Summary
This brief review focuses on the role of epigenetic mechanisms in plasticity and memory formation, and their identification as targets of activity-dependent regulation in neurons. Epigenetic modifications of chromatin, namely post-translational modifications of nuclear proteins and covalent modification of DNA, result in potent regulation of gene readout. Recent data have demonstrated that epigenetic mechanisms play a significant role in regulating synaptic plasticity and memory. In this review, we focus on this theme, describing some basic background concerning epigenetic molecular mechanisms, and describing recent results concerning plasticity and memory formation. As an understanding of these novel mechanisms of transcriptional regulation promises to invigorate many areas of investigation, we end by speculating upon some of the open questions ripe for discovery.
Keywords: memory, hippocampus, demethylation, histone, HDAC, DNA methylation, epigenetic, gene transcription, fear conditioning
Introduction – An Epigenetic Code for Memory?
Recent studies have demonstrated that nuclear targets of neuronal signaling pathways, in particular the histone proteins and DNA that comprise the core chromatin particle, are also an integral component of memory processes [reviewed in 1-3]. We and others have proposed that an epigenetic code might be involved in memory formation, whereby specific patterns of post-translational histone modifications and DNA methylation might help encode the salience of cell-surface signals and their contingencies [4-5]. This general hypothesis of an epigenetic code for memory is new and rather speculative, and stems from an earlier idea of a histone code proposed by Allis and colleagues [6].
A better understanding of the epigenetic code concept as it might apply to memory requires an appreciation of the basic biochemistry regulating chromatin structure. Here we define epigenetics as the covalent modification of chromatin that influences activity-dependent changes in gene expression necessary for cognition. These changes can be short-term and transient, or likely long-term and thus capable of perpetuating lasting changes in gene activity states. In this vein, there are two basic molecular epigenetic mechanisms that are currently studied – post-translational histone modifications and DNA methylation. For background, we will briefly review these mechanisms in the next two sections (see [1, 2] for a more detailed recent treatment).
Epigenetic histone marks
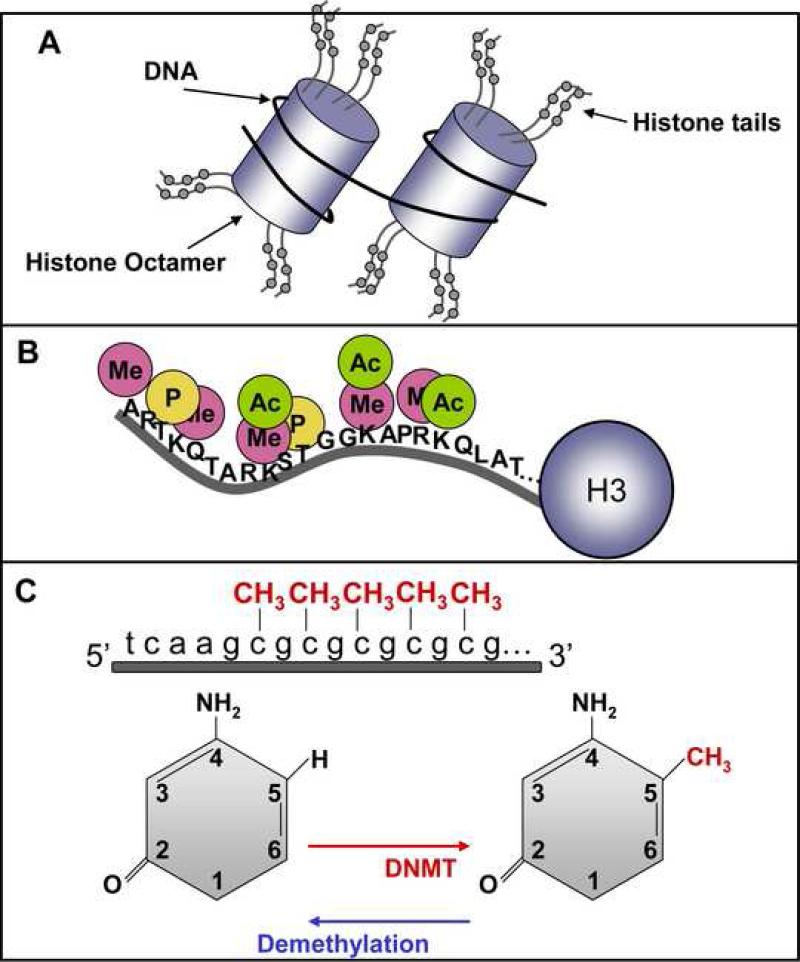
Histones are highly basic proteins that organize DNA within the nucleus. The interaction between histones, which form the core of the chromatin particle, and DNA, is mediated in part by the N-terminal tail of histone proteins. One can imagine chromatin as a core of eight histone proteins (histones 2A, 2B, 3, and 4, with two copies of each molecule) with DNA wrapped around similar to twine on a spool. Structural studies indicate the N-terminal tails of histones protrude beyond the DNA and are available for post-translational modifications [7] (Figure 1A-B).
Figure 1.
Schematic of epigenetic marks. A) In the nucleus, DNA is compressed through interactions with histones, and this DNA-protein complex is referred to as chromatin. Two copies each of histones H2A, H2B, H3 and H4 assemble, forming a histone octamer, around which DNA wraps and condenses. The DNA-histone interaction occurs at the N-terminal tail of a histone (B), where for example on the H3 N-terminal tail there are many sites for epigenetic marking via acetylation (Ac), methylation (Me), and phosphorylation (P). (C) Top - Overview of DNA methylation, where methyl groups are added to cytosine-guanine dinucleotides in an around gene regulatory regions. Bottom - DNA methylation is catalyzed by a class of enzymes known as DNA methyltransferases (DNMTs), which transfer methyl groups to the 5-position of the pyrimidine ring. DNA methylation is also a reversible process, and recent work suggests that one mechanism for active demethylation is through a DNA repair-like process.
Several sites for modifications exist within the N-terminal tails of histone proteins, and covalent chemical modification of these sites, namely acetylation, phosphorylation, methylation, ubiquitination and sumoylation, regulates the overall structure of chromatin [8]. These modifications, along with methylation of cytosine residues in DNA are the principal “epigenetic tags” impacting genome readout. As it has been best-characterized in the context of memory formation, we will focus this discussion on histone acetylation.
Acetylation of histones occurs at lysine residues, specifically on their side-chain amino group, effectively neutralizing their positive charge, decreasing the affinity between the protein tail and DNA, and thus relaxing the chromatin structure and allowing the recruitment of transcriptional machinery. Acetylated histone tails also provide a platform for the binding of additional co-activators with domains that recognize acetylated lysines. Histone acetyltransferases (HATs) catalyze the direct transfer of an acetyl group from acetyl-CoA to the ε-NH+ group of the lysine residues within a histone [9]. Histone acetylation is generally associated with transcriptional activation and is widely regarded as one of the epigenetic marks associated with active chromatin (euchromatin). CREB Binding Protein (CBP) is one known HAT regulating local chromatin structure as part of CREB-dependent activation of gene transcription in memory [10-13, 14•].
Histone acetylation is a reversible process, and the enzymes that catalyze the reversal of histone acetylation are known as histone deacetylases (HDACs). There are a total of eleven different classical HDAC isoforms, most of which are expressed in the CNS (Table 1). HDAC inhibitors are the principal way to manipulate the epigenome pharmacologically, and hold promise for therapeutic value [15-18]. Trichostatin A, sodium butyrate, valproic acid, and Suberoylanilide Hydroxamic Acid (SAHA) are commercially available HDAC inhibitors, each having varying degrees of selectivity for the classical HDAC isoforms (Table 1). Isoform-selective HDAC inhibitors are under development but are not yet broadly available. Finally, we should note that the SIR2 family of HDACs (the “Sirtuins”) is another major category of HDAC, which operate by a completely different catalytic mechanism than the classical HDAC isoforms [19].
Table 1.
Classical HDAC isoforms and their targeted HDAC inhibitors.
| Classes | HDAC Isoforms | Inhibitors |
|---|---|---|
| Class I | HDACs 1, 2, 3, 8 | Trichostatin A, Sodium Butyrate, Valproic Acid, SAHA |
| Class II | HDACs 4, 5, 6, 7, 9, 10 | Trichostatin A, Sodium Butyrate, SAHA |
| Class IV | HDAC 11 | SAHA |
Covalent modification of DNA – cytosine methylation
The second major mechanism whereby the genome can be epigenetically marked is DNA methylation (see [20, 21] for a more detailed treatment of this mechanism). Methylation of DNA is a direct chemical modification of a cytosine side-chain that adds a -CH3 group through a covalent bond (Figure 1C). Methylation of DNA is catalyzed by a class of enzymes known as DNA methyltransferases (DNMTs) [20, 21]. DNMTs transfer methyl groups to cytosine residues, specifically at the 5-position of the pyrimidine ring [20, 21]. Not all cytosines can be methylated; cytosines must be immediately followed by a guanine to be methylated [20, 21]. These CpG dinucleotide sequences are highly underrepresented in the genome, and often occur in small clusters known as CpG islands [20, 21].
There are two variants of DNMTs: maintenance DNMTs and de novo DNMTs [20, 21]. De novo DNMTs (DNMT3a and DNMT3b) methylate previously unmethylated CpG sites in DNA – sites which have no methyl-cytosine on either DNA strand. The maintenance DNMT isoform (DNMT1, though it appears to regulate de novo methylation in some cases) methylates hemi-methylated DNA – DNA which has a methylated CpG already present on one strand but no methyl-cytosine on the complementary strand. Maintenance DNMTs perpetuate methylation marks after cell division, regenerating the methyl-cytosine marks on the newly synthesized complementary DNA strand that arises from DNA replication.
DNA methylation, with its stable nature, offers an ideal substrate for the long-term cellular changes necessary for the maintenance and persistence of memory. However, DNA methylation would also have to be an active and reversible process to enable neurons to respond to physiological and environmental stimuli. To date, DNA demethylation has been viewed largely as a passive process in differentiated cells, whereby multiple rounds of cell division without DNMT-mediated remethylation is necessary to erase epigenetic marks. The idea of whether there is active DNA demethylation in post-mitotic cells, such as neurons, is controversial in the field, mainly due to the fact that the identity of a demethylase that can actively remove methyl groups remains elusive. But there is increasing evidence to suggest that there is active methylation and demethylation in mature cells [22, 23•, 24, 25••, 26•, 27]. The DNA methyltransferases DNMT3a and DNMT3b appear to have a hand in this process [23•, 26•], though recent work suggests that Gadd45b may also have a role by promoting active DNA demethylation through nucleotide-excision repair [25••].
The predominant view in the literature is that methylation of DNA is associated with suppression of gene transcription, and in many cases extensive DNA methylation triggers complete silencing of the associated gene. The precise molecular processes through which this occurs are complex, but in essence, methylation of cytosines at CpG dinucleotides recruits methyl-DNA binding proteins, at specific sites in the genome [20, 21]. Proteins binding to methylated DNA have both a methyl-DNA binding domain (MBD) and a transcription-regulatory domain (TRD). The TRD recruits adapter/scaffolding proteins, which in turn recruit HDACs to the site. The HDACs alter chromatin structure locally through removing acetyl groups from histone core proteins, leading to compaction of chromatin and transcriptional suppression. Thus, methylation of DNA triggers localized regulation of the three-dimensional structure of DNA and its associated histone proteins, resulting in a higher-affinity interaction between DNA and the histone core, and transcriptional repression by allosteric means. It is important to note that while DNA methylation is usually (and historically) associated with transcriptional suppression, recent studies have indicated that DNA methylation can also be associated with transcriptional activation [28•, 29].
Epigenetic marking of histones in memory
A diverse series of studies have demonstrated memory formation is associated with epigenetic marking of the genome. For example, contextual fear memory formation in rodents is associated with acetylation of hippocampal histone H3 [4, 30, 31••]. This epigenetic marking requires NMDA-receptor-dependent synaptic transmission and the ERK/MAPK signaling cascade in the hippocampus, as does the fear conditioning memory itself [31••, 32]. This is one specific example demonstrating epigenetic tagging of the genome during consolidation of hippocampus-dependent memory.
Considering the previous statement, one would predict disruption of HAT activity would interfere with long-term memory formation. As previously mentioned, CREB Binding Protein (CBP) is a HAT, and several studies have investigated long-term memory formation in genetically manipulated mice with impaired CBP function and have demonstrated that CBP/HAT-deficient mice have both L-LTP and long-term memory deficits [10, 11, 13, 33].
Thus, histone acetylation is regulated in long-term memory, and disruption of HAT activity impairs long-term memory. Together, these observations suggest that perturbations in the processes regulating chromatin structure can influence long-term memory formation in the behaving animal in vivo. Then, can augmentation of histone acetylation enhance memory formation? Several studies have investigated the effect of HDAC inhibitors on long-term memory formation, and found that indeed HDAC inhibition improves memory formation (4, 10, 14•, 30, 34, 35••).
Overall, there is extensive and varied data demonstrating that histone acetylation is regulated during memory consolidation, and this regulation contributes to an animal's capacity to form memories.
DNA Methylation in Memory
Studies have also begun to investigate the capacity of DNA methylation to regulate synaptic plasticity and memory in adult animals [31••, 36, 37•, 38, 39••]. Inhibitors of DNMTs alter DNA methylation in adult CNS tissue and block hippocampal Long-term Potentiation (LTP) [36, 37•, 38]. DNMT inhibition blocks hippocampus-dependent memory formation in a contextual fear conditioning paradigm [31••, 39••]. Data also demonstrate fear conditioning is associated with rapid methylation and transcriptional silencing of the memory suppressor gene Protein Phosphatase 1 (PP1), while demethylation and transcriptional activation of the synaptic plasticity gene reelin [39••]. Finally, DNA methylation controls site-specific initiation of bdnf gene transcription during memory formation [31••]. These findings have the surprising implication that both DNA methylation and demethylation are involved in long-term memory.
It is important to briefly point out here that much of what we know regarding the role of DNA methylation in synaptic physiology and memory is from pharmacological studies using drugs whose mechanisms we do not fully understand. Because both 5-aza-C and zebularine are nucleoside analogs that need to be incorporated into DNA to trap DNMT and block DNA methylation, the mechanism of how these drugs are able to alter methylation in post-mitotic neurons is not clear. They may do so by actively demethylating DNA in non-dividing cells through a replication-independent event, such as a DNA repair process, but this has not been tested. There is only one behavior study to-date that has used a non-nucleoside compound (RG108) to directly inhibit DNMT enzyme activity, and importantly, results indicated that RG108 had similar effects on memory as that of zebularine [31••].
Overall, results suggest that DNA methylation is dynamically regulated in the adult nervous system and this cellular mechanism is a crucial step in memory formation. Importantly, these studies implicating a role for altered DNA methylation in memory suggest an expansion of the original histone code concept into a broader epigenetic code concept, wherein a variety of marks associated with chromatin regulation may contribute combinatorial readout and control of memory formation.
Summary and Speculations
Thus far we have presented an emerging new view of the epigenome and its role in regulating memory formation. Indeed, this is a rapidly expanding area in neurobiology and studies are being published at a rapid pace demonstrating that epigenetic mechanisms are involved in mediating diverse experience-driven changes in the CNS. The effects of such changes are manifest at the molecular, cellular, circuit, and behavioral levels. These diverse observations support the view that the epigenome resides at the interface of the environment and the genome. Future studies geared toward understanding the role of the epigenome in experience-dependent behavioral modification will clearly be important for, and relevant to the memory field.
We would like to conclude by mentioning what we feel to be a few interesting open questions and implications raised by the emerging role of epigenetic mechanisms in memory formation.
As we have described, epigenetic molecular mechanisms are emerging as an important component of gene regulation in memory. This role in memory reprises their role as cellular information storage devices in development, and suggests an important conservation of function as molecular information storage devices. One idea we find intriguing is that utilization of epigenetic mechanisms for information storage may represent a unifying model in biology, with epigenetic mechanisms being utilized for cellular memory at levels from cellular differentiation to development to behavioral memory. However, it is important to keep in mind the only data currently available investigated the hypothesis that there are changes in epigenetic markings in response to stimuli that induce changes in cellular physiology or behavior (i.e. learning). The question of whether these epigenetic changes contribute to the maintenance and persistence of memory is still by-and-large open. The best evidence for such an idea comes from studies demonstrating that epigenetic changes contribute to the persisting effects, or “memory”, of early-life experiences [40-42, 43••, 44].
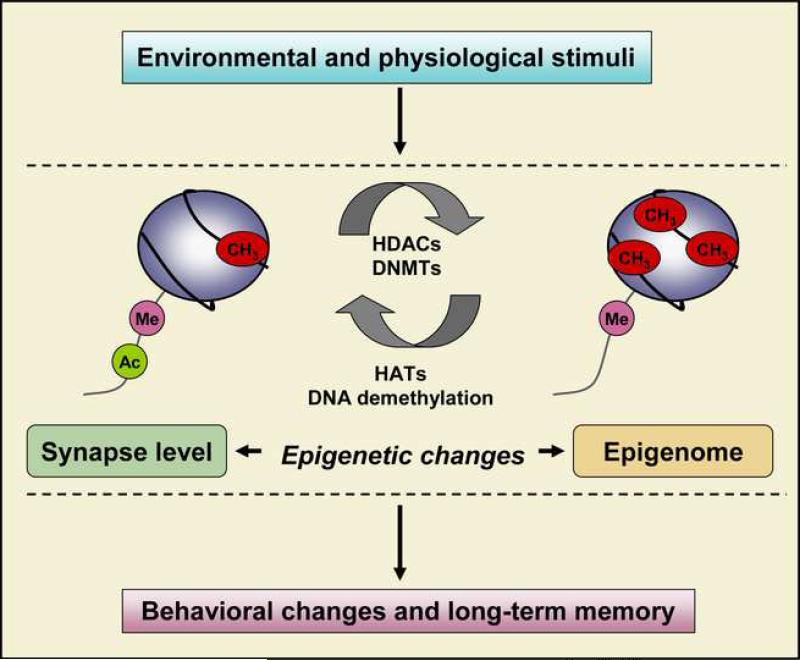
Another compelling question concerns how epigenetically driven changes in gene expression are manifest as changes in function at the synapse and elsewhere in the neuron, and on a much broader scale, across neural systems. This is completely unknown at present. However, a central organizing concept of physiological psychology, at least regarding behavioral change, is the principle of Hebbian synaptic plasticity as the central mechanism underlying behavioral modification. Recent work implicating epigenetic mechanisms in memory may necessitate a re-thinking of this fundamental assumption. A single adult neuron can have 10,000 synapses, but has only one nucleus. Within the nucleus there are only one or two copies of a given specific gene. Chemical modification of DNA through DNA methylation implies a state change for the entire neuron. Chemically modifying and completely silencing or activating a given gene must alter the genomic complement of the entire cell. If chemical modification of genes underlies behavioral plasticity, this may necessitate a reevaluation of the fundamental assumption that synaptic plasticity is the locus driving behavioral modification. At a minimum there would be two levels of mechanism in play – one residing at the individual synapse and one residing at the epigenome and operating cell- or system-wide (Figure 2). This iconoclastic concept might require a fundamental re-assessment of the role of synapse specificity in behavioral change.
Figure 2.
Epigenetic mechanisms provide a substrate for the long-term changes in gene patterns underlying behavior. Physiological cues and environmentally-relevant stimuli modulate the activity of chromatin modifying enzymes, such as histone acetyltransferases (HATs), histone deacetylases (HDACs), and DNA methyltransferases (DNMTs) that in turn modify the histone tails and methylation of CpG dinucleotides. Such changes then affect neuronal gene transcription at the synapse level to that of the epigenome and neural systems levels.
An additional thought is that epigenetics has emerged largely out of developmental biology and one of its central tenets has been that DNA methylation is immutable once laid down during cell fate determination. Recent studies have proposed DNA chemical modification is in fact plastic in non-dividing, terminally differentiated neurons. This idea being true will require a redefinition of several of the fundamental tenets of epigenetics, introducing both the idea of plasticity of DNA methylation and that epigenetic mechanisms are functionally active in terminally differentiated, non-dividing cells.
Most studies to date investigating epigenetic mechanisms in memory have utilized a target-gene approach to assessing changes at specific candidate gene loci. As an expansion beyond the target-gene approach, in the future the principal alternative is likely to be to utilize whole-epigenome approaches. These are still in development, but are likely to come online during the next few years. The capacity to use whole-epigenome high-throughput sequencing, or to use methods such as ChIPseq [45, 46] that allow comprehensive assessment of chromatin modifications across the genome, will allow investigators to identify the fraction of the genome subject to experience-driven epigenetic plasticity in the adult CNS.
Finally, we feel that there are several important missing pieces concerning the idea of a histone code in memory. While histone acetylation and phosphorylation have been more extensively examined in memory, one important missing piece is the role of histone methylation in memory. For many years the prevailing model was histone methylation was irreversible [for a review see 47]. However, histone methylation changes clearly are associated with changes in gene transcription and cognitive dysfunction [48, 49]. Furthermore, histone demethylases have been recently discovered and active regulation of histone demethylation is known to occur [48, 50]. The idea of changes in histone methylation, and sumoylation for that matter, involved in memory formation as part of a “histone code” is an open question, and evaluating this idea will test one of the critical predictions of the histone code hypothesis. On a related note, it will be important to resolve whether histone-modifying enzymes have any nonhistone functions.
In conclusion, the idea that there could be an epigenetic code operating to subserve behavioral change in the context of memory formation is beginning to gain traction. Indeed, the most recent discoveries indicate that chemical modification of DNA is also a component of the epigenetic molecular mechanisms supporting behavioral change broadly defined. While it is certainly not possible to test this epigenetic code concept thoroughly in the course of one or a few single experiments, this intriguing theory is motivating the interest of an increasing number of investigators.
Acknowledgements
We wish to thank Felecia Hester for her assistance in preparing this review. We would also like to apologize to the colleagues whose work we could not cite because of space limitations. This work was funded by grants from the National Institutes of Health, the National Alliance for Research on Schizophrenia and Depression, Civitan International, the Rotary Clubs CART fund, and the Evelyn F. McKnight Brain Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest that were published over the period of the review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behav Brain Res. 2008;192:70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 3.Sweatt JD. Experience-dependent epigenetic modifications in the central nervous system. Biol Psychiatry. 2009;65:191–197. doi: 10.1016/j.biopsych.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 5.Wood MA, Hawk J, Abel T. Combinatorial chromatin modifications and memory storage: a code for memory? Learn Mem. 2006;13:241–244. doi: 10.1101/lm.278206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 7.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 8.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 9.Marmorstein R. Structure and function of histone acetyltransferases. Cell Mol Life Sci. 2001;58:693–703. doi: 10.1007/PL00000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 11.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component on memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin KC, Sun YE. To learn better, keep the HAT on. Neuron. 2004;42:879–881. doi: 10.1016/j.neuron.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Oliveira AMM, Wood MA, McDonough CB, Abel T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn Mem. 2007;14:564–572. doi: 10.1101/lm.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB: CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [• of special interestThe authors of this study used a combination of genetic, electrophysiological, and behavioral approaches to understand how HDACs improve memory. Their data provide evidence that the enhancement of both hippocampus-dependent memory and synaptic plasticity by HDAC inhibitors is mediated by the transcription factor cAMP response element-binding protein (CREB) as well as the recruitment of the transcriptional coactivator and histone acetyltransferase CREB-binding protein (CBP)] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Current Opinion Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 18.Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009:49. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- 19.Buck SW, Gallo CM, Smith JS. Diversity in the Sir2 family of protein deacetylases. J Leukoc Biol. 2004;75:939–950. doi: 10.1189/jlb.0903424. [DOI] [PubMed] [Google Scholar]
- 20.Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 21.Miranda TB, Jones PA. DNA methylation: The nuts and bolts of repression. Journal of Cellular Physiology. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 22.Brown SE, Weaver ICG, Meaney MJ, Szyf M. Regional-specific global cytosine methylation and DNA methyltransferase expression in the adult rat hippocampus. Neurosci Letters. 2008;440:49–53. doi: 10.1016/j.neulet.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 23•.Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [• of special interestThe authors of studies [23] and [26] provide seminal data demonstrating that DNA methylation is not merely a stable epigenetic mark, but that there is dynamic DNA methylation and demethylation that supports gene transcription] [DOI] [PubMed] [Google Scholar]
- 24.Kriaucionis S, Heintz N. The nuclear DNA base 5-Hydroxymethylcytosine is present in purkinje neurons and the brain. Science. 2009:1169786. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Ma DK, Jang M-H, Guo JU, Kitabatake Y, Chang M-I, Pow-anpongkul N, Flavell RA, Lu B, Ming G-I, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [•• of outstanding interestThis land-mark study provides the first evidence that there is active DNA demethylation in mature neurons. Specifically, the authors show that Gadd45b, a stress response gene that is transiently induced by physiological and environmental stimuli, is required for activity-induced DNA demethylation at specific gene promoters supporting neurogenesis in the adult hippocampus] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [• of special interestSee [23]] [DOI] [PubMed] [Google Scholar]
- 27.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in mammalian DNA by the MLL fusion partner TET1. Science. 2009:1170116. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [• of special interestUsing genetically altered mice, the authors of this study demonstrated that methyl-CpG binding protein 2 (MeCP2) associates with the transcriptional activator CREB at the promoter of an active gene. This has broadened our understanding of the role of DNA methylation in regulation of genes, now indicating that DNA methylation can also serve as an activator of gene transcription] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen S, Zhou Z, Greenberg ME. Activating a repressor. Science. 2008;320:1172–1173. doi: 10.1126/science.1159146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of bdnf gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [•• of outstanding interestThe author's in vivo assessment of the contribution of chromatin remodeling in memory reveals that dynamic changes in methylation of bdnf DNA as well as histones modifications at specific bdnf promoters play a pivotal role in contextual fear memory formation. They also demonstrate that NMDA receptor activation supports this process. This paper begins to identify the complexity of epigenetic regulation of the bdnf gene in adult cognition] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat Neurosci. 1998;1:602–609. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- 33.Maurice T, Duclot F, Meunier J, Naert G, Givalois L, Meffre J, Celerier A, Jacquet C, Copois V, Mechti N, et al. Altered memory capacities and response to stress in p300/CBP-associated factor (PCAF) histone acetylase knockout mice. Neuropsychopharmacol. 2007;33:1584–1602. doi: 10.1038/sj.npp.1301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bredy TW, Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn Mem. 2008;15:39–45. doi: 10.1101/lm.801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai L-H. Recovery of learning and memory is associated with chromatin remodeling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [•• of outstanding interestThis paper provided the first environmental enrichment on learning and memory impairment involves histone modifications. Their data also provide evidence of the viability of HDAC therapy in reversing brain and behavioral deficits associated with neurodegeneration] [DOI] [PubMed] [Google Scholar]
- 36.Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 37•.Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [• of special interestThese studies implicate DNA methylation as a controller of presynaptic function in hippocampal neurons] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [•• of outstanding interestThese studies were the first to directly implicate active control of DNA methylation in adult behavioral memory formation] [DOI] [PubMed] [Google Scholar]
- 40.Arai JA, Li S, Hartley DM, Feig LA. Transgenerational rescue of a genetic defect in long-term potentiation and memory formation by juvenile enrichment. J. Neurosci. 2009;29:1496–1502. doi: 10.1523/JNEUROSCI.5057-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Champagne FA, Curley JP. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neurosci Biobehav Rev. 2009;33:593–600. doi: 10.1016/j.neubiorev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28:9055–9065. doi: 10.1523/JNEUROSCI.1424-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [•• of outstanding interestThis is the first evidence that abuse from a caregiver results in increases in DNA methylation that last through the lifespan and are passed-on to the next generation. Data from this study as well as others make an effective argument that epigenetic modifications contribute to the maintenance and persistence of life-long memories] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weaver ICG, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 45.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 46.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucl. Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 48.Akbarian S, Huang H-S. Epigenetic regulation in human brain--focus on histone lysine methylation. Biol Psychiatry. 2009;65:198–203. doi: 10.1016/j.biopsych.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsankova N, Berton O, Renthal W, Kumar A, Neve R, Nestler E. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 50.Xu J, Deng X, Disteche CM. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS ONE. 2008;3:e2553. doi: 10.1371/journal.pone.0002553. [DOI] [PMC free article] [PubMed] [Google Scholar]