Abstract
RNA-binding protein HuR binds U- or AU-rich sequences in the 3′-untranslated regions (UTRs) of target mRNAs, stabilizing them and/or modulating their translation. Given HuR’s links with cancer, we studied the consequences of modulating HuR levels in pancreatic cancer cells. HuR-overexpressing cancer cells, in some instances, are up to 30-fold more sensitive to treatment with gemcitabine (GEM), the main chemotherapeutic component of treatment regimens for pancreatic ductal adenocarcinoma (PDA), compared to control cells. In pancreatic cancer cells, HuR associates with deoxycytidine kinase (dCK) mRNA, which encodes the enzyme that metabolizes and thereby activates GEM. GEM exposure to pancreatic cancer cells, enriches the association between HuR and dCK mRNA and increases cytoplasmic HuR levels. Accordingly, HuR overexpression elevates, while HuR silencing reduces, dCK protein expression in pancreatic cancer cells. In a clinical correlate study of GEM treatment, we found a 7-fold increase in risk of mortality in PDA patients with low cytoplasmic HuR levels compared to patients with high HuR levels, after adjusting for other treatments and demographic variables. These data support the notion that HuR is a key mediator of GEM efficacy in cancer cells, at least in part through its ability to regulate dCK levels post-transcriptionally. We propose that HuR levels in PDA modulate the therapeutic efficacy of GEM, thus serving as a marker of the clinical utility of this common chemotherapeutic agent and a potential target for intervention in pancreatic cancer.
Keywords: pancreatic ductal adenocarcinoma, pancreatic cancer, HuR, gemcitabine, deoxycytidine kinase, mRNA binding protein, gemcitabine response, RNA binding proteins
Introduction
Pancreatic ductal adenocarcinoma (PDA) is the fourth leading cause of cancer-related deaths in the United States (1). Currently, two therapeutic options that provide the best clinical benefit are surgical resection and chemotherapy regimens that include gemcitabine (GEM) (2′,2′-difluorodeoxycytidine). For over ten years, GEM has been the reference drug for the treatment of this often fatal disease (2). GEM is also utilized to treat other malignancies including non-small-cell lung, breast, gastric, and ovarian cancers. GEM utilizes the same key metabolic enzyme for activation within the cell, deoxycytidine kinase (dCK), as does a previously developed and related nucleoside analog cytarabine (Ara-C) (3). dCK phosphorylates the prodrug, GEM, generating the active metabolites gemcitabine di- and triphosphates that inhibit DNA chain elongation and cause cellular death (4). The levels of dCK correlate with overall patient survival following GEM-based therapy in PDA specimens (p=0.0425)(4).
The stress-response protein Hu antigen R (HuR) is an RNA-binding protein that regulates gene expression post-transcriptionally. Like other related Hu/elav proteins, HuR harbors three conserved RNA recognition motifs through which it binds to target mRNAs (5) that frequently have AU- or U-rich stretches in their 3′-untranslated regions (UTRs). HuR is predominantly nuclear, but in response to various stimuli, it is mobilized to the cytoplasm, prolongs target mRNA half-life, and can modulate target mRNA translation (5). Many HuR target mRNAs encode stress-response, immune-response, cell cycle regulatory proteins, oncogenes, and tumor suppressor genes (5). HuR modulates these transcripts in response to stimuli such as therapeutic agents (i.e. tamoxifen and prostaglandin), nutrient depletion (polyamines, amino acid starvation), heat shock, immune stimuli, short-wavelength UV irradiation, oxidants, and transcriptional inhibitors (actinomycin D) (5–7).
Although HuR has never been reported to be mutated in cancer, it has been proposed to contribute to the tumorigenesis process (8–10). Elevated cytoplasmic accumulation of HuR both correlates with high-grade malignancy and serves as a prognostic factor of poor clinical outcome in some cancers (11–14). Given the extensive research examining HuR’s role in cancer and stress response over the past decade (5), we explored the role of HuR in PDA and the relationship of HuR to chemotherapeutic efficacy.
Methods
Transfection of pancreatic cancer cell lines
HuR cDNA sequence was cloned into the pcDNA 3.1.Zeo vector (Invitrogen) for stable transfection of pancreatic cancer cell lines MiaPaca2, PL-5, and Hs766t (15). Pooled cells remained under selection media containing Zeocin (Invitrogen) for several months after transfection. Mia.HuR, PL5.HuR, and Hs766t.HuR denote HuR overexpressing lines; Mia.EV, PL5.EV, and Hs766t denote empty vector or control lines. HuR and control siRNA sequences and transfection conditions were as described (7).
Immunofluorescence
Cells were plated on LabTek II™ Chamber slides (Fisher Scientific) and fixed in 3% paraformaldehyde (20 min, RT). Cells were washed with PBS and permeabilized using 0.5% Triton X-100/1% normal goat serum (Vector Laboratories) in PBS (15 min). After washes in 1% goat serum/PBS, cells were incubated (1:50 dilution, 1 hr, RT) with mouse anti-HuR (Santa Cruz) or anti-deoxycytidine kinase (dCK, Abnova) primary antibodies. After washes in PBS, cells were incubated for 1 hr with goat anti-mouse secondary antibody (1:400, Alexa Fluor 647, Molecular Probes). Nuclei were stained with DAPI and cells were evaluated under a Zeiss LSM-510 Confocal Laser Microscope.
Drug sensitivity assay
Mia.HuR, Mia.EV, Hs766t.HuR, Hs766t, and PL5.HuR, PL5.EV cells were seeded (1000 cells/well) in 96-well plates and treated with Etoposide, 5-Fluorouracil, Cis-platin, Staurosporine, Nocodazole, Colcemid, Ara-C (Sigma), and GEM (Gemzar, Eli-Lilly, Indianapolis, IN) for 6–7 days. After treatment, cells were washed with PBS and lysed with 100 μL of water/well; cell viability was quantified by staining of double-stranded DNA with Quant-iT™ PicoGreen (Invitrogen) and analyzed with a TECAN SpectraFluor.
Immunoblot
Whole-cell, cytoplasmic, and nuclear lysates were prepared as described (7), and protein was size-fractionated by SDS-PAGE (10% acrylamide). Membranes were blocked for 1 h in 5% Milk/TBS-T and incubated overnight with monoclonal antibodies (Santa Cruz) recognizing HuR, dCK, the cytoplasmic marker α-Tubulin, or the nuclear marker hnRNP. Membranes were washed with TBS-T and incubated with secondary antibodies; and the resulting signals were visualized by chemiluminescence (Millipore). Total protein was visualized with Fast Green (USB).
Cell cycle analysis and apoptosis assay
Mia.EV and Mia.HuR cell lines were either left untreated or treated with 0.03 μM GEM for 48 h. For cell cycle analysis, cells were then fixed in 100% ethanol, and stained with a propidium iodide solution containing RNAse A (Sigma Aldrich). For apoptosis assays, cells were resuspended at 106 cells/mL and incubated in Annexin V and Propidium Iodide, following the manufacturers’ protocol (FITC Annexin V, BD Pharmigen). Both assays were analyzed by flow cytometry.
RNA-binding: biotin pulldown and RNP IP assays
MiaPaCa2 cells were treated with 4 μM GEM and collected 48 hours later. For biotin pulldown analysis (7), cytoplasmic extracts were isolated using the NE-PERR® Nuclear and Cytoplasmic Extraction Reagents Kit (Pierce Biotechnology). Probes for biotin pull-down analysis were synthesized as described (7) using the following PCR primers (sense and antisense, respectively) containing the T7 RNA polymerase promoter sequence CCAAGCTTCTAATACGACTCACTATAGGGAGA (T7):(T7)GATCTTGCTGAAGACTACAGGC and TTATTAGCGT CTTTTCAATTCTACAAA for dCK 3′UTR; (T7)CTCAACGACCACTTTGTCAAGC and CACAGGGTACTTTATTGATGGTACAT for GAPDH 3′UTR (16) (see supplemental figure for depiction of UTR positions). Biotinylated probes were synthesized using the MAXIscript T7 kit (Ambion) and Biotinylated dCTP (Enzo Life Sciences).
For immunoprecipitation of endogenous RNA-protein complexes (RNP IP) from cytoplasmic (450 μg) extracts, reactions were carried out as described (7), using protein A-Sepharose beads (Sigma) that were precoated with 30 μg of either mouse immunoglobulin G1 (IgG1; BD Biosciences), or anti-HuR antibodies. After IP, the RNA in the IP materials was isolated and reverse-transcribed. GAPDH and dCK transcripts were quantified by real-time PCR analysis using each specific primers: AGCAAGGCATTCCTCTTGAA and CTACAGGCAGCCAAATGGTT for dCK, TGCACCACCAACTGCTTAGC and CTCATGACCACAGTCCATGCC for GAPDH. The relative levels of dCK product was first normalized to GAPDH product in all IP samples, then fold enrichments in HuR IP were compared with IgG IP, as described (7).
Case selection and immunohistochemistry
HuR immunostaining was performed on 32 resected PDAspecimens from the Thomas Jefferson University pathology archives after IRB approval. All patients received GEM, alone or in combination with Xeloda (2 patients), radiation therapy (8 patients) or both (2 patients). The experienced pancreatic pathologist (A.K.W.) reviewed all cases in a blinded fashion and classified the tumors as well differentiated(n=6), moderately differentiated (n=22), or poorly differentiated(n=12). For each case, representative sections were selected for immunohistochemical analysis of HuR cytoplasmic and nuclear staining patterns, which were scored using the following scale: 0 for no staining; 1 for weak and/or focal (<10% of the cells) staining; 2 for moderate or strong staining (10–50% of the cells); and 3 for moderate or strong staining (>50% of the cells). Combined scores 0 and 1 represented low expression, while combined scores 2 and 3 represented high expression.
Results and Discussion
HuR overexpression preferentially sensitized pancreatic cancer cell lines to the nucleoside analogs GEM and Ara-C
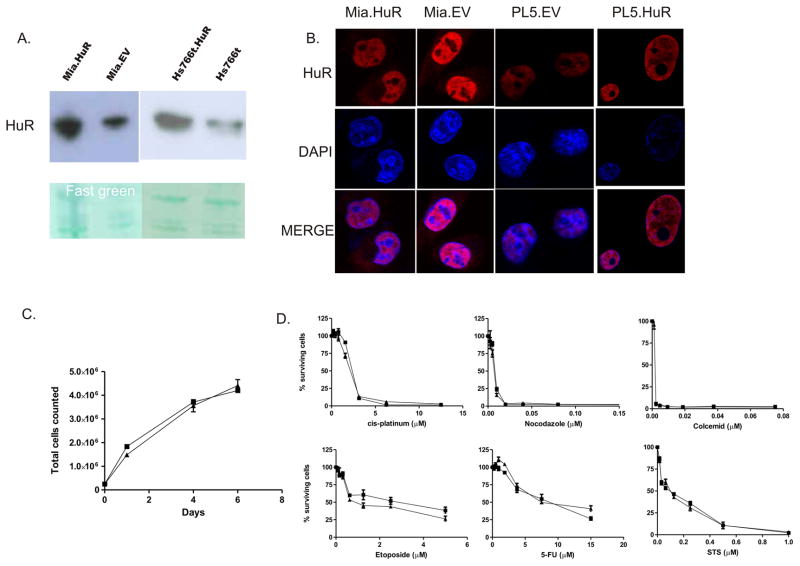
Stable HuR overexpression in the indicated pancreatic cancer cell lines was confirmed by immunoblot and immunofluorescence analyses (Figure 1A and B). Contrary to previous studies in colon cancer cell lines (9), isogenic, transfected cell lines grew roughly at the same rates (Figure 1C). Treatment with the indicated chemotherapeutic agents (but not GEM) showed no difference in sensitivity in HuR-overexpressing cells (Figure 1D).
Figure 1.
Characterization of HuR-overexpressing pancreatic cancer cell lines. (A) Immunoblot analysis of HuR expression in lysates from MiaPaCa2 (Mia.HuR and Mia.EV) and Hs766T (Hs766t. HuR and Hs766t) cells. Fast Green staining confirmed the equality of protein loading. (B) Immunofluorescence to detect HuR and nuclei (DAPI). (C) Mia.HuR and Mia.EV cell proliferation rates, as determined by direct cell counts. (D) Cell survival was measured by PicoGreen after incubation of cells for 5–7 days with the indicated compounds. Data show the means (and S.E.M.) from 3 measurements in a single experiment; each experiment is representative of at least three individual experiments. ▲, Mia.HuR cells; ■, Mia.EV cells.
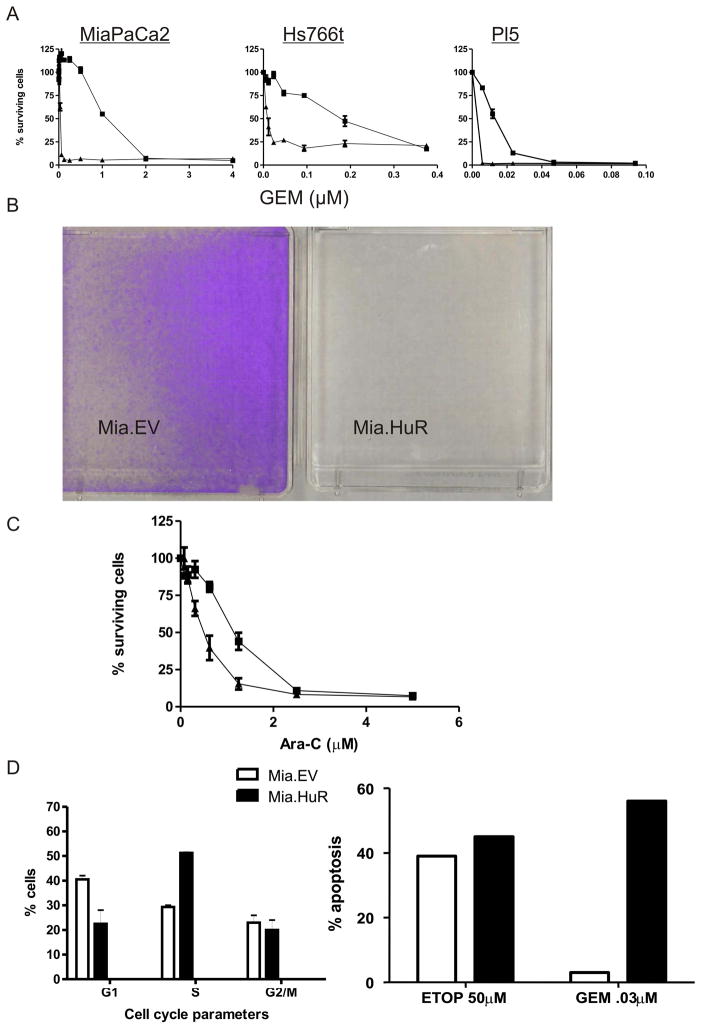
By contrast, cell lines overexpressing HuR were found to be strikingly more sensitive to GEM than were control lines, as assessed both by PicoGreen measurement (Figure 2A) and by staining with crystal violet even when cells were treated with low concentrations of GEM (Figure 2B). HuR-overexpressing cell lines were similarly selectively more sensitive to Ara-C, another anti-cancer agent that utilizes dCK (Figure 2C). After GEM treatment, HuR-overexpressing cells showed selective enrichment in the S-phase of the cell division cycle and increased apoptosis (Figure 2D) as compared to the control cells.
Figure 2.
Stable expression of HuR renders cells hypersensitive to the nucleoside analogs GEM and Ara-C. (A) Survival of MiaPaCa2, Hs766t, and PL5 cell lines was measured by the PicoGreen assay after 5–7 days of incubation with the indicated GEM doses. Graphs represent single experiments (S.E.M.); each experiment is representative of >three individual experiments. ▲, HuR expressing cells; ■, control cells. (B) Crystal violet-stained flasks of Mia.HuR and Mia.EV cultures after GEM treatment (0.1 μM, 7 days). (C) Sensitivity of MiaPaCa2 cells to Ara-C treatment was measured as explained in panel (A). (D) FACS analysis of cells treated with GEM (0.03 μM) for 48 h, depicting the percentages of cells in G1, S, and G2/M compartments (left). Measurement of apoptotic fractions (Materials and Methods) in cultures treated as explained in panel (Right).
HuR localization and association with dCK mRNA upon GEM treatment
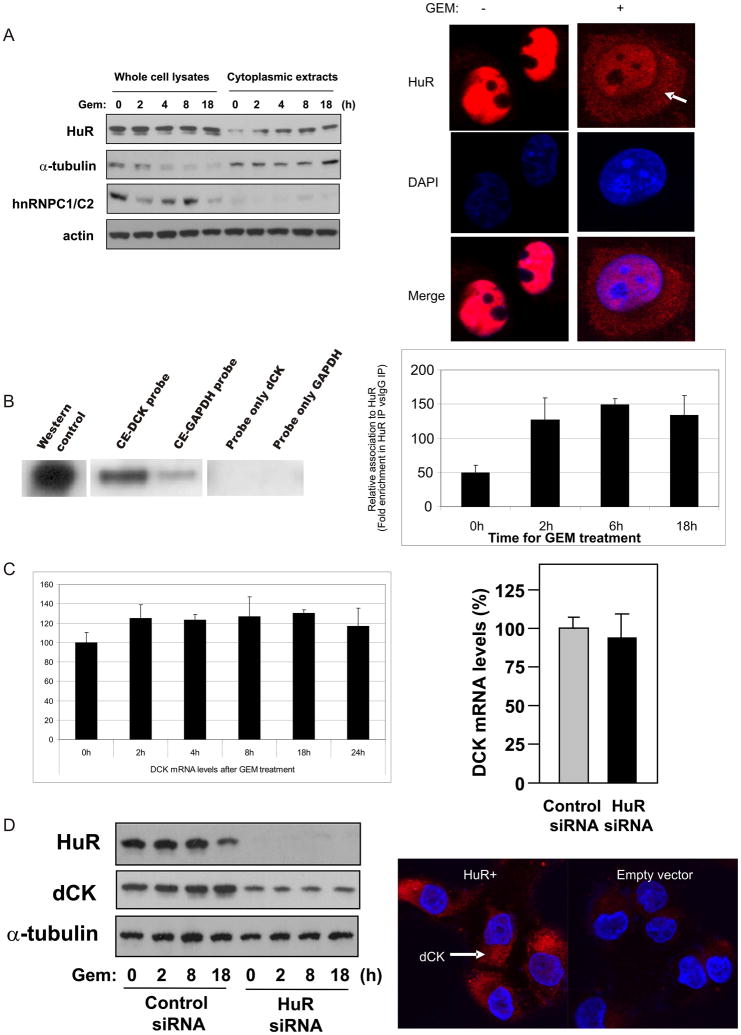
GEM treatment did not alter whole-cell HuR levels in parental MiaPaCa2 cells, but significantly increased the cytoplasmic HuR levels, as determined by immunoblot and immunofluorescence analyses (Figure 3A). Given that the dCK 3′UTR region contained 8 putative hits of an HuR recognition motif (17), we tested if HuR associates with dCK mRNA using two different RNA-binding assays. First, MiaPaCa2 cytoplasmic extracts were incubated with equimolar amounts of biotinylated transcripts spanning the dCK 3′UTR and the GAPDH 3′UTR (a control RNA, not a target of HuR); the resulting complexes were analyzed by HuR immunoblot. As shown in Figure 3B (left), HuR bound the dCK 3′UTR much more strongly than the GAPDH 3′UTR. Second, we tested the association of HuR with the endogenous dCK mRNA by using a ribonucleoprotein immunoprecipitation (RNP IP) assay. As shown, while GEM did not alter overall dCK protein or mRNA levels (Figure 3A, C), it significantly increased HuR’s association with dCK mRNA (Figure 3B, right).
Figure 3.
HuR associates with dCK mRNA and promotes dCK protein expression in MiaPaCa2 cells. (A) Western blot analysis of HuR levels in whole-cell and cytoplasmic lysates after treatment of MiaPaCa2 cells with GEM (1 μM) for the indicated times (left). Immunofluorescence analysis of HuR levels and localization (red) in cells treated with 4 μM GEM for 24 h; nuclei were distinguished by staining with DAPI (blue) (right). (B) Biotin pulldown analysis of HuR RNP complexes. Cytoplasmic extracts were incubated with biotinylated transcripts spanning the DCK or GAPDH 3′UTRs. The association of HuR with biotinylated RNAs was tested by Western blot analysis. Positive control: HuR cytoplasmic lysate. Negative controls: ‘Probe only’ lanes contain only biotinylated RNAs that were not incubated with protein lysates. Shown is a representative blot (right). HuR binding to dCK mRNA was tested by RNP IP analysis (Materials and Methods) in MiaPaCa2 cells treated with GEM for the times indicated; GEM mRNA levels in HuR and IgG IP samples were first normalized to GAPDH mRNA levels in the same IP reactions, and plotted as fold enrichment in dCK mRNA in HuR IP compared with IgG IP. Data show the means and standard deviation from 3 independent experiments (left). (C) dCK mRNA levels were measured in cells that were left untransfected (left) or were transfected with either a control siRNA or HuR siRNA(7) and tested 48 h later (right). (D) Western blot analysis of HuR, dCK, and α-Tubulin in cells expressing normal or silenced HuR levels (left). Immunofluorescence analysis of dCK levels (indicated by the arrow) and localization (red) in cells expressing normal or elevated HuR levels; nuclei were visualized by staining with DAPI (blue) (right).
Inhibition of HuR expression using small interfering (si)RNA(7) did not alter dCK mRNA levels (Figure 3C, right) but decreased dCK protein levels (Figure 3D, left) regardless of GEM treatment. Conversely, HuR-overexpressing cells displayed higher dCK signals (Figure 3D, right). Together, these data show for the first time that 1) GEM exposure to cancer cells increases cytoplasmic HuR levels (Figure 3A), 2) HuR associates with dCK mRNA (Figure 3B), and 3) HuR regulates dCK protein levels (Figure 3D).
HuR localization and expression in PDA specimens
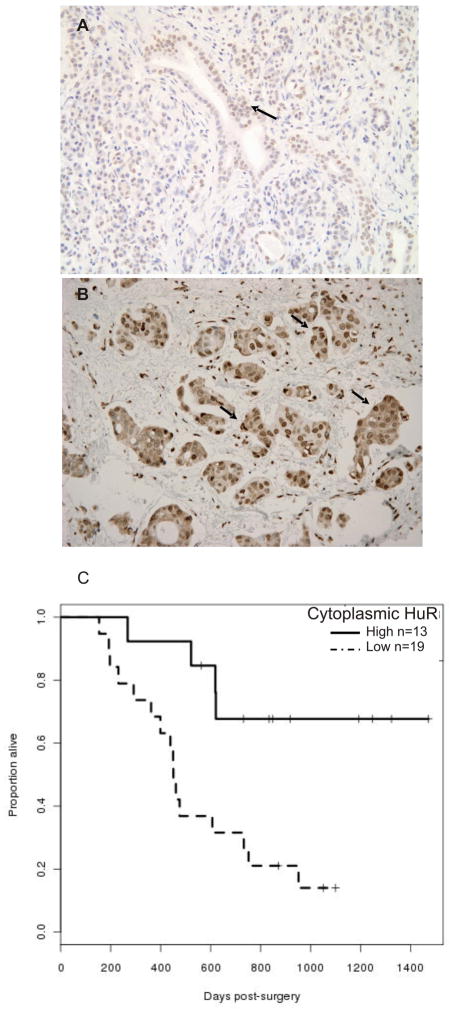
We detected primarily weak to moderate nuclear HuR expression in normal pancreatic ductal and acinarcells (Figure 4A). Strong nuclear expression of HuR was found inwell-, moderately, and poorly differentiated PDAs. Cytoplasmic HuR accumulation was associated with poorly differentiated PDAs (Figure 4B and data not shown). Figure 4C shows the Kaplan-Meier overall survival curves of patients receiving GEM, stratified by their HuR status. The median survival time for patients on GEM was 619 days, with 21 deaths out of the 32 patients who received GEM. A univariate Cox regression model gives a hazard ratio of low to high HuR of 4.48, with a 95% confidence interval of (1.49 to 13.5). Adjusting for age, sex, Xeloda use and radiation therapy in this patient group gives an adjusted hazard ratio of 7.34 (p=0.0022) with a 95% confidence interval of (2.05 to 26.22). These data indicate a greater than 7-fold increase risk of mortality in patients with low cytoplasmic HuR levels (compared to high cytoplasmic HuR levels) among patients receiving GEM, after adjusting for variables as mentioned above.
Figure 4.
HuR cytoplasmic expression correlates with GEM response in pancreatic cancer patients. (A) Arrows indicate primarily nuclear staining of HuR in normal pancreas (200X). (B) Arrows indicate high cytoplasmic expression in PDA specimen (200X). (C) Kaplan-Meier plot of overall survival among patients receiving GEM (n=32), stratified by HuR levels. The curves are significantly different (p= 0.0036 by log-rank).
Perspective
As elevated cytoplasmic HuR has been widely correlated with advanced malignancy, the finding that high cytoplasmic HuR levels were associated with an increased therapeutic efficacy of GEM in pancreatic cancer was unexpected. Our results that HuR regulates dCK protein concentration and that cytoplasmic HuR levels predict GEM response in our patient cohort lead us to hypothesize that HuR could be a key molecule involved in GEM efficacy in cancer. It is intriguing to postulate that part of HuR’s survival repertoire may be to increase dCK levels to process deoxyribonucleosides for survival, however in the presence of nucleoside analogs (such as GEM) HuR’s augmentation of dCK may be deleterious. Further studies are warranted to investigate if HuR dictates GEM effectiveness by regulating additional target mRNAs, and to assess if HuR is a suitable marker for GEM response in larger cancer patient cohorts for which GEM therapy is utilized.
Supplementary Material
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Burris HA, 3rd, Moore MJ, Andersen J, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403–13. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 3.Li ZR, Campbell J, Rustum YM. Effect of 3-deazauridine on the metabolism, toxicity, and antitumor activity of azacitidine in mice bearing L1210 leukemia sensitive and resistant to cytarabine. Cancer Treat Rep. 1983;67:547–54. [PubMed] [Google Scholar]
- 4.Sebastiani V, Ricci F, Rubio-Viqueira B, et al. Immunohistochemical and genetic evaluation of deoxycytidine kinase in pancreatic cancer: relationship to molecular mechanisms of gemcitabine resistance and survival. Clin Cancer Res. 2006;12:2492–7. doi: 10.1158/1078-0432.CCR-05-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–81. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hostetter C, Licata LA, Witkiewicz A, et al. Cytoplasmic accumulation of the RNA binding protein HuR is central to tamoxifen resistance in estrogen receptor positive breast cancer cells. Cancer Biol Ther. 2008;7:1496–506. doi: 10.4161/cbt.7.9.6490. [DOI] [PubMed] [Google Scholar]
- 7.Kuwano Y, Kim HH, Abdelmohsen K, et al. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol Cell Biol. 2008;28:4562–75. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez de Silanes I, Lal A, Gorospe M. HuR: post-transcriptional paths to malignancy. RNA Biol. 2005;2:11–3. doi: 10.4161/rna.2.1.1552. [DOI] [PubMed] [Google Scholar]
- 9.Lopez de Silanes I, Fan J, Yang X, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–54. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 10.Lopez de Silanes I, Fan J, Galban CJ, Spencer RG, Becker KG, Gorospe M. Global analysis of HuR-regulated gene expression in colon cancer systems of reducing complexity. Gene Expr. 2004;12:49–59. doi: 10.3727/000000004783992215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoo PS, Sullivan CA, Kiang S, et al. Tissue Microarray Analysis of 560 Patients with Colorectal Adenocarcinoma: High Expression of HuR Predicts Poor Survival. Ann Surg Oncol. 2008 doi: 10.1245/s10434-008-0209-3. [DOI] [PubMed] [Google Scholar]
- 12.Heinonen M, Fagerholm R, Aaltonen K, et al. Prognostic role of HuR in hereditary breast cancer. Clin Cancer Res. 2007;13:6959–63. doi: 10.1158/1078-0432.CCR-07-1432. [DOI] [PubMed] [Google Scholar]
- 13.Denkert C, Weichert W, Winzer KJ, et al. Expression of the ELAV-like protein HuR is associated with higher tumor grade and increased cyclooxygenase-2 expression in human breast carcinoma. Clin Cancer Res. 2004;10:5580–6. doi: 10.1158/1078-0432.CCR-04-0070. [DOI] [PubMed] [Google Scholar]
- 14.Denkert C, Weichert W, Pest S, et al. Overexpression of the embryonic-lethal abnormal vision-like protein HuR in ovarian carcinoma is a prognostic factor and is associated with increased cyclooxygenase 2 expression. Cancer Res. 2004;64:189–95. doi: 10.1158/0008-5472.can-03-1987. [DOI] [PubMed] [Google Scholar]
- 15.Brody JR, Witkiewicz A, Williams TK, et al. Reduction of pp32 expression in poorly differentiated pancreatic ductal adenocarcinomas and intraductal papillary mucinous neoplasms with moderate dysplasia. Mod Pathol. 2007;20:1238–44. doi: 10.1038/modpathol.3800974. [DOI] [PubMed] [Google Scholar]
- 16.Casolaro V, Fang X, Tancowny B, et al. Posttranscriptional regulation of IL-13 in T cells: role of the RNA-binding protein HuR. J Allergy Clin Immunol. 2008;121:853–9. e4. doi: 10.1016/j.jaci.2007.12.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci U S A. 2004;101:2987–92. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.