Abstract
Vibrio cholerae, the causative agent of cholera, is naturally present in the environment and autochthonous to coastal and estuarine ecosystems. V. cholerae is associated with copepods for its survival and multiplication in the natural environment. Changes in the density of its reservoir may result in modification of the bacterial population size in the environment. In this context, climate and/or environmental changes will influence the emergence of cholera in human populations. Several human pathogens are naturally occurring in the aquatic environment and can pose a threat to public health, including V. cholerae. We present results of a project, the goal of which was to improve the understanding of environmental factors associated with occurrence and distribution of the causative agent of cholera in time and space. The system that was developed provides real-time as well as short-term to seasonal forecasts of the likelihood of occurrence of V. cholerae in the Chesapeake Bay. The system, and potential future improved versions of it, may be useful to public health officials concerned with environmental factors influencing human health.
HOW CLIMATE INFLUENCES CHOLERA
Cholera is an ancient disease that during the last fifty years, has disappeared from most developed countries, but is reemerging in many parts of the world in epidemic form, especially in tropical areas. This highly contagious, dose-dependent disease is caused by the bacterium Vibrio cholerae when ingested via contaminated water or food and, most frequently, seafood.
V. cholerae is naturally present in the environment and is autochthonous in riverine, coastal, and estuarine ecosystems (1–3). The bacterium is strongly associated with plankton, forming commensal or symbiotic relationships, mainly with copepods (4–6). In the aquatic ecosystem, chitin is the most abundant polysaccharide and the principal component of the zooplankton exoskeleton. Chitinous organisms (for example, copepods, amphipods, and other crustaceans) are dominant among zooplankton populations. The copepod exoskeleton has been shown to support large populations of vibrios, including the pathogenic species, V. cholerae (7–10). Without bacterial activity that returns insoluble polysaccharide to the ecosystem in a biologically useful form, seawater would become depleted of carbon and nitrogen in a relatively short time (11). Therefore, the commensal relationship between vibrios and copepods has important consequences, as was demonstrated in studies done in the early 1980s (8, 12). Bacteria bound to plankton metabolize chitin more efficiently than free-living bacteria, thus increasing the rate of chitin mineralization in the natural environment. Attachment to copepods and, in particular, to the eggs that are dispersed in the water, provides a mechanism for extended geographic distribution of bacteria, including V. cholerae (3, 13). Based on the earlier pioneering work, plankton are now recognized as a reservoir of V. cholerae. This bacterial species, as other gram-negative bacteria, has a selective advantage in their ability to enter a dormant stage, termed the variable but nonculturable (VBNC) state, when environmental conditions are unfavorable for active growth and cell division (14–16). That also includes fully virulent strains of V. cholerae (17).
Interactions between vibrios and copepods are affected by environmental variables (16, 17). Salinity of 15‰ and temperatures ranging from 25° to 30°C have been shown to be important in influencing the attachment of V. cholerae to copepods (13). There is likely a synergistic effect between phytoplankton and zooplankton in the colonization of chitin by V. cholerae, since an alkaline pH of 8.5 is often associated with algal blooms and also has a positive effect on attachment of V. cholerae to copepods (18). Living copepods in microcosms are able to protect those bacteria that are sensitive to lower salinity (<0.5‰) and pH (13). Moreover, it has been demonstrated that V. cholerae attached to copepods, both on the surface and in their gut, survive exposure to acid pH better than unattached, free swimming bacteria, suggesting that adherence to chitin provides not only a substrate for Vibrio multiplication, but also protection from gastric acid during transit through the stomach (19).
A hierarchical model has recently been proposed, which defines the role of environmental, weather, and climate-related variables in outbreaks of cholera (20, 21) (Figure 1). At present, the main geographical regions of cholera endemicity include coastal areas surrounding the Bay of Bengal, Bangladesh, the Indian subcontinent, Africa, and coastal Latin America. In these regions, the same physical or environmental drivers most likely explain the patterns of disease. Sunlight, temperature and nutrients, in addition to affecting growth of V. cholerae, influence growth of phytoplankton and aquatic plants; these, in turn, alter the dissolved O2 and CO2 content of water and, therefore, the pH of the surrounding water. Results obtained from studies performed in a region of cholera endemicity (22) support the original hypothesis of Kaneko and Colwell (23) and Huq et al. (9) that copepods play an important role in the survival, multiplication, and transmission of V. cholerae and related vibrios in the natural aquatic environment. It is clear that V. cholerae has a close association with copepods for persistence and multiplication in the natural environment, specifically with the calanoid copepods Acartia tonsa and Eurytemora affinis, as observed recently by Rawlings et al. (24).
Fig. 1.
Hierarchical model for environmental cholera transmission. Copyright © American Society for Microbiology, Lipp et al., Clinical Microbiology Reviews, Vol. 15, No. 4, p. 757–770, Oct. 2002.
Some investigators doubted the importance of copepods in cholera transmission when the relationship was first discovered, but the correlation was conclusively demonstrated in a study in which the number of cholera cases in Bangladesh villages was reduced significantly when copepods were removed from drinking water (25). The method used was simple but ingenious: the water was filtered through several layers of the sheer but finely woven cloth of the traditional sari clothing worn by women of the region and cholera was reduced by about half (25).
The relationship between human health and climate is not a new concept, but in the existing context of global change, when most scientists now agree that our climate is changing, there is an increasing need to understand the potential outcome of such changes on human health. This can be achieved by considering how systems interact. With few exceptions, zoonotic and vector-borne diseases are readily understood as having links with the natural environment, and cholera is one of the best examples to illustrate this biocomplexity (Figure 2). Current scientific understanding of these diseases provides a basis for what is today the most complete human/natural systems model of emerging infectious diseases (26). The connection between epidemiology and ecosystem dynamics or processes is only now beginning to be appreciated.
Fig. 2.
The biocomplexity spiral is a representation of the study of complex structures and behaviors that arise from nonlinear interactions of active biological agents with their environment, which may range in scale from molecules to cells to organisms and to global scale. (Source: Rita R. Colwell)
The Chesapeake Bay, the largest estuarine ecosystem in North America, has been, and continues to be, the subject of interdisciplinary research on the ecology of V. cholerae that incorporates the fields of microbiology, ecology, physiology, physics and engineering. The Chesapeake Bay watershed, comprising eight river basins, is already showing the effects of an increase in annual mean temperature (27). The projected sea level rise in the Chesapeake region due to global warming is close to 10 inches by 2030 and up to 25 inches by 2100 (27). In addition, the human population in the Chesapeake Bay watershed will continue to increase significantly. With this background, we developed a tool for predicting the presence of V. cholerae in the Chesapeake Bay.
Cholera is no longer a threatening disease for the American population, although V. cholerae can still pose public health risks, ranging from limited outbreaks to epidemics, when it is ingested via untreated water or contaminated shellfish. But cholera was once considered “America's greatest scourge” (28), after its widespread ravages from New York to New Orleans in 1849. The disease occurred in many parts of the United States and Canada (29). Daly recently published a review of what happened when cholera came to small Midwestern towns in the United States during the “black cholera” episodes of the 19th century and later (30). In the review, he describes how populations suffered from cholera epidemics and their perceptions about and reactions to the disease, which was believed at that time to be unpreventable and incurable. However, with improvement in sanitation and public health, by the early 1900s, cholera gradually disappeared, with its first reappearance in the twentieth century reported in 1973 in Texas (31). Since then, sporadic cases are reported each year in the United States, some of which have been confirmed as indigenous in origin (32).
Because of the risk and the fact that cholera is a good model for studying water-borne diseases, it is of interest to understand the mechanisms that affect the natural populations of V. cholerae in the environment and to anticipate the potential impact of extreme climate events such as abnormally hot temperatures or floods on cholera. Changes in the number of V. cholerae reservoirs could lead to changes in the number of bacteria in the environment. Thus, climatic and/or environmental changes can potentially be responsible for the emergence of cholera in human populations (33, 34).
The Chesapeake Bay and Vibrio cholerae, a long story
In 1977, V. cholerae was isolated for the first time from water samples collected in the Chesapeake Bay (35). Pathogenic strains of V. cholerae, serotype O1, are now routinely detected in the Bay, first reported in 1981 (8). Almost 20 years later, environmental conditions have been shown to influence the appearance of V. cholerae during the warmer months of the year (36, 37). Other environmental variables, such as pH and salinity, have also been found to affect survival and multiplication of V. cholerae (18, 36, 38).
As a consequence, developing and implementing a system for predicting V. cholerae and other vibrios would be advantageous for both the prevention of this infectious disease and the mitigation of its effects in a region on both short- and long-term time scales. Short-term predictions can be employed to enhance first responder capabilities, while longer-term forecasts can be used to simulate potential consequences of climate change on this bacterium in the Bay. More generally, such a system would enhance our understanding of the connections between the oceans and human health that is crucial in addressing issues related to emerging and re-emerging diseases in the U.S. and globally.
Despite the fact that some of its general habitat preferences are known, and statistically significant empirical relationships have been established between the presence of V. cholerae and environmental factors, notably temperature and salinity affecting the growth rates of V. cholerae (36, 39), the exact mechanisms and environmental interactions giving rise to proliferation of V. cholerae are poorly understood. It is not currently possible to construct mechanistic models to predict the presence and abundance of these bacteria, but we have developed a system that generates predictions of the likelihood of V. cholerae in the Chesapeake Bay by exploiting what is known about the physical habitat of V. cholerae and taking advantage of recent advances in technology and telecommunications to retrieve, simulate and forecast relevant environmental conditions. This general approach has proven effective for nowcasting and forecasting the likelihood of encountering sea nettles (Chrysaora quinquecirrha), a stinging jellyfish (40), and is currently being developed for several harmful algal bloom species in the Bay.
The prediction system identifies geographic locations in the Bay where environmental conditions coincide with the preferred physical habitat of V. cholerae. This is accomplished by applying habitat suitability models developed previously (36) for V. cholerae in the Bay with ambient temperature and salinity fields simulated by the Regional Ocean Modeling System1 (ROMS) configured for the Bay. One of the regression models, employed by Louis et al. (36) in an early study, was used to predict the likelihood of the presence of V. cholerae in the Bay. ChesROMS, an open source Chesapeake Bay implementation of ROMS, uses historical reanalyses, near-real time observations and forecast data to provide model forcing, such as atmospheric momentum and heat fluxes, river outflow and ocean sea level, to simulate salinity, temperature and other physical (and soon biogeochemical) variables in the Bay to enable hindcasts, nowcasts and forecasts of V. cholerae.
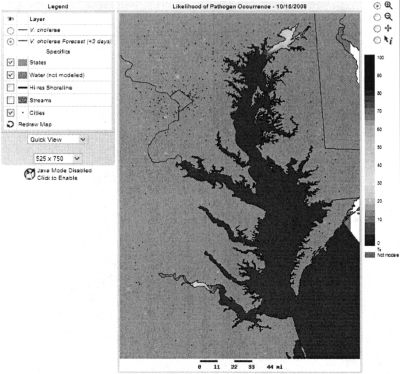
Predictions of V. cholerae probabilities are generated from the resulting daily sea-surface temperature and salinity fields of ChesROMS. Short-term predictions [i.e., nowcasts and 3-day forecasts (Figure 3)] are created daily and staged on the “Mapping Pathogens in the Chesapeake Bay” Web site, which currently has restricted access to its preliminary results.
Fig. 3.
Screenshot of the “Mapping Pathogens in the Chesapeake Bay” Web site displaying a three-day forecast of the probability of occurrence of V. cholerae in the Chesapeake Bay. The Web site supports typical capabilities, e.g. zooming, panning, and display options shown in the legend.
PERSPECTIVES
The prediction system we have produced is one of a suite of tools to identify and predict ocean-related public health risks from pathogens and represents a step toward developing a robust ecosystem modeling capability to serve current and future needs in information management. The main motivation of our project was to illustrate how an interdisciplinary research effort that includes microbiology, ecology and climatology can concretize academic research to a near-operational water-borne pathogen forecasting system. Despite current limitations, mainly due to the lack of external data for validation, the system can be of interest to a variety of users concerned with human health issues. Validation is crucial, however, in such an application and is the next step in the development of this system. Specifically, obtaining data for V. cholerae in the Chesapeake Bay will be possible through a new study that has been funded by the National Science Foundation in its “Ecology of Infectious Diseases” program. The goal of that study is to investigate the presence and abundance of three pathogenic vibrios (V. parahaemolyticus, V. vulnificus and V. cholerae) in three geographically and ecologically distinct ecosystems in the United States, one of which is the Chesapeake Bay. Sampling results will be compared to the model forecasts and will allow skill scores of assessment and either validation or model adjustment.
Future developments of the system will consist of adding information about human populations exposed to water quality to the Bay. These may include human population density data and information about activities such as recreational boating, beach bathing and sport fishing in the Chesapeake Bay and along its shoreline. Information about commercial activities that depend on the microbiological water quality of the Bay, such as shellfish harvesting, fish farms and commercial fishing, will also be included. Finally, other information, for example, locations of the occurrence of previous infections and of healthcare facilities, will be added, where available.
This operational prediction system will serve as a scientific tool to provide better understanding of in situ variability of V. cholerae by state agencies as a management tool to minimize the impact of V. cholerae on recreational activities by targeting microbial sampling efforts in the Chesapeake Bay and its tributaries. Access to the system is currently limited, but it will be customized to the needs of individuals or organizations for guidance in research and management activities, and to government agencies to guide pathogen monitoring programs and enhance first responder capabilities. In these ways, the system will provide a basic understanding and early warning of extreme weather events associated with an increase in specific microbial populations that threaten human health and, thereby, help mitigate deleterious effects on the health of both humans and the ecosystem.
ACKNOWLEDGEMENTS
This work was supported by National Oceanic and Atmospheric Administration Grant S0660009, Oceans and Human Health Initiative.
Footnotes
Potential Conflicts of Interest: None disclosed
REFERENCES
- 1.Colwell RR, Spira WM. The ecology of Vibrio cholera. In: Barua D, Greenough WB, editors. Cholera. III ed. New York: Plenum Medical Book Co.; 1992. pp. 107–27. [Google Scholar]
- 2.Faruque SM, Asadulghani, Saha MN, et al. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXPhi: molecular basis for origination of new strains with epidemic potential. Infect Immun. 1998;66(12):5819–25. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huq A, Colwell RR. Vibrios in the Marine and Estuarine Environment: Tracking of Vibrio cholerae. J Ecosystem Health. 1996;2:198–214. [Google Scholar]
- 4.Colwell RR, Huq A. Environmental reservoir of Vibrio cholerae. The causative agent of cholera. Ann N Y Acad Sci. 1994;740:44–54. doi: 10.1111/j.1749-6632.1994.tb19852.x. [DOI] [PubMed] [Google Scholar]
- 5.Shukla BN, Singh DV, Sanyal SC. Attachment of non-culturable toxigenic Vibrio cholerae O1 and non-O1 and Aeromonas spp. to the aquatic arthropod Gerris spinolae and plants in the River Ganga, Varanasi. FEMS Immunol Med Microbiol. 1995;12(2):113–20. doi: 10.1111/j.1574-695X.1995.tb00182.x. [DOI] [PubMed] [Google Scholar]
- 6.Islam MS, Draser BS, Bradley DJ. Attachment of toxigenic Vibrio cholerae O1 to various freshwater plants and survival with filamentous green algae Rhizoclonium fontanum. J Trop Med Hyg. 1989;92:396–401. [PubMed] [Google Scholar]
- 7.Colwell RR, Huq A. Global microbial ecology: biogeography and diversity of Vibrios as a model. J of App Microbiol Symp Suppl. 1999;85:134S–7S. doi: 10.1111/j.1365-2672.1998.tb05292.x. [DOI] [PubMed] [Google Scholar]
- 8.Colwell RR, Seidler RJ, Kaper J, et al. Occurrence of Vibrio cholerae serotype O1 in Maryland and Louisiana estuaries. Appl Environ Microbiol. 1981;41(2):555–8. doi: 10.1128/aem.41.2.555-558.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huq A, Small E, West P, Huq M, Rahman R, Colwell R. Ecological relationship between Vibrio cholerae and planktonic copepods. Appl Environ Microbiol. 1983;45:275–83. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl Environ Microbiol. 1990;56(6):1977–80. doi: 10.1128/aem.56.6.1977-1980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu C, Lee AM, Bassler BL, Roseman S. Chitin Utilization by Marine- Bacteria - a Physiological-Function for Bacterial Adhesion to Immobilized Carbohydrates. J Biol Chem. 1991;266(36):24260–7. [PubMed] [Google Scholar]
- 12.Sochard MR, Wilson DF, Austin B, Colwell RR. Bacteria associated with the surface and gut of marine copepods. Appl Environ Microbiol. 1979;37:750–9. doi: 10.1128/aem.37.4.750-759.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huq A. The role of planktonic copepods in the survival and multiplication of Vibrio cholerae in the aquatic environment. College Park, MD: University of Maryland; 1984. [Google Scholar]
- 14.Barcina I, Lebaron P, VivesRego J. Survival of allochthonous bacteria in aquatic systems: A biological approach. FEMS Microb Ecol. 1997;23(1):1–9. [Google Scholar]
- 15.McDougald D, Rice SA, Weichart D, Kjelleberg S. Nonculturability: adaptation or debilitation? FEMS Microb Ecol. 1998;25(1):1–9. [Google Scholar]
- 16.Kaneko T, Colwell RR. Incidence of Vibrio parahaemolyticus in Chesapeake Bay. Appl Microbiol. 1975;30(2):251–7. doi: 10.1128/am.30.2.251-257.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko T, Colwell RR. Adsorption of Vibrio parahaemolyticus onto chitin and copepods. Appl. Microbiol. 1975;29(2):269–74. doi: 10.1128/am.29.2.269-274.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huq A, West PA, Small EB, Huq MI, Colwell RR. Influence of water temperature, salinity, and pH on survival and growth of toxigenic Vibrio cholerae serovar O1 associated with live copepods in a laboratory microcosms. Appl Environ Microbiol. 1984;48:420–4. doi: 10.1128/aem.48.2.420-424.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nalin DR, Daya V, Reid A, Levine MM, Cisneros L. Adsorption and growth of Vibrio cholerae on chitin. Infect Immun. 1979;25:768–70. doi: 10.1128/iai.25.2.768-770.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colwell R, Huq A. Vibrios in the environment: Viable but Non-Culturable Vibrio cholerae. In: Wachsmuth I, Blake P, Olsvik O, editors. Vibrio cholerae and Cholera: Molecular to Global Perspectives. Washington, D.C.: ASM Press; 1994. pp. 117–33. [Google Scholar]
- 21.Lipp E, Huq A, Colwell R. Effects of global climate on infectious disease: the cholera model. Clin Microbiol Rev. 2002;15(4):757–70. doi: 10.1128/CMR.15.4.757-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiorboe T, Nielsen TG. Regulation of Zooplankton Biomass and Production in a Temperate, Coastal Ecosystem. 1. Copepods. Limnol Oceanogr. 1994;39(3):493–507. [Google Scholar]
- 23.Kaneko T, Colwell R. Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J Bacteriol. 1973;113(1):24–32. doi: 10.1128/jb.113.1.24-32.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawlings TK, Ruiz GM, Colwell RR. Association of Vibrio cholerae O1 El Tor and O139 Bengal with the Copepods Acartia tonsa and Eurytemora affinis. Appl Environ Microbiol. 2007;73(24):7926–33. doi: 10.1128/AEM.01238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colwell RR, Huq A, Islam MS, et al. Reduction of cholera in Bangladeshi villages by simple filtration. Proc Natl Acad Sci USA. 2003;100(3):1051–5. doi: 10.1073/pnas.0237386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilcox B, Colwell R. Emerging and Reemerging Infectious Diseases: Biocomplexity as an Interdisciplinary Paradigm. EcoHealth. 2005;2(4):244–57. [Google Scholar]
- 27.Boesch DF, editor. Global Warming and the Free State: Comprehensive Assessment of Climate Change Impacts in Maryland. Cambridge, Maryland: University of Maryland Center for Environmental Science; 2008. [Google Scholar]
- 28.Chambers JS. America's greatest scourge. New York: The Macmillan company; 1938. The conquest of cholera. [Google Scholar]
- 29.Barua D. History of Cholera. In: Barua D, Greenough WB, editors. Cholera. New York: Plenum Publishing; 1992. pp. 1–35. [Google Scholar]
- 30.Daly WJ. The black cholera comes to the central valley of America in the 19th century—1832, 1849, and later. Trans Am Clin Climatol Assoc. 2008;119:143–52. discussion 52–3. [PMC free article] [PubMed] [Google Scholar]
- 31.Weissman JB, DeWitt WE, Thompson J, et al. A case of cholera in Texas, 1973. Am J Epidemiol. 1974;100(6):487–98. doi: 10.1093/oxfordjournals.aje.a112061. [DOI] [PubMed] [Google Scholar]
- 32.Blake P. Endemic cholera in Australia and the Unites States. In: Wachsmuth PBIK, Olsvik O, editors. Vibrio cholerae and Cholera: Molecular to Global Perspectives. Washington, DC: Amercian Society of Microbiology; 1994. pp. 309–20. [Google Scholar]
- 33.Colwell RR. Global climate and infectious disease: the cholera paradigm. Science. 1996;274(5295):2025–31. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- 34.Pascual M, Bouma MJ, Dobson AP. Cholera and climate: revisiting the quantitative evidence. Microbes Infect. 2002;4(2):237–45. doi: 10.1016/s1286-4579(01)01533-7. [DOI] [PubMed] [Google Scholar]
- 35.Colwell RR, Kaper J, Joseph SW. Vibrio cholerae, Vibrio parahaemolyticus, and other vibrios: occurrence and distribution in Chesapeake Bay. Science. 1977;198(4315):394–6. [PubMed] [Google Scholar]
- 36.Louis VR, Russek-Cohen E, Choopun N, et al. Predictability of Vibrio cholerae in Chesapeake Bay. Appl Environ Microbiol. 2003;69(5):2773–85. doi: 10.1128/AEM.69.5.2773-2785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heidelberg JF, Heidelberg KB, Colwell RR. Seasonality of Chesapeake Bay bacterioplankton species. Appl Environ Microbiol. 2002;68(11):5488–97. doi: 10.1128/AEM.68.11.5488-5497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singleton FL, Attwell R, Jangi S, Colwell RR. Effects of temperature and salinity on Vibrio cholerae growth. Appl Environ Microbiol. 1982;44(5):1047–58. doi: 10.1128/aem.44.5.1047-1058.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipp EK, Huq A, Colwell RR. Effects of global climate on infectious disease: the cholera model. Clin Microbiol Rev. 2002;15(4):757–70. doi: 10.1128/CMR.15.4.757-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Decker MB, Brown CW, Hood RR, et al. Predicting the distribution of the scyphomedusa Chrysaora quinquecirrha in Chesapeake Bay. Mar Ecol Prog Ser. 2007;329:99–113. [Google Scholar]