic RESA peptide (Fig. 4E). With the exception of V477D, the inability to phosphorylate RESA paralleled the deficiency in GPCR phosphorylation.
The non-acidic peptide R3ASA3SA2 (RASASA) is a very poor substrate with a KM 4–5 fold higher than RESA and a Vmax 50-fold lower than RESA. Because it most likely interacts solely with active site residues, phosphorylation of RASASA is thought to be a measure of the basal rate of catalysis i.e. the fraction of GRK2 molecules that can spontaneously acquire the active form. The four mutants retained 50–70% of the WT ability to phosphorylate RASASA (Fig. 4F), suggesting that although the active site of GRK2 was not seriously perturbed by any of these point mutants, the mutant proteins still had difficulty in achieving a closed conformation capable of phosphorylating this peptide substrate. Overall, the bulk of our data indicated that the residues we found to be the most important for receptor phosphorylation also were hampered in their ability to phosphorylate soluble substrates.
We next attempted to distinguish catalytic from binding defects using Michaelis-Menten kinetics, with GRK2 purified from baculovirus-infected Sf9 cells as a source of the kinase and light-activated rhodopsin as a substrate (Fig. 5). The KM for WT GRK2 was ~8 µM, consistent with previously published values (36, 37). P473E and I485A showed 3–4 fold increases in their Michaelis constants (Table 1). V477D, but not the other mutants, exhibited a 12-fold decrease in turnover number. Thus, the inability of V477D to phosphorylate rhodopsin seemed primarily a consequence of a catalytic defect.
Figure 5. Michaelis-Menten Kinetics.
Kinase reactions were carried out with rhodopsin (2.5–20 µM), 100 nM WT GRK2, GRK2-Y281A, GRK2-P473E, and GRK2-I485A or 400 nM GRK2-V477D, and 1 mM [γ-32P] ATP (~0.2 dpm/fmol) for 2 min in the light. Rhodopsin was separated by electrophoresis, stained, and excised to quantitatively determine phosphate transferred. The data are derived from three experiments carried out in duplicate and error bars represent SEM. Initial rate data was fit to Michaelis-Menten rate equation and values for KM and kcat are given in Table 1.
Table 1.
GRK2 Kinetics with Rhodopsin
| GRK2 | kcat, sec−1 | KM, µM | Catalytic efficiency, kcat/KM, (M−1s−1) |
x-fold decrease, kcat/KM |
|---|---|---|---|---|
| WT | 0.20 ± 0.03 | 8.4 ± 2.7 | 23,800 | 1.0 |
| Y281A | 0.13 ± 0.03 | 8.0 ± 3.4 | 16,250 | 1.4 |
| P473E | 0.40 ± 0.10 | 34.5 ± 13 | 11,600 | 2.1 |
| V477D | 0.018 ± 0.002 | 11.1 ± 1.8 | 1620 | 14.7 |
| I485A | 0.37 ± 0.10 | 25.3 ± 10.9 | 14,600 | 1.6 |
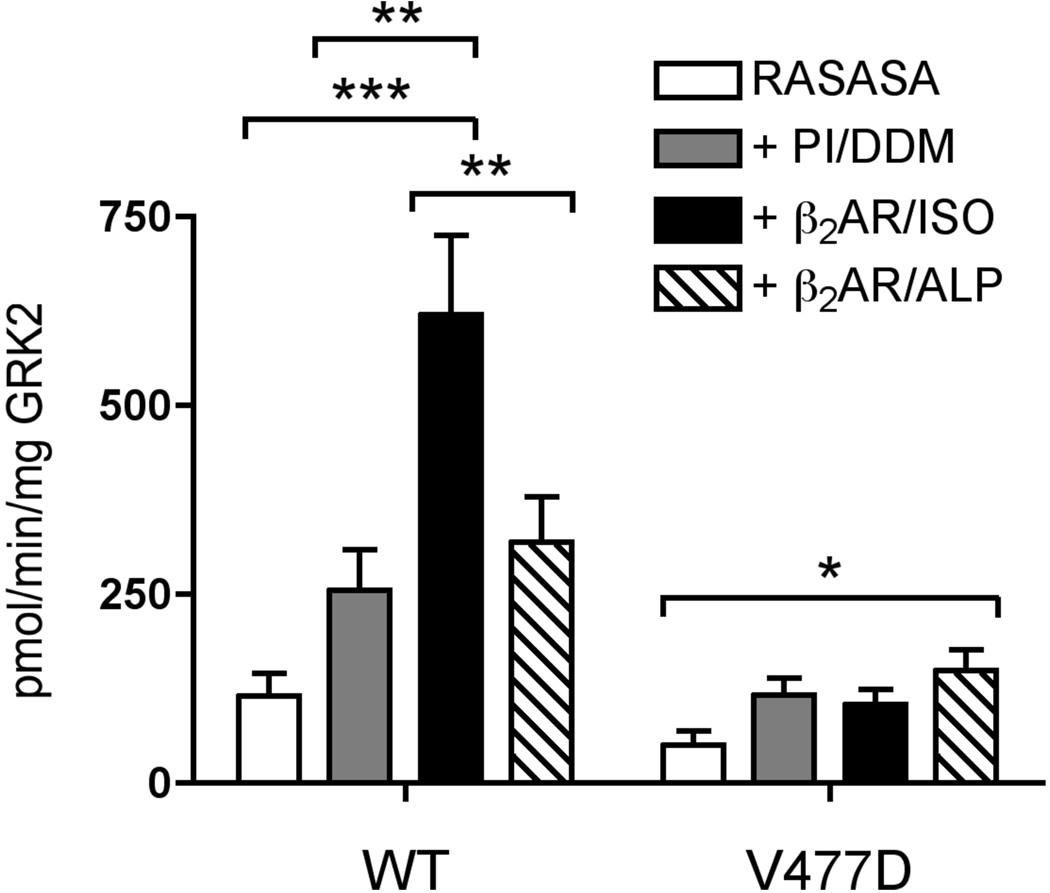
Because V477D appears to have a catalytic defect and its impairment is more severe with GPCR substrates than non-receptor substrates, we hypothesized that V477D is defective in receptor-mediated activation. While RASASA is a very poor GRK2 substrate, increased phosphorylation of this peptide can be achieved by including an activated GPCR in the kinase assay (13). Thus, phosphorylation of RASASA in the presence of ligand-treated GPCR serves as an assay of GRK activation. For our activation assays, β2AR was inserted into PI/DDM mixed micelles. Upon treatment with isoproterenol, β2AR stimulated RASASA phosphorylation by WT GRK2 6-fold (Fig. 6). However, only a ~2.5 fold stimulation can be attributed to the activated receptor, since 2–2.5 fold stimulation is observed with the addition of either PI/DDM or β2AR in PI/DDM treated with the inverse agonist, alprenolol. For comparison, Chen et al. (13) achieved a ~10-fold stimulation of RASASA phosphorylation when comparing agonist-treated and untreated β2AR. Notably when β2AR reconstituted into phospholipid vesicles was used as the activator, a much higher rate (4–7-fold) of phosphoryl transfer was achieved than in our assays where β2AR in PI/DDM mixed micelles was used. Nevertheless, in contrast with WT GRK2, isoproterenol-treated β2AR in PI/DDM did not stimulate the ability of V477D to phosphorylate RASASA, indicating that this mutant is less able to undergo receptor-stimulated allosteric activation.
Figure 6. V477D Mutant is Deficient in β2AR-mediated Activation.
Phosphorylation of 1 mM RASASA peptide by 100–200 nM GRK2 or 200 nM V477D was assessed in the absence or presence of PI/DDM mixed micelles (PI/DDM), 80 nM isoproterenol-treated β2AR in PI/DDM mixed micelles (β2AR/ISO), or 80 nM alprenolol-treated β2AR in PI/DDM mixed micelles (β2AR/ALP). Two-way ANOVA was used to test statistical significance. Error bars indicate SEM. *, p<0.05; **, p<0.01; ***, p<0.001.
DISCUSSION
Structural data suggests that the activation of a GRK requires reorientation of the small and large lobes of the kinase domain (6–8) and not other events typically associated with kinase activation. ATP binding induces only a partial closure of this domain, as well as partial ordering of the AST region of the C-tail (8). Assuming that the binding of agonist-occupied GPCRs induces full domain closure, we asked which residues of the kinase domain might be involved in this process. Our initial model of a GPCR interacting with GRK2 placed the receptor in the space between the expected membrane surface and the large lobe of the kinase domain, where phosphorylation sites in the receptor cytoplasmic loops and tail would have easy access to the kinase active site. Furthermore, by interacting with both the small and large lobes, it was envisioned that receptor binding could help pull the large lobe towards the membrane such that the kinase domain would assume an active conformation similar to that exhibited by PKA. Because activated receptors can decrease the KM for the phosphorylation of synthetic peptide substrate such as RESA (13) or peptides encompassing the phosphoacceptor sites of rhodopsin (peptide C) (12), we would expect that residues principally involved in docking to the receptor would lead to an increase in the KM for rhodopsin phosphorylation. Out of seven residues tested, the only residue in the large lobe we confirmed to be important for receptor phosphorylation was Tyr281, which is located close to the N-terminus of the αD helix (Fig. 2). The Y281A mutant only exhibited a modest reduction in kcat. Because the KM for Y281A is comparable to WT, our data suggests that this mutant harbors a deficiency in the allosteric transition that leads to full domain closure. Consistent with this hypothesis, recently reported structures of GRK1 (8) show that the residue equivalent to Tyr281 packs against the AST region, suggesting that it is primarily involved in stabilizing the closed conformation of GRK2. Although this hypothesis seems at odds with ability of Y281A mutant to phosphorylate α-synuclein at nearly WT levels, it is possible that this particular substrate can functionally substitute for the Tyr281A mutation by stabilizing the AST via a different mechanism. Indeed, it is possible that non-receptor substrates can drive kinase domain closure through unique pathways. Notably, two previously characterized mutants, D278R and D278A, are also impaired in the phosphorylation of β2ARs (38). Although initially proposed to be involved in receptor recognition, atomic structures of GRKs suggest the side chain not only caps the positive dipole at the N-terminus of αD but also bridges this helix to the ribose of bound ATP. Thus, residues in the αD helix, which immediately follows the hinge of the domain, are functionally very important in forming interactions that connect the large lobe to the active site and AST. Because no other substitutions in the large lobe altered receptor phosphorylation, our data does not support the theory that the large lobe contributes to the allosteric docking site for receptors. By analogy to AGC kinases, the large lobe is expected to contribute to the peptide channel, but studies with free peptides suggest that these interactions are of very low affinity (12, 13, 32, 33). This is common for protein kinases, which often rely on “docking sites” to achieve high affinity and selectivity for their targets (39).
A surprising result of this study is the number of conserved residues in the AST region that do not appear to play a major role in the phosphorylation of rhodopsin by GRK2. GRK2-Asp484 and GRK2-Phe488 are well conserved among all AGC kinases, whereas GRK2-Asp481 is conserved in the GRK family but not in the AGC kinase family. GRK2-Glu490 is part of a DEE490D sequence that is found in the GRK2/3 family but not the GRK1/7 or GRK4/5/6 families. GRK1 and GRK5 autophosphorylate serine and threonine residues that are similarly located (the so-called AGC kinase “turn motif”), an event that regulates receptor phosphorylation (21, 40). However, the D481A, D484A, F488D, and E490K substitutions in GRK2 did not reduce phosphorylation of rhodopsin in rod outer segments. When the autophosphorylation sites of GRK1 (Ser488 and Thr489) were converted to alanine residues, the KM for ATP was increased 20-fold while the KM for rhodopsin was relatively unperturbed (40). Thus, a potential nucleotide-binding defect in our GRK2 mutants could have went unnoticed in our assays because the ATP concentration used (200 µM) is significantly higher than the KM for ATP (60 µM) (37).
The N-terminal end of the AST region, however, does seem to play a major role in receptor phosphorylation. GRK2-Pro473 is at the N-terminus of the AST, and its conversion to glutamic acid had significant effects on receptor phosphorylation, consistent with a prior observation that this region was important for rhodopsin phosphorylation and possibly receptor binding (22). Notably, our P473A mutation did not have an effect, and thus the loss of activity appears to be due to interference caused by the glutamate side chain. Given that P473E exhibits a mild KM defect (~4-fold), it could be that the charge or length of the glutamate side chain directly interferes with receptor binding. However, our model (Fig. 2) predicts that the side-chains of Pro473 and Glu476 are juxtaposed in GRK2, and hence the P473E mutant may also introduce charge repulsion and alter the AST structure.
Ile485 is also an AST residue and the I485A mutation exhibits a phenotype similar to P473E: a small 3-fold KM defect with no decrease in kcat. The I485A mutation could therefore also be inferred to contribute to binding the loops or tails of GPCRs. However, I485A is as defective in the phosphorylation of the good non-receptor substrates, tubulin and RESA, as it is in the phosphorylation of GPCR substrates, suggesting instead that it somehow must also be involved in stabilizing the closed state of GRK2. Notably, a mutation analogous to I485A in GRK1, V484A, exhibited a 10-fold decrease in kcat and little change in KM (31).
GRK2-Val477 demonstrated the most dramatic change upon mutation by exhibiting greatly impaired receptor as well as soluble substrate phosphorylation. The equivalent residue in GRK1 points away from the small lobe and towards the membrane plane (Fig. 1A). If the AST region modeled in the partially closed structure of GRK1 is truly indicative of its position in the activated state of GRKs, then this residue would be in an ideal position to interact directly with activated receptors. However, this mutant primarily exhibited a kcat defect, and not a KM defect, suggesting that it is more important for stabilizing the active state of the kinase domain. Two aspects of our results are consistent with this conclusion. First, in the receptor activation assay (Fig. 6), the ability of WT GRK2 to phosphorylate the RASASA peptide was stimulated 2.5 fold by PI/DDM mixed micelles and 2.5 fold further by isoproterenol-treated β2AR. In contrast, V477D failed to undergo allosteric activation by agonist-treated receptor. Second, V477D demonstrates a defect in both receptor and non-receptor substrate phosphorylation. Because Val477 is positioned between Glu476 and Asp481, the V477D substitution could be imagined to disrupt structure through electrostatic repulsion. However, the analogous GRK1-V476A mutation reduced kcat for phosphorylation of both rhodopsin and its free carboxyl tail peptide, peptide C, by 12-fold while increasing KM 2-fold (31). Because the GRK1-V476A and GRK2-V477D show similar defects in catalysis, the loss of activity seems to be primarily due to the loss of the valine side chain.
In summary, we have identified three positions in GRK2: Tyr281, Pro473 and Val477 that are confined to a small region near the hinge of the kinase domain and likely participate in the mechanism of kinase domain closure and activation. A similar conclusion was reached regarding residues equivalent to Tyr281 and Val477 in GRK1 (31). Our data suggest that Ile485, which is relatively distant from the hinge, also is involved in formation of the closed state of the kinase domain. How receptor binding induces this allosteric transition is still unclear. Further characterization of these and other mutants will be necessary to dissect the mechanism of GRK2 activation.
ACKNOWLEDGMENTS
We thank Dr. Jeffrey Benovic (Thomas Jefferson University) for assistance at the early stages of this project and for providing pcDNA-GRK2, Dr. Robert Lefkowitz (Duke University) for providing β2AR, and Dr. Michel Goedert (University of Cambridge, UK) for the α-synuclein plasmid. We thank Dr. Chih-chin Huang for assistance and the communication of unpublished results, Dr. Valerie Tesmer for reagents and advice, and Siena undergraduates Nathalie Peiris, Leo Vovna, Fatima Mukadam Noorani, Kathryn Neubauer, Tim Clarke, Leslie Freeman and Becky Bussert for their contributions. Finally we are grateful to Drs. Kevin Kittredge and Jason Hofstein, and the Siena’s Department of Chemistry and Biochemistry for the use of argon, nitrogen, and specialized glassware.
The abbreviations are
- GPCR
G protein-coupled receptor
- GRK
GPCR kinase
- AGC kinases
protein kinase A/protein kinase G/protein kinase C family of protein kinases
- C-tail
carboxyl-terminal extension of the AGC kinase domain
- AST
active site tether
- RGS
regulator of G protein signaling
- RH
RGS homology
- PI
phosphatidylinositol
- DDM
dodecylmaltoside
- β2AR
β2-adrenergic receptor
- WT
wild-type
- Gα
heterotrimeric G protein α subunit
- Gβγ
heterotrimeric G protein β and γ subunits
- HSS
high-speed supernatant
Footnotes
This work was funded by the National Science Foundation grants MCB0315888 and MCB0744739 (to RSM), National Institutes of Health grants HL086865 and HL071818 and American Heart Association Scientist Development Grant 0235273N (to JJGT), and a summer undergraduate research fellowship from the Northeast Affiliate of the American Heart Association (to EDB).
REFERENCES
- 1.Neves SR, Iyengar R. Modeling of signaling networks. Bioessays. 2002;24:1110–1117. doi: 10.1002/bies.1154. [DOI] [PubMed] [Google Scholar]
- 2.Pitcher JA, Freedman NJ, Lefkowitz RJ. G protein-coupled receptor kinases. Annu Rev Biochem. 1998;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson SS. Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol Rev. 2001;53:1–24. [PubMed] [Google Scholar]
- 4.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. Faseb J. 1995;9:576–596. [PubMed] [Google Scholar]
- 5.Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci U S A. 2007;104:1272–1277. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gbetagamma. Science. 2003;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- 7.Lodowski DT, Tesmer VM, Benovic JL, Tesmer JJ. The structure of G protein-coupled receptor kinase (GRK)-6 defines a second lineage of GRKs. J Biol Chem. 2006;281:16785–16793. doi: 10.1074/jbc.M601327200. [DOI] [PubMed] [Google Scholar]
- 8.Singh P, Wang B, Maeda T, Palczewski K, Tesmer JJ. Structures of rhodopsin kinase in different ligand states reveal key elements involved in G protein-coupled receptor kinase activation. J Biol Chem. 2008;283:14053–14062. doi: 10.1074/jbc.M708974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 10.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowles C, Sharma R, Akhtar M. Mechanistic studies on phosphorylation of photoexcited rhodopsin. FEBS Lett. 1988;238:56–60. doi: 10.1016/0014-5793(88)80224-2. [DOI] [PubMed] [Google Scholar]
- 12.Palczewski K, Buczylko J, Kaplan MW, Polans AS, Crabb JW. Mechanism of rhodopsin kinase activation. J Biol Chem. 1991;266:12949–12955. [PubMed] [Google Scholar]
- 13.Chen CY, Dion SB, Kim CM, Benovic JL. Beta-adrenergic receptor kinase. Agonist-dependent receptor binding promotes kinase activation. J Biol Chem. 1993;268:7825–7831. [PubMed] [Google Scholar]
- 14.McCarthy NE, Akhtar M. Activation of rhodopsin kinase. Biochem J. 2002;363:359–364. doi: 10.1042/0264-6021:3630359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palczewski K, Buczylko J, Lebioda L, Crabb JW, Polans AS. Identification of the N-terminal region in rhodopsin kinase involved in its interaction with rhodopsin. J Biol Chem. 1993;268:6004–6013. [PubMed] [Google Scholar]
- 16.Noble B, Kallal LA, Pausch MH, Benovic JL. Development of a yeast bioassay to characterize G protein-coupled receptor kinases. Identification of an NH2-terminal region essential for receptor phosphorylation. J Biol Chem. 2003;278:47466–47476. doi: 10.1074/jbc.M308257200. [DOI] [PubMed] [Google Scholar]
- 17.Dhami GK, Dale LB, Anborgh PH, O'Connor-Halligan KE, Sterne-Marr R, Ferguson SS. G Protein-coupled receptor kinase 2 regulator of G protein signaling homology domain binds to both metabotropic glutamate receptor 1a and Galphaq to attenuate signaling. J Biol Chem. 2004;279:16614–16620. doi: 10.1074/jbc.M314090200. [DOI] [PubMed] [Google Scholar]
- 18.Narayana N, Cox S, Nguyen-huu X, Ten Eyck LF, Taylor SS. A binary complex of the catalytic subunit of cAMP-dependent protein kinase and adenosine further defines conformational flexibility. Structure. 1997;5:921–935. doi: 10.1016/s0969-2126(97)00246-3. [DOI] [PubMed] [Google Scholar]
- 19.Smith CM, Radzio-Andzelm E, Madhusudan, Akamine P, Taylor SS. The catalytic subunit of cAMP-dependent protein kinase: prototype for an extended network of communication. Prog Biophys Mol Biol. 1999;71:313–341. doi: 10.1016/s0079-6107(98)00059-5. [DOI] [PubMed] [Google Scholar]
- 20.Gan XQ, Wang JY, Yang QH, Li Z, Liu F, Pei G, Li L. Interaction between the conserved region in the C-terminal domain of GRK2 and rhodopsin is necessary for GRK2 to catalyze receptor phosphorylation. J Biol Chem. 2000;275:8469–8474. doi: 10.1074/jbc.275.12.8469. [DOI] [PubMed] [Google Scholar]
- 21.Kunapuli P, Gurevich VV, Benovic JL. Phospholipid-stimulated autophosphorylation activates the G protein-coupled receptor kinase GRK5. J Biol Chem. 1994;269:10209–10212. [PubMed] [Google Scholar]
- 22.Gan X, Ma Z, Deng N, Wang J, Ding J, Li L. Involvement of the C-terminal proline-rich motif of G protein-coupled receptor kinases in recognition of activated rhodopsin. J Biol Chem. 2004;279:49741–49746. doi: 10.1074/jbc.M407570200. [DOI] [PubMed] [Google Scholar]
- 23.Cerione RA, Strulovici B, Benovic JL, Strader CD, Caron MG, Lefkowitz RJ. Reconstitution of beta-adrenergic receptors in lipid vesicles: affinity chromatography-purified receptors confer catecholamine responsiveness on a heterologous adenylate cyclase system. Proc Natl Acad Sci U S A. 1983;80:4899–4903. doi: 10.1073/pnas.80.16.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobilka BK. Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal Biochem. 1995;231:269–271. doi: 10.1006/abio.1995.1533. [DOI] [PubMed] [Google Scholar]
- 25.Sterne-Marr R, Dhami GK, Tesmer JJ, Ferguson SS. Characterization of GRK2 RH domain-dependent regulation of GPCR coupling to heterotrimeric G proteins. Methods Enzymol. 2004;390:310–336. doi: 10.1016/S0076-6879(04)90020-1. [DOI] [PubMed] [Google Scholar]
- 26.Lodowski DT, Barnhill JF, Pyskadlo RM, Ghirlando R, Sterne-Marr R, Tesmer JJ. The role of G beta gamma and domain interfaces in the activation of G protein-coupled receptor kinase 2. Biochemistry. 2005;44:6958–6970. doi: 10.1021/bi050119q. [DOI] [PubMed] [Google Scholar]
- 27.Lodowski DT, Barnhill JF, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. Purification, crystallization and preliminary X-ray diffraction studies of a complex between G protein-coupled receptor kinase 2 and Gbeta1gamma2. Acta Crystallogr D Biol Crystallogr. 2003;59:936–939. doi: 10.1107/s0907444903002622. [DOI] [PubMed] [Google Scholar]
- 28.Shichi H, Somers RL. Light-dependent phosphorylation of rhodopsin. Purification and properties of rhodopsin kinase. J Biol Chem. 1978;253:7040–7046. [PubMed] [Google Scholar]
- 29.Der-Sarkissian A, Jao CC, Chen J, Langen R. Structural organization of alpha-synuclein fibrils studied by site-directed spin labeling. J Biol Chem. 2003;278:37530–37535. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- 30.Onorato JJ, Gillis ME, Liu Y, Benovic JL, Ruoho AE. The beta-adrenergic receptor kinase (GRK2) is regulated by phospholipids. J Biol Chem. 1995;270:21346–21353. doi: 10.1074/jbc.270.36.21346. [DOI] [PubMed] [Google Scholar]
- 31.Huang C-c, Yoshino-Koh K, Tesmer JJG. A surface of the kinase domain critical for the allosteric activation of G protein-coupled receptor kinases. doi: 10.1074/jbc.M809544200. (in revision) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benovic JL, Onorato J, Lohse MJ, Dohlman HG, Staniszewski C, Caron MG, Lefkowitz RJ. Synthetic peptides of the hamster beta 2-adrenoceptor as substrates and inhibitors of the beta-adrenoceptor kinase. Br J Clin Pharmacol. 1990;30 Suppl 1:3S–12S. doi: 10.1111/j.1365-2125.1990.tb05462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onorato JJ, Palczewski K, Regan JW, Caron MG, Lefkowitz RJ, Benovic JL. Role of acidic amino acids in peptide substrates of the beta-adrenergic receptor kinase and rhodopsin kinase. Biochemistry. 1991;30:5118–5125. doi: 10.1021/bi00235a002. [DOI] [PubMed] [Google Scholar]
- 34.Carman CV, Som T, Kim CM, Benovic JL. Binding and phosphorylation of tubulin by G protein-coupled receptor kinases. J Biol Chem. 1998;273:20308–20316. doi: 10.1074/jbc.273.32.20308. [DOI] [PubMed] [Google Scholar]
- 35.Pronin AN, Morris AJ, Surguchov A, Benovic JL. Synucleins are a novel class of substrates for G protein-coupled receptor kinases. J Biol Chem. 2000;275:26515–26522. doi: 10.1074/jbc.M003542200. [DOI] [PubMed] [Google Scholar]
- 36.Benovic JL, Mayor F, Jr, Staniszewski C, Lefkowitz RJ, Caron MG. Purification and characterization of the beta-adrenergic receptor kinase. J Biol Chem. 1987;262:9026–9032. [PubMed] [Google Scholar]
- 37.Kim CM, Dion SB, Onorato JJ, Benovic JL. Expression and characterization of two beta-adrenergic receptor kinase isoforms using the baculovirus expression system. Receptor. 1993;3:39–55. [PubMed] [Google Scholar]
- 38.Iino M, Furugohri T, Fukuzawa A, Shibano T. Asp278 of human beta-adrenergic receptor kinase 1 is essential for phosphorylation activity. Biochem Biophys Res Commun. 1997;239:548–551. doi: 10.1006/bbrc.1997.7504. [DOI] [PubMed] [Google Scholar]
- 39.Biondi RM, Nebreda AR. Signalling specificity of Ser/Thr protein kinases through docking-site-mediated interactions. Biochem J. 2003;372:1–13. doi: 10.1042/BJ20021641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palczewski K, Ohguro H, Premont RT, Inglese J. Rhodopsin kinase autophosphorylation. Characterization of site-specific mutations. J Biol Chem. 1995;270:15294–15298. doi: 10.1074/jbc.270.25.15294. [DOI] [PubMed] [Google Scholar]