Abstract
MicroRNAs (miRNAs) are short noncoding RNAs involved in post-transcriptional gene regulation via binding to mRNAs. Studies show that in a multicellular organism microRNAs (miRNAs) downregulate a large number of target mRNAs. However, predicting the target genes of a miRNA is challenging. Microarray expression profiling has been proposed as a complementary method to increase the confidence of miRNA target prediction, but it can become computationally costly or even intractable when many miRNAs and their effects across multiple tissues are to be considered. Here, we propose a statistical method, the relative R2 method, to find high-confidence targets among the set of potential targets predicted by a computational method such as TargetScanS or by microarray analysis, when expression data of both miRNAs and mRNAs are available for multiple tissues. Applying this method to existing data, we obtain many high-confidence targets in mouse.
Keywords: microRNA, microarray, regression model, TargetScanS
1. Introduction
MicroRNAs (miRNAs), which are single-stranded RNAs of ~20–23 nucleotides, posttranscriptionally regulate gene expression. Computational and molecular cloning approaches have revealed hundreds of miRNAs in a variety of organisms (Ambros et al. 2003; Houbaviy et al. 2003; Lim et al. 2003; Kim et al. 2004). Many computational methods have been developed to predict miRNA targets; for example, TargetScanS predicts the targets of a miRNA by searching for the presence of conserved 8mer or 7mer sites that match the seed region of the miRNA. A small portion of these predicted targets have been experimentally validated, showing a relatively high accuracy for target prediction (Sethupathy et al. 2006).
Computational approaches currently use sequence complementarity and most of them also use evolutionary conservation to identify potential targets. It has been suggested that the false-discovery rate for computationally predicted targets is ~50% (Lewis et al. 2005; Farh et al. 2005). Besides the sequence complementarity approach, Grimson et al. (2007) pointed out that a crux for target recognition is ~7 nt sites that match the seed region of the miRNA. Since these seed matches are not always sufficient for repression, they uncovered five general features of site context that boost binding efficacy: AU-rich nucleotide composition near the site, proximity to sites for coexpressed miRNAs, proximity to residues pairing to miRNA nucleotides 13–16, positioning within the 3’UTR at least 15 nt from the stop codon, and positioning away from the center of long UTRs.
Profiling miRNA expression is very helpful for studying the biological functions of miRNAs, so it has been used to as a complementary method for discovering miRNA targets (Lim et al. 2005). However, this method can become computationally complicated when multiple miRNAs and their effects across multiple tissues are to be considered. To overcome this difficulty, we use statistical methods to build up a network of associations between the miRNAs and their target mRNAs.
In this study, the relative R2 method is proposed to select high-confidence targets from predicted targets. The relative R2 method can be explained statistically from the degree of fitness of a model in terms of a subset of independent variables.
A method for finding miRNA targets using Bayesian variation analysis was recently proposed by Huang et al. (2007). This method is complicated and requires extensive calculations. We apply our method to the same dataset used in Huang et al. (2007) and select 448 high-confidence targets such that the relative R2 for each target reaches 0.995, which is considerably higher than those targets predicted by Huang et al. (2007), whose average relative R2 is less than 0.9.
2. Methods
We introduce our method with the usual linear regression model, before considering the general model.
2.1. The regression model
Consider n miRNAs, z1 , ……, zn, and l tissues, t1,……, t1 . Assume that the expression levels of the n miRNAs in tissue tj are z1j ,……, znj . By prediction methods, such as TargetScanS and microarray analysis, potential targets for each of these n miRNAs can be predicted. Our method is to select high-confidence miRNA targets from the set of the predicted miRNA targets, using microarray expression data.
First, for an mRNA, we can find the miRNAs, say z1 , ……, zk , from the set of potential targets such that each of the miRNAs has this mRNA as its potential target. Our goal is to find from these k miRNAs the miRNAs that have a significant effect on the expression level of this mRNA. Assume that the expression levels for the mRNA in the l tissues are y1 y2 , ,.…, yl . We fit the microarray expression data of the mRNA in terms of the microarray expression of the k miRNAs using the regression model
| (1) |
where εj is the error term.
In model (1), the best estimator of β = (b0,b1,b2,…,bk) ' is β̂ = (b̂0 b̂1,…, b̂k) '= (ZTZ)−1ZTY , where y = (y1,y2,.…,yl),Z = (zij)ix (k+1) and (z01,…,z0l) (1,.…,1). Note that b0 in (1) is the basal expression level of the mRNA and bi is the weight that miRNA zi affects the expression level of the mRNA.
Let fi = (Zβ̂)i be an estimator of yi. Define , where ȳ is the mean of y1, y2,.…,yl . The R2 for a linear regression model is defined as SS reg/SS total, which is a statistic that gives information about the goodness of fit of a model. In regression, the R2 coefficient of determination is a statistical measure of how well the regression line approximates the real data points. A R2 of 1.0 indicates that the regression line perfectly fits the data. R2 is often interpreted as the proportion of response variation explained by the regressors in the model. Thus, R2 = 1 indicates that the fitted model explains all variability in y, while R2 = 0 indicates no linear relationship between the response variable and the regressors. However, we do not directly use R2 in this study, but use the relative R2 values as a criterion to choose high-confidence targets. The definition of relative R2 is given later.
Our method of selecting miRNAs that significantly affect the level of the mRNA is first to rank the k miRNAs according to their p-values --- the smaller the p-value, the higher the rank. The p-value of miRNA zi is defined as the probability
which is the p-value used to test : H0 :bi =0, where W denotes the standard normal random variable. Note that and σ2 can be estimated by sample variance , where r denotes the rank of Z . Thus, Var(b̂i) can be approximated by the ith diagonal element of (ZTZ)−1σ̂2. Note that if the number of the tissues l is small, for obtaining more accurate probability approximation, we may use the T statistic to replace the standard normal random variable Z , where the T statistic follows the t distribution.
Rank the miRNA as the j th significant miRNA if its p-value is the j th smallest p-value. Calculate the R2 for model (1), say gk . Consider the miRNAs that have a p-value less than a critical value, say p0 . Since a p-value is an indicator of the significance of the effect of the miRNA to the mRNA, it is reasonable to require that the p-values of the selected miRNAs are not large. For example, we can choose p0 as 0.5 or less. We suggest choosing p0 near 0.5 to reduce the chance that too few miRNAs are included in the analysis. Note that p0 is not the main criterion in this approach; the main criterion is the relative R2method. Since the range of p-value is between 0 and 1, we may set the middle point 0.5 as a threshold.
Assume that there are m (m ≤ k) miRNAs, z1,……, zm , whose p-values are less than p0 . We can use the m miRNAs to fit the microarray expression data of the mRNA. The model is
| (2) |
Let c =(c0,c1,…cm), Zr = (Zij)lx(m+1) and
Denote the R2 for the regression model (2) as gm . If gm / gk ≥ S, then the m miRNAs are included in the set of the miRNAs each of which has a significant effect on the mRNA, where s can be chosen as 0.95 or larger. If gm / gk < s, then the m miRNAs are not included. Basically, the selection of the p0 and s values can be based on the proportion of high confidence targets that we intend to obtain from the set of potential targets.
We define the value gm / gk as the relative R2 . Instead of using the standard R2 , we use the relative R 2 values to evaluate the fitness of model (2). Since, from the potential target set, the k miRNAs are the only miRNAs that have significant effects on the mRNA, and the best R2 that can be derived from the linear regression model using the k miRNAs, z1 ,……, zk , is gk, it is reasonable to use the value of gk as a base to evaluate the fitness of a regression model by using some variables in the set of {z1 , ……, zk} as dependent variables. Therefore, we can use the criterion of comparing gm with gk to select high-confidence miRNAs. It is possible that gk is not high, such as the situation discussed in Section 4 later. Basically, if the correlations between an mRNA and each miRNA are not high across the l tissues, it is unlikely to find a model such that gk is high because there is no strong dependent tendency between the expression of mRNA and the expression of the miRNAs.
However, even if gk is not high, it is still possible that the mRNA is the true target for some miRNAs among these k miRNAs. Thus, we can use the relative R2 method to select a set of more significant miRNAs, which simultaneously affect the expression of the mRNA.
Using the method, for each mRNA, we can assign a set of miRNAs such that these miRNAs significantly affect the expression of the mRNA in terms of the linear model. For a miRNA, we can collect the set of mRNAs such that each mRNA in the set is a significant potential target of this miRNA by the relative R2 method. Then the mRNAs collected are the high-confidence targets of this miRNA.
Note that in order to eliminate the difference between the different tissues used in the analyses, we can first transform the expression data of mRNAs by normalizing the data in each tissue such that the scale of the expression data used in each tissue is the same. We use the normalized expression data when we apply the above approach to select high-confidence targets.
2.2. General model
The linear model used in Section 2.1 can be replaced by another kind of model, such as a nonlinear regression model. For a general model to fit the yi by using {Z1,.…, Zk}, assume that fi ' is the estimator of yi derived under this model. Define for the model. For m Zi ’s from the set {Z1,.…, Zk}, denoted as Z1,.…, Zm, we can also use the m miRNAs to derive the form of the model and the estimators for yi . Then calculate the R2 for the model based on the m miRNAs and compare it with the R2 for the model based on the k miRNAs to derive the relative R2 . Note that if the model is not a linear regression model, it may not be straightforward to derive the significant miRNAs for an mRNA by the p-value approach. It will depend on the form of the model to establish a test to select the significant miRNAs. However, to avoid the heavy calculation for deriving a test method for selecting significant miRNAs, for a set of miRNAs, we may directly calculate its relative R2 and choose the set of miRNAs corresponding to the highest relative R2 as the set of significant miRNAs. By a similar argument, the relative R2 can be used as a criterion to select high-confidence targets of a miRNA.
Furthermore, it is feasible to apply the relative R2 method to other criteria such as the adjusted R2 , etc. Since the value of adjusted R2 may be negative, a situation that requires more consideration, the application of the relative method under other criteria is currently under investigation.
3. Data Analyses
We now apply the new method to the data of Babak et al. (2004), which was used by Huang J.C. et al. (2007). The data set includes 1770 potential targets for 22 miRNAs across 17 tissues, which were predicted by Target-Scan in a dataset of 41699 mouse mRNAs in Babak et al. (2004) and Zhang et al. (2004). The 1770 potential targets are from 788 different mRNAs because some miRNAs have the same mRNA as their targets. The microarray expressions of the 41699 mRNAs across the 17 tissues can be represented by a 41699×17 matrix, and the microarray expressions of the 22 miRNAs across the 17 tissues can be represented by a 22×17 matrix. (The 22 miRNAs studied are let-7a, miR-1, miR-101, miR-107, miR-122a, miR-124a, miR-125b, miR-126, miR-133a, miR-16, miR-181a, miR-183, miR-194, miR-205, miR-22, miR-23b, miR-24, miR-26a, miR-29b, miR-34a, miR-92, and miR-93; and the 17 tissues studied are brain, femur, lung, heart, skeletal muscle, mammary gland, teeth, bladder, stomach, ES, spleen, embryo 12.5, embryo, placenta 9.5, embryo 9.5, small intestine, liver.)
To apply the relative R2 method to an mRNA in the 788 mRNAs, we first normalize the expression data of the mRNAs using the 41699 expression data for each tissue. The normalization method is first to calculate the mean and standard deviation of the 41699 expression values for each tissue. Then, for each tissue, the normalized expression data is the original expression data minus the mean and then divided by the standard deviation. Since the data set included 41699 expression data points, we can use it as a reference to make the normalization such that the scale used in the expression data of mRNA for each tissue is the same.
We find the miRNAs such that the mRNA is one of the potential targets of these miRNAs from the 1770-target dataset, and then use a regression model to fit the normalized expression data of the mRNA. Huang et al. (2007) used the Bayesian variation method to derive high-confidence targets. This method is more complicated and computationally more costly than the relative R2 method.
Using the present method, we can select high-confidence targets such that the relative R2 for each target reaches 0.995, given p0 =0.47. A total of 448 high-confidence targets are found and the average relative R2 for these 448 targets is 0.999. Here the p0 value is selected such that the number of about one-fourth targets in the 1770 targets can be selected by the method with the relative R2 reaching 0.995.
The above dataset can also be used to conduct a random permutation test of our method. We test whether our method can select more high-confidence targets from the set of the 1770 potential targets predicted by TargetScanS than from a set that is constructed by randomly assigning each one of the 1770 targets to one of the 22 miRNAs. The random permutation was repeated ten times, and the average number of selected high-confidence targets over the ten times was 336 (s.d. ≈ 27), which is significantly lower than 448, the number of high-confidence targets found by our method (see above) with a p-value of 1.4×10−10 . The p-value is derived from viewing the two proportions of the selected targets by the random permutation method and the relative R2method as the proportions of two binomial distributions and from testing the equality of the two proportions by the normal approximation. In addition, we also compare the targets shared between the random permutation case and the selected high-confidence targets. The average number of the shared targets from several comparisons is 25. So most of the selected high-confidence targets are not the same as the targets selected by random permutation. This result upholds the relative R2 method because (i) the method can select more targets than random permutation and most of the high-confidence targets are not the same as the targets selected by random permutation, and (ii) the method gives accordant results between the expression data analyses and TargetScanS analyses.
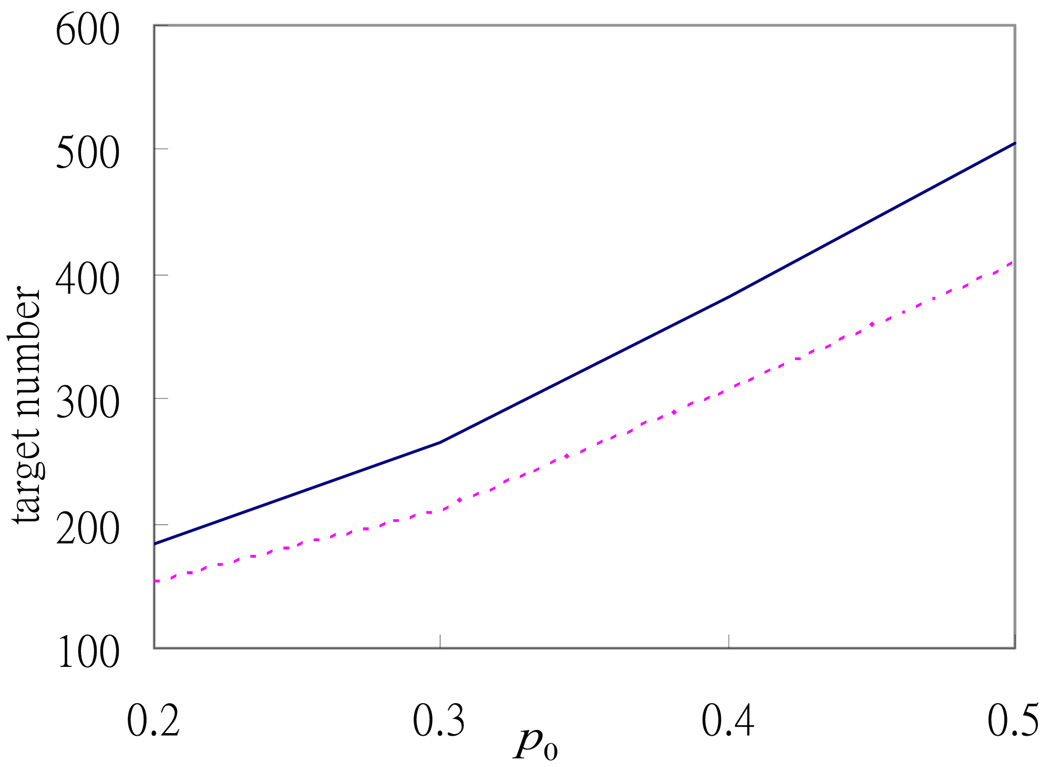
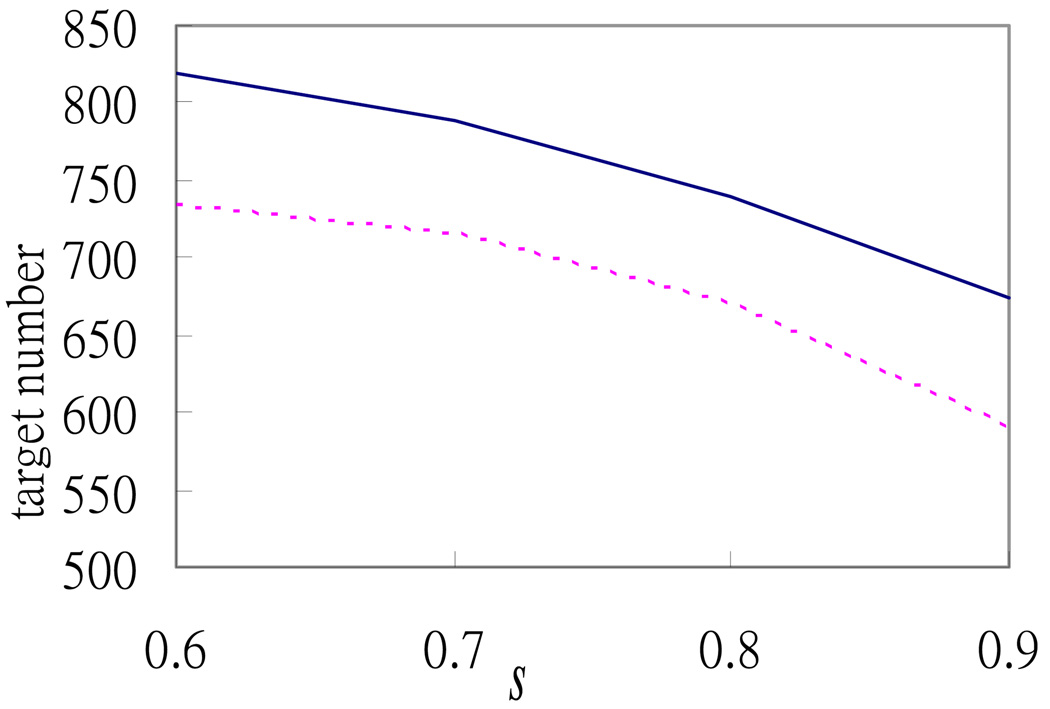
To make a more extensive comparison with the random permutation case, we conduct simulations for different cases by varying the values of p0 and s. Figure 1 and Figure 2 show that the numbers of high-confidence targets selected from the 1770 potential targets are always greater than the numbers selected from the random permutation case. Figure 1 shows that the difference between the two numbers increases with p0 , which reinforces the argument in Section 2 that the condition about the constraint of p0 should not be too strict; otherwise, the advantage of the relative R2 method is limited by this constraint.
Fig 1.
Relationship between the number of selected high-confidence targets and the p0 threshold used. The solid and dotted lines denote the numbers of high-confidence targets selected by the relative R2 method from the 1770 potential targets and from the dataset constructed by random permutation, respectively.
Fig 2.
Relationship between the number of selected high-confidence targets and the relative R2 value threshold used. The solid and dotted lines denote the numbers of high-confidence targets selected by the relative R2 method for the 1770 potential targets and the dataset constructed by random permutation, respectively, as s increases from 0.6 to 0.9, where s is the threshold of the relative R2 value as defined in the text.
In addition, in Table 1 we present several sets of p0 and s values for each of which the number of the selected targets by the relative R2 method with respect to these values is close to 450. It is seen that p0 is an increasing function of s when the proportion of the selected targets is set to be a fixed value. To obtain a fixed proportion of targets, there are more than one set of p0 and s values that can be chosen and the targets selected may be different with respect to different p0 and s . One may be interested in which set is more appropriate here. As we mentioned above, it is not recommended to choose a small p0 because it may confine the overall performance of the relative R2 method. Besides, from the correlation analysis in Section 4, the confirmed targets and miRNA may not have a high correlation when we investigate their relations individually, but can reveal the relation when we consider their overall performance by the relative R2 method. It corroborates the view that the selection of p0 should be more relaxed, while the selection of s can be more strict. Therefore, we select p0 as 0.47 and s as 0.995.
Table 1.
The 6 sets of p and s values for which the number of selected targets by the relative R2 method is close to 450.
| p | 0.47 | 0.45 | 0.4 | 0.35 | 0.3 | 0.25 |
| s | 0.995 | 0.99 | 0.97 | 0.95 | 0.85 | 0.7 |
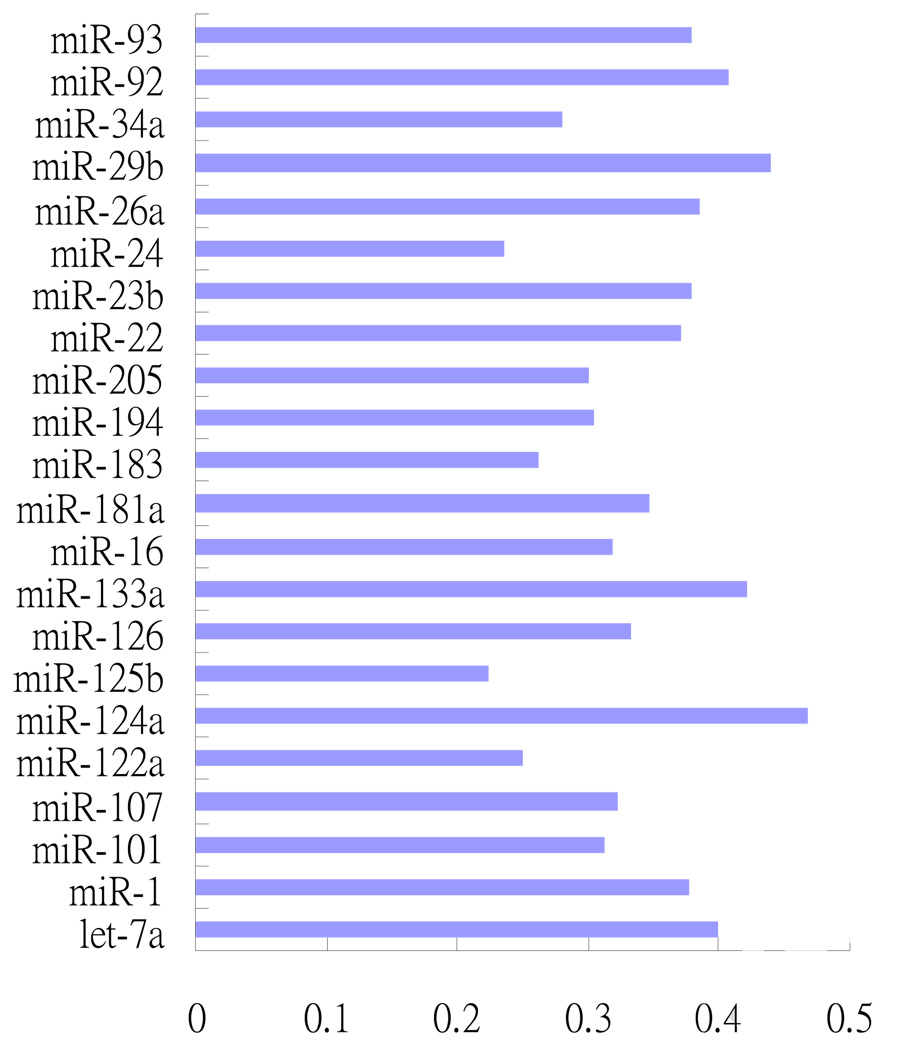
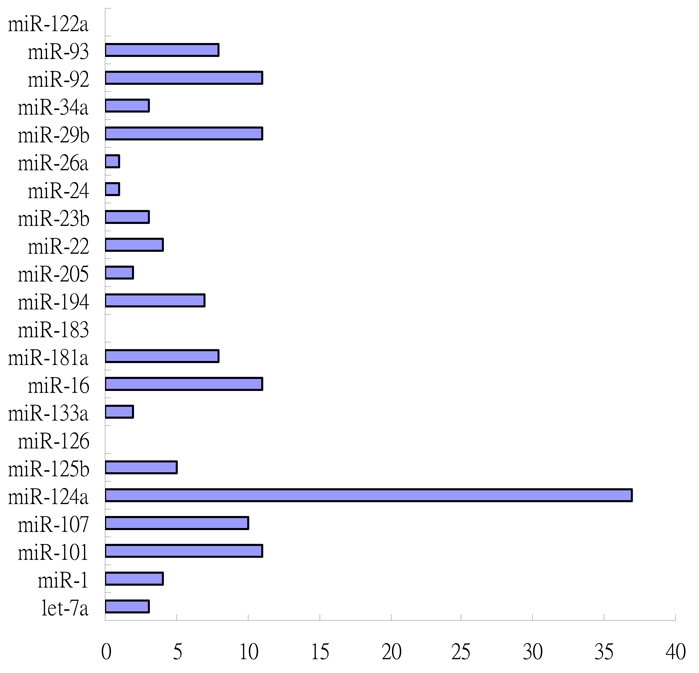
The ratio of the number of high-confidence targets found by our method to the total number of the potential targets (1770) for each of the 22 miRNAs is shown in Figure 3.
Fig 3.
The ratios of high-confidence targets for the 22 miRNAs selected by the relative R2 method to the targets for the 22 miRNAs selected by TargetScanS.
We can also check our analysis with the literature. We recover several confirmed targets from the literature (Bartel 2004, Cimmino et al. 2005, Farh et al. 2005, Lim et al. 2005, and Lewis et al. 2005). These include the relationships between miR-92 and the mitogen-activated protein kinase kinase 4 (MAP2K4) gene, between miR-16 and the B-cell CLL/lymphoma 2 (BCL2) gene, between miR-124a and the solute carrier family 15 member 4 protein (SLC15A4) gene, and between miR-124a and the homeodomain interacting protein kinase 1 (HIPK1) gene. We also recover the relationship between miR-181a and the B-cell CLL/lymphoma 2 (BCL2) from Tarbase (Sethupathy et al. 2006).
4. Discussion
The relative R2 method is proposed to analyze the data from the relative instead of from the absolute statistical point of view. If the correlation between the mRNA and miRNA is high, then we can directly adopt a standard statistical method to explore the high confidence targets. However, when the correlation between the mRNA and miRNA is not high, it is challenging to develop a statistical method to select correct targets. In such a case, if we use a standard statistical criterion, such as a high R2 to select the high-confidence targets mRNAs, then no confirmed target mRNAs may be selected. In this case, it would be better to use a variable standard that is dependent on the mRNA and miRNAs under study, rather than using a single standard for all genes. The relative R2 method can provide a variable standard to solve this problem.
We now discuss the relation between the confirmed targets and the miRNA by exploring their correlation coefficients and relative R2 values. For analyzing the data and investigating the relationship of the miRNAs and their targets, before modeling the data, a good way is to investigate the relation of the miRNA with each individual potential target. Although this study is to find the effect of multiple miRNAs on a target, rather than the effect of a single miRNA, understanding the connection between the target mRNA and a miRNA is helpful for reinforcing the validity of the proposed method.
For example, let us consider the three mRNAs, HIPK1, MAP2K4 and BCL2, mentioned in Section 3 and explore the relationship between their correlation coefficient and the R2 value.
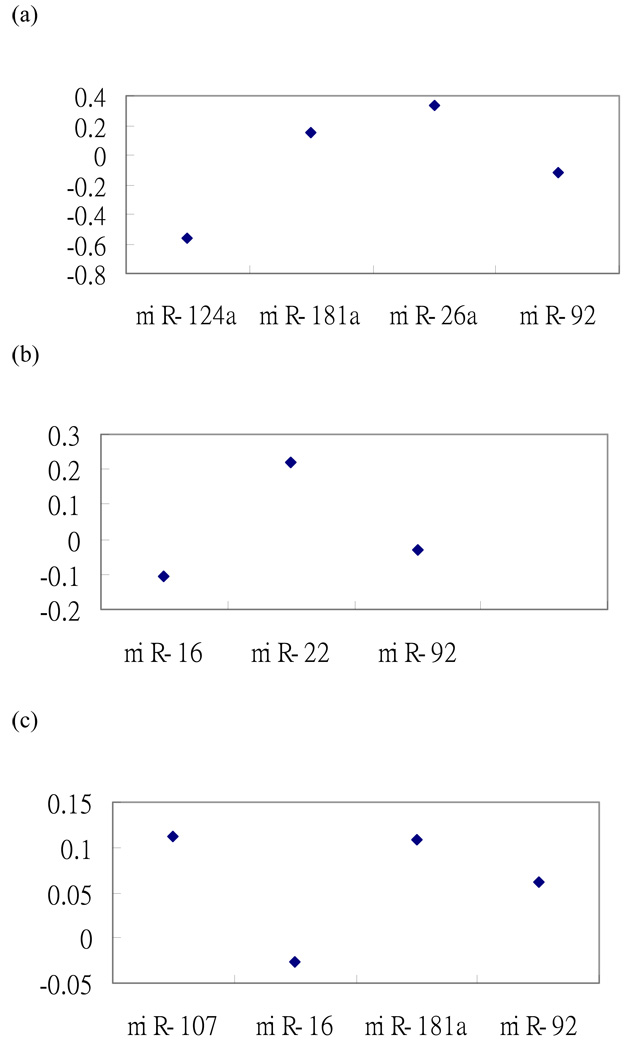
First, consider the HIPK1 mRNA, which is a potential target for the four miRNAs, miR-124a, miR-181a, miR-26a and miR-92. Although we are interested in knowing how the four miRNAs affect the expression of the mRNA, we also can investigate the relationship between the expression of each miRNA and the expression of HIPK1. The correlation coefficients of the expression for the four miRNAs with the expression of HIPK1, across the 17 tissues, are shown in Figure 4.
Fig. 4.
(a) The correlation coefficients between the homeodomain interacting protein kinase 1 ( HIPK1) mRNA expression level and the expression level of each of miR-124a, miR-181a, miR-26a and miR-92; (b) The correlation coefficients between the MAP2K4 mRNA expression level and the expression level of each of miR-16, miR-22, and miR-92; (c) The correlation coefficients of the BCL2 mRNA expression level and the expression level of each of miR-107, miR-16, miR-181a and miR-92.
By using the relative R2 method, three of the four miRNAs are selected such that its relative R2 reaches 0.995. The three selected miRNAs are miR-124a, miR-181a and miR-92 and their correlation coefficients with the HIPK1 mRNA are −0.566, 0.151 and −0.116, respectively (Figure 4(a)). Note that miR-26a, which is not selected, has the largest correlation coefficient 0.333. The correlations of two of the three selected miRNAs are negative, in agreement with the expectation that a miRNA usually downregulates its target mRNAs (Farh et al. 2005; Lim et al. 2005). The standard R2 values for the linear model of fitting the expression data of HIPK1 using the expression level of the four miRNAs and the expression level of the three selected miRNAs are 0.622 and 0.621, respectively. In this case, we can see from Figure 4 that the correlation coefficients between HIPK1 and the four miRNAs are not high. Therefore, it is hard to construct a model to fit the expression data of HIPK1 in terms of the expression data of the four miRNAs. So, if we use the standard R2 value, we may not be able to select any one of the three miRNAs, though one of the relations is confirmed in the literature. On the other hand, if we use the relative R2 method by comparing the ratio of 0.621/0.622 to 1, the confirmed relationship can be selected.
For the confirmed MAP2K4 mRNA and its corresponding miRNA miR-92, their correlation coefficient is −0.030 and the R2 is 0.198 (Fig. 4(b)). For the confirmed BCL2 mRNA and its corresponding miRNA miR-16 (Fig. 4(c)), their correlation coefficient is −0.027 and the R2 is 0.104. Therefore, if we use the correlation coefficient or R2 to select high confidence targets, these two confirmed mRNAs will not be selected. Because the two correlation coefficients are −0.030 and −0.027, we do not expect the two confirmed mRNAs to be selected by any statistical method from the absolute point of view. Instead, we need to use the relative criterion to select the targets because the coefficients of other miRNAs in the potential targets dataset are also not high. By including the other miRNAs in the potential targets dataset, we can construct the relative criterion to select the miRNAs such that the effects of the miRNAs on the expression level of the mRNA are found to be significant.
In addition, we present the overlap of the selected targets between Huang et al. 2007 and the relative R2 method in Figure 5. The total number of overlaps for the 22 miRNAs is 142. The overlap numbers for the three miRNAs, miR-126, miR-183 and miR-122a, are zero. The total overlap number is not large, perhaps because the correlation between the miRNA and their targets mRNA is not high, which was validated from several confirmed relationship as we mentioned above. This may lead to the variation between different statistical approaches.
Fig. 5.
The number of overlaps between the predictions by Huang et al. (2007) and the relative R2 method.
Besides, to estimate the accuracy of the relative R2 method, we compare the number of relationship appeared in Tarbase, but not found in the relative R2 method from the 1770 potential relationships. We found only two Tarbase interactions in the 1770 potential targets: the relationship between miR-181a and BCL2 mRNA and the relationship between miR-181a and HOXA11 mRNA. Only the relationship between miR-181a and BCL2 mRNA appeared in the 448 selected targets by relative R2method and in those selected by Huang et al. (2007). However, the other relationship between miR-181a and HOXA11 mRNA can be selected by the relative R2method if we relax the criteria by choosing 0.67 p0 and s = 0.9999 , which leads to 715 selected targets. Note that at first we set up p0 as 0.47 and s as 0.995, because we intended to obtain 25% of the targets (~ 450) among the 1770 potential targets as the high-confidence targets. But to coincide with Tarbase interactions, we can use the above new thresholds to select targets. The ratio of the number of the selected targets to the number of the potential targets is 715/1770 ≈ 0.4 . Thus, if we relax the thresholds to include 40% of the potential targets selected, then the two relationship found in Tarbase can be selected.
In summary, from the above discussions, combining results from the confirmed targets and theoretical statistical inference to develop methods for exploring the relationship between miRNA and mRNA can be more useful.
Acknowledgements
We thank Han Liang, Tsunglin Liu and Henry Lu for valuable suggestions. This study was supported by Academia Sinica, Taiwan, and by NIH grants GM30998 and GM081724.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ambros V, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farh KKH, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 6.Grimson A, Farh KKH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA Targeting Specificity in Mammals: Determinants beyond Seed Pairing. Molecular Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 8.Huang JC, Morris QD, Frey BJ. Bayesian Inference of MicroRNA Targets from Sequence and Expression Data. Journal of Computational Biology. 2007;14:550–563. doi: 10.1089/cmb.2007.R002. [DOI] [PubMed] [Google Scholar]
- 9.Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci U S A. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 11.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 16.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 17.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Supplemental Data for Lewis et al. Cell. 120:15–20. http://web.wi.mit.edu/bartel/pub/Supplemental%20Material/Lewis%20et%20al%202005%20Supp/
- 20.Zhang W, et al. The functional landscape of mouse gene expression. J Biol. 2004;3:21–43. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]