Abstract
APOBEC3G is an innate intracellular anti-viral factor which deaminates retroviral cytidine to uridine. In vivo studies of APOBEC3G (A3G) were carried out in rhesus macaques, following mucosal immunization with SIV antigens and CCR5 peptides, linked to the 70kD heat shock protein. A progressive increase in A3G mRNA was elicited in PBMC after each immunization (p<0.0002 to p≤0.02), which was maintained for at least 17 weeks. Analysis of memory T cells showed a significant increase in A3G mRNA and protein in CD4+ CCR5+ memory T cells in circulating (p=0.0001), splenic (p=0.0001), iliac lymph nodes (p=0.002) and rectal (p=0.01) cells of the immunized compared with unimmunized macaques. Mucosal challenge with SIVmac 251 showed a significant increase in A3G mRNA in the CD4+ CCR5+ circulating cells (p<0.01) and the draining iliac lymph node cells (p<0.05) in the immunized uninfected macaques, consistent with a protective effect exerted by A3G. The results suggest that mucosal immunization in a non-human primate can induce features of a memory response to an innate anti-viral factor in CCR5+ CD4+ memory and CD4+CD95+CCR7− effector memory T cells.
Keywords: Mucosal immunization, APOBEC 3G, CD4+ memory T cells
Introduction
Innate immunity is a complex system of cellular and soluble factors directed against microorganisms and foreign molecules, which act independently of an immunological memory developed by prior encounter with an agent. The lack of an immunological memory, however has recently been challenged by the demonstration that NK cells can induce antigen-specific skin hypersensitivity reaction in the absence of T cells and that priming resulted in memory mediated by NK cells (1). Furthermore, innate immunity via pattern recognition receptors, such as Toll-like receptors expressed on DC, activates antigen specific T and B cells and modulates the quantity and quality of T and B cell memory (2–5). Immunological memory has been characterised by a rapid increase and prolonged specific immune response to an antigen. Memory T cells can be characterized by a) phenotypic expression, b) cytokines profile, c) development of memory cell pathway and d) anatomical site. An emerging paradigm in the prevention of HIV-1 infection is the development of a rapid innate immune response to the virus, in view of the infection and destruction of CD4+CCR5+ memory T cells within 2–3 weeks of infection, mostly in the mucosally associated lymphoid tissue (6–9). We attempted to focus attention on some of the criteria noted above, evaluating A3G expression within CD4+ memory T cells in mucosally immunized and challenged non-human primates.
The first objective of this study was to find out if APOBEC3G (A3G) expression is elicited in rhesus macaques immunized by the mucosal route with SIV antigens and or CCR5 peptides linked to the HSP70 carrier. The second aim was to explore the possibility that immunization induces in CD4+ T cells a memory-like function for A3G. The third objective was to find out if CD4+ T cell innate and adaptive immunity are linked as exemplified by A3G expression and precursor frequency, respectively. The fourth objective was to evaluate the effect of A3G expression on the in vivo infectivity of CD4+ T cells in the SIV challenged macaques. We have found progressive upregulation of A3G mRNA after each of three immunizations. Significant levels of A3G mRNA were maintained up to 47 weeks, which raised the possibility of the development of a memory-like response. This was supported by a significantly greater increase in A3G mRNA expression in the CD4+ CCR5+ and CD95+ memory T cells, and A3G protein in CD4+ CD95+ CCR7− effector memory T cells. The A3G mRNA expression was significantly correlated with the precursor frequency of T cell proliferative response to SIVgp120 and CCR5 peptide. Furthermore, a significant increase in A3G mRNA was found in CD4+CCR5+ memory and in A3G protein in CD4+CD95+CCR7− effector memory T cells in immunized uninfected macaques.
Materials and Methods
Preparation of the vaccine antigens and peptides
HSP70 derived from Mycobacterium tuberculosis was prepared in Escherichia coli as described previously (10). It was purified by Q-sepharose followed by ATP affinity chromatography. The Q-Sepharose chromatography was repeated to remove endotoxin, which was tested by the Limulus amebocyte lysate assay and showed 1.2 pg of endotoxin per 1 μg HSP70 protein. The HSP70 preparation migrated as a doublet with molecular mass of approximately 70K, with a minor component at approximately 35K.
Recombinant SIVmac 251 gp120 was expressed in Baculovirus-infected cells and prepared by Oxford Expression Technologies. Supernatant (20L) containing SIVgp120 was purified using monoclonal antibody (KK8) immobilised onto cyanogen bromide activated sepharose 4B beads (GE Healthcare, UK). The protein was eluted with 6M guanidine and dialysed against sterile phosphate buffer. The purity of the protein was confirmed by SDS-PAGE and western blotting. Recombinant SIVmac251 p27 was prepared by pGEX-3X as a glutathione S-transferase fusion protein from E. coli. The fusion protein has a molecular weight of about 56kDa composed of SIVp27, molecular weight 27kDa and the fusion partner gst at about 28kDa. The protein was extracted from the bacterial cells by sonication and purified using gst affinity chromatography.
CCR5 peptides derived from the sequences of the N-terminal, loop 1 and loop 2 were synthesized to purity greater than 85%, as determined by HPLC and purchased from Bachem (Switzerland). The sequences of the peptides are shown below.
N-terminal (aa1-20): Met – Asp – Tyr – Gln – Val – Ser – Ser – Pro – ILe – Tyr Asp – ILe –Asp – Tyr – Tyr – Thr – Ser – Glu – Pro – Cys
Loop 1 (aa 89-102): His – Tyr – Ala – Ala – Ala – Gln – Trp – Asp – Phe – Gly- Asn –Thr – Met – Cys – Gln
Loop 2 (aa178-197): Cys – Ser – Ser – His – Phe – Pro – Tyr – Ser – Gln – Tyr- Gln – Phe–Trp – Lys – Asn – Phe – Gln – Thr – Leu– Lys
Conjugation of HSP70 to the antigens and peptides
The HSP70 was conjugated to SIV gp120, SIV p27, N terminal and 2nd loop of CCR5 by means of the SPDP reagent which is less likely to alter the immunogenicity of the vaccine components than glutaraldehyde (11). First loop was non-covalently linked to HSP70, as this peptide binds directly to the peptide binding groove, demonstrated both by surface plasmon resonance and by immunization in mice (12). At each stage of the SPDP substitution and conjugation the HSP70/protein or peptide complex was subjected to SDS-PAGE and Western blot analysis. Under non-reducing conditions high molecular weight complexes were observed which stained for both HSP70 and the conjugated protein, indicating successful conjugation. Under reducing conditions the complexes were broken down into their constituent molecules. HSP70-SIV p27 complex contained a small amount of free SIV p27.
Antibodies
The following mAb were used for immunofluorescence studies with the macaque cells: mAb to human CD95 (clone: DX, allophycocyanin or phycoerythrin conjugated) (Becton Dickinson, Oxford, UK), anti-human CCR5 mAb (CD195, 3A9, purified or phycoerythrin conjugated) (Becton Dickinson), anti-human CCR7 mAb (clone 150503, purified or phycoerythrin conjugated, all purchased from R&D), and anti-human CD4 mAb (clone L200, PerCP-Cy5.5 conjugated, BD). Rabbit anti-human A3G polyclonal antibody (IgG) or mAb to human A3G were purchased from ImmunoDiagnostics, Inc, USA.
Immunization, virus challenge and assay
Twenty rhesus macaques weighing 2.4–3.2kg were housed and maintained according to the guidelines laid down by the UK Home Office, under the Animals (Scientific Procedures) Act 1986. The animals were randomized into 4 groups of 5 animals each and treated as follows: group A HSP70+SIVgp120+SIVp27+CCR5; group B HSP70+SIVgp120+SIVp27; group C HSP70+CCR5; group D Unimmunized. The schedule of immunization at 1–6-23 weeks, challenge at 27 weeks and termination of the experiment at 47 weeks is presented in Fig. 1A. Blood samples were collected from the femoral vein before each immunization at weeks 0, 6 and 23, as well as before SIV challenge at week 27. Four weeks after the last vaccination at week 27 all macaques were challenged rectally with 3ml of 50MID50/ml of SIVmac251 stock (Robert Koch Institute, Berlin). This stock has been titrated in vitro and in vivo at PEI, (Langen Germany) and previously used in a multi-centre EU funded study. Viral infectivity was assayed for plasma viral RNA copies per ml and DNA by PCR of PBMC at intervals from 2 to 20 weeks after challenge with SIVmac251. The sensitivity of the plasma viral RNA assay was 50 SIV RNA copies per ml plasma. At the given dose all challenged naive controls were infected previously at 3 of 4 centres but failed in this study. Venous blood was withdrawn before and after each immunization at times indicated in Fig. 1.
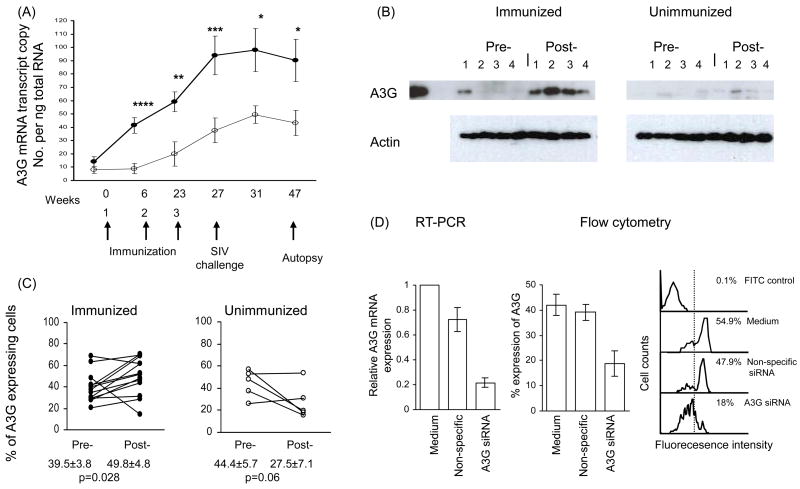
Figure 1.
(A) Sequential A3G mRNA expression in PBMC before and after (x3) immunization (●) n=15 and unimmunized control (o) n=5 rhesus macaques, which were challenged with SIVmac251 (at week 27, 4 weeks after the 3rd immunization). Blood was collected before each of the 3 immunizations and SIV challenge. The results are presented as mean ± sem; ≤0.02*, <0.01**, <0.005***, <0.0002****. (B) Western blots before and after the 3rd immunization (week 27) with lysates of PBMC from 4 macaques. (C) Intracellular expression of A3G proteins in CD4+ T cells detected by flow cytometry before and after 3rd immunization (week 27). (D) A3G mRNA and protein knock down with A3G specific and non-specific siRNA of CD4+ T cells from 4 animals and representative flow cytometry.
Preparation of PBMC and cells from post-mortem tissues
PBMC were separated from heparinsed blood following incubation with 3% gelatin (CB Gelatins UK) and density gradient centrifugation on Ficoll (GE Healthcare Ltd, UK). At autopsy spleen, iliac and axillary lymph nodes were removed and single cell suspensions were prepared by cutting the tissue into small pieces and passing through a sterile nylon sieve (Nunc UK). The cells were washed in RPMI 1640 containing 100μg/ml penicillin and streptomycin and supplemented with 2mM-L-glutamine (Sigma-Aldrich, UK). Mononuclear cells were also separated from rectal tissue as previously described (13). Briefly, the tissue was cut into small pieces and washed in hepes-buffered Hank’s balanced salt solution containing 1mM Ca Cl2 and 10mM dithiothreitol (pH 7.4). Cells were liberated from the tissue by successive rounds of digestion with Thermolysin at 4°C (Sigma-Aldrich UK Ltd) and collagenase/dispase (Calbiochem UK Ltd (as previously described (11).
Preparation of RNA, cDNA and standard A3G PCR products
Macaque PBMC (1 × 106) were thawed from cryo-perserved samples into RPMI 1640 medium supplemented with 10% FCS. After centrifugation at 500g for 5 min, the cell pellets were washed with PBS. The RNA was isolated using a Total RNA Isolation Kit (Promega, Southampton, UK), quantified using the spectrophotometer (GeneQuant II, Pharmacia Biotech), and cDNA was generated from RNA by using the Reverse Transcription System (Promega), according to the manufacturer’s instructions.
Rhesus macaques A3G and GAPDH dsDNA products were produced by PCR. The primers for rhesus macaque A3G and GAPDH were synthesized by Eurogentec S.A (Belgium). The primer sequences: for A3G, 5′-CTG TCC ACT GAC CCA AGG TT-3′ (right); 5′-ACA TGC CAC GAA GAT CA-3′ (left) and for GAPDH, 5′-GGA GCT CTC CAG AAC ATC ATC CCT-3′ (right); 5′-CCT TGA GGG GGC CCT CCG ACG CCT-3′ (left). The single amplified PCR product was verified based on size in a 3% agarose gel under UV illumination. The gel band containing the DNA target was excised and digested to recover and purify the amplified product using PureLink™ Quick Gel Extraction Kit (Invitrogen, Paisley, UK). The concentration of the amplified product was determined by spectrophotometry.
Real-time PCR for A3G mRNA
Relative amount of A3G mRNA was quantified by real-time PCR (ABI Prism 5700) using the PlatinumSYBR green qPCR SuperMix-UDG with ROX (Invitrogen Life Technologies). The primers for A3G and GAPDH were described above and used for the assays. The amplification conditions were 2 min at 50°C, 2 min at 95°C, 40 cycles of 15 s at 95°C, and 30 s at 60°C. When assaying the samples for A3G and GAPDH, the corresponding standards series was run under the same conditions and the concentration of samples was determined by reading off the standards series with the Ct values of the samples. Using the average molecular weight of the product and Avogadro’s constant, the number of copies per unit volume was calculated. The results were expressed as the copy number per ng of total RNA. Melting curve analysis was performed in every assay, and the products were also analyzed on agarose gel to confirm the specificity of amplified products.
Detection of A3G mRNA in subsets of CD4+ memory T cells
To determine A3G mRNA in different subsets of CD4+ T cell populations, T cells were enriched by the panning method using antibodies to CD8 and anti-IgG to remove B cells and monocytes. Enriched CD4+ T cells (greater than 90%, analysed by flow cytometry) were further separated into CCR5+ and CCR5− populations or CCR7+ and CCR7− populations by panning, using CCR5 and CCR7 mAbs, respectively. The adherent CCR5+ or CCR7+ cells were washed twice with medium, lysed and RNA was prepared from the lysates. The non-adherent CCR5 or CCR7 cells were also collected, washed and RNA was prepared. In the non-adherent populations, CCR5+ or CCR7+ cells were usually less than 1%. The RNA was isolated using a Total RNA Isolation Kit (Promega), and cDNA was generated from RNA by RT-PCR using the Reverse Transcription System (Promega), as described as above.
Detection of A3G protein by immunofluorescence
In order to determine A3G protein expression in different CD4+ T cell populations, intracellular A3G in combination with cell surface staining was carried out by immunofluorescence. The pre-immunized and post-immunized samples were thawed and analysed in parallel in each assay. The viability of thawed cells was checked by trypan blue exclusion and was greater than 85%. Macaque CD4+ memory and naïve cell subsets were identified using antibodies to CD95 and CCR7, as reported previously (14). Briefly, naive cells expressed CD95low and memory cells were identified by CD95high expression. Among the memory populations, central memory cells were identified as CD4+CD95+CCR7+ and effector memory cells as CD4+CD95+ CCR7− cells.
After staining with the specific fluorescence conjugated anti-cell surface marker antibodies, the cells were fixed using IC Fixation Buffer and permembilized with Permeabilization Buffer (eBioscience, UK distributer, London). The cell pellets (1 × 105 cells) were incubated for 30 min at room temperature with 5 μl (5μg/ml) of rabbit anti-human A3G antibody (Immunodiagnostics Inc), followed by incubation (20 min) with FITC-labelled secondary anti-rabbit IgG (1:100) (Sigma, Poole, UK). Cells were then washed, resuspended and analysed by flow cytometry (FACS Canto II, BD), using BD FACSDiva Software. In the side scatter and forward scatter dotplot, a live cell population was gated for subsequent analysis for either phenotypic or A3G expression. The specificity of immunofluorescence staining of A3G was verified by dose-dependent inhibition with increasing concentrations of exogenous recombinant human A3G (Immunodiagnostics Inc), MAb (Immunodiagnostics Inc.) and rabbit anti-human A3G (data not presented). Addition of recombinant human A3G (from 1–10μg/ml) produced dose-dependent decrease in intracellular A3G detected by mAb or rabbit polyclonal anti-human A3G antibodies, indicating the specificity of the A3G staining.
A3G interference by siRNA
RNA interference was used to confirm the specificity of both the PCR and flow cytometry assays for macaque A3G. Macaque CD4+ T cells were thawed and live cells were recovered by Ficoll centrifugation. A3G siRNA duplexes with the sequence r(CCAUGAAGAUCAUGAUUA)dTdT/r(UAAUUCAUGAUCUUCAUGG)dTdG (Peng et al 2006) were synthesized to specifically target macaque A3G. As the non-specific control siRNA duplex we used fluorescent labelled AllStars Neg siRNA AF488 (Qiagen, West Sussex, UK). Cell were transfected using Amaxa nucleofector and Human T Cell Nucleofector Kit (Amaxa AG, Germany). 5 × 106 cells were suspended in 100μl nucleofector solution and 5μl specific or control siRNA duplexes (40μM), or water was added. Cells were electroporated using program U-14 and recovered in 1.5ml RPMI culture medium. After cultivating cells for 18h in 24-well tissue culture plates (Costal R, Corning Inc, NY, USA), the medium was changed and the cells were incubated for further 48h before assay for A3G mRNA by PCR and protein by intracellular A3G staining.
Detection of A3G by Western blots
To detect A3G protein, 2 × 106 cells were lysed in HBSS, with 10 mM HEPES (pH 7.4) and 0.2% Nonidet P-40. Lysates were cleared by centrifugation and protein precipitated with acetone, before resuspending in a minimal volume of SDS sample buffer prior to SDS-PAGE under reducing conditions. After transfer of proteins to a polyvinylidene difluoride membrane, Western blotting was carried out with mouse mAb to beta-actin (Sigma Aldrich) or rabbit antibody to A3G (Immunodiagnostics Inc., Woburn, MA) using biotinylated anti-mouse or anti-rabbit Ig, streptavidin-peroxidase, and ECL-plus reagents (GE Healthcare, Chalfont, UK).
Precursor frequency of T cell proliferative responses
Proliferative responses of lymphocytes were measured using a flow cytometry-based assay that utilises a fluorescent membrane dye (PKH26) that is partitioned between daughter cells at division (13). Analysis software (Modfit LT, Verity Software House, Topsham, ME) was employed for mathematical deconvolution of daughter generations to provide a precursor frequency of responding cells in the parent population. The following antigens were tested in triplicates: SIVgp120, SIVgag p27 and HSP70, each at a final concentration of 5, 10 and 20μg per ml. A pool of CCR5 peptides of the N-terminal, loop 1 and loop 2 was used at final concentrations of 10, 20 and 40μg per ml., PHA at 10μg per ml and culture medium alone were employed as positive and negative controls, respectively. After 7 days of culture, the triplicates were pooled and 30,000 cells were examined by flow cytometry for precursor frequency. The results are shown as the percent of total PBMC that underwent proliferation after stimulation with the antigens. Precursor frequencies >mean+3 SD of the controls were considered to be significant. The data for SIVgagp27 are not presented as the precursor frequencies were very low and no difference was found between the groups analysed.
Statistical analysis
The results are presented as mean (±sem) and analysed by the appropriate Student’s t test; p values <0.05 were considered to be significant. Non-parametric data were analysed by the Mann-Whitney test. The Pearson’s correlation test was used to evaulate the relation between A3G and precursor frequency.
Results
Expression of A3G mRNA and protein in circulating PBMC
A3G expression was examined in rhesus macaques before and after mucosal immunization with HSP70 linked to SIV antigens or CCR5 peptides or both. As no significant difference in A3G mRNA expression was found at any of the 6 time points examined between the 3 groups of immunized macaques, they were analysed as one enlarged cohort of immunized animals, and compared with the unimmunized macaques. Mucosal immunization induced a progressive increase in A3G mRNA expression after each of the 3 immunizations; from 14.2 (±3.7) before to 41.4(±5.8) 6 weeks after 1st, 59.1(±7.4) 17 weeks after 2nd and 94.1(±14.5) 4 weeks after the 3rd immunization mRNA transcript copies per ng of total RNA (referred to as A3G mRNA copies) (Fig. 1A). There was no further increase after the macaques were challenged with SIVmac251, but 90.1 (±15.9) A3G mRNA copies were maintained at week 47 or 24 weeks following the 3rd immunization (week 23). The results suggest that A3G was maintained for at least 17 weeks without SIV challenge (between weeks 6 and 23). Significant increases in A3G mRNA expression (0.0002 to p≤0.02) were found at each time point between the immunized and unimmunized macaques (Fig. 1A). Whereas a significant increase in A3G mRNA in the immunized macaques was found after the 1st immunization at week 6 (p=0.0002), the unimmunized macaques showed no change. However, a significant increase was seen in A3G mRNA copies from 8.05 (±2.5) before to 37.7 (±9.3) in the unimmunized animals only by week 27 (p<0.05). However, all memory T cell subsets failed to show an increase in A3G expression in the unimmunized macaques between week 0 and 27 (Fig. 1, 2, 3, and 5). The small increase in A3G mRNA expression in PMBC of the controls, might be due to environmental factors.
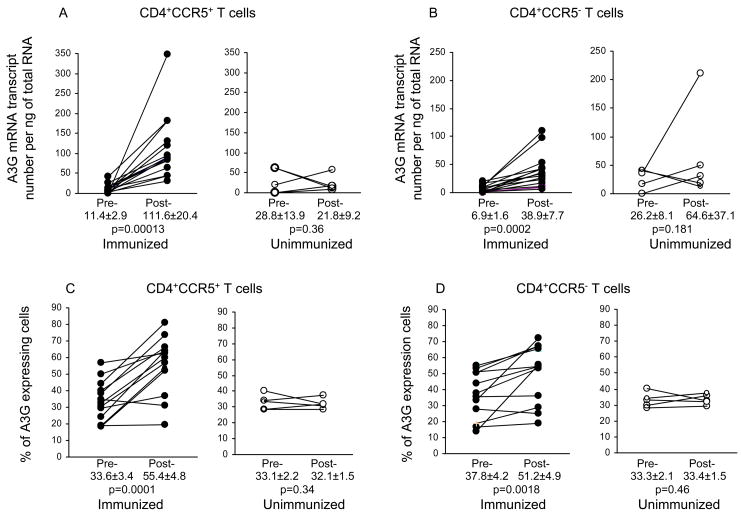
Figure 2.
Effect of immunization on A3G mRNA (A and B) and protein (C and D) expression in CCR5+CD4+ and CCR5−CD4+ T cells (n=15) as compared with unimmunized macaques (n=5); pre- and post-immunization, at week 27 (4 weeks after 3rd immunization) or at the same time point in the unimmunized animals.
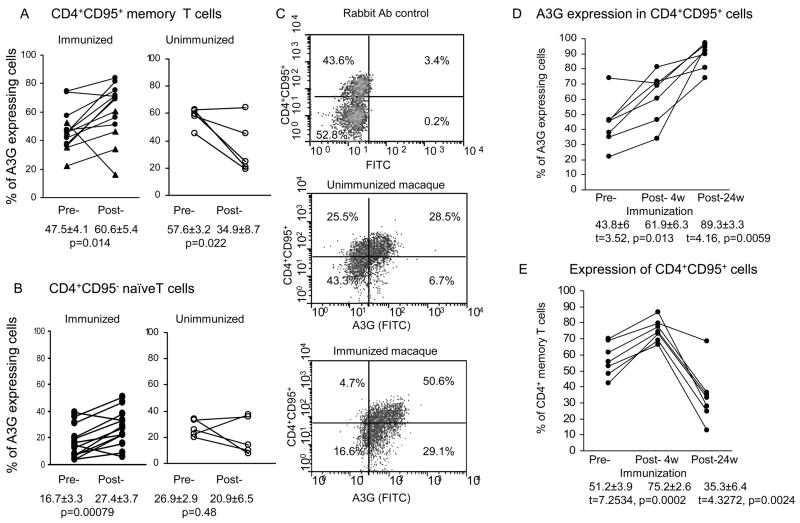
Figure 3.
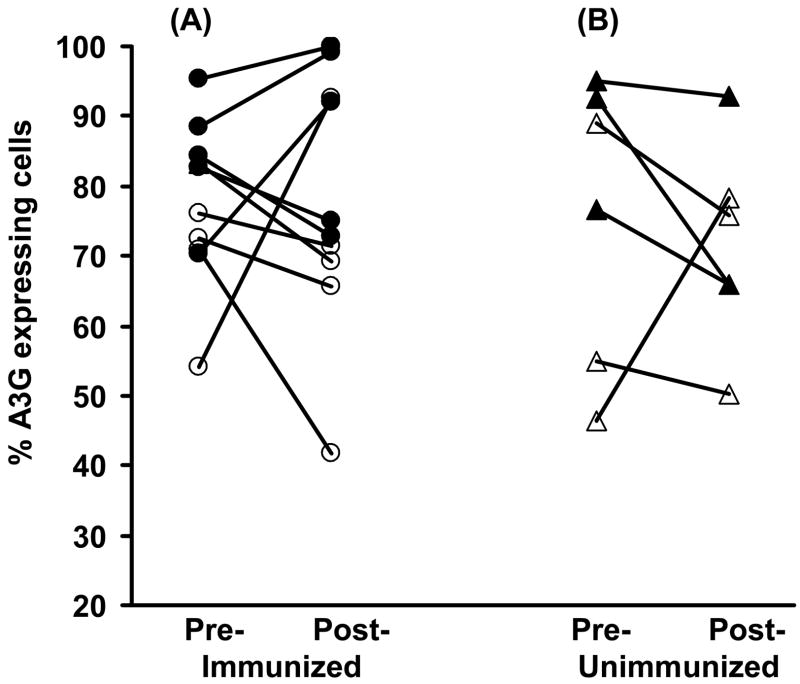
A3G expression in (A) CD4+CD95+ memory T cells and (B) CD4+CD95− naïve T cells (n=13) before and at week 27, (4 weeks after the 3rd immunization) or at the same time point in the unimmunized macaques (n=5), (C) flow cytometry profile of A3G expression in CD4+ T cells in unimmunized and immunized macaques, (D) increase in A3G in CD4+ memory T cells at weeks 27 and 47, compared with (E) increase in the proportion of these cells at week 27 but decrease at week 47.
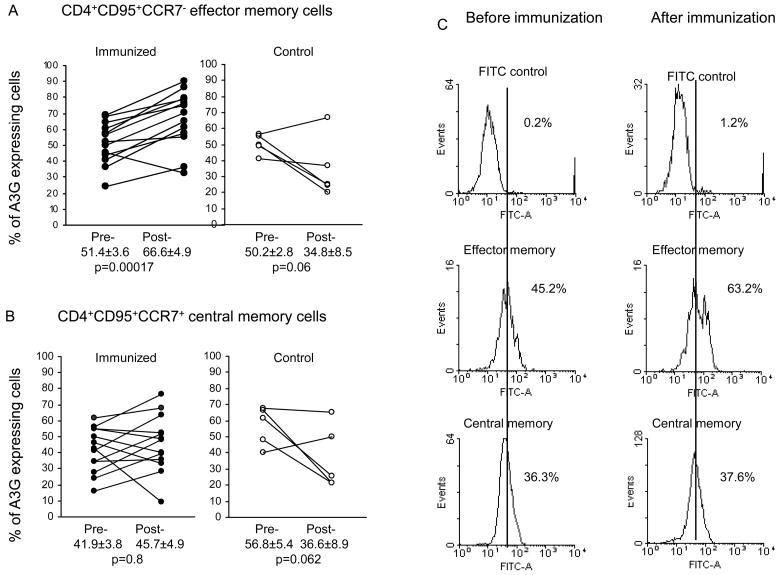
Figure 5.
Expression of A3G in CD4+CD95+CCR7− effector memory cells (A), CD4+CD95+CCR7+ central memory cells (B), before and week 27 (4 weeks after the 3rd immunization and before challenge) (n=13) or at the same time point in the unimmunized macaques (n=5), and representative profiles of A3G expression in effector and central memory CD4+ T cells before and after immunization (C).
MAb to A3G was used for Western blots of lysed PBMC before and after immunization. Representative samples of lysates from 4 macaques showed increases in A3G expression after the 3rd immunization at week 27 in all 4 samples, though one showed some A3G before immunization (Fig. 1B). In contrast, only 1 of the 4 unimmunized controls showed a slight increase in A3G band (No. 2), but the corresponding actin control was also greater in the post- than pre-sample. Increased A3G protein expression was found in all 15 macaques after immunization, including 4 of which showed detectable A3G before immunization. Surprisingly, 4 of the 5 unimmunized macaques showed weak A3G bands on Western blots before the start of immunization and by 27 weeks the 5th animal also developed an A3G protein band, however these were distinctly weaker than those of the immunized macaques. Intracellular A3G protein in CD4+ T cells was also analysed by flow cytometry (Fig. 1C) and showed a significant increase (p=0.028) from 39.5±3.8 to 49.8±4.8%, compared with a decrease from 44.4±5.7 to 27.5±7.1% in the cells from unimmunized animals which, however was not significant. Thus, the immunized macaques showed a very significant increase in A3GmRNA expression after the 1st immunization at week 6 which increased after each immunization up to week 27, and this was associated with increase in A3G protein, demonstrated both by Western blots and immunofluorescence. In contrast, the unimmunized macaques showed a small though significant increase in A3G mRNA only by week 27, and the A3G protein failed to increase when assayed by immunofluorescence and Western blots (with 1 possible exception).
The observation that A3G mRNA and protein expression was maintained for at least 17 weeks raised the possibility that immunization may have elicited A3G expression in memory T cells. Whereas the 17 week period was maintained before challenge with SIV, after the challenge A3G persisted for 24 weeks, but we cannot exclude the possibility that challenge with the virus may have boosted A3G mRNA, though SIV or HIV may not upregulate A3G (15, 16). These results demonstrate that A3G mRNA expression can be upregulated and maintained in vivo in non-human primates for at least 17 weeks, and this may play an important part in inhibiting early SIV or HIV infection.
A3G interference by siRNA
The specificity of A3G mRNA and protein expression was studied in CD4+ T cells from 4 subjects by using A3G specific and non-specific siRNA. Significant knockdown was found only with the A3G mRNA specific siRNA; untreated baseline 1, non-specific siRNA control 0.726±0.19, and specific siRNA 0.216±0.08 (Fig. 1D). The difference between the specific and non-specific siRNA was very significant (t=2.44, p=0.003). Similar results were found analysing A3G protein expression assayed by flow cytometry; untreated mean 42.1±8.65%, non-specific siRNA had no effect (mean 39.1±6.2%), whereas A3G-specific siRNA induced significant knockdown in A3G expression to a mean of 18.8±9.9% (t=3.46, p=0.01) (Fig. 1D, representive sample). Thus, both mRNA and protein expression examined were specific for A3G.
Expression of A3G mRNA in memory CD4+ T cells
Immunofluorescence data of 6 subsets of CD4+ T cells, before and at week 27 (4 weeks after the 3rd immunization) revealed significant decrease in the CD4+ CCR5+ (p=0.006) and increase in CD4+CD95+ memory T cells (p=0.013) in the immunized but no change in the unimmunized macaques sampled at the same time points (Table 1). CCR5+CD4+ T lymphocytes are the main target for SIV and primary R5 HIV infection and CCR5 is expressed especially in the CD4+ effector memory T cell population (12,13). Hence, the development of A3G in memory CD4+ T cells was examined by real-time PCR in CD4+ CCR5+ T cells at week 27 (4 weeks after the 3rd immunization) (Fig. 2). As with the PBMC, no significant difference in A3G mRNA of the CD4+CCR5+ T cells was found between the 3 groups of immunized animals, so they were analysed together (Fig. 2). This showed significantly higher A3G mRNA expression in CD4+CCR5+ T cells after (111.6±20.4) compared with (11.4±2.9) before immunization (p=0.0001). In contrast, unimmunized macaques studied at matched time points showed no change. CD4+CCR5− T cells showed a much smaller but significant increase in A3G mRNA from 6.9±1.6 to 38.9±7.7 (p=0.0002) at corresponding time points to the CD4+CCR5+ T cells (Fig. 2B).
Table 1.
Proportion of CD4+, CD4+CCR5+ and CD4+CD95+ memory T cells, CD4+CD95− naïve T cells, CD4+CD95+CCR7− effector and CD4+CD95+CCR7+ central memory T cells before and at week 27 (4 weeks after the 3rd immunization) (n=15), and at the corresponding times of sampling of the unimmunized macaques (n=5).
| Immunized group |
Unimmunized group |
|||
|---|---|---|---|---|
| Cell subset | pre- | post- | pre- | post- |
| CD4 | 23.7±1.4 | 26.1±1.8 | 35±1.6 | 37±2.7 |
| CD4+CCR5+ | 13.8±1.8 | 7.3±1.1*** | 42.2±6.1 | 47.9±8.4 |
| CD4+CCR5− | 86.1±1.8 | 92.6±2.5 | 57.8±6.1 | 52.1±8.4 |
| CD4+CD95+ | 55.9±2.4 | 65.9±4.7** | 42.2±6.1 | 47.9±8.4 |
| CD4+CD95− | 44.1±2.5 | 34.1±4.8 | 57.8±6.1 | 52.1±8.4 |
| CD4+CD95+CCR7− | 12.5±1.3 | 13.3±1.1 | 6.3±1.4 | 6.6±2.0 |
| CD4+CD95+CCR7+ | 43.4±2.0 | 52.3±4.5* | 35.1±6.0 | 41.3±7.1 |
p=0.024,
p=0.013,
p=0.006.
Expression of A3G protein in CD4+ memory T cells
We have then investigated the translation of A3G mRNA into protein by intracellular staining using antibodies to A3G in combination with those to CD4, CCR5 or CD95. A3G protein expression in CCR5+CD4+ T cells increased significantly (p=0.0001) from 33.6±3.6% before to 55.4±4.8% at week 27 (4 weeks after the 3rd immunization), unlike the unimmunized macaques which showed no change (Fig. 2C). However, A3G in the CCR5−CD4+ T cells also showed an increase from 37.8±4.2% to 51.2±4.9% after immunization (p=0.0018, Fig. 2D). A3G protein expression was then examined in CD4+CD95+ memory T cells, which showed an increase in A3G protein expression from 47.5±4.1% before to 60.6±5.4%, at week 27 (4 weeks after the 3rd immunization) (p=0.014) (Fig. 3A), but a decrease in A3G in the unimmunized macaques. A comparison between the two groups of animals showed a significantly greater expression of A3G (60.6±5.4% in the immunized than unimmunized group (34.9±8.7%; p=0.038). The CD4+ CD95− naive T cells showed a lower proportion of A3G expressing cells, but they were also significantly increased (p=0.008) after (27.4±3.7%), compared with before (16.7±3.3%) immunization (Fig. 3B). In contrast, the unimmunized control macaques again showed a decrease in A3G expression. A representative flow cytometry profile (Fig. 3C) shows greater A3G expression in CD4+CD95+ cells in the immunized (40.6%) compared with unimmunized controls (26.5%). However, whilst there was no difference in CD4+CD95+A3G+ T cells between group 1 and group 2, group 3 macaques (immunized by HSP70-CCR5) showed a significant lower levels in A3G expression because the cells from 1 animal showed a decrease from 53.0 to 16.3% and this is indicated in Fig. 3. These results suggest that a significant increase in A3G mRNA and protein is associated with immunization in CCR5+ and CD95+ CD4 memory T cells. The unimmunized control macaques failed to show a significant change in A3G expression in CCR5+ T cells, indeed a decrease in CD4+CD95+ T cells was recorded.
An increase in A3G expression in the CD4+CD95+ T cells 4 weeks after the 3rd immunization (p=0.0013) was further increased 20 weeks later (p=0.0059) (Fig. 3D). However, increase in the proportion of CD4+CD95+ memory T cells (p=0.0002) 4 weeks after immunization was followed by a significant decrease in these cells following SIV challenge (p=0.002) (Fig. 3E). These results suggest that A3G expression was raised in the CD4+CD95+ T cells despite a significant decrease in the proportion of these cells at the same time point of 20 weeks after challenge with SIV, which is consistent with a proportion of the cells being infected and killed (9).
A3G expression in DC and monocytes
To compare A3G expression in CD4+ T cells with those in the innate DC and monocytes, the latter were examined before and after immunization and this showed no consistent change (Fig. 4). Indeed, the mean ± sem values for either cell population was almost identical and this allowed the DC and monocytes to be combined for statistical purposes, which clearly showed that immunization failed to elicit significant difference in A3G expression in these cells (77.8±3.7 and 77.9±5.7, respectively). Thus, immunization elicits significant increase in A3G expression in CCR5+ or CD95+ memory CD4+ T cells, but a similar change was not found in DC and monocytes, consistent with antigen processing and presenting cells not sharing with CD4+ T cells the mechanism for cellular memory.
Figure 4.
A3G expression in DC and monocytes before and at week 27 (4 weeks after the 3rd immunization) (n=10) (A) and in corresponding cells from unimmunized macaques (n=6) (B); DC from immunized (●) and unimmunized (▲), monocytes from immunized (o) and unimmunized (△) macaques.
Central and effector memory T cells
Having established that A3G is expressed predominantly in CD4+ memory T cells, the next question was whether the central or effector memory cells are involved. We used CCR7 as a marker (17) to differentiate A3G protein expression in effector and central memory cells. The results showed a significant increase (p=0.0001) in A3G expression at week 27 (4 weeks after the 3rd immunization and before challenge 66.6±4.9%), compared to that before immunization (51.4±3.6%) in the CD4+CD95+CCR7− effector memory T cells (Fig. 5A). The corresponding data in CD95+CCR7+ central memory CD4+ T cells showed no significant difference following immunization (Fig. 5B). The unimmunized control macaques showed a decrease in A3G expression in both the effector and central memory cells, and neither reached the 5% level of significance (Fig. 5A and B). Representative flow cytometry profiles for the effector and central memory cells are shown in Fig. 5C. No significant difference was found in A3G expression of CD4+CD95+CCR7− T cells between the 3 groups of animals. It is noteworthy that RT-PCR analysis also showed significantly higher A3G mRNA expression (p=0.0003) in CD4+CCR7− cells after immunization (375.0±91.0-fold), compared with the unimmunized group (167.0±34.0 fold) (data not presented). Thus, immunization elicited a significant increase in A3G mRNA expression in the CD4+ CCR7− cells and A3G protein in CD4+CD95+CCR7− effector memory cells but not in the CD4+CD95+CCR7+ central memory cells.
Expression of A3G mRNA in splenic, lymph node and intestinal memory T cells
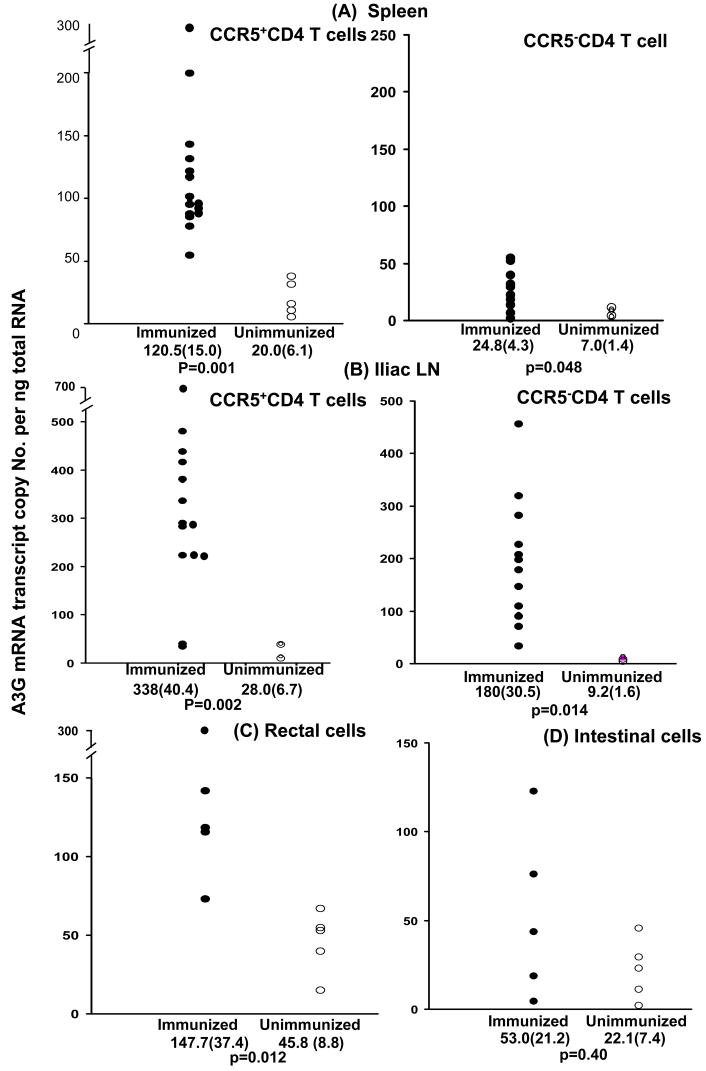
A3G mRNA was studied in the tissues after termination of the study at week 47, (24 weeks after the last immunization). Splenic CD4+ T cells showed significantly greater expression of A3G mRNA (p=0.003) in immunized (77.8±12.7) compared with unimmunized (19.2±4.6) macaques (data not presented). Splenic CD4+ T cells were then separated into CCR5+ and CCR5− cells (Fig. 6A), which showed significantly greater expression of A3G mRNA (p=0.001) in CCR5+ cells in immunized (120.5±15.0) compared with unimmunized macaques (20.0±6.1). A3G mRNA expression in CCR5−CD4+ cells was slightly higher in the immunized 24.8(±4.3) than unimmunized 7.0(±1.4) animals (p=0.048). Nonetheless, A3G mRNA expression in the immunized CCR5+ CD4+ T cells (120.5±5.2) was very significantly higher than in the CCR5− CD4 T cells (24.8±4.3; p<0.00001).
Figure 6.
A3G mRNA expression in CCR5+ and CCR5− splenic and iliac lymph node CD4 T cells and in mononuclear cells eluted from the rectal and small intestinal mucosa at the termination of the experiment (week 47); mean (±sem) and p values are given for the difference in A3G mRNA between immunized (n=15) and unimmunized macaques (n=5).
Memory cells were then studied in the iliac lymph nodes which drain the immunized and challenged rectal tissues, and axillary lymph nodes which are not part of the mucosal associated lymphoid tissue. A3G mRNA in both the CD4+CCR5+ iliac (p=0.002) and axillary (p=0.001) lymph node T cells were significantly greater in the immunized than unimmunized macaques (Fig. 6B, results of only iliac cells are presented). Furthermore, the expression of A3G mRNA was significantly greater in CCR5+ than CCR5− CD4 T cells in both the iliac (p=0.004) and axillary lymph nodes (p=0.0001).
Rectal and small intestinal lymphoid cells were eluted, but further separation into CD4+CCR5+ and CCR5− T cells was not feasible with the limited number of eluted cells. A3G mRNA was evaluated in the immunized and compared with A3G mRNA in the unimmunized macaques. A significant increase in A3G mRNA (p=0.012) was found in the rectal cells (Fig. 6C) from the immunized (147.7±37.4) compared with the unimmunized macaques (45.8±8.8). Whilst a relative increase in A3G was also observed in intestinal cells (Fig. 6D) of the immunized (53.0±21.2) compared with unimmunized macaques (22.1±7.4), this failed to reach the 5% level of significance.
Precursor frequency evaluated by the T cell proliferative response
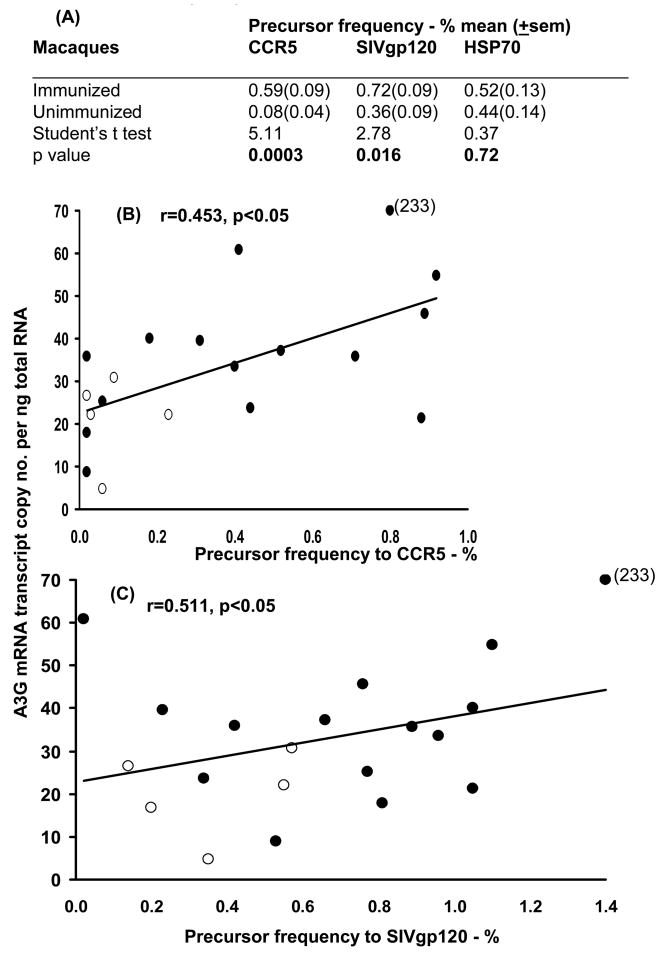
Precursor frequency to each of the 3 antigens and HSP70 was analysed at week 27 (4 weeks after the 3rd immunization); the negative control value (unstimulated cells) was subtracted. A significant increase in precursor frequency was found with the CCR5 peptides (p=0.0003) in the immunized (0.59±0.09%), compared with the unimmunized (0.08±0.04%) macaques, as well as with SIVgp120 (0.72±0.09 and 0.36±0.09, respectively, p=0.016), but not with HSP70 (Fig. 7A). The relationship between the precursor frequency and A3G mRNA expression was then analysed in CD4+ T cells, which showed a significant positive correlation (p<0.05) between A3G mRNA copies and the precursor frequency for CCR5 (r=0.453) and SIVgp120 (r=0.511) (Fig. 7B and C). The lack of a correlation with HSP70 is accounted for by the limited increase in precursor frequency of HSP70 following immunization, unlike the signficiant increase with both CCR5 and SIV (Fig. 7A).
Figure 7.
(A) Precursor frequency of CD4+ T cells, % mean (±sem), (B) relationship between A3G mRNA and precursor frequency to CCR5 and (C) to SIVgp120 of immunized (●) and unimmunized (o) macaques at week 27 (4 weeks after the 3rd immunization) or the corresponding time in the unimmunized animals (n=20).
Analysis of A3G mRNA expression between infected and uninfected immunized macaques
The expression of A3G mRNA was then evaluated in circulating CCR5+ memory T cells, according to whether the macaques were infected after challenge with SIVmac251 (Fig. 8A). Analysis of the uninfected immunized macaques showed a very significant increase in A3G mRNA (140.7±15.0), compared with that in the uninfected animals (69.2±7.9; p=0,004). In contrast, CD4+CCR5− T cells showed no significant difference in A3G mRNA between the uninfected (29.8±7.9) and infected macaques after immunization (45.1±11.7) (Fig. 8A). The viral load in the uninfected animals was <50 SIV RNA copies per ml plasma (the detection limit of the assay), as compared with that found in the infected animals (1.52±1.4 × 105) RNA copies per ml (p=0.0004, Fig. 8B). Thus, only the uninfected CCR5+ memory CD4+ T cells showed an increase in A3GmRNA, consistent with its anti-viral function.
Figure 8.
A3G mRNA transcript copy number per ng of total RNA in CCR5+ memory and CCR5− naive CD4+ T cells of SIV challenged macaques divided into uninfected (n=6) and infected groups (n=9). (A) At week 27 (4 weeks after the 3rd immunization). (B) Plasma viral load (RNA copies/ml) in the infected macaques with SIVmac251 at week 47 (n=9). (C and D) A3G mRNA in PBMC, splenic, iliac and axillary lymph nodes, rectal and small intestinal mucosal cells at necropsy; the results are presented in ■ uninfected (n=6) □ infected macaques (n=9), except in rectal and intestinal mucosa (n=2 and 3); *p<0.05, **p<0.01, NS= not significant.
Comparative analysis of A3G mRNA expression in splenic, lymph node and mucosal tissues in infected and uninfected macaques
A3G mRNA expression in the immunized macaques was significantly higher in CD4+CCR5+ T cells from uninfected than infected macaques in the iliac lymph node cells (p=0.023) in uninfected (478.0±51.0), compared with the infected (294.0±48.0) macaques (Fig. 8C), consistent with the draining pattern and homing of mononuclear cells between the iliac lymph nodes to the rectal mucosa (18). A3G expression in CD4+CCR5− cells failed to show a significant difference between the infected and uninfected macaques (Fig. 8D). Eluted cells from 5 specimens of rectal and 5 intestinal tissues were available for analysis of A3G mRNA (Fig. 8C), in which A3G mRNA copies were higher in each of the 2 rectal samples of the uninfected (141.6, 290.6) than 3 infected (72.8, 115.2, 118.1) macaques. In contrast the intestinal mucosa and the unrelated axillary lymph nodes showed no corresponding difference in A3GmRNA between the uninfected and infected animals (Fig. 8C). Furthermore, unlike the CD4+CCR5+ PBMC, those eluted from the spleen also failed to show a significant difference between the uninfected and infected macaques (Fig. 8C). The unimmunized macaques failed to show any change in CD4+CCR5+ T cells in either the uninfected (1.3±0.2) or infected macaques (3.0 and 2.75, respectively). These results suggest that rectal immunization and SIVmac251 challenge elicit an increase in A3GmRNA and prevention of SHIV infection only in the rectal mucosa and the regional iliac lymph nodes and does not involve the upper intestinal tract.
Discussion
Sequential analysis of A3G mRNA expression following rectal mucosal immunization with HSP70 linked to three extracellular CCR5 peptides, with or without SIVgp120 and gag p27, revealed a significant and progressive increase in A3G mRNA expression following each immunization of macaques. The increase in A3G mRNA was maintained for at least 17 weeks. In contrast, in vitro treatment of macrophages with IFN-α and IFN-γ upregulated A3G which was maintained only up to 1 week, after which re-exposure to the IFNs was required (19). These findings raised the question whether upregulation of A3G mRNA expression in CD4+ T cells may develop a memory-like function, despite the fact that A3G is not a part of the adaptive immune system. Indeed, examination of CCR5+ memory CD4 T cells (20, 21) clearly demonstrated that immunization significantly upregulated A3G mRNA in CD4+CCR5+ memory T cells in the circulation (p<0.0001), spleen (p=0.001), lymph nodes (p<0.02) and rectal mucosal tissue (p=0.012), compared with the corresponding cells from unimmunized macaques. A3G protein demonstrated by Western blots was also increased in PBMC after immunization in all macaques. It should be noted that both A3G mRNA and protein examined were significantly knocked down with the specific as compared with non-specific siRNA.
A3G protein was also significantly increased by immunization as demonstrated in the CD4+ CD95+ memory T cells (14) by immunofluorescence. However, unlike the persistence of A3G expression in CD4+CD95+ T cells up to 47 weeks, the proportion of these memory T cells decreased after 27 weeks, that is after they were challenged with SIVmac251, to below the pre-immunization levels. The results suggest that in CD4+ memory T cells, a large proportion of which are infected and destroyed in the mucosal associated lymphoid tissue within weeks of SIV or HIV infection (6, 8, 21), increased levels of A3G mRNA can be maintained for at least 17 weeks. This is in contrast to A3G expression in DC or monocytes in which A3G remained unchanged after immunization, probably because DC and monocytes do not manifest memory function, unlike CD4+ T cells.
Examination of central and effector memory cells showed that A3G mRNA expression was significantly increased in CD4+ CD95+ CCR7− T effector memory cells. This is consistent with the studies of protection in macaques chronically infected with the nef deleted SIVmac 239 in which CD4+ CD95+ CCR7− effector memory T cells were correlated with protection (22). However, long-term survival in macaques primed by SIVmac 239 DNA-env/gag/pol or nef and boosted with rAdeno-SIVmac 239-DNA challenged with SIVmac251 was correlated with central memory CD4+ CD28+ CD95+ T cells (23), which supports the view that central memory cells may be involved in long-term protection compared with those of effector memory T cells (24). It is noteworthy that upregulation and persistence of A3G for at least 17 weeks was elicited by rectal mucosal immunization and challenge, which has special relevance to HIV-1 vaccination and mucosal immunity at large.
The precursor frequency of the proliferative CD4+ T cells to CCR5 showed a significant increase in the immunized compared with the unimmunized macaques, and a significant direct correlation was found between A3G mRNA expression and precursor frequency of CD4+ T cells stimulated by CCR5 or SIVgp120. These results are consistent with the concept that precursor frequency is a factor controlling memory lineage development (25). A model of programmed differentiation has emerged which suggests that after primary immune stimulation the developing T cell precursor frequency is a determining factor in the differentiation of memory T cells, as well as the requirement for the CD28-CD80/86 and CD40-CD40L mediated costimulation (26–28).
As prevention of HIV or SIV infection appears to be determined within 2 weeks of exposure to the virus, and CCR5+ CD4 T cells are the prime target found predominantly in the mucosal associated lymphoid tissues (6, 8, 21), memory T cells need to be examined. Indeed, we studied the CD4+CCR5+ memory T cells and the CD4+CD95+ CCR7− T effector memory cells by mucosal immunization with HSP70 linked to CCR5 peptides or SIVgp120 or both. These elicit A3G mRNA expression predominantly in CD4+CCR5+ memory T cells in the circulation, lymph nodes, spleen and most likely mucosal tissues, though demonstrated here only in mononuclear cells.
The evidence in favour of memory-like function in CD4+ T cells for A3G, despite the innate nature of A3G is summarised below. A significant increase in A3G mRNA expression and the protein was found: (a) in PBMC following each immunization, and was maintained at least for 17 weeks, (b) in CD4+CCR5+ memory T cells in the circulation, spleen, lymph nodes, rectal and intestinal mononuclear cells, (c) CD4+ CD95+ CCR7− effector memory T cells express an increase in A3G and (d) the precursor frequency of CD4+ T cells stimulated with CCR5 peptides or SIVgp120 showed significant direct correlation with A3G mRNA expression. A3G can be upregulated in CD4+ T cells (adaptive immune cells) and DC (innate immune cells) in vitro (29), but in vivo immunization in the present study clearly demonstrated upregulation of A3G expression in CD4+ memory T cells but not in DC or monocytes. Whilst the increasingly close relationship between innate and adaptive immunity has now been well recognized (2), the boundary between the two types of immunity has been questioned by the finding that NK cells (classical innate immune cells) can induce antigen-specific skin hypersensitivity reaction in the absence of T lymphocytes (1). Further evidence has been recently published, demonstrating that repeated LPS stimulation through the TLR4 receptors on macrophages induces short-term memory for antimicrobial factors but tolerance to pro-inflammatory chemokines (30). The data presented here are consistent with CD4+ T cells expressing a memory-like response to the innate antimicrobial A3G, which however is not found in DC, monocytes or NK cells.
The potential mechanism of memory in CD4+ T cells for A3G needs to be explored, but may involve activation of CCR5 on CD4+ cells (31, 32), which signal through the MAP kinase, ERK 1/2, p38 pathway eliciting A3G expression. HSP70 also binds CCR5 and CD40 molecules on DC, eliciting maturation of DC, upregulation of the costimulatory molecules (CD40, CD80 and CD86), CC chemokines and a number of cytokines (33–35) creating an appropriate immunological milieu. Furthermore, HSP70 engaging CCR5 induces DC plasma membrane extensions and autologous or allogeneic CD4 and CD8 T cell clustering with DC within 30 seconds of stimulation, forming antigen independent immune synapses between ICAM-2 and -3 on the DC and LFA-1 on T cells (32). This may enhance cognate immune responses to CCR5 and SIVgp120, as demonstrated by increased precursor frequency of CD4+ T cells. This mechanism appears to be confined mostly to CD4+CCR5+ memory T cells in the circulation, spleen, iliac lymph nodes and the rectal mucosal tissues, and was elicited by rectal mucosal immunization. The results suggest that mucosal immunization targets CD4+CCR5+ memory T cells that may prevent their infection and destruction by the early development of A3G-mediated anti-SIV or HIV response. This is associated with CD4+ CD95+ CCR7− effector memory cells expressing A3G, which shows a significant correlation with the precursor frequency of CD4+ T cells to CCR5 and SIVgp120. The specific role of HSP70 alone in upregulation of A3G is likely to be significant, as demonstrated in vitro by stimulating the CCR5 and CD40 molecules (29). It is, however not possible to differentiate in this study the contribution that HSP70 made to upregulation of A3G, compared with those of SIVgp120, SIVp27 and the CCR5 peptides. Nonetheless, it is likely that agents, such as adjuvants that may stimulate IL-2 and IL-15 (36), IFN-α and IFN-γ (19) production or CCR5 and CD40 (29) activation will upregulate A3G. This raises the possibility that only some adjuvants may stimulate A3G expression, thus contributing to prevention of viral infection. Furthermore, as HSP70 stimulates DC and T cells to produce the CC-chemokines (CCL-3, CCL-4 and CCL-5) these will enhance upregulation of A3G by activating the CCR5 molecules (29).
The role of A3G expressing CD4+ T memory cells was examined after the macaques were challenged by the rectal mucosal route with SIVmac251. Consistently higher expression of A3G mRNA was detected in PBMC, CD4+ and CD4+CCR5+ T cells from SIV uninfected compared to infected macaques but the difference reached a significant level only with the CD4+CCR5+ memory T cells. Indeed, a significant increase in A3G mRNA expression was found in the circulating CD4+CCR5+ T cells of the uninfected compared with infected macaques (p=0.004), whereas the CD4+CCR5− T cells showed a decrease in A3G mRNA expression in the uninfected cells. Importantly, A3GmRNA expression in CD4+CCR5+ memory T cells was also increased in the rectal mucosa and the regional iliac lymph nodes of the uninfected compared with infected macaques, and demonstrates the regional nature of the immune response (37, 38). These results are unlikely to be skewed by A3G mRNA being affected by challenge with the virus, as the infected unimmunized macaques showed little or no change in A3G expression 4 and 20 weeks after challenge (73.8±10.5, and 74.4±14.8%, respectively), compared with the A3G levels before challenge (77.5±16.6%). This is consistent with the in vitro studies that HIV does not affect A3G expression (15, 16). These results suggest that upregulation of A3GmRNA in CD4+CCR5+ T cells may be important in the antiviral effect of the CCR5+ T cells targeted by the virus. However, the timing of A3G expression will need to be determined, as its optimum effect should be early in HIV or SIV infection. It is noteworthy that there was a significant correlation between the antiviral A3G (innate immunity) and the precursor frequency (adaptive immunity) to the immunizing antigens in CD4+ T cells. This supports the emerging evidence of the close association between innate and adaptive immunity, which may play an important part in the protective mechanism against SIV or HIV-1 infection.
Acknowledgments
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases, Washington, DC (No: 1-R21-A1057110-1). We thank Dr. S. Norley (Robert Koch Institute, Berlin) for the SIVmac251 stock.
We thank the referee for his comments and have revised the paper as described below.
1. The referee is raising an important point and indeed initially we have analysed all the data separately for each subset of the 3 immunization groups which did not differ significantly for A3G in CCR5+ T cells or CD4+CD95+CCR7+ T cells and this is pointed out on p.16 and 19. However, CD4+CD95+ T cells in group 3 showed lower A3G expression due to 1 outlying point (fall from 53.0 to 16.3%), and this is pointed out on p.17.
HSP70 was used as an adjuvant covalently linked with CCR5, SIVgp120 and p27 and was common to the vaccine used in the 3 groups of macaques. The lack of a significant relationship between HSP70 precursor frequency and A3GmRNA is accounted for by the limited increase in precursor frequency between the immunized (0.52±0.13%) and unimmunized animals (0.44±0.14%), unlike with the two specific antigens (2 to 7-fold increase). This is most likely due to 85% homology between the microbial HSP70 used and macaque HSP70. This point is now made on p.21.
2. We appreciate the point raised and wish to clarify the misunderstanding. The increase in A3G in the immunized macaques was shown after each of the 3 immunizations (p13). This was followed with statistical analysis of the difference between the immunized and unimmunized animals (p14). We were surprised to find that the control (unimmunized animals) showed a small increase in A3G mRNA after 27 weeks, which reached the 5% level of significance and ascribed this to environmental factors. However, A3G expression in all memory T cell subset failed to show an increase between week 0 and 27 in the unimmunized macaques (see Fig. 1C, 2C, 3C, 5A). This is now pointed out on p.14.
3. To clarify the in vivo infectivity of CD4+CCR5+ memory compared with CD4+CCR5− naïve T cells, we have modified Fig. 8A and separated it into A and B. These show that significant increase in A3GmRNA is found only in the uninfected CD4+CCR5+ memory phenotype and not in the CD4+ CCR5− naïve T cells. This is also found in CD4+CCR5+ T cells of the iliac lymph nodes draining the rectal mucosa but not in the unrelated axillary lymph nodes of the uninfected compared with the infected macaques. Furthermore, A3G expression in CD4+CD95+ T cells increases 20 weeks after challenge, but the proportion of these cells decreases (Fig. 3D and E), consistent with many of the memory cells being infected and killed (9). These points are now clarified on p.21, 22, 23, 27, 28 and 18.
The in vitro infectivity of memory cells which express high level of A3G was not examined in this investigation, as we were primarily concerned with in vivo infectivity. However, in our previous paper we have demonstrated in vitro in CCR5 transfected cells by using GFP labelled pseudovirions, as well as in primary CD4+ T cells, that upregulation in A3GmRNA is associated with inhibition of the pseudovirus HIV-1 (BaL), respectively (Pido-Lopez et al. J. Immunol. 178:1671–1679).
4. We have not tested specifically cell activation markers and/or cell cycle, which are interesting suggestions, which we will pursue in future experiments. However, effector memory CD4+ CD95+CCR7− T cells are considered to be activated cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O’Leary JGM, Goofdarzi DL, Drayton, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nature Immunol. 2006;5:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 2.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 3.Germain RN. An innately interesting decade of research in immunology. Nat Med. 2004;10:1307–1320. doi: 10.1038/nm1159. [DOI] [PubMed] [Google Scholar]
- 4.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 5.Pulendran B, Ahmed R. Translating innate immunity into immunological memory: Implications for vaccine development. Cell. 2006;124:849–863. doi: 10.1016/j.cell.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Brenchley JM, Schacker TW, Ruff LE, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Immunol. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Duan L, Estes JD, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 8.Mehandru S, Poles MA, Tenner-Racz K, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- 10.Mehlert A, Young DB. Biochemical and antigenic characterization of the Mycobacterium tuberculosis 71kD antigen, a member of the 70kD heat-shock protein family. Mol Microbiol. 1989;3:125–130. doi: 10.1111/j.1365-2958.1989.tb01801.x. [DOI] [PubMed] [Google Scholar]
- 11.Carlsson J, Drewin H, Axen R. Protein thiolation and reversible protein-protein conjugation. N-Succinimidyl 3-(2-pyridyldithio) propionate, a new heterobifunctional reagent. Biochem J. 1978;173:723–737. doi: 10.1042/bj1730723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergmeier LA, Mitchell EA, Hall G, et al. Antibody-secreting cells specific for simian immunodeficiency virus antigens in lymphoid and mucosal tissues of immunized macaques. AIDS. 1998;12:1139–1147. doi: 10.1097/00002030-199810000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Givan AL, Fisher JL, Waugh M, Ernstoff MS, Wallace PK. A flow cytometric method to estimate the precursor frequencies of cells proliferating in response to specific antigens. J Immunol Methods. 1999;230:99–112. doi: 10.1016/s0022-1759(99)00136-2. [DOI] [PubMed] [Google Scholar]
- 14.Mattapallil JJ, Douek DC, Buckler-White A, et al. Vaccination preserves CD4 memory T cells during acute simian immunodeficiency virus challenge. J Exp Med. 2006;203:1533–1541. doi: 10.1084/jem.20060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–4601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 16.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 17.Picker LJ. Immunopathogenesis of acute AIDS virus infection. Curr Opin Immunol. 2006;18:399–405. doi: 10.1016/j.coi.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell EA, Bergmeier LA, Doyle C, et al. Homing of mononuclear cells from iliac lymph nodes to the genital and rectal mucosa in non-human primates. Eur J Immunol. 1998;28:3066–3074. doi: 10.1002/(SICI)1521-4141(199810)28:10<3066::AID-IMMU3066>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Peng G, Lei KJ, Jin W, Greenwell-Wild G, Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med. 2006;203:41–46. doi: 10.1084/jem.20051512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douek DC, Picker LJ, Koup RA. T cell dynamics in HIV-1 infection. Annu Rev Immunol. 2003;21:265–304. doi: 10.1146/annurev.immunol.21.120601.141053. [DOI] [PubMed] [Google Scholar]
- 21.Veazey R, Lackner A. The mucosal immune system and HIV-1 infection. AIDS Rev. 2003;5:245–252. [PubMed] [Google Scholar]
- 22.Gauduin MC, Yu Y, Barabasz A, et al. Induction of a virus-specific effector–memory CD4+ T cell response by attenuated SIV infection. J Exp Med. 2006;203:2661–2672. doi: 10.1084/jem.20060134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letvin NL, Mascola JR, Sun Y, et al. 2006. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312:1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts AD, Ely KH, Woodland DI. Differential contributions of central and effector memory T cells to recall responses. J Exp Med. 2005;202:123–133. doi: 10.1084/jem.20050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marzo AL, Klonowski KD, Le Bon A, Borrow P, Tough DF, Lefrancois L. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat Immunol. 2005;6:793–799. doi: 10.1038/ni1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masopust D, Kaech SM, Wherry EJ, Ahmed R. The role of programming in memory T-cell development. Curr Opin Immunol. 2004;16:217–225. doi: 10.1016/j.coi.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- 28.Ford ML, Koehn BH, Wagener ME, et al. Antigen-specific precursor frequency impacts T cell proliferation, differentiation, and requirement for costimulation. J Exp Med. 2007;204:299–309. doi: 10.1084/jem.20062319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pido-Lopez J, Whittall T, Wang Y, et al. Stimulation Of Cell Surface CCR5 And CD40 Molecules By Their Ligands Or By HSP70 Upregulates APOBEC3G Expression In CD4+ T Cells And Dendritic Cells. J Immunology. 2007;178:1671–1679. doi: 10.4049/jimmunol.178.3.1671. [DOI] [PubMed] [Google Scholar]
- 30.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 31.Whittall T, Wang Y, Younson J, et al. Interaction between the CCR5 chemokine receptors and microbial HSP70. Eur J Immunol. 2006;36:2304–2314. doi: 10.1002/eji.200635953. [DOI] [PubMed] [Google Scholar]
- 32.Floto RA, MacAry PA, Boname JM, et al. Dendritic cell stimulation by mycobacterial Hsp70 is mediated through CCR5. Science. 2006;314:454–458. doi: 10.1126/science.1133515. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Kelly CG, Kartunen JT, et al. CD40 is a cellular receptor mediating mycobacterial heat shock protein 70 stimulation of CC chemokines. Immunity. 2001;15:971–983. doi: 10.1016/s1074-7613(01)00242-4. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Kelly CG, Singh M, et al. Stimulation of TH1-polarizing cytokines, C-C chemokines, maturation of dendritic cells, and adjuvant function by the peptide binding fragment of HSP70. J Immunol. 2002;169:2422–2429. doi: 10.4049/jimmunol.169.5.2422. [DOI] [PubMed] [Google Scholar]
- 35.MacAry PA, Javid B, Floto RA, et al. HSP70 Peptide Binding Mutants Separate Antigen Delivery from Dendritic Cell Stimulation. Immunity. 2004;20:95–106. doi: 10.1016/s1074-7613(03)00357-1. [DOI] [PubMed] [Google Scholar]
- 36.Kreisberg JF, Yonemoto W, Greene WC. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J Exp Med. 2006;203:865–870. doi: 10.1084/jem.20051856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehner T, Bergmeier LA, Panagiotidi C, et al. Induction of mucosal and systemic immunity to a recombinant simian immunodeficiency viral protein. Science. 1992;258:1365–1369. doi: 10.1126/science.1360702. [DOI] [PubMed] [Google Scholar]
- 38.Lehner T, Bergmeier LA, Tao L, et al. Targeted lymph node immunization with simian immunodeficiency virus p27 antigen to elicit genital, rectal and urinary immune responses in nonhuman primates. J Immunol. 1994;153:1858–1868. [PubMed] [Google Scholar]