Abstract
Pigment patterns in the integument have long-attracted attention from both scientists and non-scientists alike since their natural attractiveness combines with their excellence as models for the general problem of pattern formation. Pigment cells are formed from the neural crest and must migrate to reach their final locations. In this review, we focus on our current understanding of mechanisms underlying the control of pigment cell migration and patterning in diverse vertebrates. The model systems discussed here –chick, mouse, and zebrafish – each provide unique insights into the major morphogenetic events driving pigment pattern formation. In birds and mammals, melanoblasts must be specified before they can migrate on the dorsolateral pathway. Transmembrane receptors involved in guiding them onto this route include EphB2 and Ednrb2 in chick, and Kit in mouse. Terminal migration depends, in part, upon extracellular matrix reorganization by ADAMTS20. Invasion of the ectoderm, especially into the feather germ and hair follicles, requires specific signals that are beginning to be characterized. We summarize our current understanding of the mechanisms regulating melanoblast number and organization in the epidermis. We note the apparent differences in pigment pattern formation in poikilothermic vertebrates when compared with birds and mammals. With more pigment cell types, migration pathways are more complex and largely unexplored; nevertheless, a role for Kit signaling in melanophore migration is clear and indicates that at least some patterning mechanisms may be highly conserved. We summarize the multiple factors thought to contribute to zebrafish embryonic pigment pattern formation, highlighting a recent study identifying Sdf1a as one factor crucial for regulation of melanophore positioning. Finally, we discuss the mechanisms generating a second, metamorphic pigment pattern in adult fish, emphasizing recent studies strengthening the evidence that undifferentiated progenitor cells play a major role in generating adult pigment cells.
Keywords: melanocyte, melanophore, xanthophore, iridophore, patterning, morphogenesis, vertebrate embryos, neural crest
Introduction: What is a pigment pattern?
Many aspects of the natural world show an innate beauty, but few more so than the integumental colour patterns of vertebrates. These pigment patterns have drawn our attention for centuries, but in the last hundred years or so, pigment pattern formation –the mechanisms whereby these patterned distributions of pigments become established –has become a model system for the general problem of pattern formation. Several features make pigment patterns exciting as problems in pattern formation; these include their diversity, the relative ease of observation, the rapidity of their evolution (exemplified by the diverse patterns of closely-related groups of species), and the ecological importance of these patterns in mate choice, camouflage, communication and so on. In this review we focus on the regulation of pigment cell migration to the integument and the molecular mechanisms that guide these cells to the appropriate location. We consider both the relatively simple process in the chick and mouse embryo, as well as the more complicated fish pattern that requires the development of an early embryonic pattern and a subsequent adult, post-metamorphic pattern that is dependent on undifferentiated progenitor cells. We explore how studies of these models systems have depended on a combination of genetic screens to identify new signaling components and experimental perturbations to elucidate key molecular and cellular processes important in pigment pattern formation.
Pigment cell morphogenesis in three model systems
In all vertebrates, pigment cells, except for the pigmented retinal epithelium, arise from the neural crest. During embryogenesis, pigment cell precursors migrate away from the neuroepithelium in an extraordinary journey to contribute to one of the largest organ systems, the integument (for comprehensive review, see [1]). Variation in migration, population size, organization and differentiation of pigment cells within the integument generates the diversity of pigment patterns. Our conceptual understanding of pattern formation derives primarily from studies of three model systems: chick (and quail), mouse, and zebrafish. Although many aspects of the patterning process are shared among these organisms, there are huge differences in the timing; Table 1 compares the process of melanocyte morphogenesis in the three systems considered here.
Table 1.
Summary of melanoblast morphogenesis in the chick, mouse, and zebrafish trunk (at the axial level of the forelimb), the embryonic stages at which these morphogenetic events occur, and the known molecular determinants (or markers) involved in melanoblast migration and survival.
| Species Morphogenetic event* |
Chick | Mouse | Zebrafish*** | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Age | Determinants/Markers | References | Age | Determinants/Markers | References | Age | Determinants/Markers | References | |
| Melanoblast precursors begin emigrating from the dorsal neural tube | stage 16 | Mitf | Thomas and Erickson, unpublished | E8.5 | [7] | ||||
| Melanoblasts upregulate melanoblast specific markers | stage 18 | WRS, SLS, Mebl-1 | [60, 135–137] | E9.0 | Mitf, Kit | [42, 43] | 18-somite | mitfa, kita, dct | [48, 56, 134] |
| Melanoblasts localize to MSA | stage 19 | [27, 29] | E10.5 | Dct (Trp-2) | [45, 138, 139] | ||||
| Melanoblasts invade the dorsolateral pathway | stage 20 | EDNRB2, EphB2/ephrins | [39, 60] | Kit | [44, 45] | prim-5 stage | kita, dct | [134] | |
| Melanoblasts migrate along the dermamyotome | stage 21 | EDNRB2, EphB2/ephrins | [39, 60] | E11.5 | Ednrb | [140, 141] | |||
| Melanoblasts migrate through the lateral mesenchyme | E12.5 | Kit, AdamTS20 | [71] | ||||||
| Melanoblasts invade and disperse within the epidermis | stage 25 | Kit? | [59, 60] | E13 | Kit, E-cadherin | [75, 79, 80] | |||
| Melanoblasts invade integumental structures; in zebrafish, early larval pattern complete | E8-hatch | Kit? Fgf? | [4, 59, 87] | E15-birth | Kit | [9, 75, 142] | 5 dpf | melanin | [105] |
| Postnatal/larval pigment pattern organzation** | P4 | Kit, Foxn1, Fgf1 | [82, 84] | 14 dpf onwards | [16, 18, 23, 113] | ||||
Here we refer to all melanin producing precursor cells as melanoblasts and differentiated melanin producing cells as melanocytes. In zebrafish, however, melanin-producing pigment cells are referred to as melanophores.
In mouse this pertains to the loss of melanoblasts from the epidermis and localization to the hair follicle. In zebrafish this pertains to the change in chromatophore pattern during morphogenesis.
Due to the rostrocaudal gradient of development, we focus on the anterior trunk region here to allow clear comparison; note that melanoblast formation in the head precedes that in the trunk by c. 2 hpf.
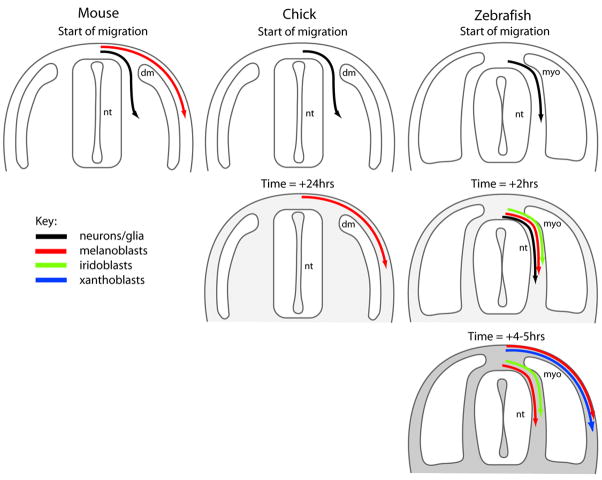
In mouse and chick, neural crest cells that migrate in the space between the somites and the neural tube (ventral pathway) give rise to neuronal and glial cells, whereas those that migrate between the somite and the non-neural ectoderm (dorsolateral pathway) differentiate into pigment cells, or melanocytes (Figure 1)[2–7]. Melanocyte precursors, or melanoblasts, initially just dorsal to the neural tube, migrate subectodermally to intermingle with the dermis, generated from the dermamyotome. Melanoblasts subsequently invade the ectoderm, and rapidly colonize the feather bud (chick) or hair follicle (mouse) where they differentiate to produce their characteristic pigment, melanin. In the adult bird, melanocytes persist primarily in the feather follicles and the epidermis [4]. In the mouse, at birth, melanocytes are observed in both the epidermis and hair follicles, but within a few days redistribute primarily to the hair follicle. As a consequence, the skin in the adult mouse is mostly unpigmented [8–10].
Figure 1. Schematic of neural crest cell migration in transverse sections.
Significant differences are observed in the timing and pathways of neural crest cell migration when comparing mouse, chick and zebrafish during early embryogenesis (see text for details).
Unlike in mouse and chick, where the pigment pattern is exclusively produced by melanocytes, the zebrafish pigment pattern is generated by a combination of different pigment cell types (or chromatophores), including melanophores, xanthophores and iridophores [11]. In zebrafish, the migratory pathways of the three cell types are differentially regulated and this contributes, in part, to their pigment pattern (Figure 1 and 2). Thus, iridoblasts utilize just the medial pathway, xanthoblasts migrate on and spread out within the lateral migration pathway [12–14], and melanoblasts migrate prominently on both ventral and lateral pathways [15]. Furthermore, many fish, including the danios (e.g. zebrafish) and medaka, develop a characteristic early larval pigment pattern before acquiring the definite adult pattern (Figure 3) [13, 16–23]. In zebrafish, this first pattern begins to be formed in the embryonic stages and consists of four melanophore stripes, three with associated iridophores, and a network of xanthophores occupying the intervening regions. During metamorphosis, a combination of embryonic and later metamorphic chromatophores contributes to the adult pattern (discussed in detail below). In further contrast to birds and mammals, fish melanophores reside in the uppermost layer of the dermis, and mostly do not invade the epidermis [24, 25].
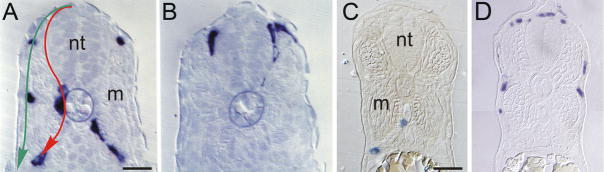
Figure 2. Chromatoblast migration in zebrafish.
Transverse sections of trunk of prim-5 (A,B) or prim-15 (C,D) stage wild-type embryos showing mRNA expression for melanoblast (dct, A, B), iridoblast (ltk, C) and xanthoblast (gch, D) markers. Note how iridoblast migration is restricted to the medial pathway, whereas xanthoblasts occupy the lateral pathway and melanoblasts migrate on both pathways (cf mouse and chick). m, muscle; nt, neural tube. Red arrow, medial pathway; green arrow, lateral pathway. Scale bar: 20 μm (A, B); 35 μm (C, D). A, B) reproduced with permission from [134].
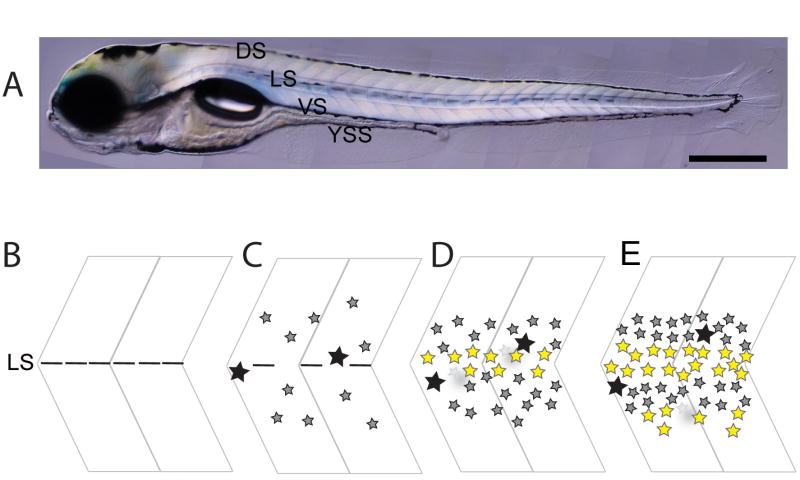
Figure 3. Wild-type zebrafish pigment patterns and pigment pattern morphogenesis.
Lateral views of wild-type early larval (A) pigment pattern. B–E) Pigment pattern metamorphosis. B) The larval pattern consists of 4 melanophore stripes, but only one, the lateral stripe (LS) is shown in this close-up of the lateral flank (horizontal black bars). C) From 14 dpf, early metamorphic melanophores (small, grey stars) differentiate in the vicinity of the horizontal myoseptum over a period of around 7 days; some larval melanophores (large black stars) emerge onto the flank from the early larval stripes (e.g. LS) and may persist. D) Melanophores aggregate dorsal and ventral to the myoseptum to form the first two stripes, while xanthophores (yellow stars) form adjacent to the horizontal myoseptum. Melanophores in the interstripe region, including those from the larval pattern’s lateral stripe in the myoseptum itself, either migrate into the nascent stripes, or die in situ (faint grey). E) Late metamorphic phase of melanophore and xanthophore production increases the pigment cell density within the stripes and interstripes; a second interstripe of xanthophores begins to form ventral to the primary melanophore stripes. Anterior to the left, dorsal up. Stages not drawn to scale. Scale bar in A): 0.5 mm. C–F) adapted from [21, 114].
How is the development of pigment cells controlled to generate the highly reproducible patterns observed in nature, and to what extent are the mechanisms shared between poikilothermic (cold-blooded) and homeothermic (warm-blooded) vertebrates? At one level, pigment pattern formation in poikilotherms must be distinctive from homeotherms, since colour differences in fish result primarily from the arrangement of the different combinations of pigment cell types in particular locations (morphological patterning), rather than from the alteration of the pigment structure and/or abundance (and thus colour) within the melanocyte (physiological patterning)(for detailed review, see [17]). However, the initial dispersal of pigment cell precursors away from the neural tube is conserved between all vertebrates, and in this process we may find some similarities.
Melanoblast specification in early pathfinding
In the chick embryo, fate specification as a melanoblast is necessary for neural crest migration on the dorsolateral pathway. This specification results in molecular changes that alter the cells’ response to signals on the dorsolateral pathway that are otherwise inhibitory to neural cell migration, but which are attractive to specified melanoblasts [26–28]. There is substantial evidence that melanocyte specification begins very early in many species, perhaps even prior to delamination from the neural tube. For example in the chick, neural tube cultures produce early migrating neural crest cells that give rise to neuronal fates, whereas late migrating cells differentiate into melanocytes [29]. Furthermore, iontophoretic labeling of these emigrating cells reveals that the early and late-migrating cells are lineage-restricted, and express distinct cell-surface markers [30, 31]. Consistent with this, almost all neural crest cells on the dorsolateral pathway express melanoblast markers [32]. Those that do not express these markers rapidly undergo apoptosis [33]. Thus, it appears that specification as a melanoblast is required to migrate along the dorsolateral pathway. Transplantation experiments in which various subpopulations of neural crest cells were isolated and then grafted into the space between the neural tube and somite support this hypothesis; only melanoblasts are able to take the dorsolateral pathway [34].
What is the molecular basis for appropriate pathway choice by these distinct populations of neural crest cells? Specifically, what prevents neural and glial precursors from migrating laterally, but permits melanoblasts to invade this space? In the chick, numerous extracellular matrix molecules and cell surface-associated signaling molecules that typically inhibit neural crest migration are found in the dorsolateral space, including chondroitin sulfate proteoglycans and PNA-binding molecules [35, 36], slits [37], semaphorins, spondins [38] and ephrins [39]. Experimentally, slits, F-spondin and ephrins have all been demonstrated to prevent early migrating crest from invading the dorsolateral pathway [37–39]. Perplexingly, there is no change in the distribution of these inhibitory molecules at the time melanoblasts enter the lateral path, with the exception of PNA-binding molecules that appear to retreat as the melanoblasts invade [36]. Given that melanoblasts can migrate in the dorsolateral space precociously when they are grafted into an early stage-12 embryo [34], the evidence suggests that autonomous molecular changes occur during melanoblast specification that permit dorsolateral migration.
What specific changes in the melanoblast allow access to the lateral pathway? Genetic studies in the mouse have made a considerable contribution to our understanding of pigment cell development, identifying over 127 genes essential to normal pigmentation [40]. Of the loci that have been cloned, over one third comprise genes that regulate melanocyte development (with the remainder involved in melanosome biogenesis and the production and deposition of melanin pigment). Mutations in these genes often manifest themselves as ‘white-spotting’ phenotypes, as a consequence of defects in melanocyte precursor proliferation, survival, migration and/or tissue invasion [41]. From these studies, several molecules involved in melanoblast pathfinding have been identified, including both receptor tyrosine kinases (e.g. Kit) and G protein-coupled receptors (e.g. Ednrb) and their ligands (KitL and Endothelin3, respectively).
Unlike in birds where neural crest subpopulations arise in temporally distinct waves [29], melanoblast precursors in the mouse appear and migrate concurrently with other non-melanogenic neural crest cells [7]. Like in birds, however, it appears that melanogenic fate is specified in mouse neural crest cells prior to migration. In the mouse, the melanoblast marker, Kit, can be identified in premigratory neural crest cells restricted to the dorsal midline of the neural tube [42]. Soon after, both Kit and the melanoblast-specific transcription factor, Mitf, colocalize in cells just dorsal to the neural tube [43]. In the mouse, it is unclear whether prior specification dictates pathway choice, however it is apparent that melanoblast survival and their subsequent dispersal into and along the dorsolateral path are dependent on the activation of Kit by KitL (also known as Steel factor) [44–47]. Melanoblast dispersal into and within the dorsolateral pathway is preceded by transient expression of Kitl by the dermatome. Kitl exists in two forms, soluble and membrane-bound, with the former implicated in melanoblast migration and the latter in survival [45]. In Nf1−/KitSl− mouse mutants, where melanoblasts survive in the absence of Kitl, melanoblasts remain sequestered in the migration staging area (the extracellular space between the dorsal neural tube and somite). This observation suggests that Kitl provides a necessary guidance cue to direct Kit+ melanoblasts laterally.
There is increasing evidence that, in fish too, pigment cell migration on both pathways may require prior specification. Early markers of melanoblast, xanthoblast and iridoblast specification have been noted to be expressed in premigratory (but not necessarily non-delaminated) neural crest in zebrafish [14, 48–50]. Indeed, it is conceivable that most pigment cells in the fish are specified prior to migration, since xanthoblasts and iridoblasts show strict localization to the lateral and medial pathways, respectively [14, 49, 50], and since most premigratory neural crest cells appear to be lineage-restricted [51, 52]. Furthermore, in zebrafish colourless/sox10 mutants, where pigment cell specification fails, there is an almost total absence of migrating neural crest cells on the lateral pathway [53–55]. Genetic studies in zebrafish also show that, as in mouse, melanophore survival and migration are also both Kit-dependent and that these two functions are genetically separable [56, 57]. Two kit orthologues, sparse/kita and kitb, as well as two kit ligands (kitla, kitlb), have been identified in zebrafish, but only Kita/Kitla have roles in melanophore development [56, 58]. kita is expressed in the melanoblasts and melanophores, whereas kitla is expressed transiently in a subset of muscle cells, some very dorsal, and others adjacent to the notochord and thus next to cells migrating in the medial pathway [58]. Using temperature-sensitive conditional mutations, Rawls and Johnson have shown that Kita signaling is needed only transiently, between 1 and 2 dpf, for melanophore migration [57]. This correlates well with the time when pigment cell precursors invade and migrate along the migration pathways and suggests that Kita signals may drive pathway invasion, but equally could have roles in continuing migration, at least on the medial migration pathway.
Interestingly, the expression patterns of Kit/Kitl do not support a homologous role for this particular receptor in early melanoblast pathfinding in the chick; Kit is detected in melanoblasts after they have already invaded the dorsolateral pathway and Kitl is upregulated in the epidermis at a similar time period [59]. Instead, recent studies have identified two other signaling systems that are required for lateral melanoblast migration in the chick. Santiago and Erickson showed that although the ventrally migrating crest cells are inhibited by ephrins in the dorsolateral pathway, melanoblasts are stimulated to migrate on ephrins [39]. The receptor mediating this migration response is EphB2; when EphB2 expression is knocked-down by short hairpin RNA, melanoblasts fail to migrate in the dorsolateral pathway [60]. These studies also provide an explanation for why melanoblasts migrate inappropriately along the ventral pathway to invade the ventromedial tissues of Silkie fowl embryos [61]. In this chicken strain ventral migration of melanoblasts correlates with the absence of the usual barrier molecules [61, 62], and also with a substantial increase in ephrins on the ventral pathway (Faraco, Harris and Erickson, unpublished data).
Endothelin signaling via the Endothelin receptor B2 (EDNRB2)/Endothelin-3 (ET-3) receptor/ligand pair is a second pathway that positively regulates melanoblast migration. Endothelins are 21-amino-acid peptides that bind to the endothelin receptors and mediate a number of physiological and cellular responses, including the development of neural crest cells (Baynash et al., 1994; Hosoda et al.,1994; Hall et al., 1999; Gether, 2000). In the chick, early migrating neural crest cells express EDNRB, but early melanoblasts upregulate a second, melanoblast-specific receptor, EDNRB2 [63]. The ligand ET-3 is expressed in the ectoderm and dermamyotome, beginning at stage 12 [64]. When EDNRB2 is knocked-down, there is a significant reduction in melanoblasts along the lateral pathway. Conversely, when EDNRB2 is misexpressed in neural and glial precursors, they migrate ectopically on the lateral pathway.
The EphB2 and EDNRB2 signaling systems appear to act additively to overcome negative cues [60]. For example, if EphB2 is knocked-down in melanoblasts at the same time that EDNRB2 is overexpressed, EDNRB2 can rescue the loss of EphB2. Thus, one model of melanoblast pathfinding is that the ability to migrate in the lateral pathway depends on the balance of stimulatory and inhibitory migratory cues. Neural crest cells and neural progenitors are sensitive to negative cues (e.g., PNA-binding molecules, chondroitin sulfate proteoglycans, ephrins, Slit) and do not bear the receptors that will positively mediate their migration (e.g., EDNRB2, EphB2). Melanoblasts, however, upregulate EDNRB2 and EphB2 just prior to their invasion, and thus become able to exploit the lateral pathway even though inhibitory molecules still remain. Another observation that supports this general model of migration being determined by the balance between stimulatory and inhibitory influences comes from overexpressing either alpha4 or alpha5 integrin (components of fibronectin-binding integrins) in sarcoma-180 cells grafted into the migration staging area of the early neural crest migratory pathways. Their ability to adhere more avidly to fibronectin allows the transfected cells to migrate in the lateral pathway [65], whereas untransfected control cells only migrate ventrally [66].
Whether this model explains early pathfinding of pigment cells in mouse and fish has not been examined directly. Where analysed, investigations of the factors mentioned above in these species have identified roles for these proteins in other aspects of pigment pattern formation. For instance, neither mouse nor zebrafish have a second endothelin receptor directly orthologous to EDNRB2 although they do express one endothelin receptor subtype, Ednrb [22, 67]. However, although Ednrb is expressed by mouse melanoblasts during their early dissemination, they are only dependent on this receptor to mediate their continued migration subectodermally once already in the lateral pathway; in Ednrb mutant embryos, melanoblasts stall at the dorsal-medial end of the dermatome (Lee et al. 2003, Shin et al. 1999). In zebrafish, rose/ednrb1 mutants have no embryonic pigment pattern defect but in the adult exhibit disrupted stripes in ventral regions due to a deficit in stripe melanocytes and interstripe iridophores during metamorphosis (see Figure 5B; [22]; E. Greenhill and RNK, unpub. data). Similarly, to our knowledge, no published studies have reported roles for ephrin signaling in pigment cell development in mouse or fish, and we postulate that this is due to redundancy in this receptor family. However, the study of Eph/ephrin signaling in these species might pay dividends, and it would be especially interesting to see whether, in zebrafish, changes in ephrin expression or Eph receptor signaling correlates with the dual pathways of migration taken by melanoblasts. Another aspect worthy of further study is how the migration pathways become shut-down. Thus, time-lapse analysis of lateral pathway migration in zebrafish indicated that invasion of the lateral pathway by melanocytes can only occur during a short time-window and then normally becomes inhibited [68], yet we have no idea of the molecular basis for this change in cell behaviour.
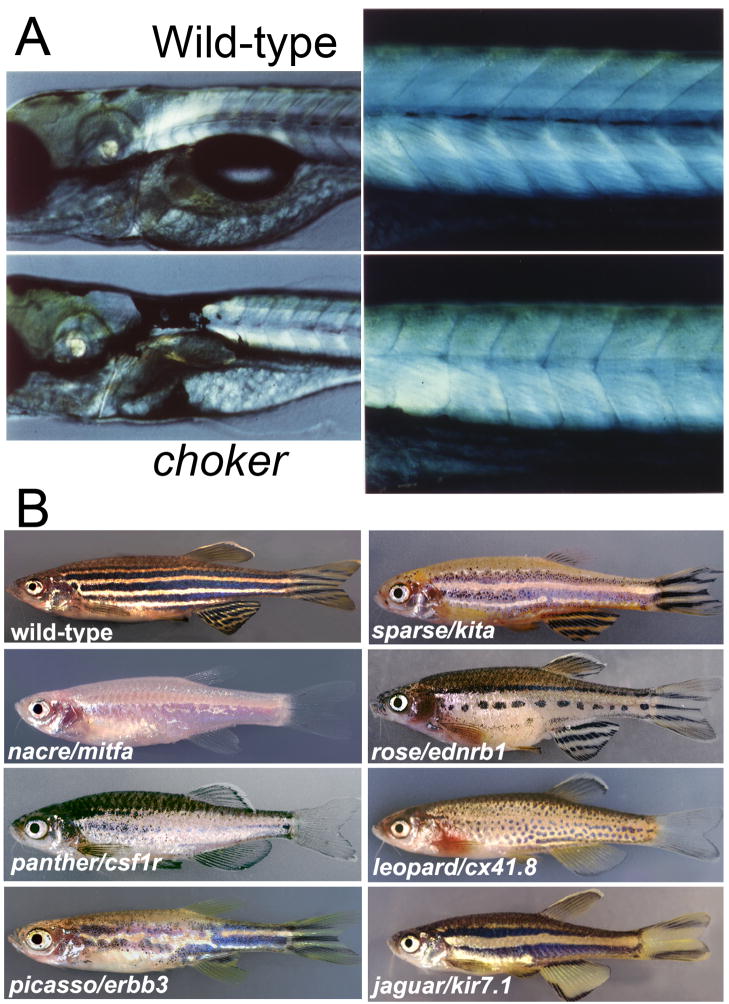
Figure 5. Pigment pattern mutants in zebrafish.
A) choker mutants at 5 dpf show ectopic melanophores in the anterior trunk region (left panels), and lack the lateral stripe melanophores of the body (right).
B) Adult pigment pattern phenotypes of wild-type and various homozygous mutant strains. See text for further details. (Adapted, with permission, from Kelsh and Parichy 2007).
Terminal migration, invasion of the epidermis and follicle, and spatial organization
Melanoblast morphogenesis after invasion of the lateral pathway is best characterized in the mouse embryo, where studies reveal how active melanoblast migration is maintained and how the invasion of the epidermis and organization of melanocytes in the integument is controlled (summarized in Figure 4).
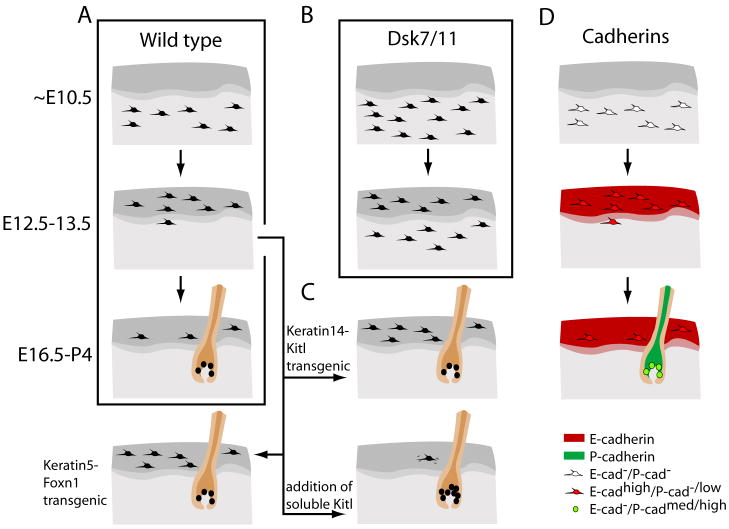
Figure 4. Regulation of melanoblast patterning in mouse skin.
Schematic of embryonic skin in wild type (A), transgenic or mutant (B, C), and cadherin-immunolabeled (C) embryos. A) Melanoblasts from the dermis (light gray) sequentially invade the epidermis (dark gray) and then the hair follicle (brown). B) In Dsk or Gnaq/11 mutants melanoblasts are produced in elevated numbers during early melanogenesis which results in ectopic pigmentation of the dermis. These results provide a basis for the regulation of cell number in maintaining normal patterning. C) Transgenic mice expressing Kitl or Foxn1 under keratin promotors results in the retention of melanocytes in the epidermis. This suggests that both play a role in the maintenance or localization of melanocytes to this tissue. Exogenous Kitl applied to skin explants causes a chemokinetic increase of melanocytes to the hair follicle. D) Cadherin expression by melanoblasts correlates with cadherin expression patterns seen in the skin and suggests an extrinsic regulation of melanoblast localization that is coincident with the development of the skin.
After melanoblasts enter the lateral pathway they migrate subectodermally at the same time as the dermatome undergoes an epithelial-to-mesenchymal transition into dermis. The recent characterization of the belted (bt) mouse mutant suggests that Kit activation is again required at this time to maintain melanoblast survival. Bt mice are identified by a white lumbar band, reminiscent of a belt, which is the consequence of nonsense or missense mutations in ADAMTS20. ADAMTS20 (a disintegrin-like and metalloproteinase domain with thrombospondin type-1 motif) is a secreted protease in the ADAM family of metalloproteinase and is expressed in the dermal mesenchyme preceding melanoblast migration [69]. Expression of ADAMTS20 is also coincident with the downregulation of Kitl mRNA, leaving only Kitl protein associated with the dermal mesenchyme [45, 70]. Melanoblast distribution is normal up to E12.5 in bt embryos, however, between E13.5–14.5 the number of melanoblasts decreases in the presumptive belt region as a result of apoptosis. ADAMTS20 is thought to participate in melanoblast survival by modulating Kit signaling. Accordingly, mutations in both bt and Kit or Kitl results in synergistic white spotting. Furthermore, in bt/bt skin explants, melanoblasts are unable to respond to soluble Kitl [71]. However, it is unknown whether ADAMTS20 acts directly on Kitl or by regulating the extracellular matrix.
Melanoblast invasion of the epidermis is coincident with its stratification and it is likely that events inherent to this process tightly regulate its colonization by melanoblasts[72]. For example, the cell adhesion molecules, E- and P-cadherin (E- and P-cad), are expressed in complementary regions of the developing skin and are required to maintain the correct organization of the epidermis, dermis and hair follicle (Figure 4). In general, E-cad is expressed by keratinocytes of the epidermis, P-cad by keratinocytes of the hair matrix, and neither in the dermis [73]. Similarly, between E12–16 when melanoblasts are invading the epidermis and then hair follicles [9, 74], they diversify into three cadherin-expressing subpopulations each of which correlates with the tissue that they colonize: dermal melanoblasts are E-cad−/P-cad−, epidermal melanoblasts are E-cadhigh/P-cad−/low and melanoblasts in the hair matrix are E-cad−/P-cadmed/high [75].
Kit signaling also appears to be important for getting melanoblasts to their appropriate position in the skin (Figure 4). Kitl has been detected in the dermis [76, 77], the epidermal basal layer and hair follicles at times that correspond with the invasion of melanoblasts into these tissues [78]. Application of Kit function-blocking antibodies (ACK2) to embryos in utero shows that Kit is required at the specific period when melanoblasts are invading the epidermis and colonizing the hair follicle [79]. Further investigations demonstrate that in hk14-SLF transgenic embryos, where ectopic expression of Kitl in the basal layer of the skin is achieved by driving Kitl under the control of human cytokeratin 14, melanoblasts are observed in regions (like the oral ectoderm) where they do not normally migrate [80]. When Jordan and Jackson [81] implanted Kitl-soaked beads into embryonic skin explants they found that the presence of Kitl did not direct melanoblasts toward the bead, but rather enhanced their migration into the hair follicles. This suggests that Kitl acts as a chemokinetic cue rather than a chemoattractant and implies that other mechanisms may be involved in directly guiding or maintaining melanoblasts in the correct location.
Until recently it was unclear why the epithelial cells of the hair matrix are the only targets for melanin deposition by melanocytes. One pigmentary unit in essence requires two types of cells—the pigment cell donor (the melanocyte) and the pigment cell receiver (the epithelial cell). A study of the Foxn1 mouse mutant may hold the key to our understanding of how these two cell types communicate [82]. Foxn1 codes for a transcription factor that is essential for the normal development of several epithelial tissues, and mutations in this gene in rodents results in the nude phenotype [83]. In the epidermis, Foxn1 expression is localized to the epithelial cells of the hair cortex, and is also required for their normal pigmentation. If Foxn1 is placed under the control of the keratin 5 promotor, targeting Foxn1 to the epidermis, melanocytes are ectopically recruited to the skin. As these transgenic mice develop, and Foxn1 distribution changes, so does the distribution of the melanocytes [82]. Interestingly, Foxn1 expression is similar to that of Kitl during development [84, 85]. Foxn1 is thought to mediate the ‘pigment cell recipient’ phenotype through the upregulation of Fgf2, so perhaps Fgf2 and Kitl together coordinate the organization of the melanocytes that make up one half of the pigmentary unit.
Significantly less is understood about how the terminal migration and organization of melanoblasts into the feather follicle is regulated in the chick. Preliminary evidence suggests that, as in mouse, migration through the ectoderm may be regulated by KIT signaling (MLH, CAE). Several studies have shown that KIT is upregulated by melanoblasts until after they invade the dorsolateral pathway. Similarly, KIT ligand is not expressed in the ectoderm until a similar stage [59, 86]. These in situ hybridization studies suggest that KIT ligand may regulate invasion into the ectoderm rather than the earlier event of migration into the dorsolateral path. In support of this notion, melanoblasts enter the dorsolateral pathway on schedule when KIT is knocked down, but are reduced in their ability to enter the ectoderm and instead accumulate in the dermis [60]. Additionally, KIT ligand may regulate the accumulation of melanoblasts in the feather germ, since at the time when melanoblasts are concentrated in the tips of the feather germ and have begun to differentiate [87], the expression of KIT ligand is intensified in the same locations [59, 87]. An alternative possibility is that KIT controls differentiation and survival at these later stages. There is also evidence from the investigation of melanoblast migration in the limb bud that FGF-2 and IGF-1 are responsible for their directed migration toward the AER [88].
The cell autonomous processes by which melanoblasts invade the ectoderm in the chick are unknown but it seems likely that proteolysis of the basal lamina is important since the ectoderm basal lamina is degraded in those regions where melanoblasts closely associate with the ectoderm [27]. However, although a number of proteases are expressed by early migrating neural crest cells, including plasminogen activator, MMP-2 and ADAM10 [89], none of these has been demonstrated to facilitate melanoblast invasion of the basal lamina.
Because the majority of pigment cells in zebrafish do not cross the epidermal basal lamina and do not invade a follicular structure that would define their final position with the embryo, the question remains in this species how the spacing of chromatophores is controlled. Contact inhibition may be sufficient to generate the relatively even spacing of xanthophores, but this remains to be tested. In contrast, other pigment-cell types, at least in the zebrafish and medaka, show more localized patterns into striped arrangements, and aspects of the embryonic/larval pattern shown by these fish can be traced to other fish and even to amphibians. As we have argued before [17], the limited evidence that we have at present indicates that both cell-cell interactions and localized positional cues determine the localized (patterned) distribution of chromatophores in zebrafish and medaka. Such mechanisms have also been deduced from studies of amphibian embryos [90–95], so these basic mechanisms are evolutionarily conserved. Recently, study of the zebrafish embryonic pigment pattern mutant choker identified a key role for localized Sdf1a, a molecule well-known for its germ cell and axonal guidance properties [96–104], in determining one part of the melanophore pattern in zebrafish [68, 105]. Three pigment pattern defects characterize choker mutants: loss of melanophores in the lateral stripe (along the horizontal myoseptum), gain of melanophores in an ectopic patch in the anteriormost trunk, and loss of xanthophores in this same area (Figure 5A; [68, 105]. Both melanocyte defects in choker mutants appear to depend, at least in part, on sdf1a; loss of expression from its normal location in the horizontal myoseptum results in loss of the embryonic/larval melanophore stripe that forms in that region, and ectopic expression of Sdf1a in the anterior somite region results in ectopic melanophores [17]. These latter cells invade the lateral pathway after it has normally been shut-down and hence Sdf1 signaling allows them to overcome the inhibitory signals that predominate in the wild-type. Interestingly, the usual Sdf1a receptor, Cxcr4b, was not detectably expressed in the melanophore lineage, so the molecular mechanism for pigment pattern formation here remains incomplete, although low-level transcriptional expression of a cxcr4 gene remains a possible explanation of the phenotype. The loss of xanthophores from the collar region was shown to be independent of melanophore invasion, but the link to Sdf1a remains to be explored. Likewise, the other three melanophore stripes must be regulated by different cues, and their elucidation will be a major goal for the future.
Pigment cell number modulates pattern
Pigment patterns are also regulated by the total number of melanoblasts, which can result in varying levels of population pressure and subsequently cell dispersion. A commonality in many white-spotting mouse mutants is the loss of pigmentation at the ventral-most portion of the trunk (belly-spot) or along a band extending dorsally from this region (belt). These characteristic pigment patterns are likely to result from a melanoblast progenitor pool that is distributed disproportionately. Specifically, regions that normally produce more melanoblast progenitors (cranial and sacral axial levels) are coincident with those areas that remain pigmented in the adult mutants, and regions with fewer melanoblast progenitors (thoracic level) are coincident with white spotting [41, 106]. Consequently, the lower number of melanoblast progenitors in the thoracic level of the trunk makes this area more sensitive to changes in cell survival or proliferation. So, how is the number of melanoblasts in the embryo determined?
Dark-skinned mouse mutants from a large-scale chemical mutagenesis screen may provide part of the answer (Figure 4; [107]. The dominant dark skin (Dsk) mutations, Dsk1, Dsk7 and Dsk10, have been identified as gain-of-function alterations in two G-protein α-subunits of the Gαq class, Gnaq (Dsk1 and Dsk10) and Gna11(Dsk7). Mice carrying these mutations are characterized by the presence of excess dermal melanin in the adult [108]. Embryos carrying both the Dsk mutation and a Dct-lacZ transgene (produces lacZ under the promoter of the melanoblast-specific enzyme, dopachrome tautomerase) demonstrate that the dark skin phenotype is not the consequence of increased pigment production or the failure of melanoblasts to invade the ectoderm, but rather results from the early expansion of melanoblast precursors. Crosses into Gnaq or Gna11-null backgrounds demonstrate that these mutations are hypermorphic, and furthermore, multiple copies of any or a combination of Dsk alleles leads to a stepwise increase in dermal pigmentation. Thus, the number of melanoblasts may be controlled directly by Gαq activity.
Gα-subunits like Gnaq and Gna11 directly mediate the transduction of signals from G protein-coupled receptors (GPCRs). One GPCR we have already discussed for its role in melanogenesis is Ednrb. Ednrb likely interacts with Dsk gene products because it is known to couple to both the Gnaq and Gna11 subunits [109]. Ednrbs-l/s-l/GnaqDsk1/+ (or Gna11Dsk7/+) double mutants are indistinguishable from Ednrbs-l/s-l single mutants, which demonstrates that the mechanism by which Dsk1 or Dsk7 affects melanogenesis depends on functional Ednrb [108]. Thus Ednrb is likely the main mediator of Gnaq and Gna11 signaling in melanoblasts. Variable amplification of the endothelin signal can therefore explain how this particular receptor might influence the number of initial melanoblast precursors, and subsequently, the final pigment pattern within the embryo. Further assessment of the mechanisms involved in defining initial melanoblast number and later cell survival (e.g. AdamTS20) will increase our understanding of how cell number affects final pigment pattern in the mouse.
The control of pigment cell number in zebrafish has been barely addressed, although a conserved role for Kit signaling in melanoblast survival is clear. In an interesting parallel to the mouse, both zebrafish and medaka mutants that result in reduced numbers of embryonic chromatophores show defects that are concentrated ventrally, reminiscent of the white belly-spot seen in mouse mutants (see [17]. Amongst these mutants are those in sparse/kita, an orthologue of the mouse mouse white-spotting gene, Kit. Kita function in melanoblast survival becomes important only from around 2 dpf [57]; upregulation of kitla transcripts in the skin at 4 dpf presumably contributes to this later survival phase [58]. At some level, Kita/Kitla signaling clearly limits melanophore number in zebrafish, since overexpression of Kitla results in a Kita-dependent increase in melanophore number, although interestingly the melanophore pattern is largely unaffected [58]. Further study of three medaka mutants with decreased melanophore numbers will be of interest in this regard [12], but an exhaustive screen for mutants affecting cell number for each of the chromatophore types is conceivable in the zebrafish and medaka systems [110, 111] and would be highly informative.
Fish have not just one, but two pigment patterns
In contrast to the highly conserved early larval pigment pattern in fish and even amphibians, the adult phenotypes are highly variable and usually very different from the larval phenotype. This suggests that the formation of these two patterns is not only evolutionarily, but also developmentally, uncoupled [21]. Since the zebrafish remains the primary fish species in which anatomical, genetic and modeling studies have been combined, we focus here on the adult pigment pattern in zebrafish.
To the eye, the striking aspect of the adult zebrafish pigment pattern is a series of blue-black stripes running along the length of the body and in most of the fins, separated by silver interstripes (see Figure 5B). Electron microscopy studies have been crucial in identifying the locations of each pigment cell-type in the adult skin and reveal a consistent pattern in the striped region of the body [24, 25]. Pigment cells contribute to the hypodermis. Xanthophores consistently occupy the most superficial position and are underlain by iridophores with relatively small reflecting platelets (type S iridophores). In the stripe regions the type S iridophores are underlain by melanophores and then iridophores with larger reflecting platelets (type L iridophores). This layer order of xanthophores/iridophores/melanophores is also seen in the dermal chromatophores unit of frogs and implies a conserved mechanism of chromatophore patterning [11, 24]. Interestingly, these cells are organized in close contact with each other, suggesting that stripe maintenance may involve cell-cell adhesion. Direct experimental support for this role is provided by studies of fms mutants, showing that genetic ablation of xanthophores results in disintegration of the stripes [112]. As we shall see, such interactions are commonly proposed to explain pattern formation too, but a major gap in our knowledge remains the physical relationships of these and other cells at stages when pigment pattern formation is occurring.
Formation of the adult stripe pattern has been described at the level of light microscopy and has been summarized before (Figure 3B)[16, 18, 23, 113]. The larval pattern remains stable from around 5 dpf until 14 dpf, but then early metamorphic melanophores differentiate in a widely scattered distribution over the dorsal and ventral myotomes for a period of around 7 days. These, plus some persisting larval melanophores, aggregate dorsal and ventral to the myoseptum to form the first two adult stripes; the appearance of xanthophores adjacent to the horizontal myoseptum marks the position of the interstripe and correlates with formation of the melanophore stripes. Melanophores in the interstripe region, including those from the larval pattern’s lateral stripe in the myoseptum itself, either migrate into the nascent stripes, or die in situ. Subsequently, between 21 and 28 dpf, a late metamorphic phase of melanophore production forms further melanophores within the stripes, contributing to melanophore density within them. As the body grows further, stripes and interstripes are added dorsal and ventral to the primary stripes. Careful study of the timing and position of appearance of xanthophores reveals differentiated xanthophores to be prominent only in the interstripe regions during these metamorphic phases [23, 113, 114]; what becomes of the early larval xanthophores that are widely distributed throughout the lateral pathway at 5 dpf has not been described. Furthermore, the localized nature of these early metamorphic xanthophores contrasts with the ubiquitous layer of xanthophores seen in striped regions of the adult skin [24, 25] and emphasizes the importance of electron microscopic study of the metamorphic fish to confirm and extend the light microscopic studies. In particular, data on the precise locations in the flank where xanthophores form, their numbers and their proximity to melanophores and iridophores is required to evaluate the interactions postulated from genetic studies (see below). Little attention has been paid to iridophores, but since homozygous viable mutant alleles of both rose/ednrb1 and shady/ltk result in adult pigment pattern defects, including severe effects on both iridophores and on melanophore stripes, and since both genes are prominently expressed in at least embryonic iridophores [14, 22], a role for iridophores in adult patterning deserves active study.
Genetic studies of adult pigment pattern formation in zebrafish
Zebrafish adult pigment pattern mutants and the natural ‘mutants’ fixed during Danio speciation provide important resources for studying Danio pigment pattern formation. These studies have been recently reviewed [17, 113] so we will just highlight specific biological characteristics of the pigment patterning process that have been revealed by these studies (Table 2). One key finding is that new chromatophores are generated from unpigmented precursors, and interestingly a reduction in the extent to which metamorphic melanophores contribute to the adult pattern seems to contribute to the distinctive pattern of D. nigrofasciatus in which secondary stripes are reduced [113, 115]. Another prominent finding is that interactions between different types of chromatophores are important to the patterning process. To date, tests have been restricted to the xanthophore and melanophore lineages[16, 18, 19, 112, 114, 116], but as noted above, consideration of iridophores as a further component is needed. Finally, even in the absence of one cell-type, hints of stripes are seen, suggesting that cues exist in the skin that contribute to the formation of stripes [114]. All these mechanisms (latent progenitors, xanthophore-melanophore interactions, environmental cues) have been postulated to be important in formation of the early larval pattern in ectothermic vertebrates, including fish and salamanders, and hence are likely to be widespread phylogenetically [90, 93, 94]
Table 2.
Key characteristics of Danio pigment patterning mechanisms
| Characteristic | Key mutants | Consequence of mutation | References |
|---|---|---|---|
| Many adult chromatophores are generated de novo from unpigmented progenitors | sparse/kit (EM) | Delayed formation of normal stripes | [16, 56, 57, 143, 144] |
| rose/ednrb1 (LM) | Fewer, broken stripes | [22] | |
| puma (EM and LM) | Reduced chromatophores, delayed formation of stripes | [23, 131] | |
| D. nigrofasciatus (EM and LM) | Fewer stripes, secondary stripes broken | [113, 115, 129] | |
| picasso/erbb3 (EM and LM) | Fewer, broken stripes | [129] | |
|
| |||
| Early and Late metamorphic melanophores are genetically distinct | sparse/kit (EM) | Delayed formation of normal stripes | [16, 56, 57, 143, 144] |
| rose/ednrb1 (LM) | Fewer, broken stripes | [22] | |
|
| |||
| Melanophore migration/death in interstripes | sparse/kit, Danio albolineatus | Melanophores remain dispersed; stripes obscured | [144] |
|
| |||
| Xanthophores provide trophic support for melanophores | panther/salz/pfeffer/fms/c sf1r | LM melanophores mostly die, so melanophore density lower | [112, 114] |
|
| |||
| Xanthophores pattern melanophores | panther/salz/pfeffer/fms/c sf1r | Melanophores form scattered small clusters, stripes obscured | [112, 114] |
| leopard/’D. frankei’ | Melanophores form scattered small clusters, stripes obscured | [16, 18, 19, 114, 116] | |
|
| |||
| Melanophores pattern xanthophores | nacre/mitfa | Xanthophores cluster | [114] |
|
| |||
| Melanophore aggregation (adhesion?) | obelix/jaguar | Melanophore stripes broadened, and xanthophores intermingled | [114, 118] |
|
| |||
| Prepattern directs xanthophore distribution | nacre/mitfa | Even in absence of melanophores, xanthophores concentrated near horizontal myoseptum | [48, 114] |
In recent years, adult pigment pattern genes have begun to be identified molecularly and this provides an exciting opportunity to understand the molecular basis of fish pigment patterns. Mutations in the gap junction protein Connexin41.8 underlie the leopard phenotype, leading to the suggestion that pigment pattern formation depends upon gap junctions likely consisting of an unidentified connexin in addition to Connexin41.8 [117]. Interestingly and importantly the authors show that connexin41.8 is expressed in melanophores and probably xanthophores in striped regions, fully consistent with the demonstration that leopard acts cell-autonomously in pigment cells [114]. Clearly, gap junctions provide a mechanism for (short-range) cell-cell communication and thus the molecular identification of leopard is fully consistent with the deduction from cell-transplantation experiments that short-range, xanthophore–melanophore interactions drive local patterning [114].
Mutations in the inwardly rectifying potassium channel 7.1 (Kir7.1) gene underlie the jaguar/obelix phenotype [118]. The broader, more diffuse stripes of jaguar/obelix/Kir7.1 mutants appear to result from changed melanophore adhesion since in mosaic fish, wild-type xanthophores intermingle with jaguar/obelix/Kir7.1 melanophores, but wild-type melanophores always form good stripes [114]. The authors suggest that reduced melanophore-melanophore adhesion results in a failure to separate fully melanophores and xanthophores and results in broader stripes. Does the identification of the jaguar/obelix gene as Kir7.1 illuminate the adult pigment pattern mechanism? Kir7.1 is expressed in melanocytes consistent with the interpretation of transplantation experiments, although intriguingly it is also expressed in xanthophores, despite no role for jaguar/obelix being identified in this cell-type [114]. When misexpressed in a cell-line, Kir7.1 proteins localize to the plasma membrane, but only wild-type protein results in detectable inwardly rectifying potassium currents [118]. These proteins affect the transport of K+ ions across the membrane, allowing depolarization of the membrane [119]. Thus, in the absence of Kir7.1 function, it might be expected that membrane depolarization is inhibited; this in turn might disrupt receptor-mediated signaling cascades. The authors provide a test of this latter idea but do not identify which signaling system underlying pigment pattern formation is affected [118]. Identification of these signaling processes will be crucial in illuminating the mechanisms of pigment cell pattern formation, and this is clearly a priority for future research.
Adult pigment pattern formation and stem cells
There is currently enormous interest in the developmental and regenerative roles and therapeutic potential of stem cells, i.e. in undifferentiated cells that can, under appropriate circumstances, both extensively self-renew and generate differentiated progeny. Neural crest-derived stem cells capable of producing melanocytes have now been described by multiple groups looking at mammalian skin [120–124], recently reviewed in [125, 126] and see also review by Fisher in this volume. To date, definitive evidence for neural crest stem cells in cold-blooded vertebrates remains to be demonstrated, but studies of pigment pattern formation and regeneration in zebrafish do reveal the production of melanophores de novo from quiescent unpigmented precursors, and a persistent population of adult melanophore stem cells is an attractive hypothesis [127, 128].
A new and exciting opportunity to identify chromatophore stem cells comes from studies of picasso/erbb3b mutants by Parichy and colleagues [129]. Erbb3b is a receptor tyrosine kinase and a zebrafish orthologue of human epidermal growth factor receptor 3 (HER3, ERBB3) which, together with ERBB2, forms a receptor for Neuregulin. Earlier studies by another group had shown that zebrafish Erbb2/Erbb3 functions in migration, proliferation and differentiation of embryonic Schwann cells from the neural crest [130]. In picasso mutants, which are likely null erbb3b alleles, the early larval pattern forms normally, but adult stripe formation at metamorphosis is defective. The authors’ analysis shows that de novo production of chromatophores is severely reduced in picasso mutants at metamorphic stages. Pharmacological Erbb kinase inhibitors were used to examine the temporal requirements for Erbb signaling, and revealed the key stage for Erbb activity to be embryonic. Although the embryonic pigment pattern is unaffected in picasso mutants or by Erbb inhibition, Erbb signaling is required in embryonic stages, in particular during neural crest migration, to allow adult pigment pattern formation to proceed normally. Thus picasso/erbb3b seems to be crucial for chromatophore progenitor/stem cell formation or maintenance. puma mutants also have defects in numbers of metamorphic chromatophore progenitors and show similar, but less severe changes, in the peripheral sox10+ glia [131]. Further work is required to identify the cellular source of the metamorphic chromatophores and how these are affected in picasso/erbb3b and puma mutants. When identification of these cells becomes possible in vivo it will provide a unique opportunity to study pigment progenitors in their endogenous niche.
Conclusion
Some of the fundamental molecular mechanisms that guide melanoblasts to the skin have now been determined. In homeothermic vertebrates, ephrins, endothelins and Kitl are known to play a critical role in directing melanoblasts into the dorsolateral pathway, but whilst Kitl promotes migration in zebrafish, the roles for these and other molecules in pathway choice remain to be tested in fish or amphibian systems. In poikilothermic vertebrates, roles for specific molecular cues in determining where pigment cells stop their migration are now beginning to be uncovered, although whether their roles are conserved throughout vertebrates remains to be established. In addition, it is likely that further signaling pathways underlying pigment pattern formation will be discovered by a combination of mutagenesis screens in zebrafish and mice, small molecule screens in zebrafish and frog [132, 133], and experimental perturbations in the chick.
An important part of pigment morphogenesis in the chick and mouse is the migration of pigment cells into the hair follicles and feather germs. The mechanisms that control this latter process are only beginning to be understood but it appears likely that Kitl and FGF’s may be critical for the migration and maintenance of melanoblasts in the hair and feather. Fish also have epidermal melanocytes and in zebrafish these cells are lost in kita mutants [56], although the cellular role for Kit signaling here remains to be investigated.
Pigment pattern formation in fish and amphibians is significantly more complex in that there are three or more chromatophores that must be assembled into the embryonic and then an adult pattern. These patterns are, in part, formed separately, with the latter dependent on stem or progenitor cells generated during embryogenesis. Analysis of these complex patterning processes is increasingly utilizing theoretical modeling, as discussed by Kondo elsewhere in this issue. Molecular identification of pigment pattern mutations is beginning to refine the patterning mechanisms involved and this will soon help to constrain the mathematical models.
Acknowledgments
We gratefully acknowledge that work in the Kelsh lab is supported by the University of Bath, MRC, BBSRC and Wellcome Trust and that in the Erickson lab by xxxx.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Robert N. Kelsh, Email: bssrnk@bath.ac.uk.
Melissa L. Harris, Email: mmlocke@ucdavis.edu.
Sarah Colanesi, Email: sc346@bath.ac.uk.
Carol A. Erickson, Email: caerickson@ucdavis.edu.
References
- 1.Le Douarin NM, Kalcheim C. The Neural Crest. 2. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- 2.Fox MH. Analysis of some phases of melanoblast migration in barred plymouth rock embryos. Physiol Zool. 1949;22:1–22. doi: 10.1086/physzool.22.1.30152023. [DOI] [PubMed] [Google Scholar]
- 3.Ris H. An experimental study of the origin of melanophores in birds. Physiol Zool. 1941;14:48–66. [Google Scholar]
- 4.Watterson RL. The morphogenesis of down feathers with special reference to the developmental history of melanophores. Physiol Zool. 1942;15:234–259. [Google Scholar]
- 5.Willier BH, Rawles ME. The control of feather color pattern by melanophores grafted from one embryo to another of a different breed of fowl. Physiol Zool. 1940;13:177–202. [Google Scholar]
- 6.Teillet MA. Recherches sur le mode de migration et la différenciation des mélanoblastes cutanés chez l’mbryon d’iseau: Etude expérimentale par la méthode des greffes hétérospécifiques entre embryons de Caille et de Poulet. Embryol Morphogen. 1971;4:95–109. [Google Scholar]
- 7.Serbedzija GN, Fraser SE, B-FM Pathways of trunk neural crest cell migration in the mouse embryo as revealed by vital dye labelling. Development. 1990;108(4):605–12. doi: 10.1242/dev.108.4.605. [DOI] [PubMed] [Google Scholar]
- 8.Hirobe T. Histochemical survey of the distribution of the epidermal melanoblasts and melanocytes in the mouse during fetal and postnatal periods. Anat Rec. 1984;208(4):589–94. doi: 10.1002/ar.1092080414. [DOI] [PubMed] [Google Scholar]
- 9.Mayer TC. The migratory pathway of neural crest cells into the skin of mouse embryos. Dev Biol. 1973;34(1):39–46. doi: 10.1016/0012-1606(73)90337-0. [DOI] [PubMed] [Google Scholar]
- 10.Silvers WK. The Coat Colors of Mice: A Model for Mammalian Gene Action and Interaction. New York: Springer.; 1979. [Google Scholar]
- 11.Bagnara JT, Matsumoto J. Comparative anatomy and physiology of pigment cells in nonmammalian tissues. In: Nordlund JJ, editor. The Pigmentary System: Physiology and Pathophysiology. Blackwell: Malden Oxford Carlton; 2006. pp. 11–59. [Google Scholar]
- 12.Kelsh RN, et al. The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech Dev. 2004;121(7–8):841–59. doi: 10.1016/j.mod.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Lamoreux LM, et al. Pigment pattern formation in the medaka embryo. Pigment Cell Res. 2005;18(2):64–73. doi: 10.1111/j.1600-0749.2005.00216.x. [DOI] [PubMed] [Google Scholar]
- 14.Lopes SS, et al. Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet. 2008;4(3):e1000026. doi: 10.1371/journal.pgen.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimmel CB, et al. Stages of Embryonic Development of the Zebrafish. Dev Dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 16.Johnson SL, et al. Genetic control of adult pigment stripe development in zebrafish. Dev Biol. 1995;167(1):27–33. doi: 10.1006/dbio.1995.1004. [DOI] [PubMed] [Google Scholar]
- 17.Kelsh RN. Genetics and evolution of pigment patterns in fish. Pigment Cell Res. 2004;17(4):326–36. doi: 10.1111/j.1600-0749.2004.00174.x. [DOI] [PubMed] [Google Scholar]
- 18.Kirschbaum F. Untersuchungen über das Farbmuster der Zebrabarbe Brachydanio rerio (Cyprinidae, Teleostei) W Roux’ Arch Dev Biol. 1975;177:129–152. doi: 10.1007/BF00848526. [DOI] [PubMed] [Google Scholar]
- 19.McClure M. Development and evolution of melanophore patterns in fishes of the genus Danio (Teleostei: Cyprinidae) J Morphol. 1999;241(1):83–105. doi: 10.1002/(SICI)1097-4687(199907)241:1<83::AID-JMOR5>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 20.Milos N, Dingle AD. Dynamics of Pigment Pattern Formation in the Zebrafish, Brachydanio rerio. I. Establishment and regulation of the lateral line melanophore stripe during the first eight days of development. J Exp Zool. 1978;205:205–216. [Google Scholar]
- 21.Parichy DM. Evolution of danio pigment pattern development. Heredity. 2006 doi: 10.1038/sj.hdy.6800867. [DOI] [PubMed] [Google Scholar]
- 22.Parichy DM, et al. Mutational analysis of endothelin receptor b1 (rose) during neural crest and pigment pattern development in the zebrafish danio rerio. Dev Biol. 2000;227(2):294–306. doi: 10.1006/dbio.2000.9899. [DOI] [PubMed] [Google Scholar]
- 23.Parichy DM, Turner JM. Zebrafish puma mutant decouples pigment pattern and somatic metamorphosis. Dev Biol. 2003;256(2):242–57. doi: 10.1016/s0012-1606(03)00015-0. [DOI] [PubMed] [Google Scholar]
- 24.Hirata M, Nakamura K, Kondo S. Pigment cell distributions in different tissues of the zebrafish, with special reference to the striped pigment pattern. Dev Dyn. 2005;234(2):293–300. doi: 10.1002/dvdy.20513. [DOI] [PubMed] [Google Scholar]
- 25.Hirata M, et al. Pigment cell organization in the hypodermis of zebrafish. Dev Dyn. 2003;227(4):497–503. doi: 10.1002/dvdy.10334. [DOI] [PubMed] [Google Scholar]
- 26.Serbedzija GN, Bronner-Fraser M, Fraser SE. A vital dye analysis of the timing and pathways of avian trunk neural crest cell migration. Development. 1989;106(4):809–16. doi: 10.1242/dev.106.4.809. [DOI] [PubMed] [Google Scholar]
- 27.Erickson CA, Duong TD, Tosney KW. Descriptive and experimental analysis of the dispersion of neural crest cells along the dorsolateral path and their entry into ectoderm in the chick embryo. Dev Biol. 1992;151(1):251–72. doi: 10.1016/0012-1606(92)90231-5. [DOI] [PubMed] [Google Scholar]
- 28.Kitamura K, et al. Avian neural crest cells express a melanogenic trait during early migration from the neural tube: observations with the new monoclonal antibody, “MEBL-1”. Development. 1992;114:367–378. [Google Scholar]
- 29.Reedy MV, Faraco CD, Erickson CA. The delayed entry of thoracic neural crest cells into the dorsolateral path is a consequence of the late emigration of melanogenic neural crest cells from the neural tube. Dev Biol. 1998;200(2):234–246. doi: 10.1006/dbio.1998.8963. [DOI] [PubMed] [Google Scholar]
- 30.Henion P, Weston J. Timing and pattern of cell fate restrictions in the neural crest lineage. Development. 1997;124(21):4351–9. doi: 10.1242/dev.124.21.4351. [DOI] [PubMed] [Google Scholar]
- 31.Luo R, et al. Molecular identification of distinct neurogenic and melanogenic neural crest sublineages. Development. 2003;130(2):321–30. doi: 10.1242/dev.00213. [DOI] [PubMed] [Google Scholar]
- 32.Erickson CA, Reedy MV. Neural crest development: the interplay between morphogenesis and cell differentiation. Current Topics In Developmental Biology. 1998;40:177–209. doi: 10.1016/s0070-2153(08)60367-1. [DOI] [PubMed] [Google Scholar]
- 33.Wakamatsu Y, et al. Avian neural crest-derived neurogenic precursors undergo apoptosis on the lateral migration pathway. Development. 1998;125(21):4205–13. doi: 10.1242/dev.125.21.4205. [DOI] [PubMed] [Google Scholar]
- 34.Erickson C, Goins T. Avian neural crest cells can migrate in the dorsolateral path only if they are specified as melanocytes. Development. 1995;121(3):915–924. doi: 10.1242/dev.121.3.915. [DOI] [PubMed] [Google Scholar]
- 35.Oakley RA, Tosney KW. Peanut agglutinin and chondroitin-6-sulfate are molecular markers for tissues that act as barriers to axon advance in the avian embryo. Dev Biol. 1991;147(1):187–206. doi: 10.1016/s0012-1606(05)80017-x. [DOI] [PubMed] [Google Scholar]
- 36.Oakley R, et al. Glycoconjugates mark a transient barrier to neural crest migration in the chicken embryo. Development. 1994;120(1):103–114. doi: 10.1242/dev.120.1.103. [DOI] [PubMed] [Google Scholar]
- 37.Jia L, Cheng L, Raper J. Slit/Robo signaling is necessary to confine early neural crest cells to the ventral migratory pathway in the trunk. Dev Biol. 2005;282(2):411–21. doi: 10.1016/j.ydbio.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 38.Debby-Brafman A, et al. F-Spondin, expressed in somite regions avoided by neural crest cells, mediates inhibition of distinct somite domains to neural crest migration. Neuron. 1999;22(3):475–88. doi: 10.1016/s0896-6273(00)80703-5. [DOI] [PubMed] [Google Scholar]
- 39.Santiago A, Erickson CA. Ephrin-B ligands play a dual role in the control of neural crest cell migration. Development. 2002;129(15):3621–32. doi: 10.1242/dev.129.15.3621. [DOI] [PubMed] [Google Scholar]
- 40.Bennett DC, Lamoreux ML. The Color Loci of Mice - A Genetic Century. Pigment Cell Res. 2003 doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 41.Baxter LL, et al. Spotlight on Spotted Mice: A Review of White Spotting Mouse mutants and Associated Human Pigmentation Disorders. Pigment Cell Research. 2004;17:215–234. doi: 10.1111/j.1600-0749.2004.00147.x. [DOI] [PubMed] [Google Scholar]
- 42.Wilson YM, et al. Neural crest cell lineage segregation in the mouse neural tube. Development. 2004;131(24):6153–62. doi: 10.1242/dev.01533. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama A, et al. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mechanisms Of Development. 1998;70(1–2):155–66. doi: 10.1016/s0925-4773(97)00188-3. [DOI] [PubMed] [Google Scholar]
- 44.Wehrle-Haller B, Meller M, Weston JA. Analysis of melanocyte precursors in Nf1 mutants reveals that MGF/KIT signaling promotes directed cell migration independent of its function in cell survival. Dev Biol. 2001;232(2):471–83. doi: 10.1006/dbio.2001.0167. [DOI] [PubMed] [Google Scholar]
- 45.Wehrle-Haller B, Weston JA. Soluble and cell-bound forms of steel factor activity play distinct roles in melanocyte precursor dispersal and survival on the lateral neural crest migration pathway. Development. 1995;121(3):731–42. doi: 10.1242/dev.121.3.731. [DOI] [PubMed] [Google Scholar]
- 46.Bernex F, et al. Spatial and temporal patterns of c-kit-expressing cells in WlacZ/+ and WlacZ/WlacZ mouse embryos. Development. 1996;122(10):3023–33. doi: 10.1242/dev.122.10.3023. [DOI] [PubMed] [Google Scholar]
- 47.Cable J, Barkway C, Steel KP. Characteristics of stria vascularis melanocytes of viable dominant spotting (Wv/Wv) mouse mutants. Hear Res. 1992;64(1):6–20. doi: 10.1016/0378-5955(92)90164-i. [DOI] [PubMed] [Google Scholar]
- 48.Lister JA, et al. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126(17):3757–67. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- 49.Minchin JE, Hughes SM. Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev Biol. 2008;317(2):508–22. doi: 10.1016/j.ydbio.2008.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parichy DM, et al. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development. 2000;127(14):3031–44. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- 51.Raible DW, Eisen JS. Restriction of neural crest cell fate in the trunk of the embryonic zebrafish. Development. 1994;120(3):495–503. doi: 10.1242/dev.120.3.495. [DOI] [PubMed] [Google Scholar]
- 52.Raible DW, Eisen JS. Regulative interactions in zebrafish neural crest. Development. 1996;122(2):501–7. doi: 10.1242/dev.122.2.501. [DOI] [PubMed] [Google Scholar]
- 53.Dutton KA, et al. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128(21):4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- 54.Elworthy S, et al. Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development. 2003;130(12):2809–2818. doi: 10.1242/dev.00461. [DOI] [PubMed] [Google Scholar]
- 55.Kelsh RN, Eisen JS. The zebrafish colourless gene regulates development of non-ectomesenchymal neural crest derivatives. Development. 2000;127:515–525. doi: 10.1242/dev.127.3.515. [DOI] [PubMed] [Google Scholar]
- 56.Parichy DM, et al. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development. 1999;126(15):3425–36. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- 57.Rawls JF, Johnson SL. Temporal and molecular separation of the kit receptor tyrosine kinase’ roles in zebrafish melanocyte migration and survival. Dev Biol. 2003;262(1):152–61. doi: 10.1016/s0012-1606(03)00386-5. [DOI] [PubMed] [Google Scholar]
- 58.Hultman KA, et al. Gene Duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. 2007;3(1):e17. doi: 10.1371/journal.pgen.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lecoin L, et al. Steel and c-kit in the development of avian melanocytes: a study of normally pigmented birds and of the hyperpigmented mutant silky fowl. Dev Dyn. 1995;203(1):106–18. doi: 10.1002/aja.1002030111. [DOI] [PubMed] [Google Scholar]
- 60.Harris ML. Patterning of neural crest-derived pigment cells in the avian embryo. University of California, Davis; Davis: 2008. [Google Scholar]
- 61.Faraco CD, et al. Hyperpigmentation in the Silkie fowl correlates with abnormal migration of fate-restricted melanoblasts and loss of environmental barrier molecules. Dev Dyn. 2001;220(3):212–25. doi: 10.1002/1097-0177(20010301)220:3<212::AID-DVDY1105>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 62.deFreitas PF, Ferreira F, de F, Faraco CD. PNA-positive glycoconjugates are negatively correlated with the access of neural crest cells to the gut in chicken embryos. Anat Rec A Discov Mol Cell Evol Biol. 2003;273(2):705–3. doi: 10.1002/ar.a.10078. [DOI] [PubMed] [Google Scholar]
- 63.Dupin E, Le Douarin NM. Development of melanocyte precursors from the vertebrate neural crest. Oncogene. 2003;22(20):3016–23. doi: 10.1038/sj.onc.1206460. [DOI] [PubMed] [Google Scholar]
- 64.Lecoin L, et al. Cloning and characterization of a novel endothelin receptor subtype in the avian class. Proc Natl Acad Sci U S A. 1998;95(6):3024–9. doi: 10.1073/pnas.95.6.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beauvais A, et al. Changes in the fibronectin-specific integrin expression pattern modify the migratory behavior of sarcoma S180 cells in vitro and in the embryonic environment. J Cell Biol. 1995;128(4):699–713. doi: 10.1083/jcb.128.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erickson CA, Tosney KW, Weston JA. Analysis of Migratory Behaviour of Neural Crest and Fibroblastic Cells in Embryonic Tissues. Dev Biol. 1980;77:142–156. doi: 10.1016/0012-1606(80)90462-5. [DOI] [PubMed] [Google Scholar]
- 67.Hosoda K, et al. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79(7):1267–76. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 68.Svetic V, et al. Sdf1a patterns zebrafish melanophores and links the somite and melanophore pattern defects in choker mutants. Development. 2007;134(5):1011–22. doi: 10.1242/dev.02789. [DOI] [PubMed] [Google Scholar]
- 69.Rao C, et al. A defect in a novel ADAMTS family member is the cause of the belted white-spotting mutation. Development. 2003;130(19):4665–72. doi: 10.1242/dev.00668. [DOI] [PubMed] [Google Scholar]
- 70.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154(1):8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 71.Silver DL, et al. The secreted metalloprotease ADAMTS20 is required for melanoblast survival. PLoS Genet. 2008;4(2):e1000003. doi: 10.1371/journal.pgen.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koster MI, Roop DR. Mechanisms regulating epithelial stratification. Annu Rev Cell Dev Biol. 2007;23:93–113. doi: 10.1146/annurev.cellbio.23.090506.123357. [DOI] [PubMed] [Google Scholar]
- 73.Hirai Y, et al. Expression and role of E- and P-cadherin adhesion molecules in embryonic histogenesis. II. Skin morphogenesis. Development. 1989;105(2):271–7. doi: 10.1242/dev.105.2.271. [DOI] [PubMed] [Google Scholar]
- 74.Yoshida H, et al. Neural and skin cell-specific expression pattern conferred by steel factor regulatory sequence in transgenic mice. Developmental Dynamics. 1996;207(2):222–232. doi: 10.1002/(SICI)1097-0177(199610)207:2<222::AID-AJA10>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 75.Nishimura EK, et al. Regulation of E- and P-cadherin expression correlated with melanocyte migration and diversification. Dev Biol. 1999;215(2):155–66. doi: 10.1006/dbio.1999.9478. [DOI] [PubMed] [Google Scholar]
- 76.Matsui Y, Zsebo KM, Hogan BL. Embryonic expression of a haematopoietic growth factor encoded by the Sl locus and the ligand for c-kit. Nature. 1990;347(6294):667–9. doi: 10.1038/347667a0. [DOI] [PubMed] [Google Scholar]
- 77.Yoshida H, et al. Distinct stages of melanocyte differentiation revealed by analysis of nonuniform pigmentation patterns. Development. 1996;122(4):1207–1214. doi: 10.1242/dev.122.4.1207. [DOI] [PubMed] [Google Scholar]
- 78.Yoshida H, et al. Review: melanocyte migration and survival controlled by SCF/c-kit expression. J Investig Dermatol Symp Proc. 2001;6(1):1–5. doi: 10.1046/j.0022-202x.2001.00006.x. [DOI] [PubMed] [Google Scholar]
- 79.Nishikawa S, et al. In utero manipulation of coat color formation by a monoclonal anti-c-kit antibody: two distinct waves of c-kit-dependency during melanocyte development. Embo J. 1991;10(8):2111–8. doi: 10.1002/j.1460-2075.1991.tb07744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kunisada T, et al. Transgene expression of steel factor in the basal layer of epidermis promotes survival, proliferation, differentiation and migration of melanocyte precursors. Development. 1998;125(15):2915–23. doi: 10.1242/dev.125.15.2915. [DOI] [PubMed] [Google Scholar]
- 81.Jordan SA, I, Jackson J. MGF (KIT ligand) is a chemokinetic factor for melanoblast migration into hair follicles. Dev Biol. 2000;225(2):424–36. doi: 10.1006/dbio.2000.9856. [DOI] [PubMed] [Google Scholar]
- 82.Weiner L, et al. Dedicated epithelial recipient cells determine pigmentation patterns. Cell. 2007;130(5):932–42. doi: 10.1016/j.cell.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 83.Nehls M, et al. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature. 1994;372(6501):103–7. doi: 10.1038/372103a0. [DOI] [PubMed] [Google Scholar]
- 84.Mak SS, et al. Indispensable role of Bcl2 in the development of the melanocyte stem cell. Dev Biol. 2006;291(1):144–53. doi: 10.1016/j.ydbio.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 85.Lee D, Prowse DM, Brissette JL. Association between mouse nude gene expression and the initiation of epithelial terminal differentiation. Dev Biol. 1999;208(2):362–74. doi: 10.1006/dbio.1999.9221. [DOI] [PubMed] [Google Scholar]
- 86.Reedy MV, Johnson RL, Erickson CA. The expression patterns of c-kit and Sl in chicken embryos suggest unexpected roles for these genes in somite and limb development. Gene Expr Patterns. 2003;3(1):53–8. doi: 10.1016/s1567-133x(02)00072-8. [DOI] [PubMed] [Google Scholar]
- 87.Niwa T, et al. Plumage pigmentation and expression of its regulatory genes during quail development--histochemical analysis using Bh (black at hatch) mutants. Mech Dev. 2002;118(1–2):139–46. doi: 10.1016/s0925-4773(02)00256-3. [DOI] [PubMed] [Google Scholar]
- 88.Schofer C, et al. The apical ectodermal ridge, fibroblast growth factors (FGF-2 and FGF-4) and insulin-like growth factor I (IGF-I) control the migration of epidermal melanoblasts in chicken wing buds. Anat Embryol (Berl) 2001;203(2):137–46. doi: 10.1007/s004290000148. [DOI] [PubMed] [Google Scholar]
- 89.Hall RJ, Erickson CA. ADAM 10: an active metalloprotease expressed during avian epithelial morphogenesis. Dev Biol. 2003;256(1):146–59. doi: 10.1016/s0012-1606(02)00133-1. [DOI] [PubMed] [Google Scholar]
- 90.Epperlein HH, Lofberg J, Olsson L. Neural crest cell migration and pigment pattern formation in urodele amphibians. Int J Dev Biol. 1996;40(1):229–38. [PubMed] [Google Scholar]
- 91.Epperlein HH, Lofberg J. The development of the larval pigment patterns in Triturus alpestris and Ambystoma mexicanum. Adv Anat Embryol Cell Biol. 1990;118(1):1–99. [PubMed] [Google Scholar]
- 92.Macmillan GJ. Melanoblast-tissue interactions and the development of pigment pattern in Xenopus larvae. J Embryol Exp Morphol. 1976;35(3):463–84. [PubMed] [Google Scholar]
- 93.Parichy DM. When neural crest and placodes collide: interactions between melanophores and the lateral lines that generate stripes in the salamander Ambystoma tigrinum tigrinum (Ambystomatidae) Dev Biol. 1996;175(2):283–300. doi: 10.1006/dbio.1996.0115. [DOI] [PubMed] [Google Scholar]
- 94.Parichy DM. Pigment patterns of larval salamanders (Ambystomatidae, Salamandridae): the role of the lateral line sensory system and the evolution of pattern-forming mechanisms. Dev Biol. 1996;175(2):265–82. doi: 10.1006/dbio.1996.0114. [DOI] [PubMed] [Google Scholar]
- 95.Tucker RP, Erickson CA. The control of pigment cell pattern formation in the California newt, Taricha torosa. J Embryol Exp Morphol. 1986;97:141–68. [PubMed] [Google Scholar]
- 96.Boldajipour B, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132(3):463–73. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 97.Chong SW, et al. The chemokine Sdf-1 and its receptor Cxcr4 are required for formation of muscle in zebrafish. BMC Dev Biol. 2007;7:54. doi: 10.1186/1471-213X-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dambly-Chaudiere C, Cubedo N, Ghysen A. Control of cell migration in the development of the posterior lateral line: antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev Biol. 2007;7:23. doi: 10.1186/1471-213X-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Doitsidou M, et al. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111(5):647–59. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- 100.Herpin A, et al. Sequential SDF1a and b-induced mobility guides Medaka PGC migration. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 101.Knaut H, et al. A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature. 2003;421(6920):279–82. doi: 10.1038/nature01338. [DOI] [PubMed] [Google Scholar]
- 102.Miyasaka N, Knaut H, Yoshihara Y. Cxcl12/Cxcr4 chemokine signaling is required for placode assembly and sensory axon pathfinding in the zebrafish olfactory system. Development. 2007;134(13):2459–68. doi: 10.1242/dev.001958. [DOI] [PubMed] [Google Scholar]
- 103.Raz E, Reichman-Fried M. Attraction rules: germ cell migration in zebrafish. Curr Opin Genet Dev. 2006;16(4):355–9. doi: 10.1016/j.gde.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 104.Valentin G, Haas P, Gilmour D. The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr Biol. 2007;17(12):1026–31. doi: 10.1016/j.cub.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 105.Kelsh RN, et al. Zebrafish pigmentation mutations and the processes of neural crest development. Development. 1996;123:369–89. doi: 10.1242/dev.123.1.369. [DOI] [PubMed] [Google Scholar]
- 106.Wilkie AL, Jordan SA, Jackson IJ. Neural crest progenitors of the melanocyte lineage: coat colour patterns revisited. Development. 2002;129(14):3349–57. doi: 10.1242/dev.129.14.3349. [DOI] [PubMed] [Google Scholar]
- 107.Fitch KR, et al. Genetics of dark skin in mice. Genes Dev. 2003;17(2):214–28. doi: 10.1101/gad.1023703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Van Raamsdonk CD, et al. Effects of G-protein mutations on skin color. Nat Genet. 2004;36(9):961–8. doi: 10.1038/ng1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85(4):1159–204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- 110.Furutani-Seiki M, et al. A systematic genome-wide screen for mutations affecting organogenesis in Medaka, Oryzias latipes. Mech Dev. 2004;121(7–8):647–58. doi: 10.1016/j.mod.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 111.Haffter P, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 112.Parichy DM, Turner JM. Temporal and cellular requirements for Fms signaling during zebrafish adult pigment pattern development. Development. 2003;130(5):817–33. doi: 10.1242/dev.00307. [DOI] [PubMed] [Google Scholar]
- 113.Parichy DM. Homology and the evolution of novelty during Danio adult pigment pattern development. J Exp Zoolog B Mol Dev Evol. 2007;308(5):578–90. doi: 10.1002/jez.b.21141. [DOI] [PubMed] [Google Scholar]
- 114.Maderspacher F, Nusslein-Volhard C. Formation of the adult pigment pattern in zebrafish requires leopard and obelix dependent cell interactions. Development. 2003;130(15):3447–57. doi: 10.1242/dev.00519. [DOI] [PubMed] [Google Scholar]
- 115.Quigley IK, et al. Pigment pattern evolution by differential deployment of neural crest and post-embryonic melanophore lineages in Danio fishes. Development. 2004;131(24):6053–69. doi: 10.1242/dev.01526. [DOI] [PubMed] [Google Scholar]
- 116.Kirschbaum F. Zur Genetik einiger Farbmustermutanten der Zebrabarbe Brachydanio rerio (Cyprinidae, Teleostei) und zum Phänotyp von Artbastarden der Gattung Brachydanio. Biol Zbl. 1977;96:211–222. [Google Scholar]
- 117.Watanabe M, et al. Spot pattern of leopard Danio is caused by mutation in the zebrafish connexin41.8 gene. EMBO Rep. 2006 doi: 10.1038/sj.embor.7400757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iwashita M, et al. Pigment pattern in jaguar/obelix zebrafish is caused by a Kir7.1 mutation: implications for the regulation of melanosome movement. PLoS Genet. 2006;2(11):e197. doi: 10.1371/journal.pgen.0020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kubo Y, et al. International Union of Pharmacology. LIV. Nomenclature and molecular relationships of inwardly rectifying potassium channels. Pharmacol Rev. 2005;57(4):509–26. doi: 10.1124/pr.57.4.11. [DOI] [PubMed] [Google Scholar]
- 120.Nishimura EK, et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416(6883):854–60. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 121.Sieber-Blum M, et al. Pluripotent neural crest stem cells in the adult hair follicle. Dev Dyn. 2004;231(2):258–69. doi: 10.1002/dvdy.20129. [DOI] [PubMed] [Google Scholar]
- 122.Lang D, et al. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433(7028):884–7. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 123.Osawa M, et al. Molecular characterization of melanocyte stem cells in their niche. Development. 2005;132(24):5589–99. doi: 10.1242/dev.02161. [DOI] [PubMed] [Google Scholar]
- 124.Wong CE, et al. Neural crest-derived cells with stem cell features can be traced back to multiple lineages in the adult skin. J Cell Biol. 2006;175(6):1005–15. doi: 10.1083/jcb.200606062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nishikawa S, Osawa M. Generating quiescent stem cells. Pigment Cell Res. 2007;20(4):263–70. doi: 10.1111/j.1600-0749.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 126.Delfino-Machin M, et al. The proliferating field of neural crest stem cells. Dev Dyn. 2007;236(12):3242–54. doi: 10.1002/dvdy.21314. [DOI] [PubMed] [Google Scholar]
- 127.Yang CT, et al. Mutations in gfpt1 and skiv2l2 cause distinct stage-specific defects in larval melanocyte regeneration in zebrafish. PLoS Genet. 2007;3(6):e88. doi: 10.1371/journal.pgen.0030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rawls JF, Johnson SL. Zebrafish kit mutation reveals primary and secondary regulation of melanocyte development during fin stripe regeneration [In Process Citation] Development. 2000;127(17):3715–24. doi: 10.1242/dev.127.17.3715. [DOI] [PubMed] [Google Scholar]
- 129.Budi EH, Patterson LB, Parichy DM. Embryonic requirements for ErbB signaling in neural crest development and adult pigment pattern formation. Development. 2008 doi: 10.1242/dev.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lyons DA, et al. erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr Biol. 2005;15(6):513–24. doi: 10.1016/j.cub.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 131.Parichy DM, Turner JM, Parker NB. Essential role for puma in development of postembryonic neural crest-derived cell lineages in zebrafish. Dev Biol. 2003;256(2):221–41. doi: 10.1016/s0012-1606(03)00016-2. [DOI] [PubMed] [Google Scholar]
- 132.Peterson RT, et al. Small molecule developmental screens reveal the logic and timing of vertebrate development [In Process Citation] Proc Natl Acad Sci U S A. 2000;97(24):12965–9. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tomlinson ML, Field RA, Wheeler GN. Xenopus as a model organism in developmental chemical genetic screens. Mol Biosyst. 2005;1(3):223–8. doi: 10.1039/b506103b. [DOI] [PubMed] [Google Scholar]
- 134.Kelsh RN, Schmid B, Eisen JS. Genetic analysis of melanophore development in zebrafish embryos. Dev Biol. 2000;225(2):277–93. doi: 10.1006/dbio.2000.9840. [DOI] [PubMed] [Google Scholar]
- 135.Reedy MV, Faraco CD, Erickson CA. Specification and migration of melanoblasts at the vagal level and in hyperpigmented Silkie chickens. Dev Dyn. 1998;213(4):476–85. doi: 10.1002/(SICI)1097-0177(199812)213:4<476::AID-AJA12>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 136.Austin LM, Boissy RE. Mammalian tyrosinase-related protein-1 is recognized by autoantibodies from vitiliginous Smyth chickens. An avian model for human vitiligo. Am J Pathol. 1995;146(6):1529–41. [PMC free article] [PubMed] [Google Scholar]
- 137.Kitamura K, et al. Avian neural crest cells express a melanogenic trait migration from the neural tube: observations with the antibody, “MEBL-1”. Development. 1992;114(2):367–378. [Google Scholar]
- 138.Mackenzie MA, et al. Activation of the receptor tyrosine kinase Kit is required for the proliferation of melanoblasts in the mouse embryo. Dev Biol. 1997;192(1):99–107. doi: 10.1006/dbio.1997.8738. [DOI] [PubMed] [Google Scholar]
- 139.Manova K, Bachvarova RF. Expression of c-kit encoded at the W locus of mice in developing embryonic germ cells and presumptive melanoblasts. Dev Biol. 1991;146(2):312–24. doi: 10.1016/0012-1606(91)90233-s. [DOI] [PubMed] [Google Scholar]
- 140.Lee HO, Levorse JM, Shin MK. The endothelin receptor-B is required for he migration of neural crest-derived melanocyte and enteric neuron precursors. Dev Biol. 2003;259(1):162–75. doi: 10.1016/s0012-1606(03)00160-x. [DOI] [PubMed] [Google Scholar]
- 141.Shin MK, et al. The temporal requirement for endothelin receptor-B signalling during neural crest development. Nature. 1999;402(6761):496–501. doi: 10.1038/990040. [DOI] [PubMed] [Google Scholar]
- 142.Jordan SA, I, Jackson J. A late wave of melanoblast differentiation and rostrocaudal migration revealed in patch and rump-white embryos. Mech Dev. 2000;92(2):135–43. doi: 10.1016/s0925-4773(99)00332-9. [DOI] [PubMed] [Google Scholar]
- 143.Rawls JF, Johnson SL. Requirements for the kit receptor tyrosine kinase during regeneration of zebrafish fin melanocytes. Development. 2001;128(11):1943–9. doi: 10.1242/dev.128.11.1943. [DOI] [PubMed] [Google Scholar]
- 144.Mills MG, Nuckels RJ, Parichy DM. Deconstructing evolution of adult henotypes: genetic analyses of kit reveal homology and evolutionary novelty during adult pigment pattern development of Danio fishes. Development. 2007;134(6):1081–90. doi: 10.1242/dev.02799. [DOI] [PubMed] [Google Scholar]