SUMMARY
Given its catalytic activity to generate diacylglycerol and inositol 1,4,5-trisphosphate (IP3), phospholipase C (PLC) is implicated in promoting cell growth. However, we found that PLC-β3-deficient mice develop myeloproliferative disease (MPD), lymphoma, and other tumors. The mutant mice have increased numbers of hematopoietic stem cells (HSC) with increased proliferative, survival, and myeloid-differentiative abilities. These properties are dependent on Stat5 and can be antagonized by the protein phosphatase SHP-1. Stat5-dependent cooperative transformation by active c-Myc and PLC-β3 deficiency was suggested in mouse lymphomas in PLC-β3−/− and in Eμ-myc;PLC-β3+/− mice and human Burkitt's lymphoma cells. The same mechanism for malignant transformation seems to be operative in other human lymphoid and myeloid malignancies. Thus, PLC-β3 is likely a tumor suppressor.
INTRODUCTION
The production and lineage commitment of hematopoietic cells is controlled by the actions of a multitude of cytokines, growth factors, and hormones (Kondo et al., 2003). Cell surface receptors bound by these ligands activate several signaling pathways including the Jak-Stat pathway. This pathway plays a crucial role in a number of biological functions by activating transcription of various target genes (Levy and Darnell, 2002; O'Shea et al., 2002; Schindler et al., 2007). Cytokine stimulation activates Jak kinases through transphosphorylation and results in tyrosine phosphorylation of receptor sites, Stats, and other substrates. Following tyrosine phosphorylation, Stats homo- or hetero-dimerize, translocate to the nucleus, and activate gene expression through sequence-specific response elements.
To date, seven mammalian Stat family members have been identified: Stats 1, 2, 3, 4, 5A, 5B, and 6; Stat5 is encoded by two recently duplicated genes, Stat5A and Stat5B, with ∼96% sequence identity (Mui et al., 1995). Stat5 plays a crucial role in early hematopoiesis - Stat5A and Stat5B doubly disrupted mice displayed a profound defect in competitive repopulation of hematopoiesis (Bunting et al., 2002; Snow et al., 2002). The commitment of embryonic stem cells to hematopoietic cells is augmented by a Stat5-mediated signal (Kyba et al., 2003). Stat5 also plays a role in myeloid cell proliferation and differentiation (Ilaria et al., 1999). Constitutive activation of Stat5 in human cord blood CD34+ cells enhances their capacity to repopulate NOD/SCID mice and promotes erythroid differentiation (Schuringa et al., 2004). Constitutively active Stat5 promotes self-renewal, proliferation, and survival of mouse HSC and induces a lethal MPD in mice (Kato et al., 2005).
Stat signals are down-regulated by three well-characterized mechanisms: dephosphorylation, nuclear export, and suppressor of cytokine signaling (SOCS) feedback inhibition (Shuai, 2000; Tanaka et al., 2005). SOCS proteins bind either activated Jak proteins or cytokine receptors to inhibit Jak activity (Alexander and Hilton, 2004). Several protein tyrosine phosphatases have been reported to dephosphorylate either Jak and/or Stat proteins. For example, Jak2 interacts with SHP-1 and may be dephosphorylated by SHP-1 (Jiao et al., 1996; Klingmuller et al., 1995).
PLC-β is a small family of enzymes that can produce diacylglycerol and IP3 downstream of heterotrimeric G proteins (Rhee, 2001). As diacylglycerol can activate protein kinase C (PKC), and IP3 can mobilize Ca2+, PLC-β is implicated in promoting cell proliferation. PLC-β directly interacts with GTP-bound Gα subunits, leading to its catalytic activation. The four isoforms of PLC-β (β1-β4) show different tissue expression specificity and heterotrimeric G protein regulation profiles. PLC-β1 and PLC-β3 are expressed in a wide range of tissues and cell types, while PLC-β2 and PLC-β4 are expressed only in hematopoietic and neuronal tissues, respectively. PLC-β2 and PLC-β3 can also be activated by βγ subunits of the Gαi/o family of G proteins (Camps et al., 1992; Katz et al., 1992; Lee et al., 1993). Consistent with their roles in G protein-coupled receptor signaling, chemokine-induced IP3 production, Ca2+ signaling, and migration are reduced in PLC-β2−/− and PLC-β2−/−;PLC-β3−/− neutrophils (Li et al., 2000) and T cells (Bach et al., 2007). However, little is known about the role of these PLC-β isoforms in hematopoiesis or tumorigenesis. Here we have studied the role of PLC-β3 in these processes using PLC-β3−/− mice.
RESULTS
PLC-β3-deficient mice develop various tumors including MPD and lymphoma
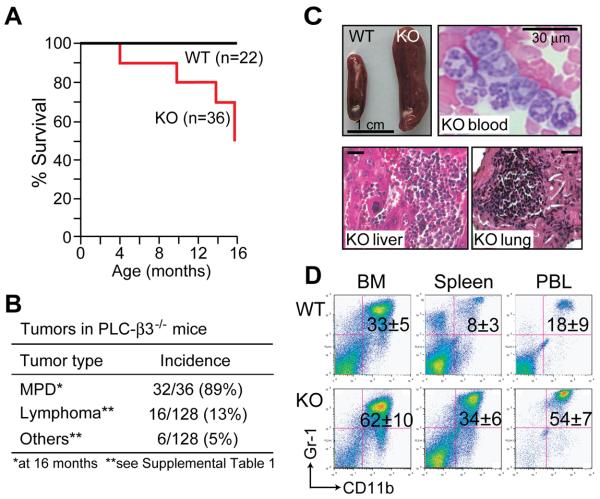
PLC-β3 deficiency led to a premature death in mice (Figure 1A). Fifty percent (18 of 36 mice) of PLC-β3−/− mice died within an observation period of 16 months, in contrast with 100% survival of wild-type (WT) mice. By the age of 16 months, most PLC-β3−/− mice in this cohort exhibited splenomegaly (Figure 1B-C), the incidence of which reached 89% when prematurely dead mice with this abnormality were included. The enlarged spleens had effaced architecture characterized by markedly increased myeloid cells and some erythroid cells, indicative of extramedullary hematopoiesis (data not shown). Livers and lungs also had foci composed of myeloid cells (Figure 1C). Dramatic increases in CD11b+Gr-1+ mature granulocytes in bone marrow (BM), spleen, and peripheral blood from these mice were observed (Figure 1C-D, Tables S1 & S2). Microbiological examinations showed no indications of bacterial infection in the diseased mice, and antibiotic treatments did not affect the number of granulocytes (data not shown). Therefore, these hematologic findings were consistent with the diagnosis of MPD (Kogan et al., 2002), unlike myelodysplastic syndrome that is frequently associated with anemia.
Figure 1. PLC-β3−/− mice develop MPD, lymphoma, and other tumors.
(A) Survival analysis. (B) Summary of tumors developed in PLC-β3−/− mice. (C) Hematologic analysis of 10-month-old PLC-β3−/− mice. Splenomegaly in PLC-β3−/− mice (top left) was associated with effaced splenic architecture (data not shown). Increased mature granulocytes in PLC-β3−/− mice were shown by blood smear (top right) and hematoxylin and eosin staining of lung and liver sections (bottom). Bars in tissue sections indicate 30 μm. (D) Flow cytometric analysis of nucleated cells in BM, spleen, and peripheral blood leukocytes (PBL) from 10 month-old mice (n=16). Granulocytes (CD11b+/Gr-1+; percentages ± SD shown) were increased in these organs of aged PLC-β3−/− mice.
During a two-year observation period, 3 in another cohort of 16 PLC-β3−/− mice with increased granulocytes developed anemia (hematocrits of 11%, 16%, and 22%) with increased numbers of blast cells in their BM (32%, 35%, and 45%, respectively). This result suggests that the MPD can evolve to accelerated and blast-crisis stages, similar to human chronic myelogenous leukemia (CML) (Sawyers, 1999). Gross and histologic examinations of a larger number (128 in total) of PLC-β3−/− mice revealed various tumors including lymphoma mostly with T cell markers and carcinomas of skin and lung (Figures 1A-B and S1 and Table S3). Unlike PLC-β3−/− mice, PLC-β2−/− mice did not develop tumors or die prematurely (data not shown).
PLC-β3-deficient mice exhibit increased numbers of HSC and myeloid progenitors as well as preferential granulocytic differentiation
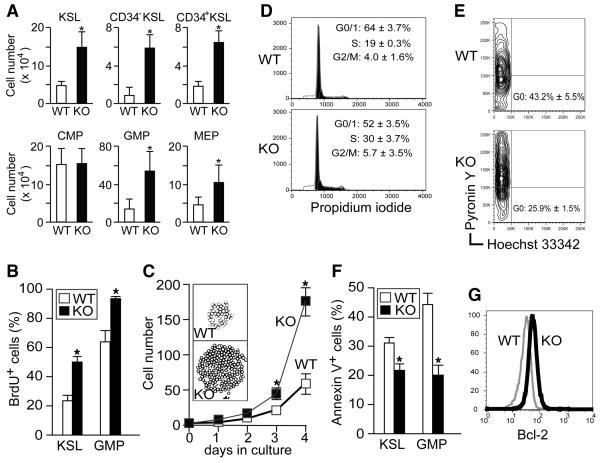
Aged PLC-β3−/− mice with splenomegaly had increased numbers of c-Kit+Sca-1+Lineage− cells (KSL cells; enriched for HSC), granulocyte-macrophage progenitors (GMP), and megakaryocyte-erythroid progenitors (MEP) in both BM (Figures 2A & S2) and spleen (data not shown), compared to age-matched WT mice. With regards to HSC subpopulations, PLC-β3−/− mice had 5-fold more CD34− KSL (enriched for long-term repopulating HSC) and 3-fold more CD34+ KSL (enriched for short-term repopulating HSC) cells (Figure 2A). Consistent with the immunophenotypic data, PLC-β3−/− BM cells and splenocytes gave rise to greater numbers of myeloid colonies than WT cells in methylcellulose medium (Figure S3A and data not shown). Purified PLC-β3−/− KSL and myeloid progenitors (CMP and GMP) generated several-fold more granulocyte (CFU-G) colonies of larger sizes than WT cells (Figure S3B-D), suggesting that PLC-β3−/− HSC and myeloid progenitors have an increased predisposition to differentiate into granulocytes, which is consistent with the MPD phenotype in PLC-β3−/− mice. Moreover, PLC-β3−/− BM and KSL cells were hypersensitive to cytokines (Figure S3E), a hallmark of human MPDs (Emanuel et al., 1991), and formed macrophage (CFU-M) and granulocyte-macrophage (CFU-GM) colonies in the absence of growth factors (Figure S3F), a feature characteristic of transformed cells.
Figure 2. Increases in KSL cells and GMP in PLC-β3−/− mice are due to increased proliferation and decreased apoptosis.
(A) BM cells from 10 month-old mice (n=16) were subjected to flow cytometric analysis of HSC and myeloid progenitors (see Figure S2 as well). Results shown are representative of at least four measurements. (B) In vivo BrdU incorporation into KSL and GMP cells in 10 month-old mice (n=4). (C) CD34− KSL cells from 10 month-old mice (n=4) were sorted into 96-well plates containing IL-3, SCF, IL-11, Flt3L, and TPO, and cultured for 4 days. The inset shows representative colonies generated at the bottom of well on day 4. Representative of 4 independent experiments. (D,E) Cell cycle analysis of propidium iodide-stained Lin− cells (D) and Hoechst 33342 and Pyronin Y-stained (E) KSL cells from 10 month-old mice (n=4). (F) Flow cytometric analysis of annexin V+ apoptotic cells in KSL and GMP cells from 10 month-old mice (n=4). Results in A-F represent mean ± SD. (G) Flow cytometric analysis for Bcl-2 expression in Lin− cells from 10 month-old mice (n=2). *, p<0.05 versus WT cells by Student's t test.
HSC-enriched populations derived from PLC-β3−/− mice exhibit increased proliferation and survival
Given the above colony-formation data, blockade of differentiation was ruled out as a contributing factor to the increase in HSC-enriched populations in PLC-β3−/−mice. Thus the increase in HSC-enriched populations could be accounted for by increased proliferation, reduced cell death, altered migration, or a combination of these factors. To dissect this point, we first performed in vivo BrdU incorporation experiments in 10-month old mice. BrdU incorporation into KSL cells was greater in PLC-β3−/− mice (Figures 2B and S4A), suggesting increased proliferation in PLC-β3−/− HSC-enriched populations. Consistent with this result, PLC-β3−/− CD34− KSL cells grew faster and formed larger colonies than WT cells in the presence of IL-3 or a cytokine cocktail (Figure 2C and data not shown). Cell cycle analysis showed increased proportions of PLC-β3−/− lineage− cells in the S phase and reduced proportions of PLC-β3−/− KSL cells in the G0 phase (Figure 2D-E). Consistent with the increased proliferation, mRNA levels of the cell cycle inhibitor p19 were reduced, but those of cyclins A2 and B2 increased, in PLC-β3−/− KSL cells (Figure S5). We then measured apoptotic cell death in KSL cells by annexin V staining. Apoptosis was less abundant in PLC-β3−/− KSL cells (Figure 2F). In line with this, expression of the anti-apoptotic protein Bcl-2 was increased in PLC-β3−/− KSL cells (Figure 2G). Finally, homing capacity of PLC-β3−/− KSL cells was not altered (Figure S6). Therefore, we conclude that the increase in HSC-enriched populations in PLC-β3−/− mice is due mainly to increased proliferation and decreased apoptosis. The same mechanisms seem operative in PLC-β3−/−GMP (Figures 2B, 2F, S4B, & S6B).
The MPD is transplantable with HSC-enriched populations derived from PLC-β3−/− mice
The MPD in PLC-β3−/− mice was BM cell-autonomous, as the irradiated Ly5.1 mice that had received PLC-β3−/− BM cells developed MPD within 6-9 months (Table S4). The increased proliferation and survival of PLC-β3−/− KSL cells suggested that HSC contain leukemic stem cells (Reya et al., 2001) that cause MPD in PLC-β3−/− mice. To test this hypothesis, purified CD34− KSL, CD34+ KSL, CMP, and GMP from PLC-β3−/− and WT mice were transferred into sublethally irradiated Rag2−/−-Ly5.1 mice. Only PLC-β3−/− CD34− KSL cells, but not other cell populations, gave rise to myeloid hyperplasia in recipient mice within only 2 months (Table S4). These results suggest that the leukemic stem cells responsible for the development of MPD in PLC-β3−/− mice are present in CD34− KSL cells.
Increased Stat5 activity is critical for increased proliferative, myeloid-differentiative, and MPD-causing capabilities of PLC-β3−/− KSL cells
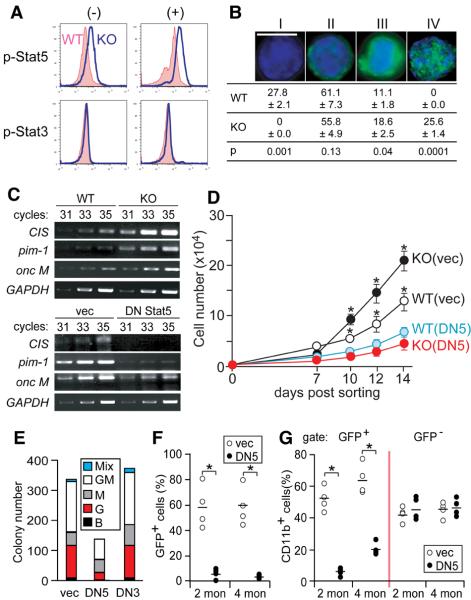
We next analyzed the signaling pathways accountable for the increased proliferation, survival, and myeloid differentiation of PLC-β3−/− KSL cells. Unlike BCR-ABL-induced CML (Sawyers et al., 1995; Skorski et al., 1997), activities of Ras, ERK, and Akt were comparable in WT and PLC-β3−/− KSL and embryonic fibroblasts (MEF) (Figure S7A and data not shown). Importantly, however, Stat5 Tyr-694 phosphorylation was constitutively increased and further induced after stimulation with IL-3 or a cytokine cocktail in PLC-β3−/− KSL cells (Figures 3A & S7B), and, compared to WT cells, more frequent and extensive nuclear localization of phospho-Stat5 was observed in PLC-β3−/− KSL cells (Figure 3B). By contrast, Stat3 phosphorylation was comparable in PLC-β3−/− and WT cells (Figures 3A & S7B). Lymphomas and a skin carcinoma from PLC-β3−/− mice also exhibited increased phosphorylation of Stat5, but not Stat3 (Figures 6C and S7C and data not shown). Consistent with Stat5 activation, mRNA expression of Stat5 target genes, e.g., CIS (Matsumoto et al., 1997), pim-1 (Lilly and Kraft, 1997; Nosaka et al., 1999), and oncostatin M (Yoshimura et al., 1996), was increased in PLC-β3−/− cells (Figure 3C).
Figure 3. Stat5 activation is required for the increased proliferation and myeloid differentiation of PLC-β3−/− KSL cells.
(A,B) Phospho-Stat5 levels were constitutively (−) higher in PLC-β3−/− KSL cells and further increased upon stimulation for 5 min with a cytokine cocktail of IL-3, SCF, IL-11, Flt3L, and TPO (+), as shown by flow cytometric analysis (A) and confocal microscopy (B) with anti-p-Stat5 (Tyr694). Nuclei were stained with DAPI (blue). A bar indicates 10 μm. The patterns of phospho-Stat5 staining in confocal microscopy were categorized into four (I-IV) and their distributions in cytokines-stimulated KSL cells (200 cells counted) from 8-10 month-old mice (n=8) are shown. Representative of 4 (A) and 2 (B) independent experiments. (C) RT-PCR analysis of mRNA expression of Stat5 target genes, CIS, pim-1, and oncostain M in KSL cells from 10 month-old mice (n=4). Numbers of PCR cycles performed are indicated. GAPDH mRNA is a house-keeping gene control. (D) CD34− KSL cells were transduced with a bicistronic retroviral vector encoding DN Stat5 (DN5) or DN Stat3 (DN3). GFP+ transduced cells were cultured in the presence of IL-3, SCF, IL-11, Flt3L, and TPO. Representative of 3 independent experiments. Results in B and D represent mean ± SD. (E) Transduced cells were cultured in methylcellulose medium containing SCF, IL-3, IL-6, and EPO. B, G, M, GM, and Mix represent BFU-E, CFU-G, CFU-M, CFU-GM, and CFU-Mix. (F,G) PLC-β3−/− CD34− KSL cells transduced with DN Stat5 or empty vector were adoptively transferred to lethally irradiated C57BL/6-Ly5.1 mice. Two and 4 months later, peripheral blood was analyzed by flow cytometry for enumeration of GFP+ cells (F) and donor-derived CD11b+ cells (G). See Table S5 as well. *, p<0.05 by Student's t test.
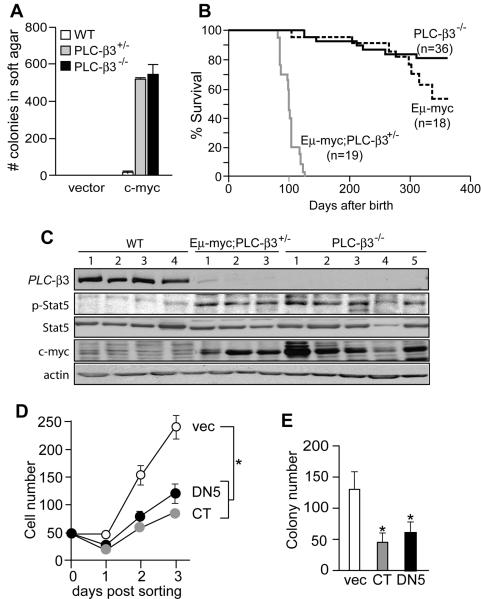
Figure 6. PLC-β3 deficiency cooperates with c-Myc to transform mouse fibroblasts and lymphocytes.
(A) MEFs from WT, PLC-β3+/−, and PLC-β3−/− mice (n=8 each) were transfected with c-myc and subjected to semisolid colony forming assays. Similar results were observed in another experiment. (B) Survival curves for the indicated mice. (C) Immunoblot analysis of PLC-β3−/− (5 independent tumors) and Eμ-myc;PLC-β3+/− (3 tumors) lymphomas and normal lymphoid tissues. WT samples 1 and 2-4 represent thymus and spleens, respectively, from WT mice. (D,E) Eμ-myc;PLC-β3+/− lymphoma cells were cultured and retrovirally transduced with the indicated genes. Their in vitro growth (D) and colony numbers in semisolid medium (E) were quantified after sorting GFP+ cells. Results represent 2 independent experiments. Results in A, D, and E represent mean ± SD.
To directly test the functional relevance of increased Stat5 activation, we introduced dominant-negative (DN) Stat5 into KSL cells, using a bicistronic retrovirus encoding DN Stat5 and green fluorescent protein (GFP). DN Stat5 induced a reduction in mRNA expression of CIS, pim-1, and oncostatin M (Figure 3C) as expected. DN Stat5 suppressed in vitro expansion of GFP-expressing PLC-β3−/− KSL cells (designated as KO/DNStat5 cells) (Figure 3D), suggesting that the increased proliferation/survival depends on the increased Stat5 activity in PLC-β3−/− HSC-enriched cells. Colony-forming assays on KO/DNStat5 cells showed a drastic reduction of granulocyte (CFU-G and CFU-GM) colonies compared to KO/vec cells harboring an empty vector (Figure 3E). By contrast, DN Stat3 had little effect on the proliferation and colony-forming abilities of PLC-β3−/− and WT cells. Furthermore, the lethally irradiated Ly5.1+ mice that had received KO/DNStat5 cells had drastically fewer donor (GFP+)-derived blood cells (Figure 3F) and KSL cells (Table S5) than the mice that had received KO/vec cells, indicating that Stat5 is required for engraftment or cell proliferation/survival of the transferred cells as shown by Bunting et al. (Bunting et al., 2002). Among the GFP+ donor cells, the former mice had much fewer CD11b+ cells than the latter mice, whereas GFP− cells in mice that had received KO/vec or KO/DNStat5 cells showed high proportions of CD11b+ cells similar to those among GFP+ cells in mice that had received KO/vec cells (Figure 3G). These results support the notion that the increased Stat5 activity is responsible for the myeloid-differentiative activities of PLC-β3−/− HSC-enriched populations. There were increases in myeloid cell percentage from 2 to 4 months after transfer in mice that received both control (vec) and DN Stat5-tranduced cells, suggesting a beginning of MPD development despite the DN Stat5 expression or retroviral inactivation or escape mechanism.
PLC-β3 suppresses the growth of hematopoietic cells through its C-terminal fragment
IL-3-dependent mouse Ba/F3 cells have been extensively used to investigate Stat5-related signaling events (Warmuth et al., 2007). Expression of PLC-β3 at 2-to 5-fold higher levels over the endogenous level inhibited IL-3-dependent proliferation of Ba/F3 cells, accompanied by reduced Stat5 phosphorylation (Figure 4B-C). Next we determined the structural requirements for growth suppression. Importantly, the C-terminal fragment (CT) corresponding to residues 809-1234 retained as strong a growth-suppressive activity as full-length PLC-β3 (Figure 4B). However, the catalytic domain of PLC-β3 was not required for suppressive activity, as two catalytically inactive mutants, E362G and del, showed a slightly stronger, if any, growth-suppressive activity (Figure 4B), consistent with the assumption that the PLC catalytic activity enhances cellular growth. The PH domain had no effect on cellular growth. The growth-suppressive function of CT was confirmed in vivo: PLC-β3−/− KSL cells transduced with CT failed to cause MPD in recipient mice (Figure S8B). KSL cells recovered from such recipient mice showed slower in vitro growth with lower levels of Stat5 phosphorylation than KSL cells from mice that had received empty vector-transduced PLC-β3−/− KSL cells (Figure S8C-D).
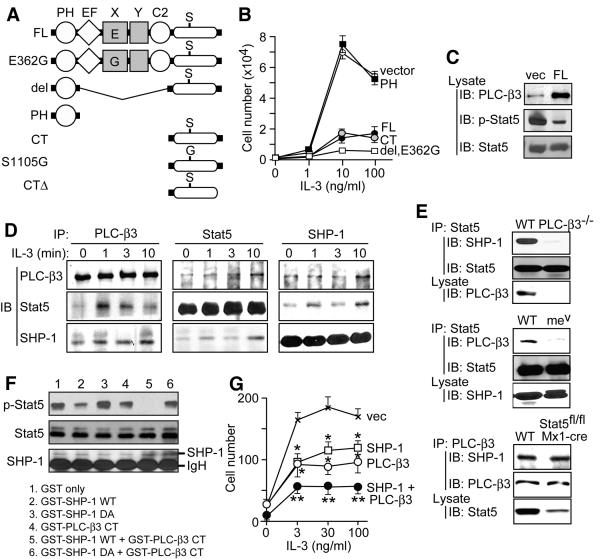
Figure 4. Functional multi-molecular SPS complexes contain PLC-β3, Stat5, and SHP-1.
(A) Scheme for a panel of PLC-β3 retroviral constructs. (B) Growth-suppressive function of various PLC-β3 constructs. Ba/F3 cells were transduced with a bicistronic retroviral vector expressing GFP alone (vector) or GFP and either full-length (FL) PLC-β3 or its fragment. Growth responses to the indicated concentrations of IL-3 are shown. S1105G and CTΔ showed almost the same inhibitory effects as CT (data not shown). Similar results were obtained in multiples experiments (at least 5 times using FL and CT constructs). (C) FL- and vector-transduced Ba/F3 cells were lysed and directly analyzed by immunoblotting. (D) Lysates of IL-3-stimulated non-transduced Ba/F3 cells were immunoprecipitated (IP) and followed by immunoblotting (IB) with the indicated antibodies. (E) Splenocytes from PLC-β3−/−, mev/mev, polyI:C-treated Stat5fl/fl;Mx1-cre, and control (WT) mice were analyzed by immunoprecipitation and immunoblotting. (F) PLC-β3 CT facilitated SHP-1-mediated dephosphorylation of phospho-Stat5 in vitro. (G) PLC-β3 and SHP-1 synergistically inhibited IL-3-dependent growth of Ba/F3 cells. *, **: p<0.05, p<0.01 versus vector control by Student's t-test. Results in B and G represent mean ± SD.
Multi-molecular interactions involve PLC-β3, Stat5, and SHP-1
We found physical interactions between PLC-β3 and Stat5. Thus, anti-PLC-β3 antibody co-immunoprecipitated Stat5 from Ba/F3 cell lysates (Figure 4D). Low-level co-immunoprecipitation before stimulation was followed by a transient increase with a peak at 1-3 min upon IL-3 stimulation. Reciprocally, PLC-β3 was co-immunoprecipitated with anti-Stat5 antibody (Figure 4D). IL-3-inducible interactions between PLC-β3 and Stat5 were also shown in mouse splenocytes (Figure S9A). Consistent with these biochemical data, confocal imaging analysis showed co-localization of PLC-β3 with phospho-Stat5 in the cytoplasm of IL-3-stimulated Ba/F3 cells (Figure S9B). By contrast, Stat3 was not co-immunoprecipitated with PLC-β3 (data not shown). Given the potential role of SHP-1 in dephosphorylating Stat5 and inhibiting IL-3-dependent cell growth (Paling and Welham, 2002), we examined whether PLC-β3 interacts with SHP-1. Indeed, co-immunoprecipitation was observed between PLC-β3 and SHP-1 in Ba/F3 and spleen cells (Figures 4D and S9A), but not between PLC-β3 and other phosphatases such as SHP-2, PP2A, and PTP-1B, despite their robust expression in Ba/F3 cells (data not shown). PLC-β3/SHP-1 interactions were largely constitutive, but increased by 30-150% at 10 min stimulation with IL-3 (Figure 4D). Stat5 also interacted with SHP-1 and this interaction slightly increased at 1 and 10 min stimulation with IL-3 (Figures 4D and S9A). The ability of PLC-β3 CT to interact with SHP-1 and Stat5 were confirmed by in vitro GST fusion protein pull-down assays (Figure S10).
Pairwise interactions among PLC-β3, Stat5, and SHP-1 raised the possibility that these molecules are present in the same multi-molecular complex. To test this hypothesis, we performed a series of immunodepletion/immunoprecipitation experiments. Briefly, depletion of one of these three molecules from Ba/F3 lysates abrogated or drastically reduced the interaction between the other two molecules (Figure S11). As these results indicate the presence of a multi-molecular complex, we propose to call it the SPS complex, which includes SHP-1, PLC-β3, and Stat5 as components. Pairwise interactions among PLC-β3, SHP-1, and Stat5 were modestly induced upon IL-3 stimulation in non-transduced Ba/F cells and splenocytes from normal mice (Figures 4D and S9A). IL-3-mediated inducibility of interactions among these molecules became more remarkable in Ba/F3 cells overexpressing PLC-β3 (Figure S9C). These results suggest that pairwise interactions among the three proteins, particularly PLC-β3/Stat5 interactions, can be inducible by IL-3 stimulation. As immunodepletion of one component from IL-3-stimulated, PLC-β3-overexpressing Ba/F3 cells completely or near completely abrogated interactions between the other two components (Figure S11), large proportions of these three molecules were interpreted to reside in SPS complexes upon IL-3 stimulation. Furthermore, PLC-β3 deficiency almost abrogated the SHP-1/Stat5 interaction and the motheaten viable (mev/mev) mutation in SHP-1 (Tsui et al., 1993) drastically reduced the PLC-β3/Stat5 interaction (Figure 4E). These results suggest that formation of stable SPS complexes require PLC-β3 and normal SHP-1 proteins. By contrast, ∼80% reduction in Stat5 expression did not affect the PLC-β3/SHP-1 interaction (Figure 4E), together with our observation of the direct interaction between PLC-β3 and SHP-1 (Figure S10F), suggesting that Stat5 may not be required for the initial assembly of SPS complexes. These results suggest the dynamic nature of the SPS complex, in which PLC-β3 and SHP-1 function as a limiting factor in its assembly.
PLC-β3 enhances SHP-1-mediated dephosphorylation of Stat5
Functional relationship among the components of the SPS complex was assessed in an in vitro phosphatase assay using recombinant SHP-1 and PLC-β3 CT (GST-PLC-β3 CT). The substrate used in this assay was phospho-Stat5 immunoprecipitated from pervanadate-stimulated Daudi cells, which expressed low levels of PLC-β3 (Figure 7A). Phospho-Stat5 levels were modestly reduced by incubation with WT, but not catalytically inactive D419A, SHP-1 (Figure 4F, lanes 2 & 3), providing the direct evidence that SHP-1 can dephosphorylate phospho-Stat5. More importantly, incubation of phospho-Stat5 with GST-PLC-β3 CT plus WT (but not D419A mutant) SHP-1 drastically reduced phospho-Stat5 levels (Figure 4F, lane 5). Incubation of phospho-Stat5 with GST-PLC-β3 CT alone slightly reduced phosphorylation levels, suggesting that the catalytic activity of the endogenous, Stat5-associated SHP-1 was enhanced with GSTPLC-β3 CT. Indeed, probing the same blot with anti-SHP-1 confirmed the presence of endogenous SHP-1 in Stat5 immunoprecipitates. We next tested the functional relevance of the SPS complex in the cellular context. Co-expression of PLC-β3 and SHP-1 had a stronger inhibitory effect on IL-3-induced Ba/F3 cell proliferation, compared to those of PLC-β3 or SHP-1 alone (Figure 4G). These results suggest that the SPS complex is functional and that PLC-β3 present in the SPS complex augments the phosphatase activity of SHP-1 toward phospho-Stat5.
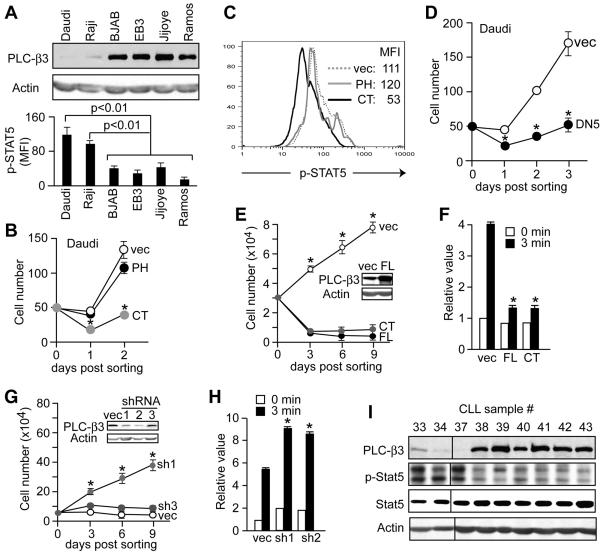
Figure 7. Growth regulation of Burkitt's lymphoma and erythroleukemia cells by PLC-β3.
(A) Daudi and Raji cells, but not other Burkitt's lymphoma cells, express very low levels of PLC-β3 (as shown by immunoblotting) and high levels of phospho-STAT5 (as shown by flow cytometry). (B-D) Daudi cells were retrovirally transduced with the indicated genes. GFP+ transduced cells were FACS-sorted and enumerated (B,D). (C) Phospho-STAT5 levels in the transduced Daudi cells were measured by flow cytometry. (E,F) TF-1 cells were transduced with full-length (FL) PLC-β3 or CT, then GFP+ cells were cultured in the presence of GM-CSF. The inset in E shows expression of PLC-β3 and panel F shows phospho-STAT5 MFI levels. (G,H) TF-1 cells were lentivirally transduced with short hairpin (sh) RNA constructs targeting the human PLC-β3 gene. The inset shows that sh1 and sh2, but not sh3, constructs efficiently inhibited expression of PLC-β3 protein. (H) Transduced TF-1 cells were stimulated by GM-CSF for 3 min before flow cytometric measurement of phospho-STAT5. (I) Immunoblot analysis of human CLL. Results in A, B, and D-H represent mean ± SD.
PLC-β3 CT cannot suppress the proliferative, myeloid-differentiative, and MPD-causing capabilities of HSC derived from motheaten viable mice
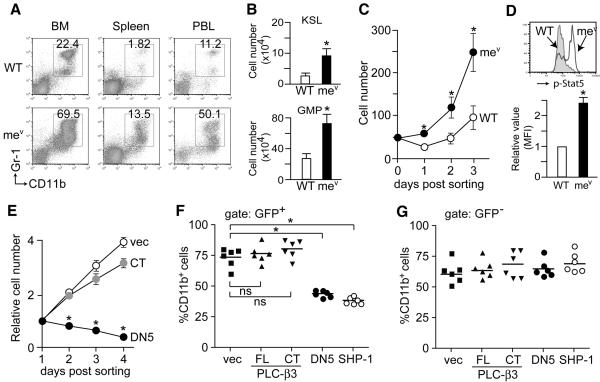
Given the probable role of the SPS complex in antagonizing Stat5 activation, we investigated whether the apparent MPD in mev/mev mice (Figure 5A) has a similar pathogenic mechanism as that in PLC-β3−/− mice. Indeed, increased numbers of KSL and GMP cells were present in the BM of mev/mev mice (Figure 5B). CD34− KSL cells from mv/mev mice grew faster in vitro with higher levels of Stat5 phosphorylation than WT cells (Figure 5C-D), and their in vitro growth was inhibited by DN Stat5 (Figure 5E). The MPD in mev/mev mice was BM cell-autonomous (Figure S12), and mev/mev CD34− KSL cells transduced with DN Stat5 failed to develop MPD in recipient mice (Figure 5F). Therefore, we concluded that constitutive Stat5 activation is a shared mechanism for MPD development in PLC-β3−/− and mev/mev mice. Interestingly, in vitro growth of mev/mev CD34− KSL cells was only marginally affected by PLC-β3 CT (Figure 5E), and transduction of mev/mev CD34− KSL cells with full-length PLC-β3 or PLC-β3 CT, did not prevent MPD development in recipient mice (Figure 5F), similar to non-transduced (GFP−) cells (Figure 5G). By contrast, mev/mev CD34-KSL cells transduced with WT SHP-1 did not cause MPD in recipient mice. Together with the reduced PLC-β3/Stat5 interaction in mev/mev cells (Figure 4E), these results suggest that aberrant SHP-1 proteins encoded by mev/mev (Shultz et al., 1993) cannot use the adaptor function of PLC-β3 to suppress Stat5 phosphorylation.
Figure 5. MPD-causing ability of HSC-enriched cells derived from motheaten viable mice depend on Stat5.
(A) Flow cytometric analysis of nucleated cells in BM, spleen, and peripheral blood (PBL) of 6-8 week-old mice (n=6). Granulocyte percentages are shown. (B) Flow cytometric analysis of KSL and GMP in BM (n=6). *, p<0.05 versus WT cells. (C) CD34− KSL cells from 6-8 week-old mice (n=4) were sorted and cultured in IL-3, SCF, IL-11, Flt3L, and TPO. Representative of 3 independent experiments. (D) Flow cytometric analysis of phospho-Stat5 levels (n=4). (E) mev/mev CD34− KSL cells were transduced with a bicistronic retroviral vector expressing DN Stat5, PLC-β3 CT, or empty vector (vec). Growth of GFP+ cells was monitored. *, p<0.05 versus vec cells. Results in B-E represent mean ± SD. (F) mev/mev CD34− KSL cells were transduced with the indicated constructs and adoptively transferred to lethally irradiated mice. Two months later, peripheral blood was analyzed by flow cytometry for enumeration of CD11b+ cells in GFP+ donor-derived cells. Similar results were observed 4 months after transfer (data not shown). *, p<0.05; ns, not significant.
PLC-β3 haploinsufficiency cooperates with c-Myc to transform fibroblasts and lymphocytes
To further analyze the transforming ability of PLC-β3-deficient cells, we transfected PLC-β3−/− MEFs with V12 H-ras or c-myc. Unlike active H-ras, which induced senescence in WT and PLC-β3−/− MEFs (data not shown), c-myc induced in vitro transformation in PLC-β3+/− and PLC-β3−/− MEFs (Figure 6A). In light of this observation, we investigated whether PLC-β3 deficiency can cooperate with c-myc to induce in vivo tumor formation, by crossing PLC-β3−/− mice with Eμ-myc transgenic mice. As reported previously (Adams et al., 1985), Eμ-myc transgenic mice developed B cell-lineage lymphomas with a long latency. Lymphoma formation in Eμ-myc transgenic mice with heterozygous PLC-β3+/− loci was dramatically accelerated (median survival: 100 days in Eμ-myc;PLC-β3+/− mice versus >365 in Eμ-myc;PLC-β3+/+ mice) (Figure 6B). These Eμ-myc;PLC-β3+/− lymphomas expressed a pre-B cell phenotype of B220+ IgM− CD43− (data not shown). These and PLC-β3−/− lymphomas showed higher levels of Stat5 phosphorylation than normal lymphoid tissues (Figure 6C). Importantly, all analyzed PLC-β3−/− lymphomas showed as high c-Myc expression as did Eμ-myc;PLC-β3+/− lymphomas. On the other hand, the expression of PLC-β3 protein in Eμ-myc;PLC-β3+/− lymphomas was drastically reduced compared to normal lymphoid tissues (Figure 6C). As extensive RT-PCR analyses covering all exon sequences of PLC-β3 mRNA gave the same results (Figure S13) in WT spleens and two Eμ-myc;PLC-β3+/− lymphomas (samples 1 and 2 in Figure 6C) and no mutations were found in the PLC-β3 exons (data not shown), the very low expression of PLC-β3 protein in these lymphomas seemed due to abnormal post-transcriptional regulation of PLC-β3 mRNA: probably stability of the PLC-β3 protein might be low. Alternatively, translation of the PLC-β3 protein might be abnormal and inefficient in Eμ-myc;PLC-β3+/− lymphomas. Another possibility is somatic inactivating point mutations in other cases of Eμ-myc;PLC-β3+/− lymphomas. However, PLC-β3 protein expression in PLC-β3+/− splenocytes is reduced by about 50% as expected (data not shown), which is consistent with the fact that PLC-β3+/− mice did not show any abnormal phenotypes. Retroviral expression of DN Stat5 or PLC-β3 CT in Eμ-myc;PLC-β3+/− lymphoma cells suppressed their in vitro growth and colony formation (Figure 6D-E). These results indicate that PLC-β3 haploinsufficiency cooperates with c-Myc to transform fibroblasts and lymphocytes.
Translocations of c-myc to immunoglobulin or other gene loci and thus abnormal expression of c-myc are causally linked to Burkitt's lymphoma (Boxer and Dang, 2001). Interestingly, two of six Burkitt's lymphoma cell lines tested, i.e., Daudi and Raji, exhibited very low levels (5-10% that in Ba/F3 cells) of PLC-β3 protein and they had higher basal levels of STAT5 phosphorylation than the other Burkitt's cells with high PLC-β3 expression (Figure 7A). Overexpression of PLC-β3 CT in Daudi and Raji cells blocked their growth (Figure 7B and data not shown) and reduced STAT5 phosphorylation (Figure 7C). Expression of DN STAT5 also blocked the growth of these Burkitt's lymphoma cells (Figure 7D and data not shown), but not that of Ramos cells expressing high levels of PLC-β3 (data not shown). These results together with observations on PLC-β3−/− and Eμ-myc;PLC-β3+/− lymphomas are consistent with the notion that reduced or abrogated expression of PLC-β3 may cooperate with active c-Myc to induce lymphoma in mice and humans. In addition to these lymphoid tumor cells, overexpression of full-length PLC-β3 or CT in GM-CSF-dependent TF-1 erythroleukemia cells suppressed GM-CSF-dependent cell growth associated with repressed STAT5 phosphorylation (Figure 7E-F); on the other hand, knockdown (to a level of 20-30% that in Ba/F3 cells) of PLC-β3 expression using lentivirus-mediated RNA interference rendered TF-1 cell growth independent of GM-CSF and associated with increased STAT5 phosphorylation (Figure 7G-H). Similarly, overexpression of full-length PLC-β3 or CT suppressed the growth factor-dependent proliferation and/or survival of other human leukemic cell lines such as MEC2 and HL-60 (data not shown). Eleven percent of chronic lymphocytic leukemia (CLL) samples showed low levels of PLC-β3 expression with high phospho-STAT5 levels (Figure 7I and data not shown). The results collectively suggest that reduced expression of PLC-β3 and thus the loss of the SHP-1-mediated Stat5 dephosphorylation mechanism cooperates with active c-myc (or an unknown oncogene) to induce lymphoid and myeloid malignancies in mice and humans.
Discussion
This study demonstrates an adaptor function of PLC-β3 that negatively regulates proliferative, survival, and myeloid-differentiative capabilities of HSC-enriched cell populations. PLC-β3 augments SHP-1-mediated deactivation of Stat5 activity (Figure 8). Loss of this regulation seems to lead to MPD development in aged PLC-β3−/− mice. Long latency suggests that an additional transforming event is required for conversion of PLC-β3−/− HSC/progenitor cells to malignant cells. Importantly, c-myc can transform PLC-β3+/− MEFs and B cell precursors. Cooperative transformation by active c-myc and PLC-β3 deficiency seems to underlie lymphomas in PLC-β3−/− and Eμ-myc;PLC-β3+/− mice and a subset of human Burkitt's lymphoma. However, c-myc expression was not increased in PLC-β3−/− KSL cells (data not shown), suggesting that an oncogene other than c-myc seems responsible for MPD development.
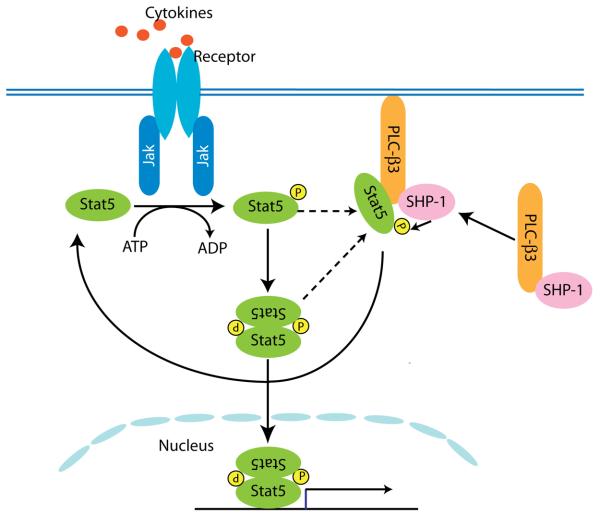
Figure 8. Proposed mechanism for PLC-β3-mediated inhibition of Stat5 overactivation.
Following cytokine stimulation, the Jak-Stat pathway is activated, leading to activation of Stat target genes. PLC-β3/ SHP-1 complexes recruit phosphorylated Stat5 to its C terminal domain, to form SPS complexes. PLC-β3 in the complexes augments the capability of SHP-1 of dephosphorylating Stat5 and inhibiting its nuclear translocation. At this point we do not know whether monomeric or dimeric phosphorylated Stat5 is recruited.
All the tested MPD, lymphoma, and other tumor cells derived from PLC-β3−/− mice had high phospho-Stat5 levels, and DN Stat5 suppressed the growth of PLC-β3−/− HSC and lymphoma cells, suggesting that Stat5 activation is part of the necessary transforming processes in these malignancies. Stat5 is frequently activated in leukemia (Benekli et al., 2003; Frohling et al., 2005). Stat5 activation was shown to be essential for MPD or myeloid leukemia induced by the activated oncogenes, such as TEL/JAK2 (Schwaller et al., 2000), TEL/PDGFRB (Cain et al., 2007), and FLT3 ITD (Choudhary et al., 2007), and by deficiencies of SHIP and Lyn/Hck (Xiao et al., 2008). An activating mutation (V617F) in JAK2 was found in human MPDs (Baxter et al., 2005; James et al., 2005; Kralovics et al., 2005; Levine et al., 2005). However, Jak2 cDNAs cloned from PLC-β3−/− KSL cells had the WT sequence around the V617 residue (data not shown). Therefore, it will be interesting to study what kinase activates Stat5 in PLC-β3−/− mice.
We provide the direct evidence that SHP-1 can dephosphorylate Stat5 to dampen Stat5 activity. This function is lost in mev/mev mice; this deficiency underlies the MPD in these mice, as transduction with DN Stat5 inhibited in vitro growth of mev/mev CD34− KSL cells and MPD development in recipient mice. Transduction of mev/mev CD34− KSL cells with SHP-1, but not PLC-β3 CT, blocked their MPD-causing capability. These results demonstrate that aberrant SHP-1 proteins generated by the mev locus cannot suppress Stat5 phosphorylation. This could be due to the loss of PLC-β3/Stat5 interactions, low enzymatic activity of the mutant SHP-1 proteins, or both.
We have provided biochemical evidence for physical interactions among PLC-β3, Stat5, and SHP-1. Pairwise interactions were modestly enhanced upon IL-3 stimulation, but they were much more strongly induced in Ba/F3 cells overexpressing PLC-β3, suggesting the dynamic nature of complex formation that is under the control of PLC-β3 levels and IL-3 stimulation. However, the structure and function of this complex remained to be defined. Our in vitro phosphatase assays showed that SHP-1 can dephosphorylate phospho-Stat5 on Tyr-694 to deactivate Stat5, and that PLC-β3 CT augments this dephosphorylation reaction. Therefore, we hypothesize that SPS complex formation enhances the activity of SHP-1 to deactivate Stat5 to prevent unchecked Stat5 activation. Further, the dysregulation of this mechanism at the level of HSC may lead to the development of MPD. It is tempting to speculate that similar dysregulation in other hematopoietic or non-hematopoietic cells may also contribute to tumorigenic processes of various malignancies.
PLC-β3 CT can specifically interact with Stat5 and SHP-1. The corresponding CT of turkey PLC-β forms a coiled-coil structure that dimerizes along its long axis, a structure for interactions with GTP-bound Gαq (Singer et al., 2002). This part of PLC-β3 has low sequence similarity (30% identity) to the corresponding region of PLC-β2. Consistent with this and biological differences between PLC-β3−/− and PLC-β2−/− mice, PLC-β2 did not co-immunoprecipitate with Stat5 or SHP-1 (data not shown). Mutations of Ser-1105 (PKC phosphorylation site) or the C-terminal residues (PDZ domain-binding site) did not affect the growth-suppressive activity. These results indicate that PLC-β3 CT contains a determinant responsible for growth suppression with interaction sites for Stat5 and SHP-1.
Information in the public domain, e.g., the Cancer Genome Anatomy Project website (http://cgap.nci.nih.gov/Chromosomes/), indicates reduced or abrogated expression of PLC-β3 in human malignancies. A recent genome-wide analysis of genetic alterations in acute lymphocytic leukemia (ALL) demonstrated that a small number of ALL patients have deletions of the region of chromosome 11 including the PLC-β3 gene (Mullighan et al., 2007). Furthermore, the Oncomine database (Rhodes et al., 2004) shows that reduced expression of PLC-β3 mRNA is associated with higher grades of bladder carcinoma (p=4.2 × 10−7). We also found loss or reduced PLC-β3 expression in human tumor cell lines (Figure 7I and data not shown). Therefore, reduced or abrogated expression of PLC-β3 may play a role in the tumorigenic process of human malignancies.
Regulation of PLC-β3 expression has not been well studied. Therefore, it is not clear how PLC-β3 expression is reduced or abrogated in tumors. One obvious possibility is deletions of the chromosomal region encompassing the PLC-β3 gene, as shown in the aforementioned ALL cases. Another possible mechanism is DNA methylation of the gene promoter. Indeed, treatment with decitabine, an inhibitor of the DNA methyltransferase activity, restored PLC-β3 expression in Daudi cells (data not shown). There might be other unknown mechanisms for reduced expression. However, simple mutations or gene fusions do not seem to be involved in reduced or abrogated expression of PLC-β3 as such mutations in the human PLC-β3 gene were not found in 136 tumor samples (COSMIC website, http://www.sanger.ac.uk/genetics/CGP/cosmic/). However, this issue warrants further investigation.
Experimental Procedures
Mice and MPD
PLC-β3−/− and PLC-β2−/− mice were described previously (Li et al., 2000). Mev, C57BL/6-Ly5.1, and the Rag2−/−-Ly5.1 mice were purchased from The Jackson Laboratory and Taconic, respectively. Stat5 floxed mice (Cui et al., 2004) with Mx1-cre will be described elsewhere (Kimura, A. et al., manuscript in preparation). For the definition of MPD, we followed the criteria adopted by the Mouse Models of Human Cancers Consortium (KOgan et al., 2002) and by Passegue et al. (Passegue et al., 2004). The Animal Care and Use Committee of the La Jolla Institute for Allergy and Immunology approved all mouse experiments.
Identification, Purification, and Proliferation of HSC
See the Supplemental Data for the identification and purification of HSC. For proliferation assays, KSL or CD34− KSL cells were sorted into a 96-well round bottom plate (one cell per well) in 200 μl of IMDM containing 5% FBS, 50 μM 2-mercaptoethanol, and either IL-3 (10 ng/ml) alone or a cocktail of stem cell factor (SCF), IL-3, Flt3L, thrombopoietin (TPO), and IL-11, and incubated at 37°C for the indicated periods. The numbers of cells per well were visually determined under an inverted microscope. In some experiments, 50 or 150 CD34− KSL cells were sorted per well and a cocktail of SCF, Flt3L, and IL-11 was added. For BrdU incorporation, cell cycle analysis, and analysis of phosphorylated signaling molecules in HSC, see the Supplemental Data.
Retroviral Transduction
Recombinant retroviruses were produced as previously described (Kato et al., 2005). Sorted CD34− KSL cells (150 cells/well) were incubated in α-MEM supplemented with 1% FBS, SCF, and human TPO for 24 h, and then transduced with a retrovirus vector (pMXGFP-DN Stat5 or DN Stat3; pMIG-SHP-1, pMIG-PLC-β3 or -CT) in the presence of protamine sulfate (10 μg/ml) and recombinant fibronectin fragment (1 μg/ml) for 24 h. Transduced cells were further subjected to liquid (in the presence of SCF, Flt-3L, and IL-11) or semi-solid cultures (MethoCult™ M3434 from StemCell Technologies). In these experiments, transduction efficiency was ∼80% as judged by GFP expression by epifluorescence microscopy.
Transplantation of Hematopoietic Cells
BM cells (2×106 cells in 400 μl PBS) were injected into the tail vein of lethally (960 rad) irradiated recipient mice (C57BL/6-Ly5.1; 8-10-week-old). For transplantation of HSC and progenitors, FACS-sorted CD34− KSL, CD34+ KSL, CMP, and GMP were injected into sublethally irradiated (450 rad) Rag2−/−-Ly5.1 recipient mice. We also transplanted retrovirally transduced HSCs into lethally irradiated C57BL/6-Ly5.1 mice together with 2.5×105 Sca-1-depleted C57BL/6-Ly5.1 helper BM cells.
Transduction, Co-immunoprecipitation, and Immunodepletion of Ba/F3 Cells
Ba/F3 cells were cultured in IL-3-containing medium and transduced with a retrovirus vector (pMIG-myc-PLC-β3 WT or mutants). Forty-eight hours later, transduced GFP+ cells were FACS-sorted into a 96-well plate at a density of 50 cells per well, and cultured in the presence of IL-3. For co-immunoprecipitation, transduced cells deprived of IL-3 were stimulated with 10 ng/ml of IL-3, and cell lysates were immunoprecipitated with anti-Stat5 (C-17), anti-PLC-β3 (C-20) (or anti-myc), or anti-SHP-1 (C-19) antibodies (all from Santa Cruz Biotechnology). For immunodepletion, cell lysates were precipitated with anti-Stat5, anti-PLC-β3, or anti-SHP-1 antibodies, and supernatants were collected for co-immunoprecipitation.
In Vitro Phosphatase Assay
In vitro phosphatase assay was performed essentially as described (Nakahira et al., 2007). In brief, phospho-Stat5 was immunoprecipitated from pervanadate-treated Daudi cells, and incubated with GST-tagged proteins in 25 mM HEPES (pH 7.5), 5 mM EDTA, 10 mM DTT at 30°C for 1 hour. GST-SHP-1 WT or D419A mutant was phosphorylated by Lyn precipitated from Ba/F3 cells in vitro.
Human Samples
Peripheral blood samples were collected from patients with human hematopoietic malignancies. Leukocytes were lysed in SDS-sample buffer and analyzed by western blotting. The study protocols were approved by the Human Subjects Research Ethics Board (HSREB) of the University of Western Ontario. Informed consent was obtained from each study subject.
Supplementary Material
Acknowledgments
We thank Federico Caligaris-Cappio, James N. Ihle, Toshio Kitamura, John Ryan, Reuben P. Siraganian, Jean Y. J. Wang, and Kirin Pharma for kindly providing reagents; Shuji Ueda for making some constructs. We are grateful to Michael Poderycki and Yu Kawakami for critical reading of the manuscript. This study was supported in part by grants from the National Institutes of Health to TK. WX was supported in part by funds from the Diabetes and Immune Disease National Research Institute. This is Publication 769 from the La Jolla Institute for Allergy and Immunology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SIGNIFICANCE
Many hematopoietic malignancies depend on the activity of Stat5 transcription factor. Here, we report a novel Stat5 suppressive mechanism by which PLC-β3 augments dephosphorylating activity of SHP-1 towards Stat5 by recruiting SHP-1 and Stat5 to its C-terminal sequence. Abrogation of this suppression leads to MPD, lymphoma, and other types of cancer in PLC-β3−/− mice. PLC-β3 deficiency or downregulation appears to cooperate with c-Myc to induce B cell lymphoma in mice and humans. The same mechanism may be operative in human myeloid and lymphoid tumors. Therefore, the adaptor function of PLC-β3 seems essential to protect the hematopoietic and non-hematopoietic systems from tumor development.
Supplemental Data
Supplemental Data include supplemental experimental procedures, 13 figures, and 6 tables and can be found with this article online.
The authors declare that they have no competing financial interests.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol. 2004;22:503–529. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- Bach TL, Chen QM, Kerr WT, Wang Y, Lian L, Choi JK, Wu D, Kazanietz MG, Koretzky GA, Zigmond S, et al. Phospholipase cbeta is critical for T cell chemotaxis. J Immunol. 2007;179:2223–2227. doi: 10.4049/jimmunol.179.4.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- Benekli M, Baer MR, Baumann H, Wetzler M. Signal transducer and activator of transcription proteins in leukemias. Blood. 2003;101:2940–2954. doi: 10.1182/blood-2002-04-1204. [DOI] [PubMed] [Google Scholar]
- Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- Bunting KD, Bradley HL, Hawley TS, Moriggl R, Sorrentino BP, Ihle JN. Reduced lymphomyeloid repopulating activity from adult bone marrow and fetal liver of mice lacking expression of STAT5. Blood. 2002;99:479–487. doi: 10.1182/blood.v99.2.479. [DOI] [PubMed] [Google Scholar]
- Cain JA, Xiang Z, O'Neal J, Kreisel F, Colson A, Luo H, Hennighausen L, Tomasson MH. Myeloproliferative disease induced by TEL-PDGFRB displays dynamic range sensitivity to Stat5 gene dosage. Blood. 2007;109:3906–3914. doi: 10.1182/blood-2006-07-036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps M, Carozzi A, Schnabel P, Scheer A, Parker PJ, Gierschik P. Isozyme-selective stimulation of phospholipase C-beta 2 by G protein beta gamma-subunits. Nature. 1992;360:684–686. doi: 10.1038/360684a0. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Brandts C, Schwable J, Tickenbrock L, Sargin B, Ueker A, Bohmer FD, Berdel WE, Muller-Tidow C, Serve H. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–374. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77:925–929. [PubMed] [Google Scholar]
- Frohling S, Scholl C, Gilliland DG, Levine RL. Genetics of myeloid malignancies: pathogenetic and clinical implications. J Clin Oncol. 2005;23:6285–6295. doi: 10.1200/JCO.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Ilaria RL, Jr., Hawley RG, Van Etten RA. Dominant negative mutants implicate STAT5 in myeloid cell proliferation and neutrophil differentiation. Blood. 1999;93:4154–4166. [PubMed] [Google Scholar]
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- Jiao H, Berrada K, Yang W, Tabrizi M, Platanias LC, Yi T. Direct association with and dephosphorylation of Jak2 kinase by the SH2-domain-containing protein tyrosine phosphatase SHP-1. Mol Cell Biol. 1996;16:6985–6992. doi: 10.1128/mcb.16.12.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Iwama A, Tadokoro Y, Shimoda K, Minoguchi M, Akira S, Tanaka M, Miyajima A, Kitamura T, Nakauchi H. Selective activation of STAT5 unveils its role in stem cell self-renewal in normal and leukemic hematopoiesis. J Exp Med. 2005;202:169–179. doi: 10.1084/jem.20042541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A, Wu D, Simon MI. Subunits beta gamma of heterotrimeric G protein activate beta 2 isoform of phospholipase C. Nature. 1992;360:686–689. doi: 10.1038/360686a0. [DOI] [PubMed] [Google Scholar]
- Klingmuller U, Lorenz U, Cantley LC, Neel BG, Lodish HF. Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, Cardiff RD, Carter JS, de Coronado S, Downing JR, Fredrickson TN, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA, Weissman IL. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- Kyba M, Perlingeiro RC, Hoover RR, Lu CW, Pierce J, Daley GQ. Enhanced hematopoietic differentiation of embryonic stem cells conditionally expressing Stat5. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11904–11910. doi: 10.1073/pnas.1734140100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Shin SH, Hepler JR, Gilman AG, Rhee SG. Activation of phospholipase C-beta 2 mutants by G protein alpha q and beta gamma subunits. J Biol Chem. 1993;268:25952–25957. [PubMed] [Google Scholar]
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, Wu D. Roles of PLC-beta2 and -beta3 and PI3Kgamma in chemoattractant-mediated signal transduction. Science. 2000;287:1046–1049. doi: 10.1126/science.287.5455.1046. [DOI] [PubMed] [Google Scholar]
- Lilly M, Kraft A. Enforced expression of the Mr 33,000 Pim-1 kinase enhances factor-independent survival and inhibits apoptosis in murine myeloid cells. Cancer Res. 1997;57:5348–5355. [PubMed] [Google Scholar]
- Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- Mui AL, Wakao H, O'Farrell AM, Harada N, Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologs. Embo J. 1995;14:1166–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui AL, Kitamura T. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. Embo J. 1999;18:4754–4765. doi: 10.1093/emboj/18.17.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002;109(Suppl):S121–131. doi: 10.1016/s0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- Paling NR, Welham MJ. Role of the protein tyrosine phosphatase SHP-1 (Src homology phosphatase-1) in the regulation of interleukin-3-induced survival, proliferation and signalling. Biochem J. 2002;368:885–894. doi: 10.1042/BJ20021054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119:431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyers CL. Chronic myeloid leukemia. N Engl J Med. 1999;340:1330–1340. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- Sawyers CL, McLaughlin J, Witte ON. Genetic requirement for Ras in the transformation of fibroblasts and hematopoietic cells by the Bcr-Abl oncogene. J Exp Med. 1995;181:307–313. doi: 10.1084/jem.181.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Levy DE, Decker T. JAK-STAT Signaling: From Interferons to Cytokines. J Biol Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- Schuringa JJ, Chung KY, Morrone G, Moore MA. Constitutive activation of STAT5A promotes human hematopoietic stem cell self-renewal and erythroid differentiation. J Exp Med. 2004;200:623–635. doi: 10.1084/jem.20041024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaller J, Parganas E, Wang D, Cain D, Aster JC, Williams IR, Lee CK, Gerthner R, Kitamura T, Frantsve J, et al. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol Cell. 2000;6:693–704. doi: 10.1016/s1097-2765(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Shuai K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene. 2000;19:2638–2644. doi: 10.1038/sj.onc.1203522. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Schweitzer PA, Rajan TV, Yi T, Ihle JN, Matthews RJ, Thomas ML, Beier DR. Mutations at the murine motheaten locus are within the hematopoietic cell protein-tyrosine phosphatase (Hcph) gene. Cell. 1993;73:1445–1454. doi: 10.1016/0092-8674(93)90369-2. [DOI] [PubMed] [Google Scholar]
- Singer AU, Waldo GL, Harden TK, Sondek J. A unique fold of phospholipase C-beta mediates dimerization and interaction with G alpha q. Nat Struct Biol. 2002;9:32–36. doi: 10.1038/nsb731. [DOI] [PubMed] [Google Scholar]
- Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi JK, Trotta R, Wlodarski P, Perrotti D, Chan TO, et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. Embo J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow JW, Abraham N, Ma MC, Abbey NW, Herndier B, Goldsmith MA. STAT5 promotes multilineage hematolymphoid development in vivo through effects on early hematopoietic progenitor cells. Blood. 2002;99:95–101. doi: 10.1182/blood.v99.1.95. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Soriano MA, Grusby MJ. SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity. 2005;22:729–736. doi: 10.1016/j.immuni.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Tsui HW, Siminovitch KA, de Souza L, Tsui FW. Motheaten and viable motheaten mice have mutations in the haematopoietic cell phosphatase gene. Nat Genet. 1993;4:124–129. doi: 10.1038/ng0693-124. [DOI] [PubMed] [Google Scholar]
- Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. Ba/F3 cells and their use in kinase drug discovery. Curr Opin Oncol. 2007;19:55–60. doi: 10.1097/CCO.0b013e328011a25f. [DOI] [PubMed] [Google Scholar]
- Xiao W, Hong H, Kawakami Y, Lowell CA, Kawakami T. Regulation of myeloproliferation and M2 macrophage programming in mice by Lyn/Hck, SHIP, and Stat5. J Clin Invest. 2008;118:924–934. doi: 10.1172/JCI34013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Ichihara M, Kinjyo I, Moriyama M, Copeland NG, Gilbert DJ, Jenkins NA, Hara T, Miyajima A. Mouse oncostatin M: an immediate early gene induced by multiple cytokines through the JAK-STAT5 pathway. Embo J. 1996;15:1055–1063. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.