Abstract
It has been found that c-Myc protein plays a critical role in controlling self-renewal versus differentiation in hematopoietic stem cells. We report that c-Myc also controls the fate of megakaryocyte-erythrocyte progenitors through regulating the differentiation of erythroid and megakaryocytic progenitors. In addition to the significant reduction of granulocytes/macrophages and B and T lymphocytes because of the reduction of their corresponding progenitors, we found significantly increased numbers of megakaryocytic progenitors and mature megakaryocytes in bone marrow and spleens of c-Myc-knockout (c-Myc−/−) mice. Differentiation of erythrocytes was blocked at the erythroid progenitor stage. This increased megakaryocytopoiesis is a cell-intrinsic defect of c-Myc-mutant hematopoietic stem cells, as shown by transplantation studies. Furthermore, we found that c-Myc is required for polyploidy formation but not for cytoplasmic maturation of megakaryocytes. Megakaryocytes from c-Myc−/− mice are significantly smaller in size and lower in ploidy than those of control mice; however, because of the dramatic increase in megakaryocyte number, although fewer platelets are produced by each megakaryocyte, a greater than 3-fold increase in platelet number was consistently observed in c-Myc−/− mice. Thus, c-Myc−/− mice develop a syndrome of severe thrombocytosis-anemia-leukopenia because of significant increases in megakaryocytopoiesis and concomitant blockage of erythrocyte differentiation and reductions in myelolymphopoiesis.
Introduction
During normal hematopoiesis, hematopoietic stem cells (HSCs) and their progeny continuously face a choice between 2 fates. The most primitive HSCs, which have long-term hematopoietic reconstitutive ability (LT-HSCs), are localized in bone marrow (BM) microenvironments (niches) where they either self-renew to maintain constant HSC numbers, ensuring life-long hematopoiesis, or differentiate to become short-term HSCs (ST-HSCs). ST-HSCs give rise to common lymphoid progenitors (CLPs) and common myeloid progenitors (CMPs), which in turn differentiate to produce T- or B-lymphocyte precursors and granulocyte/monocyte precursors (GMPs) or megakaryocyte/erythroid precursors (MEPs), respectively. Bipotent committed precursors such as MEPs and GMPs undergo further fate decisions by being directed toward either the erythroid or megakaryocytic branch or toward the granulocytic or monocytic lineage, respectively.1 It has been suggested that these dual-fate specification processes of HSCs and hematopoietic progenitors are largely controlled by fate-specific transcription factors and are regulated by stimulatory or repressive signals produced by BM niche cells.2 Despite many efforts, the transcription factors responsible for guiding the fate determination of HSCs and hematopoietic progenitors have not yet been fully identified.
c-Myc, a typical oncogene, is involved in controlling a variety of cell behaviors, including cell-cycle progression, cell proliferation, differentiation, survival, adhesion, and cell-size determination. c-Myc protein thus plays critical roles in many aspects of biologic processes, such as embryonic development, angiogenesis, and tissue regeneration.3,4 Deregulated c-Myc has been found in many malignant tissues and is critical for the pathogenesis of many types of mammalian tumors, including leukemia.5
Recent studies demonstrated that c-Myc also plays essential roles in both embryonic and adult hematopoiesis. Studies of epiblast-specific c-Myc deletion in mice suggested that c-Myc is specifically required for both yolk sac primitive and intraembryonic definitive hematopoietic development but is not necessary for the proliferation and development of nonhematopoietic tissues, such as vascular endothelia. c-Myc deficiency causes apoptotic loss of primitive erythroblasts and severely impairs definitive hematopoiesis. Although the anticipated number of phenotypically definitive HSCs was generated in c-Myc-knockout (c-Myc−/−) mice, c-Myc−/− HSCs/progenitors (HSC/Ps) were unable to expand in number and differentiate to generate a significant number of differentiated progeny. Therefore, in c-Myc−/− embryos, the percentage of HSCs was significantly increased, but the number of mature blood cells was substantially reduced. However, embryonic megakaryocyte development has not been studied in c-Myc−/− mice.6 Inducible c-Myc−/− mouse studies have demonstrated that c-Myc is also required for adult HSC function. c-Myc expression is low in quiescent LT-HSCs and is up-regulated when LT-HSCs differentiate into ST-HSCs and hematopoietic progenitors.7–10 It was found that c-Myc represses the expression of many adhesion-related molecules, including N-cadherin and integrin-β1, -α2, and -α5 in HSCs, thus regulating the process of adhesion of HSCs to the BM niche. In addition, c-Myc induces the expression of many positive regulators of the cell cycle and also represses several negative regulators of the cell cycle. These 2 sets of events are required for cell cycle progression and proliferative expansion of hematopoietic progenitors. However, the proliferation of HSCs is less dependent on c-Myc function. Because of BM niche-dependent differentiation defects, which are a consequence of enhanced BM niche adhesion, coupled with significantly impaired proliferation of hematopoietic progenitor cells, adult HSCs with c-Myc deletion fail to generate sufficient numbers of hematopoietic progenitors and mature blood cells. This results in significant HSC accumulation but also severe anemia and leukopenia in c-Myc−/− mice.7–10 Overexpression of c-Myc promotes detachment of HSCs from the BM niche and proliferation and differentiation, followed by a decline in HSC numbers.8 These studies suggest that c-Myc controls BM niche-dependent HSC self-renewal versus differentiation and fate determination, as well as hematopoietic progenitor expansion. However, the role of c-Myc in HSC/P lineage commitment has not been explored.
To study whether c-Myc is generally required for the proliferation and differentiation of all lineages of hematopoietic cells and, specifically, for the lineage commitment of HSC/Ps, we examined hematopoietic progenitors and mature blood cells in c-Myc−/− mice to determine whether certain hematopoietic lineages are less affected or may even be increased after c-Myc deletion. Consistent with findings in previous studies, we found that c-Myc−/− mice develop severe anemia and leukopenia because of impaired erythropoiesis and myelolymphopoiesis.8,10 Interestingly, we also found a 3- to 5-fold increase in platelet number in c-Myc−/− mice, which was the result of enhanced megakaryocytopoiesis.
Methods
Generation of c-Myc−/− mice
All experiments were performed in accordance with and with the approval of Loyola University Institutional Animal Care and Use Committee (protocol #06-013). c-Mycloxp mice7 were provided by Dr Frederick W. Alt of the Howard Hughes Medical Institute and Children's Hospital (Boston, MA). They were crossed with Mx1Cre mice11 to generate inducible c-Myc−/− mice. Three weeks after birth, all mice, including wild-type (WT) and heterozygous controls, were injected intraperitoneally with 25 μg of polyinosinic/polycytidylic acid (poly I:C) per gram body weight every other day for a total of 5 injections to induce c-Myc gene deletion.
Hematopoietic phenotype analysis
One month after the final poly I:C injection, mice were killed and peripheral blood (PB), BM, spleens, and thymuses were collected. Cellular components of PB of mutant mice were examined by complete blood count. Percentages and absolute numbers of HSCs and progenitor cells were examined in BM and spleen after cell surface marker staining as indicated in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) and by flow cytometric analysis, as described previously.12,13 Percentages and absolute numbers of different lineages of differentiated cells were also examined by different combinations of cell surface marker staining as indicated in the text and by flow cytometric analysis.
BM transplantation
To study whether the hematopoietic phenotype changes observed in HSCs of c-Myc−/− mice were cell-autonomous, we transplanted 2 × 106 BM nucleated cells from c-Myc−/− mice (before mutation induction) and WT mice separately into lethally irradiated recipient mice. Six weeks after transplantation, we examined white blood cell counts (WBC), hemoglobin concentration, and platelet numbers of recipient mice to confirm the complete recovery of hematopoiesis. Mice were injected with poly I:C every other day for a total of 3 injections to induce c-Myc deletions. One month hence, hematopoietic phenotypes of recipient mice, which had received BM cells from c-Myc−/− mice, were analyzed and compared with phenotypes of mice that had received WT BM cells.
CFU-S assay
One month after mutation induction, 2 × 105 BM nucleated cells from c-Myc−/− mice and WT control mice were injected separately into lethally irradiated recipient mice. Spleens of recipient mice were collected on day 12 for colony-counting and further analyses.
Colony-forming assay
Functional hematopoietic progenitors in BM of c-Myc−/− mice were analyzed and compared with those of WT controls by in vitro colony-forming assays, including granulocyte-monocyte colony-forming unit, colony-forming unit-erythroid, and burst-forming unit-erythroid, using MethoCult GF M3434 methylcellulose-based medium (StemCell Technologies) as previously described.13 Megakaryocyte progenitors were examined using MegaCult-C collagen-based medium supplemented with 50 ng/mL recombinant human thrombopoietin (TPO), 10 ng/mL recombinant mouse (rm) interleukin-3 (IL-3), 20 ng/mL rm IL-6, and 50 ng/mL rm IL-11, following the protocol provided by the vendor. Megakaryocytic colony-forming unit (CFU-Mk) numbers were counted on day 8.
RT-PCR
RNA was extracted using RNeasy Plus Mini-Kit (QIAGEN). cDNA was prepared using SuperScript First-Strand Synthesis System (Invitrogen). Quantitative polymerase chain reaction (PCR) for detecting c-Myc levels in normal hematopoietic cells and β1-tubulin, NF-E2, and PF4 levels in WT and mutant megakaryocytes was performed using SYBR Green PCR Master (Applied Biosystems). Quantitative PCRs for comparing gene expression between WT and c-Myc−/− HSCs were performed using Taqman technique. Information concerning primers used can be found in supplemental data. Triplicate reverse-transcribed (RT) PCRs were performed and confirmed in 2 independent experiments.
AChE staining
Fresh-frozen spleen sections or slides with CFU-Mk colonies were fixed in ice-cold acetone for 5 minutes. After drying at room temperature, tissue sections and colony slides were stained with acetylcholinesterase (AChE) staining solution containing 0.5 mg/mL acetylthiocholiniodide, 5 mM sodium citrate, 3 mM copper sulfate, 0.5 mM potassium ferricyanide in 0.1 M sodium phosphate buffer. After a 3-hour incubation at room temperature, the staining solution was poured off and sections were fixed in 95% ethanol for 10 minutes. Slides were counterstained with Harris hematoxylin solution for 30 seconds.14
Propidium iodide staining of CD41+ megakaryocytes
BM and spleen nucleated cells were stained with fluorescein isothiocyanate–conjugated CD41 antibody at 4°C for 30 minutes and then permeabilized using Perm/Wash Buffer (BD Biosciences). Cells were stained in propidium iodide staining solution for 40 minutes at 37°C.
Histology and immunohistochemical staining
WT and mutant mice were killed, and femurs, spleens, and livers were immediately collected. After 2 to 3 days fixation in zinc formalin, femurs were decalcified using 10% ethylenediaminetetraacetic acid (pH 7.4) for 2 weeks. BM and spleen tissues were embedded in paraffin. Sections were cut for hematoxylin and eosin staining or antibody immunostaining.
Annexin-V staining
To analyze the degree of apoptosis in CD41+ megakaryocytes and BM HSC/Ps, BM was collected from c-Myc−/− and control mice 25 days after poly I:C injections. BM cells were collected into lyse/fix buffer (BD Biosciences) for 10 minutes to fix nucleated cells and lyse red blood cells simultaneously. After 2 washes with cold phosphate-buffered saline/2% fetal bovine serum, nucleated cells were adjusted to a concentration of 5 × 106/mL in 1× Binding Buffer (BD Biosciences) and aliquoted into 5-mL staining tubes at 100 μL of cells per tube. Cells were stained with phycoerythrin-CD41 and fluorescein isothiocyanate–annexin-V (BD Biosciences) for 20 minutes at room temperature. After 2 washes in 1× Binding Buffer, cells were analyzed by flow cytometry for annexin-V–positive cell percentage in the different cell populations.12
BrdU labeling
Mice were injected with 200 μg bromodeoxyuridine (BrdU) 4 hours before being killed. The percentage of BrdU-positive cells among CD41+ megakaryocytes, Gr1+ neutrophils, and LSK-HSCs was assessed by cell-surface marker staining followed by cell permeabilization and BrdU antibody staining as previously described. BrdU in situ histologic staining was conducted as previously described.12
Statistical analyses
Student t tests were performed to assess the statistical significance of observed changes between c-Myc−/− mice and their corresponding controls. Error bars in all panels represent SEM.
Images
All pictures were taken using an Olympus Provis AX80 microscope and captured with a Q-Imaging Retiga 4000R camera. Images were acquired and processed using Adobe Photoshop 6.0.
Results
c-Myc expression in hematopoietic cells
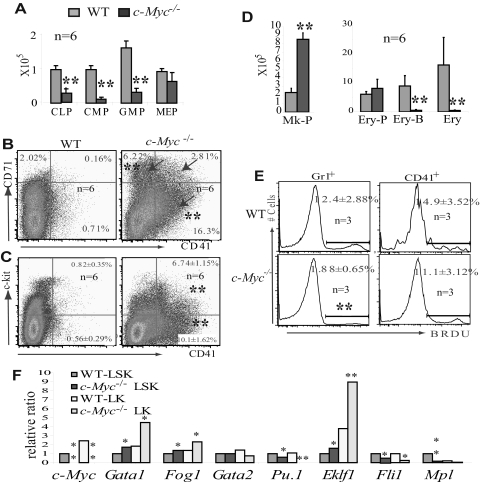
To investigate whether c-Myc is generally required for the proliferation and differentiation of all lineages of hematopoietic progenitors or whether it is selectively required for the development of certain lineages only, we first examined the expression of c-Myc in LT-HSCs, ST-HSCs, and committed progenitors (including CLPs, CMPs, GMPs, and MEPs), as well as differentiated hematopoietic cells, using quantitative RT-PCR assays. The expression levels of c-Myc in HSC/Ps and mature hematopoietic cells were normalized to c-Myc levels in LT-HSCs. As shown in supplemental Figure 1, we found low levels of c-Myc expression in LT-HSCs and significantly increased levels in ST-HSCs, CMPs, and CLPs. However, during the differentiation of CMPs to GMPs and MEPs, we found that c-Myc is differentially expressed in GMPs and MEPs, with higher levels in GMPs (similar to levels observed in CMPs) and lower levels in MEPs (similar to levels observed in LT-HSCs). Furthermore, we found that c-Myc is also differentially expressed during the differentiation of MEPs to megakaryocytes and erythrocytes. Megakaryocytic progenitors (Mk-Ps) express relatively lower levels of c-Myc than erythrocytic progenitors (Ery-Ps). Expression levels of c-Myc are significantly down-regulated during megakaryocyte maturation, are up-regulated during differentiation of Ery-Ps to erythrocytic blasts (Ery-Bs), and are significantly down-regulated during further maturation. Differences in c-Myc expression in hematopoietic cells during differentiation suggest that c-Myc may play a role in HSC/P differentiation and in lineage decisions.
Thrombocytosis in c-Myc−/− mice
To study whether c-Myc controls HSC/P lineage determination, we generated interferon-inducible c-Myc−/− mice by crossing c-MycloxP mice with Mx1Cre mice. We generated c-Myc−/− (MxCre+c-Mycloxp/loxp), c-Myc heterozygous (MxCre+c-Mycloxp/+, c-Myc+/− hereafter), and WT (MxCre−c-Mycloxp/loxp or MxCre−c-Mycloxp/+) mice in the same litter so that each c-Myc−/− mouse had proper controls. Exons 2 and 3 of the c-Myc gene (encoding almost the entire c-Myc protein) in c-MycloxP mice are flanked by 2 loxp sites.4 Deletion of these exons by Cre-mediated enzymatic recombination of loxp sites results in the complete loss of c-Myc function in target cells. Expression of the Cre enzyme, driven by the Mx1 promoter in Mx1Cre mice, can be induced by injecting mice with poly I:C,11 which can efficiently induce recombination of the 2 loxp sites and deletion of the target gene in hematopoietic cells and BM stromal cells.12
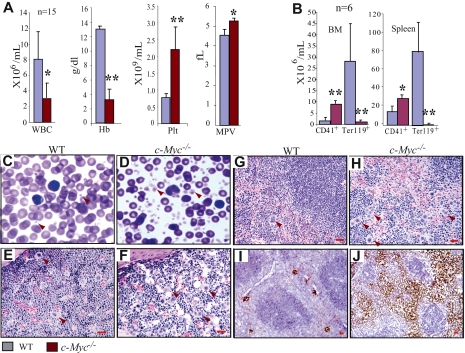
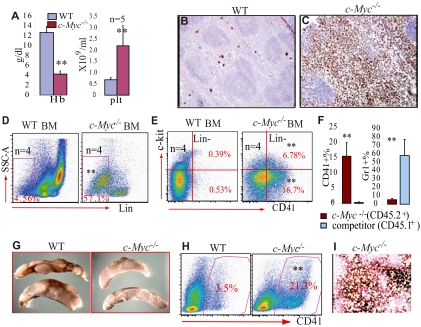
Three weeks after birth, all mice (including c-Myc+/− and WT controls) were injected with poly I:C to induce c-Myc deletion. The efficiency of c-Myc deletion in c-Myc−/− HSCs (> 99%) was determined by both quantitative PCR to detect the genomic DNA (supplemental Figure 2) and real-time RT-PCR to detect RNA expression (Figure 2F) 20 days after poly I:C injection. Hematopoietic phenotypes of the mice were analyzed one month after mutations were induced. Because the phenotype of c-Myc+/− mice is comparable with that of WT controls (data not shown), we focused on c-Myc−/− and WT in our comparative studies. First, by examining the numbers of cellular components in PB, we found significantly reduced WBC counts (Figure 1A) because of significant reductions of both neutrophils and lymphocytes (data not shown), as well as severe anemia in c-Myc−/− mice compared with their WT controls (Figure 1A). These data were consistent with what had previously been reported.8,10 However, we consistently observed a more than 3-fold increase in platelets in c-Myc−/− mice compared with their WT littermate controls (Figure 1A,C-D), which had not yet been reported. In addition, we found that platelet size (mean platelet volume) in c-Myc−/− mice was larger than that seen in WT mice (Figure 1A,C-D). Furthermore, we found that megakaryocyte numbers are significantly increased in the BM and spleens of c-Myc−/− mice compared with WT controls, as shown by both histologic examination and flow cytometric analysis (Figure 1B,E-H and supplemental Figure 3), and further confirmed by megakaryocyte-specific AChE staining (Figure 1I-J) as well as CD41 and factor-VIII–related antigen immunohistochemical staining (supplemental Figure 3E). The significant thrombocytosis, severe anemia, and grossly decreased neutrophil/monocyte and lymphocyte numbers in the PB of c-Myc−/− mice suggested that c-Myc might either be involved in the regulation of HSC/P lineage commitment or be differentially required for the proliferation and differentiation of various lineages of hematopoietic cells.
Figure 2.
HSC/Ps with c-Myc deletion are biased toward megakaryocytic differentiation at the expense of other hematopoietic lineages. (A) CMPs, CLPs, and GMPs were significantly reduced in c-Myc−/− mouse BM; however, the number of MEPs was less affected. (B) Increased percentages of CD71+ and CD41+ (arrow), 2 distinct populations of cells in BM of c-Myc−/− mice. (C) Increased percentages of both CD41+c-kit− and CD41+c-kit+ populations in c-Myc−/− BM. (D) The absolute number of Mk-Ps (Lin−Sca1−CD41+c-kit+CD9+) is significantly increased in c-Myc−/− BM (2 hind limbs), whereas the number of Ery-Ps (CD71+Ter119−) is less affected. However, the number of Ery-Bs (CD71+Ter119+) and differentiated nucleated Erys (CD71−Ter119+) is significantly reduced in c-Myc−/− BM (2 hind limbs). (E) Proliferation of CD41+ megakaryocytes is less affected, whereas the proliferation of Gr1+ granulocytes is significantly decreased in c-Myc−/− mice. (F) HSC/Ps from c-Myc−/− mice express high levels of erythrocyte and megakaryocyte-specific genes, including Gata1, Fog1, and Eklf1, but reduced lymphoid and myeloid-specific genes, such as Pu.1. The expression levels of detected genes in c-Myc−/− LSK and LK populations are shown relative to expression levels in the WT LSK cell population. The significant reduction of c-Myc levels in c-Myc−/− HSC/Ps indicates the success of the induced c-Myc deletion. Data are an average of triplicate experiments. *P < .05; **P < .01.
Figure 1.
Significant thrombocytosis in c-Myc−/− mice. (A) Decreased WBC and hemoglobin concentration but significantly increased platelets in PB of c-Myc−/− mice. Enlarged platelet size in c-Myc−/− mice is shown as mean platelet volume (MPV). (B) Significant increase in the absolute number of CD41+ megakaryocytes but reduction in Ter119+ nucleated erythrocytes in both BM (2 hind limbs) and spleens of c-Myc−/− mice compared with their age- and gender-matched WT controls. (C-D) PB smears (Wright-Giemsa staining 100×/1.3) show increased platelets in c-Myc−/− mice. (E-J) Increase in megakaryocyte numbers in c-Myc−/− BM (F) and spleens (H,J), compared with WT BM (E) and spleens (G,I), as shown by hematoxylin and eosin staining (40×/0.8 air; E-H) and confirmed by megakaryocyte-specific AChE staining (20×/0.7 air) oil (I,J). *P < .05; **P < .01.
The sensitivities of various lineages of hematopoietic progenitors to c-Myc deletion differ
To investigate whether the combined thrombocytosis, anemia, and leukopenia phenotype of c-Myc−/− mice is the result of lineage commitment defects in HSC/Ps, we analyzed hematopoietic progenitors in c-Myc−/− BM. Consistent with previous reports, the Lin−Sca1+c-kit+ (LSK) population is significantly increased because of the dramatic elevation of LT-HSCs and the notable increase in ST-HSCs in c-Myc−/− mice (supplemental Figure 4B,G). However, CLP, CMP, and GMP cell populations are significantly reduced in the BM of c-Myc−/− mice, which is consistent with the significant reduction in mature neutrophils/monocytes and lymphocytes (Figure 2A, supplemental Figure 4C-D).8,10 However, we found that the MEP population is less affected in c-Myc−/− mice (Figure 2A, supplemental Figure 4C). In addition, significant increases in CD71+, CD41+, and CD71+/CD41+ cell populations were observed in both BM and spleens of c-Myc−/− mice (Figure 2B, supplemental Figure 4). Further studies demonstrated that the percentages of Lin−Sca1−CD41+c-kit+CD9+ Mk-Ps (supplemental Figure 4E) and CD41+c-kit− differentiated megakaryocytes (Figure 2C), as well as CD71+Ter119− erythrocytic progenitors (Ery-Ps; supplemental Figure 4F), are significantly increased in the BM of c-Myc−/− mice compared with WT controls. However, the percentages of CD71+Ter119+ Ery-Bs and CD71−Ter119+ differentiated erythrocytes (Erys) are significantly reduced in c-Myc−/− mice (supplemental Figure 4F). Despite the hypocellularity of c-Myc−/− mouse BM (supplemental Figure 3B), the absolute number of Mk-Ps is still significantly increased in c-Myc−/− BM compared with WT; however, the number of Ery-Ps in c-Myc−/− BM is comparable to that seen in WT controls, whereas the number of Ery-Bs and Erys is reduced more dramatically (Figure 2D). These data suggest that the differentiation of HSCs to MEPs is less dependent on c-Myc function than is the differentiation of HSCs to GMPs and CLPs. MEPs from c-Myc−/− mice can differentiate to both Mk-Ps and Ery-Ps. However, because of the differentiation blockage of Ery-Ps to Ery-Bs in c-Myc−/− mice, more MEPs differentiate to Mk-Ps and further mature to Mks, and finally to platelets. The significant increase in expression levels of the MEP lineage-specific transcription factor Gata1 and its partner gene Fog1, as well as the erythrocyte-specific gene Eklf1, together with the significant reduction in the myeloid/lymphocyte lineage-specific transcription factor Pu.1 in LSK-HSC/Ps, suggested that the lineage commitment defects of c-Myc−/− HSCs might already be determined at the HSC stage (Figure 2F). The further increase or decrease in these transcription factors in the c-Myc−/− lin−c-kit+sca1− (LK) progenitor cell population might be the consequence of the increased MEP percentage within the LK cell population in c-Myc−/− mice compared with WT controls. Interestingly, the sensitivity of hematopoietic cells to c-Myc deletion is well correlated to c-Myc expression levels (supplemental Figure 1). BrdU pulse-labeling indicated that the proliferation of Gr1+ granulocytes, which express relatively higher levels of c-Myc, is significantly reduced in c-Myc−/− mice, whereas the proliferation of CD41+ megakaryocytes, which express low levels of c-Myc, is less affected by c-Myc deletion (Figure 2E). However, despite the obvious increase in phenotypic Mk-Ps in c-Myc−/− BM, the colony-forming ability of these cells is compromised, as shown by the significant reduction in megakaryocytic colonies, CFU-Mk (supplemental Figure 4H), which may be the result of the increased apoptotic cell death of the mutant megakaryocytes (Figure 3B-C,F) or in vivo microenvironment-dependent growth of the mutant megakaryocytes.
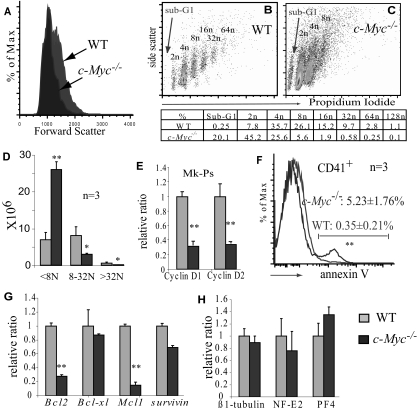
Figure 3.
c-Myc is required for endomitosis and polyploidy differentiation but is not essential for megakaryocyte maturation. (A) c-Myc−/− megakaryocytes are smaller than WT megakaryocytes as shown by flow cytometry forward scatter. (B,C) c-Myc−/− megakaryocytes have lower ploidy, as shown by propidium iodide staining. The appearance of the sub-G1 population in the c-Myc−/− CD41+ population suggests increased apoptosis in c-Myc−/− megakaryocytes. A typical representative of 3 separate experiments is shown. (D) Comparison of the absolute number of low-ploidy (< 8N), medium ploidy (8-32N), and high ploidy (> 32N) in BM plus spleens of c-Myc−/− and WT control mice. (E) Cylin D1 and D2 expression is down-regulated in c-Myc−/− MK-Ps (lin−Sca1−c-kit+Cd41+) compared with WT counterparts. (F) Increased apoptosis of CD41+ megakaryocytes as shown by annexin-V staining. (G) Down-regulation of pro-survival genes Bcl-2 and Mcl1 in CD41+ megakaryocytes of c-Myc−/− mice compared with WT controls. (H) Expression of mature megakaryocytic markers in c-Myc−/− megakaryocytes (CD41+c-kit−) compared with that of WT counterparts. **P < .01.
c-Myc is required for megakaryocyte endomitosis and polyploidy formation but is not essential for megakaryocyte maturation
Mk-Ps undergo endomitosis to become large polyploid cells. At the same time, the cytoplasm matures, resulting in the formation of proplatelets. We found that CD41+ megakaryocytes from c-Myc−/− mice are smaller than megakaryocytes from WT mice, as shown by both histologic study (Figures 1E-J, 4) and flow cytometric analysis (Figure 3A,D). This might be the result of a reduction in the formation of polyploidy (Figure 3B-C). Previous studies suggested that polyploid formation in megakaryocytes might be dependent on the expression of cyclin D isotypes. We found that, in c-Myc−/− MK-Ps, cyclin D1 and cyclin D2 are significantly down-regulated (Figure 3E). In addition, we found a significant increase in apoptosis in c-Myc−/− megakaryocytes, as shown by a significant increase in the sub-G1 population (Figure 3C). This was confirmed by annexin-V staining (Figure 3F) and was probably the result of the down-regulation of expression of pro-survival genes such as Bcl-2 and Mcl-1 (Figure 3G). The comparable levels of mature markers such as β1-tubulin, NF-E2, and PF4 in c-Myc−/− CD41+c-kit− megakaryocytes and WT megakaryocytes suggested that c-Myc may not be essential for the cytoplasmic maturation of megakaryocytes (Figure 3H).
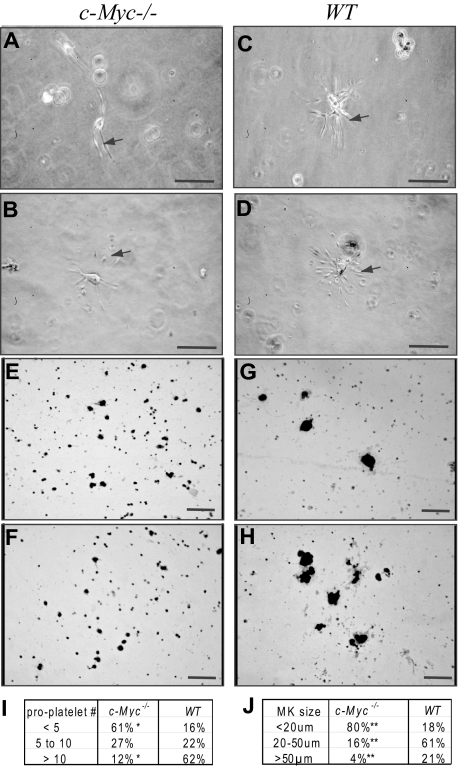
Figure 4.
The percentage of small-sized megakaryocytes, which produce fewer numbers of proplatelets, was increased in c-Myc−/− mice compared with WT controls. BM nucleated cells from c-Myc−/−and WT control mice were seeded at a final concentration of 5 × 105/mL in MegaCult-C Collagen-Medium (StemCell Technologies) supplemented with 50 ng/mL rh TPO,10 ng/mL rm IL-3, 20 ng/mL rh IL-6, and 50 ng/mL rh IL-11. Cells were incubated at 37°C, 5% CO2. The megakaryocyte maturation process was observed 48 hours later. We found that significant numbers of megakaryocytes had matured, as indicated by the development of proplatelet projections and, later (A,C) by platelet production (B,D). Shown are representative photomicrographs of megakaryocytes from c-Myc−/−mice (A-B) and WT control mice (C-D). Based on the number of proplatelet projections, megakaryocytes were classified into 3 groups as summarized in panel I. We found that the percentage of megakaryocytes producing fewer proplatelets (< 5) was significantly increased, whereas the percentage of megakaryocytes producing larger numbers of proplatelets (> 10) was significantly decreased in c-Myc−/− mice compared with WT controls. A total of 83 and 95 megakaryocytes were counted in c-Myc−/− and WT control mice, respectively. Megakaryocytes were confirmed by AChE staining as shown in panels E to H. Megakaryocytes cultured as described in panels A to D were harvested 72 hours after incubation and stained with AChE. Shown are representative photomicrographs of AChE-positive megakaryocytes from c-Myc−/− (E-F) and WT control mice (G-H). The size of AChE-positive megakaryocytes was measured. Based on diameter, megakaryocytes were classified into 3 groups as summarized in panel J. *Significant difference compared with WT control mice. Bar represents 100 μm. Panels A-D, 40×/0.8 air; panels E-H, 20×/.7 air.
Small-sized low-ploidy megakaryocytes are able to mature and produce platelets
It is normally accepted that large polyploid megakaryocytes produce platelets. In c-Myc−/− mice, only small-sized low-ploidy megakaryocytes are significantly increased, whereas medium- and large-sized high-ploidy megakaryocytes are reduced (Figures 3D, 4). Therefore, we predicted that the small megakaryocytes might produce platelets, which contribute to the thrombocytosis phenotype of c-Myc−/− mice. To address this hypothesis, we incubated the BM cells from c-Myc−/− mice and WT mice in MegaCult-C Collagen-Medium supplemented with proper amounts and types of growth factors. Two days after incubation, proplatelet-producing megakaryocytes were analyzed by counting the number of proplatelet projections of each megakaryocyte. After AChE staining, the sizes of megakaryocytes were measured. We found that small megakaryocytes, which produce fewer numbers of proplatelets, were significantly increased in c-Myc−/− mice compared with WT controls. These data demonstrated that low-ploidy megakaryocytes from c-Myc−/− mice have the ability to produce proplatelets, although fewer proplatelets are produced by each of these megakaryocytes compared with their WT counterparts (Figure 4).
The thrombocytosis phenotype of c-Myc−/− mice is the result of HSC cell-intrinsic defects
Mx1Cre-mediated rearrangement induces deletion of c-Myc in both hematopoietic cells and their BM niche stromal cells. To study whether the megakaryocytic lineage differentiation bias of c-Myc−/− HSC/Ps is cell-intrinsic or the result of BM microenvironmental defects, we examined whether the thrombocytosis phenotype of c-Myc−/− mice is transplantable. We found that c-Myc−/− BM transplantation was able to effectively regenerate HSC accumulation and thrombocytosis in the WT BM environment. This suggested that cell-autonomous defects in c-Myc−/− HSC/Ps were causative of the thrombocytosis-anemia-leukopenia phenotype (Figure 5A-E). This was further confirmed by competitive transplantation experiments (Figure 5F), as well as by studying colony-forming units in spleen (CFU-S; Figure 5G-I). In competitive transplantation experiments, the contribution of c-Myc−/− HSCs to BM hematopoiesis in recipient mice is significantly lower than that of competitor HSCs (< 6%, data not shown), which is consistent with what was observed in previous studies.2 However, the percentage of CD41+ megakaryocytes in hematopoietic cells derived from c-Myc−/−HSCs (CD45.2+) is significantly higher than that in hematopoietic cells derived from competitor HSCs (CD45.1+; Figure 5F). In CFU-S experiments, transplantation of c-Myc−/− BM cells generated relatively smaller CFU-Ss in recipient mouse spleens, which makes counting colony numbers difficult (Figure 5G). Histologic analysis indicated that the CFU-Ss generated by c-Myc−/− HSC/Ps contain significantly more megakaryocytes, as shown by flow cytometric analysis of CD41 expression and AChE staining (Figure 5H-I).
Figure 5.
Cell-intrinsic defects in c-Myc−/− HSC/Ps. (A-E) A total of 2 × 106 BM cells from c-Myc−/− mice (before c-Myc deletion was induced) and WT control mice were transplanted into lethally irradiated recipient mice separately. Six weeks after transplantation, at which point hematopoiesis was completely regenerated in the recipient mice by donor HSCs, recipient mice were injected with poly I:C to induce c-Myc deletion. The hematopoietic phenotypes of recipient mice were analyzed one month after poly I:C injection. (A) Thrombocytosis and anemia developed in recipient mice receiving c-Myc−/− BM hematopoietic cells. (B-C) Increased megakaryocytosis in the spleens of recipient mice receiving c-Myc−/− BM cells, as shown by AChE staining (20×/.7 air). (D-E) Increased Lin− cell population (D), as well as Lin−CD41+c-Kit+ and Lin−CD41+c-Kit− megakaryocytes (E) in BM of mice receiving c-Myc−/− BM hematopoietic cell transplantation, as shown by flow cytometric analysis. (F) Equal numbers of c-Myc−/− BM cells (before c-Myc deletion was induced, CD54.2+ background) and competitor BM cells (CD54.1+ background) were transplanted into lethally irradiated recipient mice (CD54.1+ background). One month after transplantation, recipient mice were injected with poly I:C to induce c-Myc deletion. The percentages of CD41+ and Gr1+ cells in CD45.1+ population (competitor BM cell-derived) or CD45.2+ (c-Myc−/− BM-derived) population were examined, respectively, by flow cytometry. (G-I) Colony-forming units in spleen (CFU-S) generated by c-Myc−/− HSC/Ps (G) are enriched for CD41+ megakaryocytes, as shown by flow cytometric analysis (H) and AChE staining (I). **P < .01.
Discussion
The role of c-Myc in regulating embryonic and adult HSCs has been well demonstrated.6,8,10 Here, we report an as yet undefined role for c-Myc in megakaryocytic development. We found significant increases in megakaryocytopoiesis and reductions in erythropoiesis/myelolymphopoiesis, resulting in a thrombocytosis-anemia-leukopenia phenotype in c-Myc−/− mice. Two potential mechanisms can explain these selective changes: (1) c-Myc controls the lineage switch of HSC/Ps; and (2) different lineages of hematopoietic cells differ in their requirement for c-Myc to regulate their proliferation and differentiation. Our data support the latter explanation. We found that the transition of CMPs to MEPs as well as the proliferation and differentiation of MK-Ps seem less dependent on c-Myc function than is the case for other lineages. This increased megakaryocytopoiesis seems not to be a consequence of an anemia-induced feedback reaction (megakaryocytes and erythrocytes share a common progenitor cell) because the number of MEPs is not increased in c-Myc−/− mice and also because, in competitive transplantation experiments, c-Myc−/− HSC/Ps generated more megakaryocytes than their WT counterparts in control mice in which no anemia was observed (Figure 5F).
c-Myc is an oncogene known to be a target gene and effector molecule of many leukemogenic fusion proteins. Up-regulation of c-Myc gene has been found in patient BM samples of most types of leukemia, including acute lymphocytic leukemia, acute myeloid leukemia, and acute erythroid leukemia, suggesting that c-Myc might be critical in the pathogenesis of these diseases.15–17 The involvement of c-Myc in these types of leukemia has also been confirmed by transgenic animal model studies.18,19 However, the role of c-Myc in acute megakaryocytic leukemia (AMkL) has not been explored. Interestingly, c-Myc has been found to be the major target of Rbm15, an AMkL-related protein, which mediates the role of Rbm15 in HSC-BM niche interactions and megakaryocytopoiesis in mice. The AMkL fusion protein Rbm15/Mkl1 functions in a dominant negative fashion to down-regulate c-Myc expression in HSC/Ps (C.N., J.Z., P.B., S.W.M. et al, unpublished data). The significant increase in CD41+CD71+ cell populations in our c-Myc−/− mice is reminiscent of increases in the same cell populations in some AMkL in Down syndrome persons.20 We propose that, in contrast to other types of leukemia, down-regulation of c-Myc might play a role in the pathogenesis of AMkL.
Selective requirement for c-Myc in proliferation of different populations of hematopoietic cells
c-Myc has been shown to be a critical cell cycle mediator promoting cell cycle progression from G0/G1 to S phase by up-regulating positive cell cycle regulators and down-regulating negative cell cycle regulators.3,21,22 In vitro studies have suggested that c-Myc expression is necessary and sufficient to induce quiescent cells to enter S phase in most cells.22 Deletion of c-Myc leads to a markedly prolonged cellular doubling time and severely impaired cellular proliferation in response to mitogenic stimulation.22,23 However, recent studies demonstrated that the role of c-Myc in cell cycle progression and proliferation might be cell-type specific.6,24
During early embryonic development, c-Myc is highly expressed in hematopoietic cells and is specifically required for proliferation and differentiation of HSC/Ps. However, proliferation in most nonhematopoietic tissues is not affected by c-Myc deletion at this developmental stage.6,25 Conditional c-Myc knockout studies suggested that the proliferation of a subset of cells in adult tissues is c-Myc-independent.26,27 Indeed, the requirement of c-Myc for oncogenesis can sometimes be replaced by other oncogenic events.28 All of these studies suggested that the requirement for c-Myc in cell cycle progression and proliferation might be cell context-dependent. Whether this is the result of compensation by other Myc family members such as N-Myc or L-Myc requires further study in Myc compound-mutant mice.9,29
We found that the proliferation of HSCs is independent of c-Myc, consistent with previous reports.8 In addition, we found that megakaryocyte proliferation is also not affected by c-Myc deletion. However, the proliferation of neutrophils is significantly impaired in c-Myc−/− mice. Why c-Myc is selectively required for the proliferation of other hematopoietic cells but not for HSCs and megakaryocytes is an important topic to be addressed in future studies. Currently, we cannot rule out the possibility of compensation by other Myc family members in HSCs and megakaryocytes, although we found no alterations in the expression of either N-Myc or L-Myc in HSCs and megakaryocytes. Nor are these genes altered in neutrophils after c-Myc is deleted (supplemental Figure 5).
Selective requirement for c-Myc in differentiation of certain types of hematopoietic progenitors
Previous transgenic studies suggested that c-Myc might be involved in regulating lineage specification in epidermal stem cells.30 The molecular mechanism underlying this process is not yet known. It is commonly accepted that overexpression of c-Myc promotes cell proliferation and blocks full maturation in tissue progenitor cells, thereby inducing abnormal growth of tissue cells and tumor formation.31 However, high levels of c-Myc might also induce apoptosis in certain cell types because of elevated p53 levels via activation of the Arf-Mdm2-p53 pathway.32 Interestingly, some types of cells might be relatively more resistant to c-Myc-induced apoptosis. This could explain why c-Myc transgenic mice tend to develop certain histologic tumor types but not others.18,33
Whether c-Myc is physically involved in cell lineage decisions is unknown. We found that, compared with GMPs and CLPs, the number of MEPs is less affected in c-Myc−/− mice. Furthermore, Mk-Ps and mature Mks are significantly increased, whereas Ery-Ps are less affected; Ery-Bs and mature Erys are significantly reduced. These data indicate that c-Myc is more important for the development of certain lineages of hematopoietic cells (such as CMP to GMP to granulocytes and Ery-Ps to Ery-Bs to mature erythrocytes) than other lineages (such as MEP to Mk-Ps to mature Mks). We do not think that this is because other lineages of hematopoietic cells are more heavily reliant on c-Myc for their survival than megakaryocytes because we found that, in c-Myc−/− mice, apoptosis is even greater in megakaryocytes than in other lineages. Our data support the idea that megakaryocytes are less reliant on c-Myc for their proliferation and differentiation than other lineages. Actually, the effect of c-Myc deletion on hematopoietic cells correlates well with c-Myc expression; cells expressing higher levels of c-Myc (such as GMPs, CLPs, and Ery-Bs) are significantly reduced, whereas cells expressing lower levels (HSCs, MEPs and megakaryocytes) are less affected or are even increased in number.
The role of c-Myc in regulating bifate differentiation of MEPs to megakaryocytes or erythrocytes has not been explored. In vitro studies using cell lines demonstrated that c-Myc is required for proliferation of both megakaryocyte and erythrocyte progenitors; c-Myc expression is down-regulated during this process.34,35 Transgenic overexpression of c-Myc in MEPs, driven by the Gata1 promoter, leads to erythroid leukemia development.36 However, megakaryocytic lineage-specific c-Myc overexpression under the control of the platelet factor-4 promoter yields only a slight increase in the frequency of low-ploidy megakaryocytes because of enhanced proliferation and survival along with the blocking of differentiation.37 We found that c-Myc−/− MEPs can differentiate to both Ery-Ps and Mk-Ps. However, c-Myc is required for Ery-Ps to differentiate to Ery-Bs. Therefore, erythrocyte differentiation is blocked at the Ery-P stage in c-Myc−/− mice, whereas further maturation of megakaryocytes and platelet production might not rely on c-Myc. The role of c-Myc in the fate determination of MEPs was further confirmed by c-Myc overexpression and in vitro culture studies (supplemental Figure 6). Furthermore, our data suggest that c-Myc is necessary for polyploidy development and increased cell size in megakaryocytes but is not required for cytoplasmic maturation or platelet production in megakaryocytes.
Several hematopoietic transcription factors, including Gata11/Fog1, Fli-1, and Eklf, have been shown to be critical for the commitment of HSC/Ps to megakaryocytic or erythrocytic lineages. Fog1 forms a complex with Gata1, which is essential for both megakaryocytic and erythrocytic differentiation.38,39 Overexpression of Gata1 converts CLPs and GMPs to the megakaryocyte/erythrocyte lineages.40 Mice with Gata1 and Fog1 deletions develop significant thrombocytopenia and anemia.41,42 Eklf is expressed in MEPs and erythrocytes, being essential for erythrocyte maturation but limiting megakaryocyte differentiation.43 Fli-1 functions conversely to Eklf; the former promotes megakaryocytic lineage differentiation while restricting commitment to the erythroid lineage.44 Interestingly, in our c-Myc−/− HSC/Ps, Gata1, Fog1, and Eklf are all up-regulated, whereas Fli-1 is significantly down-regulated (Figure 2F), contrary to what we expected. We propose that c-Myc might function downstream and independently of these hematopoietic transcription factors in the lineage commitment and differentiation process.
c-Myb also plays a critical role in regulating HSC self-renewal and lineage commitment.45,46 Inactivation of c-Myb activity results in significant accumulation of HSCs in BM and a differentiation bias toward megakaryocytes over erythrocytes and lymphocytes. The significant thrombocytosis and severe anemia phenotype of c-Myb mutant mice are reminiscent of the phenotype of our c-Myc−/− mice. We found that c-Myb regulates c-Myc expression in HSCs/Ps (supplemental Figure 7), consistent with previous observations in cell line studies.47 We propose that c-Myc might be a downstream target of c-Myb, mediating c-Myb functions in HSC/Ps.
TPO has been shown to be a key hematopoietic cytokine for megakaryocytopoiesis.48 TPO signaling is also essential for the maintenance of hematopoietic homeostasis by restricting HSCs within a quiescent state in the BM niche and promoting HSC self-renewal after transplantation.49,50 Interestingly, in contrast to other hematopoietic cytokines that induce c-Myc expression, TPO down-regulates c-Myc expression in HSC/Ps49 (and our unpublished data). Thus, we speculate that down-regulation of c-Myc expression might be one aspect of lineage specification of this cytokine.
Supplementary Material
Acknowledgments
The authors thank Dr Ignacio Moreno de Alborán of Department of Immunology and Oncology, Centro Nacional de Biotecnología/CSIC, Universidad Autónoma de Madrid, Madrid, Spain, for his kindness in providing the c-Mycloxp mouse line. We appreciate the excellent animal care services provided by the staff of the Department of Comparative Medicine at Loyola University Medical Center. Ms Patricia Simms helped with flow cytometric sorting of HSCs and progenitors, as well as in the analysis of flow cytometry data. We appreciate the ongoing professional collaboration of Drs Nancy Zeleznik-Le and Andrew Dingwall, whose scientific suggestions and discussions improved the present studies. We acknowledge Dr John Crispino of the Division of Hematology/Oncology, Northwestern University (Chicago, IL) for his scientific suggestions and discussions which improved our data presentation.
This work was partially supported by a Stem Cell Research Grant from the Illinois Department of Public Health through Loyola University Medical Center, and also by Cancer Center Core grant NCI CA21765 and a grant from The American Lebanese Syrian Associated Charities (ALSAC), both through St. Jude Children's Research Hospital. P.B. is supported in part by a grant from the Jimmy Burns Foundation.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.G., C.N., P.B., W.W., M.T., and S.Z. performed research and analyzed data; A.R.K., G.P.P., and S.A. contributed vital new reagents or analytical tools; S.W.M., M.D., and P.J.S. were involved in research design and in paper writing; J.Z. designed the research, performed research, analyzed data, and wrote the paper; and P.B. was also involved in writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jiwang Zhang, Oncology Institute, Cardinal Bernardin Cancer Center, Pathology Department, Loyola University Medical Center, 2160 S 1st Ave, Maywood, IL 60153; e-mail: jzhang@lumc.edu.
References
- 1.Shizuru JA, Negrin RS, Weissman IL. Hematopoietic stem and progenitor cells: clinical and preclinical regeneration of the hematolymphoid system. Annu Rev Med. 2005;56:509–538. doi: 10.1146/annurev.med.54.101601.152334. [DOI] [PubMed] [Google Scholar]
- 2.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 4.de Alboran IM, O'Hagan RC, Gärtner F, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman B, Amanullah A, Shafarenko M, Liebermann DA. The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene. 2002;21:3414–3421. doi: 10.1038/sj.onc.1205400. [DOI] [PubMed] [Google Scholar]
- 6.Dubois NC, Adolphe C, Ehninger A, Wang RA, Robertson EJ, Trumpp A. Placental rescue reveals a sole requirement for c-Myc in embryonic erythroblast survival and hematopoietic stem cell function. Development. 2008;135:2455–2465. doi: 10.1242/dev.022707. [DOI] [PubMed] [Google Scholar]
- 7.Murphy MJ, Wilson A, Trumpp A. More than just proliferation: Myc function in stem cells. Trends Cell Biol. 2005;15:128–137. doi: 10.1016/j.tcb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Wilson A, Murphy MJ, Oskarsson T, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurenti E, Varnum-Finney B, Wilson A, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baena E, Ortiz M, Martinez AC, Moreno de Alboran I. c-Myc is essential for hematopoietic stem cell differentiation and regulates Lin(-)Sca-1(+)c-Kit(-) cell generation through p21. Exp Hematol. 2007;35:1333–1343. doi: 10.1016/j.exphem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Niu C, Ye L, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Grindley JC, Yin T, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 14.Mercher T, Cornejo MG, Sears C, et al. Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell. 2008;3:314–326. doi: 10.1016/j.stem.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cairo S, De Falco F, Pizzo M, Salomoni P, Pandolfi PP, Meroni G. PML interacts with Myc, and Myc target gene expression is altered in PML-null fibroblasts. Oncogene. 2005;24:2195–2203. doi: 10.1038/sj.onc.1208338. [DOI] [PubMed] [Google Scholar]
- 16.Schreiner S, Birke M, García-Cuéllar MP, Zilles O, Greil J, Slany RK. MLL-ENL causes a reversible and myc-dependent block of myelomonocytic cell differentiation. Cancer Res. 2001;61:6480–6486. [PubMed] [Google Scholar]
- 17.Sawyers CL, Callahan W, Witte ON. Dominant negative MYC blocks transformation by ABL oncogenes. Cell. 1992;70:901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- 18.Luo H, Li Q, O'Neal J, Kreisel F, Le Beau MM, Tomasson MH. c-Myc rapidly induces acute myeloid leukemia in mice without evidence of lymphoma-associated antiapoptotic mutations. Blood. 2005;106:2452–2461. doi: 10.1182/blood-2005-02-0734. [DOI] [PubMed] [Google Scholar]
- 19.Smith DP, Bath ML, Harris AW, Cory S. T-cell lymphomas mask slower developing B-lymphoid and myeloid tumours in transgenic mice with broad haemopoietic expression of MYC. Oncogene. 2005;24:3544–3553. doi: 10.1038/sj.onc.1208399. [DOI] [PubMed] [Google Scholar]
- 20.Langebrake C, Creutzig U, Reinhardt D. Immunophenotype of Down syndrome acute myeloid leukemia and transient myeloproliferative disease differs significantly from other diseases with morphologically identical or similar blasts. Klin Padiatr. 2005;217:126–134. doi: 10.1055/s-2005-836510. [DOI] [PubMed] [Google Scholar]
- 21.Wanzel M, Herold S, Eilers M. Transcriptional repression by Myc. Trends Cell Biol. 2003;13:146–150. doi: 10.1016/s0962-8924(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 22.Amati B, Alevizopoulos K, Vlach J. Myc and the cell cycle. Front Biosci. 1998;3:250–268. doi: 10.2741/a239. [DOI] [PubMed] [Google Scholar]
- 23.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 24.He C, Hu H, Braren R, et al. c-myc in the hematopoietic lineage is crucial for its angiogenic function in the mouse embryo. Development. 2008;135:2467–2477. doi: 10.1242/dev.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douglas NC, Jacobs H, Bothwell AL, Hayday AC. Defining the specific physiological requirements for c-Myc in T cell development. Nat Immunol. 2001;2:307–315. doi: 10.1038/86308. [DOI] [PubMed] [Google Scholar]
- 26.Bettess MD, Dubois N, Murphy MJ, et al. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol Cell Biol. 2005;25:7868–7878. doi: 10.1128/MCB.25.17.7868-7878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baena E, Gandarilla A, Vallesphino M, et al. c-Myc regulates cell size and ploidy but is not essential for postnatal proliferation in liver. Proc Natl Acad Sci U S A. 2005;102:7286–7291. doi: 10.1073/pnas.0409260102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oskarsson T, Essers MA, Dubois N, et al. Skin epidermis lacking the c-Myc gene is resistant to Ras-driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes Dev. 2006;20:2024–2029. doi: 10.1101/gad.381206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Habib T, Park H, Tsang M, et al. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J Cell Biol. 2007;179:717–731. doi: 10.1083/jcb.200704173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koster MI, Huntzinger KA, Roop DR. Epidermal differentiation: transgenic/knockout mouse models reveal genes involved in stem cell fate decisions and commitment to differentiation. J Investig Dermatol Symp Proc. 2002;7:41–45. doi: 10.1046/j.1523-1747.2002.19639.x. [DOI] [PubMed] [Google Scholar]
- 31.Janz S. Uncovering MYC's full oncogenic potential in the hematopoietic system. Oncogene. 2005;24:3541–3543. doi: 10.1038/sj.onc.1208473. [DOI] [PubMed] [Google Scholar]
- 32.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vousden KH. Switching from life to death: the Miz-ing link between Myc and p53. Cancer Cell. 2002;2:351–352. doi: 10.1016/s1535-6108(02)00186-1. [DOI] [PubMed] [Google Scholar]
- 34.Kirsch IR, Bertness V, Silver J, Hollis GF. Regulated expression of the c-myb and c-myc oncogenes during erythroid differentiation. J Cell Biochem. 1986;32:11–21. doi: 10.1002/jcb.240320103. [DOI] [PubMed] [Google Scholar]
- 35.Brewer G. Regulation of c-myc mRNA decay in vitro by a phorbol ester-inducible, ribosome-associated component in differentiating megakaryoblasts. J Biol Chem. 2000;275:33336–33345. doi: 10.1074/jbc.M006145200. [DOI] [PubMed] [Google Scholar]
- 36.Skoda RC, Tsai SF, Orkin SH, Leder P. Expression of c-MYC under the control of GATA-1 regulatory sequences causes erythroleukemia in transgenic mice. J Exp Med. 1995;181:1603–13. doi: 10.1084/jem.181.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson A, Zhang Y, Kamen D, Jackson CW, Cardiff RD, Ravid K. Deregulated expression of c-myc in megakaryocytes of transgenic mice increases megakaryopoiesis and decreases polyploidization. J Biol Chem. 1996;271:22976–22982. doi: 10.1074/jbc.271.38.22976. [DOI] [PubMed] [Google Scholar]
- 38.Kuhl C, Atzberger A, Iborra F, Nieswandt B, Porcher C, Vyas P. GATA1-mediated megakaryocyte differentiation and growth control can be uncoupled and mapped to different domains in GATA1. Mol Cell Biol. 2005;25:8592–8606. doi: 10.1128/MCB.25.19.8592-8606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Z, Richmond TD, Muntean AG, Barber DL, Weiss MJ, Crispino JD. STAT1 promotes megakaryopoiesis downstream of GATA-1 in mice. J Clin Invest. 2007;117:3890–3899. doi: 10.1172/JCI33010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwasaki H, Mizuno S, Wells RA, Cantor AB, Watanabe S, Akashi K. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 2003;19:451–462. doi: 10.1016/s1074-7613(03)00242-5. [DOI] [PubMed] [Google Scholar]
- 41.Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang AN, Cantor AS, Fujiwara Y, et al. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc Natl Acad Sci U S A. 2002;99:9237–9242. doi: 10.1073/pnas.142302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouilloux F, Juban G, Cohet N, et al. EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood. 2008;112:576–584. doi: 10.1182/blood-2007-07-098996. [DOI] [PubMed] [Google Scholar]
- 44.Kostyak JC, Naik UP. Megakaryopoiesis: transcriptional insights into megakaryocyte maturation. Front Biosci. 2007;12:2050–2062. doi: 10.2741/2210. [DOI] [PubMed] [Google Scholar]
- 45.Carpinelli MR, Hilton DJ, Metcalf P, et al. Suppressor screen in Mpl−/− mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc Natl Acad Sci U S A. 2004;101:6553–6558. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandberg ML, Sutton SE, Pletcher MT, et al. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 47.Cogswell JP, Cogswell PC, Kuehl WM, et al. Mechanism of c-myc regulation by c-Myb in different cell lineages. Mol Cell Biol. 1993;13:2858–2869. doi: 10.1128/mcb.13.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaushansky K, Broudy VC, Lin N, et al. Thrombopoietin, the Mp1 ligand, is essential for full megakaryocyte development. Proc Natl Acad Sci U S A. 1995;92:3234–3238. doi: 10.1073/pnas.92.8.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshihara H, Arai F, Hosokawa K, et al. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 2007;1:685–697. doi: 10.1016/j.stem.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 50.Qian H, Buza-Vidas N, Hyland CD, et al. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1:671–684. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.