Abstract
Endometriosis is a common gynecologic disorder characterized by pain and infertility. In addition to estrogen dependence, progesterone resistance is an emerging feature of this disorder. Specifically, a delayed transition from the proliferative to secretory phase as evidenced by dysregulation of progesterone target genes and maintenance of a proliferative molecular fingerprint in the early secretory endometrium (ESE) has been reported. MicroRNAs (miRNAs) are small noncoding RNAs that collectively represent a novel class of regulators of gene expression. In an effort to investigate further the observed progesterone resistance in the ESE of women with endometriosis, we conducted array-based, global miRNA profiling. We report distinct miRNA expression profiles in the ESE of women with versus without endometriosis in a subset of samples previously used in global gene expression analysis. Specifically, the miR-9 and miR-34 miRNA families evidenced dysregulation. Integration of the miRNA and gene expression profiles provides unique insights into the molecular basis of this enigmatic disorder and, possibly, the regulation of the proliferative phenotype during the early secretory phase of the menstrual cycle in affected women.
Keywords: endometriosis, endometrium, microRNA, microarray
Introduction
Endometriosis is a benign, estrogen-dependent disorder associated with pelvic pain and infertility and is characterized by the ectopic distribution of endometrial tissue, primarily on the pelvic peritoneum and ovaries. It occurs in 6–10% of women in the general population and in 35–50% of women with pain and/or infertility (Eskenazi and Warner, 1997). The retrograde displacement of eutopic endometrium into the pelvis and its subsequent implantation on peritoneal surfaces is a leading theory as to the etiology of this condition. A heritable or acquired molecular aberration within the endometrium may impart a selective survival advantage to refluxed endometrial tissue in women predisposed to the development of endometriosis. The identification of molecular differences in the eutopic endometrium of women with endometriosis is an important step toward understanding the pathogenesis of this condition and toward developing novel strategies for the treatment of associated infertility and pain.
Previously, we demonstrated maintenance of a proliferative fingerprint in the transcriptome of early secretory endometrium (ESE) from women with endometriosis, consistent with attenuated progesterone action or progesterone resistance on the part of the endometrium in the setting of endometriosis (Burney et al., 2007). Functional genomic analysis of ESE revealed a signature of enhanced cellular survival and persistent expression of genes involved in DNA synthesis and cellular mitosis in women with this disorder.
MicroRNAs (miRNAs) are endogenous 22 nucleotide-long, highly conserved, noncoding RNAs which, in general, negatively regulate gene expression. Functional analysis of miRNAs has revealed their significant regulatory influence on the expression of target genes involved in both physiologic and pathologic conditions (Ambros, 2004; Li et al., 2007). Cell-cycle progression, proliferation and differentiation are among the biologic processes regulated by miRNAs via altered expression of target genes. Our finding of incomplete transitioning from proliferative to secretory phase endometrium in women with endometriosis, as evidenced by dysregulation of numerous genes in the cell-cycle pathway, suggested the potential for miRNA involvement. Consequently, we targeted the early secretory (ES) phase eutopic endometrium from women with versus without moderate–severe stage endometriosis for global miRNA profiling.
Herein, we demonstrate that ESE from women with endometriosis is characterized by a miRNA expression profile that differs from that of healthy ESE. Among the miRNAs underexpressed in ESE in the setting of endometriosis are members of the miR-9 and miR-34 families. Recent studies support direct reduction of target mRNA with antisense complementarity of their 3′ untranslated region (UTR) to the hexameric ‘seed’ region of the miRNA (Lim et al., 2005). The effect of miRNA on mRNA degradation may result in changes to transcript levels detectable by array-based gene expression analysis. By using the same RNA specimens that were used in our previously reported gene expression analysis (Burney et al., 2007), the current study provided the opportunity for parallel miRNA–mRNA expression profiling in the identification of molecular dysregulation. These findings highlight new candidates within an emerging dimension of genetic regulation that may be involved in the incomplete transitioning of ESE in women with endometriosis.
Materials and Methods
Human subject characteristics
All patients included in this study provided informed consent for participation under a protocol approved by the University of California, San Francisco, Committee on Human Research and the Stanford University Committee on the Use of Human Subjects in Medical Research. Samples were obtained through the UCSF NIH Human Endometrial Tissue and DNA Bank. Endometriosis is a phenotypically heterogeneous disorder and the inaccuracy in its visual diagnosis, particularly in cases of minimal/mild stages, is well documented (Marchino et al., 2005). Accordingly, endometrial biopsies were obtained from women with histologically confirmed, moderate–severe endometriosis at laparoscopy (n = 4) and from women found to be free of endometriosis at surgery (n = 3). Moderate–severe endometriosis (Stage III–IV disease) was defined in accordance with the Revised American Fertility Society (rAFS) classification system. All subjects were normo-ovulatory with regular menstrual cycles, were between the ages of 23 and 50, and had not received steroid hormone medications within 3 months of endometrial sampling. Women without endometriosis at surgery were undergoing hysterectomy for uterine leiomyomata, none of which were submucosal in location. Review of pathology reports revealed no evidence of inflammation within the endometrium of women without endometriosis. Samples were collected using a Pipelle catheter or curettage. Endometrial biopsies were dated as ES (days 15–18) by menstrual dating and by histologic dating according to the Noyes criteria (Noyes et al., 1975) upon independent review by up to four pathologists. Importantly, the total RNA samples used in this study were the same as those used in our previous reported global gene expression profiling study (Burney et al., 2007) with the exception of one sample in each group (607 and 389). These substitutions were necessary due to the lack of available RNA from two specimens analyzed in the prior study. The clinical data of samples used for miRNA profiling is provided in Table I.
Table I.
Subject characteristics
| Patient ID | Cycle phase | Age (years) | Distribution | Other diagnoses | Ethnicity | Medications |
|---|---|---|---|---|---|---|
| Moderate–severe stage endometriosis | ||||||
| 489 | ES | 39 | R,PI | Leiomyomata | Asian | Levothyroxine |
| 27A | ES | 22 | R,O,PI | None | Caucasian | None |
| 575 | ES | 26 | O,PI | None | Unknown | None |
| 607 | ES | 24 | R,O,PI | None | Asian | None |
| No endometriosis | ||||||
| 629 | ES | 46 | N/A | Leiomyomata | Caucasian | Celexa |
| 650 | ES | 48 | N/A | Leiomyomata | Caucasian | None |
| 389 | ES | 41 | N/A | Leiomyomata | Caucasian | None |
PI = peritoneal endometriosis, defined as biopsy-proven serosal implant; O = ovarian endometriosis, defined as biopsy-proven endometrioma; R = rectovaginal endometriosis, defined as posterior cul de sac obliteration due to endometriotic lesions. Endometrial specimens from subjects with endometriosis were surgically staged with moderate–severe stage of disease in accordance with the rAFS criteria.
RNA isolation and miRNA microarray analysis
Total RNA from whole endometrial specimens was isolated using Trizol reagent (Invitrogen) as described previously (Burney et al., 2007). Immediately prior to sample inclusion in the miRNA microarray analysis, the quality, yield and size of the miRNA fractions were validated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA, USA). The nanodrop instrument (Thermo Fisher Scientific Inc., Waltham, MA, USA) was used to verify minimal contamination from protein (ratio abs260/abs280) and from organic compounds such as Trizol (ratio abs260/abs230). Following validation of RNA sample quality, two micrograms of total RNA from each sample and from common reference pooled samples were labeled with Hy3™ and Hy5™ fluorescent label, respectively, using the miRCURY™ Labeling kit (Exiqon, Vedbaek, Denmark) following the procedure described by the manufacturer. The Hy3™-labeled samples and Hy5™-labeled reference RNA sample were mixed pair wise and hybridized to the miRCURY™ LNA Array (version 10.0, Exiqon). This platform consists of Tm-normalized capture probes for 1488 distinct miRNAs, including all known miRNAs registered in miRBASE version 10.1 at the Sanger Institute and 151 proprietary miRPlus sequences not yet annotated in miRBase. Each sample was evaluated in duplicate. Microarray slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies) and image analysis was carried out using the ImaGene 7.0 software (BioDiscovery, Inc., USA). Signal intensities were normalized using the global Lowess (LOcally WEighted Scatterplot Smoothing) regression algorithm. Analysis was performed using the log2 of the background-subtracted, normalized median spot intensities of ratios from the two channels.
miRNA real-time PCR
For validation of microarray data by real-time PCR, cDNA was synthesized from total RNA using commercially available miR-specific (for miR-34c-5p, miR-34b* miR-9, and miR-9*) or endogenous control-specific reverse primers according to the miRCURY LNA™ microRNA PCR System protocol instructions (Exiqon). Each reverse transcriptase (RT) reaction contained 10 ng of RNA samples, 2 µl of specific RT primer, 1× RT buffer, 500 µM of dNTP mix, 20 U of RT enzyme and 10 U of RNase inhibitor (all included in First-Strand cDNA Synthesis kit), with total volume of 10 µl. Reactions were incubated in Mastercycler gradient (Eppendorf, Hamburg, Germany) for 30 min at 50°C, followed by heat-inactivation of RT for 10 min at 85°C and held at 4°C.
Real-time PCR was performed using the Stratagene Mx3005 system (Stratagene, La Jolla, CA) with 20-µl PCR reaction mixture including 4-µl RT product, 1× SYBR Green Master Mix and 1 µl of LNA™ microRNA primer and Universal primer and 4-µl nuclease-free water (Exiqon). Reactions were incubated in a 96-well optical plate at 95°C for 10 min, followed by 40 cycles of 95°C for 20 s and 60° for 1 min with subsequent melting curve analysis. Expression of target miRNAs was normalized to the levels of an endogenous standard housekeeping miRNA, miR-5S. The variability of miR-5S expression was low and not significant. The expression of each miRNA relative to 5S miRNA was determined using the 2−ΔCT method (Livak and Schmittgen, 2001). Data are presented as fold change relative to the control group. Nonparametric Mann–Whitney U-test was used for statistical evaluation of miRNA expression differences with significance accepted at P-values ≤0.05.
Pathway analysis
Previous global gene expression data comparing endometrium from women with versus without moderate–severe endometriosis identified a number of differentially expressed genes in the ESE (Burney et al., 2007). This list of genes was compared with the list of TargetscanS (Lewis et al., 2005) predicted gene targets of the three miRNAs (miR-9, miR-34b* and miR-34c-5p) validated to be differentially expressed in the current study. The union of these gene lists was analyzed for biologic pathway relationships using Ingenuity Pathway Analysis software (IPA, version 7.5, Ingenuity Systems, Redwood City, CA). This platform provides a literature-curated analysis of networks, canonical pathways and biological processes involved in gene expression data. An association between predicted miRNA gene targets and biologically relevant pathways identified by this analysis was deemed statistically significant at a P-value of <0.05.
Results
A total of six miRNAs were differentially expressed with a fold change of >1.5 and a false discovery rate of <0.05 in the ESE from women with versus without endometriosis. Down-regulated miRNAs included miR-9, miR-9*, miR-34b*, miR-34c-5p, miR-34c-3p and the unannotated miRPlus_42 780. MiR-9 represented the most significantly dysregulated miRNA (P = 0.0032). The majority of miRNAs were unchanged or not expressed in endometrium, in agreement with the previous data demonstrating spatiotemporal-specific expression of a high percentage of miRNAs.
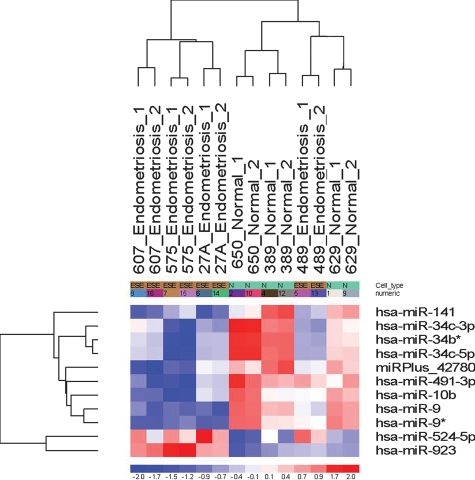
Unsupervised hierarchical clustering analysis was conducted using the miRNA expression profiles of the seven endometrial samples (four with endometriosis and three without endometriosis) based on the combined list of miRNAs showing differential expression in the ES phase of the menstrual cycle (Fig. 1). The samples demonstrate self-segregation into normal and disease clusters with the exception of the endometrial specimen obtained from subject 489, which clustered with specimens taken from women without endometriosis. Subject 489 was a 39-year-old woman noted to have leiomyomata at surgery and was the only subject in the endometriosis cohort who was not affected with an endometrioma. Variables correlating with segregation of this specimen with those of the no endometriosis cohort include older subject age, presence of intramural/subserosal leiomyomata and absence of ovarian endometriosis/endometrioma.
Figure 1.
Unsupervised hierarchial clustering of differentially expressed miRNAs in ESE from women with versus without endometriosis (fold change of ≥1.5).
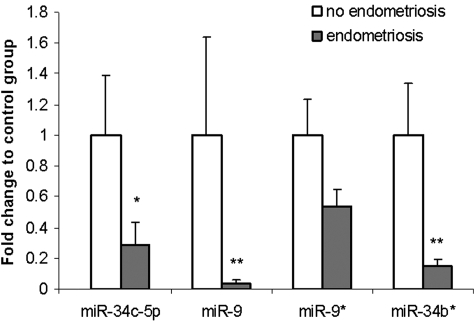
To validate our array expression findings, four of the six differentially expressed miRNAs (miR-34c-5p, miR-9, miR-9* and miR-34b*) were chosen for quantitative real-time PCR (qRT–PCR) analysis (Fig. 2). The trends for down-regulation of miRNA expression were consistent in all four qRT–PCR measurements, and significant for three of the four miRNAs (miR-34c-5p, miR-9 and miR-34b). MiR-9* did not demonstrate statistically significant difference in expression between ESE from women with versus without endometriosis (Table II).
Figure 2.
Validation by qRT–PCR analysis of miRNA expression. Data are presented as fold change of expression in eutopic endometrium from women with endometriosis relative to expression in endometrium from women without endometriosis after normalization to miR-5S. For comparative analysis, the expression values for each miRNA were set as 1 in ESE from women without endometriosis. Significant fold changes are marked by *P = 0.05; **P < 0.05. Data are mean ± SEM.
Table II.
Differentially expressed miRNAs in ES phase endometrium
| miRNA | Accession | Seed 7-mer | Microarray fold |
qRT–PCR fold |
||
|---|---|---|---|---|---|---|
| Change | P-value | Change | P-value | |||
| miR-34c-5p | MIMAT0000686 | GGCAGUG | −2.96 | 0.015 | −3.57 | 0.05 |
| miR-34b* | MIMAT0000685 | AGGCAGU | −2.84 | 0.019 | −6.80 | 0.03 |
| miR-34c-3p | MIMAT0004677 | AUCACUA | −2.54 | 0.025 | ||
| miR-9 | MIMAT0000441 | CUUUGGU | −1.90 | 0.0032 | −25.64 | 0.03 |
| miR-9* | MIMAT0000442 | UAAAGCU | −1.90 | 0.0152 | −1.90 | 0.11 |
| miRPlus_42 780 | Unannotated | −1.79 | 0.038 | |||
Fold changes provided compare the endometrium from women with versus without moderate–severe endometriosis.
We previously reported significant dysregulation in gene expression signatures during the proliferative, ES and mid-secretory phases of the menstrual cycle in the endometrium from women with versus without endometriosis (Burney et al., 2007). Of the three phases of the menstrual cycle investigated, the ES phase involved the greatest number of statistically significant and differentially expressed genes, with 747 up-regulated and 1741 down-regulated with fold change of ≥1.5 and P ≤ 0.05. The complete gene lists for all cycle phases in women with disease versus no disease are published as supplemental data on The Endocrine Society's Journals Online website at http://endo.endojournals.org. The data were submitted to the Gene Expression Omnibus database under the identifier GSE6364.
To explore the biologic relationship between the differentially expressed messenger- and miRNAs identified in our analysis of ESE from women with versus without endometriosis, we performed in silico analysis using the TargetScanS algorithm (Lewis et al., 2005) for identification of predicted messenger RNA targets for each of the three miRNAs that we identified and validated to be significantly differentially expressed. Predicted mRNA gene targets of each miRNA were then cross-referenced against the 2488 genes that we found to be differentially expressed between normal ESE and that from women with endometriosis in our microarray gene expression study (Table III and for full lists see supplementary table). Of interest is the finding that the down-regulation of several miRNAs correlated with the up-regulation of cell-cycle target genes, which may contribute to the maintenance of the proliferative fingerprint in ESE from women with endometriosis.
Table III.
miRNAs differentially expressed by microarray and qRT–PCR and their mRNA targets (P < 0.05)
| Down-regulated miRNA | miRNA Ct valuea | mRNA targets |
Target mRNA gene ID | Gene name | Fold change | |
|---|---|---|---|---|---|---|
| Up | Down | |||||
| miR-9 | 30.29 ± 0.38 versus 26.94 ± 1.04 | 31 | 63 | CALB2 | Calbindin 2 | +3.77 |
| CCNE2 | Cyclin E2 | +3.29 | ||||
| TGFBI | TGF, β induced | +3.14 | ||||
| FBN1 | Fibrillin 1 | +2.03 | ||||
| BCL2 | B cell CLL/lympoma 2 | +1.72 | ||||
| FGFR2 | Fibroblast growth factor receptor 2 | +1.65 | ||||
| miR-34c-5p | 29.09 ± 0.57 versus 27.88 ± 0.46 | 18 | 20 | CCNE2 | Cyclin E2 | +3.29 |
| BCL2 | B cell CLL/lympoma 2 | +1.72 | ||||
| CDK6 | Cyclin-dependent kinase 6 | +1.51 | ||||
| miR-34b* | 26.22 ± 0.29 versus 24.21 ± 0.36 | 10 | 14 | MAPK4 | Mitogen-activated protein kinase 4 | +1.98 |
| BRD4 | Bromodomain containing 4 | +2.20 | ||||
| CDK6 | Cyclin-dependent kinase 6 | +1.51 | ||||
These were identified by cross-referencing the predicted mRNA gene targets of each miRNA against the gene transcripts that were found to be differentially expressed in endometrium from women with versus without moderate–severe endometriosis in previous global gene expression analysis (Burney et al., 2007).
aMiRNA Ct value from RT–PCR validations expressed as mean ± SEM in endometriosis versus nonendometriosis specimens.
A total of 156 mRNA genes were both differentially expressed between endometrium from women with versus without endometriosis and predicted mRNA target genes of differentially expressed miRNAs (miR-9, miR-34b* and miR-34c-5p). Forty-seven of these genes were eligible for analysis of network relationships using Ingenuity Pathway Analysis software (IPA version 7.5, Ingenuity Systems, Redwood City, CA). This analysis generated a network most significantly associated with the biological processes of cell death, cell cycle and cellular assembly and organization and the canonical pathways of molecular mechanisms of cancer, semaphorin signaling in neurons and cell-cycle regulation (P < 0.05). Eighteen of the 47 genes (38%) were represented in this network, each demonstrating statistically significant association (P < 0.05), and included cell-cycle genes CCNE, CCNE2, CDK4, CDK6, CDC25A and the anti-apoptosis gene, BCL2.
Discussion
Previously, we reported differences in the global gene expression profile of endometrium from women with versus without surgically confirmed moderate–severe endometriosis. Evaluation of the transcriptome differences were most striking in the ES phase, during which incomplete transitioning from the proliferative to secretory phase was observed on a molecular level in women with endometriosis. Interestingly, genes related to the biological processes of cell cycle and DNA replication were enriched in the profile of dysregulated genes. Given their central role in the regulation of processes such as cellular proliferation, miRNAs are biologically plausible contributors to the maintenance of the proliferative fingerprint in ESE of women with endometriosis. To our knowledge, this study is the first to evaluate the cycle phase-specific differential expression of miRNAs in the eutopic endometrium from women with and without endometriosis, and integrate the findings with the gene expression signature from the same specimens.
Our array-based global miRNA profiling demonstrated differential expression of two miRNA families, miR-9 and miR-34, in the ESE of women with endometriosis. Both the 3′-end (miR-9) and 5′-end (miR-9*) forms of miR-9 were down-regulated in the ESE of women with endometriosis. MiR-9 represented the most significantly dysregulated miRNA in our microarray data set, and qRT–PCR revealed a 25-fold reduction in miR-9 in the setting of endometriosis. In the human genome, three distinct genes encode different primary miR-9 transcripts (pri-miR-9) generating identical mature miR-9. Among the predicted mRNA targets of miR-9 is BCL2, a gene encoding an anti-apoptosis protein found to be over-expressed in the ESE from women with endometriosis in our transcriptome study. Down-regulation of miR-9 and miR-9* has been reported in clear cell and endometrioid histotypes of ovarian cancer, for which patients with endometrioma are at increased risk (Iorio et al., 2007). MiR-9 was the most significantly down-regulated miRNA in a study of recurrent versus primary ovarian cancers (Laios et al., 2008). Further integration with transcriptome findings suggested a tumor suppressor role for miR-9 and the reliable confirmation of its deregulation in training cohorts suggested utility as a biomarker for recurrent ovarian cancer.
Three down-regulated miRNAs were members of the miR-34 family. Functional studies have concluded miR-34b and miR-34c to act as mediators of p53-dependent suppression of proliferation (Corney et al., 2007). Hexamer analysis of these down-regulated miRNAs against the cohort of up-regulated genes in the mRNA profile demonstrated enrichment of predicted target genes involved in the cell-cycle pathway. The down-regulation of miR-34c and miR-34b in the ESE of women with endometriosis may be involved in the maintenance of the proliferative fingerprint observed in our transcriptome analysis.
It is important to recognize that the relationship between the expression of miRNA and the expression of target genes is complex and evolving. Most miRNAs are thought to repress expression of target genes by post-transcriptional repression or degradation of transcripts. This would suggest an inverse relationship between miRNA and target gene mRNA expression. However, translational activation of target genes by miRNAs has also been reported (Vasudevan et al., 2007), rendering the relationship between miRNA and mRNA less generalizable. Equally important to recognize is the putative nature of the relationship between miRNA and mRNA expression, as this relationship is currently more based on computational biology than experimental validation. Several algorithms designed to predict target genes of miRNA sequences exist, each predicated on the degree of sequence complementarity between the miRNA and a target UTR. On the basis of these computational algorithms, each miRNA is predicted to regulate an average of 200 genes (Krek et al., 2005). Several or even multiple miRNAs, including ones without significant changes in the expression level, may share target genes in a given cell or tissue, thereby adding to the complexity of proposed relationships between miRNA and mRNA expression.
Hierarchial clustering of differentially expressed miRNAs in ESE samples showed segregation not only by endometriosis status, but also correlated with the presence of leiomyomata. All women in the no endometriosis cohort and one in the affected cohort (subject 489) were found to have leiomyomas at surgery. Importantly, none of these leiomyomas was submucosal in position, as this was a contraindication to study inclusion and confirmed upon review of operative reports. Endometrial HOXA10 and HOXA11 gene expression was reported to be significantly decreased in endometrium from women with submucosal leiomyomas but unchanged in women with intramural leiomyomata (Rackow and Taylor, 2008). While a recent microarray study demonstrated that intramural leiomyomas may alter the expression pattern of some endometrial genes, those involved in implantation were unaffected (Horcajadas et al., 2008). This is corroborated by our work with cultured endometrial stromal fibroblasts from women with intramural fibroids revealing normal decidualization after exposure to decidualizing stimuli (Aghajanova et al., 2009). Global miRNA profiling of leiomyomas versus matched normal myometrium revealed differential expression of a number of miRNAs (Wang et al., 2007), yet none of these were common to those found to be dysregulated in the current study. Collectively, these lines of evidence suggest that intramural leiomyomata are unlikely to affect the endometrial miRNA profile observed in the current study.
More plausible in the explanation of the clustering profile of specimens by differential expression of miRNAs are the variables of subject age and/or presence of endometrioma. Though clinical studies involving a donor oocyte model suggest that endometrial age is not as significant a factor as oocyte age in determining fertility, experiments using a mouse system support age-related changes in cycling endometrium (Archer 2008). More recently, HOXA10 gene expression was demonstrated to be inversely related to uterine age in women (Fogle et al., 2009). These studies highlight a potential role for patient age in endometrial molecular expression signatures. Interestingly, the endometrial specimen from the subject with endometriosis, which clustered with the normal cohort, was the only endometriosis subject without endometrioma(s) present at surgery. In view of the molecular data demonstrating that miR-9 is deregulated in endometrioid ovarian cancer (Laios et al., 2008) and clinical data suggesting an increased risk of endometrioid ovarian cancer in women with endometrioma (Korner et al., 2006), the miRNA-based clustering of endometrial specimens by endometrioma status may be reflective of fundamental differences in endometrial biology.
The current study is strengthened by the stringency of the surgical confirmation of presence or absence of disease and by the inclusion of only biopsy confirmed, moderate–severe (rAFS III–IV) stage endometriosis among the affected cohort. Additionally, our study is strengthened by the use of microarray technology employing locked nucleic acid (LNA) probes. Mature miRNAs are approximately 22 nucleotides in length, and this short length presents problems of specificity in their detection and localization. Microarray analysis using LNA probes allow very sensitive detection of these short-coding sequences (Neely et al., 2006).
In contrast to the current study, the differential expression of miRNAs in eutopic compared with ectopic endometrium has been evaluated by two studies previously (Pan et al., 2006; Ohlsson Teague et al., 2009). These studies highlight molecular pathways that may be associated with the development of endometriosis as well as the changes in expression signature that exist in ectopically located endometrial tissue. Decreased expression of miR-34c was observed in ectopic versus eutopic endometrium in the study by Ohlsson Teague et al. Our study focused on the differences in miRNA expression in eutopic endometrium from women with versus without endometriosis matched for a specific cycle phase, and this focus may explain the comparatively finite number of dysregulated miRNAs observed herein. Functional relationships between differentially expressed miRNAs and putative mRNA targets and their cellular co-localization require further study.
In summary, we report for the first time an integrated transcriptome-miRNA analysis of eutopic endometrium from women with versus without endometriosis, identifying potential miRNA mediators in the delayed proliferative to secretory transition of endometrium observed in women with advanced endometriosis. These data illustrate an altered miRNA expression profile in women with endometriosis, and provide candidates within a novel layer of genetic regulation in the pathogenesis of this enigmatic disorder.
Supplementary Data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Funding
This work was supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)/National Institutes of Health (NIH) through cooperative agreement [U54 HD-055764-02 (L.C.G.) and NICHD HD-35041-12 (B.A.L.)] as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research. Tissues were obtained through the UCSF NIH U54 Human Endometrial Tissue and DNA Bank [HD055764 (L.C.G.)].
References
- Aghajanova L, Hamilton A, Kwintkiewicz J, Vo KC, Giudice LC. Steroidogenic enzyme and key decidualization marker dysregulation in endometrial stromal cells from women with versus without endometriosis. Biol Reprod. 2009;80:105–114. doi: 10.1095/biolreprod.108.070300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Archer D. Aging of the endometrium. In: Alpin JD, Fazleabas AT, Glasser SR, Giudice LC, editors. The Endometrium. 2nd edn. Informa Healthcare; 2008. p. 761. [Google Scholar]
- Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezhat CR, Lessey BA, Giudice LC. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology. 2007;148:3814–3826. doi: 10.1210/en.2006-1692. [DOI] [PubMed] [Google Scholar]
- Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- Eskenazi B, Warner ML. Epidemiology of endometriosis. Obstet Gynecol Clin North Am. 1997;24:235–258. doi: 10.1016/s0889-8545(05)70302-8. [DOI] [PubMed] [Google Scholar]
- Fogle RH, Li A, Paulson RJ. Modulation of HOXA10 and other markers of endometrial receptivity by age and human chorionic gonadotropin in an endometrial explant model. Fertil Steril. 2009 doi: 10.1016/j.fertnstert.2008.11.002. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Horcajadas JA, Goyri E, Higón MA, Martínez-Conejero JA, Gambadauro P, García G, Meseguer M, Simón C, Pellicer A. Endometrial receptivity and implantation are not affected by the presence of uterine intramural leiomyomas: a clinical and functional genomics analysis. J Clin Endocrinol and Metab. 2008;93:3490–3498. doi: 10.1210/jc.2008-0565. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu C-G, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Korner M, Burckhardt E, Mazzucchelli L. Higher frequency of chromosomal aberrations in ovarian endometriosis compared to extragonadal endometriosis: a possible link to endometrioid adenocarcinoma. Mod Pathol. 2006;19:1615–1623. doi: 10.1038/modpathol.3800699. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Laios A, O'Toole S, Flavin R, Martin C, Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008;7:35–48. doi: 10.1186/1476-4598-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li SC, Tang P, Lin WC. Intronic microRNA: discovery and biological implications. DNA. Cell Biol. 2007;26:195–207. doi: 10.1089/dna.2006.0558. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marchino GL, Gennarelli G, Enria R, Bongioanni F, Lipari G, Massobrio M. Diagnosis of pelvic endometriosis with use of macroscopic versus histologic findings. Fertil Steril. 2005;84:12–15. doi: 10.1016/j.fertnstert.2004.09.042. [DOI] [PubMed] [Google Scholar]
- Neely LA, Patel S, Garver J, Gallo M, Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S, Rooke J. A single-molecule method for the quantitation of microRNA gene expression. Nat Methods. 2006;3:41–46. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- Noyes RW, Hertig AT, Rock J. Dating the endometrial biopsy. Am J Obstet Gynecol. 1975;122:262–263. doi: 10.1016/s0002-9378(16)33500-1. [DOI] [PubMed] [Google Scholar]
- Ohlsson Teague EMC, Van der Hoek KH, Van der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, Print CG, Hull ML. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23:265–275. doi: 10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13:797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- Rackow BW, Taylor HS. Submucosal uterine leiomyomas have a global effect on molecular determinants of endometrial receptivity. Fertil Steril. 2008 doi: 10.1016/j.fertnstert.2008.03.029. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Wang T, Zhang X, Obijuru L, Laser J, Aris V, Lee P, Mittal K, Soteropoulos P, Wei J. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes, Chromosomes Cancer. 2007;46:336–347. doi: 10.1002/gcc.20415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.