Abstract
Ubiquitin (Ub) receptor proteins as a group must contain a diverse set of binding specificities to distinguish the many forms of polyUb signals. Previous studies suggested that the large class of ubiquitin associated (UBA) domains contains members with intrinsic specificity for lysine 63-linked polyUb (K63-polyUb) or K48-polyUb, thus explaining how UBA-containing proteins can mediate diverse signaling events. Here we show that previously observed K63-polyUb selectivity in UBA domains is the result of an artifact in which the dimeric fusion partner, glutathione-S-transferase (GST), positions two UBAs for higher affinity, avid interactions with K63-polyUb, but not K48-polyUb. Freed from GST, these UBAs are either non-selective or prefer K48-polyUb. Accordingly, NMR experiments reveal no K63-polyUb specific binding epitopes for these UBAs. We re-examine previous conclusions based on GST-UBAs and present an alternative model for how UBAs achieve a diverse range of linkage-specificities.
Ubiquitination, the covalent linkage of the small protein ubiquitin (Ub) to a substrate protein, is involved in nearly every aspect of eukaryotic cell biology1. Substrates can be modified with a single Ub unit or polymeric chains of Ub, with the Ub-Ub linkages typically occurring through one of the seven Ub lysine sidechains2. The prevailing model holds that the diversity in the forms of the signal is partly responsible for the diversity in the outcomes associated with ubiquitination3. Mono and differently-linked polyUb chains are associated with distinct outcomes for the proteins they modify. PolyUb formed through Ub lysine 48 (K48-polyUb) is predominantly associated with proteolysis of the substrate4. In contrast, K63-polyUb is associated with non-proteolytic roles in DNA repair, DNA damage tolerance, NF-κB signaling, and translation3-5. Likewise, cargo monoubiquitination is associated with endocytosis, trafficking, and transcriptional control6,7. This model predicts that diverse (poly)Ub binding preferences should exist among Ub receptor proteins to promote the proper downstream consequences.
A key finding in support of this model was that a large class of ubiquitin binding domains known as ubiquitin associated (UBA) domains contains a diverse set of ubiquitin specificities8. Glutathione-S-transferase (GST) fusions of UBA domains from more than 30 proteins, including all but one of the UBAs from budding yeast, were evaluated by quantitative pull-down assays for mono- and polyUb binding preferences. K48- and K63-polyUb selectivities were observed, as well as tight binding to monoUb that was associated with little polyUb linkage preference. Although K48-specific UBAs were known9, this was the first report of K63-linkage selectivity for isolated UBA domains. This study indicated that UBAs could present a diverse range of linkage-specific epitopes, and that linkage selectivity was achieved mainly at the level of these small, modular domains.
We expected that the reported K63-specific UBA interactions would be explained by binding at a linkage-specific epitope on K63-polyUb, which we set out to identify. Here we report that the apparent K63-selectivity of some UBAs is actually due to avid interactions that are artificially promoted in the dimeric GST-fusions used to classify the domains. We demonstrate that UBAs formerly considered K63-selective based on the GST-UBA fusions lose or reverse selectivity as free domains. Accordingly, those domains individually exhibit no K63-selective contacts with polyUb. We re-examine previous studies of UBAs in the light of this linkage-preference artifact to resolve some functional and mechanistic inconsistencies. We also examine how this artifact suggests an additional level of linkage specificity that could arise from multivalent arrangements of UBA domains in nature.
Results
GST-Ede1 UBA preferentially binds K63-linked polyUb
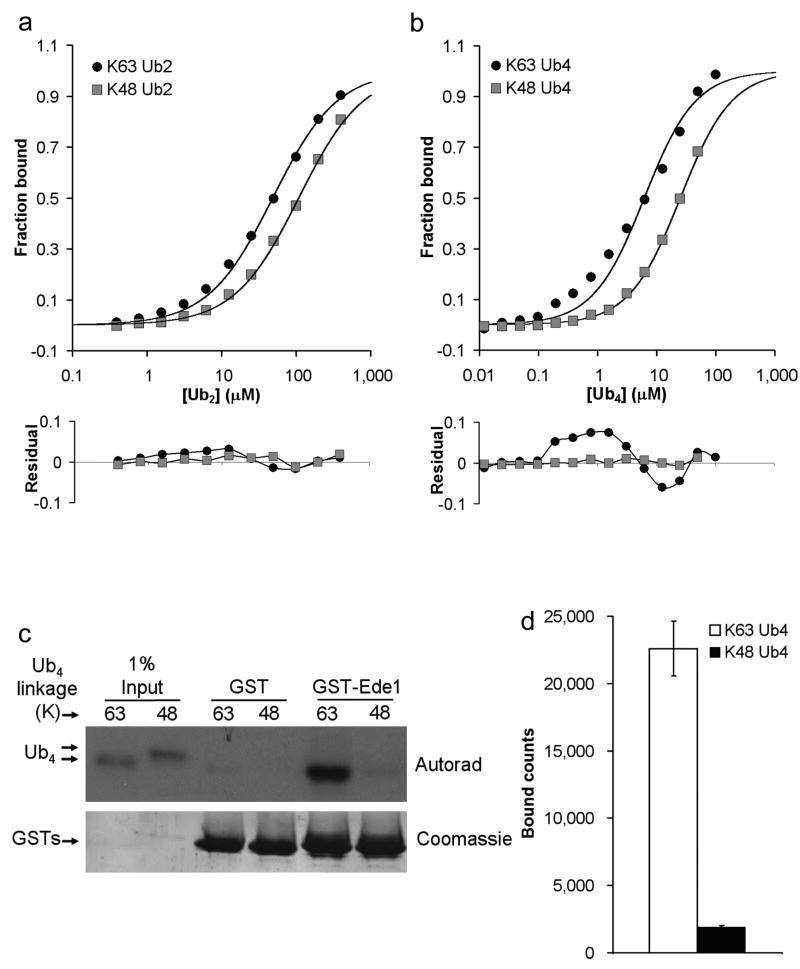
To identify a model K63-selective UBA domain for structural studies, we examined 4 of the 7 UBA domains classified as K63-selective by Raasi et al 8. We used the GST-fused minimal UBA domain constructs from their original study to assay binding to K63- and K48-Ub2 using surface plasmon resonance (SPR). The equilibrium dissociation constants (Kds) determined are shown in Table 1, and representative binding curves [for GST-Ede1 UBA (yeast) binding to K63-Ub2 and K48-Ub2] are shown in Figure 1a. All four GST-UBA fusion proteins bound K63-Ub2 more tightly than K48-Ub2. Next we examined binding to Ub4 for the two UBAs with the highest affinity, GST-Ede1 UBA and GST-hHR23A UBA1 (human). GST-Ede1 UBA was more linkage selective, and so became our model K63-selective UBA domain (4.2-fold selective for K63-Ub4; Table 1 and Fig. 1b). The magnitude of this selectivity is comparable to other linkage-selective UBA domains8,10 [e.g., ∼5-fold K48 preference by hHR23A UBA2].
Table 1.
Dissociation equilibrium constants (Kd) for GST-UBA domains interacting with polyubiquitins.
| GST-UBA | K63-Ub2 | K48-Ub2 | K63-Ub4 | K48-Ub4 |
|---|---|---|---|---|
| Ede1 | 48 ± 9 | 110 ± 16 | 6.0 ± 1.9 | 25 |
| hHR23A UBA1 | 26 | 36 | 4.7 | 13 |
| Ubc1 | 130 | 460 | ND | ND |
| DRM2 UBA1 | 89 | 140 | ND | ND |
Kd values (μM) were determined by SPR. Measurements with 3 or 4 replicates are presented as (Kd ± standard deviation). ND, not determined.
Figure 1.
GST-Ede1 UBA is selective for K63-linked polyUb. SPR analysis of GST-Ede1 binding to K63- and K48-linked (a) Ub2 and (b) Ub4 reveals preference for K63-linked polyUb (upper panels). The residuals (lower panels) indicate a systematic deviation from the 1:1 binding model used to fit the K63-linked polyUb data. These data are the averages of three or four independent experiments, except K48-Ub4, which is the average of two trials. (c) GST-Ede1 was used to pull-down radiolabeled Ub4 chains (upper panel). The lower panel shows even loading of GST-Ede1 or control GST onto the glutathione beads. The averages of three such experiments are quantified in (d). The error bars indicate standard deviations for the three trials.
The differences between the measured and theoretical SPR values (residuals) for the Ub4 ligands are plotted in Figure 1b (lower panel). For K48-Ub4, the small and random residuals indicate that the data are well described by the 1:1 interaction model used in the fit. For K63-Ub4 binding, however, large and non-random residuals are evident, potentially indicating more complex binding modes. SPR data for GST-Ede1 UBA vs. the Ub2 ligands (Fig. 1a) and GST-hHR23A UBA1 vs. Ub2 and Ub4 exhibit similar patterns of residuals, although the magnitude of the systematic deviations is smaller. We note that this pattern of non-random error in the 1:1 fit is frequently a feature of SPR data for UBA·polyUb binding, particularly when the UBA domain is the immobilized binding partner (J.J.S. and R.E.C., unpublished; references10,11).
Because GST pull-down assays are the most common technique used to evaluate polyUb linkage specificity, we also performed a quantitative version of a pull-down assay using GST-Ede1 UBA to capture radiolabeled Ub4 chains for comparison to the SPR data. By this method, the preference of GST-Ede1 for K63-Ub4 over K48-Ub4 appeared to be even larger (12-fold K63 selective by pull-down, Fig. 1c,d).
The mode of Ede1 UBA binding is not linkage-specific
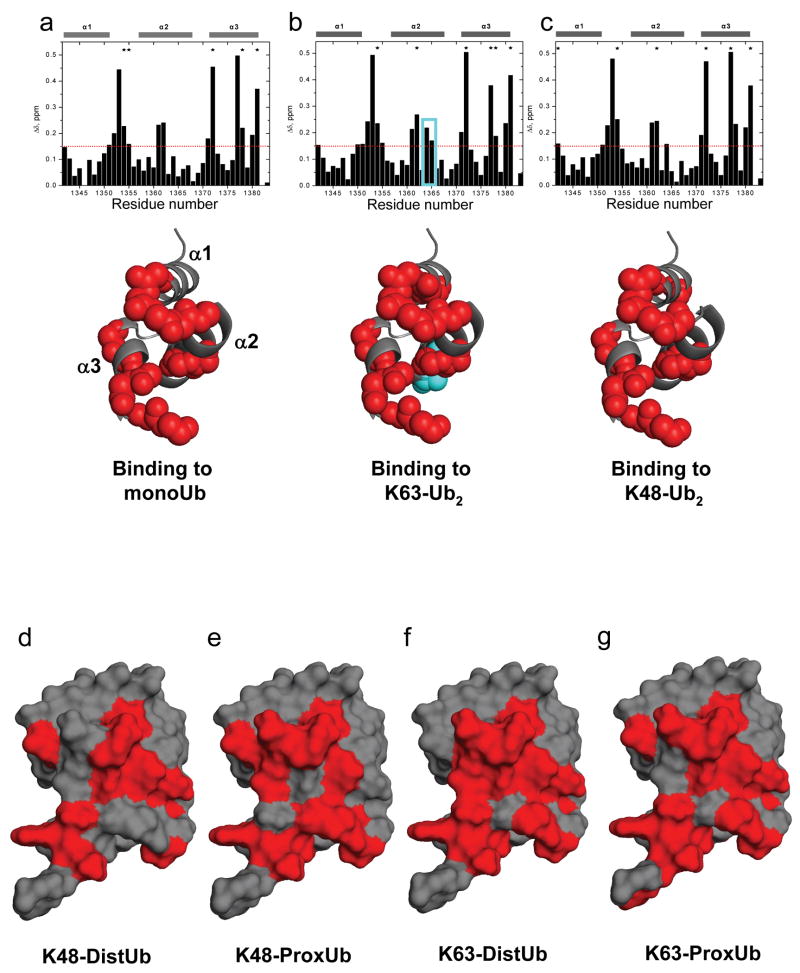
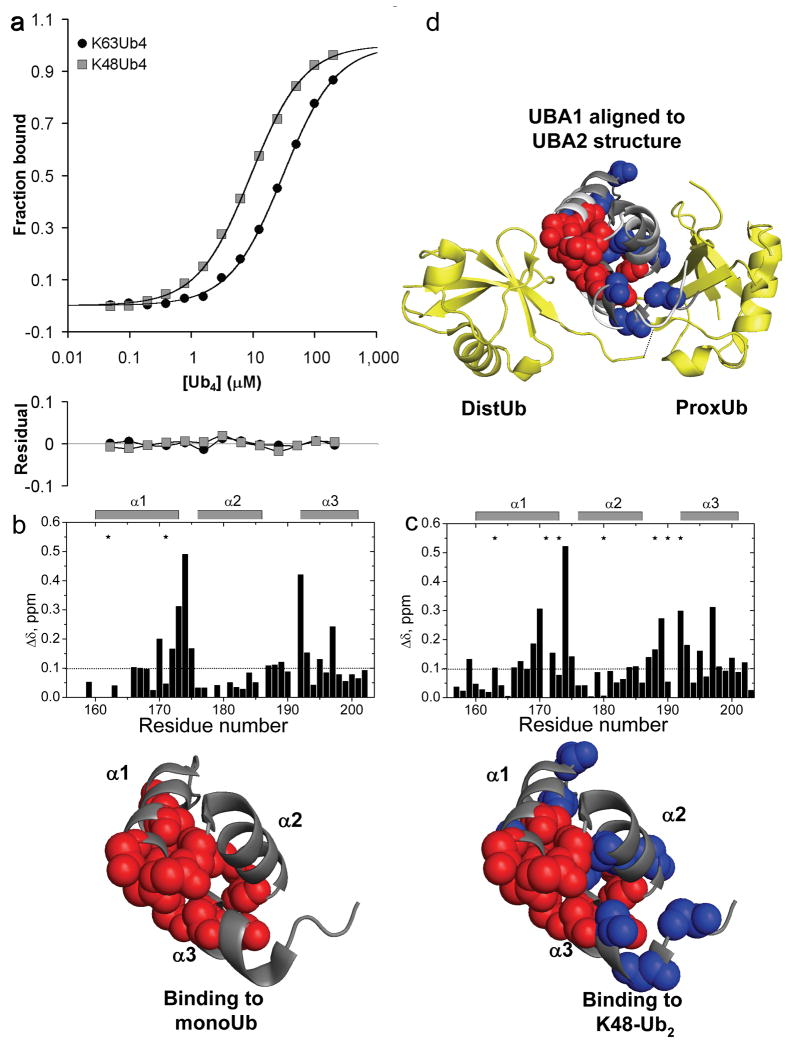
Next we performed NMR backbone amide (1H, 15N) chemical shift perturbation (CSP) studies to gain insight into the molecular basis of K63-selective binding. We titrated 15N-labeled Ede1 with monoubiquitin, K63-Ub2, or K48-Ub2 to determine the residues responsible for each of these interactions (Fig. 2a-c). The observed CSPs for monoUb·Ede1 UBA agree well with published data for this interaction12. However, to our surprise, the Ede1 CSP maps with all three ligands were nearly identical, with the primary interaction surface composed of residues on helix 1 and helix 3 of the UBA domain. Though the chemical shifts for residues E1364 and K1365 are slightly more perturbed in the complex with K63-Ub2 than with K48-Ub2 or monoUb (Fig. 2b), binding studies with E1364A and K1365A UBA domains indicate that these residues are not important determinants of binding or selectivity (Supplementary Fig. 1). This result is in contrast to the interaction of hHR23A UBA2 with its preferred binding partner, K48-Ub2 13. In this model of selective polyUb binding, an additional, distinct interface on UBA2 was evident from the titration with K48-Ub2 but not with K63-Ub2 or monoUb.
Figure 2.
CSP mapping of the interactions of Ede1 UBA with monoUb, K63-polyUb and K48-polyUb reveals no linkage-specific mode of interaction. Amide CSPs at titration endpoints are shown as a function of residue number for the titrations of 15N-Ede1 UBA with (a) monoUb, (b) K63-Ub2, and (c) K48-Ub2 (upper panels). Residues that were significantly perturbed upon binding (Δδ>0.15 ppm, or signal attenuation >60%) are mapped to the Ede1 UBA structure12 (2g3q.pdb) in red spheres below each plot. E1364 and K1365 are shown in cyan in (b). Likewise, Ub2 molecules were segmentally labeled with 15N and titrated with unlabeled Ede1 UBA. Ub residues that were significantly perturbed (Δδ>0.1 ppm, or signal attenuation >60%) are mapped to the surface of Ub12 (2g3q.pdb) in red for the (d) distal and (e) proximal K48-linked Ubs, and for the (f) distal and (g) proximal K63-linked Ubs. The CSP values are shown in Supplemental Figure 3, along with 15N monoUb CSPs, which are highly similar to both polyUb measurements.
Next we performed the inverse experiments, using versions of Ub2 with either the proximal (free C-terminus) or distal (free lysine 48 or 63) Ub selectively labeled with 15N and titrated with unlabeled Ede1 UBA. We collected (1H, 15N) monoUb CSPs for comparison to the polyUb data (Fig. 2d-g). These data reveal that, contrary to our expectations for a K63-selective UBA, both the proximal and distal Ub CSPs from the K63-Ub2·Ede1 UBA interaction are essentially the same as those observed from monoUb binding. For Ede1·K48-Ub2, the distal Ub CSPs were like those for both Ubs in K63-Ub2 and for monoUb. Significant differences were seen only in resonances from the proximal Ub of the K48-Ub2 titration, where the overall magnitude of CSPs was lower than in the other titrations (Supplementary Fig. 2). This phenomenon has been observed for other K48-Ub2 interactions and may relate to the opening-closing dynamics of the K48-Ub2 chain13,14. These results indicate that Ede1 UBA interacts with polyUb in a non-linkage-selective manner, in contrast to the SPR and pull-down results with GST-Ede1 UBA.
Free Ede1 UBA domain is not linkage-selective
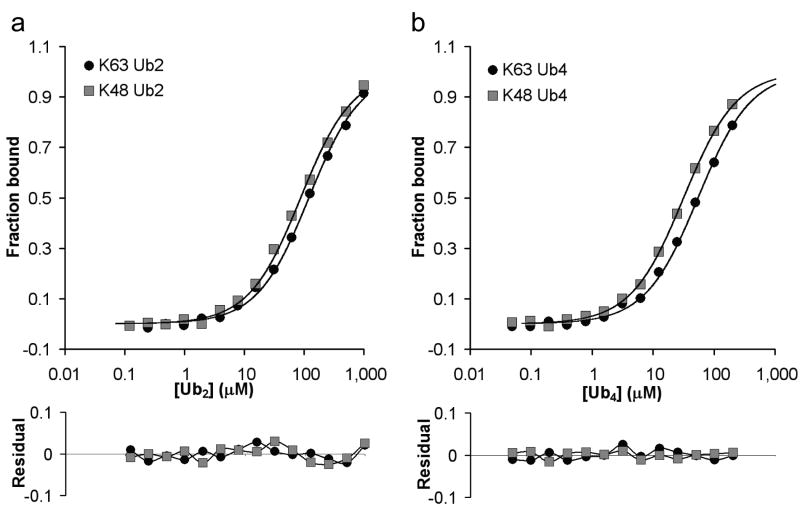
To examine polyUb specificity of the free Ede1 UBA domain, we produced a fluorescent version of the minimal domain free of affinity tags by expressed protein ligation15 (Ede1_rhodamine), and measured binding to polyUb by fluorescence anisotropy. In contrast to the >4-fold K63 selectivity of the GST-Ede1 UBA, Ede1_rhodamine is not linkage-selective (Fig. 3a,b and Table 2). This is in agreement with our CSP-mapping result that Ede1 UBA binds to K63-polyUb, K48-polyUb, and monoUb with virtually identical interfaces. The small preference (<2-fold) of Ede1_rhodamine for K48-polyUb has been observed for other UBDs that bind in non-linkage-specific configurations16,17. In K48-linked diUb, the residues on the Ub surface that are required for UBA interaction can face each other in a deep pocket18. For a UBA bound to a K48-linked Ub, non-specific contacts with an adjacent Ub in the chain might facilitate rebinding after dissociation, explaining the small observed preference for K48-Ub2. Another possibility is that the compact, closed form of K48-Ub2 presents a slightly smaller entropic barrier to binding than the relatively flexible K63-linked Ub dimer.
Figure 3.
Free Ede1-UBA is not linkage selective. Binding to K63- and K48-linked (a) Ub2 and (b) Ub4 were detected by monitoring the fluorescence anisotropy of Ede1_rhodamine (upper panels). These data reveal no substantial (i.e., >2-fold) linkage selectivity for the free UBA domain. The results from a single experiment are presented here; replicates in similar assays typically varied by <10%. Small, random residuals were observed for the free UBA domain binding to chains (lower panels).
Table 2.
Dissociation equilibrium constants (Kd) for fluorescent UBA domains interacting with polyubiquitins.
| UBA | K63- Ub2 | K48- Ub2 | K63- Ub4 | K48- Ub4 |
|---|---|---|---|---|
| Ede1 | 120 | 86 | 54 | 31 |
| hHR23A UBA1 | ND | ND | 30 | 9.0 |
| Ubc1 | ND | ND | 170 | 37 |
| Dsk2 | ND | ND | 2.2 | 1.4 |
| Ddi1 | ND | ND | 92 | 50 |
Kd values (μM) were determined by fluorescence anisotropy. ND, not determined.
K48-polyUb Kds agreed closely between GST-Ede1 and Ede1_rhodamine measurements (Tables 1 and 2). The large deviations in K63-polyUb affinities between the two constructs (9-fold for Ub4) suggested that GST-fusion can artificially promote UBA·K63-polyUb interactions. We suspected that the dimeric GST moiety of the GST-fusion could bring together two UBA domains in a configuration that promotes simultaneous or avid binding to a single K63-linked chain, but not to K48-polyUb. Since avid interactions are potentially more favorable, this could lead to the apparent linkage selectivity observed for some GST-UBAs. We previously showed that this mechanism, termed ‘linkage-specific avidity’, can determine the polyUb linkage preference for sets of ubiquitin interacting motifs (UIMs) that are held close in space by a short linking sequence17. Modeling suggests that two GST-fused UBAs could interact avidly with adjacent Ubs in a chain (Supplementary Fig. 3).
For GST-UBAs, a tighter, avid binding mode would contribute to binding at ligand (Ubn) concentrations below the intrinsic UBA·Ub Kd. At ligand concentrations nearer to the intrinsic Kd, each UBA could bind a separate chain. This mixed mode of binding would explain the systematic deviations from the 1:1 model that we have observed for some GST-UBAs interacting with K63-polyUb (Fig. 1a,b), i.e. more binding at low ligand concentrations than can be accounted for by the 1:1 binding model. In support of this hypothesis, we note that the free UBA binding data do not exhibit these same systematic deviations (Fig. 3a,b).
Bivalency accounts for linkage selectivity in GST-Ede1 UBA
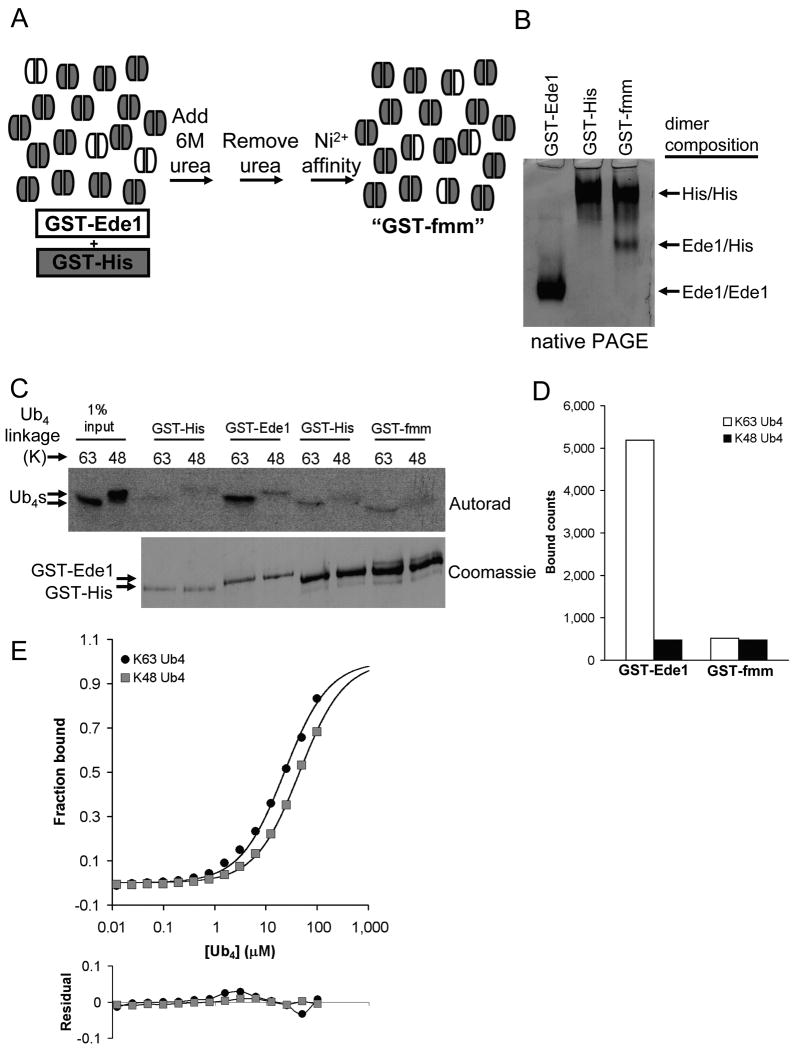
To test whether the bivalency of the GST-Ede1 UBA is responsible for its higher K63-polyUb affinity, we created a GST dimer that contained only one UBA fusion polypeptide. We did this by taking advantage of the slow exchange of subunits between GST dimers15,19. First, we produced a GST protein with a 6-His affinity tag, but no UBA fusion (GST-His). We mixed GST-His with GST-Ede1 at a 12:1 mole ratio, unfolded the mixture in 6 M urea, and then refolded the mixture by rapid 10-fold dilution into urea-free buffer. In a final step, dimers with a His tag were purified on Ni2+-NTA agarose, thus removing any reformed GST-Ede1/GST-Ede1 homodimers (Fig. 4a). The mixture obtained, termed GST-fmm for ‘functionally monomeric mixture,’ contained GST-His homodimers and GST-His/GST-Ede1 heterodimers at a ratio of about 6:1. The GST-Ede1/GST-Ede1, GST-His/GST-His, and GST-His/GST-Ede1 dimers could be separated by native PAGE because of charge differences between the two component polypeptides (Fig. 4b). The large excess of GST-His homodimers in the mixture was intended to reduce artificial multivalency between adjacent, non-dimerized UBAs in assays such as pull-downs and SPR where proteins are immobilized at high density on a solid surface.
Figure 4.
The bivalency of GST-Ede1 UBA preferentially promotes binding to K63-polyUb over K48-polyUb. (a) The process of making the functionally monomeric version of GST-Ede1 (GST-fmm) from a mixture of GST-His (gray subunits) and GST-Ede1 (white subunits) is shown schematically. (b) GST subunit exchange can be seen by comparing the starting materials to the final GST-fmm mixture after separation by native PAGE. The heterodimer has an intermediate pI and migrates between the positions of the two homodimers. (c) GST-Ede1 and GST-fmm were used to pull-down 125I-labeled Ub4 chains. The upper panel shows an autoradiogram of SDS-PAGE-separated Ub4 chains. To resolve GST-Ede1 and GST-His by SDS-PAGE, smaller amounts of the same samples were separated for a longer time on a second gel (lower panel). The amounts of GST-Ede1 and GST-fmm used in the assay were adjusted to contain similar amounts of the GST-Ede1 subunit (upper band). Separate negative controls (GST-His) were performed to account for different amounts of total protein on the GST-Ede1 and GST-fmm beads. The results of two assays are averaged in (d). (e) SPR analysis confirms this result. K63-Ub4 affinity and selectivity were reduced with GST-fmm (upper panel) relative to the GST-Ede1 homodimer. Data shown are an average from two independent trials that differed by <10%. Deviations from the fits are shown in the lower panel.
We compared GST-Ede1 to GST-fmm by pull-down assay with radiolabeled Ub4 chains (Fig. 4c). GST-Ede1 and GST-fmm samples were loaded onto glutathione-agarose so that equal amounts of GST-Ede1 polypeptides were on the beads. Because the resin with GST-fmm contained more total protein due to the excess GST-His, separate negative controls with GST-His alone were performed. Similar to previous pull-down assay results (Fig. 1c,d), GST-Ede1 was >11-fold selective for K63-Ub4 over K48-Ub4. In contrast, GST-fmm bound Ub4 chains with no linkage preference (Fig. 4d). Note that the GST-fmm and GST-Ede1 bound nearly identical amounts of K48-Ub4. This suggests that affinity for K48 chains is largely independent of the oligomeric state of the Ede1 UBA, and supports the idea that the linkage selectivity of dimeric GST-Ede1 arises from avid interactions that are only possible with K63-polyUb.
Using the SPR assay, GST-fmm bound K63-Ub4 more weakly than GST-Ede1 and had reduced linkage selectivity (Fig. 4e; KdK63Ub4 = 23 μM, KdK48Ub4 = 44 μM). The small difference between these SPR Kds and those derived from the fluorescence assays with free UBA (Table 2) may be the result of some inevitable multivalency generated by neighboring heterodimers on the surface of the SPR chip. We note that the systematic error associated with SPR measurements of GST-Ede1·K63-Ub4 was reduced but not eliminated for GST-fmm (Fig. 4e). If this indicates a link between multivalent binding and deviations from the 1:1 fit, then it indeed seems that the GST-fmm mixture reduced but did not totally eliminate multivalency in the context of the SPR assay. Nonetheless, it is clear from these measurements that the additional valency in the GST-Ede1 homodimer is responsible for most of the artifactual K63-polyUb affinity.
GST-fusion inflates K63-polyUb binding for other UBAs
We suspected that this artifactual K63-selectivity may apply to GST-hHR23A-UBA1 as well. We produced a fluorescein-labeled hHR23A-UBA1 domain free of affinity tags and measured its interactions with Ub4 chains (Fig. 5a). In contrast to the K63-selectivity observed for GST-UBA1, the free domain actually has a small but significant preference for K48-linked polyUb (Fig. 5a, Table 2). As with Ede1 UBA, K48-polyUb affinities agree for the two constructs, but K63-polyUb affinities do not (the GST-UBA1 is >6-fold tighter for K63-Ub4 than the free domain, Tables 1 and 2).
Figure 5.
hHR23A-UBA1 is a K48-selective UBA domain. (a) Fluorescence anisotropy binding data for hHR23A-UBA1 interacting with K63-Ub4 or K48-Ub4 indicate a preference for K48-polyUb. CSP mapping was used to identify the UBA1 surface responsible for binding to (b) monoUb and (c) K48-Ub2. Upper panels show the amide CSPs as a function of 15N-UBA1 residue number. Residues that were significantly affected by binding (CSP Δδ>0.10 ppm, or signal attenuation >60%) are mapped to the UBA1 structure20 (1IFY.pdb) below (spheres). Blue spheres indicate the residues that were only perturbed upon K48-Ub2 binding. UBA1 interacts with K48-Ub2 with an expanded set of residues in a configuration that is similar to the linkage-specific binding of UBA2 from the same protein. (d) To show this similarity, we aligned UBA1 (contacts indicated as before by spheres) to the UBA2 coordinates from its bound complex with K48-Ub2 (1ZO6.pdb)13. Ub2 is in yellow ribbons; UBA2 is in white ribbons; the aligned UBA1 is in dark gray ribbons. The putative interface of UBA1 with the distal Ub is similar to the monoUb interface (red spheres), whereas additional residues on the other side of UBA1 (blue spheres) that are specifically perturbed in the K48 interaction could make linkage-specific interactions with the isopeptide region and proximal Ub.
We again mapped NMR CSPs to investigate the molecular determinants of this preference. We titrated 15N-labeled UBA1 with unlabeled monoUb or K48-Ub2. These experiments revealed that, as with most UBA domains, the interaction with monoUb is mediated by residues primarily on one face of the domain, comprising helix 1 and helix 3 (Fig. 5b). The interaction with K48-Ub2, however, involves an extended surface that contains residues on the helix 2-helix 3 side of the domain in addition to the helix 1-helix 3 interface (Fig. 5c). This observation closely matches the published results for UBA2 from the same protein, which is K48-selective13. For UBA2, the helix 2-helix 3 contacts are part of a K48-specific epitope that is fully engaged only when the UBA is ‘sandwiched’ between the Ubs of K48-Ub2. Our results suggest that UBA1 achieves K48-linkage preference in the same way, with an extended interface that can be completely occupied only in the complex with K48-Ub2. Aligning the coordinates for free UBA120 to the K48-Ub2·UBA2 complex13 shows how the K48-specific contacts on UBA1 could engage K48-Ub2 along the isopeptide bond and proximal Ub (Fig. 5d). As with Ede1 UBA, the structural details of the interaction are consistent with the binding preference of the free domain, and not the GST-fused construct. Thus, it is likely that the smaller K63-selectivity we observed for GST-fused hHR23A-UBA1 (Table 1) is the result of competition between the intrinsic K48-selectivity of the domain and the avidity-driven K63-selectivity of the GST-fusion.
Another UBA from the K63-selective GST fusions, Ubc1 UBA (yeast), bound with an even larger K48-preference when expressed as a free domain (Table 2). However, we found that GST fusion does not necessarily result in the overestimation of K63-polyUb affinity for all GST-UBAs. Both GST-Dsk2 UBA (yeast) and GST-Ddi1 (yeast) UBA were originally shown to bind polyUb without a linkage preference8; our experiments indicate that the free UBAs are indeed non-selective (<2-fold difference in Kd for K63- and K48-Ub4, Table 2). In support of these results, the human homolog of Dsk2, ubiquilin-1, was found to bind without linkage-selectively to K63- and K48 polyUb in a manner similar to its interaction with monoUb16.
One possible difference between the UBA domains prone to the K63-GST artifact and those that are not may be the degree or nature of UBA self-association. Forced proximity from a dimerized GST (Supplementary Fig. 3) could promote even weak or non-specific self-association of some UBAs, resulting in a conformation that favors avid binding to adjacent, K63-linked Ubs. Several UBA domains have been shown to self-associate21-23, and in at least one instance this UBA·UBA interaction promotes polyUb binding24. We have collected some evidence that Ede1 UBA domain may self-associate at high concentrations and in the context of the GST-fusion (data not shown), though our studies were not conclusive about the contribution this property makes to apparent linkage selectivity.
Intriguingly, Dsk2 oligomerization has been suggested to play a part in K48-polyUb selectivity25. Full-length Dsk2 self-associates26 and, by inference, has shown an in vivo binding preference for K48-linked polyUb27. However, no linkage selectivity for the isolated domain has been observed under conditions that favor dimerization (i.e., as a GST fusion8) or conditions that should prevent UBA self-association (i.e., use of low UBA concentrations in our fluorescence binding assays). It is likely that the precise configuration of self-associated UBAs would influence linkage selectivity. This property of some UBAs could be either functionally relevant or an artifact of some assays. Detailed biophysical studies will be required to determine the contributions of UBA domain self-association to linkage-selective binding.
Discussion
A widely cited conclusion is that isolated, minimal UBA domains contain a broad range of intrinsic Ub binding specificities, including K48- and K63-polyUb preference8. Here we report that the assays used to reach those conclusions have artificially promoted K63-polyUb binding for some UBA domains, and thus may have overestimated the range of UBA·polyUb specificities attributable to the minimal UBA domain. Accordingly, we see no structural or biophysical evidence for K63-selective binding in isolated, free UBA domains. Of the seven UBA domains originally identified as K63-selective, we have shown that one is non-selective (Ede1 UBA from yeast) and two are actually K48-selective in isolation (human hHR23A UBA1 and yeast Ubc1). By extension, we expect that the homologs of these UBAs, yeast Rad23 UBA1 and human E2-25k UBA, respectively, are also K48-selective in isolation. We have not examined the remaining two domains, both from the Arabidopsis protein DRM2.
The apparent source of the GST-fusion artifact that we have observed is a type of linkage-specific avidity in which the dimeric GST moiety can position two UBAs close in space to make simultaneous contacts with K63-polyUb, but not K48-polyUb. Previously, we showed that closely-spaced tandem UIM domains can achieve polyUb linkage preference through the same mechanism17. This work extends the range of configurations that can result in linkage-specific avidity, as well as the types of UBDs that can be involved.
Why then should some multivalent arrangements promote binding to a K63-polyUb chain over a K48-polyUb chain that has an equivalent number of Ub binding sites? As we have shown previously for tandem UIM domains, the orientation and spacing of two UBDs can promote avid binding to one linkage, but not another, and thus provide an effective means of linkage selectivity17. Considering the flexibility that probably exists in the GST-UBA linking sequence of the constructs we have examined (Supplementary Fig. 3), it seems unlikely that the GST and linker alone could exert much influence over the orientation and spacing of UBA fusions. However, as mentioned above, UBA self-association in the context of a dimeric GST-fusion could provide control over the orientation of the UBAs. Alternatively, the different conformations of K48- and K63-polyUb chains in solution28,29 could either promote or restrict access to avid binding modes by multiple UBAs. Careful structural and biophysical studies will be required to determine the precise mechanisms of the linkage-specific avidity we have observed.
Pull-down assays with immobilized Ub-binding proteins are widely applied to assess linkage specificity, particularly since small amounts of K48- and K63-polyUb have been made commercially available. Our results suggest that any immobilization of UBD proteins on a solid surface such as glutathione-coated beads or SPR chips may result in artificial multi-valency that can profoundly influence polyUb binding properties such as chain length preference, linkage preference, and affinity. Multivalent interactions allow individual sites to re-bind after dissociation and therefore slow observed off-rates; non-equilibrium wash steps in pull-down assays can exaggerate these differences in off-rates, particularly for weak receptors. In fact, for typically weak Ub receptors, most of the retained polyUb chains in a pull-down assay are probably bound avidly. Thus, conclusions about intrinsic linkage-preference drawn from such experiments should be re-examined. Another complication is that, to conserve chains, polyUb pull-downs often employ (typically) non-linear western-blots to achieve sensitive chain detection, and use anti-Ub antibodies that can differentially stain polyUb chains of different linkages. As well as being an additional source of error, western-blotting can also have the effect of reducing subtle differences in linkage preference to “all or nothing” conclusions.
The SPR assay is closer to an equilibrium binding measurement, but because immobilization is achieved with divalent anti-GST antibodies, the commonly used GST-fusion based version of this assay may add a double layer of valency. Indeed, it seems that we could not eliminate all traces of multivalency from our SPR assay (Fig. 4e). Ideally, polyUb binding studies would be performed in the solution phase with full-length proteins near their physiological concentrations, or in the context of a relevant protein complex. Since this standard is impractical or impossible for most polyUb receptors, a reasonable compromise may be to at least avoid a known source of experimental artifact (GST fusion or immobilization) in favor of solution-based assays with a UBD or UBD-protein to determine linkage selectivity.
Avidity artifacts in previous polyUb binding studies may have led to some confusion about the functional significance of polyUb selectivity in UBD proteins30. Human hHR23A UBA1 and its yeast homolog were originally classified8 as K63-selective but occur in proteasomal ubiquitin receptor proteins, where K48-polyUb binding is presumed to be more relevant. Likewise, two other members of the original K63-selective class, yeast Ubc1 and its human homolog E2-25K, have stronger functional connections to K48-polyUb pathways. Consistent with our finding of K48-selectivity, both are involved in endoplasmic reticulum-associated degradation (ERAD), a pathway that requires K48-polyUb31, and E2-25K assembles K48-linked chains exclusively in vitro 32. In fact, the earlier study8 identified just one K48-selective UBA in yeast (UBA2 from Rad23), although K48-linked chains are likely the most common type of polyUb33. Our study resolves these inconsistencies and recognizes the intrinsic K48-polyUb selectivity of many more UBA domains. Nonetheless, conclusions about the role of these relatively modest linkage preferences will require studies that directly examine the link between selectivity and function.
Previous work has explained intrinsic K48-polyUb linkage-selective binding by human hHR23A UBA213; and fission yeast Mud1 UBA11. These UBAs meet the expectations for intrinsically linkage-selective domains because they present similar, K48-specific epitopes on their surfaces. UBA2 binds K48-Ub2 at a contiguous interface that includes the isopeptide bond and both Ub hydrophobic patches, the sites of all known UBA·Ub interactions. Our study indicates a similar arrangement for UBA1 of hHR23A interacting with K48-Ub2. This is because K48 is adjacent to the hydrophobic patch, and K48-Ub2 adopts a structure that brings both hydrophobic patches into close proximity18. However, it is unclear how the small UBA domain could achieve an analogous, linkage-specific interface with K63-Ub2 because lysine 63 is not close to the hydrophobic patch, and K63-linked ubiquitins adopt an elongated, open structure in solution28. We therefore speculate that intrinsic K63 selectivity, at least in the mold of UBA2·K48-Ub2 recognition, may not exist among UBA domains or any of the other small UBDs that require contacts with the Ub hydrophobic patch (e.g., CUE, UIM, or NZF domains)(unpublished results, D.F.). In contrast, larger and more extended UBDs appear to be capable of intrinsic K63-polyUb selectivity, as recently shown for the CC2-LZ domain of NEMO34. CC2-LZ engages linear or K63-linked diUb along an extended surface that includes both Ub units and the junction between them.
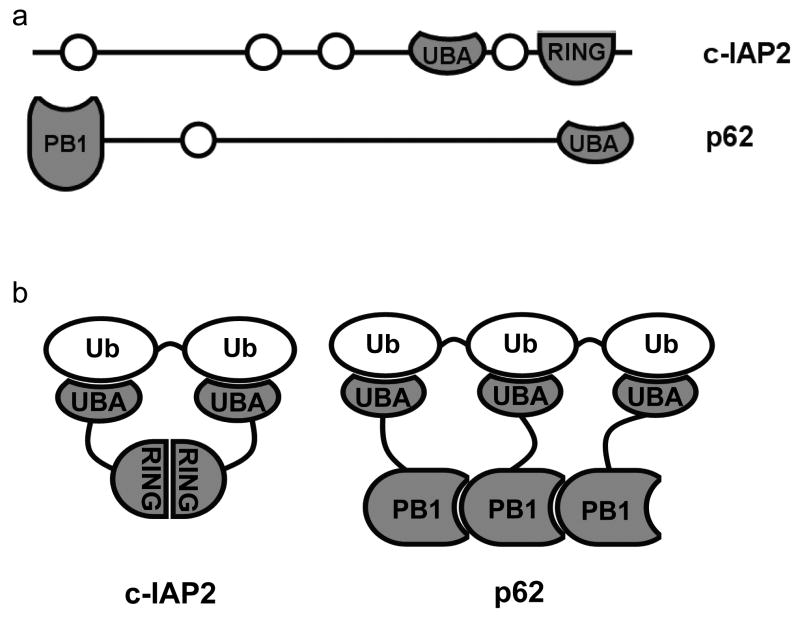
If the range of signaling functions accomplished by UBA proteins requires a similarly diverse range of polyUb linkage preferences, our study indicates that the origins of linkage selectivity are more complex than the intrinsic specificities of minimal domains. The GST effect described here suggests how UBA proteins can use two mechanisms to diversify polyUb linkage preferences: some UBAs are intrinsically K48-specific, and K63-polyUb selectivity can arise from certain avid combinations of intrinsically non-specific UBA interactions. Unfortunately, few measurements of UBA protein binding specificity have considered the influence of multiple domains in a complex or the oligomeric state of a single-UBA protein. Nonetheless, a survey of the literature yields several cases where oligomeric proteins that contain UBA domains achieve K63-selectivity. One example is the IAP (inhibitor of apoptosis) proteins, a family of anti-apoptotic proteins that are involved in NF-κB signalling, in which UBA-mediated IAP interaction with K63-linked polyUb is critical for function35. One recent study showed that c-IAP2 is K63-selective and that polyUb binding required not only the UBA domain, but also an adjacent, dimerizing RING domain36 (Fig. 6). The c-IAP2 RING domain may determine the selectivity for polyUb in the same way that GST modulates selectivity of GST-Ede1 UBA, i.e., through linkage-specific avidity.
Figure 6.
Oligomeric UBA proteins may achieve K63-selectivity through linkage-specific avidity. (a) Domain maps for two UBA proteins, c-IAP2 (top) and p62/SQSTM1 (bottom), that are functionally linked to K63-polyUb binding; the UBAs and the oligomerizing domains are highlighted (dark gray). (b) In their oligomeric forms, these proteins may present arrays of UBAs that are specific for K63-polyUb.
In another example, the highly oligomeric p62/SQSTM1 is a multi-functional scaffolding protein with links to K63-polyUb signaling in NF-κB and autophagy pathways37-39 (Fig. 6). Solution-phase measurements of linkage selectivity have not been published for p62 UBA, though there is some evidence that the isolated domain binds Ub weakly and without regard to linkage8. One study showed that full-length p62 binds K63-polyUb preferentially in vivo40. In another41, localization to autophagosomes, also likely to be signaled by K63-polyUb39,42,43, was abrogated by point mutations in the PB1 domain that prevented p62 self-association as well as point mutations in the UBA domain that eliminated Ub binding. Interestingly, another oligomeric protein with a similar architecture to p62, NBR1, was recently shown to localize to sites of autophagy, though the role of oligomerization in Ub binding is less clear44.
With regard to the Ede1 UBA, we note that yeast Ede1 may be effectively oligomerized when a group of endocytic network proteins including Ede1 gather at high density around ubiquitinated cargo to recruit oligomerized clathrin to the sites of endocytosis45-47, a process that in some cases may involve K63-polyUb. Careful biophysical and structural studies will be required to determine whether the linkage-specific avidity that we observe for some artificially oligomerized UBA domains relates to a functionally relevant mechanism of K63-selective binding by UBA proteins.
Methods
Plasmids and proteins
We have listed the sequences of all the proteins used in this study in the Supplementary Materials, along with the details of their cloning, expression, purification, and (in some cases) fluorescent or radioisotopic labeling.
Pull-down assays
We added GST-UBA protein to 15 μl of glutathione-agarose and then washed the beads with binding buffer [25 mM phosphate pH 7.4, 150 mM NaCl, 10 mM β -mercaptoethanol, 1 mM EDTA, 0.05% (v/v) Brij35]. We then added 125I-labeled K63 or K48-Ub4 (1 μM) in 100 μl of binding buffer plus 1 mg ml-1 BSA, and agitated the beads gently for 20 min at room temperature. We normalized the specific radioactivities of 125I-labeled K63-and K48-Ub4 beforehand with unlabeled chains. We then washed the beads quickly 2 or 3 times with binding buffer. We eluted the bound chains with SDS-PAGE sample buffer and resolved them by SDS-PAGE. We excised the Ub4 bands from the gel and quantified them with a gamma-counter. ‘Bound counts’ in Figure 1d and Figure 4d is the radioactivity (cpm) for a particular band minus the counts in the corresponding gel slice in the negative-control lane.
Surface plasmon resonance
We performed SPR analyses on a Biacore 3000 instrument at 25 °C in HBS-EP buffer (Biacore). We immobilized anti-GST antibody (Biacore) by amine coupling on a Biacore CM5 chip. We captured GST-UBA proteins on a measurement surface at a density of 150-400 RU; an antibody-coupled surface served as the reference. We applied Ubn chains to the chip with a 5 μl/min flow rate and recorded the response; we used 50 mM glycine pH 1.8 or 15 mM NaOH to remove GST-UBA proteins and renew the surface. We fit the data with a single-site binding model as described48.
NMR
We performed all NMR studies on a cryoprobe-equipped Bruker 600 MHz spectrometer at 23 °C. We prepared NMR samples of Ede1 UBA, hHR23A UBA1, monoUb and Ub2 (protein concentrations 0.35–0.8 mM) in the appropriate buffers containing 20 mM sodium phosphate at pH 6.8, 7% (v/v) D2O, and 0.02% (w/v) NaN3. In addition, all Ede1 UBA samples contained 5 mM β-mercaptoethanol. We synthesized segmentally 15N-labeled Ub2 chains as described18,28. NMR signal assignments for monoUb and Ub2 at pH 6.8 were from our previous studies18,28. NMR signal assignments for the Ede1 UBA and hHR23A UBA1 domains were from the literature12,20. We processed NMR data using XWINNMR and analyzed using the program CARA and in-house software.
We achieved binding-interface mapping in a series of NMR titration experiments in which 2D 1H-15N HSQC or SOFAST spectra of a 15N-labeled species of interest (e.g. Ede1 UBA) were recorded as a function of the increasing amount of unlabeled binding partner (e.g., Ub2). To map the binding surface on a specific Ub unit in Ub2, we performed a similar assay in which unlabeled UBA was added to segmentally 15N-labeled Ub2. We monitored binding through accompanying changes in the peak positions in 2D 1H-15N HSQC spectra and quantified using combined amide chemical shift perturbation (CSP) calculated as Δδ = [(ΔδH)2 + (ΔδN/5)2]1/2, where ΔδH and ΔδH are the observed chemical shift changes for 1H and 15N, respectively. To monitor site-specific changes in NMR signal intensities due to line broadening (as a result of intermediate or slow exchange), we uniformly scaled the NMR spectra obtained in the course of titration to compensate for the higher molecular weight of the complex. We then calculated the signal attenuation for each residue as the ratio of peak intensities in the corresponding spectra of the free and bound protein. We have included additional details about the NMR experiments in the Supplementary Materials.
Fluorescence anisotropy binding assays
We performed fluorescence anisotropy measurements as described17 using excitation and emission maxima of 492 nm/520 nm (fluorescein), 556 nm/569 nm (Alexa Fluor 546), or 555 nm/578 nm (rhodamine), in binding buffer at 25 °C. We calculated the concentrations of the fluorescent proteins using published extinction coefficients (Invitrogen-Molecular Probes). We assessed polyUb concentrations from absorbance at 280 nm using the Ub extinction coefficient of 0.16 (mg per ml)-1cm-1.49 We fit the data with a single-site binding model48.
Supplementary Material
Acknowledgments
We thank Phil Cole (Johns Hopkins School of Medicine) for materials and advice for fluorescent-labeling by expressed protein ligation. Work in this study was supported in part by NIH grant GM065334 (to D.F.) and NIH Roadmap grant RR020839.
Footnotes
Author Contributions: J.J.S. and R.E.C. conceived of the study; J.J.S., D.F., and R.E.C. designed the experiments; J.J.S., R.E.C., A.H., and B.C.D. prepared the proteins; J.J.S. performed all the cloning and binding assays, and interpreted the data with R.E.C.; A.H. and B.C.D. performed the NMR experiments and analyzed the data with D.F.; J.J.S., D.F., and R.E.C. wrote the paper.
References
- 1.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 7.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 8.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 9.Raasi S, Pickart CM. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- 10.Raasi S, Orlov I, Fleming KG, Pickart CM. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J Mol Biol. 2004;341:1367–1379. doi: 10.1016/j.jmb.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 11.Trempe JF, et al. Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J. 2005;24:3178–3189. doi: 10.1038/sj.emboj.7600797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swanson KA, Hicke L, Radhakrishnan I. Structural basis for monoubiquitin recognition by the Ede1 UBA domain. J Mol Biol. 2006;358:713–724. doi: 10.1016/j.jmb.2006.02.059. [DOI] [PubMed] [Google Scholar]
- 13.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Haririnia A, D'Onofrio M, Fushman D. Mapping the interactions between Lys48 and Lys63-linked di-ubiquitins and a ubiquitin-interacting motif of S5a. J Mol Biol. 2007;368:753–766. doi: 10.1016/j.jmb.2007.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheibner KA, Zhang Z, Cole PA. Merging fluorescence resonance energy transfer and expressed protein ligation to analyze protein-protein interactions. Anal Biochem. 2003;317:226–232. doi: 10.1016/s0003-2697(03)00087-3. [DOI] [PubMed] [Google Scholar]
- 16.Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377:162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan W, et al. Conformational stability of pGEX-expressed Schistosoma japonicum glutathione S-transferase: a detoxification enzyme and fusion-protein affinity tag. Protein Sci. 1997;6:399–406. doi: 10.1002/pro.5560060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller TD, Feigon J. Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions. J Mol Biol. 2002;319:1243–1255. doi: 10.1016/S0022-2836(02)00302-9. [DOI] [PubMed] [Google Scholar]
- 21.Bayrer JR, Zhang W, Weiss MA. Dimerization of doublesex is mediated by a cryptic ubiquitin-associated domain fold: implications for sex-specific gene regulation. J Biol Chem. 2005;280:32989–32996. doi: 10.1074/jbc.M507990200. [DOI] [PubMed] [Google Scholar]
- 22.Kozlov G, et al. Structural basis for UBA-mediated dimerization of c-Cbl ubiquitin ligase. J Biol Chem. 2007;282:27547–27555. doi: 10.1074/jbc.M703333200. [DOI] [PubMed] [Google Scholar]
- 23.Prag G, et al. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003;113:609–620. doi: 10.1016/s0092-8674(03)00364-7. [DOI] [PubMed] [Google Scholar]
- 24.Peschard P, et al. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol Cell. 2007;27:474–485. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 25.Lowe ED, et al. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr D Biol Crystallogr. 2006;62:177–188. doi: 10.1107/S0907444905037777. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki T, Funakoshi M, Endicott JA, Kobayashi H. Budding yeast Dsk2 protein forms a homodimer via its C-terminal UBA domain. Biochem Biophys Res Commun. 2005;336:530–535. doi: 10.1016/j.bbrc.2005.08.126. [DOI] [PubMed] [Google Scholar]
- 27.Matiuhin Y, et al. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell. 2008;32:415–425. doi: 10.1016/j.molcel.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varadan R, et al. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 29.Ryabov Y, Fushman D. Interdomain mobility in di-ubiquitin revealed by NMR. Proteins. 2006;63:787–796. doi: 10.1002/prot.20917. [DOI] [PubMed] [Google Scholar]
- 30.Kim I, Rao H. What's Ub chain linkage got to do with it. Sci STKE. 2006;2006:pe18. doi: 10.1126/stke.3302006pe18. [DOI] [PubMed] [Google Scholar]
- 31.Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z, Pickart CM. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J Biol Chem. 1990;265:21835–21842. [PubMed] [Google Scholar]
- 33.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo YC, et al. Structural basis for recognition of diubiquitins by NEMO. Mol Cell. 2009;33:602–615. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broemer M, Meier P. Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol. 2009 doi: 10.1016/j.tcb.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Gyrd-Hansen M, et al. IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-kappaB as well as cell survival and oncogenesis. Nat Cell Biol. 2008;10:1309–1317. doi: 10.1038/ncb1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seibenhener ML, Geetha T, Wooten MW. Sequestosome 1/p62--more than just a scaffold. FEBS Lett. 2007;581:175–179. doi: 10.1016/j.febslet.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2006;2:138–139. doi: 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- 39.Tan JM, Wong ES, Dawson VL, Dawson TM, Lim KL. Lysine 63-linked polyubiquitin potentially partners with p62 to promote the clearance of protein inclusions by autophagy. Autophagy. 2007;4 [Google Scholar]
- 40.Seibenhener ML, et al. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan JM, et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17:431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- 43.Olzmann JA, Chin LS. Parkin-mediated K63-linked polyubiquitination: a signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy. 2008;4:85–87. doi: 10.4161/auto.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 45.Aguilar RC, Watson HA, Wendland B. The yeast Epsin Ent1 is recruited to membranes through multiple independent interactions. J Biol Chem. 2003;278:10737–10743. doi: 10.1074/jbc.M211622200. [DOI] [PubMed] [Google Scholar]
- 46.Gagny B, et al. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J Cell Sci. 2000;113(Pt 18):3309–3319. doi: 10.1242/jcs.113.18.3309. [DOI] [PubMed] [Google Scholar]
- 47.Maldonado-Baez L, et al. Interaction between Epsin/Yap180 adaptors and the scaffolds Ede1/Pan1 is required for endocytosis. Mol Biol Cell. 2008;19:2936–2948. doi: 10.1091/mbc.E07-10-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkinson KD. Quantitative analysis of protein-protein interactions. Methods Mol Biol. 2004;261:15–32. doi: 10.1385/1-59259-762-9:015. [DOI] [PubMed] [Google Scholar]
- 49.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.