Abstract
It is often difficult to produce eukaryotic membrane proteins in large quantities, which is a major obstacle for analyzing their biochemical and structural features. To date, yeast has been the most successful heterologous overexpression system in producing eukaryotic membrane proteins for high-resolution structural studies. For this reason, we have developed a protocol for rapidly screening and purifying eukaryotic membrane proteins in the yeast Saccharomyces cerevisiae. Using this protocol, in 1 week many genes can be rapidly cloned by homologous recombination into a 2 μGFP-fusion vector and their overexpression potential determined using whole-cell and in-gel fluorescence. The quality of the overproduced eukaryotic membrane protein-GFP fusions can then be evaluated over several days using confocal microscopy and fluorescence size-exclusion chromatography (FSEC). This protocol also details the purification of targets that pass our quality criteria, and can be scaled up for a large number of eukaryotic membrane proteins in either an academic, structural genomics or commercial environment.
INTRODUCTION
One way to improve the success in obtaining pure membrane proteins for biochemical and structural analysis is to use GFP-based fusion technology. As the C-terminal GFP folds and becomes fluorescent only if the upstream membrane protein integrates into the membrane, the resultant fluorescence is a fast and accurate measure of membrane-integrated expression1. Fluorescence is easy to measure directly in liquid culture, standard SDS-gels and detergent-solubilized membranes2,3. Detergent-solubilized membranes can also be subjected to fluorescence size-exclusion chromatography (FSEC) to measure the ‘monodispersity’ of the sample4. FSEC was recently used to facilitate the purification of the chicken acid-sensing ion channel which crystallized and diffracted to 1.9-Å resolution5. In short, although membrane-integrated expression is no guarantee of function, the GFP-tag, nevertheless, speeds up the empirical process that all membrane proteins are required to go through on the way to obtain stable and homogeneous material for functional and structural work.
On the basis of a recent publication6, we have constructed a reliable protocol for screening the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. This system was adapted from a GFP-based Escherichia coli pipeline2, because yeast possess salient features absent in E. coli that are often essential for producing functional eukaryotic membrane proteins (e.g., post-translational modification and a more suitable lipid composition). With the exception of the human lipoxygenase structure to 4-Å resolution7, these differences could explain why functional eukaryotic membrane proteins in E. coli have failed to give high-resolution structures. With similar costs to E. coli and the possibility to clone by homologous recombination into standard vectors, S. cerevisiae is a convenient and efficient expression host to use. Although more structures of eukaryotic membrane proteins have been determined using material isolated from heterologous overexpression in the methylotrophic yeast Pichia pastoris, this difference is likely due to the popularity of P. pastoris because of its ability to produce a large biomass during fermentation with methanol8. However, recent work has demonstrated that S. cerevisiae is also a suitable host for obtaining enough material that will give high-resolution membrane protein structures9,10.
Using GFP-fusions, we can quickly identify membrane proteins that are highly overexpressed in S. cerevisiae. From our analysis of ~ 150 eukaryotic membrane protein-GFP fusions, approximately one-quarter can be overexpressed to > 1 mg l−1. The reason we prefer to screen in S. cerevisiae, rather than P. pastoris, is that cloning in P. pastoris is more time-consuming, as genes must be cloned into vectors, typically in E. coli, before their genomic integration. S. cerevisiae also has the added advantage that numerous localization studies have been carried out with functional GFP-fusions, and because S. cerevisiae has a highly regulated and extensively characterized quality-control systemin the endoplasmic reticulum11,12, assessing localization of fusions under overexpression conditions can be used as a good indication of functional expression. Of the highly expressed eukaryotic membrane proteins in S. cerevisiae, we find that more than half of those tested are targeted to the correct organelle and are monodisperse in more than one detergent6. Many eukaryotic membrane proteins have been purified to high levels (> 5 mg from 10 l) in our laboratory, and for those tested, functional data (Sonoda, Y. et al., manuscript in preparation) are consistent with other studies13. However, we also anticipate that, due to an absence of certain lipids (e.g., cholesterol), membrane proteins that express well in yeast may in some cases need to be produced in insect or mammalian cells for structural work. Nonetheless, because of its speed and comparatively low cost, S. cerevisiae remains a valuable prescreening host even in such situations.
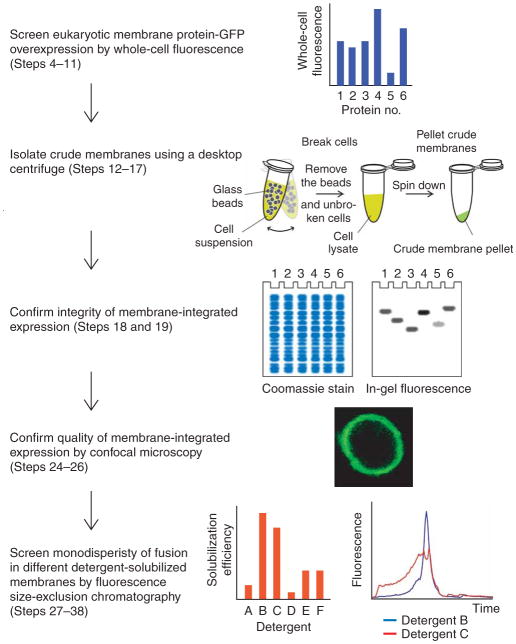
Here, we describe in detail the practical steps that constitute our S. cerevisiae GFP-based pipeline. This comprises (i) cloning by homologous recombination, (ii) whole-cell and in-gel fluorescence for estimating expression, (iii) confocal microscopy for confirming subcellular localization, (iv) FSEC for selecting detergents for purification, (v) GFP fluorescence-assisted immobilized metal-affinity chromatography (IMAC) and (vi) removal of GFP by His-tagged tobacco etch virus (TEV) protease and reverse IMAC (Fig. 1).
Figure 1.
Flowchart illustrating the screening process for the overexpression and purification of eukaryotic membrane protein-GFP fusions in Saccharomyces cerevisiae.
MATERIALS
REAGENTS
pep4 Deletion yeast strain, for example, FGY217 (MATa, ura3-52, lys2Δ201, pep4Δ)14
N,N-Dimethyldodecylamine N-oxide (LDAO; Anatrace, cat. no. D360)
n-Decyl-β-D-maltopyranoside (DM; Anatrace, cat. no. D322)
n-Dodecyl-β-D-maltopyranoside (DDM; Anatrace, cat. no. D310)
Fos-choline 12 (Anatrace, cat. no. F308)
Polyethylene(9)dodecyl ether (C12E9; Anatrace, cat. no. AP0129)
Pure yeast-enhanced green fluorescent protein (yEGFP) (S65G; S72A)15
Cholesterol hemisuccinate Tris salt (CHS; Sigma, cat. no. C6013)
Yeast nitrogen base without amino acids (BD, cat. no. 291920)
Yeast synthetic drop-out medium supplement without Ura (Sigma, cat. no. Y1501)
D-(+)-Glucose (Sigma, cat. no. G7021)
D-(+)-Galactose (Sigma, cat. no. G0625)
D-Sorbitol (Sigma, cat. no. S1876)
Bicinchoninic acid (BCA) protein assay kit (Pierce, cat. no. 23225)
Imidazole, minimum 99% (Sigma, cat. no. I2399)
Ni-NTA superflow resin (Qiagen, cat. no. 30430)
DTT (Sigma, cat. no. 43815)
Complete protease inhibitor cocktail tablets (Roche, cat. no. 04 693 132001)
TEV protease16, His-tagged stored at −80 °C in buffer containing 50% glycerol, 20 mM Tris–HCl pH 7.5, 0.3 M NaCl and 1 mM DTT
Coomassie brilliant blue R-250 (Sigma, cat. no. B-7920)
Fluorescent protein standard (Invitrogen, cat. no. LC 5928)
Protein ladder (Invitrogen, cat. no. LC 5625)
PEG 3350 (Sigma, cat. no. P3640)
Lithium acetate (Sigma, cat. no. L4158)
Single-stranded carrier DNA, salmon sperm (Sigma, cat. no. D1626)
Bacteriological agar (Sigma, cat. no. A5306)
Yeast peptone dextrose media (YPD media; Sigma, cat. no. P7750) (see REAGENT SETUP)
DMSO (Sigma, cat. no. D2438)
Novex 12% Tris–Gly gels (Invitrogen)
–URA media (see REAGENT SETUP)
Yeast suspension buffer (YSB) (see REAGENT SETUP)
PBS (see REAGENT SETUP)
Solubilization buffer (SB) for in-gel fluorescence (see REAGENT SETUP)
Cell resuspension buffer (see REAGENT SETUP)
Equilibration buffer (EB) (see REAGENT SETUP)
Membrane resuspension buffer (MRB) (see REAGENT SETUP)
Dialysis buffer (DB) (see REAGENT SETUP)
EQUIPMENT
Äkta FPLC system (GE Healthcare)
Frac-950 fraction collector with Rack C (GE Healthcare)
Peristaltic pump P-1 (GE Healthcare)
5-ml His-trap columns (GE Healthcare)
Constant Systems TS series cell disruptor (Constant Systems)
Dialysis tubing, 12–14 kDa molecular weight cut-off (Spectrumlabs)
LAS-1000–3000 charge-coupled device (CCD) imaging system (Fujifilm)
Nunc 96-well black optical bottom plate (Nunc)
Poly-Prep/glass Econo-Column chromatography columns (Bio-Rad)
Shaking incubator with temperature control (New Brunswick Scientific)
SpectraMax M2e microplate reader (Molecular Devices)
Superose 6 10/300 GL Tricorn gel filtration column (GE Healthcare)
Superdex 200 10/300 GL Tricorn gel filtration column (GE Healthcare)
Acid washed glass beads, 500 μm (Sigma)
Universal laboratory mixer-mill disruptor, for example the TissueLyser mixer (Qiagen)
50-ml Aerated capped tubes (TPP)
Tunair 2.5-l full baffled shaker flasks (VWR)
Preparative ultracentrifuge, Beckman Coulter Optima L-XP series with Beckman 45Ti rotor (Beckman)
Benchtop ultracentrifuge, Beckman Coulter Optima MAX series with TLA-55 and TLA 120.1 rotors (Beckman)
1.5-ml Polyallomer microcentrifuge tubes (Beckman)
Centrifugal filter devices (Millipore/Vivascience)
Confocal microscope (Leica TCS SP2 upright confocal microscope; Leica)
15- and 50-l Fermenter reaction vessels (Applikon)
Prominence HPLC (Shimadzu)
REAGENT SETUP
URA media
2.0 g l−1 of yeast synthetic drop-out medium without Ura, 6.7 g l−1 yeast nitrogen base without amino acids and either 2% glucose (for pre-culture) or 0.1% glucose (expression culture). For plates, use 2% glucose and add bacteriological agar 20 g l−1.
YSB
50 mM Tris–HCl (pH 7.6), 5 mM EDTA, 10% glycerol, 1× complete protease inhibitor cocktail tablets.
PBS
1.44 g Na2HPO4 · 2H2O (8.1 mM phosphate), 0.25 g KH2PO4 (1.9 mM phosphate), 8.00 g NaCl, 0.2 g KCl in 1,000 ml H2O; adjust pH to 7.4 using 1 M NaOH or 1 M HCl.
SB for in-gel fluorescence
50 mM Tris–HCl (pH 7.6), 5% glycerol, 5 mM EDTA (pH 8.0), 0.02% bromophenol blue to make aliquots of 700 μl and keep at −20 °C. Before use, add 200 μl of 20% SDS and 100 μl of 0.5M DTT. Working solution can be kept for 1 month at room temperature (20–25 °C).
CRB
50 mM Tris–HCl (pH 7.6), 1 mM EDTA, 0.6 M sorbitol.
EB
1× PBS, 10 mM imidazole (pH 8.0), 150 mM NaCl, 10% glycerol (wt/vol), 3× CMC detergent of choice.
MRB
Mix 20 mM Tris–HCl, (pH 7.5), 0.3 M sucrose, 0.1 mM CaCl2.
DB
20 mM Tris–HCl, (pH 7.5), 0.15 M NaCl, 3× CMC detergent of choice.
YPD media
10 g l−1 of bacto-yeast extract, 20 g l−1 bacto-peptone, 20 g l−1 dextrose.
PROCEDURE
Construction of GFP-8His-containing vector ● TIMING 2–3 weeks
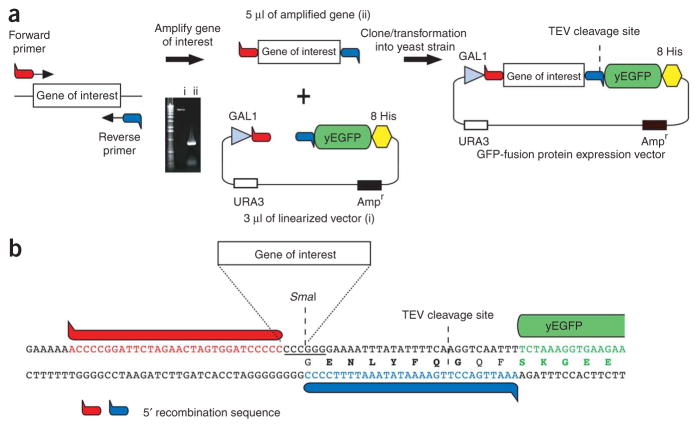
1. Clone the sequence for yEGFP15 (less start Methionine) with a C-terminal octa-His tag and a TEV protease site upstream of GFP-8His into a 2 μ vector that harbors a GAL1 promoter and URA selection marker (p424GAL1)17. In addition, introduce a SmaI site preceding the TEVp-GFP-8His sequence (Fig. 2a). Digestion of vector with SmaI gives rise to blunt ends that eliminate background colonies (typically <5) caused by the re-annealing of linearized vector during transformation by homologous recombination (problem when restriction enzymes that produce sticky ends are used). Note that although we recommend a GAL1 promoter, in some cases, a constitutive promoter gives higher expression. For propagation of yeast vector, use E. coli with appropriate antibiotic. As there are examples where an N-terminal GFP-fusion is more suitable for functional18 or structural work5 than a C-terminal GFP fusion, an N-terminal GFP fusion vector should also be considered. Note, however, that an N-terminal GFP fusion may reduce expression for Nout–membrane protein topologies19, and as the upstream GFP can be translated more efficiently than the downstream membrane protein, the GFP fluorescence from whole cells is no longer a reliable reporter of membrane-integrated expression.
Figure 2.
Cloning by homologous recombination into 2 μ Saccharomyces cerevisiae GFP-fusion vector. (a) The amount of PCR and vector used in cloning (as stated in Box 1) is depicted in the UV-exposed agarose-gel inset: i, SmaI linearilized vector; ii, amplified gene with overhangs required for homologous recombination. (b) Cloning site used in our 2μ GFP-fusion vector pDDGFP-2 (ref. 6). TEV, tobacco etch virus; yEGFP, yeast-enhanced green fluorescent protein.
Cloning of membrane protein gene(s) into GFP-8His-containing vector by homologous recombination ● TIMING5 d
2. Amplify the gene of interest with primers that include 5′ overhangs (35 bp) complementing the upstream and downstream sequences to either side of the SmaI site in the GFP-8His vector (Fig. 2b).
▲ CRITICAL STEP Because we find that only a quarter of mammalian membrane proteins are heterologously overexpressed to 1 mg l−1 (see ANTICIPATED RESULTS), initial screening of at least 20 targets is recommended in order to implement this screening protocol effectively.
3. Transform with 5 μl of PCR product and 3 μl of SmaI-linearized vector solution into S. cerevisiae competent cells as described in Box 1. Typical number of colonies is ~ 80–150. We recommend using a S. cerevisiae strain with the vacuolar pep4 deletion6. Check that the genotype of the yeast strain is compatible with the selection marker used in the vector.
BOX 1. TRANSFORMATION OF GENE-VECTOR CONSTRUCT INTO SACCHAROMYCES CEREVISIAE.
Preparation of competent cells ● TIMING2 d
1. Using a sterile loop, inoculate 5 ml of yeast peptone dextrose (YPD) medium with yeast strain in a 50-ml capped tube. Shake in an orbital shaker at 280 r.p.m. overnight at 30 °C.
2. Dilute overnight culture into 50 ml of YPD in a 250-ml shaker flask to an OD600 of 0.1. Grow cells in an orbital shaker at 280 r.p.m. at 30 °C until an OD600 of 0.5–0.6.
3. Centrifuge cells at 3,000g for 5 min at 4 °C, discard supernatant and resuspend the cell pellet in 25 ml of sterile dH2O. Centrifuge cells at 3,000g for 5 min at 4 °C, discard supernatant and resuspend cell pellet in 1 ml of 100 mM lithium acetate (LiAc).
4. Transfer suspension into a 1.5-ml tube and centrifuge cells at 8,000g for 15 s. Discard supernatant and resuspend cell pellet in 400 μl of 100 mM LiAc.
Transformation ● TIMING90 mi
5. Prepare new 1.5-ml tubes containing 240 μl of 50% (wt/vol) PEG 3350 for each transformation. Put tubes on ice.
6. Add 50 μl of cell suspension from Step 4 into each tube and vortex for 5 s. Add 25 μl of 2mgml−1 single-stranded carrier DNA into each tube and vortex for 5 s.
7. Add 50 μl of DNA mix (3 μl of 25 ng μl−1SmaI-digested vector, 5 μl of 150 ng μl−1 PCR product and 42 μl of sterile dH2O) into each tube and vortex for 5 s.
8. Incubate cells for 30 min at 30 °C, and heat shock cells for 25 min at 42 °C.
Plating ● TIMING30 min
9. Centrifuge cells at 8,000g for 15 s at room temperature, discard supernatant and resuspend the cell pellet with 100 μl of sterile dH2O.
10. Plate cell suspension onto a –URA selective plate and incubate for 2–3 d at 30 °C.
■ PAUSE POINT Yeast plates stored at 4 °C can be kept for 1 month with no reduction in the level of overexpression.
Screening membrane protein overexpression using whole-cell and in-gel fluorescence ● TIMING4 d
4. Inoculate a colony of the yeast transformant in an aerated 50-ml tube containing 10 ml –URA medium with 2% glucose.
▲CRITICAL STEP Aeration-capped tubes should be used, as they produce a more reliable large-scale culturing estimate than nonaerated tubes.
5. Incubate the culture overnight in an orbital shaker at 280 r.p.m. set at 30 °C.
6. Spot 10 μl of each overnight culture onto a fresh –URA plate, allow to dry at room temperature and transfer to a 30 °C incubator for 1–2 d.
7. Dilute the overnight culture (from Step 5) to an OD600 of 0.12 into two 50-ml aerated tubes, each containing 10 ml –URA medium with 0.1% glucose.
▲CRITICAL STEP Make sure that 0.1% glucose is used and not 2% glucose in the expression media because high levels of glucose repress the GAL1 promoter, while the former helps to maintain cell growth.
8. Incubate the cultures from Step 7 in an orbital shaker at 280 r.p.m. set at 30 °C. At an OD600 of 0.6 (after ~ 7 h), induce expression of membrane protein-GFP fusion by adding 20% (wt/vol) galactose (final 2%).
▲CRITICAL STEP In order to avoid diluting culture medium, prepare the 20% galactose stock in –URA medium. Induction before reaching an OD600 of 0.6 (typically) causes a reduced biomass that lowers protein production. Although the whole-cell GFP fluorescence can be higher if the cells are induced at a later OD600 than 0.6, we find that there is proportionately greater degradation, so that there is no linear gain in the amount of membrane-integrated material produced (as measured in Step 32).
9. Twenty-two hours after induction, centrifuge the cells at 3,000g for 5 min, remove the supernatant and resuspend the cell pellet in 200 μl of YSB.
▲CRITICAL STEP It is important to culture for 22 h after galactose addition, as, although OD600 is constant after 12 h, membrane protein production is maximal at 12–20 h after induction6. Because different final volumes can affect the level of whole-cell fluorescence measured, it is important to remove all of the supernatant. We recommend removal of supernatant by vacuum suction, or if it is removed by hand, to pat dry the tubes using absorbent paper.
10. Transfer 200 μl of the cell suspension to a black Nunc 96-well optical bottom plate.
▲CRITICAL STEP Because yeast cells settle to the bottom of the plate, for accurate measurements, proceed to Step 11 within 5 min of transfer.
11. Measure GFP fluorescence emission at 512 nm by excitation at 488 nm in a microplate spectrofluorometer. For plate readers with bottom read option, choose this setting. Estimate membrane protein overexpression levels (in mg l−1) from the yeast whole-cell fluorescence by applying the methodology detailed in Box 2 (Table 1). As previously illustrated, E. coli and S. cerevisiae whole-cell GFP fluorescent measurements (from samples with differing final OD600 densities) correlate linearly (R2 > 0.9) to expression levels as measured by the quantification of in-gel fluorescent band intensities2,6.
BOX 2. ESTIMATE OF MEMBRANE PROTEIN OVEREXPRESSION LEVELS.
Membrane protein overexpression estimates from whole cells
1. Harvest 10 ml of yeast cells that have been cultured with and without galactose addition (to estimate background fluorescence), remove supernatant and resuspend in 200 μl of yeast suspension buffer (small-scale culture) or cell resuspension buffer (CRB) (from large-scale culture).
2. Measure GFP fluorescence as outlined in Steps 9–11 of the main procedure. For example, no addition of galactose addition (MP-GFP – GAL)= 3,000 relative fluorescence units (RFU). Addition of galactose (e.g., Rattus norvegicus sugar transporter, Table 1) (MP-GFP + GAL) = 31,291 RFU.
3. To correlate the whole-cell fluorescence with the amount of GFP produced, measure the fluorescence of a set concentration of yeast-enhanced green fluorescent protein (yEGFP) (in a final volume of 200 μl). For example, fluorescence of pure yEGFP at a concentration (conc.) of 0.03 mg ml−1 is 11,300 RFU. Drew et al.2 have described the purification of GFP previously.
Numerical estimate of whole-cell overexpression
-
4. Calculate GFP concentration in mg ml−1 in whole cells as follows:
For example using the above measurements:
5. To determine the concentration of GFP in 200 μl of cell culture, divide by 40 [8,000 μl (cell culture)/200 μl (resuspension volume)]. Although the initial cell culture volume was 10 ml, there is an effective 2-ml loss by the transfer of only 200 μl of the resuspended cells (200 μl buffer + cell pellet = 250 μl) to the 96-well plate.
6. Using above example: (0.075 mg ml−1)/40 = 0.0018 mg ml−1, which equates to an expression yield of 1.8 mg l−1 (upscale correlation between 10 ml screening volume versus 1 l is reliable6).
7. As the typical recovery of GFP counts from 1-l culture into membranes is 60% (Fig. 3c,d), multiply this number by 0.6.
8. Using above example: 1.8 mg l−1 × 0.6 = 1.1 mg l−1 of GFP fusion.
-
9. Calculate amount of membrane protein expressed as follows:
Equation 1:Using above example:
Numerical estimate of membrane protein from solution
-
10. Calculate GFP concentration in mg ml−1 in 100 μl of solution (membranes or buffer) as follows:
(1)
Using R. norvegicus sugar transporter example (Table 1)
Membranes RFU (see Step 32) = 31,879 (resuspended in 6 ml CRB per l)
From 10 l of cell culture, the resuspended membrane volume was 60 ml
Amount of GFP-fusion:
(0.12 mg ml−1) × 60 ml = 7.2 mg
Amount of membrane protein (Eq. 1)
Immobilized metal-affinity chromatography elution RFU (see Step 46) = 34,622 (in 35 ml of EB)
Amount of GFP-fusion in elution:
(0.13 mg ml−1) × 35 ml = 4.6 mg
Amount of membrane protein in elution:
Note: GFP fluorescence (in our instrument setup) does not linearly increase with the volume in the 96-well plate; that is, 100 μl of pure GFP (0.03 mg ml−1) = 8,000 RFU and 200 μl of pure GFP (0.03 mg ml−1) = 11,300 RFU. For this reason, it is important to use the same volume of GFP standard as the experimental sample volume in the 96-well plate.
TABLE 1.
Example data-sheet showing information to be recorded throughout this protocol.
| Mammalian membrane protein | Step | Sugar transporter | Nucleotide sugar transporter | ABC transporter |
|---|---|---|---|---|
| MW (kDa) | 54 | 35 | 174 | |
| Small-scale expression | ||||
| Normal | 11 | 31,291 | 32,038 | 23,412 |
| 2.5% DMSO | 23 | 21,479 | 38,050 | 20,882 |
| 0.004 mg m−1 His | 23 | 27,422 | 34,929 | 21,108 |
| Large-scale expression | ||||
| 2.5-l Flask culture (10 l total) | 29 | 31,879** | 37,500 | 20,000** |
| Fermenter culture (15 l total) | 29 | 30,754 | 38,000** | 19,800 |
| Membrane preparation | ||||
| Breakage efficiency (%) | 30 and 31 | 78 | 85 | 78 |
| Membrane protein expression in membrane (mg l−1) | 32 | 1.4 | 1.4 | 1.2 |
| Purification | ||||
| Solubility (%) | 39 and 40 | 89 | 100 | 55 |
| Binding (%) | 43 | 71 | 86 | 58 |
| GFP count of elution | 46 | 34,622 | 53,000 | 2,919 |
| Amount of GFP-fusion in elution (mg) | 46 | 4.6 | 10 | 0.3 |
| Amount of membrane protein in elution (mg) | 46 | 8.9 | 13 | 2.1 |
| Amount of membrane protein after reverse immobilized metal-affinity chromatography (IMAC) (mg) | 51 | 7 | 9 | 1 |
Large-scale culture for which purification data are shown.
12. Transfer the cell suspension from the 96-well plate into a 1.5-ml capped tube.
■ PAUSE POINT The cell pellets can be stored at −20 °C for several days.
13. Add glass beads so that the final volume with the cell suspension is 500 μl. Add an additional 500 μl of YSB. Note that yeast cells are not solubilized directly by SB and need to be broken by other means for SDS-PAGE analysis.
14. Break the yeast cells with a mixer-mill disruptor set at 30 Hz for 7 min at 4 °C Alternatively, a vortex can be used, but we recommend using a heavy-duty lyser, as the cell breakage is more efficient, reproducible and easier to scale up.
15. Remove unbroken cells by centrifugation at 22,000g in a desktop centrifuge for 5 s at 4 °C. Transfer 500 μl of the supernatant into a new tube. Add 500 μl of YSB to the mixture of unbroken cell pellet and glass beads. Repeat Step 14 and transfer the supernatant to the 500-μl batch obtained from the first round of cell breakage.
16. To pellet crude membranes, centrifuge the 1 ml of supernatant at 20,000g in a desktop centrifuge at 4 °C for 1 h. Alternatively, to collect membranes, the supernatant can be pelleted using a desktop ultracentrifuge (120,000g) for 1 h. However, we find the recovery from centrifugation in a desktop centrifuge is sufficient for this analysis, and as the final pellet is less compact, it is easier to resuspend (Step 17).
17. Resuspend crude membranes in 50 μl of YSB, and transfer 15 μl into a tube containing 15 μl of SB. Load 10 μl for SDS-PAGE. Include nonfluorescent and fluorescent protein standards, such as Benchmark Fluorescent and SeeBlue Plus Prestained standard, respectively. For this step, we recommend our SB recipe with standard SDS denaturing cast gels for in-gel fluorescence. We have also tested Pre-cast Criterion (Bio-Rad) and Tris–Gly gels with equal success. We have found that the NuPAGE gels (Invitrogen) are not compatible with in-gel fluorescence.
▲CRITICAL STEP Do not boil samples for SDS-PAGE, as this denatures GFP and often causes membrane protein samples to aggregate.
18. Rinse the SDS gel with dH2O and detect the fluorescent bands with a CCD camera system. Expose the gel to blue light (EPI source) set at 460 nm and cut-off filter of 515 nm. Capture images and increase the exposure time until fluorescent bands are clearly visible (Fig. 3, panel a).
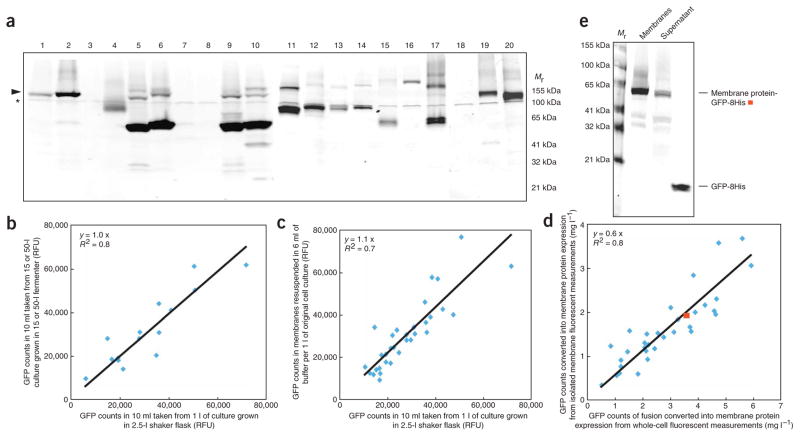
Figure 3.
Estimating reliable expression levels of eukaryotic membrane protein-GFP fusions from S. cerevisiae whole-cell fluorescent measurements. (a) In-gel fluorescence of crude yeast membrane samples isolated after the overexpression of 20 different eukaryotic transport proteins (Steps 6–18): arrowhead, membrane protein-GFP fusion; asterisk, endogenous fluorescent ‘background’ protein. (b) Comparison of whole-cell GFP counts (Steps 4–11) for 15 yeast and mammalian membrane protein-GFP fusions grown in either a 2.5-l shaker flask (Steps 27–29) or a 15-/50-l fermenter. (c) Whole-cell GFP counts from 10-ml culture volume for 10 yeast and 24 mammalian membrane proteins versus their GFP counts from membranes isolated from 10-l cultures; membrane pellet was resuspended in 6 ml of membrane resuspension buffer (MRB) per 1 l of initial cell culture volume (Steps 27–32). (d) Calculated membrane protein overexpression levels (as outlined in Box 2 with the exception that Step 7 was omitted) in whole-cells versus membranes for fusions presented in panel c. Red square, data for a membrane protein-GFP fusion of ‘typical’ recovery, which is analyzed further in panel e. (e) In-gel fluorescent SDS-PAGE gel for fusion depicted as red square in panel d: left lane, 10 μl of membranes after resuspension into prespin volume; middle lane, 10 μl of high-speed spin supernatant; right lane, pure GFP standard. Using quantification software band intensities were measured. RFU, relative fluorescence units.
▲CRITICAL STEP Blue light is recommended over UV light, as it is closer to the excitation wavelength of GFP. In addition, we do not recommend detecting GFP-fusion expression by western-blotting, as the transfer of membrane protein-GFP fusions is often inconsistent between membrane protein samples.
? TROUBLESHOOTING
19. Stain the SDS gel with Coomassie, and after sufficient time, transfer to destain. Note the intensity of Coomassie staining is a poor indication as to the levels of overexpression, as some membrane proteins bind Coomassie better than others6.
Optimization of membrane protein overexpression ● TIMING3 d
20. Select the fusions that give the best overexpression as established in Steps 4–19. Using the spotted yeast culture (Step 6), inoculate 10 ml of –URA medium containing 2% glucose in aerated capped 50-ml tubes. Incubate the culture overnight in an orbital shaker set at 30 °C at 280 r.p.m.
21. Dilute the overnight culture to an OD600 of 0.12 into six 50-ml aerated tubes containing 10 ml –URA medium with 0.1% glucose. Label two tubes each with the following: no chaperone addition, DMSO, His. Incubate at 30 °C at 280 r.p.m.
22. Monitor the OD600 of the cultures. Upon reaching an OD600 of 0.6 (after ~ 7 h), induce with 20% (wt/vol) galactose (final 2%). Add DMSO to a final concentration of 2.5% (vol/vol) and His to final concentration of 0.04% (wt/vol) to the tubes with the relevant labels (see Step 21). Note that, for the initial optimization, we recommend adding either DMSO or His, as we find that the presence of these chemical chaperones can afford an additional 30% improvement to the whole-cell expression levels6. Until now it was unclear as to why these chemical chaperones can improve membrane protein production levels20.
23. Harvest cells after 22 h of induction and measure whole-cell GFP fluorescence as outlined in Steps 9–11. Using Steps 20–23, one can also test other culture conditions that have shown to be successful to improve membrane protein production, for example, the addition of a ligand to the culture medium20, or to switch the culture to 20 °C after galactose induction. However, in the latter case, we have found that only 5% of the targets we have tested give an additional improvement in expression compared with standard growth at 30 °C (ref. 6).
Confocal microscopy ● TIMING2 d
24. Repeat the membrane protein overexpression culturing condition that gave the highest level of whole-cell fluorescence (Steps 20–22). Resuspend harvested cells in 1 ml of –URA media containing 50% glycerol. Glycerol slows yeast mobility to facilitate the capture of quality images.
25. Add 1 μl of cell suspension to a microscope slide and place coverslip on top. Focus on the sample using Köhler illumination with a × 10 magnification lens. Set the focal plane to zero.
▲CRITICAL STEP Before setting any additional parameters on the microscope, make sure that you can see your cells in transmitted light. This is achieved through Köhler illumination, resulting in an even illumination of the sample with minimal flare. At this point, you can see whether your cells are granular, which can help interpret fusion localization, for example, punctuated internal spots are often seen as fluorescent vacuoles in Step 26.
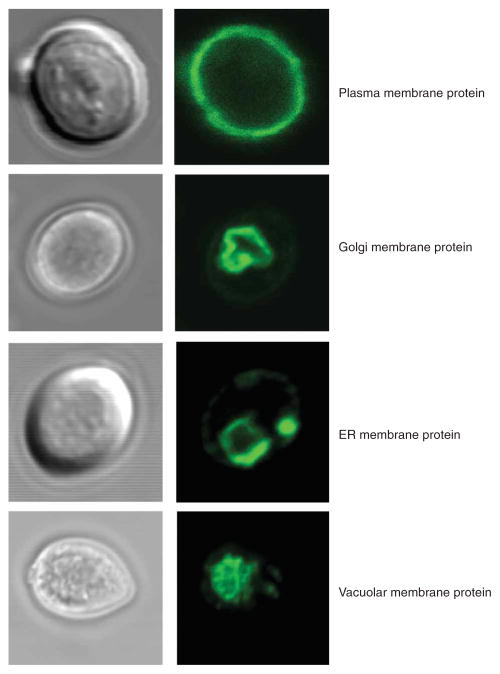
26. Add a drop of lens oil, and change to the higher magnification oil-immersion lens. Turn off the brightfield lamp. Turn the blue light on, and estimate gross localization of the membrane protein-GFP fusion. Turn the blue light off. Use the argon laser and excite at 488 nm to capture a detailed localization image of the GFP-fusion (emission detection at 505–535 nm) (Fig. 4). Note that in contrast to E. coli, in which GFP is inactively fused to periplasmic segments of inner-membrane proteins21, C-terminal membrane protein-GFP fusions are fluorescent in all yeast subcellular compartments22.
Figure 4.
Examples of membrane protein GFP-fusion localization in S. cerevisiae as monitored by confocal microscopy. Top, plasma membrane protein Ctr1p; top-middle, Golgi membrane protein Hut1p; middle-bottom, Endoplasmic reticulum (ER) membrane protein Isc1p; bottom, vacuolar membrane protein Uga4p (examples have been published in core ref. 6).
? TROUBLESHOOTING
Isolation of large-scale membranes ● TIMING5 d
27. Inoculate 10 ml of –URA media with spotted yeast culture as outlined in Step 20. Next day, add overnight culture to a 500-ml shaker flask containing 150 ml –URA medium and 2% glucose. Incubate the culture overnight in an orbital shaker at 280 r.p.m. set at 30 °C.
28. Dilute the 150-ml overnight culture to an OD600 of 0.12 into 1 l of –URA containing 0.1% glucose in a 2.5-l baffled shaker flask. Incubate the culture in an orbital shaker at 280 r.p.m. set at 30 °C. Induce at OD600 of 0.6 using the parameters established in the overexpression optimization screen (Steps 20–23). Note that the volume of the overnight culture in an appropriate shaker flask depends on the number of 1 l cultures to be inoculated (~ 20 ml l−1). For detergent screening and FSEC analysis, we recommend the preparation of 2 l of yeast culture per clone. For purification, we recommend 10–15 l of yeast culture. Surprisingly, 1 l cultures grown in baffled 2.5-l shaker flasks give comparable expression per liter to that of 15- and 50-l fermentations (Fig. 3b).
29. Harvest the cells after 22 h of incubation by centrifugation at 4,000g at 4 °C for 10 min. Decant the supernatant and resuspend the cell pellet in 25 ml of CRB per liter of original cell culture.
▲CRITICAL STEP From this step onward, even when not indicated, material should be kept on ice or at 4 °C.
■ PAUSE POINT Cell suspensions can be rapidly frozen in liquid nitrogen and stored at −80 °C for up to 6 months. Use screw-capped tubes for storage.
30. Break the cells using heavy-duty cell disruptor for four passes at incremental pressures of 25, 30, 32 and 35 kpsi (~ 1.7–2.4 × 103 atm) at 4–15 °C. Remove 100 μl of cells, transfer into a 96-well plate and measure GFP fluorescence as outlined in Step 11.
▲CRITICAL STEP It is advisable not to break the cells immediately at the higher pressure, as the suspension is more viscous at this stage and leads (often) to a blocked inlet valve. We recommend cell disruption using disruptors from Constant Systems, which typically break 75% of the yeast cells (as measured by GFP fluorescence). On the other hand, glass beads with a BeadBeater can be used to break yeast cells, but typically with lower efficiency with large culture volumes.
31. Remove the unbroken cells and debris by centrifugation at 10,000g at 4 °C for 10 min and collect the supernatant containing membranes. Transfer 100 μl of supernatant to a 96-well plate and measure GFP fluorescence as outlined in Step 11. Calculate the yeast cell breakage efficiency by comparing GFP fluorescence to that measured in Step 30.
? TROUBLESHOOTING
32. To collect the membranes, centrifuge the cleared supernatant at 150,000g at 4 °C for 120 min. Discard the supernatant and resuspend the pellet to a final volume of 6 ml l−1 of original cell culture with MRB using a disposable 10-ml syringe with a 21-gauge needle. Transfer 100 μl of the membrane suspension into a 96-well plate and measure GFP fluorescence as outlined in Step 11. Calculate the amount of overexpressed membrane protein (Box 2). Calculate the amount of total protein using the BCA protein assay kit following the manufacturer’s instructions. If the membranes isolated from 1 l of cells are resuspended in 6 ml of MRB, the GFP fluorescent counts typically match the original whole-cell fluorescent counts (Fig. 3c). This corresponds to ~ 60% of the amount of GFP measured in whole cells actually recovered into membranes (Fig. 3d). Analysis of the supernatant from the high-speed spin, shown here for a typical membrane protein, indicates that ~ 12% of the GFP-fusion is not pelleted (Fig. 3e). Given cell-breakage efficiency of ~ 75% means that the average fraction of ‘free GFP’ is ~ 15%.
■ PAUSE POINT Membrane suspensions can be rapidly frozen in liquid nitrogen and stored at −80 °C for up to 6 months. Although this is the routine in our laboratory, some membrane protein crystallographers avoid freezing and storing membranes and continue with purification immediately.
Detergent screen ● TIMING2 h
33. Adjust the membrane suspension to a protein concentration of 3.5 mg ml−1 in PBS. Transfer 900 μl aliquots of the membrane suspension into 1.5-ml Beckman polyallomer microcentrifuge tubes.
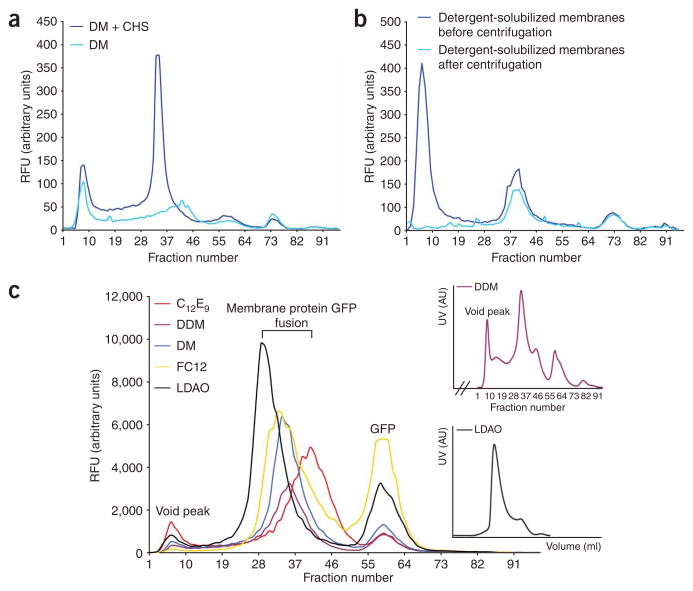
34. Add 100 μl of each freshly prepared detergent listed in the table below (final 1%) to each of the 1.5-ml tubes containing 900 μl of membrane suspension (final 3 mg ml−1). Incubate the mixtures at 4 °C for 1 h with mild agitation. We also recommend testing the addition of CHS to the detergent mixture (final 0.2%), as this can be essential for the isolation of monodisperse mammalian membrane proteins (Fig. 5a).
Figure 5.
Determining the monodispersity of membrane protein-GFP fusions in different detergents using fluorescence size-exclusion chromatography (FSEC). (a) FSEC traces showing the effect of cholesterol hemisuccinate salt addition on the detergent extraction efficiency and monodispersity of a Homo sapiens metal transporter. (b) FSEC traces of membranes containing Rattus norvegicus ABC transporter-GFP fusion after adding C12E9: before ultracentrifugation, dark blue line; after ultracentrifugation, light-blue line. (c) FSEC traces obtained for a yeast trafficking chaperone protein in the five recommended initial screening detergents (Steps 33–38). Aggregated protein in void peak, membrane protein-GFP fusion and ‘free’ GFP peaks are as highlighted. Inset (top), the SEC trace for the n-dodecyl-β-D-maltopyranoside (DDM)-solubilized total membranes of the sample shown in panel c, main. Inset (bottom), the SEC trace of yeast chaperone purified in N,Ndimethyldodecylamine N-oxide (LDAO) using Steps 36–51. CHS, cholesterol hemisuccinate Tris salt; DM, n-Decyl-β-D-maltopyranoside; RFU, relative fluorescence units.
▲CRITICAL STEP Other detergents should be screened. This selection is considered a starting point only.
| FSEC screening detergents | Stock concentration (% wt/vol) |
|---|---|
| DDM | 10 |
| DM | 10 |
| LDAO | 10 |
| Fos-choline 12 | 10 |
| C12E9 | 10 |
35. Transfer 100 μl of the detergent-solubilized membranes into a 96-well plate and measure GFP fluorescence as outlined in Step 11. To pellet nonsolubilized material, centrifuge the remaining 900 μl in a benchtop ultracentrifuge at 100,000g at 4 °C for 45 min.
36. Transfer the supernatant into a new 1.5-ml tube. Transfer 100 μl into a 96-well plate and repeat the GFP fluorescent measurement as outlined in Step 11. Calculate the detergent solubilization efficiency by comparing to the GFP fluorescence measured in Step 35. In the example shown in Figure 5b, before ultracentrifugation, detergent solubilized membranes harbor a fusion in an aggregated and monodisperse state. In this example, all aggregation is removed by ultracentrifugation. This indicates that nondetergent solubilized fusion is protein in an aggregated state, rather than nonaggregated protein in detergent-resistant membranes (monodisperse peaks were equivalent before and after ultracentrifugation). However, because aggregation is still evident in many detergent-solubilized samples after ultracentrifugation6 (e.g, Fig. 5c), these aggregates are often solubilized with detergent to varying degrees. For this reason, the solubilization efficiency estimate is an unreliable indicator to decide the detergent for purification, and as such, samples should be analyzed by FSEC (see Steps 37–38) and, if possible, also for activity.
■ PAUSE POINT The detergentsolubilized supernatant can be rapidly frozen in liquid nitrogen and stored at −80 °C. Note, however, that some membrane protein crystallographers do not freeze and analyze by FSEC directly (Step 37).
? TROUBLESHOOTING
FSEC ● TIMING1 d
37. Inject 0.5 ml of the detergent-solubilized sample onto a Superose 6 10/300 column equilibrated in 20 mM Tris–HCl (pH 7.5), 0.15 M NaCl and 0.03% DDM. After 6 ml, fractionate row-by-row 0.2 ml into a 96-well plate. Note that the use of a low percentage of DDM (0.03%) in the buffer used for separation of detergent-solubilized membranes by SEC does not rescue membrane protein that has aggregated in a different detergent4.
38. Set the 96-well parameters in the plate reader to read wells row-by-row. To improve the signal-to-noise ratio, measure the GFP fluorescence emission at 512 nm by excitation at 470 nm (this wavelength is used instead of 488 nm, as it produces a lower background fluorescent signal6). Plot the GFP fluorescence in each well against the fraction number (Fig. 5c). Alternatively, as originally outlined by Kawate and Gouaux4, the fluorescence can be measured with higher sensitivity using an in-line detector e.g., with a Prominence HPLC (Shimadzu).
? TROUBLESHOOTING
Purification of membrane protein-GFP fusions ● TIMING2 d
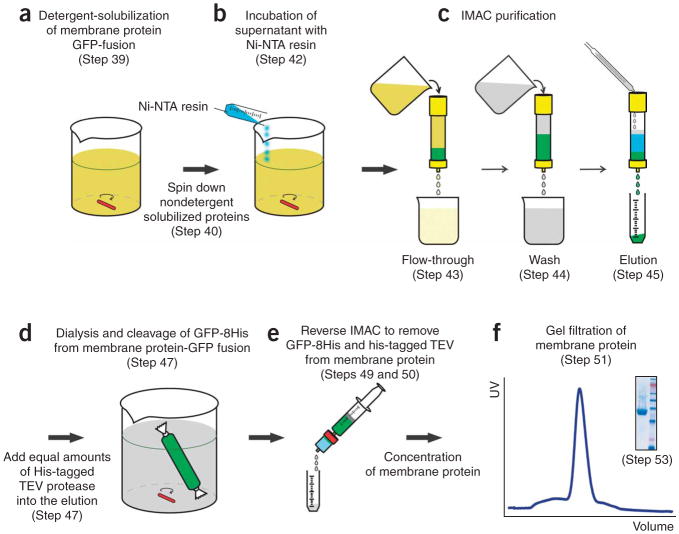
39. Dilute the membrane suspension isolated from 10–15 l of culture into a 500-ml beaker at a final protein concentration of 3 mg ml−1 using EB. Add the detergent powder, as outlined in Steps 33–38, that produced the most homogeneous sample. Incubate mixture at 4 °C for 1 h with mild agitation (Fig. 6). Note that the EB composition should be considered a starting point only, and inhibitors should be added at this point if necessary.
Figure 6.
Flowchart illustrating the purification of eukaryotic membrane proteins from GFP-fusions. (a) Detergent solubilization of membrane protein- GFP fusion (Step 39). (b) Incubation of supernatant with Ni-NTA resin (Step 42). (c) Immobilized metal-affinity chromatography (IMAC) purification (Steps 43–45). (d) Dialysis and cleavage of GFP from membrane protein-GFP fusion (Step 47). (e) Reverse IMAC to remove GFP and tobacco etch virus (TEV) from membrane protein (Steps 49 and 50). (f) Gel filtration of membrane protein (Step 51): example shown here is for Homo sapiens sugar transporter with the pooled fraction analyzed by SDS-PAGE (Step 53) as shown in inset.
40. Pellet the unsolubilized material by centrifugation at 100,000g at 4 °C for 45 min. Transfer supernatant into an appropriately sized glass beaker. Transfer 100 μl of the detergent-solubilized membranes to a 96-well plate and measure GFP fluorescence as outlined in Step 11. Calculate the amount of the GFP fusion as detailed in Box 2.
41. Use 1 ml of Ni-NTA resin (2 ml of 50% slurry) per 1 mg of GFP fusion to be purified (Step 40), and equilibrate with five column volumes of EB.
▲CRITICAL STEP As the His tag is C-terminal to GFP, the amount of GFP that will bind as a fusion is calculated at this step rather than the amount of membrane protein. Although the level of resin used is much higher than for soluble proteins, we have empirically found that this amount is necessary to ensure good binding efficiency.
42. Add equilibrated Ni-NTA resin to detergent-solubilized membranes. Use a magnetic stirrer to mix the solution at 4 °C for 2–3 h.
▲CRITICAL STEP Batch binding of yeast-solubilized membranes improves the protein recovery compared with flow loading. One can pellet a 1 ml sample to compare fluorescence to that measured in Step 40. Usually binding efficiency reaches a plateau after 2 h, with only a modest gain of 10–15% by overnight incubation.
43. Pour the slurry into a glass Econo-Column (BioRad). Transfer 100 μl of the flowthrough into a 96-well plate and measure GFP fluorescence as outlined in Step 11. Compare measurement to Step 40 and calculate the binding efficiency of the fusion to the Ni-NTA resin.
? TROUBLESHOOTING
44. Wash the column with 10-column volumes of EB. Add 1 M imidazole to EB to a final concentration of 30 mM and wash the column for 35-column volumes.
45. Add 1 M imidazole to EB to a final concentration of 250 mM and elute the protein from column using 2–3 column volumes.
46. Transfer 100 μl of the eluate into a 96-well plate and measure GFP fluorescence as outlined in Step 11. Determine the amount of GFP fusion (Box 2). The amount of GFP-fusion in the eluate should be determined by measuring GFP fluorescence as the BCA assay measures total protein content (including contamination).
Removal of GFP-8His tag from the membrane protein-GFP fusion ● TIMING16 h
47. Add to the GFP fusion an equal amount of His-tagged TEV protease as calculated in Step 46. Transfer into dialysis tubing and dialyze overnight at 4 °C against ~ 3 l of DB. In this protocol, the TEV protease constructed by Doudna and co-workers was used16, which describes how to overexpress and isolate His-tagged TEV protease. Conveniently, GFP and TEV protease have approximately the same Mr, which means a quick calculation for the amount of GFP-fusion is a molar equivalent of TEV protease. For commonly used detergents, such as DDM, equimolar amounts of TEV protease is typically sufficient for a complete overnight digest at 4 °C (ref. 2). The DB chosen here is only a starting point.
48. Optional: After overnight dialysis, add 10 μl of digest to 10 μl SB and process as described in Steps 17–19. Analyze the TEV digest reaction for completeness by in-gel fluorescence. Usual cleavage under these conditions is > 98% complete. Note that we do not add any additional DTT to the TEV protease from that already contained in its storage buffer.
49. For the reverse Ni-NTA binding, equilibrate a 5-ml His-Trap column in fresh DB containing 15 mM imidazole using a 20-ml syringe or a peristaltic pump. Remove the dialyzed sample from the tubing and add 1 M imidazole to a final concentration of 15 mM.
? TROUBLESHOOTING
50. To remove the His-tagged TEV protease, cleaved GFP-8His tag and co-eluting contaminating proteins, pass the dialyzed sample through the His-Trap column at a flow rate of 2 ml min−1. Collect the flowthrough containing membrane protein into a 50-ml Falcon tube. After collecting the flowthrough, the bound material (GFP-8His, His-tagged TEV protease and contaminating proteins) can be eluted using buffer containing 250 mM imidazole. Elution can be analyzed by SDS-PAGE to confirm that the cleaved membrane protein did not inadvertently bind to the His-Trap column.
51. Concentrate the flowthrough from Step 50 using a centrifugal concentrator to 0.5 ml. Calculate the amount of protein using the BCA assay and lower to 20 mg ml−1 if necessary. Inject 0.5 ml onto a Superdex 200 10/300 column pre-equilibrated with buffer used for dialysis at a flow rate of 0.5 ml min−1. Alternatively, if crystallization screening is the final step, inject only half of the sample onto the size-exclusion column, as it has been found that lipid removal by SEC can influence the quality of the membrane protein crystals23. Another strategy shown to be successful is to equilibrate the column in buffer that also contain lipids24. To use the sample without SEC, remove imidazole by re-diluting and concentrating sample several times.
52. Collect 0.5 ml-fractions. Analyze the UV-trace and collect and pool the membrane protein peak.
53. Analyze the eluate (membrane protein-GFP fusion), the flowthrough (membrane protein before SEC) and the protein peak (membrane protein after SEC) by SDS-PAGE (Fig. 7a). Note: For further examples to those presented in this protocol, refer to core ref. 6. Cloning and handling of S. cerevisiae was based on ref. 25.
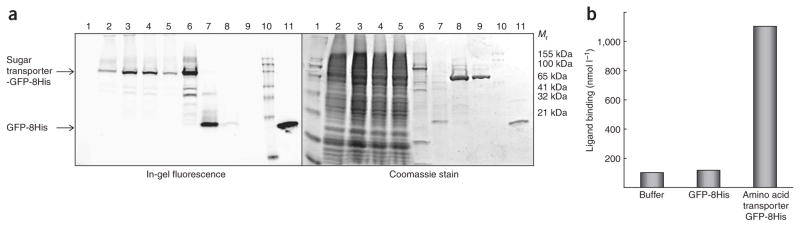
Figure 7.
Examples of purified eukaryotic transport proteins. (a) SDS-PAGE gel as analyzed by in-gel fluorescence and Coomassie staining for the purification of a Homo sapiens sugar transporter: 1, prestained molecular weight (MW) protein ladder; 2, total membranes; 3, n-dodecyl-β-D-maltopyranoside (DDM)-solubilized membranes before ultracentrifugation; 4, DDM-solubilized membranes after ultracentrifugation; 5, Ni-NTA flowthrough; 6, Ni-NTA elution; 7, elution of reverse Ni-NTA after GFP cleavage; 8, flowthrough of reverse Ni-NTA after GFP cleavage; 9, purified membrane protein after SEC; 10, fluorescent MW protein ladder; 11, pure GFP standard. (b) Assay for the binding of a specific inhibitor (nanomole affinity) to a Rattus norvegicus amino acid transporter, left lane, buffer solution; middle lane, GFP; right lane, amino acid transporter-GFP fusion.
^ TIMING
Construction of GFP-8His containing vector: 2–3 weeks
Cloning of membrane protein gene(s) into GFP-8His containing vector by homologous recombination: 5 d
Preparation of competent cells: 2 d
Transformation: 90 min
Plating: 30 min
Screening membrane protein overexpression using whole-cell and in-gel fluorescence: 4 d
Optimization of membrane protein overexpression: 3 d
Confocal microscopy: 2 d
Isolation of large-scale membranes: 5 d
Detergent screen: 2 h
FSEC: 1 d
Purification of membrane protein-GFP fusions: 2 d
Removal of GFP-8His tag from the membrane protein-GFP fusion: 16 h
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
TABLE 2.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 18 | No bands are visible | Incorrect setup or there is no membrane-integrated product | Check the in-gel fluorescence setup with pure yeast-enhanced green fluorescent protein (yEGFP). Blue light is needed for best sensitivity. Bands should be visible down to 5 ng (ref. 2). If still no signal is visible for the experimental sample, increase exposure time. If GFP counts are low (<5,000 relative fluorescence units), the protein may not be expressed or was expressed but has degraded so that only ‘free GFP’ is left6 (see Fig. 3 for average fusion recovery into membranes from whole cells) |
| 26 | Incorrectly localized protein | Protein is incorrectly folded or has been re-routed to vacuoles for degradation | If a non-endoplasmic reticulum (ER) membrane protein is retained in the ER, it may be misfolded and unable to exit. Check localization under different culture condition, for example, growth at 20 °C after adding galactose. If it is located in vacuoles, then the protein may have been initially targeted correctly, but has been downregulated for degradation in vacuoles, a common problem for transporters in the presence of their substrates26 (e.g., Ura yeast transporter Fur4 when Ura is abundant27). Try using a specialized media, and/or a constitutive promoter, and/or different yeast strain28 |
| 31 | Low breakage efficiency, that is, <70% | Sample is too viscous | If final OD600 was high (>7), cell suspension may need to be diluted further with cell resuspension buffer to achieve better breakage efficiency. If no improvement is detected, then increase breaking pressure to 40 kpsi (2.4 × 103 atm) |
| 36 | Solubilization efficiency <45% | Total protein concentration is too high | Make sure total protein concentration is at 3 mg ml−1. Solubilization efficiency can also be increased (typically) by 10–15% by the dilution of total protein to 1.5 mg m−1 in the same detergent concentration |
| 38 | Free-GFP peak in fluorescence size-exclusion chromatography trace | This is a sign of protein degradation | If free GFP-to-fusion peak is <10%, this is not usually problematic and one should carry on with the protocol, provided that the protein peak is monodisperse |
| If free GFP-to-fusion peak is 20–50%, then try different detergent, and/or add glycerol, ligand, inhibitor, etc., to improve protein stability | |||
| If free GFP-to-fusion peak >50%, severe proteolysis has occurred. Try parameters suggested for lowering the amount of protein degradation, but also consider modification of protein, for example, make N- and/or C-terminal truncation(s), adding N-terminal tag, for example, Flag-tag | |||
| Only aggregation peak is apparent | Protein is unstable in this detergent | Try different detergents, additions to buffer (e.g., glycerol, ligand/inhibitor), or homologs to the protein of interest | |
| 43 | Not all of the protein binds to the column as evident by GFP fluorescence measured in flow through | Saturation of Ni-NTA resin or detergent lowers binding | Typically binding efficiency is ~50–60% for GFP-fusions in yeast purifications. Efficiency may increase in a differ- ent detergent, but it is not predictable and often difficult to improve. The addition of more Ni-NTA resins or overnight batch incubation is worth testing, but in our hands such modifications affords only modest improvements (10–15%) |
| 49 | The solution appears slightly cloudy | A small fraction of tobacco etch virus (TEV) protease can precipitate during removal of glycerol by dialysis | Filter sample through 0.22-mM filter. To confirm precipitant is due to TEV protease, repeat dialysis with pure TEV only. In almost all such cases, TEV cleavage has been >98% complete, and the target protein has not aggregated |
| 49 | The appearance of a heavy precipitant is observed | The protein is unstable without GFP | To distinguish between general instability and instability due to removal of GFP-8His tag, repeat dialysis of eluate without adding His-tagged TEV protease. In either scenario (aggregation in general or aggregation due to GFP-8His removal), try dialysis in a buffer with a higher or lower pH (e.g., 6 and 8), add glycerol, and/or repeat in a different detergent, and/or add inhibitor, etc. Note, that GFP-fusions can still be completely functional as fusions |
ANTICIPATED RESULTS
To date, we have cloned 110 eukaryotic transport proteins into a GFP-fusion vector: 30 from yeast and 80 from higher eukaryotic organisms. After obtaining colonies (Steps 2 and 3), we tested the expression (Steps 4–11) of two colonies from each transformation. In 90% of the cases, whole-cell GFP counts in both colonies were similar. In cases where whole-cell GFP fluorescence over background was low (<5,000 relative fluorescence units (RFU)), correct clones could be confirmed by PCR and/or DNA sequencing. Thus, error-free clones should be obtained quickly by homologous recombination.
Analyzing the overexpression levels for these clones (Box 2), we find that only 25% of mammalian membrane proteins, compared with 75% of yeast membrane proteins, were produced > 1 mg l−1 (ref. 6). These data emphasize the importance of initially screening the overexpression potential of many membrane proteins. Typical in-gel fluorescence of isolated crude membranes (Steps 12–19) for 20 targets is illustrated in Figure 3a. As shown in Figure 3b, 10 ml whole-cell screening provides reliable estimates for large-scale production up to 50-l fermentations. Importantly, there is a good correlation (R2 = 0.8) between whole-cell and membrane-integrated expression, Figure 3c,d. This correlation means that overexpression culture conditions can be confidently optimized by whole-cell GFP fluorescence measurements. Modest improvement (typically 30%) could be obtained for a third of transport proteins tested by adding DMSO or His to the culture medium (Steps 20–23)6.
So far, analysis of membrane protein localization by confocal microscopy (Steps 24–26) is consistent with all previously reported functional localization data for membrane protein-GFP fusions6. Overall, 70% of membrane proteins tested were targeted to the correct organelle. Typical localization patterns are shown in Figure 4 (for further examples, see core reference article6). We found no correlation between overexpression levels and mistargeting.
Detergent screening and FSEC analysis (Steps 33–38) has been carried out for 33 eukaryotic transporters: 15 from S. cerevisiae and 18 from Rattus norvegicus, Mus musculus or Homo sapiens. Our criterion for monodispersity is that a protein peak is symmetric and is equal to or larger than any ‘free GFP’ or aggegration peak(s). A typical detergent screen for a yeast membrane protein-GFP fusion is shown in Figure 5c. In this example, the detergents DDM, DM and LDAO produce a monodisperse peak, whereas Fos-choline 12 and C12E9 detergents do not (for further examples see refs. 4,6). We have found no correlation between the efficiency of the detergent to extract the fusion protein and the quality of the FSEC traces6. We attribute this poor correlation to the ability of detergents to solubilize fusion protein that has aggregated (Fig. 5b). Eleven of fifteen yeast and seventeen of eighteen mammalian transporters tested were monodisperse in at least one detergent. Nine of these yeast membrane proteins and all mammalian membrane proteins produced monodisperse profiles after purification by IMAC (Steps 39–46) (results not shown). After GFP-8His cleavage by His-tagged TEV protease and reverse IMAC (Steps 47–50), all yeast proteins and 15 of the 17 mammalian transporters were monodisperse as untagged proteins (2 mammalian transporters aggregated after the removal of the GFP-8His tag). Overall, 60% of the yeast and 85% of the mammalian transporters that expressed > 1 mg l−1 could be purified as homogeneous samples (Steps 51–53) (see Fig. 6f, and core reference article ref. 6). So far, we have monitored the transport activity for four mammalian transporters. Activity is consistent with the activity of these proteins recorded either in vivo or after reconstitution into lipids (Sonada, Y. et al., manuscript in preparation). Specific binding of a inhibitor to one of these proteins in detergent solution is illustrated in Figure 7b. An example data-sheet encompassing most steps outlined in this protocol is shown in Table 1. We have found this information important for troubleshooting and for maintaining reproducibility in a large-group setting.
Acknowledgments
We thank Jan-Willem de Gier, Mitsunori Shiroishi, Shuichiro Goda and the reviewers for critically reading the manuscript and for useful comments. D.D. was a recipient of an European Molecular Biology Organization (EMBO) long-term fellowship. Funded by grants from the European Membrane Protein Consortium (E-MEP), the Membrane Protein Structure Initiative (MPSI) and the Biotechnology and Biological Sciences Research Council (BBSRC) (to S.I.). The project was also supported by grant number R01GM081827 from the National Institute of General Medical Sciences (to G.v.H. and H.K.); the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Drew DE, von Heijne G, Nordlund P, de Gier JW. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 2001;507:220–224. doi: 10.1016/s0014-5793(01)02980-5. [DOI] [PubMed] [Google Scholar]
- 2.Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW. Optimization of membrane protein overexpression and purification using GFP fusions. Nat Methods. 2006;3:303–313. doi: 10.1038/nmeth0406-303. [DOI] [PubMed] [Google Scholar]
- 3.Drew D, et al. A scalable, GFP-based pipeline for membrane protein overexpression screening and purification. Protein Sci. 2005;14:2011–2017. doi: 10.1110/ps.051466205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 6.Newstead S, Kim H, von Heijne G, Iwata S, Drew D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:13936–13941. doi: 10.1073/pnas.0704546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferguson AD, et al. Crystal structure of inhibitor-bound human 5-lipoxygenaseactivating protein. Science. 2007;317:510–512. doi: 10.1126/science.1144346. [DOI] [PubMed] [Google Scholar]
- 8.Nyblom M, et al. Exceptional overproduction of a functional human membrane protein. Protein Expr Purif. 2007;56:110–120. doi: 10.1016/j.pep.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Jidenko M, et al. Crystallization of a mammalian membrane protein overexpressed in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:11687–11691. doi: 10.1073/pnas.0503986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- 11.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 12.Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niebauer RT, Robinson AS. Exceptional total and functional yields of the human adenosine (A2a) receptor expressed in the yeast Saccharomyces cerevisiae. Protein Expr Purif. 2006;46:204–211. doi: 10.1016/j.pep.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 14.Kota J, Gilstring CF, Ljungdahl PO. Membrane chaperone Shr3 assists in folding amino acid permeases preventing precocious ERAD. J Cell Biol. 2007;176:617–628. doi: 10.1083/jcb.200612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cormack BP, et al. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology. 1997;143 (Pt 2):303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 16.Lucast LJ, Batey RT, Doudna JA. Large-scale purification of a stable form of recombinant tobacco etch virus protease. Biotechniques. 2001;30:544–546. 548. doi: 10.2144/01303st06. 550 passim. [DOI] [PubMed] [Google Scholar]
- 17.Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 18.Sato K, Sato M, Nakano A. Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J Cell Biol. 2001;152:935–944. doi: 10.1083/jcb.152.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner S, Bader ML, Drew D, de Gier JW. Rationalizing membrane protein overexpression. Trends Biotechnol. 2006;24:364–371. doi: 10.1016/j.tibtech.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 20.André N, et al. Enhancing functional production of G protein-coupled receptors in Pichia pastoris to levels required for structural studies via a single expression screen. Protein Sci. 2006;15:1115–1126. doi: 10.1110/ps.062098206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drew D, et al. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc Natl Acad Sci USA. 2002;99:2690–2695. doi: 10.1073/pnas.052018199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 23.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci USA. 2007;104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K. channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 25.Burke D, Dawson D, Stearns T. Cold Spring Harbor Laboratory Course Manual. 2000. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2000. Methods in Yeast Genetics. [Google Scholar]
- 26.Stimpson HE, Lewis MJ, Pelham HR. Transferrin receptor-like proteins control the degradation of a yeast metal transporter. EMBO J. 2006;25:662–672. doi: 10.1038/sj.emboj.7600984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galan JM, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Sitaram A, Burd CG. Regulation of copper-dependent endocytosis and vacuolar degradation of the yeast copper transporter, Ctr1p, by the Rsp5 ubiquitin ligase. Traffic. 2007;8:1375–1384. doi: 10.1111/j.1600-0854.2007.00616.x. [DOI] [PubMed] [Google Scholar]