Abstract
Polychlorinated biphenyls (PCBs) are toxic industrial chemicals, complete carcinogens and efficacious tumor promoters. However, the mechanism(s) of PCB-mediated carcinogenicity remains largely undefined. One likely pathway by which these agents may play a role in carcinogenesis is the generation of oxidative DNA damage by redox cycling of dihydroxylated PCB metabolites. We have now employed a new 32P-postlabeling system to examine novel oxidative DNA lesions induced by Cu2+-mediated activation of PCB metabolites. 32P-Postlabeling of DNA incubated with various PCB metabolites resulted in over a dozen novel polar oxidative DNA adducts that were chromatographically similar for all active agents. The most potent metabolites tested were the hydroquinones (hydroxyl groups arranged para to each other) yielding polar oxidative adduct levels ranging from 55 to 142 adducts/106 nucleotides. PCB catechols, or ortho-dihydroxy metabolites, were up to 40% less active than their corresponding hydroquinone congeners while mono hydroxylated and quinone metabolites did not produce detectable oxidative damage over that of vehicle. With the exception of 2,4,5-Cl-2′,5′-dihydroxybiphenyl, this oxidative DNA damage appeared to be inversely related to chlorine content: no chlorine ≈ mono- > di- > tri-chlorinated metabolites. Importantly, copper, but not iron, was essential for activation of the PCB metabolites to these polar oxidative DNA adducts since in its absence or in the presence of the Cu+-specific scavenger, bathocuproine, no adducts were detected. Intervention studies with known reactive oxygen species (ROS) modifiers suggested that H2O2, singlet oxygen, hydroxyl radical and superoxide may also be involved in this PCB-mediated oxidative DNA damage. These data indicate a mechanistic role of several ROS, in addition to copper, in PCB-induced DNA damage and provide further support for oxidative DNA damage in PCB-mediated carcinogenesis.
Keywords: PCBs, oxidative DNA adducts, 32P-postlabeling
Introduction
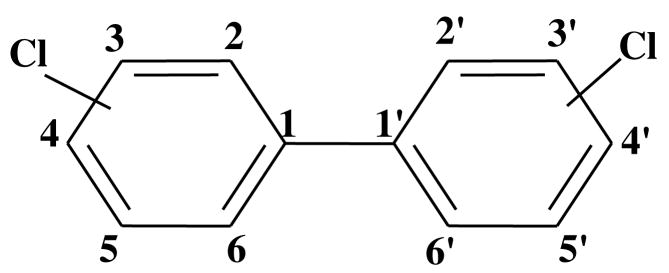
Polychlorinated biphenyls (PCBs)1 (Figure 1) are widespread environmental contaminants once produced commercially for a variety of diverse applications including dielectric, heat transfer, and hydraulic fluids, flame retardants, lubricating and cutting oils, and additives in paints, adhesives, sealants, plastic and pesticides (1). PCBs were mass produced from 1929 up until the early 1980s resulting in an estimated worldwide production of 1.5 million tons (2). Relatively large amounts, estimated to be as high as 31% of the total production, of these stable compounds have been released into the environment due to inappropriate disposal practices, accidents and leakages from industrial facilities. Sixty-five percent of the total environmental burden is still in use or in storage while only 4% is believed to have been destroyed (2, 3). Environmental sampling has shown that PCBs are almost always present as complex mixtures and found in just about every environmental compartment or matrix including human and animal tissues (4). The environmental impact of PCBs is further confounded by their ability to bioaccumulate and biomagnify in higher trophic organisms due to their high stability and lipophilicity.
Figure 1.
General structure and numbering system of polychlorinated biphenyls (PCBs).
Concerns regarding potential health detriments of PCBs arose in the early 1970s following the Yusho incident of 1968 during which approximately 1800 Japanese men and women ingested relatively large amounts of a complex mixture of PCBs and polychlorinated dibenzofurans from accidental contamination of rice oil (5). Reports of chloracne, follicular accentuation and pigmentation, eye discharge and swelling and peripheral neuropathy soon followed this disaster. A second contamination of rice oil occurred in Tiawan in 1978 (Yucheng incident). Similar effects of PCB toxicity were observed in the more than 2000 people affected by this latter incident. In a 40 year follow-up study of the Yusho accident examining the mortality of 1,664 patients, men showed an elevated mortality, compared to the general population, from all types of cancer, particularly lung and liver, while in women the mortality ratio was only increased for liver cancer (6). A similar follow-up study of the Yucheng accident did not find any increase in cancer mortality in either sex, as compared to the general Taiwanese population, but did find an early increase in liver mortality in men and a delayed increase in systemic lupus erythematosus mortality was found in women (7). In addition to these accidental poisonings by PCBs and dibenzofurans, epidemiologic, wildlife and experimental studies of the effects of PCBs have revealed reproductive toxicity, birth defects, abnormal growth and development in children of exposed mothers, dermatolgical toxicity, neurocognitive deficiencies and increased risk to immunological and infectious diseases (8). Evidence has also linked PCB exposure to cardiovascular disease (9) and a variety of cancers (10–12).
Several routes of exposure to PCBs have been identified, however, ingestion and dermal are believed to be the most common in humans. Due to their high lipophilicity, PCBs are easily absorbed through these avenues (13). Although many congeners of PCBs have been found to be quite stable in the body and tend to accumulate in fatty tissues, metabolism and excretion does occur to some extent, albeit at a slower rate than would be expected for less lipophilic compounds. In the first step of metabolism, P450 enzymes catalyze epoxidation or hydroxylation of the PCB. Subsequent glucuronidation, sulfation, or binding to glutathione may then result in excretion of these metabolites. If not excreted, the hydroxylated and epoxide metabolites can be further metabolized by P450 or epoxide hydrolase to dihydroxylated compounds or dihydrodiols, respectively. The dihydrodiols, through the action of dihydrodiol dehydrogenase, can also be metabolized to dihydroxylated species (13). Several studies have suggested that formation of dihydroxylated metabolites in the form of catechols (hydroxyl groups are ortho) and hydroquinones (hydroxyl groups are para) may play a significant role in PCB toxicity and carcinogenicity due to their redox cycling capacity (10, 14, 15).
Studies from our laboratory as well as others have shown that catechols and hydroquinones can be oxidized to semiquinones and ultimately quinones both non-enzymatically and enzymatically (14, 16–18). The resulting (semi-) quinones can bind to DNA and protein producing DNA (16, 17, 19–21) and protein (22) adducts. Studies from this laboratory with lower chlorinated PCB congeners, found that both human and rodent hepatic enzymes were able to metabolize these compounds to quinone metabolites resulting in direct DNA adduction (19). Subsequent studies suggested the involvement of intermediary semiquinone radicals in the binding of benzoquinones and hydroquinones to DNA (20). Further, Lin et al. (22) provided some of the earliest evidence that quinone metabolites are a reactive species formed during PCB metabolism in vivo. Using human hepatocytes, Borlak et al. (21) were able to establish a link between PCB exposure and DNA damage. In recent reports from our laboratory and others, redox cycling of the PCB (semi) quinones have also been shown to increase oxidative DNA damage as measured by DNA strand breaks, 8-oxodeoxyguanosine (8-oxodG) and 3-(2′-deoxy-beta-D-erythro-pentofuranosyl)-pyrimido[1,2-a]-purin-10(3H)-one (M1dG) adducts (15, 23–25).
In these current studies we have employed a new 32P-postlabeling system (26) coupled with Cu2+-mediated activation to evaluate the oxidative DNA-damaging potential of several dihydroxylated PCBs and PCB quinones. By using this model system we were able to detect numerous PCB catechol- and hydroquinone-induced novel polar oxidative DNA adducts. In order to study the reactive oxygen species (ROS) which may mediate the formation of these DNA adducts we have also employed several ROS modifiers.
Experimental Procedures
Chemicals
Hydroxylated PCBs and PCB quinones were synthesized and characterized as previously described (15, 27, 28). All other chemicals were purchased from Sigma Chemical Co. (St. Louis, MO) or Aldrich (Milwaukee, WI) unless stated otherwise. Sources of chemicals used for 32P-postlabeling analysis were as described previously (29).
DNA Adduction
Salmon testis DNA (300 μg/ml), freed from RNA by treatment with RNases and solvent extractions (29), was incubated (37°C, 1 h) with either vehicle or various PCB metabolites (100 μM) in the presence or absence of CuCl2 (100 μM) in 10 mM Tris HCl, pH 7.4. Intervention with ROS scavengers was carried out as described above in the presence of either 600 U/ml superoxide dismutase (SOD), 200 U/ml catalase, 100 mM sodium azide, 10 mM tiron, 10 mM bathocuproine, 100 mM mannitol or 10% dimethylsulfoxide. Modified DNA was purified by solvent extractions and ethanol precipitation of DNA as previously described (29). Concentrations of reactants and ROS scavengers were chosen based on earlier studies from this laboratory in which oxidation of PCB dihydroxy metabolites, in the presence of CuCl2, increased 8-oxodG levels (23).
32P-Postlabeling Analysis
Analysis of polar oxidative DNA adducts was as described elsewhere (26). Briefly, DNA (6 μg) was hydrolyzed to 3′-monophosphates with micrococcal nuclease and spleen phosphodiesterase and adducts were enriched by treatment with nuclease P1. Adducts were 5′-32P-labeled in the presence of molar excess of [γ-32P]ATP (< 3 μM; 550 Ci/mmol) and T4 polynucleotide kinase. The labeled nucleotides were treated with a mixture of nuclease P1 and potato apyrase to convert labeled bisphosphates of the residual normal nucleotides to monophosphates and unused [γ32P]ATP to 32Pi, respectively. Adducts were resolved by two-directional polyethyleneimine (PEI)-cellulose TLC (D1 = 50 mM sodium phosphate, pH 5.8/1 M formic acid onto a 6 cm Whatman no. 17 paper wick; D2 = 80 mM sodium phosphate, pH 6.0/10% acetonitrile (v/v).
Total nucleotides were analyzed by labeling dilute DNA digest (2 ng) in parallel with adducts followed by one-directional PEI-cellulose TLC in 0.5 M acetic acid/2 M formic acid (30). Adducts were visualized and quantified by a Packard InstantImager. Adduct levels were determined by relative adduct labeling (RAL), i.e., RAL = [cpm of adducts/cpm of normal nucleotides] × 1/dilution factor. Levels are expressed as adducts/106 nucleotides.
Statistical Analysis
Statistical comparisons were made using the two-way ANOVA followed by Fisher’s Least-Significant-Difference procedure. Values were considered significantly different when P ≤ 0.05.
Results
Detection of Novel Oxidative DNA Adducts
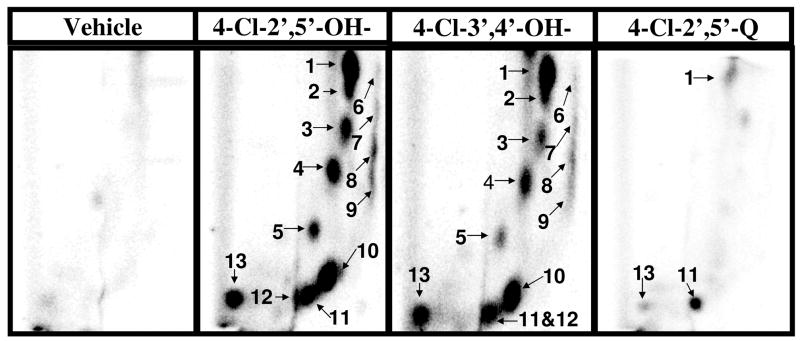
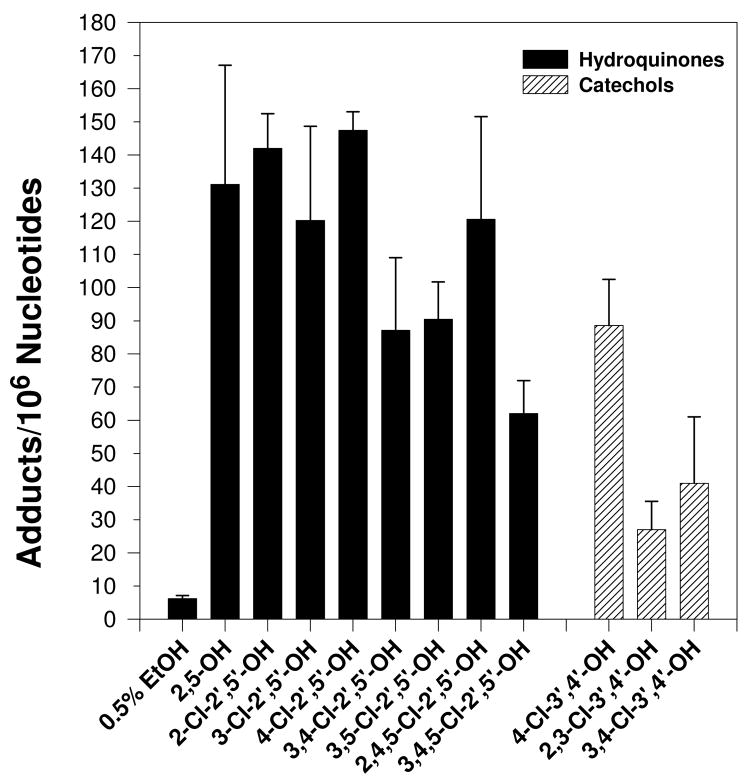
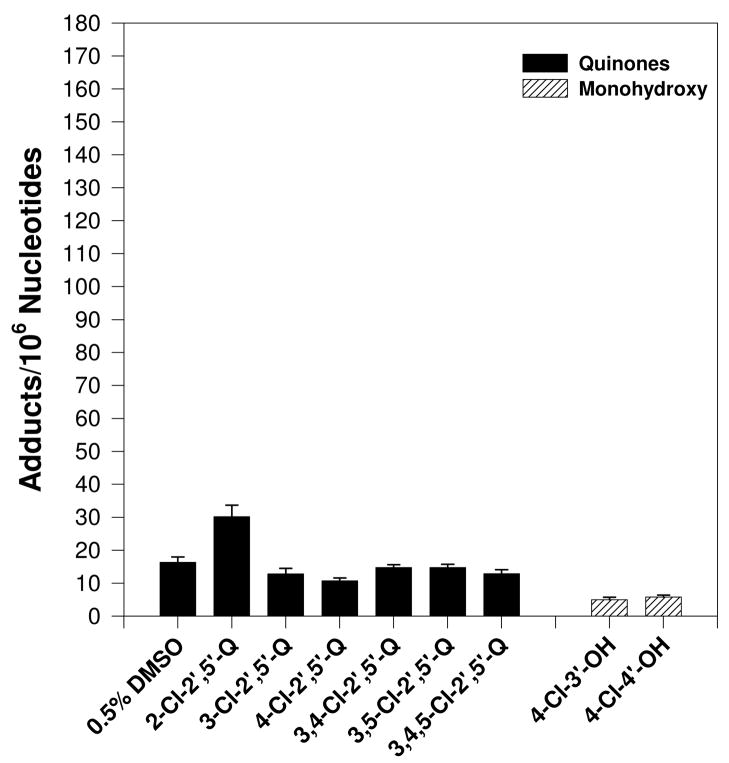
Cu2+ (100 μM)-mediated activation of PCB catechols and hydroquinones (100 μM), but not PCB quinones or monohydroxylated metabolites, in the presence of salmon testis DNA (300 μg/ml) resulted in the formation of nearly a dozen novel polar oxidative DNA adducts (Figure 2). Trace amounts of adduct #11 were also detected in several vehicle treated samples (data not shown). The characterization or these adducts as polar oxidative adducts is based on their ability to migrate under low-salt conditions, while adducts formed from the covalent binding of PCB quinones to DNA require high-salt and high-urea concentrations to displace them from the origin during thin-layer chromatography (19, 20). No adducts were detected in the absence of CuCl2 (data not shown). All dihydroxy metabolites tested resulted in essentially the same adduct pattern. DNA adduct levels induced by the dihydroxy metabolites ranged from 25 to 150 adducts/106 nucleotides with 2- and 4-chloro-2′,5′-dihydroxybiphenyl resulting in the highest adduct levels (Figure 3). Cu2+-activation of PCB monohydroxylated metabolites and quinones did not result in adduct levels significantly above that of vehicle treated DNA (Figure 4).
Figure 2.
Representative autoradiographs of 32P-labeled DNA adducts resulting from Cu2+ (100 μM)-mediated activation of indicated PCB metabolites (100 μM) or vehicle (0.5% ethanol) only. Specific conditions are described in text. Adduct numbers 6–9 can be viewed with longer exposure times.
Figure 3.
Effect of various dihydroxylated PCB metabolites (100 μM) on polar oxidative DNA adduct levels following incubation with DNA (300 μg/ml) and CuCl2 (100 μM). Vehicle used was 0.5% ethanol. Adduct levels were determined as the mean of 3 to 4 replicates ± S.E. Asterick (*) indicates values significantly different from vehicle at P ≤0.05. Specific conditions are described in text.
Figure 4.
Effect of various PCB monohydroxylated and quinone metabolites (100 μM) on polar oxidative DNA adduct levels following incubation with DNA (300 μg/ml) and CuCl2 (100 μM). Vehicle used was 0.5% DMSO for the quinones and 0.5% ethanol for the monohydroxylated metabolites. Adduct levels were determined as the mean of 3 to 4 replicates ± S.E. Specific conditions are described in text.
In order to determine if Cu2+ was the only transition metal which could catalyze dihydroxylated PCB-induction of these polar oxidative DNA adducts, 2-Cl-2′,5′-dihydroxybiphenyl was reacted with either Cu+ (CuCl), Cu2+ (CuCl2), Fe2+(FeSO4), or Fe3+ (FeCl3) (100 μM) in the presence of DNA (300 μg/ml) and 10 mM Tris HCl, pH 7.4. While all transition metals tested resulted in a chromatographically-identical DNA adduct pattern, both Cu+- and Cu2+-activation of 2-Cl-2′,5′-dihydroxybiphenyl resulted in similar adduct levels, but Fe2+ and Fe3+ resulted in adduct levels 3 to 5-fold lower than found in the presence of copper (data not shown).
Effect of ROS Modifiers on 2-Chloro-2′,5′-dihydroxybiphenyl-Mediated Polar Oxidative DNA Adduction
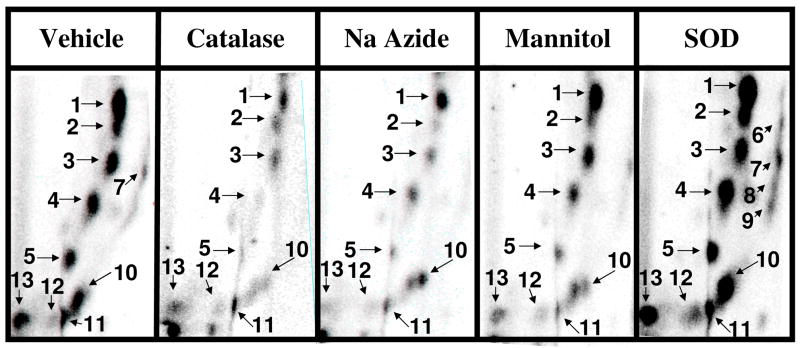
In order to determine the specific ROS involved in dihydroxy-PCB-mediated oxidative DNA adduction several ROS modifiers were added to the standard reaction described above. No differences in the polar oxidative DNA adduct pattern were observed following intervention with the modifiers tested (Figure 5). However, significant differences in adduct levels were observed (Figure 6). Tiron and bathocuproine, a scavenger of superoxide and Cu+-specific chelator, respectively, were the most potent adduct inhibitors (>99%) essentially preventing 2-chloro-2′,5′-dihydroxybiphenyl-induced oxidative DNA damage. Other agents tested, including catalase - an H2O2 scavenger, sodium azide – a singlet oxygen scavenger, and sodium benzoate, mannitol and dimethylsulfoxide – hydroxyl radical scavengers, were also effective inhibitors (30–85%) of this oxidative DNA damage. Interestingly, superoxide dismutase (SOD), a catalyst for the dismutation of superoxide to hydrogen peroxide, increased 2-chloro-2′,5′-dihydroxybiphenyl-mediated oxidative DNA adduction up to 230%. However, SOD in the presence of Cu2+ alone resulted in adduct levels (69 ± 10 adducts/106 nucleotides) significantly above that of vehicle (6 ± 1 adducts/106 nucleotides) indicating that the combination of SOD and Cu2+, in the absence of the PCB metabolite, can induce oxidative DNA damage (data not shown). Thus, it appears that SOD and Cu2+ have an additive, not synergistic, effect of 2-chloro-2′,5′-dihydroxybiphenyl-mediated polar oxidative DNA adduction.
Figure 5.
Representative autoradiographs of 32P-labeled DNA adducts resulting from Cu2+ (100 μM)-mediated activation of 2-chloro-2′,5′-dihydroxybephenyl (100 μM) in the presence of the indicated ROS modifiers or vehicle (HPLC water). Adduct numbers 6–9 can be viewed with longer exposure times.
Figure 6.
Effect of ROS modifiers or vehicle (HPLC water) on Cu2+(100 μM)-mediated oxidative DNA damage by 2-chloro-2′,5′-dihydroxybiphenyl (100 μM). Adduct levels were determined as the mean of 3 to 4 replicates ± S.E. Asterick (*) indicates values significantly different from vehicle at P ≤ 0.05. Specific conditions are described in text.
Discussion
PCBs can be metabolized to both mono- and dihydroxy metabolites which can be further metabolized to redox cycling (semi-) quinones (16–18). In previous studies from our laboratory and others we have shown that these quinones can induce DNA damage directly by covalently binding to DNA (16) or indirectly through redox cycling and subsequent attack of ROS on DNA resulting in 8-oxodG formation (23) and DNA strand breaks (15). We have extended these studies here and have detected numerous novel polar oxidative DNA adducts following Cu2+-mediated metabolism of the dihydroxylated, but not monohydroxylated or quinone, PCB metabolites. These polar adducts were chromatographically similar for all dihydroxy metabolites tested. Moreover, essentially the same DNA adduct pattern was found following transition metal activation of numerous structurally diverse dihydroxylated compounds, including estrogen catechols (unpublished data) and polycyclic aromatic hydrocarbon diols (31) as well as the Fenton reaction (Fe3+/H2O2) (32). These findings strongly support the role of common redox cycling intermediates in the formation of these DNA adducts.
Although the chemical structure of these adducts has not been determined, their characterization as polar oxidative DNA lesions is based on several observations: 1) ability to migrate under low-salt conditions, while adducts formed from the covalent binding of PCB quinones to DNA require high-salt and high-urea concentrations to displace them from the origin during thin-layer chromatography (19, 20); 2) chromatographic similarity to polar oxidative adducts produced from typical Fenton chemistry (32); 3) proximity to known oxidative markers including 5-hydroxy-deoxyuridine, 5,6-dihydrothymidine, thymidine glycol and 8-oxo-dG (26); 4) ability to be inhibited in the presence of antioxidants (33); and 5) chromatographically similar adduct patterns have also been found with numerous structurally diverse redox-active agents, including estrogen catechols (unpublished data), PAH diols (31) and catecholic dopamine analogs (unpublished data).
The level of oxidative damage induced by dihydroxylated PCB metabolites appeared to be structurally dependent. Chlorine was not required for oxidative DNA damage as Cu2+-mediated activation of 2,5-dihydroxybiphenyl resulted in relatively high levels of oxidative DNA adduction (131 adducts/106 nucleotides). In fact, with the exception of 2,4,5-Cl-2′,5′-dihydroxybiphenyl, the lower chlorinated congeners resulted in greater levels of oxidative DNA damage than the higher chlorinated congeners. This is supported by a recent study in which lower chlorinated dihydroxy PCB metabolites produced higher levels of superoxide than higher chlorinated congeners, although no structure-activity relationship was found for DNA strand breaks (15). The most distinguishing characteristic, with respect to oxidative DNA damage, of the dihydroxy metabolites tested was the position of the hydroxyl groups, either ortho (catechols) or para (hydroquinones) to each other. The hydroquinones produced, on average, 40% more oxidative DNA damage than their respective catechol counterparts following Cu2+-mediated activation. Similar findings were found for both dihydroxy PCB-mediated DNA strand breaks and 8-oxodG formation (15, 23).
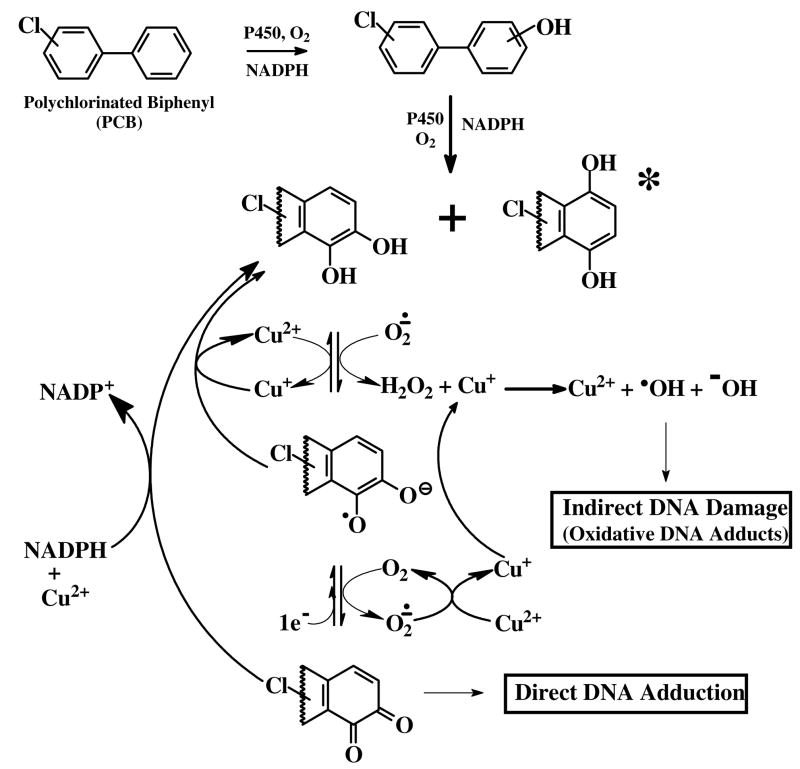
The proposed mechanism of this oxidative DNA damage is believed to occur via redox cycling of PCB catechols/hydroquinones and quinones via semiquinone intermediates (Figure 7). Oxidation of these dihydroxy metabolites to quinones can occur both enzymatically, via oxidases and peroxidases, and non-enzymatically, in the presence of transition metals such as copper (34). Our studies have indicated that Cu+ and/or Cu2+ are required for induction of the polar oxidative DNA adducts from PCB catechols and hydroquinones since in its absence no DNA adducts were detected. In further support of copper’s role in PCB-mediated polar DNA adduction, the addition of bathocuproine, a specific chelater of Cu+1, to the Cu2+-mediated activation of 2-chloro-2′,5′-dihydroxybiphenyl completely inhibited (>99%) the formation of polar oxidative DNA adducts. The replacement of Cu2+ with Cu+ in the PCB activation system resulted in similar adduct levels, however, the use of Fe2+ or Fe3+ was resulted in substantially lower levels of polar oxidative DNA adducts.
Figure 7.
Proposed mechanism of PCB catechol/hydroquinone-mediated oxidative DNA damage. Lower halogenated PCB’s can be metabolized by CYP450s to hydroquinones, which in the presence of transition metals such as copper, can be further oxidized to semiquinones and quinones that can bind directly to DNA forming covalent DNA adducts. These semiquinones and quinones are also capable of redox cycling producing superoxide which can be readily converted to H2O2. Superoxide can also reduce a metal ion, such as Cu+ to Cu2+, which in the presence of hydrogen peroxide can then be converted to the highly reactive hydroxyl radical. The hydroxyl radical can attack nucleophiles such as DNA producing oxidative DNA adducts and can generate additional reactive products including peroxyl radicals.
We have also determined that a peroxidase/H2O2 system is not sufficient to induce PCB dihydroxy metabolite-mediated polar oxidative DNA damage. This is in contrast to a previous report from our laboratory in which a low (2-fold) increase in 8-oxodG levels, over that of vehicle, was observed following lactoperoxidase/H2O2-mediated metabolism of 3,4-dichloro-2′,5′-dihydroxybiphenyl (23). It should be noted, however, that the latter study employed a non-adduct enrichment version of the 32P-postlabeling method for measuring 8-oxodG and thus had an increased susceptibility for artifactual production of 8-oxodG during the work up as unadducted dG was not removed from the DNA digest prior to labeling. We hypothesize that copper mediates oxidative DNA damage by reacting with hydrogen peroxide, produced from dismutation of superoxide - a byproduct of redox cycling of the PCB quinones and semiquinones - resulting in the production of hydroxyl radicals, singlet oxygen and metal-peroxide complexes previously shown to damage DNA (35).
Copper is an essential element in human diet and is required for normal enzymatic function (36). It is localized in human and animal tissues and circulated in the blood bound to protein carriers and transporters in both +1 and +2 valence states. The total body burden of copper has been estimated to be between 80–120 mg in a healthy adult with approximately 20% of this amount stored in the nuclei of cells (36, 37). In the nucleus, Cu2+ is found bound to nucleosomes with a binding constant of 4–5 × 104 M−1 (38). Studies have also shown that Cu2+ has a high affinity to DNA, preferentially binding to guanine residues at the N7 position (36). Further, this Cu2+-DNA interaction has been shown to promote DNA oxidation and the resulting damage enhanced by packaging of DNA as a nucleosome (36, 39). Thus, the availability and reactivity of copper coupled with its close proximity to DNA makes this transition metal a likely participant in ROS-mediated DNA damage.
In order to support the role of various ROS, in addition to Cu2+, in PCB-mediated oxidative DNA damage we tested several known ROS modifiers for their effect on 2-chloro-2′,5′-dihydroxybiphenyl/Cu2+-mediated polar oxidative DNA adduction. Known scavengers of singlet oxygen – sodium azide, superoxide - tiron, H2O2 – catalase and hydroxyl radicals –sodium benzoate, mannitol and dimethysulfoxide reduced, 30 to 99%, oxidative DNA damage induced by this PCB metabolite. Thus, these polar oxidative DNA adducts are mediated, at least in part, by several ROS including hydroxyl radical, superoxide, H2O2 and singlet oxygen. Interestingly, SOD, responsible for the dismutation of superoxide to H2O2, induced oxidative DNA damage in the presence of Cu2+ alone and had an additive effect on PCB-mediated oxidative DNA damage. Although some of the DNA damaging activity of SOD was independent of enzyme activity (heat inactivated SOD yielded adduct levels 5-fold above vehicle), this did not account for the entirety of the DNA damage. Mechanisms of increased oxidative damage by SOD in the presence of dihydroxy PCBs include release of copper ions from damaged SOD, peroxidase activity exhibited by SOD that has been shown to result in self-inactivation and the formation of enzyme bound oxidants such as SOD-Cu2+-•OH and induction of Cu+ accumulation (40).
This study coupled with several previously reported in vitro studies supports the role of ROS and DNA damage in PCB-mediated carcinogenesis, however, very few studies have addressed the in vivo effects of PCBs on oxidative DNA damage. In a recent study, the carcinogenicity of a mixture of PCBs was associated with chronic exposure but not acute exposure. Further, the incidence of preneoplastic lesions and subsequent cancer development, cholangio carcinoma and hepatocellular adenoma, correlated with the accumulation of the [3-(2′-deoxy-B-D-erythro-pentofuranozyl)-pyrimido[1,2-a]-purin-10(3H)-one (M1dG) oxidative DNA adduct in both a dose and time dependent manner following PCB exposure in rats (25). The inability of acute PCB exposure to induce oxidative DNA damage is also supported by an earlier study in male rats in which investigators were unable to detect any measurable induction of 8-oxodG levels following a single oral dose (20 mg/kg b.wt.) of Aroclor 1242, a PCB mixture composed predominantly of di-, tri-, and tetrachlorobiphyenyls (18). Studies from our own laboratory indicated a significant increase (up to 2 fold) in 8-oxodG levels following i.p. treatment (300 μmol/kg b.wt.) of male rats pretreated with a mixture of phenobarbital and 3-methylcholanthrene - P450 inducers - with either 4-chloro- or 3,4-dichlorobiphenyl (41). However, in a parallel study, 8-oxodG levels did not increase over that of vehicle following treatment of uninduced male rats with either 3,4-dichloro- or 3,4,5-trichlorobiphenyl. These results further suggest that that exposure to various agents - via dietary, lifestyle, environmental and other pathways - which may modify basal metabolic function, such as P450 inducers, may have a significant impact on disposition of toxic and carcinogenic environmental contaminants such as PCBs.
In recent unpublished studies from our laboratory we have also found that i.p. treatment (100 μmols/kg b.wt., single injection every third day) of female rats with 3,4,5-trichloro-, 3,3′,4,4′-tetrachloro- and 2,2′,4,4′,5,5′-hexachlorobiphenyls resulted in the enhancement of numerous polar endogenous oxidative DNA adducts from 2 to 3 fold over that of vehicle-treated animals. These endogenous oxidative adducts were chromatographiclly similar, although not identical, to those found by Cu2+-mediated activation of PCB catechols and hydroquinones. In a separate study employing the same PCBs with a modified dosing protocol (50 μmols/kg b.wt, single injection on alternate days), however, we did not find any induction of the endogenous polar oxidative DNA adducts for any of the PCBs. Neither study indicated any increase in 8-oxodG levels over that of vehicle-treated animals. Thus, the ability of PCBs to induce oxidative DNA damage in vivo is not conclusive and appears to be greatly dependent on dosing regimen and length of exposure. Therefore, these data clearly warrant further in-depth investigations into the oxidative activity and DNA damaging effects of PCBs using animal models.
In conclusion, we have shown that Cu2+-mediated activation of PCB catechols and hydroquinones can induce oxidative DNA damage as detected by the formation of numerous novel polar DNA adducts. This oxidative damage is mediated by copper as well as several ROS, including singlet oxygen, H2O2, superoxide and hydroxyl radical. These studies suggest a causal role of oxidative DNA damage in PCB-mediated cancer development and may contribute PCB-mediated toxicity.
Footnotes
Work was performed at the Department of Preventive Medicine and Environmental Health and Graduate Center for Toxicology, University of Kentucky, Lexington, Kentucky 40536
Abbreviations: PCBs, polychlorinated biphenyls; ROS, reactive oxygen species; 8-oxodG, 8-hydroxydeoxyguanosine; M1dG, 3-(2′-deoxy-beta-D-erythro-pentofuranosyl)-pyrimido[1,2-a]-purin-10(3H)-one; PEI, polyethyleneimine; RAL, relative adduct level
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Erickson MD. Introduction: PCB properties, uses, occurrence, and regulatory history. In: Robertson LW, Hanson LG, editors. PCBs: Recent Advances in Environmental Toxicology and Health Effects. The University Press of Kentucky; Lexington, KY: 2001. [Google Scholar]
- 2.Tanabe S. PCB problems in the future: foresight from current known knowledge. Environ Pollut. 1988;50:5–28. doi: 10.1016/0269-7491(88)90183-2. [DOI] [PubMed] [Google Scholar]
- 3.Holoubek I. Polychlorinated biphenyl (PCB) contaminated sites worldwide. In: Robertson LW, Hanson LG, editors. PCBs: Recent Advances in Environmental Toxicology and Health Effects. The University Press of Kentucky; Lexington, KY: 2001. pp. 17–26. [Google Scholar]
- 4.Fiedler H. Global and local disposition of PCBs. In: Robertson LW, Hanson LG, editors. PCBs: Recent Advances in Environmental Toxicology and Health Effects. The University Press of Kentucky; Lexington, KY: 2001. pp. 11–15. [Google Scholar]
- 5.Guo YL, Hsu CC. Yucheng and Yusho: The effects of toxic oil in developing humans in Asia. In: Robertson LW, Hanson LG, editors. PCBs: Recent Advances in Environmental Toxicology and Health Effects. The University Press of Kentucky; Lexington, KY.: 2001. pp. 137–141. [Google Scholar]
- 6.Onozuka D, Yoshimura S, Furue M. Mortality after exposure to polychlorinated biphenyls and polychlorinated dibenzofurans: a 40-year follow-up study of Yusho patients. Am J Epidemiol. 2009;169:86–95. doi: 10.1093/aje/kwn295. [DOI] [PubMed] [Google Scholar]
- 7.Tsai PC, Ko YC, Huang W, Liu HS, Guo YL. Increased liver and lupus mortalities in 24-year follow-up of the Taiwanese people highly exposed to polychlorinated biphenyls and dibenzofurans. Sci Total Environ. 2007;374:216–222. doi: 10.1016/j.scitotenv.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 8.Safe SH. Polychlorinated biphenyls (PCBs): Environmental impact, biochemical and toxic responses, and implications for risk assessment. Critical Rev Toxicol. 1994;24:87–149. doi: 10.3109/10408449409049308. [DOI] [PubMed] [Google Scholar]
- 9.Hennig B, Slim R, Toborek M, Hammock B, Robertson LW. PCBs and cardiovascular disease. In: Robertson LW, Hanson LG, editors. PCBs: Recent Advances in Environmental Toxicology and Health Effects. The University Press of Kentucky; Lexington, KY: 2001. pp. 211–220. [Google Scholar]
- 10.Ludewig G. Cancer initiation by PCBs. In: Robertson LW, Hanson LG, editors. PCBs: Recent Advances in Environmental Toxicology and Health Effects. The University Press of Kentucky; Lexington, KY: 2001. pp. 337–354. [Google Scholar]
- 11.Glauert HP, Robertson LW, Silberhorn EM. PCBs and tumor promotion. In: Robertson LW, Hanson LG, editors. PCBs: Recent Advances in Environmental Toxicology and Health Effects. The University Press of Kentucky; Lexington, KY: 2001. pp. 355–371. [Google Scholar]
- 12.Sargent LM. Role of polychlorinated biphenyl exposure in the progression of neoplasia. In: Robertson LW, Hanson LG, editors. PCBs: Recent Advances in Environmental Toxicology and Health Effects. The University Press of Kentucky; Lexington, KY: 2001. pp. 373–379. [Google Scholar]
- 13.James MO. Polychlorinated biphenyls: Metabolism and metabolites. In: Robertson LW, Hanson LG, editors. PCBs: Recent Advances in Environmental Toxicology and Health Effects. The University Press of Kentucky; Lexington, KY: 2001. pp. 35–46. [Google Scholar]
- 14.Robertson LW, Gupta RC. Metabolism of polychlorinated biphenyls (PCBs) generates electrophiles and reactive oxygen species that damage DNA. In: Williams GM, Aruoma OI, editors. Molecular Drug Metabolism and Toxicology. OICA International; 2000. pp. 16–32. [Google Scholar]
- 15.Srinivasan A, Lehmler H-J, Robertson LW, Ludewig G. Production of DNA strand breaks in vitro and reactive oxygen species in vitro and in HL-60 cells by PCB metabolites. Toxicol Sci. 2001;60:92–102. doi: 10.1093/toxsci/60.1.92. [DOI] [PubMed] [Google Scholar]
- 16.McLean MR, Robertson LW, Gupta RC. Detection of PCB adducts by the 32P-postlabeling technique. Chem Res Toxicol. 1996;9:165–171. doi: 10.1021/tx9500843. [DOI] [PubMed] [Google Scholar]
- 17.Oakley GG, Robertson LW, Gupta RC. Analysis of polychlorinated biphenyl-DNA adducts by 32P-postlabeling. Carcinogenesis. 1996;17:109–114. doi: 10.1093/carcin/17.1.109. [DOI] [PubMed] [Google Scholar]
- 18.Amaro A, Oakley G, Bauer U, Spielmann H, Robertson L. Metabolic activation of PCBs to quinones: reactivity toward nitrogen and sulfur nucleotides and influence of superoxide dismutase. Chem Res Toxicol. 1996;9:623–629. doi: 10.1021/tx950117e. [DOI] [PubMed] [Google Scholar]
- 19.Pereg D, Robertson LW, Gupta RC. DNA adduction by polychlorinated biphenyls: adducts derived from hepatic microsomal activation and from synthetic metabolites. Chem Biol Interact. 2002;139:129–144. doi: 10.1016/s0009-2797(01)00292-7. [DOI] [PubMed] [Google Scholar]
- 20.Arif JM, Lehmler HJ, Robertson LW, Gupta RC. Interaction of benzoquinone- and hydroquinone-derivatives of lower chlorinated biphenyls with DNA and nucleotides in vitro. Chem Biol Interact. 2003;142:307–316. doi: 10.1016/s0009-2797(02)00141-2. [DOI] [PubMed] [Google Scholar]
- 21.Borlak J, Hock A, Hansen T, Richter E. DNA adducts in cultures of polychlorinated biphenyl-treated human hepatocytes. Toxicol Appl Pharmacol. 2003;188:81–91. doi: 10.1016/s0041-008x(02)00075-3. [DOI] [PubMed] [Google Scholar]
- 22.Lin PH, Sangaiah R, Ranasinghe A, Upton PB, La DK, Gold A, Swenberg JA. Formation of quinonoid-derived protein adducts in the liver and brain of Sprague-Dawley rats treated with 2,2′,5,5′-tetrachlorobiphenyl. Chem Res Toxicol. 2000;13:710–718. doi: 10.1021/tx000030f. [DOI] [PubMed] [Google Scholar]
- 23.Oakley GG, Devanaboyina U-S, Robertson LW, Gupta RC. Oxidative DNA damage induced by activation of Polychlorinated biphenyls (PCBs): Implications for PCB-induced oxidative stress in breast cancer. Chem Res Toxicol. 1996;9:1285–1292. doi: 10.1021/tx960103o. [DOI] [PubMed] [Google Scholar]
- 24.Schilderman PAEL, Maas LM, Pachen DMFA, de Kok TMCM, Kleinjans JCS, van Schooten FJ. Induction of DNA adducts by several polychlorinated biphenyls. Environ Mol Mut. 2000;36:79–86. doi: 10.1002/1098-2280(2000)36:2<79::aid-em1>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 25.Jeong YC, Walker NJ, Burgin DE, Kissling G, Gupta M, Kupper L, Birnbaum LS, Swenberg JA. Accumulation of M1dG DNA adducts after chronic exposure to PCBs, but not from acute exposure to polychlorinated aromatic hydrocarbons. Free Rad Biol Med. 2008;25:585–591. doi: 10.1016/j.freeradbiomed.2008.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravoori S, Vadhanam MV, Davey DD, Srinivasan C, Nagarajan B, Gupta RC. Modulation of novel DNA adducts during human uterine cervix cancer progression. International journal of oncology. 2006;29:1437–1443. [PubMed] [Google Scholar]
- 27.Bauer U, Amaro AR, Robertson LW. A new strategy for the synthesis of polychlorinated biphenyl metabolites. Chem Res Toxicol. 1995;8:92–95. doi: 10.1021/tx00043a012. [DOI] [PubMed] [Google Scholar]
- 28.McLean MR, Bauer U, Amaro AR, Robertson LW. Identification of catechol and hydroquinone metabolites of 4-monochlorobiphenyl. Chem Res Toxicol. 1996;9:158–164. doi: 10.1021/tx950083a. [DOI] [PubMed] [Google Scholar]
- 29.Gupta RC. 32P-Postlabeling for detection of DNA adducts. In: Pfeifer GP, editor. Technologies for Detection of DNA Damage and Mutations. Plenum Press; New York: 1996. pp. 45–61. [Google Scholar]
- 30.Gupta RC, Arif JM. An improved 32P-postlabeling assay for the sensitive detection of 8-oxodeoxyguanosine in tissue DNA. Chem Res Toxicol. 2001;14:951–957. doi: 10.1021/tx000131d. [DOI] [PubMed] [Google Scholar]
- 31.Vadahnam MV, Krzeminski J, Amin S, Gupta RC. Cu2+-mediated oxidative DNA adducts from aromatic hydrocarbon catechols and diones. Proc Am Assoc Cancer Res. 2002;43 abstract 4289. [Google Scholar]
- 32.Gupta RC, Ravi Kumar MNV, Vadhanam MV, Arif JM. Detection of oxidatively damaged polar DNA adducts by 32P-postlabeling. Proc Am Assoc Cancer Res. 2001;42 abstract 2521. [Google Scholar]
- 33.Aiyer HS, Vadhanam MV, Gupta RC. Efficacy of cancer chemopreventive agents in protecting against oxidative DNA damage from Cu2+-mediated activation of 4-hydroxy estradiol. Proc Am Assoc Cancer Res. 2002;43 abstract 5682. [Google Scholar]
- 34.McLean M, Twaroski T, Robertson LW. A new mechanism of toxicity for PCBs: redox cycling and superoxide generation. Organo-Halogen Compounds. 1998;37:59–62. [Google Scholar]
- 35.Yamamoto K, Kawanishi S. Hydroxyl free radical is not the main active species in site-specific DNA damage induced by copper(II) ion and hydrogen peroxide. J Biol Chem. 1989;264:15435–15440. [PubMed] [Google Scholar]
- 36.Theophanides T, Anastassopoulou J. Copper and carcinogenesis. Crit Rev Oncol. 2002;42:57–64. doi: 10.1016/s1040-8428(02)00007-0. [DOI] [PubMed] [Google Scholar]
- 37.Agarwal K, Sharma A, Talukder G. Effects of copper on mammalian cell components. Chem Biol Interact. 1989;69:1–16. doi: 10.1016/0009-2797(89)90094-x. [DOI] [PubMed] [Google Scholar]
- 38.Gelagutashvili ES, Sigua KI, Sapojnikova NA. Binding and the nature of Cu(II) ion interaction with nucleosomes. J Inorg Biochem. 1998;70:207–210. doi: 10.1016/s0162-0134(98)10016-8. [DOI] [PubMed] [Google Scholar]
- 39.Liang Q, Dedon PC. Cu(II)/H2O2-induced DNA damage is enhanced by packaging of DNA as a nucleosome. Chem Res Toxicol. 2001;14:416–422. doi: 10.1021/tx0002278. [DOI] [PubMed] [Google Scholar]
- 40.Midorikawa K, Kawanishi S. Superoxide dismutases enhance H2O2-induced DNA damage and alter its site specificity. FEBS Letters. 2001;495:187–190. doi: 10.1016/s0014-5793(01)02383-3. [DOI] [PubMed] [Google Scholar]
- 41.Oakley GG, Robertson LW, Gupta RC. Oxygen free-radical mediated DNA damage in polychlorinated biphenyl (PCB)-treated male Sprague-Dawley rats. Proc Am Assoc Cancer Res. 1997;38 abstract 131. [Google Scholar]