Abstract
The study of how transcriptional control and cell signaling influence neurons and glia to acquire their differentiated properties is fundamental to understanding CNS development and function. The Drosophila CNS midline cells are an excellent system for studying these issues because they consist of a small population of diverse cells with well-defined gene expression profiles. In this paper, the origins and differentiation of midline neurons and glia were analyzed. Midline precursor (MP) cells each divide once giving rise to two neurons; here, we use a combination of single-cell gene expression mapping and time-lapse imaging to identify individual MPs, their locations, movements and stereotyped patterns of division. The role of Notch signaling was investigated by analyzing 37 midline-expressed genes in Notch pathway mutant and misexpression embryos. Notch signaling had opposing functions: it inhibited neurogenesis in MP1,3,4 and promoted neurogenesis in MP5,6. Notch signaling also promoted midline glial and median neuroblast cell fate. This latter result suggests that the median neuroblast resembles brain neuroblasts that require Notch signaling, rather than nerve cord neuroblasts, the formation of which is inhibited by Notch signaling. Asymmetric MP daughter cell fates also depend on Notch signaling. One member of each pair of MP3–6 daughter cells was responsive to Notch signaling. By contrast, the other daughter cell asymmetrically acquired Numb, which inhibited Notch signaling, leading to a different fate choice. In summary, this paper describes the formation and division of MPs and multiple roles for Notch signaling in midline cell development, providing a foundation for comprehensive molecular analyses.
Keywords: CNS midline, Drosophila, Glia, Neuroblast, Neuron, Notch
INTRODUCTION
The central nervous system (CNS) consists of a diverse collection of neurons and glia that differ in both morphology and function. These properties arise during a sequence of developmental events that require numerous gene regulatory and signaling processes. The cells that lie along the midline of the Drosophila CNS provide a useful system for the comprehensive study of neurogenesis and gliogenesis. The mature, embryonic CNS midline cells consist of a functionally diverse group of ~22 cells, including midline glia (MG), local interneurons, projection neurons, peptidergic motoneurons and neuromodulatory motoneurons (Wheeler et al., 2006). The embryonic expression patterns of nearly 300 midline-expressed genes have been identified (Kearney et al., 2004), and transcriptional maps permit detailed genetic analysis of the entire process of midline cell development (Bossing and Brand, 2006; Wheeler et al., 2006). Thus, the Drosophila midline cells combine cellular diversity with extensive molecular genetic characterization for the study of CNS development.
The Drosophila midline cells originate from about eight precursor cells/segment that undergo synchronous cell division (δ1414) at stage 8 (Foe, 1989) to give rise to ~16 cells (Bossing and Technau, 1994). These cells are characterized by expression of the single-minded (sim) gene (Crews, 2003; Thomas et al., 1988). By late stage 11, the midline cells consist of about ten MG, comprising two populations, the anterior midline glia (AMG) and posterior midline glia (PMG), two midline precursor 1 (MP1) neurons, two MP3 neurons, six ventral unpaired median (VUM) neurons (two VUM4s, two VUM5s and two VUM6s) and the median neuroblast (MNB) (Wheeler et al., 2006). The PMG die during embryogenesis along with about half of the AMG. The remaining three AMG ensheathe the axon commissures. Whereas the two MP1 neurons appear to be identical, the MP3 neurons differentiate into the dopaminergic H-cell and glutamatergic H-cell sib. Each VUM precursor (MP4–6) divides once, giving rise to a GABAergic VUM interneuron (iVUM4–6) and a glutamatergic/octopaminergic VUM motoneuron (mVUM4–6). Thus, MPs can give rise to either two identical neurons (MP1) or two non-identical neurons (MP3–6). The MNB stem cell divides asymmetrically to generate about eight GABAergic neurons during embryogenesis, and a much larger number postembryonically (Truman et al., 2004). Despite the small number of embryonic midline cells, the origins of midline neurons and glia remain largely unknown. In this study, for the first time, we identified each MP and described their patterns of cell division. This information was then utilized to reveal multiple roles of Notch signaling in midline neuronal and glial cell development.
MATERIALS AND METHODS
Drosophila strains and genetics
Drosophila strains included w1118 (used as wild type), Dl3, Dl7, numb2 (Uemura et al., 1989), numb4 (Skeath and Doe, 1998), spdoG104 and spdoZ143 (Skeath and Doe, 1998), N55e11, Nts1, P[12xSu(H)bs-lacZ] (Go et al., 1998) and Gbe-lacZ (Furriols and Bray, 2001). Gal4 and UAS lines used were: sim-Gal4 (Xiao et al., 1996), UAS-numb (Wang et al., 1997), UAS-spdo (O’Connor-Giles and Skeath, 2003), UAS-Su(H).VP16 (Kidd et al., 1998) and UAS-tau-GFP (Brand, 1995). For N temperature-shift experiments, N55e11/Nts1 embryos were collected for 2 hours at 18°C, further incubated for 2 hours at 18°, then shifted to the restrictive temperature (30°C) for 6 hours, followed by fixation (approximately stage 14).
In situ hybridization and immunostaining
Embryo collection, in situ hybridization and immunostaining were performed as previously described (Kearney et al., 2004). Primary antibodies used were: mouse (Promega) and rabbit (Cappel) anti-β-galactosidase, rabbit anti-Cas (Kambadur et al., 1998), mouse and rat anti-Elav [Developmental Studies Hybridoma Bank (DSHB)], mouse anti-En MAb 4D9 (Patel et al., 1989), anti-Futsch MAb 22C10 (DSHB), guinea pig anti-Hb [East Asian Distribution Center (EADC)] (Kosman et al., 1998), chicken anti-GFP (Upstate), rabbit anti-GFP (Abcam), guinea pig anti-Lim1 (Broihier and Skeath, 2002), guinea pig anti-Numb (O’Connor-Giles and Skeath, 2003), rabbit anti-Odd (Ward and Skeath, 2000), rabbit anti-Period (Per) (Liu et al., 1992), rabbit antiphosphohistone H3 (Millipore), guinea pig anti-Runt (EADC), guinea pig and rat anti-Sim (Ward et al., 1998), rabbit anti-Spdo (O’Connor-Giles and Skeath, 2003), mouse anti-Tau (Sigma) and rat anti-Tup (Broihier and Skeath, 2002). Alexa Fluor-conjugated secondary antibodies were used (Molecular Probes). The Tyramide Signal Amplification System (Perkin Elmer) was employed for some immunostaining.
Microscopy and image analysis
In situ hybridization and immunostaining were carried out as previously described (Kearney et al., 2004; Wheeler et al., 2006). Midline cells were examined in abdominal segments A1–8. Owing to the three-dimensional structure of the midline cells, it was difficult to represent all relevant cells in a single focal plane, so, for clarity, irrelevant portions of single images within a stack of confocal images were subtracted and projections were generated. Thus, a single composite image is made from different focal planes that each contained relevant data.
Live imaging of midline cells
Time-lapse imaging of midline cell development was carried out in sim-Gal4 UAS-tau-GFP and sim-Gal4 UAS-tau-GFP; Dl3/Dl3 embryos by visualizing GFP. Embryos were collected for 1 hour, aged for an additional 4 hours, dechorionated, mounted on a glass coverslip, and immersed in halocarbon oil 700 on slides containing an oxygen-permeable membrane. GFP-fluorescent images were captured using a Nikon Eclipse TE300 equipped with a Perkin Elmer Ultraview confocal scanner and 40× or 60× oil-immersion objectives. Embryos were visualized for ~4 hours with an image captured every 30 seconds. Movies were assembled from images of single focal planes using MetaMorph software (Molecular Devices). Ten movies of wild-type embryos viewing 29 segments, and five movies of Dl mutant embryos viewing 14 distinct groups of cells, were analyzed.
RESULTS
Identification of midline precursors and their pattern of division
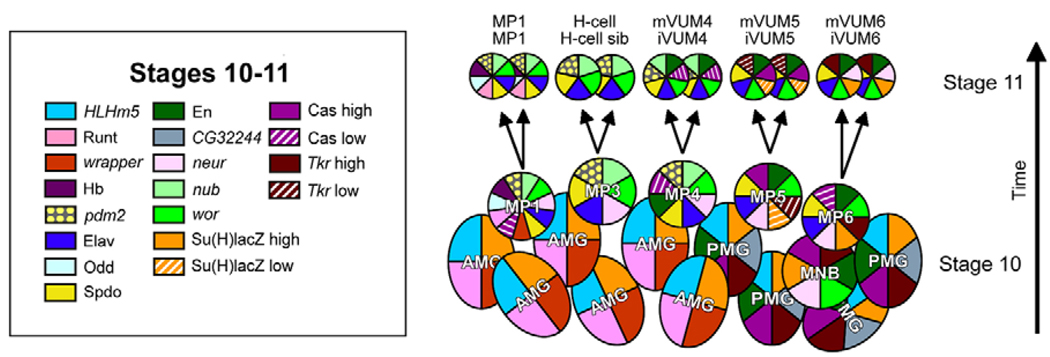
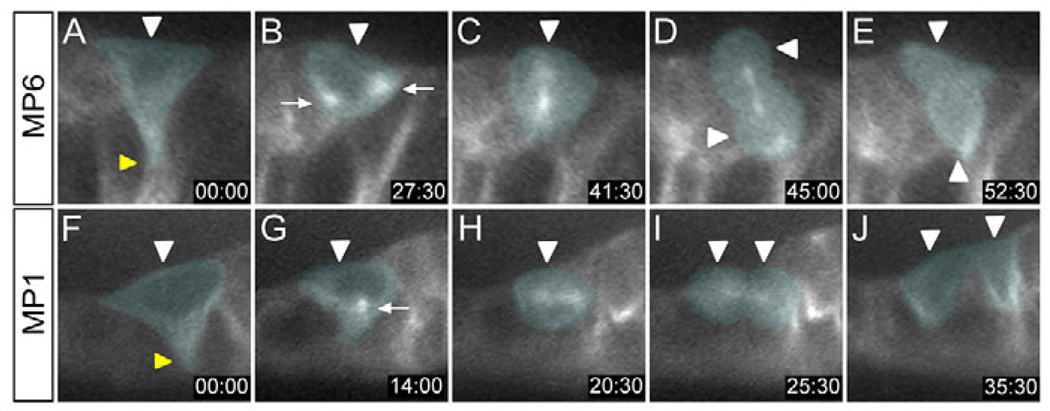
As a prelude to studying the molecular mechanisms that control MP neuronal cell fate decisions, it was important to identify the MPs and to determine when these cells divide. Previously, we generated molecular maps of stages 9, late 11, 13 and 17 (Wheeler et al., 2006), which allowed identification of individual midline cells. In this paper, we mapped the midline cell expression of 16 genes (Fig. 1 and see Fig. S1 in the supplementary material) at multiple periods during stages 10–11 of embryogenesis. Each of the MPs, the MNB, and their progeny, were defined and distinguished from each other by gene expression differences, position, size and visualization of cell division. These data provided strong evidence that MP divisions occur during stage 11, as confirmed by time-lapse imaging of midline cells in sim-Gal4 UAS-tau-GFP embryos (Fig. 2 and see Movie 1 in the supplementary material).
Fig. 1. Formation of midline precursors (MPs) and MP neurons in Drosophila.
Molecular map of stage 10 and 11 MPs and midline neurons (circles) and glia (ovals) shown in sagittal view. One segment is shown, with anterior to the left and interior (basal) at top. Each cell is depicted in terms of its pattern of gene expression as indicated by colors (the corresponding genes as listed on the left). The five MPs are shown at late stage 10, and the arrows indicate MPs dividing into their neuronal progeny at stage 11. The number of midline glia does not change appreciably from stage 10 to 11.
Fig. 2. Time-lapse imaging of sequential MP delamination and division.
Images in sagittal view, internal (basal) up, from time-lapse imaging of an (A–E) MP6 division and an (F–J) MP1 division. GFP fluorescence was visualized in sim-Gal4 UAS tau-GFP embryos during stage 11. Time is displayed as minutes:seconds. Relevant cells in each panel are pseudocolored. (A) Prior to division, the MP6 (white arrowhead) delaminates from the apical surface and takes on a triangular shape. The tip of the retracting cell is indicated by the yellow arrowhead. (B–D) During mitosis, (B) the centrosomes (arrows) move toward opposite poles, (C) the spindle fibers have an apical-basal orientation, and (D) the MP6 divides (arrowheads) along this axis. (E) Two MP6 neurons (arrowheads) are produced. (F) The MP1 (white arrowhead) delaminates from the apical surface, also acquiring a triangular shape (retraction point, yellow arrowhead). (G) The centrosomes (arrow) can be seen just before they separate and begin their migration. (H) The MP1 spindle maintains an orientation perpendicular to the apical-basal axis. (I,J) Cytokinesis results in the formation of two MP1 neurons (arrowheads).
At early stage 10, the midline cells constitute a monolayer along the anterior-posterior axis. However, beginning at late stage 10, MPs began to delaminate, and migrated basally (internally). As the cells migrated, they retracted a cytoplasmic process from the apical surface. The MP1,3,4 precursors acquired a flattened shape, resided internal to the MG, and were separated from other MPs by MG. The five MPs were arranged in a defined order, MP1→MP3→MP4→MP5→MP6 (anterior to posterior), within the segment. However, they delaminated and divided in the order MP4→MP3→MP5→MP1→MP6. The MP divisions were characterized by loss of an apical projection, retraction of the MG that separate the MPs, and the subsequent juxtaposition of neuronal progeny. The MP3–6 divisions were along the apical-basal axis, whereas the MP1 division was perpendicular to the apical-basal axis. After the MPs divided, the MNB delaminated posterior to the MP6 progeny and began dividing to generate ganglion mother cells (GMCs).
Notch signaling promotes midline glia, MNB and MP5,6 formation and inhibits MP1,3,4 formation
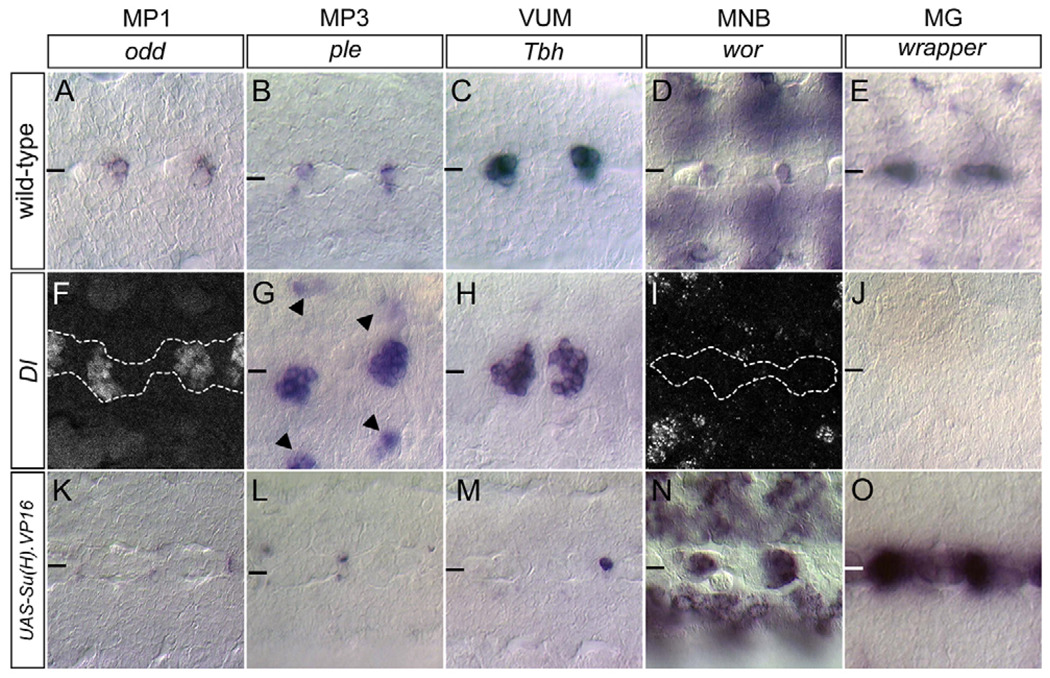
Based on the important roles of Notch signaling in CNS development, Delta (Dl) and Notch (N) mutants were screened for midline phenotypes, including alterations in expression of midline-expressed genes. In both Dl3 homozygotes and Dl3/Dl7 transheterozygotes, an increase was observed in the number of midline neurons at the expense of MG (Fig. 3). At stage 14, the number of MP1 neurons increased from two cells/segment to 9.3±1.6 (n=14 segments) cells (Fig. 3A,F). The number of H-cells increased from one cell/segment to 9.6±1.1 (n=17) (Fig. 3B,G), and the number of mVUMs increased from three cells/segment to 11.5±1.7 (n=51) (Fig. 3C,H). H-cell sib- and iVUM-specific gene expression was absent in Dl mutants (not shown). As described below, in the absence of Notch signaling, all MP3 neurons are H-cells and all VUMs are mVUMs owing to cell fate defects. Both MP1 and MP3 neurons increased ~5-fold in Dl mutant embryos. The VUM neurons, by contrast, increased only 2-fold.
Fig. 3. Notch signaling influences midline cell fate.
Ventral views of (A–E) wild-type, (F,G,I,J) Dl3/Dl3, (H) Dl3/Dl7 and (K–O) sim-Gal4 UASSu(H).VP16 stage 14 Drosophila embryos. Cell types are listed at the top of each column, and the gene or protein assayed that identifies each cell type is listed below. Horizontal bars indicate the location of the midline. (F,I) To differentiate (F) Odd+ and (I) wor+ midline cells from lateral CNS cells, embryos were double-stained with anti-Sim [not shown, but outlined (dashed line) to show location of midline cells]. In Dl mutants, there was an (F–H) increase in MP1, MP3 (H-cell) and mVUM neurons, and an absence of the (I) MNB and (J) MG. (G) Ectopic ple+ cells (arrowheads) were present off the midline; double-staining with anti-Sim indicated that these are not midline-derived (not shown). (K–O) sim-Gal4 UAS-Su(H).VP16 embryos showed the opposite phenotype to Dl mutants: (K–M) strong reduction of MP1, MP3 and mVUM neurons, and increases in (N) MNB and (O) MG.
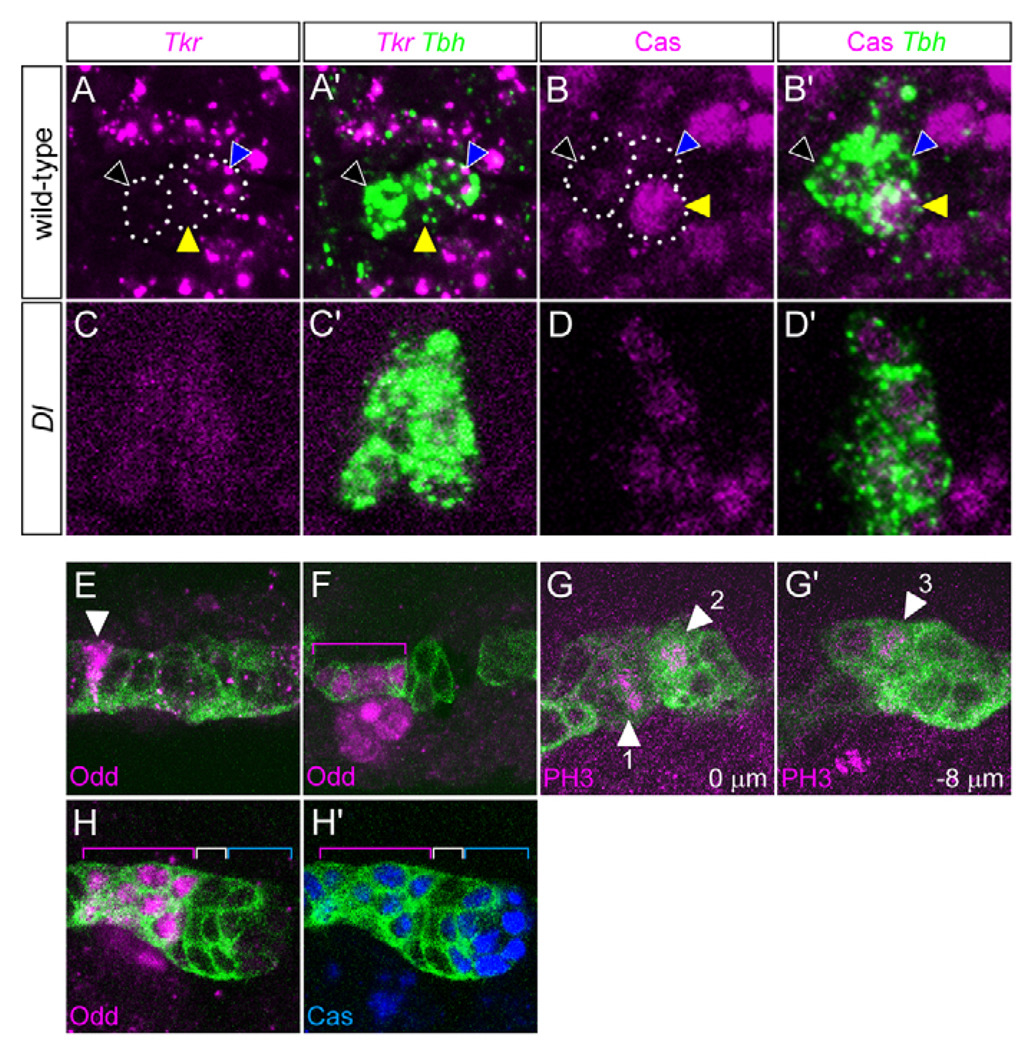
This disparity led us to investigate the identity of the mVUM neurons observed in Dl mutants. All mVUMs can be uniquely identified in the midline by Tyramine β hydroxylase (Tbh) expression, and mVUM4–6 can be distinguished from each other based on Tyrosine kinase-related protein (Tkr) and Castor (Cas) levels. The wild-type mVUM4 and mVUM5 neurons are Tkr−, whereas mVUM6 is Tkr+ (Fig. 4A). The expanded Tbh+ mVUMs in Dl mutants were Tkr− (Fig. 4C), indicating that none was mVUM6. The one significant difference between wild-type mVUM4 and mVUM5 is that mVUM4 has low levels of Cas (Caslo) and mVUM5 has high levels of Cas (Cashi) (Fig. 4B). Quantitation of Cas staining intensity was measured using the Mean Gray Value (MGV) function of ImageJ (Abramoff et al., 2004). In wild type (n=4 segments), mVUM4, mVUM5 and mVUM6 showed MGVs of 67, 155 and 22, respectively. In Dl mutants (n=6 segments), all Tbh+ cells showed similar MGVs with an average of 67, identifying them as mVUM4s. These results, together with the observation that the 11.5 mVUMs/segment observed in Dl mutants was close in number to the approximately nine MP1s and ten H-cells observed, suggested an expansion of a single VUM precursor, probably the MP4.
Fig. 4. MP number increases in the absence of Notch signaling.
(A–D′) Ventral views of (A–B′) wild-type and (C–D′) Dl3/Dl3 mutant stage 14 Drosophila embryos. (A,A′,C,C′) Single segments stained for Tkr (magenta) and Tbh (green). (A,A′) The mVUM6 (blue arrowhead) was Tkr+, whereas mVUM4 (black arrowhead) and mVUM5 (yellow arrowhead) were Tkr−. Dotted ovals outline the mVUMs. (C,C′) In Dl, Tkr expression was absent indicating that the excess Tbh+ cells were not mVUM6s. (B,B′,D,D′) Single segments stained for Cas (magenta) and Tbh (green). (B,B′) There are three Tbh+ mVUMs in each segment: mVUM4 (black arrowhead) was Caslo, mVUM5 (yellow arrowhead) was Cashi, and mVUM6 was Cas− (blue arrowhead). (D,D′) Excess Tbh+ cells in Dl mutants were Caslo, indicating that they were mVUM4s. (E–H′) Sagittal views of single segments of (E) wild-type and (F–H′) Dl3/Dl3 mutant embryos. Midline cells are defined as MPs based on their presence at stage 10 and relatively large size. (E) At mid-stage 10, there is a single Odd+ (magenta) MP1 (arrowhead). (F) In Dl, the number of Odd+ MP1s (bracket) was increased. (G,G′) Dl mutant embryo at two focal planes, 8 µm apart, showing three dividing cells (arrowheads 1–3) in close proximity, stained with anti-phosphohistone H3 (PH3, magenta). (H,H′) In Dl, there is an increase in Odd+ (magenta) Cas+ (blue) MP1 neurons (magenta bracket), Odd− Cas− MP3 neurons (white bracket), and Odd− Cas+ MP4 neurons (blue bracket).
The expanded numbers of MP1,3,4 neurons in Dl mutant embryos (~30) could be due to either: (1) a transformation of all of the ~16 midline cells to MPs1,3,4, followed by a single division of each MP; or (2) an overproliferation of one or a few MP1,3,4 cells, accompanied by the death or unrecognizable fate change of the other midline cells. This was tested by assaying stage 10–11 Dl mutant embryos for gene expression and positions and timing of cell division. Late stage 10 mutant embryos had an increased number of Odd-skipped (Odd)+ MP1s (4.1±1.2; n=17) (Fig. 4E,F). Live imaging of Dl mutant embryos during stage 11 indicated that the observable MP divisions occurred within a relatively short time interval (88±16 minutes) (see Movie 2 in the supplementary material). Divisions of closely juxtaposed cells were frequently observed to occur in close temporal sequence in both live imaging and fixed embryos stained for phosphohistone H3 (Fig. 4G). There was no evidence of cell death. Confocal imaging of stage 11 Dl embryos, after division, revealed 7.9±2.1 (n=19) Odd+ Cas+ MP1 neurons, 6.9±1.4 (n=12) Odd− Cas− MP3 neurons, and 10.0±2.2 (n=7) Odd− Cas+ MP4s (Fig. 4H). These data are most consistent with a model in which there is a transformation of ~16 midline cells into approximately five MP1s, five MP3s and six MP4s, followed by a single division of each MP.
In contrast to the expansion of MP1,3,4-derived neurons in Dl mutants, there was an absence of MG and of the MNB. MG gene expression was reduced from 10.0±1.3 (n=15) cells/segment in the wild type to 0.1±0.2 (n=176) cells/segment in Dl mutants (Fig. 3E,J). The wild-type MNB has prominent expression of three genes: worniu (wor) (Fig. 3D), miranda (mira) (not shown), and sanpodo (spdo) (not shown), which are specific to the MNB after stage 11. In stage 14 Dl mutant embryos, wor (n=84) (Fig. 3I), mira (n=56) and spdo (n=47) expression was absent from the midline. Involvement of the Notch receptor was confirmed by analysis of a N mutant combination, Nts1/N55e11, that showed similar phenotypes to Dl mutants, although at a reduced frequency (not shown).
Notch activation converts MPs to midline glia
In experiments complementary to Notch and Dl mutant analyses, sim-Gal4 was used to misexpress constitutively-active Suppressor of Hairless.VP16 [Su(H).VP16] (Kidd et al., 1998) in all midline cells. Stage 14 sim-Gal4 UAS-Su(H).VP16 embryos were examined because MG undergo apoptosis beginning at stage 15. At stage 14, these embryos showed a 3-fold increase in the number of MG (30±5.5 cells/segment, n=19) compared with wild type (10.0±1.3, n=15) (Fig. 3E,O, and see Fig. S2A–H in the supplementary material). The expanded MG had wild-type properties: they underwent apoptosis, both AMG and PMG were present, and they wrapped commissural axons. In addition, there was a near complete absence of midline axons (see Fig. S3A,B in the supplementary material) and fewer than one MP-derived neuron/segment was present (Fig. 3K–M). The larval and adult phenotypes of these midline neuron-less animals were assessed: 62% of embryos survived to adulthood, but were female sterile (see Fig. S3C in the supplementary material), and larvae had reduced motility (see Fig. S3D in the supplementary material). When sim-Gal4 UAS-Su(H).VP16 embryos were stained for the MNB markers wor (Fig. 3N), mira and spdo, there was an increase in cell number from one cell/segment in the wild type to 4.9±1.8 (n=12). These wor+ cells also had MG gene expression. The expansion of MNB gene expression was consistent with the Dl mutant data indicating that Notch signaling was required for MNB formation. By contrast, there was no evidence that Su(H).VP16 misexpression resulted in additional MP5,6 progeny.
To further understand the spatial and temporal dynamics of midline Notch signaling, the expression of two reporters of Su(H) activity was examined: P[12xSu(H)bs-lacZ] (Go et al., 1998) and Gbe-lacZ (Furriols and Bray, 2001). Reporter expression was observed in AMG and PMG during stage 10, and was maintained through to the end of embryogenesis, although levels were low by stage 17 (see Fig. S2I–L in the supplementary material). Expression was dependent on Notch signaling, as it was absent in the CNS midline cells in Dl mutant embryos (see Fig. S2M,N in the supplementary material). In addition to MG, expression of P[12xSu(H)bs-lacZ] was present in MP5,6 and in the MNB (see Fig. S2I–K in the supplementary material) during stage 11, prior to their division. MP5 expresses a low level of P[12xSu(H)bs-lacZ], MP6 an intermediate level, and the MNB higher levels. After division, the MP5,6 and MNB progeny express P[12xSu(H)bs-lacZ] at the same relative levels as the precursors (see Fig. S2L in the supplementary material). The neuronal expression is maintained throughout embryogenesis. No expression of the reporter was observed in MP1,3,4 or their progeny. The expression pattern of Gbe-lacZ was similar, although levels of lacZ expression were reduced compared with P[12xSu(H)bs-lacZ]. These data indicate that Notch signaling is occurring in MG, MP5, MP6 and the MNB during stages 10–11, consistent with genetic requirements for Notch signaling in these cells.
numb and spdo direct sibling neuronal fates in MP asymmetric divisions
MPs either divide symmetrically (MP1) or asymmetrically (MP3–6). A possible mechanism for generating MP asymmetric cell fates is asymmetric localization of Numb in conjunction with Notch signaling. To assess cell fate in Dl, numb and spdo mutant and overexpression embryos, the MP1, MP3 and VUM neurons were analyzed for changes in the expression of 37 genes, which encode transcription factors, signaling molecules, neurotransmitter biosynthetic enzymes, neurotransmitter receptors and neuropeptide receptors. Additionally, axonal trajectories were analyzed based on sim-Gal4 UAS-tauGFP visualization.
MP3 neurons
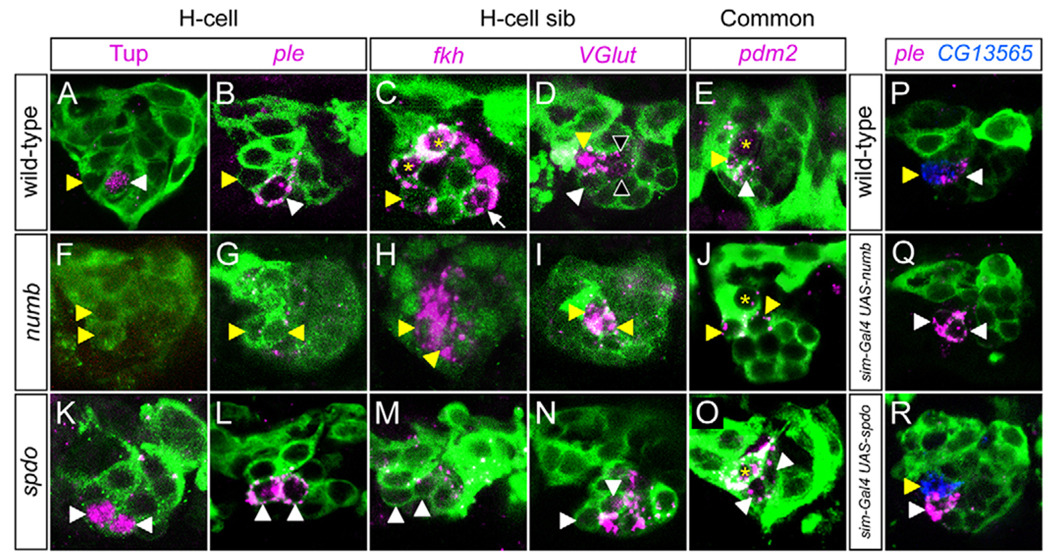
Analysis of 19 genes expressed in the H-cell and H-cell sib neurons showed that H-cell-specific gene expression was absent in numb mutant embryos (Fig. 5A,B,F,G), but was present in both neurons in spdo mutants (Fig. 5K,L). The opposite results were observed for H-cell sib-specific gene expression (Fig. 5C,D,H,I,M,N). Another indicator of neuronal cell fate is axonal trajectory. Consistent with the gene expression results, numb mutants showed an absence of H-cell axons and the presence of H-cell sib axons, whereas spdo mutants showed the opposite phenotype (see Fig. S4A–C in the supplementary material). These results were confirmed by analysis of H-cell gene expression in numb-overexpression and Dl mutant embryos. When numb was overexpressed in all midline cells, there were two pale (ple)+ cells (H-cells), an absence of CG13565+ H-cell sib, and a duplication of H-cell axons (Fig. 5P,Q; see Fig. S4D in the supplementary material). Overexpression of spdo did not result in cell fate defects (Fig. 5R). Analysis of Dl mutant embryos revealed an expansion of neurons derived from the MP3. Only ple+ H-cells (Fig. 3G), and not CG13565+ H-cell sibs (data not shown), were present. Four genes, including POU domain protein 2 (pdm2) (Fig. 5E,J,O), that are expressed in both cell types had no alterations in expression in either numb or spdo mutant embryos, indicating that numb and spdo affect cell type-specific gene expression, but not expression present in both cells. Thus, assays of both neuronal morphology and gene expression indicated that Notch controls all of the divergent aspects of H-cell versus H-cell sib cell fate.
Fig. 5. numb and spdo control MP3 neuronal cell fate.
Confocal images of stage 14–15 Drosophila embryos in sagittal view. (A–E,P) Wild type, (F–J) numb4/numb4, (K–O) spdoG104/spdoG104, (Q) sim-Gal4 UAS-numb and (R) sim-Gal4 UAS-spdo. All embryos had sim-Gal4 UAS-tau-GFP (green) in the background, except H, which shows anti-Sim (green) staining. To identify the MP3 neurons, numb mutants were double-labeled with Vesicular glutamate transporter (VGlut) (not shown, except in I); spdo mutants were double-labeled with ple (not shown, except in L). In A–O, white arrowheads denote cells expressing H-cell genes and yellow arrowheads indicate cells expressing H-cell sib genes. (A,B) In wild type, Tailup (Tup) protein and ple were present in the H-cell, and absent from H-cell sib. (F,G) In numb, Tup and ple were absent from both MP3 neurons. (K,L) In spdo, Tup and ple were present in both MP3 neurons. (C) In wild type, fork head (fkh) was expressed in H-cell sib, the two MP1 neurons (*) and iVUMs; only one iVUM (arrow) is present in this focal plane. (H) In numb, fkh was expressed in two Sim+ MP3 neurons, and was absent (M) from spdo MP3 neurons. (D) In wild type, VGlut was expressed in H-cell sib and at a lower level in mVUMs (black arrowheads), whereas (I) in numb, VGlut was expressed in two MP3 neurons and absent from VUM neurons. By contrast, (N) in spdo, the two MP3 neurons (arrowheads) lacked VGlut, whereas it was present in all VUM neurons. (E) pdm2 was expressed in the MP1 (*) neurons and in both MP3 neurons in wild type (only one MP1 neuron is present in this focal plane). The expression of pdm2 was unaltered in (J) numb and (O) spdo. (P–R) Overexpression of numb, but not spdo, causes an MP3 cell fate change. The H-cell is marked by ple expression (magenta) and H-cell sib by CG13565 expression (blue). (P) Wild-type expression of ple and CG13565. (Q) In sim-Gal4 UAS-numb, H-cell sib was transformed into an H-cell, as shown by the presence of two ple+ cells and the absence of CG13565-expressing cells. (R) sim-Gal4 UAS-spdo showed a wild-type pattern of gene expression with a single ple+ cell and a single CG13565+ cell.
VUM neurons
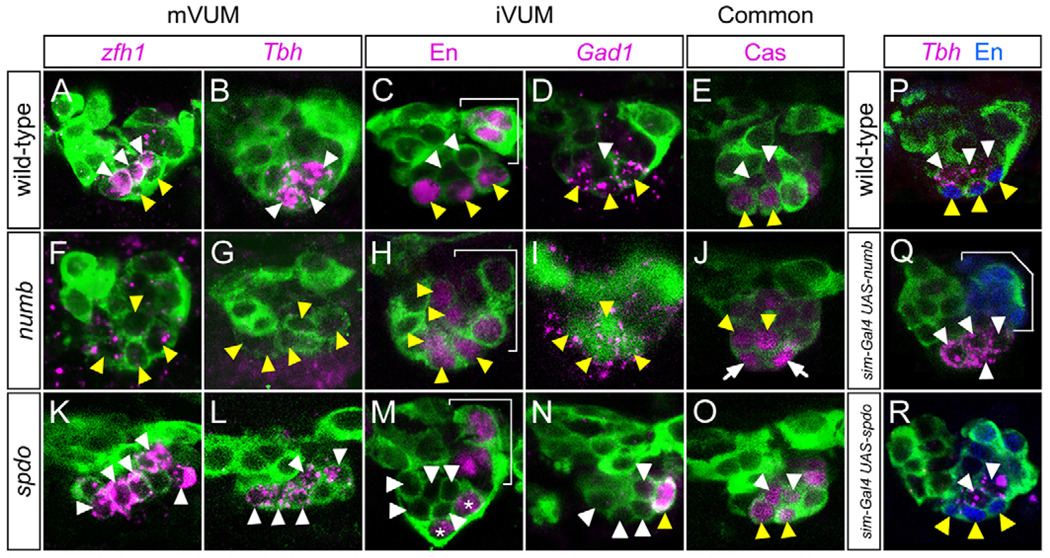
The expression of 21 VUM neuron-expressed genes (see Table S1 in the supplementary material) was examined in numb and spdo mutants. mVUM-specific gene expression was absent in numb mutant embryos and expanded in spdo mutants (Fig. 6A,B,F,G,K,L). By contrast, iVUM-specific gene expression was expanded in numb mutants and absent from spdo mutants (Fig. 6C,D,H,I,M,N). In numb mutant embryos, the mVUM axons were absent and the iVUM axons appeared thickened, suggesting a duplication; spdo mutants had the opposite phenotype (see Fig. S4E–G in the supplementary material). Embryos mutant for Dl showed an increase in Tbh+ mVUMs (Fig. 3H), but lacked CG15236+ iVUMs (data not shown). In sim-Gal4 UAS-numb embryos, Tbh expression (mVUMs) was expanded to six cells and Engrailed (En) (iVUMs) was absent (Fig. 6P,Q). Furthermore, mVUM but not iVUM axons were present (see Fig. S4H in the supplementary material). Analysis of sim-Gal4 UAS-spdo did not show alterations in VUM cell fate (Fig. 6R). Genes expressed in both iVUMs and mVUMs showed no alterations in expression in either numb or spdo mutants (Fig. 6E,J,O). In conclusion, Notch signaling, in conjunction with numb and spdo, controls iVUM/mVUM asymmetric cell fate choices.
Fig. 6. numb and spdo control VUM neuronal cell fate.
Confocal images of stage 14–15 Drosophila embryos in sagittal view. (A–E,P) Wild-type, (F–J) numb4/numb4, (K–O) spdoG104/spdoG104, (Q) sim-Gal4 UAS-numb and (R) sim-Gal4 UAS-spdo. All embryos had sim-Gal4 UAS-tau-GFP (green) in the background, except I, which shows anti-Sim (green) staining. To identify the VUM neurons, numb mutants were double-labeled for En (not shown, except in H); spdo mutants were double-labeled with Tbh (not shown, except in L). In A–O, white arrowheads indicate cells expressing mVUM genes and yellow arrowheads indicate cells expressing iVUM genes. (A) In wild type, Zn finger homeodomain 1 (zfh1) was present in all three mVUMs and not in the iVUMs. (F) In numb, zfh1 expression was absent. (K) In spdo, zfh1 expression was expanded to five VUMs. (B) Tbh was expressed in three mVUMs in wild type. (G) In numb, Tbh was not expressed. (L) In spdo, five VUMs expressed Tbh. (C) In wild type, En was present in three iVUMs as well as other cell types, including the PMG (bracket). (H) In numb, En was present in five VUMs in addition to the PMG (bracket). (M) In spdo, En was absent from VUMs, but was present in the PMG (asterisk and bracket). (D) Glutamic acid decarboxylase 1 (Gad1) was expressed in three iVUMs in wild type. (I) Gad1 expression was expanded to six cells in numb (four of the six VUMs can be seen in this focal plane). (N) In spdo, Gad1 expression was present in only one VUM. (E) In wild type, Cas was present in two iVUMs (iVUM4,5) and two mVUMs (mVUM4,5). In (J) numb and (O) spdo mutant embryos, Cas was also present in iVUM4,5 and mVUM4,5. (P–R) Overexpression of numb causes a VUM cell fate change. (P) Wild-type expression of Tbh (magenta) in three mVUMs and of En protein (blue) in three iVUMs. (Q) In sim-Gal4 UAS-numb, six ventral Tbh+ En− mVUMs (two of the six cells are absent in this focal plane) were present. En in PMG (bracket) was unaffected. (R) sim-Gal4 UAS-spdo had a wild-type Tbh and En pattern (two of three Tbh+ mVUMs are present in this image).
MP1 neurons
The MP1 neurons are unique among MP progeny in that they appear identical. Consequently, their development might be independent of numb and spdo regulation. This was addressed by examining mutant and overexpression embryos for ten MP1-expressed genes (see Table S1 and Fig. S5 in the supplementary material). There were no alterations in MP1 neuronal gene expression in numb, spdo or sim-Gal4 UAS-numb embryos (see Fig. S5 in the supplementary material), nor were there alterations in MP1 neuronal axonal trajectories (see Fig. S5A–E,G in the supplementary material). These data indicate that numb and spdo do not play a role in the cell fate specification of MP1 neurons. In Dl mutant embryos, we observed an expanded set of neurons that are Hunchback+ and Odd+ (Fig. 3F); within the midline, these genes are specific for the MP1 neurons. Taken together, the Dl, numb and spdo mutant results suggest that Notch signaling is not important for MP1 cell fate determination.
Numb and Spdo are localized asymmetrically during MP3–6, but not MP1, divisions
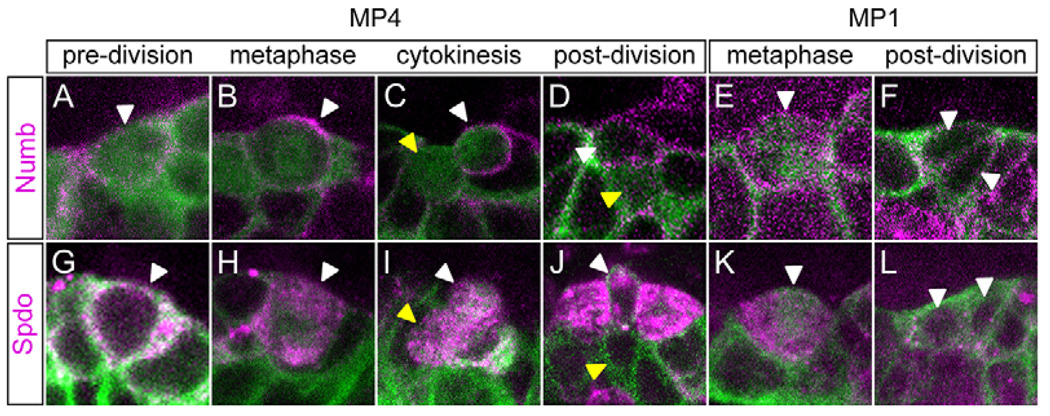
Analysis of the numb and spdo mutant phenotypes suggested that Numb and Spdo proteins would be asymmetrically localized during MP divisions. Our analysis showed that Numb localization was regulated in a cell-cycle-dependent manner in MP3–6. Prior to mitosis, Numb was localized uniformly around the MP cell membrane (Fig. 7A), then became enriched along the basolateral surface (Fig. 7B), and finally segregated into only the basal H-cell daughter (Fig. 7C,D). During mitosis (Fig. 7G–I), Spdo was localized around the MP membrane and in puncta throughout the cytoplasm. Immediately after division, Spdo was localized uniformly around the membrane of the Numb− daughter cell at a low level (Fig. 7J), whereas in the Numb+ cell the membrane localization of Spdo was reduced, being instead found in intracellular puncta (Fig. 7J). These puncta are likely to be intracellular vesicles (Hutterer and Knoblich, 2005; O’Connor-Giles and Skeath, 2003). In summary, MPs asymmetrically generate a Numb+ intracellular Spdo+ neuron (H-cell, mVUM) and a Numb− cortical Spdo+ neuron (H-cell sib, iVUM).
Fig. 7. Numb and Spdo localization during MP divisions.
Confocal images of MP1 and MP4 divisions in sim-Gal4 UAS-tau-GFP (green) stage 11 Drosophila embryos stained with (A–F) anti-Numb (magenta) and (G–L) anti-Spdo (magenta). Sagittal views with anterior left and internal (basal) up. White arrowheads indicate: (A,B,G,H) MP4, (C,D) Numb+ VUM4 neuron, (I,J) basal VUM4 neuron with cytoplasmic punctate Spdo, (E,K) MP1 and (F,L) MP1 neurons. Yellow arrowheads indicate: (C,D) Numb− VUM4 neuron, (I) apical VUM4 neuron with cytoplasmic punctate Spdo and (J) membranous Spdo VUM4 neuron.
What happens in the MP1, which generates two identical neurons? In this case, Numb was uniformly localized to the membrane prior to, during and after MP1 cell division (Fig. 7E,F). Spdo was found at the membrane and in cytoplasmic puncta prior to and during division, and in both progeny after division (Fig. 7K,L). Although Numb is present in both MP1 neurons, other mechanisms must cause these cells to be refractory to Notch signaling because numb mutants do not exhibit changes in MP1 gene expression.
DISCUSSION
Patterns of stage 10–11 midline cell divisions and gene expression
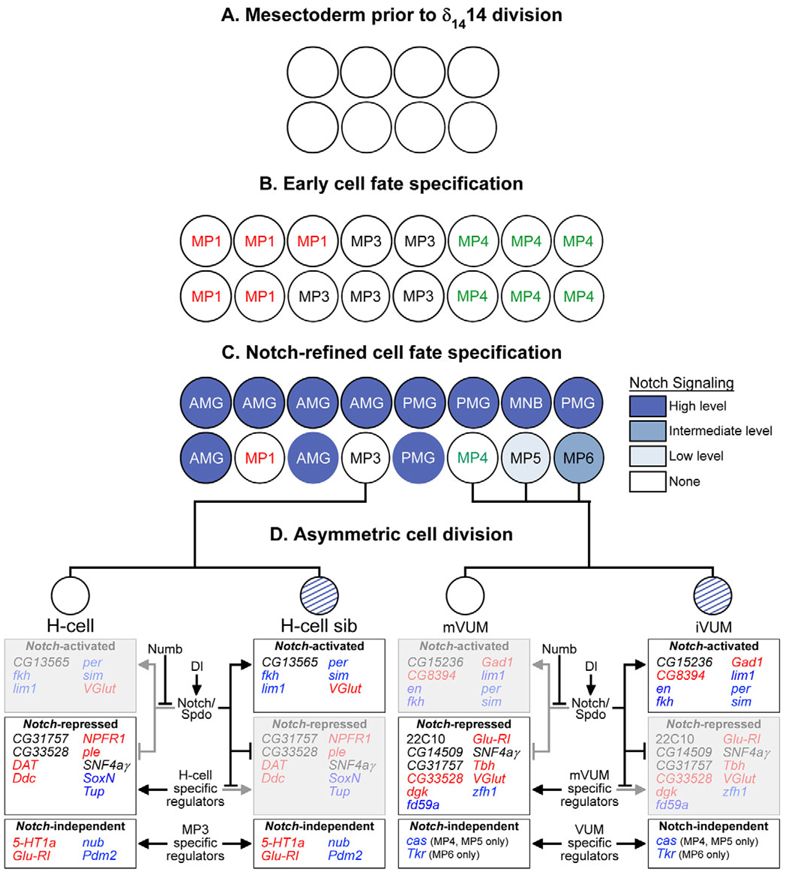
The Drosophila MPs form at specific positions and divide in a reproducible sequence. Descriptive work in grasshopper indicated that MPs each undergo a single division (Bate and Grunewald, 1981; Goodman et al., 1981; Jia and Siegler, 2002). We propose that the Drosophila cells described here are homologous, and that MP4 gives rise to the anterior pair of VUMs (VUM4s), MP5 to the medial VUM pair (VUM5s), and MP6 to the posterior VUM pair (VUM6s). This picture of Drosophila stage 11 MP divisions runs counter to the prevailing Drosophila models, which propose that the MP divisions occur at stage 8 during the δ1414 synchronous cell division (Bossing and Technau, 1994; Jacobs, 2000; Klambt et al., 1991). Instead, we propose that the precursors dividing at stage 8 give rise to glial-glial, neuronal-neuronal and mixed glial-neuronal lineages (Fig. 8). In general, this new model fits DiI-labeling data from previous reports in which mixed clones were noted (Bossing and Technau, 1994; Schmid et al., 1999), but no compelling arguments put forward for how they arose.
Fig. 8. Model of Notch regulation of midline cell fate in Drosophila.
(A) Mesectodermal cells meet at the ventral midline before the stage 8 δ1414 division. Little is known regarding influences on midline cell development at this stage. (B) After the stage 8 division, but before Notch signaling, the 16 midline cells can be considered as three equivalent groups of cells: MP1s, MP3s and MP4s. (C) After Notch signaling, the 16 cells acquire specific fates, and differ in their levels of Notch signaling as indicated by the expression of Su(H)-lacZ reporter. For simplicity, midline cells are shown as paired cells along the anterior-posterior axis. The precise anterior-posterior and left-right positions of individual cells are unknown, except that AMG, MP1 and MP3 tend to reside in the anterior half, and PMG, MP5, MP6 and MNB in the posterior half. Different shades of blue indicate relative levels of Notch signaling. (D) Asymmetric cell division. Notch signaling is required for MP3–6 asymmetric cell fates. Notch signaling is active (blue diagonals) in H-cell sib and iVUMs, and inhibited in H-cell and mVUMs. Assayed genes expressed in the MP3 and VUM lineages are shown below each neuron: bold text indicates expression, whereas the light, shaded text indicates repression. Genes are categorized as either Notch-activated, Notch-repressed or Notch-independent. Functional classes of genes are color-coded: transcription factors (blue), neural function genes (red) and others (black).
Notch signaling directs the formation of midline glia and inhibits neurogenesis
Dl mutant and Su(H) misexpression experiments indicated that: (1) Notch signaling is required for the formation of both AMG and PMG; (2) Dl is a ligand for N; and (3) transcriptional output involves Su(H) beginning at stage 10. Consistent with these results, analysis of a Nts mutant showed changes in expression in MG and neuronal enhancer-trap lines, but lacked specific markers to fully characterize the phenotype (Menne and Klambt, 1994). Genes of the Enhancer of split-Complex [E(spl)-C] are commonly activated by Notch signaling and repress transcription. We note that the HLHm5 E(spl)-C gene is expressed specifically in MG at stages 10–11 (Fig. 1), and other E(spl)-C members are also expressed in midline cells (Kearney et al., 2004; Wech et al., 1999). While E(spl)-C genes could be direct targets of Su(H) and repress midline neuronal gene expression in MG, what activates MG gene expression? The sim gene was previously shown to activate MG gene transcription (Ma et al., 2000; Wharton et al., 1994), and could be a direct target of Su(H).
Dl mutants not only showed a complete lack of MG gene expression, but also an expansion of anterior midline neurons (MP1,3 and VUM4) and absence of posterior neurons (VUM5,6 and MNB). Expanded MP1s have also been noted in work describing the role of Notch signaling in MP2 development (Spana and Doe, 1996). Do the expanded Dl mutant MP1,3,4 neurons result from transformation of MG precursors to MPs, or from excessive division of a small number of MPs? Analysis of Dl mutants at stages 10–11 suggests that the midline cells at these stages consist of approximately five MP1s, five MP3s and six MP4s. If each divided once, this would equal the ten MP1 neurons, ten H-cells and 12 mVUM4s observed in Dl mutant embryos at later stages. In this model (Fig. 8), Notch signaling promotes MG development, while MP1, MP3 and MP4 are selected from their respective MP fields. This midline role for Notch parallels known functions of Notch in both Drosophila and vertebrates, in which it promotes gliogenesis and inhibits neurogenesis (Gaiano et al., 2000; Morrison et al., 2000; Udolph et al., 2001).
Notch signaling promotes MNB and MP5,6 formation
The progeny of MP5,6 and the MNB were absent from Dl mutants, indicating that Notch signaling is required for the formation of the MNB as well as for VUM5,6. This was a surprising result for the MNB because in the ventral nerve cord, Notch signaling inhibits NB formation early in development (Campos-Ortega, 1993) and plays no apparent role in the asymmetric division of postembryonic nerve cord NBs (Almeida and Bray, 2005). However, Notch signaling controls central brain NB number (Lee et al., 2006; Wang et al., 2007), indicating a parallel between the MNB and brain NBs. Thus, the MNB has a number of properties distinct from other nerve cord NBs in that it is not part of a neural/epidermal equivalence group and does not utilize the Hunchback>Krüppel>Pdm>Cas cascade (Isshiki et al., 2001). Similarly, it is unusual that VUM5,6 require Notch function, as Notch signaling inhibited MP1,3 and MP4 neurogenesis. Consistent with the genetic data, P[12xSu(H)bs-lacZ] expression is restricted to MP5,6 and the MNB. This suggests that the different responses to Notch signaling might reflect anterior-posterior location. However, there might also be differences with respect to cell type, because sim-Gal4 UAS-Su(H).VP16 embryos have expanded MNB-like cells, but the MP5,6 cells were not expanded. One potential model involves successive waves of signaling, by Notch or other signaling molecules, to generate the MNB, MP5,6 and MG, similar to what happens during development of the Drosophila retina (e.g. Doroquez and Rebay, 2006). Bossing and Brand have proposed an equivalence group in which Notch signaling would inhibit cells from becoming a MNB, and instead promote the VUM cell fate (Bossing and Brand, 2006). However, our Dl mutant and Su(H).VP16 misexpression data indicate that Notch signaling promotes, not inhibits, MNB formation. Another view is that the presence of PMG is required for MP5,6 and MNB formation, and that the absence of PMG in Dl mutants also results in the loss of the neural precursors. In summary, alterations in Notch signaling have revealed its requirement in the formation of MP5,6 and the MNB, but additional work will be required for mechanistic insight.
Notch signaling and numb generate asymmetric midline neuronal cell fates
Asymmetric neuronal cell fates of MP3–6 progeny are determined by Numb and Spdo asymmetric localization in one of the two daughter cells (Fig. 8), similar to asymmetric cell fate determination of the non-midline MP2 cell and GMCs (O’Connor-Giles and Skeath, 2003; Spana and Doe, 1996; Spana et al., 1995). In the H-cell sib and iVUMs, Numb is absent, and Notch signaling, in combination with cortical Spdo, activates H-cell sib- and iVUM-specific gene expression and represses H-cell and mVUM gene expression. Genes that are expressed in both siblings are not dependent on Notch signaling. The MP1 progeny are identical by gene expression and morphological criteria. Numb is present in both MP1 neurons, but the significance of this is unclear because MP1 gene expression and morphology were unaffected in numb mutants; nor were defects observed in Dl mutants. This suggests that Notch signaling does not influence MP1 development.
Another difference between MP1 and the other MPs is that MP1 divides perpendicular to the apical-basal axis, whereas MP3–6 rotate their spindles during cell division along the apical-basal axis. The basal cell is always the Numb+ cell, which is the Notch-independent H-cell or mVUM. The orientations of the divisions might aid in positioning the cells towards their final locations in the CNS. In the mature CNS, the iVUMs are apical to the mVUMs, and during MP divisions the iVUM is the more apical sibling. In the case of the MP1s, it might be important that both cells are in the same position along the basal/apical axis.
Kuwada and Goodman examined the development of grasshopper MP3 (Kuwada and Goodman, 1985). Their data suggested a model in which the two MP3 neurons are born equivalent with an H-cell sib dominant fate, and, within 5 hours, signaling between the two cells generates different fates. These data appear inconsistent with the Drosophila results, as the Drosophila MP3 neurons asymmetrically localize Numb and are inherently different at birth. However, it is important to recognize that the grasshopper and Drosophila results are based on different types of experiments (genetic versus experimental ablation), and the grasshopper data might be revealing additional levels of regulation or different mechanisms for generating cell fates.
Towards a molecular basis for neuronal and glial cell fate determination
Nearly 300 genes are known to be expressed in the developing Drosophila CNS midline cells, and many have been mapped at the single-cell level by confocal microscopy. The work described here examined the role of Notch signaling in the expression of 37 MG-and neuronal-expressed genes (Fig. 8). Molecular analysis can now be carried out on these genes to identify direct targets of Notch action. Additional studies are also beginning to identify transcription factors that regulate the Notch-independent neuronal pathways (our unpublished results). The large number of genes identified, in combination with the utility of Drosophila molecular and genetic tools, will facilitate a detailed understanding of the regulatory pathways controlling midline neurogenesis and gliogenesis.
Supplementary Material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/135/18/3071/DC1
Acknowledgments
We thank Amaris Guardiola, Catarina Homem, Joe Kearney, Tony Perdue, Nasser Rusan and Ferrin Wheeler for valuable advice and assistance; Stephanie Freer for investigation of larval phenotypes; and Joe Pearson for helpful comments on the manuscript. We are grateful to Spyros Artavanis-Tsakonas, Nipam Patel and Francois Schweisguth for contributing antibodies and fly stocks; and are particularly appreciative of the endless advice and supplies from Jim Skeath and Beth Wilson. We also thank the Developmental Studies Hybridoma Bank for monoclonal antibodies and the Bloomington Drosophila Stock Center for fly stocks. This work was supported by NIH grant R37 RD25251 to S.T.C., an NRSA postdoctoral fellowship to S.R.W., and UNC Developmental Biology NIH training grant support to S.B.S.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with Image. J. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Almeida MS, Bray SJ. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 2005;122:1282–1293. doi: 10.1016/j.mod.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Bate CM, Grunewald EB. Embryogenesis of an insect nervous system II: a second class of neuron precursor cells and the origin of the intersegmental connectives. J. Embryol. Exp. Morphol. 1981;61:317–330. [PubMed] [Google Scholar]
- Bossing T, Technau GM. The fate of the CNS midline progenitors in Drosophila as revealed by a new method for single cell labelling. Development. 1994;120:1895–1906. doi: 10.1242/dev.120.7.1895. [DOI] [PubMed] [Google Scholar]
- Bossing T, Brand AH. Determination of cell fate along the anteroposterior axis of the Drosophila ventral midline. Development. 2006;133:1001–1012. doi: 10.1242/dev.02288. [DOI] [PubMed] [Google Scholar]
- Brand A. GFP in Drosophila. Trends Genet. 1995;11:324–325. doi: 10.1016/s0168-9525(00)89091-5. [DOI] [PubMed] [Google Scholar]
- Broihier HT, Skeath JB. Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron. 2002;35:39–50. doi: 10.1016/s0896-6273(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA. Early neurogenesis in Drosophila melanogaster. In: Bate M, Arias AM, editors. The Development of Drosophila melanogaster. vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1091–1129. [Google Scholar]
- Crews ST. Drosophila bHLH-PAS developmental regulatory proteins. In: Crews ST, editor. PAS Proteins: Regulators and Sensors of Development and Physiology. New York: Kluwer Academic Publishers; 2003. pp. 69–108. [Google Scholar]
- Doroquez DB, Rebay I. Signal integration during development: mechanisms of EGFR and Notch pathway function and cross-talk. Crit. Rev. Biochem. Mol. Biol. 2006;41:339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- Foe VE. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 1989;107:1–22. [PubMed] [Google Scholar]
- Furriols M, Bray S. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 2001;11:60–64. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Go MJ, Eastman DS, Artavanis-Tsakonas S. Cell proliferation control by Notch signaling in Drosophila development. Development. 1998;125:2031–2040. doi: 10.1242/dev.125.11.2031. [DOI] [PubMed] [Google Scholar]
- Goodman CS, Bate M, Spitzer NC. Embryonic development of identified neurons: origin and transformation of the H cell. J. Neurosci. 1981;1:94–102. doi: 10.1523/JNEUROSCI.01-01-00094.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer A, Knoblich JA. Numb and alpha-Adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep. 2005;6:836–842. doi: 10.1038/sj.embor.7400500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Jacobs JR. The midline glia of Drosophila: a molecular genetic model for the developmental functions of glia. Prog. Neurobiol. 2000;62:475–508. doi: 10.1016/s0301-0082(00)00016-2. [DOI] [PubMed] [Google Scholar]
- Jia XX, Siegler MV. Midline lineages in grasshopper produce neuronal siblings with asymmetric expression of Engrailed. Development. 2002;129:5181–5193. doi: 10.1242/dev.129.22.5181. [DOI] [PubMed] [Google Scholar]
- Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney JB, Wheeler SR, Estes P, Parente B, Crews ST. Gene expression profiling of the developing Drosophila CNS midline cells. Dev. Biol. 2004;275:473–492. doi: 10.1016/j.ydbio.2004.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S, Lieber T, Young MW. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klambt C, Jacobs JR, Goodman CS. The midline of the Drosophila central nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- Kosman D, Small S, Reinitz J. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev. Genes Evol. 1998;208:290–294. doi: 10.1007/s004270050184. [DOI] [PubMed] [Google Scholar]
- Kuwada JY, Goodman CS. Neuronal determination during embryonic development of the grasshopper nervous system. Dev. Biol. 1985;110:114–126. doi: 10.1016/0012-1606(85)90069-7. [DOI] [PubMed] [Google Scholar]
- Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 2006;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zwiebel LJ, Hinton D, Benzer S, Hall JC, Rosbash M. The period gene encodes a predominantly nuclear protein in adult Drosophila. J. Neurosci. 1992;12:2735–2744. doi: 10.1523/JNEUROSCI.12-07-02735.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Certel K, Gao Y, Niemitz E, Mosher J, Mukherjee A, Mutsuddi M, Huseinovic N, Crews ST, Johnson WA, et al. Functional interactions between Drosophila bHLH/PAS, Sox, and POU transcription factors regulate CNS midline expression of the slit gene. J. Neurosci. 2000;20:4596–4605. doi: 10.1523/JNEUROSCI.20-12-04596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menne TV, Klambt C. The formation of commissures in the Drosophila CNS depends on the midline cells and on the Notch gene. Development. 1994;120:123–133. doi: 10.1242/dev.120.1.123. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000;101:499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- O’Connor-Giles KM, Skeath JB. Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev. Cell. 2003;5:231–243. doi: 10.1016/s1534-5807(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Patel NH, Martin-Blanco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Doe CQ. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development. 1998;125:1857–1865. doi: 10.1242/dev.125.10.1857. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Spana EP, Kopczynski C, Goodman CS, Doe CQ. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development. 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- Thomas JB, Crews ST, Goodman CS. Molecular genetics of the single-minded locus: a gene involved in the development of the Drosophila nervous system. Cell. 1988;52:133–141. doi: 10.1016/0092-8674(88)90537-5. [DOI] [PubMed] [Google Scholar]
- Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131:5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
- Udolph G, Rath P, Chia W. A requirement for Notch in the genesis of a subset of glial cells in the Drosophila embryonic central nervous system which arise through asymmetric divisions. Development. 2001;128:1457–1466. doi: 10.1242/dev.128.8.1457. [DOI] [PubMed] [Google Scholar]
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- Wang H, Ouyang Y, Somers WG, Chia W, Lu B. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature. 2007;449:96–100. doi: 10.1038/nature06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Younger-Shepherd S, Jan LY, Jan YN. Only a subset of the binary cell fate decisions mediated by Numb/Notch signaling in Drosophila sensory organ lineage requires Suppressor of Hairless. Development. 1997;124:4435–4446. doi: 10.1242/dev.124.22.4435. [DOI] [PubMed] [Google Scholar]
- Ward EJ, Skeath JB. Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development. 2000;127:4959–4969. doi: 10.1242/dev.127.22.4959. [DOI] [PubMed] [Google Scholar]
- Ward MP, Mosher JT, Crews ST. Regulation of bHLH-PAS protein subcellular localization during Drosophila embryogenesis. Development. 1998;125:1599–1608. doi: 10.1242/dev.125.9.1599. [DOI] [PubMed] [Google Scholar]
- Wech I, Bray S, Delidakis C, Preiss A. Distinct expression patterns of different enhancer of split bHLH genes during embryogenesis of Drosophila melanogaster. Dev. Genes Evol. 1999;209:370–375. doi: 10.1007/s004270050266. [DOI] [PubMed] [Google Scholar]
- Wharton KA, Jr, Franks RG, Kasai Y, Crews ST. Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation. Development. 1994;120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Kearney JB, Guardiola AR, Crews ST. Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells. Dev. Biol. 2006;294:509–524. doi: 10.1016/j.ydbio.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Hrdlicka LA, Nambu JR. Alternate functions of the single-minded and rhomboid genes in development of the Drosophila ventral neuroectoderm. Mech. Dev. 1996;58:65–74. doi: 10.1016/s0925-4773(96)00559-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/135/18/3071/DC1