Abstract
Several metabolic abnormalities are associated with relative excess or deficiency of adipose tissue. Identifying the regulators of adipogenic differentiation is critical for its successful manipulation. Ad36, a human adenovirus, is a novel factor that promotes adipogenesis. We exploited the adipogenic potential of Ad36 to reveal exogenous modifiers of adipogenesis in rodent preadipocyte cell line in presence or absence of differentiation inducers methyl-isobutyl-xanthine, dexamethasone, and insulin (M, D, and I). A non-adipogenic human adenovirus Ad2 was used as a negative control for viral infection. First, we confirmed that, Ad36, but not Ad2, increases lipid accumulation in presence or absence of MDI. Time course studies for expression of key genes of adipogenic cascade showed that Ad36, but not Ad2 down regulated preadipocyte marker gene Wnt10b, and up regulated expression of early (C/EBPδ and C/EBPβ), intermediate (PPARγ2) and late genes (aP2 and G3PDH) of adipogenic cascade even in the absence of MDI. In presence of MDI, onset of expression of adipogenic genes coincided for Ad36 and control groups, but the expressions were significantly greater for the Ad36 group. Next, we observed that attenuation of Ad36 mRNA expression by an anti-adenoviral agent reduced 3T3-L1 differentiation, indicating that viral mRNA expression is required for the process. Furthermore, with or without MDI or its components, Ad36 significantly increased lipid accumulation in 3T3-L1 cells. Cell confluency at the time of Ad36 infection positively influenced lipid accumulation. The results reveal that Ad36 is an MDI-independent exogenous regulator of the adipogenic process. Elucidating the molecular pathways involved may reveal novel regulatory controls of adipogenesis.
Keywords: adenovirus, Ad36, obesity, adiposity, 3T3-L1, adipocytes, lipogenesis, adipogenesis
INTRODUCTION
Excessive or impaired adipogenesis may lead to obesity or lipodystrophy, respectively, and can play a major role in dysregulation of insulin secretion and action, glucose and lipid metabolism, energy balance, immune functions or reproduction(1). This underscores the need to determine various intra- and extra-cellular regulators of the adipogenic process.
Most of our understanding about adipogenesis comes from studies using cell culture models. While it does not fully resemble mammal primary preadipocytes, the best-studied model is 3T3-L1 cells–a murine preadipocyte cell line. Upon reaching confluence, the proliferative preadipocytes become growth arrested by contact inhibition. Upon stimulation by a combination of adipogenic inducers, including a glucocorticoid agonist (Dexamethasone, D), an agent to increase intracellular cAMP (methyl-isobutyl-xanthene, M) and high concentrations of insulin (I) to stimulate insulin like growth factor-I receptors, confluent cells re-enter the cell cycle and undergo a few rounds of cell division, before differentiating terminally. The last stage is characterized by the coordinated expression of specific genes that will finally determine the specific adipocyte phenotype of the cells(2). Adipogenesis in vitro follows a highly ordered and well-characterized temporal sequence, including a reduction in expression of preadipocyte specific genes such as Pref-1 and Wnt10b and increased expression of the peroxisome proliferator-activated receptor(PPAR) family (a subgroup of nuclear hormone receptors) and the CCAAT/enhancer binding protein (C/EBP) family of genes (3, 4). Detailed insight about adipogenic process in 3T3-L1 cells enables in understanding the role of various adipogenic agents.
Human Adenovirus Ad36 is such an adipogenic agent, which increases adiposity in experimentally infected animal models such as chickens, mice, rats and marmosets(5–9) increases commitment, differentiation and lipid accumulation in adipocyte progenitors(10, 11) and improves glucose disposal in cell culture and animal models(9, 12–14). In the United States, Ad36 shows about 15 – 17% prevalence in adult population (15). About 30% of the obese, but only 11% of the non-obese adults are naturally infected with the virus(15) and the natural infection is associated with greater BMI(15). Ad36 DNA can be isolated from human adipose tissue(10). Human adenoviruses are non-enveloped DNA viruses. Ad36 belongs to serotype D of 51 human adenoviruses identified and is antigenically unique(16). Ad36 was first isolated from fecal sample of a girl suffering from enteritis(16) Subsequently, additional adipogenic (Ad5 and Ad37)(17, 18) or non-adipogenic (Ad2, Ad31)(17) human adenoviruses were identified.
To comprehensively determine adipogenic effect of Ad36, we studied various aspects of adipogenic process, including the requirement of viral gene expression and the pattern of cellular gene modulation, role of adipogenic inducers, and the degree of cellular confluency on Ad36 induced lipid accumulation. Ad2 was used as a negative control for viral infection. 3T3-L1 cells are non-permissive for Ad2 entry. Therefore, for experiments using Ad2, 3T3-L1 cells which overexpress “coxsackie virus adenovirus receptor” (3T3ΔCARL1) were used. As previously described, 3T3ΔCARL1 cells are permissive to Ad36 and Ad2 (19) and were a gift from Dr. David Orlicky (20).
Collectively, the experiments presented below indicate that without the presence of adipogenic inducers, Ad36 can induce adipogenesis. In presence of adipogenic media, the effect of Ad36 on lipid accumulation is enhanced. Viral mRNA expression is required for this effect.
MATERIALS AND METHODS
Media used
Growth media
Dulbeco’s Minimum Essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic-antimycotic.
Differentiation media
DMEM, 10% FBS, 1% antibiotic –antimycotic, 10μg/mL insulin, 39μg/mL dexamethasone, and 115μg/mL 3-isobutyl-1-metylxanthine.
Maintenance media
DMEM, FBS, antibiotic-antimycotic, 2.5 μg/mL insulin.
1) To determine the effect of Ad36 on adipogenic pathway in presence or absence of MDI
a) Gene expression
Two days post confluence, 3T3ΔCARL1 cells were incubated for 1h with growth media (Control) or maintenance media + 3.8 PFU(plaque forming units)/cell of either Ad36 or Ad2. Next, the media were replaced with either differentiation media (MDI group) or growth media (No MDI group). After 48h, the differentiation media were replaced with maintenance media. Media in all groups were refreshed every 2d. Expressions of key genes of adipogenic cascade- Wnt10b, C/EBPβ, C/EBPα, C/EBPδ, PPARγ2, aP2 and G3PDH were determined by qRT-PCR on days 0, 1, 3, 5, 7, and 9 using β-actin as a control for normalization and quantification. Expression of viral gene E1A was determined for Ad36 and Ad2 by qRT-PCR, to confirm successful viral entry in cells.
b) Lipid accumulation
Confluent 3T3ΔCARL1 cells treated with or without MDI and inoculated with Ad36, Ad2 or media as described above in experiment 1a were used for quantifying lipid accumulation using the Oil Red O (ORO) assay described below. To pictorially demonstrate specificity of lipogenic potential of Ad36, we compared lipid accumulation by Ad36 vs Ad2. 3T3ΔCARL1 cells were either mock infected(control) or infected with 3.8 MOI of Ad36 or Ad2 and incubated without the differentiation media or in presence of MDI, as described above and stained with ORO 12 d or 14 d post inoculation, respectively.
2) Is Ad36 mRNA expression required to induce adipogenesis
3T3-L1 preadipocytes support Ad36 mRNA expression, but not the replication of virus particles, suggesting that Ad36 mRNA is sufficient to induce adipogenesis in these cells (21). Here we determined if the attenuation of Ad36 mRNA expression by an anti-adenoviral agent may reduce Ad36 induced proliferation, differentiation and lipid accumulation in 3T3-L1 cells. Cidofovir (CF; a gift from Gilead Sciences, Inc.), an antiviral agent that successfully reduces Ad36 enhanced lipid accumulation in presence of MDI (21), was used as follows:
Confluent 3T3-L1 preadipocytes (ATCC#CCL-92-1, Mannassas, VA) cultured in growth medium were divided in 4 groups (Con, Con+CF, Ad36 and Ad36+CF). The two Ad36 groups were inoculated with 3.8 PFU/cell of Ad36 and the remaining 2 groups were inoculated with media (control). One group each from Ad36 and control received 0 or 10μM CF. Dose of CF that is effective but not cytotoxic was previously determined (21). RNA was extracted on days 1 and 2, treated with deoxyribonuclease I(Cat#18068-015, Invitrogen, Carlsbad, CA) and reverse-transcribed to cDNA. Expression of C/EBPβ, E4-orf1 and E1A (early genes of Ad36) was determined by qRT-PCR using β-actin as a control for normalization and standard curve quantification. Lipid accumulation was measured by ORO staining(21). Cells were counted using a hemocytometer. Total and phosphorylated PKB (protein kinase B) was determined by western blot analysis.
3) What is the effect of individual components or the combination of M,D, and I on lipid accumulation promoted by Ad36
a) Time course of lipid accumulation and cell proliferation
Confluent 3T3-L1 cells treated with or without MDI and inoculated with Ad36 or media as described above in experiment 1a were used for cell counts and lipid accumulation determination using hemocytometer and ORO assay, respectively.
b) Effect of M, D, and I combinations on lipid accumulation
3T3-L1 cells were inoculated with Ad36 (3.8 PFU/cell) or media (control), 2d post confluence in 6-well plates. After 1h of incubation, the media from all the plates were removed. One control plate and one infected plate received differentiation media (MDI group). A second set of control and infected plates received growth media (No-MDI group). The next set of control and infected plates received differentiation media without insulin(MD group), without Dexamethasone(MI group) or without Isobutyl-methyl-xanthene(DI group). Forty-eight hours post inoculation, the groups without insulin (“No-MDI” and MD) received growth media, while the others received maintenance media. Five days post inoculation, lipid accumulation was determined by ORO in the groups MDI, MD, DI and MI, and on day-8, for the ‘No-MDI’ group.
4) Does the level of confluency of 3T3-L1 cells modify the lipogenic effect of Ad36
To better detect small changes in lipid accumulation, 3T3-L1 cells were infected with a low viral dose (1PFU/cell) on the day of confluence or 1 or 2d earlier and exposed to MDI at confluency. Lipid accumulation was determined by BODIPY staining(10), either 5d post-infection, or 5d post-confluence. Briefly, cells were fixed with 4% paraformaldehyde and stained with BODIPY at 10μg/ml overnight, washed, and the fluorescence was quantitated using Flexstation-196 at 485–538 nm. This provided the cells of different baseline confluence, an opportunity to accumulate lipid for a constant number of days either post Ad36 infection or post confluence. This design was repeated in absence of MDI, and the lipid accumulation was determined 9d post Ad36 infection or post-confluence.
Techniques and Assays
Virus Preparation
Ad36 was obtained from American Type Culture Collection(ATCC) and propagated in A549 cells after plaque purifying three times as described(5, 6). Viral titers were determined by plaque assay and cell inoculations were expressed as plaque forming units per cell(5, 6).
Lipid Content
Lipid accumulation was assessed by either ORO staining(22) or by BODIPY staining(10),
Cell counts
Cells were trypsinized and counted using a hemocytometer.
qRT-PCR
Real-time-quantitative PCR was conducted using AB-PRISM-7700 sequence detector(Applied Biosystems, Branchburg, NJ) and a SYBR-Green detection system(Bio-Rad). A standard was generated using cDNA pooled from the experimental samples. Relative expression levels were determined by normalization to β-actin and expressed as arbitrary units(AU). The following primer sequences were used.
Mouse β-actin primers
Forward Primer: 5′-ACGTTGACATCCGTAAAGAC-3′
Reverse Primer: 5′-GATCTTCATGGTGCTAGGAG-3′
Product size: 125 bp
Mouse C/EBPα primers
Forward: 5′-TGGACAAGAACAGCAACGAG-3′
Reverse: 5′-TCACTGGTCAACTCCAGCAC-3′
Product size: 126 bp
Mouse C/EBPβ primers
Forward: 5′-GAGCGACGAGTACAAGATGCGG-3′
Reverse: 5′-TTGTGCTGCGTCTCCAGGTTG-3′
Product size: 95 bp
Mouse C/EBPδ primers
Forward: 5′-CTGAACGACCTATACCTCAGACC-3′
Reverse: 5′-AGCTTCTCTCGCAGTCCAGT-3′
Product size: 121 bp
Mouse PPARγ2 primers
Forward: 5′-CTCCGTGATGGAAGACCACT-3′
Reverse: 5′-AACCATTGGGTCAGCTCTTG-3′
Product size: 120 bp
Mouse G3PDH primers
Forward: 5′-GATGCTAAATGGGCAGAAGC-3′
Reverse: 5′-CTGGCCCTCATAGCACACTT-3′
Product size: 129 bp
Mouse aP2 primers
Forward: 5′-GCG TGG AAT TCG ATG AAA TCA -3′
Reverse: 5′-CCC GCC ATC TAG GGT TAT GA -3′
Mouse Wnt10b primers
Forward: 5′-CCACTGGTGCTGTTATGTGC-3′
Reverse: 5′-CAGTGCTTCTCCTCCTCGTC-3′
Product size: 108 bp
Ad-36 E1A 13S
Forward: 5′-TGAGCAGCAGATGGCTCTAATCTC-3′
Reverse: 5′-GGTCTTCTTCTGAGGGTGATGACTC -3′
Ad-36E4orf1
Forward: 5′-GGCATACTAACCCAGTCCGATG -3′
Reverse: 5′-AATCACTCTCTCCAGCAGCAGG -3′
Ad2 E1A 13S
Forward: 5′-CTGTGGCATGTTTGTCTACAGTCC -3′
Reverse: 5′-ATGTCGGGCGTCTCAGGATAG -3′
Western blot assay
Proteins were quantitated using standard bicinchoninic-acid assay. The samples were run on gradient gels (Cat#161–1159, BioRad, Hercules, CA) along with a molecular weight standard (Cat#sc-2035, Santa Cruz Biotech, Santa Cruz, CA). Total protein measurements were normalized to Mitogen-Activated Protein Kinase(MAPK). Protein-Kinase B(PKB) phosphorylation was normalized to total PKB. Antibodies were purchased from Cell Signaling, (Danvers, MA) as follows: phospho-PKB(ser473), (Cat#9271S); total PKB(Cat#9272) MAPK(Cat#9212).
Statistical analysis
At least triplicate samples were used for all assays. Mock infected (control), Ad2, and Ad36 groups were compared using one-way ANOVA followed by Tukey comparison for all groups using GraphPad InStat software (GraphPad Software, Inc., San Diego, CA, version 3.06). Student’s T-Test determined significance(p<.05) when two groups were compared (Ad36 vs control).
RESULTS
1) In the absence of MDI, Ad36 induces adipogenesis, and enhances it in the presence of MDI
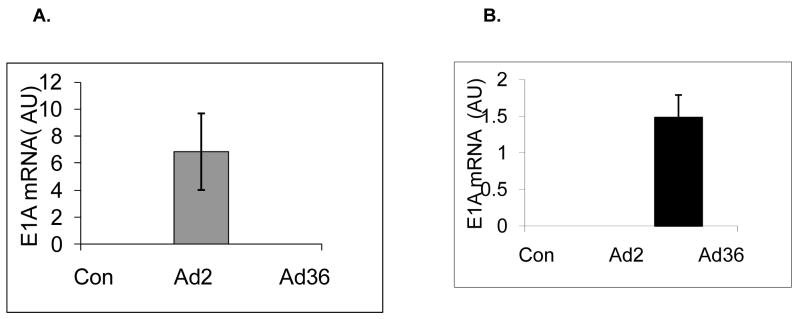
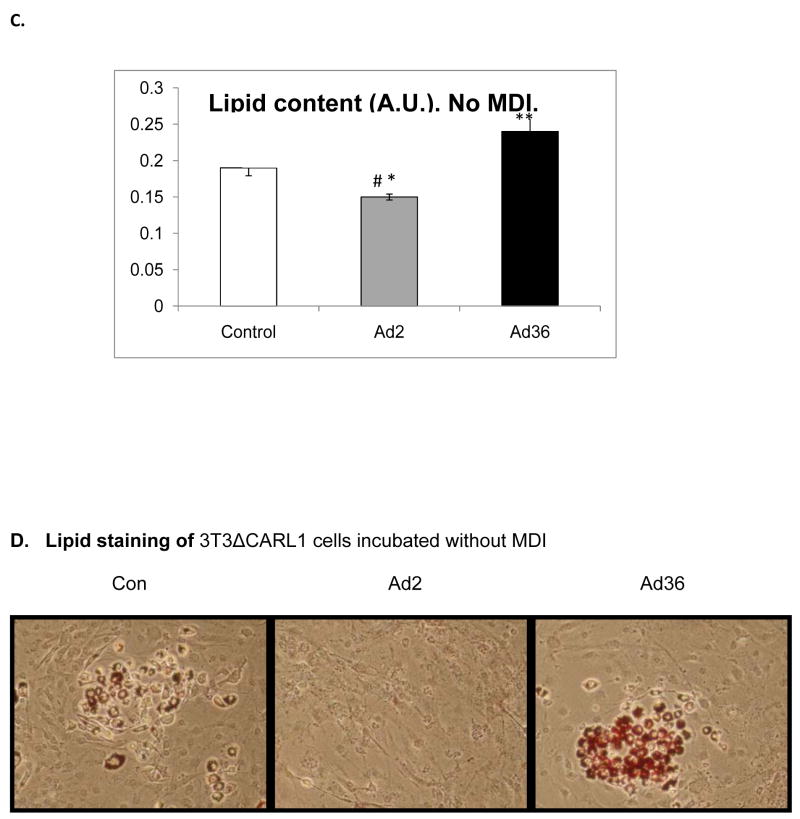
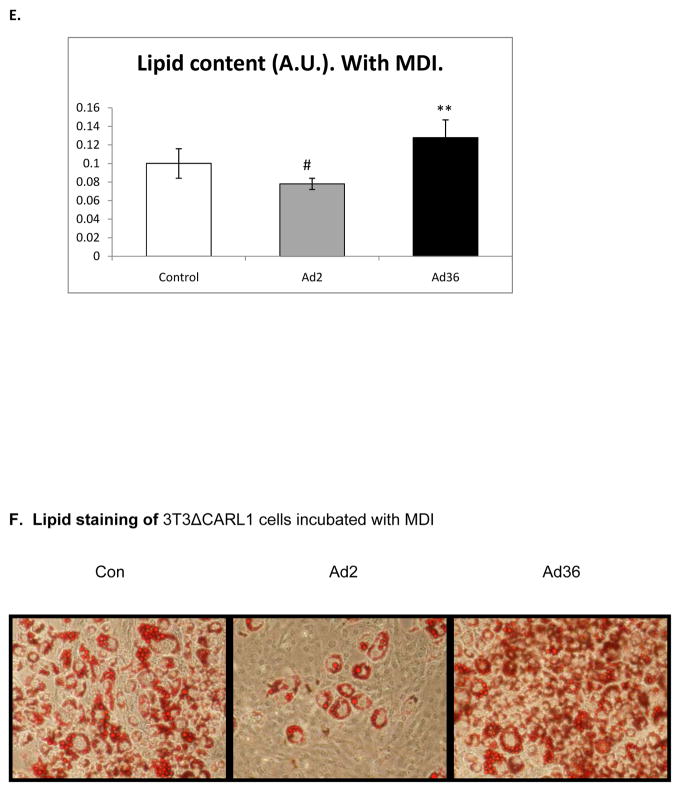
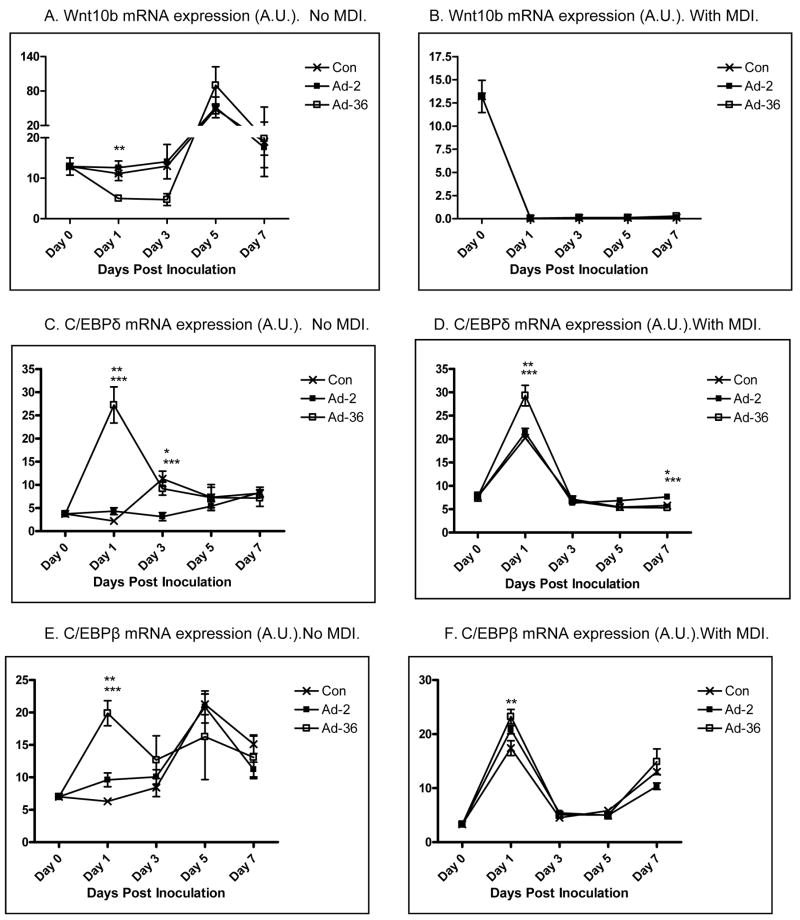
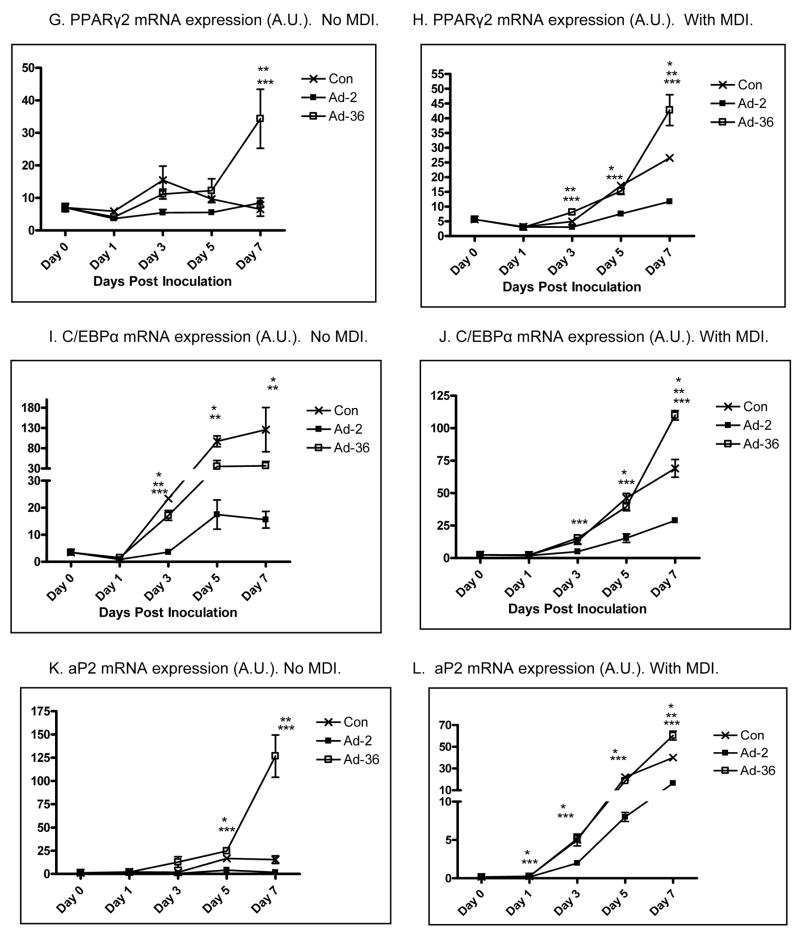
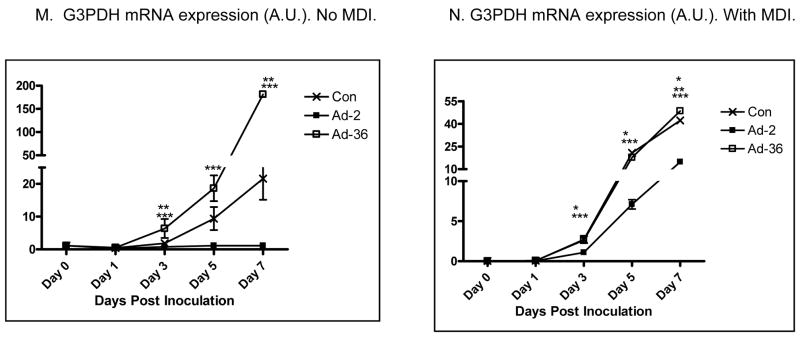
We first confirmed that Ad36 and Ad2 infect 3T3ΔCARL1 cells as indicated by viral mRNA expression in the respective groups (Figure 1A & B). Despite the infection, Ad36, but not Ad2 infected groups showed greater lipid accumulation compared to uninfected control (Figure 1C–F). This also confirmed that Ad2 is an appropriate negative control for viral infection. The effect of Ad36 on time course for expression of key genes of adipogenesis and lipid accumulation was extensively characterized in presence or absence of MDI (Figure 2). Ad36 significantly induced the adipogenic cascade even in the absence of MDI, as indicated by a reduction in preadipocyte gene marker (Wnt10b), and increase in differentiation promoters (C/EBPδ, C/EBPβ and PPARγ2) and lipogenic markers (aP2, G3PDH). As expected, not much activation of the cascade was observed in Ad2 or control groups. When induced by MDI, the control group responded predictably, by increasing the expressions of adipogenic genes. This adipogenic effect of MDI was further enhanced by Ad36, but not by Ad2. In fact, even in presence of MDI, the Ad2 group had significantly lower induction of adipogenic genes. This is in congruence with lower lipid accumulation observed in Ad2 infected groups (Figure 1, C–F).
Figure 1. Ad36 but not Ad2 increases lipid accumulation.
3T3ΔCARL1 cells were either mock infected (control) or infected with 3.8 MOI of Ad36 or Ad2. Expression of Ad36 and Ad2 E1A genes was determined in all groups. The cells were incubated with or without MDI, and stained with ORO 12 d or 14 d post inoculation for quantifying lipid accumulation. Mean ± SD. * Ad2 significantly different from control at p<0.05. ** Ad-36 significantly different from control at p<0.05. # Ad-2 significantly different from Ad-36.
Figure 2. Time course of Ad36-induced adipogenic gene expression presence or absence of MDI.
Time course of mRNA expression of key genes of adipogenic cascade in 3T3ΔCARL1 cells infected with Ad36 or Ad2 (3.8PFU/cell either virus) or mock infected and incubated with or without MDI. Mean±SD. Values from different group for a given day were compared to determine significant difference, by one-way ANOVA, followed by Tukey comparison.
* Ad-2 significantly different from control at p<0.05. ** Ad-36 significantly different from control at p<0.05. *** Ad-2 significantly different from Ad-36 at p<0.05.
Some interesting patterns were observed in individual gene expressions. C/EBPβ expression showed a second peak for the Ad36 group in presence or absence of MDI (Figure 2 & F). Possibly, the second peak in the Ad36 group was contributed by the uninfected cell population in the group, as it corresponded with the peak in the Control and Ad2 groups. Similarly, in no-MDI induction, Wnt10b expression showed initial and significant reduction in Ad36 group, followed by a second peak corresponding to the peaks in Ad2 and Control groups (Figure 2A). Taken together, these second peaks suggest rounds of replication and new cell formation. It appears that the MDI induction sharply reduced preadipocyte cell type as indicated by a dramatic reduction in Wnt10b expression in all groups (Figure 1B). Moreover, C/EBPα was modulated differentially by Ad36 in presence or absence of MDI induction (Figure 2I–J). C/EBPα expression increased by day 7 in presence or absence of MDI in the Control groups, but remained significantly lower in Ad36 or Ad2 groups in absence of MDI.
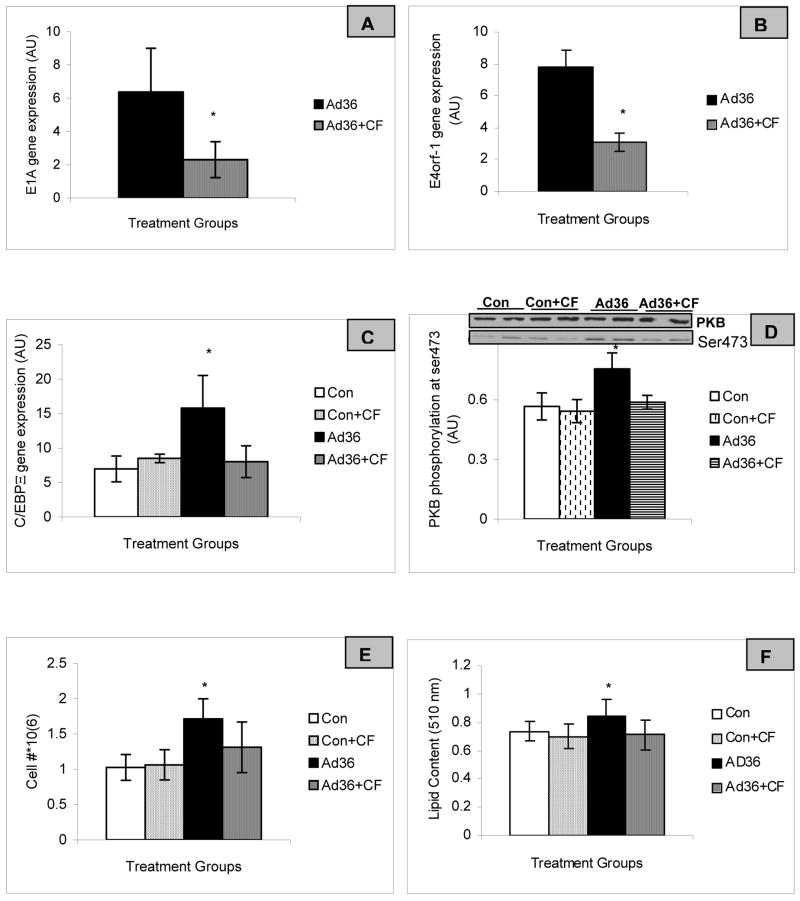
2) Ad36 mRNA expression is required to induce adipogenesis
After establishing non-adipogenic potential of Ad2 above, we focused on determining the adipogenic effect of Ad36 in the subsequent experiments. Viral entry of Ad36 in 3T3-L1 cells was confirmed by the presence of Ad36 E1A and E4orf1 mRNA expression (Figure 3). Predictably, Ad36 induced adipogenic changes as suggested by increased C/EBPβ mRNA expression, phosphotidyl inositol 3–kinase(PI3K) pathway activation as indicated by PKB phosphorylation (19), lipid accumulation and cell proliferation, even in the absence of MDI. Reduction of Ad36 mRNA expression by CF appears to attenuate the adipogenic potential of the virus. The results indicate that Ad36 mRNA expression is required for its adipogenic effect.
Figure 3. Active Ad36 is required for inducing differentiation in 3T3-L1 cells.
3T3-L1 cells were inoculated with Ad36 (3.8 PFU/cell) or media with 0 or 10μM cidofovir in the absence of MDI on day 1 post inoculation. One day later, E1A-13S, E4orf1 and C/EBPβ mRNA levels were determined by qRT-PCR and normalized to β-actin. Total and phospho-PKB protein abundance were determined by Western blot. Lipid accumulation was measured by ORO 11d post inoculation and cell were counted using a hemocytometer. Mean±SD from n=3. * Values are significantly different at p<.05 compared to the rest of the groups.
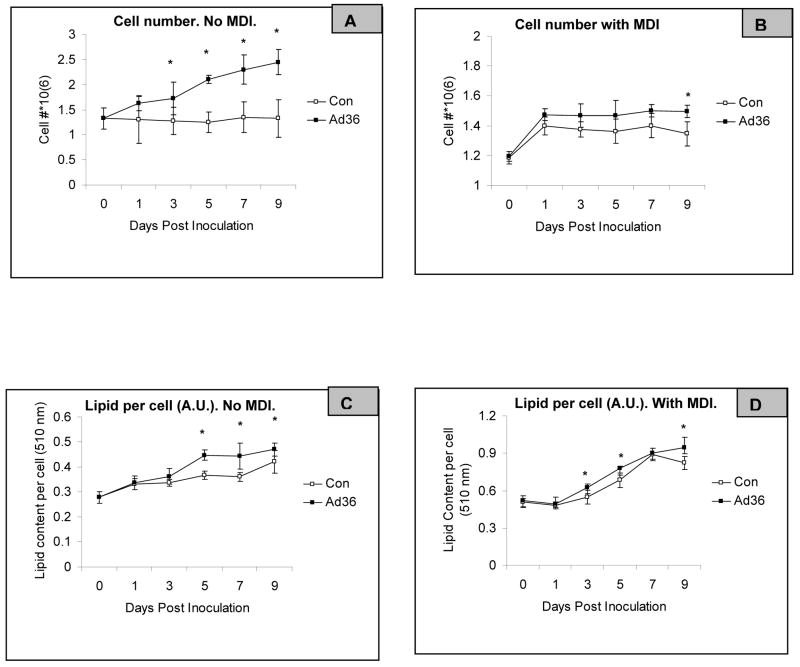
3) Ad36 can increase lipid accumulation in presence of various combinations of M,D, and I
While Ad36 significantly increased cell number regardless of MDI presence, the effect was most pronounced in the absence of MDI, suggesting a stronger suppressive effect of MDI on cell proliferation, in favor of differentiation (Figure 4A–B). To adjust for the increasing cell number, lipid accumulation was expressed on per cell basis. In the absence of MDI, Ad36 induced cellular lipid accumulation is delayed (Figure 4C). In the presence of MDI, while the control group predictably increases cellular lipid, Ad36 has greater lipogenic effect (Figure 4D).
Figure 4. Time course of Ad36 induced cell proliferation and lipid accumulation in presence or absence of MDI.
A–D: Time course of cell count and lipid accumulation in 3T3-L1 cells infected with Ad36 (3.8PFU/cell) or mock infected and incubated with or without MDI. Cells were counted by hemocytometer. The lipid accumulation was determined by ORO. Mean±SD. *P<.05 between the two groups for respective days.
Based on the different rates of lipid accumulation observed in the time course above in presence or absence of MDI, days 5 and 8 were selected to determine the effects of individual components of M, D, I treatments. As expected, the MDI induction increased lipid accumulation by 4.5-fold in uninfected cells (Table 1) compared to the group without MDI. However, the presence of Ad36 further increased MDI induced lipid accumulation by 23% per cell. Of the three constituent inducers of MDI, Ad36 induced significantly greater lipid accumulation in the absence of I or D, but not without M, compared to cells exposed to similar inducers. This suggests the significance of M, a cyclic-AMP promoter, in Ad36 induced lipid accumulation.
Table 1.
Cellular lipid content in presence of different inducers. Mean±SE
| Group | Control Absorbance/μg DNA | Ad36 Absorbance/μg DNA | P |
|---|---|---|---|
| MDI | 0.48 ± 0.029 | 0.59 ± 0.020 | < 0.02 |
| No MDI | 0.11 ± 0.005 | 0.19 ± 0.020 | < 0.02 |
| MD | 0.07 ± 0.005 | 0.14 ± .009 | < 0.0003 |
| MI | 0.35 ± 0.012 | 0.41 ± 0.017 | < 0.02 |
| DI | 0.27 ± 0.017 | 0.30 ± 0.020 | NS |
3T3-L1 cells were mock infected (Control) or infected with Ad36 (3.8PFU/cell). Lipid accumulation was determined by ORO 8d post infection for the “No-MDI” group and 5d post infection for other groups.
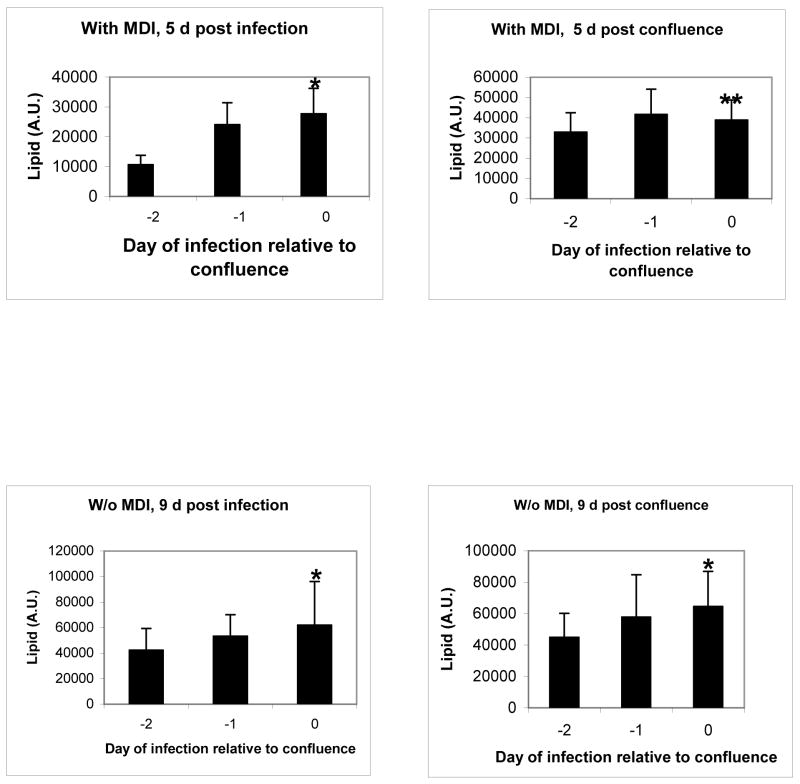
4) Confluency of 3T3-L1 cells contributes to their lipogenic response to Ad36
Considering the robust promotion of cell proliferation by Ad36 (Figure 4A–B), we determined if the infecting proliferating cells before reaching confluence would enhance their lipogenic response to Ad36. Compared to the cells infected by Ad36 2d before confluence, those infected upon confluence accumulated significantly greater lipid, either 5d post infection or 5d post confluence and in presence of MDI(Figure 4). Similarly, in absence of MDI, cells infected at confluence had significantly greater lipid compared to their uninfected counterparts 9d post infection or post confluence. These results indicated that while 3T3-L1 cells proliferate in response to Ad36 infection, infecting the confluent cells was important for greater Ad36 induced lipid accumulation.
Discussion
Preadipocyte differentiation is extensively studied in 3T3L1 and 3T3F442A cells as surrogate models for deciphering the role of fat cell differentiation in human obesity. In this study, preadipocyte differentiation induced by Ad36 can be compared to conventional induction of differentiation by MDI. Overall, the adipogenic program induced by Ad36 was very similar to that induced by MDI with respect to expression of key adipogenic genes, followed by lipid accumulation, indicating the ability of Ad36 to simulate the MDI effect and induce adipogenesis. A noticeable difference was the relative down-regulation of C/EBPα by Ad36 in absence of MDI. C/EBPα suppresses cell cycle progression (23), therefore, considering the increased cell proliferation by Ad36 in absence of MDI, its down-regulation by Ad36 would be expected. Down regulation of C/EBPα by Ad36 agrees well with our earlier report that showed Ad36 down regulated expression and secretion of leptin (13), a gene under the transcriptional control of C/EBPα (24). Down regulation of leptin may further promote adipogenesis in autocrine/paracrine way as previously discussed (13).
While C/EBPα and PPARγ coordinately upregulate adipocyte differentiation, Ad36 induced differentiation appears to proceed without upregulation of C/EBPα but with upregulation of PPARγ2 and the downstream genes G3PDH and aP2. Effect of Ad36 was perhaps most remarkable for PPARγ2, compared to uninfected control, in presence or absence of MDI. Probably due to concomitantly increasing gene expression in MDI treated control group, statistical significance between the infected vs the uninfected groups was reached later (day 5) compared to that for the no-MDI group (day 3).
A virus may not infect every cell in a plate. Ad36 infects estimated 30% of 3T3-L1 cells at 3.8 MOI (unpublished observations). Thus, it should be noted that even within the Ad36 infected group, there are two cell populations, one infected with the virus and other uninfected. Presence of these two cell populations in Ad36 groups was evident from two peaks of gene expression Wnt10 and C/EBPβ, an early peak due to Ad36 infection and a late peak coinciding with normal gene expression of the uninfected cell population. Moreover, unlike human primary adipocyte progenitors, 3T3-L1 cells do not support Ad36 viral particle formation ((10, 21) and unpublished observations). Therefore, the direct effect of Ad36 is limited to only the cells it enters. This also implies that the effect of Ad36 on infected cells is so robust that the infection of only a fraction of cells significantly increases average response for the entire group.
Infection of cells by a viral agent is likely to induce non-specific changes in cell metabolism. We used Ad2 to control for such non-specific effect of Ad36 infection. As we observed before, Ad2 is not adipogenic (19) and in fact may reduce adipocyte differentiation. Despite successful cellular entry, it did not modulate adipogenic program. However, caution should be used in interpreting the effects of a negative control virus. While it is reasonable in principle, in practice it is difficult to find an idea negative control virus. An ideal negative control virus for Ad36 would be the one that exactly mimics the effects of Ad36, but is non-adipogenic. A virus that enters a cell will induce its own set of modulation of cellular machinery, which may need another control. Hence, finding a perfect negative control for Ad36 is extremely difficult and perhaps, a proper control for “infection” is “no infection”. Nonetheless, it could be concluded that not all human adenoviruses posses the adipogenic effect like Ad36.
Subsequent to observing the modulation of cellular mRNA expression by Ad36, we wished to determine how the virus modulates cellular response. In a permissive cell, Ad36 attaches to a cell receptor, next it is internalized, which is followed by viral mRNA expression, DNA replication and virus assembly followed by the release of newly formed virus particles. A cellular change may be induced by attachment of a virus to a cell-receptor, or more actively, by viral mRNA, DNA or protein interaction with cellular counterparts. Broadly, Ad36 genes are grouped under early, and late genes, based on their initiation in virus replication cycle. Early genes are involved in inducing viral gene expressions, promoting cell cycle progression and blocking apoptosis, and viral DNA replication. The expression of late genes is involved in completing assembly of new virus particles. Despite the expression of early or late viral genes, 3T3-L1 cells do not support assembly of new Ad36 particles as indicated by a decrease in viral DNA(21) or a lack of viral structural protein formation (unpublished observations).
This narrowed the focus to cellular attachment of Ad36, or to subsequent mRNA expression as mediators of cellular modulations. Sufficient time was allowed for cellular attachment and internalization of Ad36 before the CF treatment. Once the virus is internalized, CF interferes with its replication as evident by a reduction in viral mRNA expression of early viral genes E1a and E4orf1. This was accompanied with attenuation of adipogenic effects induced by Ad36. This suggests the process requires viral mRNA (more likely viral proteins) and that the cellular attachment of Ad36 is not adequate for inducing cellular modulations. These findings also agree with our recent data showing a lack of adipogenic effect of ultra-violet light (UV) inactivated Ad36(25). UV irradiation induces DNA damage, but preserves surface proteins of a virus particle for cellular attachment, which was not adequate for inducing adipogenic effects of Ad36. Another approach to determine the effect of receptor engagement is to block internalization of the attached virus, which is particularly challenging because the exact receptor used by Ad36 is unknown.
Adipogenic inducers, such as M,D and I are required to experimentally stimulate differentiation of confluent preadipocyte cell lines. Lipid accumulation by Ad36 was 3-fold greater when MDI was added, compared to that without MDI (Table 1), which may be due to a synergistic effect of Ad36 and MDI or an additive effect of Ad36 through a novel pathway. Greater upregulation of adipogenic gene expression by Ad36 + MDI compared to MDI alone (Figure 2) supports a synergistic effect. Also, M,D and I can potentially induce adipogenesis in most cells in a plate, whereas, the effect of Ad36 may be limited to only the infected cells. This, coupled with lack of spread of virus infection in 3T3-L1 cells can limit the lipid accumulation attained by Ad36. Despite the limitations, even in the absence of MDI, Ad36 nearly doubled lipid accumulation compared to uninfected control. This was further assessed by observing Ad36 induced differentiation in selective presence of inducers. Percent increase in lipid accumulation in Ad36 group compared to respective uninfected control varied greatly between groups (MDI 27.5%, No-MDI 72%, MD 100%, MI 18% and DI 11%). Overall, those groups without insulin stimulation had much lower levels of lipid accumulation, suggesting the significance of insulin in preadipocyte differentiation and lipid accumulation. Interestingly, Ad36 was unable to increase lipid accumulation (DI group) in absence of M – which is required for increasing cAMP levels. The critical role of cAMP in Ad36 induced lipid accumulation is also supported by our previous findings showing marked increase in its levels by Ad36 in adipocyte progenitors(19). Overall, it is well documented that adipogenic induction is required for differentiation of preadipocytes in vitro. Ability of Ad36 to induce adipogenic program without conventional inducers suggests the presence of exogenous modulators of the program. It is likely that Ad36 interacts with an unknown upstream target, which initiates an adipogenic program similar to that induced by MDI.
Ad36 appears to strongly promote cell proliferation. Due to increasing cell number in Ad36 infected group, lipid expression was normalized to cell count. The total lipid unadjusted for cell number was even greater for Ad36 group, compared to control. Thus, Ad36 increases accumulation of lipid in an infected cell and the cell number. This led us to determine the lipogenic ability of Ad36 in proliferating vs confluent cells. Reaching confluence before adipogenic induction is important for adequate differentiation of 3T3-L1 cells. Thus, the two potential variables, number of days post infection as well as post confluence, may influence lipid accumulation. Ad36 infection of confluent cells induced greater lipid accumulation vs that of subconfluent proliferating cells, determined after similar days post infection or confluence in presence or absence of MDI. Possibly, the lower lipogenic response of Ad36-infected subconfluent 3T3-L1 cells was due to fewer cells present for viral infection compared to those infected at confluence and because the infection does not spread in 3T3-L1 cells. Future experiments will determine if infection of proliferating human preadipocytes carry Ad36 in daughter cells, which may increase the adipogenic impact of the virus.
In summary, Ad36 is an inducer of adipogenic program in preadipocytes, and this single factor appears to qualitatively mirror the combined adipogenic effects of M, D, and I. This phenomenon is of particular relevance since Ad36 increases adiposity in experimentally infected animal models(5–7, 9) and preadipocytes of humans naturally infected with the virus show greater potential to differentiate and accumulate lipid(10). Adipogenic potential of Ad36 has been recently re-confirmed in vivo by others(26) and several other adipogenic adenoviruses have been reported (27). Considering very high prevalence of adenovirus infection in population, it would be important to determine the adipogenic effect, if any, of certain adenovirus serotypes on preadipocyte differentiation. Importantly, the exact cellular target(s) of Ad36 that initiate the adipogenic response is(are) unclear. Identifying hitherto unknown regulators of cellular adipogenesis may provide additional tools to better understand and effectively manipulate preadipocyte differentiation.
Figure 5. Confluent 3T3-L1 cells respond better to lipogenic effect of Ad36.
3T3-L1 cells were infected with Ad36 (1PFU/cell) on the day of confluence or 1 or 2d earlier and exposed to MDI at confluency. Lipid accumulation was determined by BODIPY(10), either 5d post infection, or 5d post-confluence. The same design was repeated in absence of MDI, and the lipid accumulation was determined 9d post Ad36 infection or post confluence. *p<.007 (at least), **p<.03, vs lipid accumulation on −2d.
Acknowledgments
Funded by NIH1R01DK066164 awarded to NVD.
References
- 1.Otto TC, Lane MD. Adipose development: from stem cell to adipocyte. Crit Rev Biochem Mol Biol. 2005;40:229–42. doi: 10.1080/10409230591008189. [DOI] [PubMed] [Google Scholar]
- 2.Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol. 2000;16:145–71. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- 3.Farmer SR. Regulation of PPARgamma activity during adipogenesis. Int J Obes (Lond) 2005;29 (Suppl 1):S13–6. doi: 10.1038/sj.ijo.0802907. [DOI] [PubMed] [Google Scholar]
- 4.Ntambi JM, Young-Cheul K. Adipocyte differentiation and gene expression. J Nutr. 2000;130:3122S–6S. doi: 10.1093/jn/130.12.3122S. [DOI] [PubMed] [Google Scholar]
- 5.Dhurandhar NV, Israel BA, Kolesar JM, Mayhew G, Cook ME, Atkinson RL. Transmissibility of adenovirus-induced adiposity in a chicken model. Int J Obes Relat Metab Disord. 2001;25:990–6. doi: 10.1038/sj.ijo.0801668. [DOI] [PubMed] [Google Scholar]
- 6.Dhurandhar NV, Israel BA, Kolesar JM, Mayhew GF, Cook ME, Atkinson RL. Increased adiposity in animals due to a human virus. Int J Obes Relat Metab Disord. 2000;24:989–96. doi: 10.1038/sj.ijo.0801319. [DOI] [PubMed] [Google Scholar]
- 7.Dhurandhar NV, Whigham LD, Abbott DH, et al. Human adenovirus Ad-36 promotes weight gain in male rhesus and marmoset monkeys. J Nutr. 2002;132:3155–60. doi: 10.1093/jn/131.10.3155. [DOI] [PubMed] [Google Scholar]
- 8.Kapila M, Khosla P, Dhurandhar NV. Novel short-term effects of adenovirus Ad-36 on hamster lipoproteins. Int J Obes Relat Metab Disord. 2004;28:1521–7. doi: 10.1038/sj.ijo.0802710. [DOI] [PubMed] [Google Scholar]
- 9.Pasarica M, Shin AC, Yu M, et al. Human adenovirus 36 induces adiposity, increases insulin sensitivity, and alters hypothalamic monoamines in rats. Obesity (Silver Spring) 2006;14:1905–13. doi: 10.1038/oby.2006.222. [DOI] [PubMed] [Google Scholar]
- 10.Pasarica M, Mashtalir N, McAllister EJ, Kilroy GE, Koska J, Permana P, de Courten B, Yu M, Ravussin E, Gimble JM, Dhurandhar NV. Adipogenic human adenovirus Ad-36 induces commitment, differentiation and lipid accumulation in human adipose-derived stem cells. Stem Cells. 2008;26:969–78. doi: 10.1634/stemcells.2007-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vangipuram SD, Sheele J, Atkinson RL, Holland TC, Dhurandhar NV. A human adenovirus enhances preadipocyte differentiation. Obes Res. 2004;12:770–7. doi: 10.1038/oby.2004.93. [DOI] [PubMed] [Google Scholar]
- 12.Wang ZQCW, Zhang XH, Yongmei Y, Qin J, Son L, Rogers PM, Mashtalir N, Bordelon JR, Ye J, Dhurandhar NV. Human adenovirus type 36 enhances glucose uptake in diabetic and non-diabetic human skeletal muscle cells independent of insulin signaling. Diabetes. 2008;57:1805–13. doi: 10.2337/db07-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vangipuram SD, Yu M, Tian J, et al. Adipogenic human adenovirus-36 reduces leptin expression and secretion and increases glucose uptake by fat cells. Int J Obes (Lond) 2007;31:87–96. doi: 10.1038/sj.ijo.0803366. [DOI] [PubMed] [Google Scholar]
- 14.Rogers PMMN, Rathod MA, Dubuisson O, Wang ZQ, Dasuri K, Babin S, Gupta A, Markward N, Cefalu WT, Dhurandhar NV. Metabolically Favorable Remodeling of Human Adipose Tissue by Human Adenovirus Ad-36. Diabetes. 2008;57:2321–31. doi: 10.2337/db07-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkinson RL, Dhurandhar NV, Allison DB, et al. Human adenovirus-36 is associated with increased body weight and paradoxical reduction of serum lipids. Int J Obes (Lond) 2005;29:281–6. doi: 10.1038/sj.ijo.0802830. [DOI] [PubMed] [Google Scholar]
- 16.Wigand R, Gelderblom H, Wadell G. New human adenovirus (candidate adenovirus 36), a novel member of subgroup D. Arch Virol. 1980;64:225–33. doi: 10.1007/BF01322702. [DOI] [PubMed] [Google Scholar]
- 17.Whigham LD, Israel BA, Atkinson RL. Adipogenic potential of multiple human adenoviruses in vivo and in vitro in animals. Am J Physiol Regul Integr Comp Physiol. 2006;290:R190–4. doi: 10.1152/ajpregu.00479.2005. [DOI] [PubMed] [Google Scholar]
- 18.So PW, Herlihy AH, Bell JD. Adiposity induced by adenovirus 5 inoculation. Int J Obes Relat Metab Disord. 2005;29:603–6. doi: 10.1038/sj.ijo.0802917. [DOI] [PubMed] [Google Scholar]
- 19.Rogers PM, Fusinski K, Rathod MA, et al. Human adenovirus Ad-36 induces adipogenesis via its E4 orf-1 gene. International Journal of Obesity. 2007;32:397–406. doi: 10.1038/sj.ijo.0803748. [DOI] [PubMed] [Google Scholar]
- 20.Orlicky DJ, DeGregori J, Schaack J. Construction of stable coxsackievirus and adenovirus receptor-expressing 3T3-L1 cells. J Lipid Res. 2001;42:910–5. [PubMed] [Google Scholar]
- 21.Rathod M, Vangipuram SD, Krishnan B, Heydari AR, Holland TC, Dhurandhar NV. Viral mRNA expression but not DNA replication is required for lipogenic effect of human adenovirus Ad-36 in preadipocytes. Int J Obes (Lond) 2007;31:78–86. doi: 10.1038/sj.ijo.0803358. [DOI] [PubMed] [Google Scholar]
- 22.Ramirez-Zacarias JL, Castro-Munozledo F, Kuri-Harcuch W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry. 1992;97:493–7. doi: 10.1007/BF00316069. [DOI] [PubMed] [Google Scholar]
- 23.Johnson PF. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J Cell Sci. 2005;118:2545–55. doi: 10.1242/jcs.02459. [DOI] [PubMed] [Google Scholar]
- 24.Miller SG, De Vos P, Guerre-Millo M, et al. The adipocyte specific transcription factor C/EBPalpha modulates human ob gene expression. Proc Natl Acad Sci U S A. 1996;93:5507–11. doi: 10.1073/pnas.93.11.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasarica M, Loiler S, Dhurandhar NV. Acute effect of infection by adipogenic human adenovirus Ad36. Archives of Virology. 2008 doi: 10.1007/s00705-008-0219-2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas GCR, Tilkorn D, Keramidaris E, Morrison W, Penington A, Blick T. Effect of human adenovirus −36 in a localized in vivo model of neoadipogenesis. Obesity. 2008;16:S74. [Google Scholar]
- 27.Pasarica M, Dhurandhar NV. Infectobesity: obesity of infectious origin. Adv Food Nutr Res. 2007;52:61–102. doi: 10.1016/S1043-4526(06)52002-9. [DOI] [PubMed] [Google Scholar]