Summary
Substrates of the E3 ubiquitin ligase CRL4Cdt2, including Cdt1 and p21, contain a PCNA-binding motif called a PIP box. Upon binding of the PIP box to PCNA on chromatin, CRL4Cdt2 is recruited and the substrate is ubiquitylated. Importantly, a PIP box cannot be sufficient for destruction, as most PIP box proteins are stable. Using Xenopus egg extracts, we identify two sequence elements in CRL4Cdt2 substrates that promote their proteolysis: a specialized PIP box which confers exceptionally efficient PCNA binding, and a basic amino acid four residues downstream of the PIP box, which recruits CRL4Cdt2 to the substrate-PCNA complex. We also identify two mechanisms which couple CRL4Cdt2-dependent proteolysis to the chromatin-bound form of PCNA, insuring that this proteolysis pathway is active only in S phase or after DNA damage. Thus, CRL4Cdt2 recognizes an unusual degron, which is assembled specifically on chromatin via the binding of a specialized PIP box to PCNA.
Introduction
Cells use ubiquitin-mediated proteolysis to regulate many processes, including cell cycle progression, transcription, signaling, DNA replication, and DNA repair. During ubiquitin-mediated proteolysis, an E3 ubiquitin ligase attaches a polyubiquitin chain to the substrate, which targets it for destruction by the 26S proteasome (Nakayama and Nakayama, 2006; Petroski and Deshaies, 2005). Many ubiquitin ligases recognize substrates via a short amino acid motif called a “degron” (Nakayama and Nakayama, 2006; Petroski and Deshaies, 2005). Because the binding of the ligase to its cognate degron is frequently a key point of regulation, ligase-degron interactions have been the subject of intense investigation.
Several E3 ubiquitin ligases control cell cycle progression by binding to specific degron motifs. One of these, the Anaphase Promoting Complex (APC), is active from mitosis until the end of the G1 phase. The APC binds the activating subunits Cdc20 or Cdh1 (Peters, 2006). These activators function as `substrate receptors' by binding directly to a degron motif within the substrate. APCCdc20 is positively regulated via cyclin-dependent kinase 1 (CDK1)-dependent phosphorylation of Cdc20, and it recognizes proteins with a `D box' degron, whose consensus is R-X-X-L (Glotzer et al., 1991). In contrast to APCCdc20, the activity of APCCdh1 is negatively regulated by CDK, such that it only functions from anaphase until late G1, when CDK activity is low. APCCdh1 recognizes proteins that contain either a D box or a `KEN' box, which comprises the motif K-E-N (Pfleger and Kirschner, 2000).
Another example of a cell cycle regulated E3 ubiquitin ligase is SCF, now known as CRL1 (Cullin-ring ligase 1). The CRL1F-box complex is composed of Cul1, a scaffolding protein; Skp1, an adapter protein; Rbx1, a ring protein that interacts with the E2; and one of numerous substrate receptors called F-box proteins that bind Skp1 (Nakayama and Nakayama, 2006; Petroski and Deshaies, 2005). Recognition by CRL1F-box often requires substrate phosphorylation at serine or threonine residues, which creates a `phosphodegron' that interacts with the F-box protein. For example, proteins destroyed by CRL1β-TRCP typically employ the degron D-pS-G-X-X-pS (Ang and Wade Harper, 2005; Nakayama and Nakayama, 2006), while the degron for CRL1Fbw7 is L-X-pT-P-P-X-pS (Ang and Wade Harper, 2005).
Recently, CRL4DCAF has emerged as another E3 ubiquitin ligase that is an essential regulator of cell cycle progression and genome stability. In this E3 ligase, the Cul4 scaffold links to the Ddb1 adaptor, which in turn binds to different putative substrate receptors called DCAFs (Jin et al., 2006; Lee and Zhou, 2007; O'Connell and Harper, 2007). Within this family, CRL4Cdt2 targets the replication licensing factor Cdt1 for destruction in S phase of the cell cycle in all metazoans (Arias and Walter, 2006; Jin et al., 2006; Zhong et al., 2003). Cdt1 is required to recruit the MCM2-7 complex to origins in the G1 phase of the cell cycle (“licensing”), and CRL4Cdt2-mediated destruction of Cdt1 prevents de novo licensing in S phase, thereby limiting DNA replication to one round per cell cycle (Jin et al., 2006; Kim and Kipreos, 2007; Lovejoy et al., 2006; Sansam et al., 2006; Zhong et al., 2003). CRL4Cdt2 likely also targets the Xenopus CDK inhibitor Xic1 (Chuang and Yew, 2001, 2005; Chuang et al., 2005; You et al., 2002), and it was recently shown to target p21 (Abbas et al., 2008; Kim et al., 2008; Nishitani et al., 2008), CKI-1 (Kim et al., 2008), E2F (Shibutani et al., 2008), and DNA polymerase η (Kim and Michael, 2008) for destruction. Interestingly, Cdt1 and Xic1 destruction in S phase is coupled directly to DNA replication (Arias and Walter, 2005; Chuang and Yew, 2001; May et al., 2005; Nishitani et al., 2006). Cdt1 and p21 are also destroyed after DNA damage in a manner that depends on PCNA and CRL4Cdt2 (Abbas et al., 2008; Arias and Walter, 2006; Higa et al., 2003; Hu et al., 2004; Jin et al., 2006; Kim et al., 2008; Lovejoy et al., 2006; Nishitani et al., 2008; Ralph et al., 2006; Sansam et al., 2006). CRL4Cdt2 and PCNA-dependent proteolysis of two substrates (Cdt1 and Spd1) after DNA damage have also been detected in S. pombe but not in S. cerevisiae (Liu et al., 2005; Ralph et al., 2006). Currently, there is no evidence that CDK, ATR, ATM or any other protein kinase is required for Cdt1 destruction in S phase or after DNA damage, and all the data suggest that the mechanism of destruction in these two contexts is identical.
Many proteins involved in DNA metabolism interact with PCNA, including DNA polymerases, ligases, endonucleases, chromatin remodeling factors and CDK inhibitors (Moldovan et al., 2007). PCNA is a homotrimeric ring that encircles DNA and thereby tethers interacting proteins to the DNA. Most PCNA binding proteins contain a PCNA Interaction Protein motif, or PIP box, through which they interact with PCNA. PIP boxes are 8 amino acids long, and are thought to contain 4 essential residues (Q-x-x-ψ-x-x-ϑ-ϑ, where ψ is any moderately hydrophobic amino acid L, V, I, or M, and ϑ is an aromatic residue, Y or F). The PIP box interacts with the Inter Domain Connector Loop (IDCL) and a hydrophobic pocket of PCNA (Moldovan et al., 2007). Importantly, all known substrates of CRL4Cdt2 contain a PIP box, and in each case, mutation of the PIP box abolishes destruction by CRL4Cdt2. An unresolved puzzle in the field is why the vast majority of PIP box proteins are not targeted for destruction by CRL4Cdt2.
Another important question is how PCNA promotes CRL4Cdt2-dependent proteolysis. Our results in Xenopus egg extracts showed that Cdt1 ubiquitylation occurs exclusively on chromatin in a PIP-box, PCNA, and replication or damage dependent manner, suggesting that chromatin-bound PCNA (PCNAChromatin) serves as a platform for Cdt1 ubiquitylation (Arias and Walter, 2005, 2006). Consistent with this idea, CRL4Cdt2 binds to chromatin during replication and in a fashion that depends on Cdt1's PIP box (Arias and Walter, 2006; Jin et al., 2006). These data suggest that Cdt1 docks onto PCNAChromatin, and that the PCNA-Cdt1 complex recruits CRL4Cdt2, after which ubiquitin transfer occurs (Figure 1A). This model explains why Cdt1 is destroyed only in S phase and after DNA damage, since PCNA is bound to chromatin under both circumstances. However, the model does not address why unbound PCNA (PCNAFree) is not competent to support Cdt1 destruction, a critical feature that prevents aberrant Cdt1 destruction in G1. One idea is that Cdt1 does not bind efficiently to PCNAFree. In apparent support of this explanation, we failed to detect an interaction between Cdt1 and PCNAFree in extracts lacking DNA, although we were unable to verify binding to PCNAChromatin under the same conditions [unpublished results cited in (Arias and Walter, 2006; Jin et al., 2006)]. Another possibility is that only the complex of PCNAChromatin and Cdt1 is competent to interact with CRL4Cdt2.
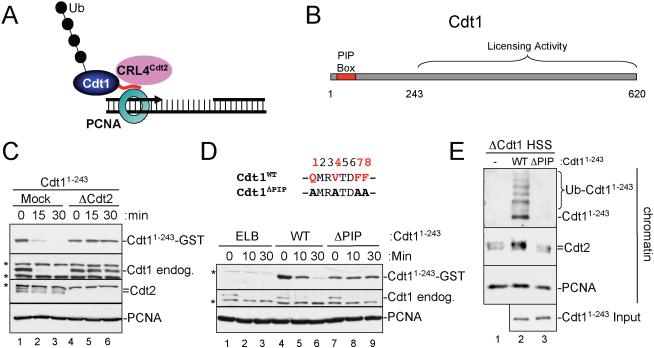
Figure 1. Cdt11-243 is destroyed in a CRL4Cdt2- and PCNA-dependent manner and interacts with chromatin-bound PCNA.
(A) Model of PCNA-Cdt1-CRL4Cdt2 complex on DNA
(B) Schematic of PCNA binding and licensing regions of Xenopus Cdt1.
(C) Cdt2-dependent destruction of endogenous Cdt1 and recombinant Cdt11-243 in HSS. Mock-depleted or Cdt2-depleted HSS was supplemented with 50 nM recombinant Cdt11-243-3xNLS-GST-Flag (Cdt11-243) and 5 ng/μl MMS plasmid. At different times, samples were blotted for the indicated proteins. Asterisks indicate background bands.
(D) Cdt11-243 destruction is PIP box-dependent. HSS was supplemented with 1 kb MMS DNA and Cdt11-243 or Cdt11-243/ΔPIP. At different times, samples were blotted for the indicated proteins. The sequence of the ΔPIP mutant is shown.
(E) Cdt1-depleted HSS was supplemented with immobilized, 1 kb MMS DNA and 2 mg/ml methyl ubiquitin. 50 nM Cdt11-243 or Cdt11-243/ΔPIP was added, as indicated, and after 10 minutes, chromatin was recovered from the extract, washed, and blotted for the indicated proteins.
To define the sequence elements that comprise the CRL4Cdt2 degron, and to understand why Cdt1 destruction depends on PCNAChromatin, we studied the DNA-damage induced proteolysis of Cdt1 in Xenopus egg extracts. Here, we demonstrate that Cdt1 binds much more efficiently to PCNAChromatin than to PCNAFree. In addition, an artificial CRL4Cdt2 substrate that binds to PCNAFree and PCNAChromatin alike only recruits CRL4Cdt2 in the context of PCNAChromatin. These results suggest that Cdt1 destruction is limited to S phase and damaged cells because Cdt1 only binds to PCNAChromatin and because DNA is a necessary co-factor for CRL4Cdt2 recruitment. We also show that the degron of Cdt1 contains a TD motif at positions 5 and 6 of the PIP box, which confers high affinity PCNAChromatin binding, and a positively charged amino acid four residues downstream of the PIP box that recruits CRL4Cdt2 to the Cdt1- PCNAChromatin complex. When transferred to Fen1, a PIP box protein that is normally not destroyed, these sequence elements induce CRL4Cdt2-dependent proteolysis. Together, the data support the idea that a select number of proteins in eukaryotic cells contain a specialized PIP box that assembles on PCNAChromatin to form the degron for CRL4Cdt2.
Results
Selective binding of Cdt1 to PCNAChromatin
We wanted to directly test whether Cdt1 has a higher affinity for PCNAChromatin than PCNAFree (Arias and Walter, 2006; Jin et al., 2006). Our goal was complicated by the fact that Cdt1 can associate with chromatin in two modes, via PCNAChromatin (Arias and Walter, 2006) and via ORC (Ferenbach et al., 2005; Maiorano et al., 2000), which obscures binding to PCNAChromatin (Figure S1). To circumvent this problem, we exploited the fact Cdt1's PIP box resides at the N-terminus of the protein (Arias and Walter, 2006; Nishitani et al., 2006; Senga et al., 2006), while the ORC-binding domain is located in residues 243-620 (Figure 1B; (Ferenbach et al., 2005)). We therefore prepared Xenopus Cdt11-243 with a GST and Flag tag on the C-terminus. Like endogenous Cdt1 (Jin et al., 2006), this protein was destroyed when added to a high-speed supernatant of egg cytoplasm (HSS) containing plasmid that had been damaged with Methyl methanesulfonate (MMS) (Figure 1C, top panel, lanes 1-3). As expected, Cdt11-243 was stable in Cdt2-depleted extract (Figure 1C, lanes 4-6). In addition, a protein in which the four consensus PIP box residues were mutated (Cdt11-243/ΔPIP) was stable (Figure 1D, top panel, compare lanes 4-6 and 7-9), as was Cdt11-243 after addition of a competitor PIP box peptide derived from p21 (Figure S2, lanes 4-6). Therefore, Cdt11-243 is destroyed in a PIP box and CRL4Cdt2-dependent manner, similar to endogenous Cdt1.
To examine the binding of Cdt11-243 to chromatin, we coupled MMS-treated 1 kb linear DNA to magnetic beads and added these to Cdt1-depleted HSS. The DNA fragment caused efficient destruction of the endogenous Cdt1, as well as recombinant Cdt11-243 (Figure 1D, lanes 4-6). In all experiments where chromatin was isolated, methyl-ubiquitin was added to help visualize Cdt1 ubiquitylation on chromatin (Arias and Walter, 2005). Cdt11-243, but not Cdt11-243/ΔPIP was recovered on the DNA beads (Figure 1E, top panel, compare lanes 2 and 3), and recovery was inhibited after PCNA depletion (Figure S3). Together, these results show that this assay detects a direct interaction between the Cdt11-243 PIP box and PCNAChromatin. Importantly, Cdt11-243 but not Cdt11-243/ΔPIP was ubiquitylated on chromatin and induced loading of Cdt2 (Figure 1E, compare lanes 2 and 3). The Cdt2 protein that bound to chromatin in the absence of added Cdt1 (Figure 1E, lane 1) does not reflect a non-specific background, since it disappears upon inhibition of PCNA loading by pre-incubation with a competitor PIP box peptide (Figure S1, compare lanes 5 and 7). Therefore, this basal Cdt2 is likely recruited to DNA via unknown, endogenous CRL4Cdt2 substrates other than Cdt1. Since we have eliminated the binding of Cdt1 to ORC, these results clearly show that Cdt1 interacts directly with PCNAChromatin during CRL4Cdt2-mediated ubiquitylation.
We next asked whether Cdt1 interacts more efficiently with PCNAChromatin than with PCNAFree. In the presence or absence of damaged DNA, Cdt11-243 or Cdt11-243/ΔPIP was added to HSS and then immunoprecipitated (IP'd) from the extract using the C-terminal Flag-tag. Equal amounts of Cdt11-243 were recovered from the extract under all conditions (Figure 2A, lanes 2, 3, 5, and 6). As expected, Cdt11-243 was only ubiquitylated in the presence of MMS DNA and a PIP box (Figure 2A, top panel, compare lane 2 with lanes 3 and 5). Strikingly, Cdt11-243 co-IP'd with PCNA only in the presence of damaged DNA (Figure 2A, PCNA panel, compare lanes 2 and 5) and a PIP box (Figure 2A, PCNA panel, compare lanes 2 and 3). Considering that the large majority of PCNA is soluble (Figure 2A, compare lanes 7-9 and 11-13), these results demonstrate that Cdt1 binds much more efficiently to PCNAChromatin than to PCNAFree. Importantly, CRL4Cdt2 only co-IP'd with Cdt11-243 in the presence of PCNAChromatin (Figure 2A, Cdt2 and Ddb1 panels, compare lanes 2 and 5), while Cdt11-243/ΔPIP did not interact with Cdt2 (lanes 3 and 6). Furthermore, it was possible to extract DNA from the anti-Flag IP only when Cdt11-243 contained a PIP box (Figure 2B, lane 2). In contrast to Cdt1, Fen1, a PIP box protein that is not destroyed by CRL4Cdt2 (Figure S4), was able to bind efficiently to PCNAFree (Figure 2C, lane 5), suggesting that not all PIP box proteins exhibit selective binding for PCNAChromatin. Together, these results demonstrate that Cdt1 interacts efficiently only with PCNAChromatin, providing one explanation for why Cdt1 is destroyed only in S phase and after DNA damage.
Figure 2. Specific assembly of the PCNA-Cdt1-CRL4Cdt2 complex on chromatin.
(A) 10 μl aliquots of HSS supplemented with methyl ubiquitin were mixed with 350 ng of 1 kb MMS DNA (lanes 1-3), no DNA (lanes 4-6), or 350 ng immobilized 1 kb MMS DNA template (lanes 7-9), as well as 250 nM Cdt11-243-Flag or Cdt11-243/ΔPIP-Flag for 10 minutes. For lanes 1-6, Cdt11-243 was precipitated with flag antibody and the IPs were analyzed. For lanes 7-10, the beads were recovered and associated proteins analyzed. In lanes 11-13, input extract was analyzed. The material recovered from the equivalent of 2 μl of HSS was loaded in lanes 1-10, whereas 1 μl of HSS was loaded in lanes 11-13. HC, heavy chain; LC, light chain.
(B) For reactions described in lanes 1-3 of Panel (A), DNA was extracted from the Flag IP, 50% was analyzed on an agarose gel and then stained with SYBER gold alongside the total input DNA, which was also extracted, but not exposed to extract.
(C) 10 μl aliquots of HSS supplemented with methyl ubiquitin were mixed with 350 ng of 1 kb MMS DNA (lanes 1-3) or 350 ng immobilized 1 kb MMS DNA template (lanes 7-9), as well as 250 nM Flag-Fen1 or Flag-Fen1ΔPIP for 10 minutes. For lanes 1-6, Flag-Fen1 was precipitated with flag antibody and the IPs were analyzed. For lanes 7-10, the DNA was recovered and associated proteins analyzed. In lanes 11-13, input extract was analyzed. The material recovered from the equivalent of 2 μl of HSS was loaded in lanes 1-10, whereas 0.5 μl of HSS was loaded in lanes 11-13.
A PIP box peptide is sufficient to recruit CRL4Cdt2 to chromatin
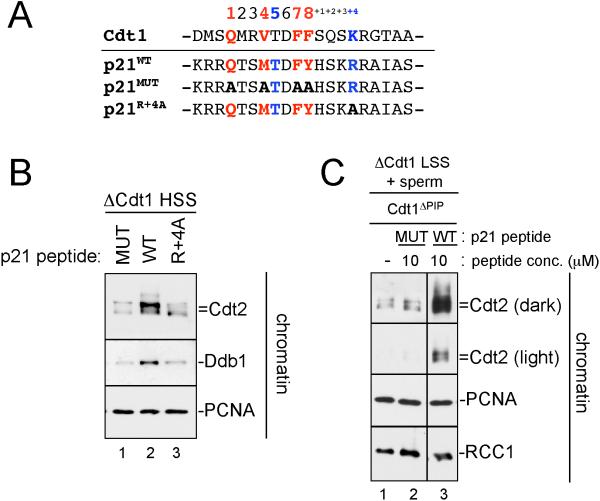
Previous results showed that a short peptide derived from Cdt1 promotes DNA damage-induced destruction (Nishitani et al., 2006; Senga et al., 2006). We wanted to determine whether a PIP box peptide is sufficient to recruit CRL4Cdt2 to PCNAChromatin, as predicted by these results. We prepared a synthetic peptide containing the Cdt1 PIP box, but it was highly insoluble (data not shown). We therefore used a 20 amino acid peptide containing the PIP box of p21 ((Mattock et al., 2001), Figure 3A), as this protein was recently identified as a target of CRL4Cdt2 (Abbas et al., 2008; Kim et al., 2008; Nishitani et al., 2008). When added to Cdt1-depleted HSS, the p21 peptide but not a mutant peptide lacking core PIP box residues was able to recruit Cdt2 and Ddb1 onto immobilized MMS DNA above basal levels (Figure 3B, lanes 1 and 2).
Figure 3. The p21 PIP box peptide is sufficient to recruit CRL4Cdt2 to chromatin.
(A) Sequence of human p21 PIP box peptides used in panels (B) and (C) aligned with Xenopus Cdt1.
(B) HSS was mixed with methyl ubiquitin and immobilized 1 kb MMS DNA. Three minutes after the addition of DNA, 10 μM p21ΔPIP peptide (MUT), p21 peptide (WT), or p21R+4A peptide (R+4A) were added to the reaction. Samples were stopped with sample buffer after 10 minutes and blotted for the indicated proteins.
(C) LSS was supplemented with methyl ubiquitin, sperm chromatin and buffer, p21ΔPIP peptide (Mut) or p21 peptide (WT). After 45 minutes, chromatin was isolated, washed, and blotted for the indicated proteins. All samples were run on the same gel, but some irrelevant lanes were removed between lanes 2 and 3.
We wanted to determine whether a PIP box is also sufficient to recruit CRL4Cdt2 to PCNAChromatin in the context of chromosomal DNA replication. To this end, we employed a low-speed supernatant of egg cytoplasm (LSS), which assembles added sperm chromatin into nuclei that undergo chromosomal DNA replication and support Cdt1 destruction (Arias and Walter, 2006). Endogenous Cdt1 was depleted from LSS and replaced with Cdt1ΔPIP to allow DNA replication but prevent Cdt1-mediated CRL4Cdt2 loading. Addition of p21 peptide but not mutant peptide to this extract induced efficient CRL4Cdt2 recruitment (Figure 3C, compare lanes 2 and 3). Together with previous data (Chuang et al., 2005; Nishitani et al., 2006; Senga et al., 2006; You et al., 2002), our results demonstrate that a short, PIP box containing peptide is sufficient to recruit the CRL4Cdt2 ubiquitin ligase to PCNAChromatin.
Sequence alignment identifies a potential `PIP degron' motif
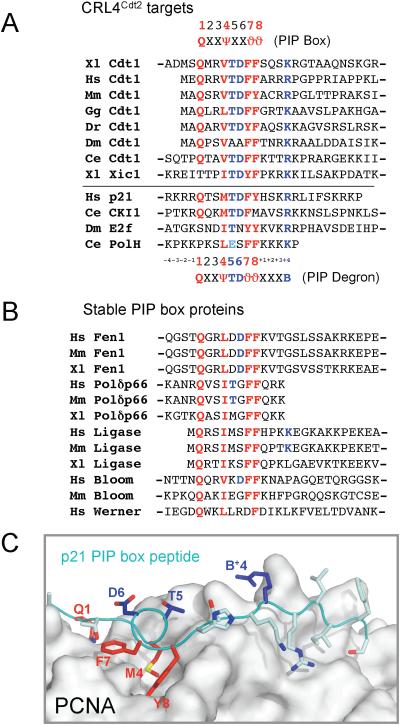
A critical question is why some PIP box proteins, such as Cdt1, Xic1, p21, and E2F are destroyed by CRL4Cdt2, whereas the vast majority of PIP box proteins are stable. Given that a short PIP box peptide is sufficient to recruit CRL4Cdt2, we reasoned that the PIP boxes of CRL4Cdt2 substrates might contain sequences that distinguish them from the PIP boxes of stable PCNA binding proteins. We therefore aligned the PIP boxes of Cdt1 from multiple species with that of Xic1 (Figure 4A; other CRL4Cdt2 targets had not yet been identified), and compared this set to the PIP boxes from numerous proteins, which are presumably stable (Figure 4B). For example, under conditions that induce Cdt1 destruction, endogenous Fen1 is completely stable (Figure S4). When comparing PIP boxes of different proteins, we refer to the position of an amino acid relative to the PIP box, rather than its absolute position within the amino acid sequence of the protein (Figure 4A). The alignment revealed that Cdt1 and Xic1 contain two unique features. First, with the exception of Drosophila Cdt1, all the proteins in this group contain a TD motif at positions 5 and 6 of the PIP box (Figure 4A, in blue). In addition, all the proteins contain a positively charged residue four amino acids downstream of the PIP box (Figure 4A; “K/R+4”). Importantly, we found no examples of canonical PIP boxes that contain both of these features (Figure 4B). Together, these observations suggested that CRL4Cdt2 might recognize a degron with the following consensus: Q-x-x-Ψ-T-D-ϑ-ϑ-x-x-x-B (Ψ = I, L, M or V, ϑ = Y or F, B = K or R). In support of this idea, the recently identified CRL4Cdt2 substrates (human p21, worm CKI, fly E2F, and worm DNA pol η) also conform closely to this consensus (Figure 4A).
Figure 4. Sequence alignment of PIP box proteins.
(A) Alignment of PIP boxes from proteins that are known to be targeted for destruction by CRL4Cdt2. Canonical PIP box residues are shown in red and putative “degron-specific” residues are shown in blue. The PIP box consensus, and the putative PIP degron consensus are shown (ψ = I/L/M/V, J = Y/F, B=K/R). The absence of a dash indicates the N or C terminus.
(B) Alignment of PIP boxes from proteins that are not likely targets of CRL4Cdt2.
(C) An image of the PCNA-p21 peptide co-crystal structure (Gulbis et al., 1996) was generated using PDB accession number 1AXC and PyMOL (www.pymol.org).
Cdt1 T5 and K+4 residues are essential for Cdt1 destruction
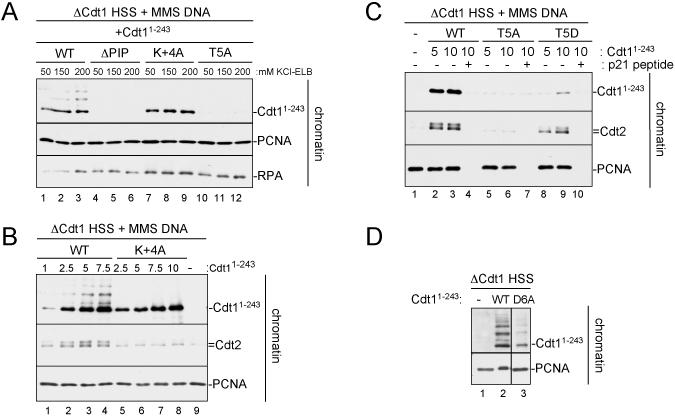
The above sequence alignment predicts that T5, D6, and B+4 are required for PCNA-dependent protein destruction. Indeed, Dutta and colleagues previously showed that mutation of T5 to alanine in human Cdt1 reduced damage-dependent destruction (Senga et al., 2006). Consistent with these data, mutation of T5 to alanine in Xenopus Cdt11-243 (Cdt11-243/T5A) abolished destruction of the protein in response to DNA damage (Figure 5A, compare lanes 1-3 and 7-9). Similar results were obtained when the T5A mutation was made in the context of full-length Cdt1 and assayed for DNA damage or replication dependent destruction (Figure S5A and S5B). Dutta and colleagues also reported that mutation of D6 to alanine had no effect on Cdt1 stability (Senga et al., 2006). We found that while Cdt11-243/D6A was still degraded, the rate was reduced compared to the WT protein (Figure 5B, compare lanes 1-4 to 9-12).
Figure 5. Mutational analysis of the Cdt1 PIP box.
(A) HSS was mixed with MMS plasmid and 50 nM recombinant Cdt11-243, Cdt11-243/ΔPIP, Cdt11-243/T5A, or Cdt11-243/K+4A. Reactions were stopped at different times and blotted for Cdt1 or Cdt11-243-GST (anti-GST antibody).
(B) HSS was mixed with MMS plasmid and 50 nM recombinant Cdt11-243, Cdt11-243/ΔPIP, or Cdt11-243/D6A. Reactions were stopped at different times and blotted as in (A).
(C) HSS was mixed with MMS plasmid and 50 nM recombinant Cdt11-243, Cdt11-243/ΔPIP, Cdt11-243/R+5A or Cdt11-243/T5D. Reactions were stopped at the indicated times and blotted as in (A).
More recently, simultaneous mutation of positions +3, +4, and +5 in human p21 (KRR to AAA) was shown to prevent destruction (Nishitani et al., 2008), suggesting that at least one positively charged residue beyond the core PIP box is important for PCNA-dependent destruction. Consistent with the sequence alignment shown above, we found that mutation of only the lysine at position +4 (Cdt11-243/K+4A) resulted in a completely stable protein (Figure 5A). In contrast, Cdt11-243/R+5A was still degraded, albeit at a reduced rate (Figure 5C, lanes 5-8). Together, these data argue that the T5 and K+4 residues are essential for Cdt1 destruction. In addition, D6 and R+5, while not essential, enhance the rate of destruction.
The TD motif confers highly efficient PCNAChromatin binding, whereas the K+4 residue is required for CRL4Cdt2 recruitment
We next wished to determine what contribution the TD and K+4 residues make to Cdt1 PCNAChromatin binding and CRL4Cdt2 recruitment. The co-crystal structure of soluble PCNA with p21's PIP box reveals that the core PIP box resides (Figure 4C, red) interact directly with the hydrophobic pocket of PCNA (Gulbis et al., 1996). Interestingly, T5, D6 and B+4, the residues that are specific to CRL4Cdt2 substrates, all protrude from the surface of PCNA into solution (Figure 4C, blue). Although this structure does not contain DNA, it suggests that T5, D6, and B+4 might be dispensable for PCNA binding, while being specifically required to recruit CRL4Cdt2 to the PIP box- PCNAChromatin complex. Consistent with this prediction, Cdt11-243/K+4A bound as efficiently to PCNAChromatin as Cdt11-243 (Figure 6A, compare lanes 1-3 and 7-9). However, unlike Cdt11-243, Cdt11-243/K+4A was not ubiquitylated (Figure 6A), consistent with it being completely stable (Figure 5A). Notably, we found no concentration at which Cdt11-243/K+4A was able to recruit Cdt2 above the basal level seen in the absence of added Cdt1 (Figure 6B, compare lanes 5-8 with lane 9). Similarly, when the basic residue at +4 in the p21 peptide was changed to an alanine, it could no longer recruit Cdt2 to chromatin (Figure 3B, lane 3). In contrast, Cdt11-243/R+5A bound PCNAChromatin and recruited Cdt2, but not quite as efficiently as Cdt11-243 (Figure S6, compare lanes 1, 2 and 4, 5), consistent with Cdt11-243/R+5A being destroyed at a slightly reduced rate (Figure 5C). We conclude that K+4 is not essential for efficient PCNAChromatin binding, but rather is specifically required for the recruitment of CRL4Cdt2 to the Cdt1-PCNAChromatin complex.
Figure 6. PCNAChromatin binding and Cdt2 recruitment of Cdt11-243 mutants.
(A) Cdt1-depleted HSS was supplemented with immobilized 1 kb MMS DNA, methyl ubiquitin, and 50 nM Cdt11-243, Cdt11-243/ΔPIP, Cdt11-243/K+4A or Cdt11-243/T5A. After 10 minutes, the beads were recovered, washed with buffer containing different concentrations of salt, and blotted for the indicated proteins.
(B) Cdt1-depleted HSS was mixed with immobilized 1 kb MMS DNA, methyl ubiquitin and 50 nM (“1x”) to 500 nM (“10x”) Cdt11-243 or Cdt11-243/K+4A. After 10 minutes, the beads were recovered and blotted for the indicated proteins.
(C) Cdt1-depleted HSS was mixed with immobilized 1 kb MMS DNA, methyl ubiquitin, and 250 (5x) or 500 nM (10x) Cdt11-243, Cdt11-243/T5A, or Cdt11-243/T5D. Chromatin-bound proteins were analyzed as in (B).
(D) Cdt1-depleted HSS was mixed with immobilized 1 kb MMS DNA, methyl ubiquitin, and 50 nM Cdt11-243 or Cdt11-243/D6A. Chromatin-bound proteins were analyzed as in (B). All samples were run on the same gel, but some irrelevant lanes were removed between lanes 2 and 3.
Strikingly, Cdt11-243/T5A did not bind detectably to PCNAChromatin (Figure 6A, lanes 10-12), which explains why it failed to be destroyed (Figure 5A) or recruit Cdt2 (Figure 6C). Like Cdt11-243/T5A, full length Cdt1T5A also failed to bind efficiently to PCNA on chromatin or to recruit Cdt2 (data not shown). We considered the possibility that an alanine is not well tolerated at this position of Cdt1's PIP box. Indeed, alanine is significantly under-represented at position 5 of the PIP box in Xenopus proteins. In contrast, a considerable number of these proteins contain an aspartic acid at this position. We therefore prepared Cdt11-243/T5D and found it was destroyed, albeit slowly (Figure 5C, lanes 9-12), and it still bound poorly to PCNAChromatin (Figure 6C, lanes 8-9). When we titrated Cdt11-243/T5D into the extract, we found that at high levels, it was able to recruit Cdt2 efficiently (Figure 6C, lanes 8-9). Based on these results, we conclude that T5 does not form a critical interaction with CRL4Cdt2, but instead, mediates tight binding of Cdt1 to PCNAChromatin (see also below). Similar to Cdt11-243/T5D and Cdt11-243/R+5A, Cdt11-243/D6A did not bind PCNAChromatin as well as Cdt11-243 (Figure 6D, compare lanes 2 and 3), but it was able to recruit Cdt2 when added at higher concentrations (data not shown).
In summary, in order to be destroyed, Cdt1 requires a canonical PIP box and a basic residue at position +4 to recruit CRL4Cdt2. In addition, T5, D6, and a basic residue at position +5 each enhance the binding of Cdt1 to chromatin-bound PCNA, with T5 providing the most important and essential contribution.
Binding of Cdt1 to PCNAChromatin does not require Cdt2
We have shown that the K+4A mutant of Cdt1 binds to PCNAChromatin, but does not interact with CRL4Cdt2 (Figure 6A and 6B), indicating that the ligase is not required for efficient binding of Cdt1 to PCNA. In agreement with this conclusion, Cdt11-243 bound to PCNAChromatin with similar efficiency in control-depleted and Cdt2-depleted extracts (Figure S7, compare lanes 1 and 3). These results establish a hierarchical assembly pathway, in which Cdt1 first docks onto chromatin-bound PCNA, followed by recruitment of CRL4Cdt2.
PCNA- and CRL4Cdt2-dependent destruction of a canonical PIP box protein
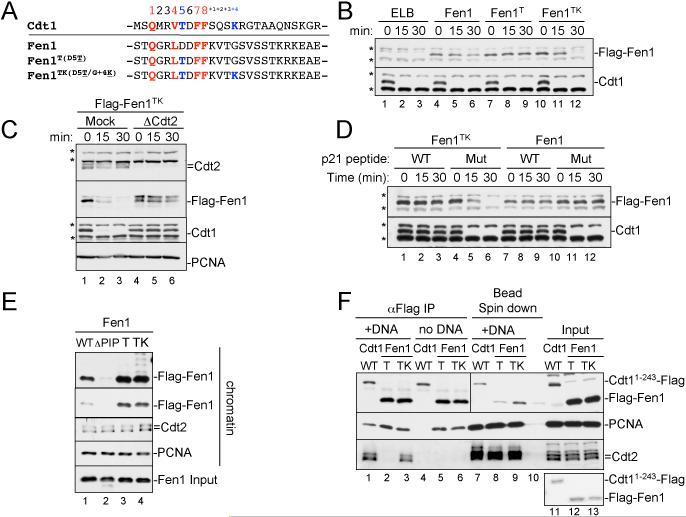
To determine whether the degron motifs we defined in Cdt1 are sufficient to induce CRL4Cdt2-dependent proteolysis, we examined Fen1, a PIP box protein which is normally not destroyed (Figure S4). To this end, the aspartic acid in position 5 of Fen1's PIP box (Figure 7A) was changed to a threonine (“Fen1T”), the glycine in the +4 position was changed to a lysine residue (“Fen1K”), or both residues were altered (“Fen1TK”). Like Fen1, Fen1 K was not destroyed in response to damaged DNA (data not shown). Fen1T was also not destroyed (Figure 7B, compare lanes 4-6 and 7-9). However, Fen1T (but not Fen1K) bound more efficiently than Fen1 to PCNAChromatin (Figure 7E, compare lanes 1 and 3 and data not shown), consistent with the results from Cdt1, which indicate that PIP boxes containing T5 bind exceptionally well to PCNAChromatin.
Figure 7. The PIP degron is portable.
(A) Sequence comparison of Xenopus Cdt1's PIP box with that of Xenopus Fen1 and various mutants of Fen1.
(B) HSS was mixed with MMS plasmid and 50 nM recombinant Fen1, Fen1T, or Fen1TK. Reactions were stopped at the indicated times and blotted for Cdt1 and the Flag peptide, to visualize Flag-Fen1.
(C) Mock-depleted or Cdt2-depleted HSS was supplemented with 5 ng/μl MMS plasmid and 50 nM Fen1 or Fen1TK. Samples were blotted for the indicated proteins.
(D) HSS was incubated with 50 μM mutant or WT p21 PIP peptide and then supplemented with 50 nM recombinant Fen1TK or Fen1 and 5 ng/μl MMS plasmid. At different times, samples were blotted for the indicated proteins.
(E) HSS was mixed with immobilized 1 kb MMS DNA, methyl ubiquitin, and 200 nM Fen1, Fen1ΔPIP, Fen1T or Fen1TK. After 10 minutes, the beads were recovered and blotted for the indicated proteins.
(F) 10 μl aliquots of HSS supplemented with methyl ubiquitin were mixed with 350 ng of 1 kb MMS DNA (lanes 1-3) or 350 ng immobilized 1 kb MMS DNA (lanes 7-9), as well as 250 nM Cdt11-243-Flag, Flag-Fen1T or Flag-Fen1TK for 10 minutes. Lane 10 contained all three proteins, HSS and magnetic beads, but no DNA. For lanes 1-6, Flag-tagged proteins were precipitated with flag antibody and the IPs were analyzed. For lanes 7-10, the beads were recovered and associated proteins analyzed. In lanes 11-13, input extract was analyzed. The material recovered from the equivalent of 2 μl of HSS was loaded in lanes 1-10, whereas 0.5 μl of HSS was loaded in lanes 11-13. All samples were from the same experiment, but for the anti-Flag panel, a darker exposure of lanes 7-13 is shown. Lower panel, coomassie gel showing that equal amounts of recombinant proteins were used. Note that Cdt11-243-Flag contains a single C-terminal Flag tag, whereas Flag-Fen1 contains 2 N-terminal Flag tags.
Unlike Fen1, Fen1TK was destroyed in the presence of DNA damage (Figure 7B, lanes 10-12) and in a manner that depended on Cdt2 (Figure 7C, compare lanes 1-3 and 4-6). In addition, Fen1TK destruction was PCNA-dependent, since addition of a competing PIP box peptide (but not a mutated peptide) also stabilized the protein (Figure 7D, compare lanes 1-3 and 4-6). Finally, Fen1TK was ubiquitylated, and it recruited Cdt2 to the chromatin above basal levels (Figure 7E, compare lanes 1 and 4). Therefore, the specific sequence elements we identified in Cdt1 as being important for PCNA binding and CRL4Cdt2 recruitment are transferable to another substrate. We propose to call these sequence elements a `PIP degron' (see discussion).
CRL4Cdt2 only binds to the Cdt1-PCNAChromatin complex
Having defined the CRL4Cdt2 degron, we revisited the question of why Cdt1 destruction is critically dependent on PCNAChromatin. We were curious whether a PIP degron protein that is able to bind PCNAFree would also recruit CRL4Cdt2. Unlike Cdt1, Fen1 binds efficiently to PCNAFree (Figure 2). We therefore examined the binding of Fen1TK, which contains a PIP degron, to Cdt2 in the presence of PCNAFree or PCNAChromatin. Like Fen1 (Figure 2C), Fen1TK bound efficiently to PCNAFree, demonstrating that a PIP degron does not necessarily impose selective binding to PCNAChromatin (Figure 7F, compare lanes 3 and 6). In addition, Fen1T and Fen1TK both bound PCNAFree and PCNAChromatin more efficiently than Fen1 (Figure 7E and data not shown), demonstrating that the T5 position of the PIP box enhances binding to PCNAFree and PCNAChromatin alike. Most importantly, although Fen1TK bound stably to PCNAFree, it did not recruit Cdt2 in this context (Figure 7F, compare lanes 3 and 6). Therefore, binding of a PIP degron to PCNAFree is not sufficient to promote assembly of the Cdt1-PCNA-CRL4Cdt2 complex.
Discussion
In this paper, we identified the cis-acting sequences within Cdt1 that target it for destruction via CRL4Cdt2. Two elements are important to promote efficient Cdt1 destruction: a PIP box that binds PCNA with high affinity, and a basic residue at the +4 position relative to the PIP box that recruits CRL4Cdt2. In addition we identify two mechanisms that insure that CRL4Cdt2 targets are destroyed only in the presence of chromatin-bound PCNA (PCNAChromatin), limiting the activity of this pathway to S phase and damaged cells. Thus, our data show that CRL4Cdt2 recognizes a highly unusual degron that is formed on chromatin via the interaction between a specialized PIP box and PCNA.
Coupling of CRL4Cdt2-dependent proteolysis to PCNAChromatin
In this paper, we identify two mechanisms which explain our previous observation that Cdt1 destruction is strictly dependent on PCNAChromatin (Arias and Walter, 2006; Jin et al., 2006). First, we showed that Cdt1 binds efficiently to PCNAChromatin, whereas there is no detectable interaction with PCNAFree (Figure 2). Consistent with this observation, Cdt1 only co-IP'd with CRL4Cdt2 in the presence of PCNAChromatin. The highly selective binding of Cdt1 to PCNAChromatin is not a general property of PIP box proteins, as illustrated by Fen1, which also binds to PCNAFree. Selective PCNAChromatin-binding is not an obligatory property of the PIP degron, since Fen1TK, which was engineered to contain the PIP degron, still binds efficiently to PCNAFree (Figure 7). The mechanistic basis of Cdt1's selectivity for PCNAChromatin is presently unclear. It could involve a conformational change in PCNAChromatin, chromatin-specific modification of PCNA, or cooperative binding of Cdt1 to PCNA and DNA. In support of the latter model, mouse Cdt1 is reported to bind DNA (Yanagi et al., 2002), and the destruction of Drosophila E2F by CRL4Cdt2 was recently reported to require the DP protein (Shibutani et al., 2008), which binds E2F and recruits it to DNA.
We discovered a second mechanism that couples CRL4Cdt2-dependent ubiquitylation to PCNAChromatin when we examined Fen1TK. This mutant of Fen1, which contains a PIP degron, binds efficiently to PCNAFree but is only able to recruit CRL4Cdt2 in the context of PCNAChromatin (Figure 7F). Thus, even if a PIP degron protein should inadvertently bind to PCNAFree, it will not be destroyed. We speculate that the conformations of PCNAFree and PCNAChromatin differ, and that Cdt2 is able to interact with the PIP degron only when it is displayed on PCNAChromatin. However, another explanation is that other, unknown DNA-binding proteins cooperate with PCNAChromatin to recruit CRL4Cdt2, although such factors would have to be shared between the replication and repair machineries. The two mechanisms we have described, which couple CRL4Cdt2-dependent proteolysis to PCNAChromatin, together insure that this pathway is only active in S phase and after DNA damage. It remains to be seen whether bona fide CRL4Cdt2 substrates other than Cdt1 employ both mechanisms.
Interestingly, it has been reported that in the absence of DNA, Cdt1 and p21 co-IP with PCNAFree (Abbas et al., 2008; Arias and Walter, 2006; Hu and Xiong, 2006; Kim et al., 2008; Nishitani et al., 2008; Nishitani et al., 2006). However, the experiments were performed with purified proteins or in lysates of cells overexpressing these proteins, calling into question whether such interactions really occur in vivo. Even if they do, our results show that a PIP degron-PCNAFree complex does not support CRL4Cdt2 recruitment. In addition, Cdt1 and p21 were found to bind to CRL4Cdt2 in the absence of PCNA or a PIP box (Abbas et al., 2008; Higa et al., 2006; Hu et al., 2004; Kim et al., 2008; Nishitani et al., 2008). These observations are difficult to reconcile with the fact that these substrates are only destroyed in S phase and after DNA damage, unless PCNAChromatin is not only required to join CRL4Cdt2 with its substrates, but also to facilitate ubiquitin transfer.
Anatomy of the PIP degron
In this study, we identify two functional elements that are necessary and sufficient to create a degron for CRL4Cdt2. The first is a basic residue four amino acids downstream of the PIP box. Every known CRL4Cdt2 substrate contains a K or R residue at the +4 position (Figure 4A). This residue is dispensable for substrate binding to PCNAChromatin but is essential to recruit CRL4Cdt2 to the substrate-PCNAChromatin complex. In support of this conclusion, we show that Cdt11-243/K+4A is completely stable, binds PCNAChromatin normally, but fails to recruit Cdt2 to chromatin. Moreover, Fen1 (Figure 7) and p21 peptide (Figure 3) only recruit CRL4Cdt2 if a positively charged amino acid is present at the +4 position. Although numerous CRL4Cdt2 targets contain multiple positive charges downstream of the PIP box, only the +4 position is essential. This is apparent from the fact that some targets have only a single positive charge, which is always at the +4 position. In addition, several proteins that were engineered to contain only the positive charge at the +4 position are destroyed (Cdt11-243/R+5A and Fen1TK). Although Cdt11-243/R+5A is destroyed slowly, this is not due to defective CRL4Cdt2 recruitment, but rather due to poor binding to PCNAChromatin (see below). Thus, the basic residue at +4 is an indispensable determinant of the degron that enables CRL4Cdt2 recruitment.
The second element required for efficient destruction by CRL4Cdt2 is a PIP box. Importantly, for the PIP box to promote destruction, it appears that it must have a very high affinity for PCNAChromatin. For most CRL4Cdt2 substrates, high affinity PCNAChromatin binding is conferred by a TD motif, which is located at positions 5 and 6 of the PIP box (Figure 4A). Thus, we found that mutation of T5 to A dramatically decreases the binding of Cdt1 to PCNAChromatin, thereby preventing destruction, and that insertion of a T5 in Fen1 strongly enhances PCNAChromatin binding and enables proteolysis when a basic residue is also present at +4. These results are further supported by data from Xic1, a likely CRL4Cdt2 target, where a T5A mutation blocked proteolysis and greatly attenuated PCNA binding (Chuang et al., 2005). Interestingly, Cdt11-243/T5D was still destroyed, albeit with reduced kinetics. Although this raised the possibility that T5 might be phosphorylated, numerous lines of evidence in diverse systems argue against this idea (see Supplemental Discussion). Instead, it seems that D5 simply allows better binding to PCNAChromatin than A5 (Figure 6C). Importantly, we also found that mutation of D6 to A in Cdt1 significantly reduced PCNAChromatin binding and slowed, but did not eliminate, Cdt1 destruction. Together, the data argue that a TD motif confers high affinity binding to PCNAChromatin, and that in most CRL4Cdt2 substrates, this motif is essential for destruction.
Notably, three proteins that are destroyed by CRL4Cdt2 (worm DNA pol η, fly Cdt1, and fly E2F) lack the TD motif (Figure 4A). We speculate that these proteins are destroyed efficiently despite the absence of a TD motif because they achieve high affinity PCNAChromatin binding by other means. In the case of fly Cdt1, it is conceivable that an AA motif at positions 5 and 6 is just as adept at binding PCNAChromatin as TD. However, we consider it more likely that other sequence elements compensate for the absence of TD motifs in these proteins. Specifically, it is conspicuous that all three proteins contain many positively charged amino acids downstream of the PIP box (at positions +1 to +5), and in the case of pol η, immediately upstream of the PIP box (at positions -1 to -4). Interestingly, the co-crystal structure of p21 and PCNA shows that the analogous basic residues in p21 form salt bridges with acidic residues in PCNA, and PIP residues -1 through -4 form poorly ordered ionic interactions with the C-terminus of PCNA (Gulbis et al., 1996). In support of these interactions being relevant for proteolysis, we found that Cdt11-243/R+5A exhibits reduced PCNAChromatin binding and destruction kinetics. Importantly, this was not due to defective CRL4Cdt2 recruitment, because at equivalent levels of chromatin-binding, Cdt11-243/R+5A and Cdt11-243 recruited the same amount of ligase (Figure S6). Further, in p21, mutation of the -4RKRR-1 motif inhibited destruction (Nishitani et al., 2008). Together, these data suggest that positively charged residues might be able to compensate for the absence of a TD motif and/or an imperfect core PIP box in some CRL4Cdt2 substrates.
Although the -4RKRR-1 motif appears to play a role in p21 destruction (assuming that mutation of these residues to alanines did not grossly disorder the protein's structure), a cluster of positive charges upstream of the PIP box is clearly not a universal feature of the PIP degron. Thus, although numerous CRL4Cdt2 substrates contain positively charged amino acids in this area (or they are located at the extreme N-terminus, which provides a positive charge), several substrates (frog and worm Cdt1, Fen1TK) lack positive charges (Figure 4A). Therefore, although positively charged residues upstream of the PIP box might enhance PCNAChromatin binding, they are not essential elements of the PIP degron.
In summary, our data suggest that as long as a protein binds to PCNAChromatin with sufficient affinity, contains a basic residue at +4 and has an accessible degron, it will be a target of CRL4Cdt2. We speculate that high affinity PCNAChromatin binding is essential for proteolysis because it allows processive ubiquitylation of Cdt1 by CRL4Cdt2 and thereby mediates synthesis of polyubiquitin chains. Although the basic residue at the +4 position is the only residue that is essential for recruitment of CRL4Cdt2 to the PCNAChromatin-PIP box complex, the binding must involve other interactions. CRL4Cdt2 might recognize other general features of the PIP box (such as the hydrophobic residues at 4, 7, and 8 positions) and/or specific residues that lie on the surface of PCNAChromatin.
PCNA binding affinity and canonical PIP box proteins
A large number of stable proteins that function in chromosome metabolism bind to PCNAChromatin via a PIP box (Moldovan et al., 2007), but it appears that most of these have non-optimal PCNA binding affinity. This conclusion is based on the fact that the vast majority of PIP box proteins lack a TD motif, and the finding that in all cases where the TD motif has been mutated, binding to PCNAChromatin or PCNAFree is diminished (Cdt1 in this paper, (Chuang et al., 2005; Nakanishi et al., 1995; Warbrick et al., 1995)). Moreover, when we introduced a TD motif into Fen1's PIP box, its binding to PCNAChromatin increased substantially (Figure 7E). We therefore conclude that the TD motif can enhance PCNAChromatin binding in the context of many and perhaps all PIP boxes, and the question arises why more PCNA-interacting proteins do not contain this motif. Given the number of proteins that need to access PCNAChromatin in the course of chromosomal DNA replication (polymerases, ligases, nucleases, chromatin assembly factors), and the requirement for frequent exchange between these factors, we speculate that replication would be adversely affected if the affinity of these proteins for PCNAChromatin was very high. In contrast, high affinity PCNAChromatin binding by CRL4Cdt2 substrates is not deleterious since they are rapidly destroyed upon binding to PCNAChromatin.
A new paradigm for temporally regulated proteolysis?
Most E3 ubiquitin ligases recognize short degron motifs, and regulation of proteolysis usually involves post-translational modifications of the substrate or the ligase, most notably by phosphorylation. The degron for CRL4Cdt2 departs from this paradigm in two ways. First, there is currently no evidence that post-translational modifications regulate the binding of CRL4Cdt2 to its substrates. Rather, binding requires the assembly of a cell-cycle regulated structure, PCNAChromatin, and this insures that substrates are only destroyed in S phase and after DNA damage. Second, the degron is formed via the interaction of two proteins. Since the only unique determinant for ligase recruitment in the substrate is a single basic residue, it is almost certainly true that other essential determinants for CRL4Cdt2 recognition are provided by residues on PCNA. Thus, CRL4Cdt2 is an example of a ubiquitin ligase that appears to recognize a specific surface created by two different proteins. We will be surprised if this principle is not exploited repeatedly by cells to couple proteolysis to the formation of other transient protein-protein interactions.
Experimental Procedures
Egg extract and immunological methods
HSS, LSS (Walter and Newport, 2000; Walter et al., 1998) and chromatin spin downs (Arias and Walter, 2005) were performed as described. We used previously described antibodies against Orc2 (Walter and Newport, 1998), Cdt1 (Arias and Walter, 2005), RCC1 (Dasso et al., 1992), Ddb1 (Arias and Walter, 2006), RPA (Walter and Newport, 2000), Cdt2 (Jin et al., 2006), Fen1 (Cell Signaling Technology), GST (New England Bio Labs), M2 and Rabbit Flag (Sigma), and PCNA (Santa Cruz sc-056). Whenever a Western blot was performed with anti-Flag or anti-GST antibody, the protein being detected is indicated with its tag to the right of the panel.
For Cdt11-243-Flag and Flag-Fen1 IPs, HSS extract was first precleared with anti-Flag-agarose beads (Sigma), then recombinant proteins and DNA were added and 10 min. later, Flag-tagged proteins were IP'd using anti-Flag-agarose resin. The resin was washed 3 × in 2.5 mM MgCl2, 150 mM KCl, 250 mM sucrose, 10 mM HEPES, pH 7.7, 0.6% Triton and then resuspended in sample buffer for SDS-PAGE. DNA from the Flag IP was extracted, run on an agarose gel and stained with SYBER gold (Molecular Probes). The p21 WT and ΔPIP peptides were previously published (Arias and Walter, 2006), and the p21R155A peptide (CKRRQTSMTDFYHSKARAIAS) was synthesized by the Tufts University Core Facility, Boston, MA.
Plasmid construction and protein purification
Recombinant full length Xenopus Cdt1 was purified as described (Arias and Walter, 2005). Recombinant Xenopus Cdt11-243-3xNLS-GST-Flag was cloned into pDONR201 and produced in SF9 cells using the BaculoDirect Baculovirus Expression System (Invitrogen). All Cdt1 mutants were cloned using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). See supplemental data for primers and cloning information. For purification of Cdt11-243, insect cell pellets were lysed in lysis buffer (50 mM Tris pH 7.5, 300 mM NaCl, 50 mM KCl, 5 mM MgCl2, 0.5% Triton X-100 and 10 mM Imidazole), sonicated, and spun for 30 min at 30,000 × g. The supernatant was loaded onto Ni-NTA beads (Qiagen), washed in lysis buffer with 30 mM Imidazole and Cdt11-243 was eluted in lysis buffer with 250 mM Imidazole. The eluted Cdt11-243 was incubated with GST-sepharose (Amersham) which was washed to remove Imidazole and Triton (50 mM Tris pH 7.5, 300 mM NaCl, 50 mM KCl, 5 mM MgCl2). Cdt11-243 was eluted with 10 mM Tris pH 8.0, 10 mM reduced glutathione, 250 mM NaCl, 10% glycerol.
See suppelemental data for details on Xenopus Fen1 cloning. For purification of Fen1, pET28b-2xFlag-Fen1-GST was transformed into BL-21, grown at 37°C to an OD600 nm of 0.6 and induced with 1mM IPTG overnight at 19°C. Pellets were frozen at - 80°C and then purified in buffer A plus protease inhibitors (500 mM NaCl, 20 mM Hepes pH 8, 10% glycerol, 2 mM ß-mercapto-ethanol, 0.1% NP-40) (Hohl et al., 2007), except that the lysate was loaded onto Ni-NTA resin (Qiagen) in buffer A containing 7.5 mM Imidazole. The resin was washed with buffer A containing 20 mM Imidazole and Fen1 eluted in 250 mM Imidazole. A second purification step using Glutathione Sepharose™ 4 Fast Flow was used to remove the Imidazole, transfer Fen1 into freezing buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM DTT), and cleave the GST-tag from the C-terminus using PreScission Protease (GE Healthcare).
MMS DNA preparation and bead spin-down assay
Methylated DNA was generated as described (Stokes and Michael, 2003). A 2 x biotinylated double stranded 1 kb linear DNA was generated by PCR (see supplemental data for primer sequences). The DNA product was then purified using a PCR purification kit (Qiagen), MMS treated and then coupled to M-280 Streptavidin Dynabeads (Invitrogen). 100 ng of Biotinylated MMS treated PCR product was used per 10 μg of Dynabeads. DNA and beads were bound for 2 hr at 25°C in 10 mM Tris 8.0, 100 mM NaCl, 1 mM EDTA, then incubated overnight at 4°C in the presence of excess streptavidin to block any free biotin ends and finally washed 3 times. 60-70% of DNA bound to beads. To spin down MMS-DNA beads, a modification of our standard chromatin spin down protocol (Arias and Walter, 2006) was used, in which the ELB wash step was supplemented with 0.6% Trition X-100.
Supplementary Material
Acknowledgements
We are grateful to Yoshihiro Matsumoto for the gift of Xenopus Fen1 cDNA; Yi Zhang and Jarrod Marto for the mass spectrometry analysis of Cdt1 phosphorylation sites; Puck Knipscheer for generating Figure 4C and helpful discussions; and Anna Kochaniak for the gift of the Xenopus PCNA antibody. We thank Emily Arias, Xiaolu Ang, David Long & Randall King for critical reading of the manuscript. This research was funded by NIH-F32-GM082014 to C.G.H. and NIH-R01-GM080676 to J.C.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang XL, Wade Harper J. SCF-mediated protein degradation and cell cycle control. Oncogene. 2005;24:2860–2870. doi: 10.1038/sj.onc.1208614. [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 2005;19:114–126. doi: 10.1101/gad.1255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2006;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- Chuang LC, Yew PR. Regulation of nuclear transport and degradation of the Xenopus cyclin-dependent kinase inhibitor, p27Xic1. J Biol Chem. 2001;276:1610–1617. doi: 10.1074/jbc.M008896200. [DOI] [PubMed] [Google Scholar]
- Chuang LC, Yew PR. Proliferating cell nuclear antigen recruits cyclin-dependent kinase inhibitor Xic1 to DNA and couples its proteolysis to DNA polymerase switching. J Biol Chem. 2005;280:35299–35309. doi: 10.1074/jbc.M506429200. [DOI] [PubMed] [Google Scholar]
- Chuang LC, Zhu XN, Herrera CR, Tseng HM, Pfleger CM, Block K, Yew PR. The C-terminal domain of the Xenopus cyclin-dependent kinase inhibitor, p27Xic1, is both necessary and sufficient for phosphorylation-independent proteolysis. J Biol Chem. 2005;280:35290–35298. doi: 10.1074/jbc.M506430200. [DOI] [PubMed] [Google Scholar]
- Dasso M, Nishitani H, Kornbluth S, Nishimoto T, Newport JW. RCC1, a regulator of mitosis, is essential for DNA replication. Mol Cell Biol. 1992;12:3337–3345. doi: 10.1128/mcb.12.8.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenbach A, Li A, Brito-Martins M, Blow JJ. Functional domains of the Xenopus replication licensing factor Cdt1. Nucleic Acids Res. 2005;33:316–324. doi: 10.1093/nar/gki176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle. 2006;5:1675–1680. doi: 10.4161/cc.5.15.3149. [DOI] [PubMed] [Google Scholar]
- Higa LA, Mihaylov IS, Banks DP, Zheng J, Zhang H. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat Cell Biol. 2003;5:1008–1015. doi: 10.1038/ncb1061. [DOI] [PubMed] [Google Scholar]
- Hohl M, Dunand-Sauthier I, Staresincic L, Jaquier-Gubler P, Thorel F, Modesti M, Clarkson SG, Scharer OD. Domain swapping between FEN-1 and XPG defines regions in XPG that mediate nucleotide excision repair activity and substrate specificity. Nucleic Acids Res. 2007;35:3053–3063. doi: 10.1093/nar/gkm092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- Hu J, Xiong Y. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J Biol Chem. 2006;281:3753–3756. doi: 10.1074/jbc.C500464200. [DOI] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Kim SH, Michael WM. Regulated Proteolysis of DNA Polymerase eta during the DNA-Damage Response in C. elegans. Mol Cell. 2008;32:757–766. doi: 10.1016/j.molcel.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kipreos ET. The Caenorhabditis elegans replication licensing factor CDT-1 is targeted for degradation by the CUL-4/DDB-1 complex. Mol Cell Biol. 2007;27:1394–1406. doi: 10.1128/MCB.00736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell. 2007;26:775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Liu C, Poitelea M, Watson A, Yoshida SH, Shimoda C, Holmberg C, Nielsen O, Carr AM. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J. 2005;24:3940–3951. doi: 10.1038/sj.emboj.7600854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Lock K, Yenamandra A, Cortez D. DDB1 maintains genome integrity through regulation of Cdt1. Mol Cell Biol. 2006;26:7977–7990. doi: 10.1128/MCB.00819-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D, Moreau J, Mechali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- Mattock H, Jares P, Zheleva DI, Lane DP, Warbrick E, Blow JJ. Use of peptides from p21 (Waf1/Cip1) to investigate PCNA function in Xenopus egg extracts. Exp Cell Res. 2001;265:242–251. doi: 10.1006/excr.2001.5181. [DOI] [PubMed] [Google Scholar]
- May NR, Thomer M, Murnen KF, Calvi BR. Levels of the origin-binding protein Double parked and its inhibitor Geminin increase in response to replication stress. J Cell Sci. 2005;118:4207–4217. doi: 10.1242/jcs.02534. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Nakanishi M, Robetorye RS, Pereira-Smith OM, Smith JR. The C-terminal region of p21SDI1/WAF1/CIP1 is involved in proliferating cell nuclear antigen binding but does not appear to be required for growth inhibition. J Biol Chem. 1995;270:17060–17063. doi: 10.1074/jbc.270.29.17060. [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem. 2008;283:29045–29052. doi: 10.1074/jbc.M806045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell BC, Harper JW. Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr Opin Cell Biol. 2007;19:206–214. doi: 10.1016/j.ceb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Ralph E, Boye E, Kearsey SE. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep. 2006;7:1134–1139. doi: 10.1038/sj.embor.7400827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, Hopkins N, Lees JA. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 2006;20:3117–3129. doi: 10.1101/gad.1482106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem. 2006;281:6246–6252. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- Shibutani ST, de la Cruz AF, Tran V, Turbyfill WJ, 3rd, Reis T, Edgar BA, Duronio RJ. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell. 2008;15:890–900. doi: 10.1016/j.devcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Michael WM. DNA damage-induced replication arrest in Xenopus egg extracts. J Cell Biol. 2003;163:245–255. doi: 10.1083/jcb.200306006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Warbrick E, Lane DP, Glover DM, Cox LS. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol. 1995;5:275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- Yanagi K, Mizuno T, You Z, Hanaoka F. Mouse geminin inhibits not only Cdt1-MCM6 interactions but also a novel intrinsic Cdt1 DNA binding activity. J Biol Chem. 2002;277:40871–40880. doi: 10.1074/jbc.M206202200. [DOI] [PubMed] [Google Scholar]
- You Z, Harvey K, Kong L, Newport J. Xic1 degradation in Xenopus egg extracts is coupled to initiation of DNA replication. Genes Dev. 2002;16:1182–1194. doi: 10.1101/gad.985302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–889. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.