Abstract
There has been increasing evidence pointing to the mitochondrial respiratory chain (MRC) as a novel and important target for the actions of 17β-estradiol(E2) and estrogen receptors (ER) in a number of cell types and tissues that have high demands for mitochondrial energy metabolism. This novel E2-mediated mitochondrial pathway involves the cooperation of both nuclear and mitochondrial ERα and ERβ and their co-activators on the coordinate regulation of both nuclear DNA- and mitochondrial DNA-encoded genes for MRC proteins. In this paper, we have: 1) comprehensively reviewed studies that reveal a novel role of estrogens and ERs in the regulation of MRC biogenesis; 2) discussed their physiological, pathological and pharmacological implications in the control of cell proliferation and apoptosis in relation to estrogen-mediated carcinogenesis, anticancer drug resistance in human breast cancer cells, neuro-protection for Alzheimer’s disease and Parkinson’s disease in brain, cardiovascular protection in human heart and their beneficial effects in lens physiology related to cataract in the eye; and 3) pointed out new research directions to address the key questions in this important and newly emerging area. We also suggest a novel conceptual approach that will contribute to innovative regimines for the prevention or treatment of a wide variety of medical complications based on E2/ER-mediated MRC biogenesis pathway.
Keywords: Anti-cancer drug resistance; Alzermer’s Diseases; Cataract; Estrogens; Estrogen carcinogenesis in breast cancer; Estrogen Protection of Cardiovascular Diseases; Estrogen receptors; Mitochondrial DNA replication; transcription and translation; Mitochondrial Mitochondrial Estrogen Receptors; Respiratory Chain Biogenesis; Mitochondrial Reanscription Factor A; Nuclear Respiratory Factors, Parkinsopn’s Disease
1. Introduction
Estrogens, notably 17β-estradiol (E2), have a wide variety of physiological and pathological effects on a number of cell types and organs, including primary and secondary reproductive, cardiovascular, central nerve, immune, bone, gastrointestinal, and respiratory systems as well as oral and lens epithelial cells. The classical genomic mechanisms underlying the regulation of nuclear gene transcription by estrogens via the nuclear estrogen receptors alpha and beta (ERα and ERβ) have been well established (for review see [1, 2]) Recently, great interest has also been drawn to the non-genomic pathways involving plasma membrane-associated ERs that activate a number of intracellular protein kinase-mediated phosphorylation signaling cascades and the subsequent physiological activities (for review see [3–8]). In recent years, there has been increasing evidence pointing to the mitochondrial respiratory chain (MRC) as a novel and important target for the actions of E2 and ERs in a number of cell types and tissues that have high demand for mitochondrial energy metabolism for their biological activities. This novel E2-mediated mitochondrial pathway involves the cooperation of the nuclear ERα and ERβ with mitochondrial localized ERs and their co-activators on the coordinate regulation of both nuclear DNA (nDNA)-encoded genes and mitochondrial DNA (mtDNA)-encoded genes for MRC proteins. In this paper, we have: 1) reviewed recent studies that reveal a novel role of estrogens and ERs in the regulation of MRC biogenesis; 2) discussed their potential physiological, pathological and pharmacological implications in the control of cell proliferation and apoptosis in relation to estrogen-mediated carcinogenesis in human breast cancer cells, cardiovascular protection in heart, neuro-protection for Alzheimer’s disease and Parkinson’s disease in brain and their beneficial effects in lens physiology related to cataract in the eye; and 3) pointed out new research directions to address the key questions in this important and newly emerging area.
2. MRC Structure, Functions and Biogenesis
2.1. MRC Structure and Functions
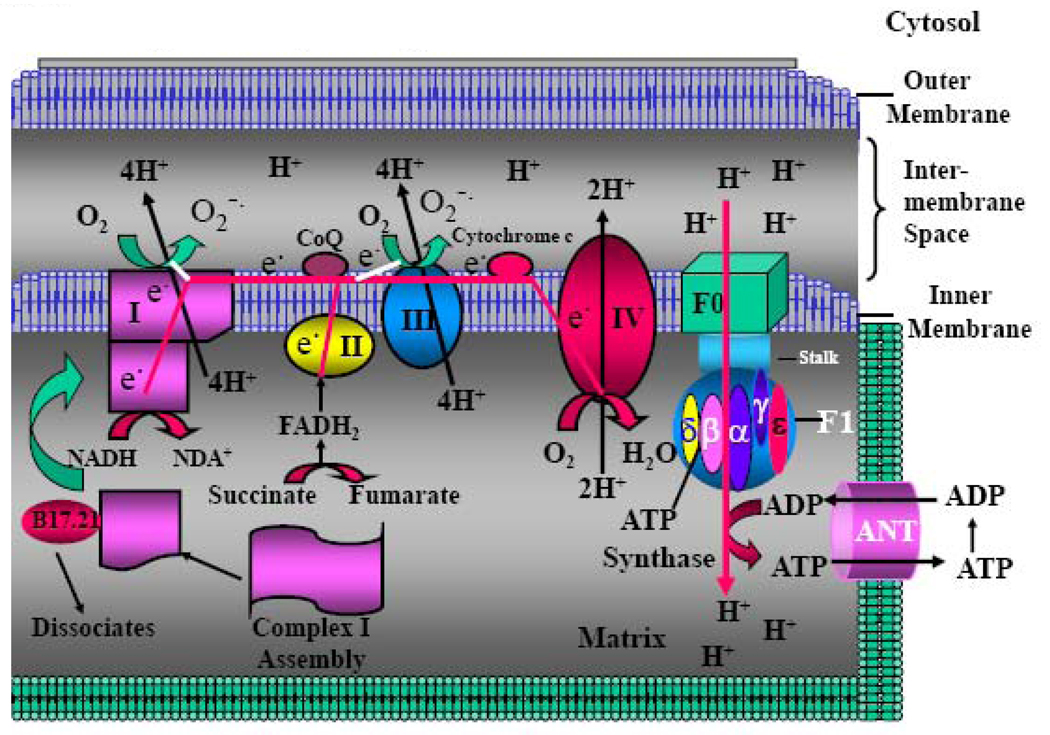
MRC (also called electron transport chain)(Figure 1) is one of the most important structural and functional parts of mitochondria. MRC consists of a series of metalloproteins bound to the mitochondrial inner membrane (also named cristea). There are four large protein complexes (designated complex I to complex IV) that are associated with mitochondrial electron transport. These complexes cooperate in electron transfer and proton pumping across the inner mitochondrial membrane. Electron transfer between these complexes is accomplished by the mobile coenzymes ubiquinone (CoQ) in the lipid membrane, from complexes I and II to complex III and cytochrome c in the inter-membrane space, from complex III to complex IV. The mitochondrial F0–F1 ATP synthase couples the proton gradient across the mitochondrial inner membrane to the synthesis of ATP from ADP + Pi. The best-known function of MRC is its ability to generate the vast majority (more than 90%) of cellular energy in the form of ATP, which is essential to drive and maintain the majority of the physiological activities. In addition, MRC also generates a large quantity of reactive oxygen species (ROS) as by-product, which is the major source of cellular ROS. ROS play important role in redox regulation of gene expression, the control of cell proliferation and apoptosis, as well as serving as second messenger involved in regulation of a number of physiological and pathological activities (see below). The ROS also have the potential for causing oxidative damage to DNA and other critical molecules within the mitochondria. Together, the MRC-generated ATP and ROS play crucial roles in regulating the large majority of physiological and pathological activities of the cells.
Fig. 1. MRC, H+-ATP Synthase and ANT.
Schematic picture showing MRC complexes, H+-ATPase, and ANT. MRC: Mitochondrial respiratory chain; ANT: Adenosine Nucleotide Translocase.
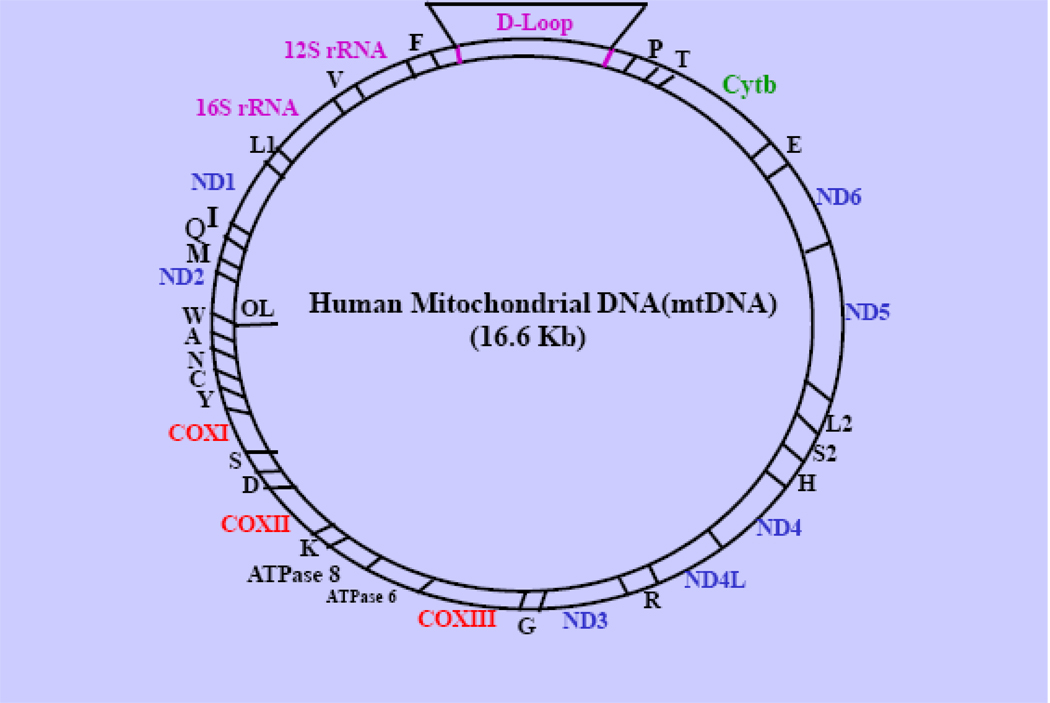
Human/mammalian mitochondria contain their own genetic system, which undergoes a unique mode of cytoplasmic inheritance. Each organelle has multiple copies of a covalently closed circular, 16.6 kD DNA genome (mitochondrial DNA, mtDNA), which encodes mRNAs for 13 essential subunits of MRC, 2 rRNAs (12S and 16S), and 22 tRNAs that are needed for mitochondrial protein synthesis (9) (Figure 2). The majority of MRC proteins and all of the other protein factors that are involved in replication-transcription-translation of mtDNA, assembly of MRC complexes, heme biosynthesis, mitochondrial protein import machinery and other mitochondrial functions (see sections 2 and 3) are encoded by nuclear genes. Coordination of the expression of the nDNA-and mtDNA-encoded genes is thus essential for maintaining normal mitochondrial functions and responses to paphological and environmental factors. The structural components, functions and assembly/biogenesis of the individual MRC complexes are briefly described as follows.
Fig. 2. Human Mitochondrial Genome.
The relative locations of genes encoding 13 MRC proteins for complex I (ND1, ND2, ND3, ND4, ND4L, ND5 and ND6); Complex III (cytb); Complex V (COX I, COX II and COX III) and H-ATP synthase (ATP6/8), two rRNAs (12S rRNA and 16S rRNA) and 22 tRNAs for specific amino acids indicated by the letters are shown. F: Phe, V: Val, L: Leu, I: Ile, Q: Gln; M: Met; W: Trp; A: Ala; N: Asn; C: Cys; Y: Tyr; S1: Ser, D: Asp; K: Lys; G: Gly; R: Arg; H: His; S2: Ser-2; L2: Leu-2; E: Glu; P: Pro; T: Thr. D-loop: Displace loop.
2.2.1. Complex I - NADH-ubiquinone Oxidoreductase
Complex I (NADH-ubiquinone oxidoreductase, EC 1.6.5.3), the largest among the MRC complexes, has two domains that form a L-shape: one hydrophobic domain lies within and is oriented parallel to the membrane, while a hydrophilic arm extends into the mitochondrial matrix. This complex transports two electrons from tricarboxyl acid (TCA) cycle-derived NADH in the mitochondrial matrix to quenone (Q) within the membrane, via flavin momonucleotide (FMN) and a series of seven iron-sulphur clusters. FMN can break up the simultaneous transfer of two electrons from NADH along with a proton, transferring H+ into two separate one-electron steps, as required for electron transfer through the iron-sulphur clusters. Complex I is also a major site of superoxide (O−˙) generation as a by-product due to the direct reaction of electrons that leak from this site with dioxygen (Figure 1).
Complex I consists of 46 protein subunits with seven subunits being encoded by mtDNA and the remaining 39 subunits being encoded by nuclear DNA (nDNA)(Table 1A). The mtDNA-encoded subunits play the major catalytic role of the enzyme. The majority of the nDNA-encoded subunits provide the regulatory functions. They are sub-classed into flavoprotein (FP), iron-sulfur protein (IP), and hydrophobic protein (HP) subclasses. The FP contains three subunits with molecular masses of 51, 24, and 9 kDa, respectively. The 51-kDa subunit contains FMN and a tetranuclear iron-sulfur cluster and is the principal site of entry for electrons donated by NADH into the MRC. The 24-kDa subunit contains a binuclear iron-sulfur cluster [9]. IP subclass contains seven subunits, designated NDUFS1 to NDUFS7, with molecular masses of 75, 49, 30, 18, 15, 13, and 11 kDa, respectively[10]. This subclass is important because it contains important prosthetic groups highly conserved among species. It is noted that the NDUFS2 contains a highly conserved protein kinase C phosphorylation site and the NDUFS3 subunit contains a highly conserved casein kinase II phosphorylation site [10]. This makes them strong candidates to be regulated by phosphorylation/dephosphorylation, an important regulatory mode that can controls their activity [11]. FP and IP make contact through the 51- and the 75-kDa subunits. The hydrophobic protein (HP) subclass, which includes more than thirty subunits, is largely membrane-intercalated and contains two iron-sulfur clusters apparently in a 23-kDa subunit and possibly another in a 20-kDa subunit [9]. Stoichiometrically, per mole of complex I, there are 2 mol of the 15-kDa subunit and 1 mol each of the FP and the four largest IP subunits [9].
Table 1.
| Table 1A: Properties of mtDNA-amn nDNA-encoded Complex I Subunits and complex I Assembly Proteins | ||||||
|---|---|---|---|---|---|---|
| Properties |
NRF-1 & NRF-2 Binding Sites |
Other Factors Binding Sites |
Regulated by E2/ERs |
References | ||
| Complex I Subunits | ||||||
| mtDNA- Encoded |
ND1 | via Tfam | TFBIM & TFBIM | + | 48, 79 | |
| ND2 | via Tfam | TFBIM & TFBIM | ||||
| ND3 | via Tfam | TFBIM & TFBIM | ||||
| ND4L | via Tfam | TFBIM & TFBIM | ||||
| ND4 | via Tfam | TFBIM & TFBIM | ||||
| ND5 | via Tfam | TFBIM & TFBIM | ||||
| ND6 | via Tfam | TFBIM & TFBIM | ||||
| nDNA- Encoed |
FP | NDUFV1 | NRF-2 | Sp1 & YY1 | 143 | |
| NDUFV2 | ||||||
| NDUFV3 | ||||||
| IP | NDUFS1 | |||||
| NDUFS2 | ||||||
| NDUFS3 | ||||||
| NDUFS4 | ||||||
| NDUFS5 | ||||||
| NDUFS6 | ||||||
| NDUFS7 | ||||||
| NDUFS8 | NRF-1 | Sp1 & YY1 | + | 116 | ||
| NDUFA8 | + | 116, 142 | ||||
| HP | 31 subunits | |||||
| Nuclear- Encoded Assembly Proteins |
CIA30 NDUFAF1 C20orf7 C6orf66 |
|||||
| Table 1B: Properties of nDNA-encoded complex II subunits | |||||
|---|---|---|---|---|---|
| Properties |
NRF-1 & NRF-2 Binding Sites |
Other Factors |
Regulated By E2/ERs |
References | |
| Genes | |||||
| nDNA- Encode |
SDHA | NRF-1 | NA | 122 | |
| SDHB | NRF-1; NRF-2 | N/A | + | 116, 120 | |
| SDHC | NRF-1; NRF-2 | N/A | 18 | ||
| SDHD | NRF-1; NRF-2 | N/A | 121 | ||
| CytC | NRF-1, NRF-2 | + | 67, 118 | ||
| Table 1C: Properties of mtDNA and nDNA encoded Complex III Subunits and Assembly Proteins | |||||
|---|---|---|---|---|---|
| Properties |
NRF-1/NRF-2 Binding Sites |
Other Factors Binding Sites |
Regulated by E2/ERs |
References | |
| Complex III Subunits | |||||
| mtDNA- Encoded |
Cytb | via Tfam | TFBIM & TFBIIM | + | 48 |
| nDNA- Encoded |
QCR6 | N/A | N/A | ||
| QCR8 | N/A | N/A | |||
| QCR9 | N/A | N/A | + | 116 | |
| QCR10 | N/A | N/A | |||
| nDNA- Encoded Assembly Proteins |
QCR7 | N/A | N/A | ||
| BCSIL | N/A | N/A | |||
| Table 1D: Properties of mtDNA and nDNA encoded Complex IV Subunits and complex IV Assembly Proteins | |||||
|---|---|---|---|---|---|
| Properties |
NRF-1 & NRF-2 Binding Sites |
Other Factors Binding Sites |
Regulated By E2/ERs |
References | |
| Complex IV Subunits | |||||
| mtDNA- Encoded |
COXI | via Tfam | TFBIM & TFBIIM | + | 48, 51, 53,55, 67, 69 |
| COXII | via Tfam | TFBIM & TFBIIM | + | 48, 51, 53, 55 | |
| COXIII | via Tfam | TFBIM & TFBIIM | + | 48, 51, 53, 55 | |
| nDNA- Encoded |
COXIV | NRF-1; NRF-2 | Sp1 | + | 67, 76, 79,130, 135, 136 |
| COXVa | NRF-1; NRF-2 | + | 115, 130, 135, 136 | ||
| COXVb | NRF-1; NRF-2 | Sp1, Ets; NF-E1 | + | 126, 127, 130, 135, 136 | |
| COXVIb | NRF-1; NRF-2 | + | 130, 135, 136 | ||
| COXVIc | NRF-1, NRF-2 | + | 130, 135, 136 | ||
| COXVIIb | NRF-1, NRF-2 | + | 130, 135, 136 | ||
| COXVIIc | NRF-1, NRF-2 | + | 130, 135, 136 | ||
| COXVIII | NRF-1, NRF-2 | + | 128, 133, 134 | ||
| COXVIa(L) | NRF-1; NRF-2 | Sp1, YY-1,Est | 124 | ||
| COXVIa(H) | MEF2A | ||||
| COXVIIa(L) | NRF-1; NRF-2 | Sp1 | |||
| COXIIa(H) | MEF2A | + | 116, 137 | ||
| COXVIII(H/L) | |||||
| nDNA- Encoded Assembly Proteins |
COX17p | NRF-1; NRF-2 | Sp1 | 128 | |
| SCO1 | |||||
| SCO2 | |||||
| SURF1 | |||||
| COX11 | |||||
| COX15 | |||||
| Table 1E: Properties of mtDNA and nDNA encoded ATP Synthase Subunits and Assembly Proteins | ||||||
|---|---|---|---|---|---|---|
| Properties | NRF-1/NRF-2 Binding Sites |
Other Factors Binding Sites |
Regulated By E2/ERs |
References | ||
| F0-F1 ATP Synthase Subunits | ||||||
| mtDNA- Encoded |
ATP6 | via Tfam | TFBIM & TFBIIM | + | 48 | |
| ATP8 | via Tfam | TFBIM & TFBIIM | + | 48 | ||
| nDNA- Encoded |
F0 | Inhibitor | ||||
| protein | ||||||
| a | ||||||
| b | ||||||
| c | ||||||
| d | + | 116 | ||||
| e | + | 116 | ||||
| f | + | 115 | ||||
| g | + | 115 | ||||
| OSCP | ||||||
| J | + | 114 | ||||
| F6 | ||||||
| A6L | ||||||
| F1 | Alpha(α) | ATPF1, YY-1 | ||||
| Beta(β) | + | 116 | ||||
| Gamma(γ) | NRF-1 | 142 | ||||
| Delta(δ) | ||||||
| Epsilon(ε) | + | 114 | ||||
| Stalk | 45-A | |||||
| nDNA- Encoded Assembly Proteins |
Oxal ATP12 TMEM70 |
|||||
The biogenesis of intact complex I requires the correct assembly of both mtDNA-and nDNA-encoded subunits. The mtDNA-encoded subunits first assemble into intermediate complexes and require significant chase times for their integration into the holoenzyme. In contrast, a set of newly imported nDNA-encoded subunits integrates with preexisting complex I subunits to form intermediates and/or the fully assembed holoenzyme. One of the intermediate complexes represents a subassembly associated with the chaperone protein B17.2L. Recent studies (for review see [12]) have pointed to a mechanism of complex I biogenesis involving two complementary processes: (i) synthesis of mtDNA-encoded subunits to seed de novo assembly and (ii) exchange of preexisting subunits with newly imported ones to maintain complex I homeostasis. Subunit exchange may also act as an efficient mechanism to prevent the accumulation of oxidatively damaged subunits that would otherwise be detrimental to mitochondrial oxidative phosphorylation and have the potential to cause disease.
Several proteins including mtDNA-encoded ND6[13] and the nDNA-encoded CIA30 (complex I intermediate associated protein)[14], NDUFAF1[15], C20orf7[16] and C6ORF66[17] have been known to play essential roles in complex I assembly. CIA30 associates with the newly translated mtDNA-encoded complex I subunits at early stages in their assembly before dissociation at a later stage. CIA30-deficiency, which causes reduced levels and activity of complex I, was detected in a patient with cardioencephalomyopathy. Genetic analysis has revealed that the patient had mutations in both alleles of the NDUFAF1 gene that encodes CIA30. Complex I assembly in patient’s cells is defective with subunits being degraded at early stages of development. Complementing the deficiency in patient fibroblasts with normal CIA30 restores steady-state complex I levels [14]. C20orf7 peripherally associates with the matrix face of the mitochondrial inner membrane. Silence of its expression with small interference RNA (siRNA) decreases complex I activity. Furthermore, mutation of C20orf7 disruptes complex I assembly and causes lethal neonatal mitochondrial disease[16]. NDUFAF1 has been demonstrated to be an important protein for the assembly and stability of complex I. NDUFAF1 is associated to two intermediate complexes of 600 and 700 kDa in size. The 700 kDa complex appears to represent a key step in the complex I assembly process. The relative distribution of these two complexes is altered in two complex I-deficient patients. Knock down of NDUFAF1 expression with siRNA led to a reduced amount and activity of complex I [15]. Homozygosity mapping in five patients from a consanguineous family presented with infantile mitochondrial encephalomyopathy led to identification of a missense mutation in a conserved residue of the C6ORF66 gene, which encodes a 20kDa mitochondrial protein [17]. The same mutation has also been detected in a patient who had antenatal cardiomyopathy. In muscle of two patients, the levels of the C6ORF66 protein and the fully assembled complex I were markedly reduced. Transfection of the patient's fibroblasts with wild-type C6ORF66 cDNA restored complex I activity. These data suggest that C6ORF66 is an assembly factor of complex I. More importantly, the C6ORF66 gene product was found to promote invasiveness of breast cancer cells [17].
2.2.2. Complex II - Succinate Dehydrogenase
Complex II (succinate dehydrogenase, EC 1.3.5.1) oxidizes succinate to fumarate and reduces quenone (Q) to QH2 within the membrane. The electron transfer from FDAH2 proceeds initially via an FAD cofactor, and then through a series of three iron-sulphur clusters. The first is a [2Fe2S] cluster where one of the Fe atoms is bound by aspartate oxygen. The second and third ones are the [4Fe4S] and [3Fe4S] clusters. All these groups are in the hydrophilic protein chains external to the membrane. Finally, electrons pass through the home iron of cytochrome b560 (within the hydrophobic protein inside the membrane), and then outside the protein to ubiquinone, which binds near the heme group.
Complex II contains four nDNA-encoded subunits but no mtDNA-encoded subunits: the flavoprotein (FP) and iron-sulfur protein (IP) of the dehydrogenase, and two integral membrane proteins referred to as C (II-3) and D (II-4). Their respective genes in mammals are SDHA, SDHB, SDHC and SDHD [18](Table 1B). Mutations in any of these may disrupt complex II enzymatic activity. In fact, defects in SDHA produce bioenergetic deficiency while defects in SDHB, SDHC, or SDHD induce tumor formation, suggesting that Complex II has tumor suppressor functions. It has been demonstrated that loss of the SDHB, but not the SDHA, subunit triggers ROS-dependent hypoxiainducible factor activation and tumorigenesis [19]. SDHB, SDHC, and SDHD germline mutations are prevalently detected in patients with head and neck paragangliomas [20–22]. These observations suggest that the dysfunction and loss of these genes due to mutations can have an important role in the malignant transformation of the paragangliomas.
2.2.3. Complex III - Cytochrome bc1 Complex
Complex III (ubiquinol cytochrome c reductase, EC 1.10.2.2) catalyzes electron transfer from succinate via FDAH2− and NADH-linked dehydrogenases to cytochrome c, a mobile electron carrier located outside the mitochondrial membrane, shuttling one electron at a time from complex III to complex IV.
Complex III can be divided into two halves. Each half has a binding site for the lipid-mobile carrier ubiquinol/ubiquinone (QH2/Q). Quinol oxidation takes place near the inter-membrane space, while quinone reduction takes place near the matrix side of the membrane. Half the electrons delivered at Qo pass through the complex to Qi. Because the redox of Q/QH2 also involves the transfer of protons, this process results in the release of protons into the inter-membrane space and the uptake of protons from the cell matrix. In effect, H+ has been transported from one side of the membrane to the other, even though it is not actually the same proton coming and going. The other half is delivered to another intermembrane-mobile carrier, cytochrome c, which contains a heme group with two axial ligands, one hisN and one metS, giving octahedral coordination to the iron atom. The two iron oxidation states are Fe(III) d5 and Fe(II) d6, both low spin. The transfer of a non-bonding t2g electron doesn't dramatically affect the coordination geometry of the iron atom, making for kinetically facile electron transfer. There are two pathways for electrons to pass through Complex III from ubiquinol at Qo. One is via two heme iron centers, cytochromes b562 (or bL, with lower potential) and b566 (or bH, with higher potential), and then to ubiquinone at Qi. The other is via a Rieske iron-sulphur cluster to cytochrome c1, and then to cytochrome c. Complex III is another site for generation of superoxide within MRC (see Figure 1).
Complex III is made up of 11 subunits, of which all but one (cytochrome b) (cytb), are encoded by nDNA (Table 1C). The biogenesis of the yeast MRC complex III has been described (for review see [23]) Several studies have indicated that the mtDNA-encoded Cytb plays an important role not only in assembly of complex III but also in the maintenance of the complex I stability. Mutations in the cytb gene are associated either with only complex III deficiency or with combined complex I and III deficiency. In the processes of the assembly of the entire MRC, the complexes I and III form a stable core respirasome to which complex IV binds. The formation of respirasomes is essential for the assembly and stability of complex I. The dependence of complex I stability on the Cytb-mediated assembly of complex III has been demonstrated by a study using mouse and human cultured cell models harboring cytb mutations with a combined complex I and complex III defects. In both, complex III assembly was impeded and causes a severe reduction in the amount of complex I, not observed when complex III activity was pharmacologically inhibited. Metabolic labeling in mouse cells revealed that complex I was assembled, but its stability was severely hampered. Conversely, complex III stability was not influenced by the absence of complex I. This structural dependence among complexes I and III was confirmed in a muscle biopsy of a patient harboring a nonsense cytb mutation [24, 25]. In addition to Cytb, a nuclear gene named BCS1L, which encodes a member of the AAA family of ATPases, is required for the assembly of complex III and for the expression of functional Rieske iron-sulfur protein. Mutations in this gene cause disruption of Complex III assembly, associated with reduced activity of the MRC and increased ROS production. A number of functional mutations in BCS1L have been reported in patients with tubulopathy, encephalopathy, liver failure and the Björnstad syndrome [26, 27]. More recently, pathogenic mutations in the 5′ untranslated region of BCS1L mRNA have been identified, which were associated with decreased BCS1L mRNA and protein levels, and a complex III assembly impairment [28].
2.2.4. Complex IV - Cytochrome c Oxidase (COX)
Complex IV [Cytochrome c Oxidase (COX), EC 1.9.3.1],the major component of the MRC complex localized within the mitochondrial inner membrane, acts as the terminus of mitochondrial electron transport in all aerobic life. This reaction is coupled with the transfer of four protons across the mitochondrial inner membrane, driving the synthesis of ATP via mitochondrial F0–F1 ATP synthase (section 2.2.5). In each half of COX, there are three metal sites involved in electron transfer: bimetallic CuA, monometallic cytochrome a, and bimetallic cytochrome a3/CuB. In addition, there are non-redox Mg2+ and Zn2+ ions, which, along with the CuB/cyt a3 site, may be involved in the proton-pumping mechanism. Cytochrome c initiates electron flow through the enzyme by delivering one electron at a time to CuA, which sits just outside the membrane. Electrons flow from CuA through cyt a to the a3/CuB site, which is where O2 is bound and reduced to water. With four one-electron redox centers (CuA, cyt a, CuB, and cyt a3), the enzyme can hold the four electrons needed for the complete reduction of a single O2 molecule.
Complex IV consists of thirteen subunits (Table 1D). Of which, the largest subunits, COXI, COXII, and COXIII, are encoded by the mtDNA and synthesized within the mitochondria. They represent the catalytic core of the enzyme; the rest of the smaller subunits are encoded by nDNA and imported into mitochondria following their synthesis in the cytosol. The nDNA-encoded subunits are implicated in the regulatory functions. Thus, its biosynthesis involves a coordinate interplay between nuclear and mitochondrial genomes. The subunits IV, Va, Vb, VIb, VIc, VIIb, VIIc, and VIII(L) are ubiquitously expressed in all the tissues, although the mRNA levels for the individual subunits vary in different tissues. Several nDNA-coded subunits are expressed in a tissue- and development-specific manner. For example, the subunits VIa (H), VIIa (H), and VIII (H) are exclusive to heart and skeletal muscle. COX biogenesis includes a variety of steps starting from translation to the formation of the mature complex. Each step involves a set of specific factors that assists translation of subunits, their translocation across membranes, the insertion of essential cofactors, assembly and final maturation of the enzyme. The organization and biogenesis of COX have been described (for details see [29, 30]).
The COX IV assembly/biogenesis relies on a number of proteins, including SCO1, SCO2, COX11, COX15, Surf1p, COX17 and SURF1 (Table 1D), which are essential for the correct assembly and stability of this complex. SCO1, SCO2 and COX17 are also responsible for delivery of copper ions to the mitochondrion and for insertion of these ions into the enzyme and maintenance of cell copper homeostasis [31–33]. Mutations in SCO2 have been reported in infants with early onset of fatal cardioencephalomyopathy and who have a severe COX deficiency in striated muscle [33–35]. More interestingly, SC02 has been known to couple with p53 within mitochondria in modulating the balance between the utilization of MRC and glycolytic pathways [36]. Maturation of the heme a (3)-Cu(B) center is a step that limits the association of subunits I and II in the COX assembly. Surf1p plays a role in facilitating the insertion of heme into the active site of COX. Numerous mutations in human Surf1p lead to severe mitochondrial disease with defective complex IV functions [37]. Surf1 appears to be involved in an early step of heme insertion into subunit I [38]. In humans, the loss of Surf1 function due to mutations is associated with Leigh syndrome, a fatal neurodegenerative disorder which is highly prevalent in Poland [39].
2.2.5. Complex V-mitochondrial F0–F1 ATP synthase
Complex V (the mitochondrial F0–F1 ATP synthase) is a membrane protein complex, consisting of catalytic sector, F1; the membrane sector, F0, and a long stalk that connects F1 to F0 (Figure 1). F0–F1 ATP synthase couples the proton gradient across the mitochondrial inner membrane to the synthesis of ATP from ADP + Pi. The complex is composed of essential subunits for its motor functions and supernumerary subunits. Two subunits of ATP synthase, subunits 6 and 8 (ATP6/8), are encoded by mtDNA and the remaining subunits are encoded by nDNA (Table 1E). The well characterized subunits of the bovine ATP synthase complex are the subunits alpha (α), beta (β), gamma (γ), delta (δ), and epsilon (ε) of F1; the ATPase inhibitor protein; and subunits a, b, c, and d, OSCP (oligomycin sensitivity-conferring protein), F6, and A6L are present in F0, and the 45-A-long stalk. Bovine ATP synthase preparations also contain three small polypeptides, designated e, f, and g, with respective molecular masses of 8.2, 10. 2 and 11.3 kDa, respectively. It has been shown that (i) e, f, and g could be immunoprecipitated with anti-OSCP from a fraction of bovine submitochondrial particles enriched in oligomycin-sensitive ATPase;(ii) the NH2 termini of subunits f and g are exposed on the matrix side of the mitochondrial inner membrane and can be curtailed by proteolysis; (iii) the COOH termini of all three polypeptides are exposed on the cytosolic side of the inner membrane; and (iv) f cross-links to A6L and to g, and e cross-links to g and appears to form an e-e dimer. Thus, the bovine ATP synthase complex appears to have 16 subunits [10].
The assembly of mitochondrial F0–F1 ATP synthase is a complicated process, which is not fully understood. However, several proteins including Oxa1, ATP12, subunit ε and TMEM70 are involved in the assembly and biogenesis of this complex. The Oxa1 protein is involved in assembly of the COX complex by facilitating the co-translational membrane insertion of mtDNA-encoded COX subunits. It has been demonstrated [40] that Oxa1 also directly supports the assembly of the inner membrane embedded F0-sector of the ATP synthase via its physical interaction with the newly synthesized mtDNA-encoded ATP8 protein. In the absence of Oxa1, ATP8 is observed to assemble into an oligomeric complex containing F1-subunits, but its further assembly with subunit 6 (ATP6) of the F0-sector is perturbed. By directly interacting with newly synthesized ATP8 in a posttranslational manner, Oxa1 is required to maintain the assembly competence of the ATP8-F1-subcomplex for its association with ATP6. Subunit ATP12 is also involved in assembly for F0-F1 ATP synthase. A mutation in the ATP12 gene has been identified in one patient, which is believed to be the cause of the impaired ATP synthase activity[41]. Bisetto et al. [42] have determined that integral inner membrane subunit ε is essential for self-association of F0–F1 ATP synthase. Whole-genome homozygosis mapping, gene expression analysis and DNA sequencing in individuals with isolated mitochondrial ATP synthase deficiency led to identification of disease-causing mutations in TMEM70. Complementation of the cell lines derived from these individuals with wild-type TMEM70 restored biogenesis and metabolic function of the enzyme complex. These results indicate that TMEM70 is involved in mitochondrial ATP synthase biogenesis [43].
3. Regulation of mtDNA-encoded MRC proteins by E2 and ERs
Mitochondria integrate a large number of signal transduction pathways for a wide variety of biologically active molecules. There is increasing evidence that mtDNA is one of the major targets for the direct actions of steroid and thyroid hormones and their respective receptors (for reviews see [44–47]). The regulation of mtDNA-encoded MRC protein synthesis by E2 and ERs is described as follows.
3.1. Localization of ERβ and ERα in Mitochondria of Target Cells/Tissues
In addition to their presence in nuclei and plasma membrane, ERα and ERβ are also localized in mitochondria of a number of cell types and tissues. While several earlier studies have provided several clues suggesting the association of ERs with non-nuclear/cytoplasmic compartments in target tissues (for review see [46]), definitive demonstration for the presence of ERα and ERβ in mitochondria in target cells comes from more recent studies. The studies examining the effects of 17β-ethinyl estradiol plus and minus ER antagonist ICI182780 on MRC gene expression and MRC-mediated superoxide generation in human liver cancer HepG2 cells provided the first line of evidence suggesting the presence of ERs in their mitochondria [48, 49]. Monje and Boland reported the first direct detection of ERβ in the mitochondria in rabbit uterus and ovary [50]. Using subcellular fractionation/Western blot, immunochemical localization with confocal fluorescent microscopy and immunogold microscopy, Chen et al. [51–53] demonstrated that a substantial fraction (approximately 20%) of total cellular ERβ are localized within the mitochondrial matrix of human breast cancer MCF-7 and that E2 enhanced the import of ERβ into MCF-7 cell mitochondria in a time- and dose-dependent manner. Pedram et al. [54] independently confirmed the presence of functional ERβ in MCF-7 and endothelial cells. Chen et al. [55] observed that ERβ is predominantly localized in mitochondria of normal, immortalized human breast epithelial cells (MCF-10F) and the E2-induced transformed subline of MCF-10F cells (trMCF). More importantly, they demonstrated that silence of ERβ expression with ERβ-specific siRNA markedly diminished both nuclear and mitochondrial ERβ in MCF-10F and MCF-7 cells. In addition, they observed a progressive shift of ERβ from its predominant localization in mitochondria in MCF-10F and trMCF to the nuclei of the more invasive and tumorigenetic MCF-10F sub-lines [55]. The mitochonrial localization of ERβ has been extended to a number of cell type and tissues. Yang et al. [56] demonstrated the mitochondrial localization of ERβ in primary neurons, primary cardiomyocytes, murine hippocampus cell lines and human heart cells. Moreover, they verified, using a proteomic approach, tat a protein with molecular weight between 50 to 60 kDa within mitochondria of human heart cells was ERβ. Furthermore, ERβ has been detected in mitochondria of human lens epithelial cells[57–59], human liver cancer HepG2 cells[53, 60], osteosarcoma SaOS-2[60, 61], sperm [62], periodontal ligament cells [63], and rat brain rostral ventrolateral medulla and the hippocampal CA1 region where silver grains indicative of 125I-estradiol binding are localized [64, 65].
It should be noted that the wild-type human ERβ (designated ERβ1) coexists with several isoforms (designated ERβ2 to ERβ5) in a wide range of tissues. These isoforms are generated via alternative splicing of exons encoding the carboxyl terminus. Using antibodies specific for ERβ1, ERβ2 and ERβ5, Cammarata et al. [58] observed that ERβ-1 immunostaining was prominently present in the mitochondria along with weaker staining in the nucleus of normal cultures of human lens epithelial cells (nHLE), virally transformed HLE-B3 and MCF-7 cells, as determined by co-localization with Mitotrack-633. However, ERβ2 staining was distributed throughout the cytosol and was associated with the nucleus of these cells. ERβ5 staining was diffuse in the cytosol and also associated with their nuclei. The differential subcellular distribution of ERβ1 to the mitochondria and ERβ2 to the nucleus suggests a new aspect of regulation and function of the ER signaling systems. Indeed, ERβ1 has been shown to play an important role in mitochondrial cytoprotection from oxidative stress in cultured normal male and female human lens epithelial cells [59](see section 7.5. for more details).
The presence of ERβ1 but not of ERβ2 and ERβ5 in mitochondria of many types of cells and tissues indicates that, like other mitochondrially localized proteins, ERβ1 is imported into mitochondria via mitochondrial protein import machinery and that its import likely requires the mitochondrial targeting polypeptide signal (mTPs) and information encoded within the carboxyl terminus of ERβ1. Indeed, a putative mTPs has been identified within the internal amino acid sequence of human ERβ1[14, 51]. It is likely that ERβ is imported into the mitochondria of target cells, through tethering to cytosolic chaperone protein and/or through direct interaction with mitochondrial import proteins [66]. The mechanisms underlying the mitochondrial import of ERβ deserves further investigation.
While ERα is mainly present in the nucleus, several studies have demonstrated its presence in mitochondria of several types of cells, including cerebral blood vessels obtained from ovariectomized (Ovx) female rats [67], MCF-7 cells [51, 54, 68], HepG2 cells [53, 60], and 2C12 murine skeletal muscle cell line[69].
The presence of ERα and ERβ in mitochondria of breast, heart, brain, bone, eyes, sperm, and periodontal ligament tissues, which have high demand on energy supply from mitochondria for their proper functions, indicates that mitochondrial ERα and ERβ may have important roles in regulation of mitochondrial energy metabolism in these systems (see section 7).
3.2. Induction of mtDNA Transcription by E2 and ERs
E2 has been known to up-regulate the transcript levels of several mtDNA-genes, which encode MRC complex proteins, and to have direct and indirect effects on MRC activity [48, 51, 53, 70–72]. The previous studies on the effects of estrogens on the transcript levels of mtDNA genes encoding MRC proteins in a number of cell types and tissues have been reviewed ([46, 73] and references therein) and summaried[74]. A number of recent studies have provided strong additional support for an important role of E2 in regulation of the expression of mtDNA-encoded MRC proteins.
It was reported [67] that treatment of Ovx female rats in vivo with E2 increased the levels of a number of specific proteins in cerebrovascular cell mitochondria, including mtDNA-encoded COX1, the nDNA-encoded cytochrome c and COX IV of complex IV, ERα and manganese superoxide dismutase (MnSOD), associated with increased complex IV activity. Irwin et al. [75] investigated the effects of progesterone (P) and E2 on mitochondrial functions. They treated ovx rats with P, E2, or E2 + P and then isolated whole-brain mitochondria for functional assessment and observed that brain mitochondria from E2-treated rats exhibited increased expression and activity of complex IV coupled to MRC functions, and that this increased MRC activity was coupled with a decreased rate of reactive oxygen leakage and reduced lipid peroxidation, representing a systematic enhancement of brain mitochondrial efficiency. Hsieh et al. [76] treated male rats underwent trauma-hemorrhage (T-H) with either ERα selective agonist propylpyrazole triol (PPT) or ERβ selective agonist diarylpropionitrile (DPN) or E2 or vehicle and another group of rats with Complex IV inhibitor sodium cyanide (SCN) ± DPN. Their results indicated that 24h after T-H, cardiac functions were depressed in the vehicle-treated but were normal in DPN-treated rats. Either E2 or DPN treatment after T-H normalized cardiac mitochondrial ERβ expression and increased mitochondrial ERβ DNA-binding activity, accompanied by an increase in complex IV gene expressions and activity. Moreover, inhibition of complex IV in DPN-treated T-H rats by SCN abolished the DPN-mediated cardioprotection, ATP production, mitochondrial cytochrome c release, caspase-3 cleavage, and apoptosis. These results indicate that E2 and ERβ-mediated cardioprotection following T-H appears to be mediated via mitochondrial ERβ-dependent complex IV activity and inhibition of mitochondrial apoptotic signaling pathways. Human periodontal ligament (PDL) cells express mainly ERβ protein. Jonsson et al. [63] demonstrated ERβ in mitochondria of PDL cells, in which E2 induced attenuation the expression of COXI of complex IV, an effect that was blocked by ER antagonist, ICI182780. Chen et al. [55] reported that treatment of immortalized human breast epithelial cells that are negative to ERα but contain ERβ in their mitochondria with E2 and DPN for 24h significantly stimulated the expression of subunits COX I and COX II of complex IV, and ND1 subunit of complex I and that knock down of ERβ expression with siRNA diminished the mitochondrial expression of ERβ in these cells and blocked E2-induced expression of COXII protein. These results demonstrated that the mitochondrial ERβ in MCF-10F cells is directly involved in E2-induced expression of mtDNA-encoded MRC proteins. Consistent with these observations, Nilson et al. [77] reported increased mRNA and protein levels of COXI, COXII and COXIII in mitochondria of rat brain neurons and commitant increase in COX activity.
3.3. Activation of Nuclear Respiratory Factors (NRFs) and Mitochondrial Transcription Factor A (Tfam) by E2/ERs
3.3.1. Activation of NRFs by E2 and ERs
While the molecular mechanisms underlying the above-described E2/ER-mediated effects on expression of mtDNA-encoded MRC proteins are not completely understood, several recent studies have pointed to an important role of nuclear respiratory factors 1 and 2 (NRF1 and NRF-2) in mediating these effects. NRF-1 and NRF-2 are primary transcription factors of nDNA-encoded mitochondrial proteins, e.i. the majority of MRC complex proteins and mitochondrial transcription factor A (Tfam), which control transcription of the mtDNA, are regulated by NRF-1 and NRF-2 (see below). Stirone et al. [67] observed that long-term treatment of ovx female rats with E2 increased NRF-1 protein in their cerebral blood vessels. Similar stimulatory effects of E2 on NRF-2 protein levels were also seen in heart cells of male rats [78]. Mattingly et al. [79] revealed an important role of NRFs in regulation of E2/ER-mediated biogenesis. These investigators reported that E2 increased NRF-1 mRNA and protein in both MCF-7 breast and H1793 lung adenocarcinoma cells in a time-dependent manner. These effects were inhibited by ER antagonist ICI 182,780 and transcription inhibitor, actinomycin D but not by phosphoinositide-3 kinase and MAPK inhibitors, indicating that up-regulation of NRF-1 by E2 is mediated via ERs at the transcriptional level. Importantly, NRF-1 promoter contains an estrogen response element (ERE) that specifically binds to ERα and ERβ in vitro as demonstrated by gel electrophoresis mobility shift assays (EMSA). E2 induced the recruitment of both ERα and ERβ to this ERE in vivo in MCF-7 cells as revealed by in Chip assays. Furthermore, the ERE of NRF-1 has been shown to activate expression of a reporter gene in transfected cells. Knock down of the expression of ERα and ERβ with respective siRNAs has revealed that it is ERα that mediates E2-induced NRF-1 transcription in MCF-7 cells.
3.3.2. Regulation of Tfam and PGC by E2 via NRFs
Mitochondrial transcription factor A (Tfam, also named MTFA or mtTFA) is an nDNA-encoded, mitochondrially-localized protein that is crucial for replication, transcription and maintenance of mtDNA [80, 81]. Human Tfam binds to mtDNA in a sequence-independent manner and is abundant enough to cover the entire mtDNA genome, thereby stabilizing mtDNA through formation of a nucleoid (mitochromosome corresponding to chromosome of nDNA) and regulates the amount of mtDNA[81]. Over-expression of human Tfam in mice increased the amounts of mtDNA and dramatically ameliorated the cardiac dysfunction caused by myocardial infarction [82]. Tissue-specific knock out of Tfam in mouse heart resulted in dramatic reduction in mtDNA content. This effect was associated with abnormal mitochondrial functions[83]. Knock down of Tfam expression with siRNA in HeLa cells resulted in a reduction of Tfam levels to 15% of control, associated with a closely parallel reduction in amount of mtDNA[84]. Thus, Tfam may play a crucial role in maintaining mtDNA as a main component of the nucleoid. Tfam also interacts with the mitochondrial p53, protecting mtDNA from oxidative damage [85, 86]. As a key transcription factor, Tfam plays an essential role in activation of mtDNA transcription. Transient over-expression of Tfam stimulated mtDNA transcription. This effect was more evident when Tfam was transiently over-expressed in cells having less mtDNA caused by pre-treatment with ethidium bromide [87].
Because of its important role within mitochondria, the expression and activities of Tfam must be linked to cellular energy needs in response to physiological, pathological changes and environmental insults. Its expression is under the control of a number of transcriptional factors, including NRF-1, NRF-2, specificity protein 1 (Sp1) and hStaf/ZNF143. Indeed, NRF-1 and NRF-2 binding sites are present in the proximal promoter of the human Tfam gene [80]. The promoter of the human Tfam gene is highly dependent on both NRF-1 and NRF-2 binding sites for activity. The affinity-purified factors from HeLa cells were shown to specifically bind to these NRF-1 and NRF-2 sites. Mutations in these contact sites eliminated NRF-1 and NRF-2 binding and dramatically reduced promoter activity in transfected cells. NRF-1 binding appears to be the major determinant of promoter function. This dependence on NRF-1 activation was confirmed by in vitro transcription using highly purified recombinant proteins that displayed the same binding specificities as the HeLa cell factors. The activation of the Tfam promoter by both NRF-1 and NRF-2 therefore provides a link between the expression of nuclear and mitochondrial genes and suggests a mechanism for their coordinate regulation during organelle biogenesis [80]. The role of NRF-1 in the regulation of Tfam expression is further demonstrated by the observation [79] that the E2-induced increase in NRF-1 is followed by increased Tfam and the transcription of Tfam-regulated mtDNA-encoded COXI and NDI genes, and increased mitochondrial biogenesis. Knock down of NRF-1 with siRNA blocked E2 stimulation of mitochondrial biogenesis and activity, indicating a mechanism by which estrogens regulate mitochondrial function by increasing NRF-1 expression.
In addition to NRFs, two cross-species conserved binding sites for the transcription factor hStaf/ZNF143 have been identified within the promoter region of human Tfam gene[88]. Promoter binding assays, transient expression of mutant Tfam reporter gene constructs and Chip assay had revealed that hStaf/ZNF143 was involved in promoter activity and that the promoter of the human Tfam gene harbored a complex organization with at least six transcriptional regulatory elements[88].
Three binding sites for Sp1 and one for NRF-2 but none for NRF-1 are present in the proximal upstream region within the 5'-flanking region of the rat Tfam gene [89]. Transfection of a reporter gene linked to this region into rat skeletal muscle cells has demonstrated that the promoter activity was 10 times higher than that of control vectors. Despite the absence of a NRF-1 binding site, co-transfection of human NRF-1 expression vector increased the Tfam promoter activity by 2-fold. An EMSA assay for NRF-1 has shown that NRF-1 binds to the proximal region of the promoter between −112 and +49 [89]. Consistent with this observation, Dong et al. [90] reported that the mRNA levels for NRF-1 and NRF-2 were 5- and 3-fold higher, respectively, in the rapidly growing rat hepatoma relative to the host liver, associated with more than a 10- fold increase in levels of Tfam, and 10-, 8-, 5-, and 3-fold increases in mtDNA-encoded ND5, ND6, COXI, COXII, respectively, the downstream targets of Tfam. Furthermore, the DNA binding activity of Sp1 in the hepatoma nuclear extract was 4-fold greater than that in the liver extract.
Several studies have indicated that the expression of Tfam is regulated by E2 via ERs. It has been reported [76, 78] that E2 increased the expression of rat cardiac PGC-1, NRF-2, Tfam and that these effects were associated with increased in COX IV and mtDNA-encoded COXI and ATP synthase β-subunit, mitochondrial ATP, and COX activity in rats that underwent trauma-hemorrhage (T-H). These effects were totally abolished by the ER antagonist ICI-182780, indicating the involvement of ER in mediating these effects. Furthermore, treatment of T-H animals with ERβ-selective agonist, DPN, mimicked E2’s effects [76, 78]. Similar stimulatory effects of E2 on Tfam were seen in Kupffer cells following T-H [91] and in human breast MCF-7 cells [79]. In contrast, E2 was reported to inhibit the expression of Tfam and NRF-1 in cultured rat brown adipocytes [92].
The effects of E2 on the expression of Tfam are most likely mediated via NRF-1 and NRF-2 because the promoter of human Tfam contains functional NRF-1 and NRF-2 binding sites[80] and E2-mediated up-regulation of NRF-1 in MCF-7 cell [79](see above). In addition, the effects of E2 on Tfam in heart cells may be mediated via the interaction of NRFs with PGC-1α because it has been shown that administration of antisense PGC-1α prevented both E2 and DPN-mediated cardioprotection and increased levels of ATP and Tfam following T-H and that the expression of PGC-1α was up-regulated by E2 and DPN [76, 78]. These findings suggest that the effects of E2 on cardiac function following T-H are mediated via ERβ up-regulation of PGC-1α through Tfam-dependent pathway [76, 78]. The effects of E2 on Tfam could be mediated via the cooperation of NRF-2 and Sp1. Sp1, a ubiquitous transcription factor, has been shown to be involved in regulation of a number of estrogen-responsive genes [93, 94]. Indeed, both NRF-2 and Sp1 have been shown to occupy the NRF-2 and Sp1 binding sites on the promoter of rat Tfam gene as revealed by in vivo genomic footprinting [90].
3.4. Regulation of Protein Factors Involved in Replication, Transcription and Translation of mtDNA by NRF
The replication, transcription and translation of mtDNA into these MRC proteins within mitochondria require the involvement of a number of nDNA-encoded protein factors that are synthesized on cytosol ribosomes and imported into mitochondria.
3.4.1. Regulation of Protein Factors Involved in mtDNA Replication by NRFs
Mammalian mtDNA replication usually occurs by asymmetric synthesis of the two strands, starting at the multiple origins in the displacement loop (D-loop). Replication of mtDNA is carried out by several proteins, including mitochondrial-specific DNA polymerase, mitochondrial RNA polymerase (POLRMT), the DNA helicase TWINKLE, and mitochondrial single strand binding proteins (mtSSB). DNA polymerase γ consists of a catalytic (POLG) subunit and an accessory (POLG2) subunit. Replication in the leading-strand origin is coupled to transcription via the formation of an RNA-DNA hybrid known as an R-loop [95]. POLRMT acts as lagging-strand primase in mammalian cells and highly processes on double-stranded DNA, but synthesizes RNA primers on a single-stranded template. The short RNA primers synthesized by POLRMT are used by the mitochondrial DNA polymerase γ to initiate DNA synthesis in vitro. Addition of mtSSB reduces overall levels of primer synthesis, but stimulates primerdependent DNA synthesis. When combined, POLRMT, DNA polymerase γ, DNA helicase TWINKLE, and mtSSB are capable of performing simultaneous leading- and lagging-strand DNA synthesis in vitro [96]. An RNA processing enzyme, Ribonucleoprotein endonuclease (RNase MRP), is also involved in primer maturation. In vitro initiation of leading-strand mtDNA synthesis requires the actions of RNA polymerase and RNase MRP for the generation of replication primers. Mutations of the RNase MRP gene are known to cause a recessively inherited developmental disorder, cartilage-hair hypoplasia [97]. Tfam, acting as a DNA binding stimulatory factor, coordinates the assembly of multiple DNA molecules into nucleoid-like structures and thus, is required for mtDNA maintenance[98]. Another major replication origin is present at position 57 in the D-loop of mtDNA in several human cell lines and immortalized lymphocytes. The nascent chains starting at this origin do not terminate prematurely at the 3' end of the D-loop but proceed well beyond this control point, behaving as "true" replicating strands. This origin is mainly responsible for mtDNA maintenance under steady-state conditions, whereas mtDNA synthesis from other D-loop origins may be more important for recovery after mtDNA depletion and for accelerating mtDNA replication in response to physiological demands [99].
The expression of several proteins involved in mtDNA replication appears to be controlled by NRFs. As described above, the expression of Tfam is under the control of NRF-1, NRF-2 and Sp1. Furthermore, a functional NRF-1 site in the promoter of Rnase MRP gene [100], functional Sp1 and NRF-1 binding sites in genes encoding the mtSSB [101] and the human hSUV3 gene that encodes ATP-dependent RNA and DNA helicase [102] have also been identified (Table II).
Table 2.
Protein Factors Involved Replication, Transcription and Transclation of mtDNA
| Processes | Protein Factors |
NRF-1 & NRF-2 Binding Sites |
Other Factors | Regulated By E2/ERs |
References |
|---|---|---|---|---|---|
| Involved in Replication & Maintenance of mtDNA |
POLG1 | N/A | |||
| POLG2 | N/A | ||||
| RNase MRP | NRF-1; NRF-2 | 100 | |||
| Tfam | NRF-1; NRF-2 | Sp1 | + | 76–80, 89–92 | |
| Mtssb | NRF-1 | Sp1 | 101 | ||
| hSUV3 | NRF-1 | Sp1 | 102 | ||
| Involved in In mtDNA Transcription |
POLRMT | ||||
| Tfam | NRF-1; NRF-2 | Sp1 | + | 76–80, 89–92 | |
| TFB1M | NRF-1; NRF-2 | 104 | |||
| TFB2M | NRF-1; NRF-2 | 104 | |||
| mtERF | |||||
| Involved in mt mRNA Translation |
MTIF2 | NRF-1; NRF-2 | Sp1, ERs | 109, 110 | |
| EF-TU | N/A | N/A | |||
| EF-TS | N/A | N/A | |||
| EF-G | N/A | N/A | |||
| HMRFIL | N/A | N/A | |||
| mtRF1a | N/A | N/A | |||
3. 4. 2. Regulation of Protein Factors Involved in mtDNA Transcription by NRFs
Using the heavy strand as the template under the control of heavy strand promoter (HSP) within the D-loop regulatory region, mtDNA is first transcribed to a large mitochondrial transcript precursor. 12 out of 13 mature mRNAs, two rRNAs and 22 tRNAs are derived from the processed transcript precursor. The other mRNA that encodes for a complex I subunit, ND4L, is transcribed using the light strand of mtDNA as the template under the control of light strand promoter (LSP). Transcription of mtDNA by POLRMT is initiated bidirectionally from closely spaced promoters, HSP and LSP, within the regulatory region (D-loop) and requires assembly of several transcriptional factors, including Tfam, mitochondrial transcription factors B1 (TFB1M) and B2 (TFB2M). Termination factor (mTERF) is involved in termination of mtDNA transcription (see Table II).
As a key transcription factor, Tfam is essential for activation of mtDNA transcription. TFB1M and TFB2M, acting as auxiliary factors for promoter recognition, are necessary for basal transcription of mtDNA. They markedly enhance mtDNA transcription in the presence of Tfam and POLRMT[103–105]. Human TFB1M and TFB2M are expressed ubiquitously and can each support promoter-specific mtDNA transcription in a pure recombinant in vitro system containing POLRMT and Tfam. Both TFB1M and TFB2M interact directly with POLRMT, but TFB2M is at least one order of magnitude more active in promoting transcription than TFB1M. The presence of two proteins that interact with mammalian POLRMT may allow flexible regulation of mtDNA gene expression in response to the complex physiological demands of mammalian metabolism [103].
It has been established [104] that the expression of human TFB1M and TFB2M is governed by NRF-1 and NRF-2. NRF binding sites are present within both TFB1M and TFB2M promoters, which have been shown to be required for maximal trans activation by the PGC-1 family co-activators, PGC-1α and PRC. The evidence includes: a) The physiological induction of these co-activators has been associated with the integration of NRFs and other transcription factors in a program of mitochondrial biogenesis; 2) the TFB1M and TFB2M genes are up-regulated along with Tfam and either PGC-1α or PRC in cellular systems where mitochondrial biogenesis is induced; and 3) Ectopic expression of PGC-1α is sufficient to induce the coordinate expression of all three nucleus-encoded mitochondrial transcription factors along with nuclear and mitochondrial respiratory subunits. Thus, the coordinate regulation of nDNA-encoded mitochondrial transcription factors by NRFs and PGC-1 family coactivators is essential to the control of mitochondrial biogenesis. Because the expression of NRFs and PGC-1α is induced by E2 via ERα and ERβ [76, 79], it is likely that the expression of these mitochondrial transcription factors is regulated by E2 and ERs via NRFs and PCG-1α and, perhaps, other protein factors.
3.4. 3. Regulation of Proteins Factors involved in Mitochondrial Protein Translation by NRFs
Translation of 13 mRNAs into MRC proteins occurs within mitochondria. A number of protein factors are involved in the steps of initiation, elongation, and termination of mitochondrial translation. The properties of some of these factors are summarized in Table II. The nDNA-encoded mitochondrial translation initiation factor 2(MTIF2) functions to initiate the translation. The mitochondrial elongation factor EF-Tu is a GTPase that delivers amino-acylated tRNAs to the ribosome during the elongation step of translation. EF-Tu/GDP is recycled by the guanine nucleotide exchange factor EF-Ts. Mitochobdrial release factor 1α and human mitochondrial release factor 1L (HMRF1L) have been identified as mitochondrial translation release factors involved in the decoding of the termination codons UAA and UAG [106, 107]. HMRF1L is methylated in the GGQ motif in vivo by the human mitochondrial methyltransferase (HMPrmC). The methylation of HMRF1L by HMPrmC is involved in the control of the translation termination process, probably by preventing the undesired suppression of termination codons and/or abortive termination events at sense codons [107].
Bot1p is also required for mitochondrial translation. Bot1p localizes to the mitochondria and cofractionates with purified mitochondrial ribosomes. Reduced levels of Bot1p lead to mitochondrial fragmentation, associated with decreased mitochondrial protein translation, and in cell respiration. Over-expression of Bot1p resulted in cell cycle delay, accompanied by increased cell size and cell length and enhanced cell respiration rate. These results indicate that Bot1p has a novel function in the control of cell respiration by acting on the mitochondrial protein synthesis machinery [108].
The 5' flanking regions of MTIF2 gene have been isolated and mapped to 296 bp upstream from the translation initiation site in human heart tissue. The promoter contains binding sites for NRF-2, Sp1, ERs and enhancer binding sites upstream. NRF-2 binds to the NRF-2 site in the MIF2mt promoter. Reporter assays in HEK293T cells co-transfected with an NRF-2-expressing vector and/or a MIF2 promoter reporter vector have revealed that NRF-2 trans-activated the MIF2 promoter [109, 110]. Importantly, NRF-2 sites are also present in the promoters of several other mitochondrial translation factors, suggesting that NRF-2 may play a key role in the regulation of mitochondrial protein synthesis [110]. The presence of Sp1 and ER binding sites in the promoter of MTIF2 gene suggests that the expression of this gene can be directly activated by E2 via ERs, Sp1 and NRFs.
3.5. Regulation of mtDNA Transcription and Translation by Estrogens/ERs
The presence of ERα and ERβ in mitochondria suggests that these mitochondrial ERs are directly involved in E2-induced mtDNA transcription. The D-loop, the major regulatory region, of mtDNA contains four putative estrogen responsive elements (mtEREs) [111, 112]. Using EMSA, supershift assays and surface plasman resonance (SPR), Chen et al. [52] have demonstrated that ERα, ERβ and mitochondrial proteins that contain ERβ specifically bind to these mtEREs. E2 dose- and time-dependently enhanced the binding of MCF-7 cell mitochondrial proteins to these mtEREs. It is likely that E2 stimulates the interaction of ERα and/or ERβ with Tfam, TFB1M, TFB2M and other factors within mitochondria, and form active transcription complex, thus enhancing mtDNA transcription. Because E2 stimulates the expression of Tfam, and possibly TFB1M and TFB2M via activation of NRF-1 and NRF-2, it is likely that E2 and ERs stimulate the transcription via activation the expression of these mitochondrial transcriptional factors. On the other hand, E2 may activate the expression of several key proteins involved in mitochondrial protein translation and thus, enhance the rate of mitochondrial proteins synthesis. The direct effects of E2 and ERs on the control of the expression of protein factors involved in mitochondrial protein translation and the mechanisms involved deserve further investigation.
4. Regulation of nDNA-Encoded MRC proteins by E2 via NRFs
As mentioned above, the majority of MRC proteins are encoded by nDNA, synthesized on cytosol ribosomes and then imported into mitochondria where they are assembled with 13 mtDNA-encoded proteins into the four MRC complexes and mitochondrial ATP synthase. Previous studies that suggest that estrogens are involved in regulation of nDNA-encoded MRC proteins have been reviewed [46, 113]. A number of recent studies have provided a strong support for a key role of ERα and ERβ in regulation of E2-induced expression of nDNA genes encoding MRC proteins in a variety of cells and tissues.
4.1. Regulation of nDNA-encoded MRC proteins by estrogens/ERs
Chen et al. [48, 114] reported that treatment of female rats with EE and HepG2 cells with E2 enhanced the transcript levels of a nuclear gene encoding H+-ATP synthase subunit ε. Gene expression profiling [115] of prostate from E2 plus testosterone-treated Noble rats revealed the enhanced expression of several nuclear genes encoding MRC complex subunits, including COXVa, COXVIa and mitochondrial H+-ATP synthase subunits b and f. O’Lone et al. [116] performed gene expression profiling in mouse aorta of ERα knockout (ERαKO) and ERβKO mice to identify comprehensive gene sets whose expression was regulated by long-term E2 treatment. They treated ERαKO and ERβKO mice with E2 for 1 week and then performed microarray analysis for identification of ER subtype-dependent gene expression. Their data revealed that ERα- and ERβ-dependent pathways regulated distinct and largely non-overlapping sets of genes, i.e. ERα was essential for most of the E2-mediated increase in gene expression in wild-type aortas whereas ERβ mediated nearly 90% of E2-mediated decreases in gene expression. The E2-regulated genes include those encoding for MRC complexes (see Table 1A, 1B, 1C, 1D, 1E), for proteins involved in ROS pathways and extracellular matrix synthesis. Importantly, the E2/ERβ pathway mediated down-regulation of mRNAs for nuclear genes encoding subunits for all five MRC complexes in mouse aorta. Using a combined proteomic and functional biochemical approach, Nilsen et al. [77] determined the overall impact of E2 on mitochondrial protein expression and functions in neurons of rat brain. They treated ovx adult female rats with a single injection of E2 for 24h, purified neuron mitochondria and then conducted two-dimensional gel electrophoresis and liquid chromatography-tandem mass spectrometry to screen the E2-mediated changes in mitoproteome. The proteomic analyses revealed a twofold or greater change in expression of 66 proteins. Of these, 28 proteins were increased in expression after E2 treatment whereas 38 proteins were decreased in expression relative to control. E2 enhanced several key metabolic enzymes, including the subunits for MRC complex IV and ATP-synthase subunits (Table 1D and 1E) as well as the TCA cycle enzymes. Consistent with the increased gene expression is the increased respiratory control ratio, elevated complex IV activity, in association with simultaneous reduction of free radical generation in brain. Araújo et al. [117] examined E2 effects on MRC gene expression and the MRC activity of rat cortical and mesencephalic astrocytes. They observed that MRC structural and functional properties were regulated dependent on the E2 exposure time and the brain region, but independent of the nuclear ERs. They demonstrate that long-term E2 exposure increases the expression of genes for MRC complex subunits and the mtDNA content, thereby indicating an up-regulation of the amount of mitochondria per cell together with an increase of MRC production. This could represent an important indirect mechanism by which long-term estrogen exposure protects neurons from cell death under neurotoxic conditions. On the other hand, they observed short-term effects of estrogen on the activity of mitochondrial, proton-pumping MRC complexes. In astrocytes from the cortex, MRC activity was decreased, whereas it was increased in astrocytes from the mesencephalon. An increased production of ROS would be the consequence of an increased MRC activity in mesencephalic astrocytes. This could explain the different efficiencies of estrogen-mediated short-term protection in distinct brain regions, but also indicates the limitations for a therapeutic short-term application of estrogen. Razmara et al. [118] observed that treatment of cultured human brain microvascular endothelial cells with E2 for 24h increased mitochondrial cytochrome c protein and mRNA. These effects were mediated via ERα as demonstrated by their being blocked by knocking down ERα expression with ERα siRNA but not by knock down of ERβ expression with ERβ siRNA. Direct measurement of mitochondrial superoxide with MitoSOX Red showed that 17β-estradiol, but not 17α-estradiol, significantly decreased superoxide production, which was blocked by the ER antagonist, ICI182780. Use of selective ER agonists also demonstrated that the decrease in mitochondrial superoxide was mediated by ERα but not ERβ. Stimulation of nDNA-encoded MRC genes by E2 was also seen in human breast MCF-7 cells [79, 119].
4.2. Role of NRFs and Other Transcription Factors in E2-Induced Transcription of nDNA-encoded MRC Proteins
Transcriptional coordination of nDNA-encoded subunits for four MRC complexes and mitochondrial ATP synthase is likely accomplished by transcription factors responding to upstream signals. The transcription activity of the promoters in most of the ubiquitous genes encoded for MRC proteins is regulated by factors binding to the 5' upstream NRF-1, NRF-2, Sp1 and YY1 sites. Among these, NRF- and NRF-2 have been extensively shown to play an important role in coordinate regulation of the expression of MRC complexes and F0–F1 ATP synthase.
4.2.1. Role of NRFs in Control of Complex II protein expression
The promoters of nDNA-encoded genes (SDHA, SDHB, SDHC and SDHD) of complex II have been cloned and analyzed. NRF-1 and NRF-2 binding sites have been identified in the promoter regions of all four genes (Table 1B)[18, 19, 120–122]. Reporter gene analysis, site-directed mutagenesis and EMSA have revealed that both NRF-1 and NRF-2 were required for SDHB gene expression [120]. It ha been also demonstrated that NRF-1 bound to the promoters of SDHA and SDHD genes and that the activity of complex II was dynamically regulated through the catalytic SDHA flavoprotein [122].
4.2.2. Role of NRFs in the Control of nDNA-encoded genes for Complex IV and Its assembly
As mentioned above, complex IV consists of proteins that are bigenomically encoded. Of its thirteen subunits, three are encoded in the mtDNA and ten are encoded in the nDNA. Transcriptional coordination of ten nuclear-encoded COX subunit genes is likely accomplished by transcription factors responding to upstream signals.
The functional nDNA-encoded genes for the COX subunits, including IV, Vb, VIa 'L' & 'H', VIIa 'L' & 'H', VIIc and VIII (H), from different mammalian species and their 5' flanking putative promoter regions have been sequenced and extensively characterized [123–127]. The promoters of several genes for COX assembly (e.g. COX17P and SURF1) have also been characterized [128, 129]. While the ubiquitous COX genes and assembly genes show extensive heterogeneity in their promoter regions, one common feature is that the majority of these genes contain multiple transcription initiation sites for both general as well as the specialized transcription factors NRF-1, NRF-2, Sp1 and YY1 (Table 1D). A number of studies [123–134] have demonstrated that NRF-1, NRF-2, Sp1 and YY1 bind to the corresponding sites in gene promoters and activate the transcription of these genes.
Since the nuclear genes encoding 10 subunits for COX are localized in different chromosomes, several studies [130, 135, 136] were performed to address whether the 10 nuclear genes are transcribed simultaneously and whether a single transcription factor binds to all ten of COX subunit promoters. Dhars et al. [136] performed in silico analysis of murine nDNA-encoded COX subunit promoters and identified NRF-1 binding sites in all ten promoters, that are highly conserved among mice, rats, and humans. Using EMSA, supershift assays and site-directed mutagenesis, they demonstrated that NRF-1 binds to all ten promoters. Moreover, using in vivo ChIP assays, they also showed in murine neuroblastoma cells that NRF-1 bound to all ten promoters. Furthermore, they observed that silencing NRF-1 expression with siRNA reduced all ten COX subunit mRNAs and mRNAs of other genes involved in mitochondrial biogenesis. These results indicate that NRF-1 plays a significant role in coordinating the transcriptional regulation of all ten nuclear-encoded COX subunits in neurons.
Putative NRF-2 binding sites were also identified on all ten nDNA-encoded COX gene promoters in the rat genome. A study by Ongwijitwat et al. [135] provided several lines of evidence demonstrating that NRF-2 is also an important mediator for coordinated regulation of all ten nuclear-encoded COX subunit genes: 1) NRF-2 binds in vivo to six of the ten nDNA-encoded COX subunit promoters as revealed by in vivo ChIP assay; 2) NRF-2 also binds to the other four subunits as shown by EMSA, super shift assays, and promoter mutation study; 3) COX expression is significantly reduced by transfection of dominant-negative constructs of NRF-2 proteins; and 4) more importantly, NRF-2 has been shown to sense changing cellular energy demands in rat neurons and silence of its expression down-regulates cytochrome oxidase and other target gene mRNAs [130].
It is interesting to note that the promoters of the heart- and muscle-specific COXVIα (H) and COXVIIα (H) do not contain NRF-1 and NRF-2 sites but, instead, have conserved myocyte enhancer factor 2 (MEF2) elements. It has been shown [137, 138] that the expression and functional activity of MEF2 is under the control of NRF-1. First, over-expression of NRF-1 induced MEF2A mRNA levels. The MEF2A 5'-regulatory region contains an evolutionarily conserved canonical element that binds endogenous NRF1 in ChIP assays; Second, NRF-1 regulated MEF2A promoter-reporters as demonstrated by the stimulatory effects of its over-expression, the suppressive effects of its under-expression with siRNA, and by promoter mutation studies; and third, ChIP assays using isoform-specific antibody to four mammalian MEF2 isotypes confirmed specific MEF2A binding to the COX6A(H) promoter. These findings support a role for MEF2A as an intermediary in coordinating respiratory chain subunit expression in heart and muscle through a NRF1 --> MEF2A --> COX(H) transcriptional cascade. MEF2A also binds the MEF2A and PPARGC1A promoters in ChIP assays, placing it within a feedback loop with PGC-1α in controlling NRF1 activity. Interruption of this cascade and loop may account for striated muscle mitochondrial defects in mef2a null mice [137, 138].
The PGC-1 family of regulated coactivators (PGC-1α, PGC-1β and PRC) plays an important role in directing respiratory gene expression in response to environmental signals. PRC and PGC-1α differ in their interactions with nuclear hormone receptors but are highly similar in their direct binding to several nuclear transcription factors implicated in the expression of the MRC proteins. Vercauteren et al. [139] observed that the NRF-2 subunits and PRC were co-immunoprecipitated from cell extracts, indicating that the two proteins exist in a complex in vivo. The association between PRC and NRF-2 is mediated by HCF-1 (host cell factor 1), a major chromatin component. Both PRC and NRF-2 bind to HCF-1 in vitro and invo and their association was required for PRC trans-activation through promoter-bound NRF-2.
NRF-1 and NRF-2 are known to activate several mitochondrial transcription factors including Tfam, TFB1M and TFB2M (see above). NRF-2, PRC and HCF associate with NRF-2-dependent nuclear genes that direct the expression of TFB1M and TFB2M. Short hairpin RNA-mediated knock down of PRC protein levels led to reduced expression of TFB2M mRNA and mitochondrial transcripts for COXII and cytb and these changes in gene expression coincided with a marked reduction in COX activity [139].
NRF-1 and NRF-2 in cooperation with co-activators such as PGC-1α and related factors HCF-1 are directly involved in regulation of 10 nDNA-encoded subunits and indirectly regulate, via activating mitochondrial transcription factors, e.g. Tfam, TFB1M and TFB2M, and the expression of the three mtDNA-encoded COX subunits. Thus, NRF-1 and NRF-2 prove to be the two key bigenomic coordinators for transcriptional regulation of all COX subunits in neurons. Because the transcription of NRF-1, NFR-2, PGC-1α are up-regulated by E2 via ERs [67, 78, 79], the effects of E2/ER on the transcript levels of both nDNA- and mtDNA-encoded subunits of COX are likely mediated through these factors.
4.2.3. Role of NRFs in Control of nDNA-Encoded Genes for Complexes I, III and Mitochondrial ATP Synthese and Their assembly
While much less is known about the role of NRFs, Sp1 and other transcription factors in the control of the expression of the nDNA-encode genes for complexes I, III and V and their assembly proteins, NRF-1, NRF-2, Sp1 and YY-1 binding sites have been identified in the promoters of a few genes for Complex I (e.g. NDUFV1 and NDUFS8)(Table 1A)[140–143], and mitochondrial ATP synthase (e.g. ATPγ)(Table 1E)[144]. There are no reports, to date, for the presence of binding sites for NRF-1, NRF-2, Sp1 and YY-1 in the promoters of nDNA-encoded complex III genes. Several studies have demonstrated that the NRF-1 and/or NRF-2 are capable of binding to these sites in the promoters of NDUFV1, NDUFS8 and ATPγ genes and activate their transcription [141, 143, 144]. It has been demonstrated [140, 142] that Sp1 and YY-1also play an important role in the transcriptional activation of NDUFV1 and NDUFS8 genes.
5. Regulation of Mitochondrial import Machinery by NRFs
The majority of nuclear encoded, mitochondrially localized proteins, including the nDNA-encoded MRC proteins and the protein factors that are involved in mtDNA replication, transcription and translation and MRC assembly proteins (Table I and Table II), are synthesized on cytosol ribosomes with N-terminal signal sequences termed pre-sequences or internal targeting signals, and then imported into mitochondria. Precise targeting of mitochondrial precursor proteins to mitochondria requires Translocase of the Outer mitochondrial Membrane (designated Tom), a multi-subunit complex containing specific import receptors. Three important Toms, namely Tom20, Tom22, and Tom70, are anchored in the outer membrane by a single transmembrane α-helix, located at the N-terminus in the case of Tom20 and Tom70 (signal-anchored) or in the C-terminal portion in the case of Tom22 (tail-anchored), where they serve as receptors for specific recognition and membrane translocation of nuclear-encoded preproteins [145]. Each Tom binds to individual mitochondrial preproteins with different specificity. Tom20 recognizes preferentially mitochondrial pre-sequences, and preproteins with internal targeting signals. Generally, preproteins with presequences are initially recognized by Tom20 and, subsequently, by Tom22. It has been shown that the cytosolic domains of Tom20 and Tom22 also function to maintain their substrate preproteins unfolded and prevent them from aggregation on the mitochondrial surface. The cytosolic domain of Tom22 appears to function as a receptor in cooperation with Tom 20. It has been shown that Tom20 and Tom22 are apparently involved in the same step or sequential steps along the same pathway of targeting signal recognition in import [145]. The hydrophobic preproteins with internal targeting signals are first recognized by Tom70, which then associates with molecular chaperones, thereby maintaining their substrate preproteins in an import-competent state [146–148]. In addition to these Toms, the human Tom34 gene encodes a cytosolic protein with chaperone-like activity that helps import some preproteins to the mitochondria by keeping them in an unfolded, import-compatible state.
All of the Tom proteins are encoded by nuclear genes, synthesized as precursors on the cytosol ribosomes and then translocated to the outer mitochondrial membrane. The channel-forming β-barrel protein Tom40 is targeted to mitochondria via Tom receptors and inserted into the outer membrane by the sorting and assembly machinery (SAM complex). A further outer membrane protein, Mim1, plays a less defined role in assembly of Tom40 into the Tom complex. These Toms are anchored in the outer membrane: insertion of the precursor of Tom22 into the outer membrane requires pre-existing Tom receptors. Mim1 is required for efficient membrane insertion and assembly of Tom20 and Tom70, but not Tom22. Mim1 associates with SAM (core) components to a large SAM complex, explaining its role in late steps of the assembly pathway of Tom40. Thus, Mim1 is not only required for biogenesis of the beta-barrel protein Tom40 but is also required for membrane insertion and assembly of signal-anchored Tom receptors. Thus, Mim1 plays an important role in the efficient assembly of the mitochondrial Tom complexes.
Because Tom complexes act as the central entry gate for nDNA-encoded mitochondrial precursor proteins, the biogenesis of the mitochondrial Tom complex is subjected to strict control. NRF-1 and NRF-2 are involved in the regulation of the expression of the mitochondrial Tom20, Tom22, Tom70 and Tom34.
NRF-2 has been shown to be critical for maintaining normal transcriptional levels of the Tom20 gene. Blesa et al. [149] identified a NRF-2 and two NRF-1 binding motifs in the promoter of human Tom20 gene. Using Chip assays and reporter assays in HeLa S3 and A204 cells, they demonstrated that only NRF-2 and the proximal NRF-1 motifs were involved in the expression of the gene. The NRF-2 binding site was required to activate transcription. The proximal NRF-1 site cooperated with NRF-2 in regulating the expression of the gene whereas the distal NRF-1 binding site was not functional.
Blesa et al.[150] identified two NRF-2 binding motifs in the 5'-flanking region of the human Tom70 gene. This region contained thirteen potential CpG methylation sites, three of which occur in the sequence 5'-CCGG-3'. Interestingly, each NRF-2 site contained one CCGG sequence. Reporter assays in HeLa cells, Chip assays, site-directed mutagenesis, and EMSA assays demonstrated that NRF-2 bound in vivo to the Tom70 promoter and that interactions between these sites and a CpG island contributed to expression. Furthermore, the two NRF-2 binding sites of the promoter have different functional contributions in promoting TOM70 expression, i.e. they worked in an additive manner as single sites [151]. Site-specific methylation of the NRF-2 binding sites in the Tom70 promoter reduced the expression of a luciferase reporter in HeLa S3 cells. Abrogation of NRF-2 binding at the methylated sites was confirmed by EMSA assays. These results have established essential role of the NRF-2 binding sites for promoter activity of Tom70[152]. However, studies of methylation on DNAs from different sources found no methylation in the promoter regions of Tom70 and other Tom/TIM family genes. Thus, although in vitro methylation inactivates the expression of Tom70, these results suggest that this is not the mechanism modulating its expression in vivo. Since a number of nuclear genes encoding mitochondrial translocases have NRF-2 binding sequences containing CpG methylation sites, it is possible that methylation/demethylation of the NRF-2 binding sites might be a regulatory mechanism of mitochondrial biogenesis [152].
Blesa et al. [153] reported the existence of binding sites for NRF-1, NRF-2 and Sp1 in the 5' region of the human Tom34 gene. Site-directed mutations at these sites on promoter activity in HeLa S3 and A204 cells, in conjunction with Chip assays, EMSA assays, and in vivo methylation analysis of the promoter region have demonstrated that NRF-1 is the main transcription factor regulating the expression of Tom34. Sp1 interacts with NRF-1 to stimulate the promoter's full activity. Tom34 is over-expressed frequently in colorectal tumors, suggesting that it has a role in the growth of cancer cells. In this context, Tom34 is a potential target for novel anticancer drugs, and it might also be used in the diagnosis of colorectal cancer.
In summary, NRF-1 and NRF-2 are involved in the regulation of the expression of nuclear genes encoding: 1) MRC complex subunits, ATP synthase and their assembly proteins; 2) proteins required for replication, transcription and translation of mtDNA; and 3) Proteins involved in mitochondrial protein machinery. Therefore, NRFs coordinate the expression of nuclear and mitochondrial genes involved in mitochondrial biogenesis and respiration. The nucleo-mitochondrial interactions depend on the interplay between NRF-1, NRF-2 and other transcriptional factors including Sp1, MEF2, and other members of the PGC-1 family of regulated coactivators (e.g. PGC-1α and PRC). The transcription factors target genes that specify the MRC, the mitochondrial transcription, translation and replication machinery, and protein import and assembly apparatus among others. These transcriptional paradigms provide a basic framework for understanding the integration of mitochondrial biogenesis and function with signaling events that dictate cell- and tissue-specific energetic properties. Because the expression of NRF-1 and NRF-2 is activated by E2 via ERs, and Sp1 acts as a transcription factors in regulating the expression of a number of estrogen responsive genes, ERs, Sp1 and other transcription factor are likely involved in E2-mediated coordinate regulation of 1) MRC complex subunits and their assembly proteins; 2) proteins required for replication, transcription and translation of mtDNA; and 3) proteins involved in mitochondrial protein import machinery and 4) mtDNA-encoded MRC proteins.
6. MRC as an important regulator of apoptosis and cell proliferation
In addition to playing important roles in intracellular energy generation and redox-dependent intracellular signaling, mitochondria are the focal points of a variety of key events in apoptosis, including the release of caspase activators (e.g. cytochrome c), changes in electron transport, loss of mitochondrial transmembrane potential, altered cellular oxidation-reduction, and regulation of pro- and antiapoptotic Bcl-2 family proteins. The different signals that converge on mitochondria to trigger or inhibit these events and their downstream effects delineate several major pathways in physiological cell death [154]. As an important structural and functional part of mitochondria, the MRC plays a key role in regulation of cell proliferation and apoptosis, and thus may be involved in tumor progression, as supported by the several lines of genetic and pharmacological evidence.
6.1.1. Genetic Evidence
As described in section 3.2.2, Tfam is essential for replication, transcription and maintenance of mtDNA. It has been observed [83] that tissue-specific knockout of Tfam in mouse heart caused dramatic reduction in mtDNA and severe MRC deficiency. These effects are associated with increased in vivo apoptotic cardiocytes. Over-expression of Tfam in transgenic mice ameliorated the decreased mtDNA copy number and MRC enzyme activities in post-myocardial infarction hearts and significantly increased survival rate and decreased apoptosis [82]. Kwong et al. [155] has clearly demonstrated a correlation between MRC composition and apoptotic responses. These researchers investigated the role of MRC dysfunction in the control of apoptosis, using mtDNA mutations as genetic models. Six genetic models were used: 1) wild type cell with full complements of the MRC complexes I, II, III, IV and V; 2) ρo cells that are missing complexes I, III, IV, V but retain complex II; 3) COX cells that are missing complex IV and have reduced complex I activities; 4) CYTb cells that are missing complex III; 5) MERRF cells that are missing complexes I, III, IV, but have some residual ATP hydrolytic complex V activity; and 6) NAPP cells that have all the MRC complexes in place but have reduced amounts and activities. The responses of these cells to both mitochondrial (MT)- and endoplasmic reticulum (ER) stress-induced apoptosis are summarized in Figure 3A. There are significant differences among these cell models in responses to apoptotic stimuli: cells lacking MRC are protected against both MT- and ER stress–induced apoptosis. Cells with MRC, but unable to generate electron flux, are protected against MT-apoptosis, although they have increased sensitivity to ER stress and cells with a partial reduction in electron flux have increased apoptosis under both conditions. These results have shown that the MRC modulates apoptosis in a context-dependent manner independent of ATP production and that apoptotic responses are the result of the interplay between mitochondrial functional state and environmental cues.
Fig. 3. Genetic and pharmacological evidence for MRC as Modulator of Apoptosis 3A. Correlation between MRC Composition and Apoptotic Response in mtDNA Mutant Cybrids.
The RC components of the cybrids are depicted. WT cells have a full complement of RC complexes. ρ0 cells are missing complexes I, III, IV, and V. COX cells are missing complex IV and have reduced complex I (depicted as a smaller circle). CYTB cells are missing complex III and have reduced complex I. MERRF cells are missing complexes I, III, IV, and V, detectable by BN gel, but have some residual ATP hydrolytic complex V activity. NARP cells have all of the complexes in place, but have reduced amounts and activities. Normal response to apoptosis induction is represented by +, high response by +++, and low response by −. (Adapted from Kwong JQ et al. (153) with permission granted by J. Cell. Biology).
3B. Pharmacological Evidence for MRC as Modulator of Apoptosis: effects of specific inhibitors on individual complexes
Induction of apoptosis by inhibitors specific for individual MRC complexes is shown. Inhibition of individual complex is represented by X; Normal response to apoptosis induction is represented by +.
Several studies have also demonstrated that over-expression of individual MRC proteins led to suppression of apoptosis. Bcl-2-associated X protein (Bax) and its related protein, Bak, are pro-apoptotic proteins involved in cell death. Both proteins are essential for maximum apoptotic response induced by several apoptotic inducers including tumor necrosis factor-α (TNF-α)[156], curcumin (a polyphenolic compound and cancer chemopreventive agent) [157] and sulforaphane (a cruciferous vegetable-derived cancer chemopreventive agent)[158]. Human MRC complex IV subunit VIa (COX6A1) was identified as a novel suppressor of Bax-mediated cell death in yeast [159]. Over-expression of COX6A1 significantly suppressed Bax- and N-(4-hydroxyphenyl) retinamide (4-HPR)-induced apoptosis in yeast and human glioblastoma-derived U373MG cells. The ROS generation in response to Bax or 4-HPR was inhibited in yeast and U373MG cells expressing COX6A1, indicating that COX6A1 exerts a protective effect against ROS-induced cell damage. 4-HPR-induced mitochondrial translocation of Bax, mitochondrial cytochrome c release, and activation of caspase-3 were markedly attenuated in U373MG cells that stably expressed COX6A1. These results demonstrate that over-expression of MRC proteins, e.g. COX6A1, has a potential role in protecting mammalian cells from 4-HPR-induced apoptosis.
6.1.2. Pharmacological Evidence
A large number of studies with MRC complex-specific inhibitors and chemotherapy reagents that specifically target individual MRC complexes have provided strong evidence for the MRC as an important regulator of apoptosis (Figure 3B). For example, it has been shown that inhibition of Complex I with rotenone[160–163] and chemotherapeutic reagents e.g. arsenic trioxide[162, 164], and TNF-α [165], and ceramide[166] causes increased MRC-derived ROS generation, leading to apoptosis characterized by induction of mitochondrial membrane permeability transition (MPT), cytochrome c release, activation of various caspases and DNA fragmentation. Inhibition of Complex II with specific inhibitors, e.g. 3-nitroproprionic acid [167], and alpha-tocopheryl succinate (a vitamin E analogue)[168–172], also causes apoptosis. Benzyl isothiocyanate (BITC) is a dietary cancer chemopreventive agent that causes apoptosis in breast cancer cells[173]. The BITC-induced apoptosis was initiated by MRC-derived ROS via regulation by Bax and Bak in breast cancer cells [173, 174]. Xiao et al. [175] demonstrated that the BITC-induced apoptosis in these cells was initiated by ROS due to its inhibition of Complex III. Inhibition of complex III with Antimycin A induced apoptotic effects in lung cancer cells [176]. Complex III is a target of C2-ceramide [177], which induced apoptosis in lung cancer cells[178].
Lyssaviruses are highly neurotropic viruses. The matrix proteins of some lyssaviruses strongly induced neuronal apoptosis. Gholamic et al. [179] reported that for Mokola virus (MOK), the M(M-MOK), targeted mitochondria, disrupted the mitochondrial morphology, and induced apoptosis by its physical association with the COXI of complex IV. N-retinyl-N-retinylidene ethanolamine (A2E) is a compound suspected to cause age-related macular degeneration. A2E has been shown to interfere with the binding of cytochrome c to COX, detaching proapoptotic proteins from mitochondria, and inducing apoptosis in mammalian retinal pigment epithelial cells[180, 181].
Most human carcinomas contain reduced levels of the catalytic subunit of the mitochondrial ATP synthase. In colon and lung cancers, this alteration correlates with a poor patient prognosis. Down-regulation of the H+-ATP synthase has been shown to link to the resistance of the cells to chemotherapy[182]. It has been shown that inhibitors of the ATP synthase delayed staurosporine-induced apoptosis in liver cells that are dependent on oxidative phosphorylation for energy provision [182]. Roy et al. [183] observed that 3,3'-Diindolylmethane (DIM) inhibited mitochondrial ATP synthase of Leishmania donovani and induced apoptosis in protozoan parasites. DIM-induced inhibition of ATP synthase activity caused depletion of mitochondrial ATP levels and significant stimulation of mitochondrial ROS production, followed by depolarization of mitochondrial membrane potential (Deltapsim). Since Deltapsim is the driving force for mitochondrial ATP synthesis, loss of Deltapsim results in depletion of cellular ATP level. The loss of Deltapsim caused the cellular ROS generation and in turn led to the oxidative DNA lesions followed by DNA fragmentation. On the other hand, loss of Deltapsim led to release of cytochrome c into the cytosol and subsequent activation of the caspase-like proteases, which led to oligonucleosomal DNA cleavage. They have also shown that mtDNA depleted cells were insensitive to DIM to induce apoptosis. Therefore, mitochondria are necessary for cytotoxicity of DIM. These results indicate that DIM-induced mitochondrial dysfunction by inhibition of ATP synthase activity leads to apoptosis in Leishmania parasites.
The genetic and pharmacological studies described above strongly support the notion that up-and down-regulation of the MRC biogenesis and functions is closely related to the control of apoptosis: up-regulation of the MRC biogenesis/function inhibits apoptosis and stimulate cell growth and proliferation whereas their down-regulation and/or inhibition induces apoptosis and inhibition cell growth and proliferation. Physiological, pathological and environmental factors that cause dramatic and persistent changes in the MRC biogenesis and functions may alter the balance of the cell growth and cell death.
6.2. Mitochondrial Permeability Transition Pore as Mediator of Apoptosis and Its Regulation by E2/ERs
Cellular stresses, such as ischemia, hypoxia, oxidative stress, and cytotoxic drugs induce a cascade of events at the level of the mitochondrion that, if left unchecked, will kill the cell. Initially this involves excessive production of ROS, and Ca2+ overload of the mitochondrial matrix [184–187]. This in turn causes permeabilization of the inner mitochondrial membrane. This phenomenon, termed the mitochondrial permeability transition (MPT), dissipates the proton electrochemical gradient (ΔΨm) that drives many mitochondrial functions, leading to ATP depletion, further ROS production, and ultimately swelling and rupture of the organelle [184–187]. This in turn releases pro-apoptotic intermembrane space proteins, most notably cytochrome c, Smac/DIABLO, htrA2/Omi protease, and endonuclease-G [184, 188]. The coordinated action of these proteins subsequently results in the death of the cardiomyocyte by apoptosis. However, if the stress is severe and/or prolonged, ATP will be depleted and the cell will instead undergo necrosis. Indeed, the relative contribution of MPT to apoptosis versus necrosis is still the subject of debate[189].
The mitochondrial permeability transition pore (MPTP), a non-specific channel thought to span both mitochondrial membranes, mediates these lethal increases in inner mitochondrial membrane permeability [184–187]. The pore itself is permeable to solutes up to 1.5kDa. This causes equilibration of H+ across the inner membrane, which dissipates ΔΨm and inhibits ATP production. A concomitant influx of water causes swelling of the mitochondria, which stretches the membranes to the point where the outer membrane fails. The mitochondrial pore is redox, Ca2+, voltage, adenine nucleotide, Pi, and pH sensitive [184–187]. Most importantly, increases in matrix Ca2+ and ROS induce pore opening, whereas adenine nucleotides inhibit the pore; and many diseases are associated with increases in mitochondrial pore activators (Ca2+, ROS) and reductions in mitochondrial pore inhibitors (ATP/ADP). Indeed, studies have shown that inhibition of the MPTP blunts ischemia-reperfusion injury in multiple organs [190–193], and can also protect against the cell loss that underlies heart failure [194, 195], muscular dystrophy [196, 197], cancer drug-induced toxicity [198, 199], and several neurodegenerative diseases [199–201].
Unfortunately, the precise molecular architecture of the MPTP remains unknown. Based upon biochemical and pharmacological studies, the pore was proposed to consist of the voltage-dependent anion channel (VDAC) in the outer membrane, the adenine nucleotide translocase (ANT) in the inner membrane, plus CypD in the matrix (1–4). VDAC, ANT, and CypD interact at membrane contact sites and reconstitution of this complex in vesicles yields a Ca2+-sensitive channel reminiscent of the mitochondrial pore [202]. However, recent genetic studies have seriously questioned the validity of this paradigm. While several studies have shown that mice lacking CypD are resistant to Ca2+- and ROS-induced MPT and cell death in the heart and other tissues [190, 203, 204], mice lacking ANT still exhibit a classical permeability transition phenomenon and respond normally to death-inducing stimuli[205]. Baines et al. have also shown that deletion of all 3 VDAC isoforms does not inactivate the mitochondrial pore nor prevent cell death [206]. Therefore, CypD is the only defined molecular component of the MPTP, where it functions in an enzymatic capacity to induce pore formation and cell death by binding and regulating unknown proteins. Consequently, there is now a concerted effect to identify novel components of the mitochondrial pore by a variety of proteomic and genetic screening methodologies.
The ability of estrogens and mitochondrial ERs to modulate MPTP has not been extensively studied. However, evidence that estrogen and its receptors can directly (i.e., non-genomically) influence MPT is beginning to accumulate. Using the well-established technique of mitochondrial swelling, Morkuniene et al demonstrated that direct application of physiological concentrations of either E2 or 4-hydroxytamoxifen to rat cardiac mitochondria could suppress Ca2+-induced permeability transition [207]. Similarly, Custudio and colleagues reported that tamoxifen could prevent Ca2+-induced mitochondrial swelling, ΔΨm collapse and matrix Ca2+− release, all indices of MPT, in isolated liver mitochondria [208, 209]. Comparable effects have also been observed in brain mitochondria[210].
These exciting findings have been extended to intact cells. E2 has been shown to prevent MPT in CHO cells (Miyaguchi) [211], neurons [210, 212], cervical epithelia [213], and lens epithelia [59]. Antithetically, withdrawal of E2 induces MPT in human breast cancer cells [214]. The one caveat with these studies is that they primarily used changes in ΔΨm as their index of MPT. While opening of the mitochondrial pore will indeed dissipate ΔΨm, alterations in ΔΨm can still occur independently of pore opening, and therefore are not the most reliable way of observing MPT in intact cells. Instead, the calcein/CoCl2 method developed by Bernardi’s group is a more reliable measurement of permeability transition in intact cells[215], and studies examining the anti-permeability transition effects of E2 using this technique are needed.
The most obvious question is how does E2 and its analogues modulate the mitochondrial pore. Although in intact cells this is most likely a multi-factorial process involving both genomic and non-genomic pathways, the fact that E2 and tamoxifen can have direct effects in isolated mitochondria suggests that specific intra-mitochondrial mechanisms must exist. Lobaton et al have demonstrated an ER-dependent inhibition of the mitochondrial Ca2+ uniporter [216]. Reduced Ca2+ influx would certainly account for an inhibitory effect of estrogens on permeability transition. As has already been extensively discussed in this review, mitochondrial ER-dependent changes in mtDNA transcription and MRC complex formation has been shown to reduce mitochondrial oxidant production. Alternatively, activation of ER has also been reported to increase MnSOD activity in mitochondria, independent of changes in protein[217]. Either way a reduction in ROS would decrease the probability of mitochondrial pore opening. Off-ER estrogen targets may also exist. For example, tamoxifen may be able to directly affect the ANT[218], inhibition of which can block mitochondrial permeability transition [219]. Of course the easiest way for E2 to inhibit the pore would be for it to bind, either directly or through the ER complex, to components of the mitochondrial pore. However, until the true components of the pore are identified, the existence of such a mechanism remains untestable. In regard to a role of ERs in regulation of MPTP, Yang et al. [220] recently demonstrated that ERβ was mainly localized within mitochondria of immortal hippocampal cells and primary hippocampal neurons and that knocking down ERβ expression in these cells resulted in a lower resting mitochondrial membrane potential ΔΨm and increase in resistance to hydrogen peroxide-induced ΔΨm depolarization, in association with changes in ATP concentrations and reduced levels of mitochondrial superoxide in these cells. Similar mitochondrial phenotype changes were observed in primary hippocampal neurons derived from ERβKO mice. These data support the notion that mitochondrial ERβ functions as a mitochondrial vulnerability factor involved in ΔΨm maintenance, potentially through a mitochondrial transcription dependent mechanism.
In addition to playing a regulatory role in MPTP, ANT is involved in the mediation of ATP/ADP shuttle cross the mitochondrial membrane, thereby transporting the majority of MRC-generated ATP out of mitochondria in exchange of ADP and Pi (Figure 1). There are several tissue-specific isoforms of ANT, namely ANT-1, ANT-2, ANT-3 and ANT-4. It has been reported that the expression of the heart-specific ANT isoform (ANT-1) is enhanced by E2 [221]. Interestingly, the anti-apoptotic protein, Bcl-2, is an important component of mitochondrial ANT system and is involved in the regulation of ANT shuttle. Bcl-2 was shown to enhance ANT-dependent ADP/ATP exchange activity by direct protein-protein interaction with ANT [222, 223]. Its expression is enhanced by E2 in a number of cells [72, 224–229]. It has been suggested that Bcl-2 family members may regulate important mitochondrial and cellular functions and serve as sentinels to detect abnormalities in these pathways and, when the abnormalities are severe enough, to initiate or facilitate cell death. Understanding the physiological processes controlled by the Bcl-2 and Bcl-2 family will be important in understanding cell regulation, and may provide new insights into the regulation of apoptosis [230].
6.3. E2/ER-mediated Mitochondrial Effects in Regulation of Cell Proliferation and Anti-Apoptosis
6.3.1. Implications of E2/ER-mediated MRC biogenesis in Cell Proliferation
E2 stimulates cell proliferation in a number of cells including breast epithelial cells. MRC plays an important role in the control of cell proliferation. It is likely that the E2/ER-mediated the MRC biogenesis has important implications in regulation of cell proliferation via generating the large majority of cellular energy in the form of ATP and ROS. The contribution of the MRC to cell proliferation is most evidenced based on the several studies on the effects of down-and up-regulation of MRC protein expression/functions on cell cycle progression and cell growth.
Down-regulation of the MRC protein expression/function is related to cell cycle arrest and reduced cell growth. Mandal et al. [231] observed that a mutation in COXVa gene caused a drop in intracellular ATP to levels sufficient to maintain cell survival, growth, and differentiation, but not to enable progression through the cell cycle. Analysis of this gene revealed that a specific pathway involving AMPK and p53 was activated that eliminated Cyclin E, resulting in cell cycle arrest. This study demonstrates that mitochondria have a direct and specific role in enforcing a G1-S cell cycle checkpoint during periods of energy deprivation. Schauen et al.[232] demonstrated that loss of a functional MRC by depletion of mtDNA inhibited cell-cycle progression through decreasing ROS levels, leading to down-regulation of p21CIP1/WAF1. Sustained inhibition of MRC oxidative phosphorylation (OXPOP) with antimycin A or a partial uncoupling of OXPOP with uncoupler carbonylcyanide trifluoromethoxyphenylhydrazone (FCCP) impaired cell proliferation and induced premature senescence in human fibroblasts [233, 234]. Treatment of human lung cancer cells with either antimycin A or 2,4-dinitrophenol, another uncoupler of OXPOP, prevented the cell growth via inducing apoptosis[176, 235].
Over expression of MRC proteins, on the other hand, has been found to promote cell proliferation. For example, in a poorly differentiated, rapidly growing Morris hepatoma 3924A, the mRNA and protein levels of Tfam are elevated more than 10 fold. The mRNA levels for COX I, II and ND5 and ND6, the downstream targets of Tfam, were increased 10, 8, 5, and 3 fold, respectively. The mRNA levels for NRF-1, NRF-2, were 5 and 3 fold higher, respectively, in the hepatoma relative to the host liver [90]. Up-regulation of the MRC proteins and cellular proliferating regulators were seen in thyroid oncocytomas compared with normal thyroid tissue[236]. In proliferating cells, transcription and replication of mtDNA is highly enhanced by parkin, a RING protein located within the mitochondria, via its association with Tfam [237]. Increased mitochondrial ATP synthase was seen to be involved in liver metastasis of colorectal cancer [238].
The contribution of enhanced MRC biogenesis and functions to cell proliferation can be attributed, in part, to its generation and supply of energy in form of ATP and ROS for a number of cellular processes. The large number of cellular activities, including cell cycle progression, synthesis of proteins, DNA, RNA, carbohydrates, lipids and other molecules, active transport across membrane by various ion channels/transporters, kinase-dependent signaling pathways, are all driven by ATP. ROS at low to moderate levels serve as second messenger for redox regulation of proteins involved in growth-related processes. By up-regulating the MRC biogenesis and functions, E2 and ERs contribute, at least in part, to the stimulation of cell growth and proliferation.
6.3.2. Implications of E2/ER-mediated MRC Biogenesis in Inhibition of Apoptosis
Apoptosis is a fundamental cellular activity to protect the cells against neoplastic development by eliminating genetically damaged cells or those cells that have been improperly induced to divide by mitotic stimuli. Like a number of tumor promoters [239], E2 normally inhibits spontaneous and induced-apoptosis in a number of cell types including human breast cancer cells[225, 240, 241], brain neurons[242, 243], heart endothelial cells [244, 245], skeletal muscle cells[246], human lens epithelial cells [247], rat hepatocytes[248] and other cells [249, 250]. The mechanisms underlying E2-mediated inhibition of apoptosis are not clearly understood. Several mechanisms/pathways may be involved in these effects, depending on the cell types. A number of signaling pathways, including mitogen-activated protein kinase and phosphoinositide 3-kinase signaling that are mediated by the membrane-associated ERα, are implicated in the anti-apoptotic effects of E2 in human breast cells, neurons and cardiomyocytes [240, 241, 251, 252]. The anti-apoptotic effects of E2 in other cells may be mediated by other events, including induction of the expression of baculoviral inhibitors of apoptosis repeat-containing 1 (Birc1) in uterine cells [249]. Apoptosis is triggered by increasing levels of the MRC-derived ROS induced by a number of anti-cancer drugs (see section 7.2). Inhibition of ROS-induced apoptosis can be achieved by increasing mitochondrial antioxidants, which detoxify ROS. It has been shown that E2 activated the mitochondrial ERα to increased mitochondrial Mnsuperoxide dismutase (MnSOD) activity, which is involved in inhibition of UV-induced apoptosis in MCF-7 cells [54, 253]. It has also been reported that MnSOD activity was activated by E2 in mitochondria of cerebral vessels of rat brain [67] and human lens epithelial cells [217] (for more detail see section 7.5). In addition, E2 treatment of rat hepatocytes caused increased mitochondrial distribution of glutathione (GSH), another major mitochondrial anti-oxidant [72]. The increased mitochondrial MnSOD activity and GSH levels within mitochondria can detoxify ROS. Furthermore, it has been known that estrogens up-regulate the expression of the anti-apoptotic proteins, e.g. Bcl-2 [72, 225, 254, 255]. The combination of these effects may partially account for E2’s anti-apoptotic effects.
In addition, E2/ER-mediated MRC biogenesis/functions may contribute to E2-mediated inhibition of apoptosis. It has shown that the inhibition of TFGβ-induced apoptosis by EE in rat hepatocytes was accompanied by EE induction of transcript levels of MRC proteins and ATP levels [72, 248]. ERβ has been known to play a major role in mediating E2’s beneficial effects on cardiac function following trauma-hemorrhage (T-H). Treatment of male rats that underwent T-H with E2 or DPN (an ERβ-selective agonist) enhanced the expressions and activity of complex IV and ATP production. These effects were blocked by co-treatment with complex IV specific inhibitor. Importantly, these effects are required for inhibition of mitochondrial apoptotic signaling pathways [76]. Estrogens may up-regulate the expression of human ATP-dependent RNA and DNA helicase (SUV3) that is involved in mtDNA replication because the promoter of this gene contains NRF-1, Sp1 and NF-κB binding sites. Interestingly, this protein has been found to be associated with HBXIP that was identified as a cofactor of survivin in suppression of apoptosis. The HBXIP binding domain is important for mitochondrial import and stability of the Suv3 protein in vivo [102, 256].
The genetic and pharmacological studies described in section 6.1 indicate that up- or down-regulation of the MRC biogenesis/functions may determine the fate of cell death via apoptosis or cell survival by inhibition of apoptosis. Thus, persistent stimulation of MRC biogenesis/functions by E2 and ERs should contribute to inhibition of apoptosis, causing the normal balance of cell growth/cell death toward survival and enhanced cell growth. Inhibition of apoptosis is one of important mechanisms of cancinogenesis.
7. Physiological and pathological implications
The MRC is crucial to the large majority of cellular functions. The regulation of the MRC biogenesis and functions by E2/ERs confers a large number of physiological effects. The fact that E2 and ERs stimulate MRC biogenesis and functions in such organs/tissues as breast, brain, heart, and eyes that have high demand for energy supply for their proper functions reflects the physiological importance of these effects. MRC-derived ROS also play important roles in the regulation of a number of cellular functions. However, over-abundance and/or deficiency of these E2/ER-mediated mitochondrial effects could lead to pathological consequences and diseases, depending on the types and age of the target cells and tissues. The energy demand, availability/levels of E2 and ER isoforms, subcellular localization of ERs and the duration of their actions, and the vulnerability of the cells to ROS stress will determine the physiological and pathological consequences in particular target cell types and tissues.
7.1. Implications in Estrogen Carcinogenesis in Breast Cancer Cells
Breast cancer is the second leading cause of cancer-related death among women in U.S. and other developed countries. Prolonged exposure to excess E2 is a key etiological factor for the development of breast cancer. Because human breast cells are exposed to relatively high levels of E2 due to the in situ E2 synthesis by local aromatase activity [257, 258] and estrogen replacement therapy, their growth and development are under persistent control by E2. The carcinogenic effects of E2 are mediated via ERα- and ERβ-mediated genomic pathways and by the genotoxic effects caused by oxidative estrogen metabolites [259]. As described above, over-abundance of E2/ER-mediated MRC biogenesis and functions may contribute to stimulation of cell proliferation and inhibition of apoptosis. It is likely that persistent stimulation of the MRC biogenesis and functions by E2 and ERs exists in breast cells, which can lead to alterations in energy metabolism, abnormal cell growth and inhibition of apoptosis in breast cells. All of these may contribute to estrogen carcinogenesis of breast cancer. The potential implications and relevance of E2/ER-mediated effects on mitochondria to estrogen carcinogenesis have been described elsewhere [46, 113]. Several lines of new evidence are presented here to further support this notion. First, altered mitochondrial functions are becoming crucial to understanding cancer mechanisms. For example, Putignani et al. [260] observed distinct differences in mitochondrial proteins between normal and breast-infiltrating ductal carcinoma (IDC) cells. They detected depressed expression levels for all MRC complexes in human mammary carcinoma, particularly for NDUFS3 (complex I), SDHB subunits (complex II), 2 UQCRC2 (complex III), COX 1 (Complex IV) and ATPβ (complex V). These changes were associated with reduction in COX and ATP synthase activity, deterioration in mitochondria morphology and changes in mitochondria membrane. These data described bioenergetic and phenotypic alterations of IDC cell mitochondria. Similarly, Abril et al. [261] detected altered expression of 12S/RNR1, COX1/COX2 (complex IV), and ATP6 (complex V) in prostate cancer cells. Furthermore, a proteomic analysis of brain metastasis of human breast cancer cells[262] revealed increased expression of proteins involved in several energy metabolism pathways including glycolysis, tricarboxylic acid cycle, oxidative phosphorylation, the pentose phosphate pathway and the glutathione system. These metabolic alterations were associated with strongly enhanced tumor cell survival and proliferation in the brain microenvironment. Second, other carcinogens can cause alterations in mitochondrial functions. For example, the carcinogenic polycyclic aromatic hydrocarbon DNA adducts caused multiple alterations in gene expression. A proteomic analysis revealed altered expression of several MRC proteins of MCF-7 cells treated with benzo[a]pyrene [263]. Third, persistent enhanced generation of MRC-derived ROS induced by E2/ERs may cause progressive oxidative damage to mtDNA and mitochondrial proteins, leading to mutations in mtDNA and malfunction of MRC. Indeed, several types of mtDNA mutations have been detected in breast cancer tissue and in matched nipple aspirate fluid [264–266]. It has been shown that mitochondrial genetic background modified breast cancer risk [267, 268].
In addition, altered MRC functions could lead to retrograde regulation that has been implicated in carcinogenesis [269]. Mitochondrial retrograde signaling is a pathway of communication from mitochondria to the nucleus that influences many cellular and organismal activities under both normal and pathological conditions [269–271]. This signaling pathway is triggered by mitochondrial dysfunction and is not a simple on-off switch, but rather it responds in a continuous manner to the changing metabolic needs of the cell. Communication between mitochondria and the nucleus is important for a variety of cellular processes such as carbohydrate and nitrogen metabolism, cell cycle and proliferation, cell growth and morphogenesis [269]. By altering the levels of MRC-derived ATP, ROS and other intermediate metabolites, E2/ER-mediated MRC biogenesis and functions can trigger mitochondrial retrograde signaling. For example, it has been shown [73, 272, 273] that the E2-mediated MRC-derived ROS are involved in the regulation of the nuclear redox-sensor kinases, e.g. A-Raf, Akt and PKC, which in turn, activate transcription factors NF-κB, CREB, AP-1 via MEK/ERK pathways, that are involved in cell growth. Depletion of mtDNA has been shown to cause mitochondrial impairment in breast cancer cells, leading to altered expression of nuclear genes involved in signaling, cellular architecture, metabolism, cell growth/differentiation, and apoptosis [274]. Proteomic analysis of mitochondria-to-nucleus retrograde response in human cancer identified quantitative changes in expression of several proteins including subunits of complex I and complex III, molecular chaperones, and a protein involved in cell cycle control were downregulated and inosine 5'-monophosphate dehydrogenase type 2 (IMPDH2) involved in nucleotide biosynthesis was upregulated in mtDNA-depleted rho (0) cells, suggesting that these proteins play key roles in retrograde response. Interestingly, further analysis of expression of UQCRC1 gene (encoding ubiquinol cytochrome-c reductase core protein I) in breast and ovarian tumors revealed that UQCRC1 was highly expressed in breast (74%) and ovarian tumors (34%) and that its expression positively correlated with mtDNA-encoded COXII [275].
7. 2. Implications in Anti-Cancer Drug Resistance
The majority of anti-cancer drugs confer their anti-cancer activities by inducing apoptosis and/or inhibiting cell growth and proliferation. Cancer cells acquire drug resistance as a result of selection pressure dictated by unfavorable microenvironments. This survival process is facilitated through efficient control of oxidative stress originating from mitochondria that typically initiates apoptosis. Several anti-cancer drugs have been shown to target MRC and alter MRC biogenesis and functions. Alterations in MRC protein expression/functions are frequently seen in a number of cancer cells that are resistant to anticancer drugs.
7.2.1. Implications in Tamoxifen Resistance in Breast Cancer Cells
Tamoxifen (TAM), an ER antagonist that blocks E2-mediated cell growth and provokes apoptosis, is widely used for treatment of ER-positive breast cancer patients. TAM and its metabolite, 4-hydroxytamoxifen (4-OH-TAM), can induce apoptosis through both ER-dependent and ER-independent pathways[276]. Several lines of evidence have indicated that MRC are important targets of TAM via ER-dependent and ER-independent pathways.
TAM has acute inhibitory effects on oxidative phosphorylation in isolated rat liver mitochondria [209, 218, 277, 278]. It has been observed [279] that treatment of TAM-sensitive MCF-7 and MAB-MB-231 cells with TAM induced apoptosis and that these effects were inhibited by bongkrekic acid, an inhibitor of MPT. The pure anti-estrogen ICI 182780 partly inhibited the effect of TAM in ER-positive MCF-7 cells, but not in ERα-negative MDA-MB-231 cells. Pre-culturing MCF-7 cells in the absence of E2 or in the presence of a low TAM concentration made the cells even more susceptible to rapid death induced by TAM. These results suggest that disruption of mitochondrial function has a primary role in the acute death response of the cells and that E2 via ERs has protective effects on mitochondrial functions. Moreira et al. [280] demonstrated that TAM interacted with the flavin mononucleotide site of complex I, leading to mitochondrial failure. Ridaifen-B, a novel TAM derivative, has been shown to induce apoptosis in ER-negative breast cancer cells by reducing mitochondrial membrane potential [281]. Pancratistatin (PST), a natural compound obtained from the Hawaiian spider lily, has been known to be specific and selective in inducing apoptosis in multiple cancer cell lines. It targets mitochondria specifically in cancer cells to induce increased levels of ROS, decreased ATP and mitochondrial membrane permeabilization [282]. When combined with TAM, PST caused synergic effects on breast cancer cells in inducing apoptosis by targeting mitochondria [283]. In addition, TAM has been shown to indirectly influence MRC functions via increasing intramitochondrial ionized Ca(2+) levels and stimulating mitochondrial NO synthase (mtNOS) activity in MCF-7 cells. By stimulating mtNOS, TAM hampered mitochondrial respiration, releases cytochrome c, elevates mitochondrial lipid peroxidation, increased protein tyrosine nitration of certain mitochondrial proteins, decreased activity of succinyl-CoA:3-oxoacid CoA-transferase, and induced mitochondrial aggregation [284].
A major clinical problem for TAM therapy is that a high percentage of breast cancer patients ultimately become TAM resistant (TAM-R). While the mechanisms underlying TAM-R are not completely understood, altered MRC protein expression and deregulation of MRC functions, which have been observed in TAM-R cells, may be a contributing factor. It has been reported [285] that TAM induced apoptosis in normal human mammary epithelial cells (HMECs) that have lost of the tumor suppressor p53 function (p53−). However, these cells rapidly developed resistance to TAM-induced apoptosis within 10 passages in vitro. Resistance to TAM in late passages of p53−/− HMEC-E6 cells correlated with an increase in mitochondrial mass, a lack of mitochondrial depolarization and caspase activation following TAM treatment, suggesting altered mitochondrial functions in TAM-R cells. Indeed, a proteomic comparison of TAM-S versus TAM-R xenografted human breast cancer revealed an enhanced expression of MRC proteins in association with altered expression of proteins involved in cell-cell adhesion/interaction, signal transduction, DNA and protein synthesis machinery, oxidative stress processes and apoptosis[286]. Similar alterations in the mitochondrial proteome were also detected in adriamycin resistant MCF-7 cells [287].
7.2.2. Implications in Other Anti-cancer Drug Resistance
Alterations in MRC protein expression and MRC activities are also observed in other cancer cells resistant to several widely used anti-cancer drugs, including 5-luorouracil (5-FU), cisplatin and docetaxel.
5-FU is the mainstay of chemotherapy for the treatment of patients with advanced colon cancers. However, the acquisition of resistance to 5-FU is a prominent obstacle to successful chemotherapy. Several studies reported altered expression of MRC proteins, ATP synthase and proteins involved in MRC protein synthesis in 5-FU resistant cells. For example, Tanaka et al. [288] identified enhanced levels of Tfam in association with a heterogeneous nuclear ribonucleoprotein G histone H2B, histone H4 and ribosomal protein L3 in 5-FU resistant cells. These proteins are potentially related to 5-FU resistance by protecting the cells from mtDNA and nDNA damage. Shin et al. [289] detected lower levels of the ATPα subunit and ATP synthase d-subunit, reduced ATP synthase activity and ATP contents in 5-FU-resistant cells compared with parent cells. Furthermore, suppression of ATP synthase d-subunit expression by siRNA increased cell viability in the presence of 5-FU. Down-regulation of ATP synthase β-subunit expression was also seen in breast, liver, kidney, colon, squamous oesophageal, and lung carcinomas and gastric adenocarcinomas, suggesting that ATP synthase down-regulation may lead to cellular events responsible for 5-FU resistance.
Cisplatin is a platinum drug used as a cornerstone of current chemotherapy regimens. Development of resistance to this drug by tumors significantly decreases its usefulness in the clinic. It has been reported that the expression of SIRT1, which belongs to the family of type III histone deacetylase and is implicated in diverse cellular processes, was markedly up-regulated in androgen-refractory PC3 and DU145 cells compared with androgen-sensitive LNCaP cells and that its expression level was correlated with cell growth in PC3 cells. Treatment with a SIRT1 inhibitor, sirtinol, inhibited cell growth and increased sensitivity to cisplatin. Silence of SIRT1 expression by siRNA also suppressed cell proliferation and reduced resistance in PC3 cells, mimicking the effect caused by sirtinol. Also in DU145 cells, sirtinol treatment enhanced sensitivity to cisplatin [290]. SIRT1 contributes in part to cisplatin resistance by reducing glucose use and altering mitochondrial metabolism [291]. Qian et al. [292] observed that mitochondrial density in intestinal epithelium determined the cellular sensitivity to cisplatin-induced cell death.
Docetaxel is an effective chemotherapeutic agent against cancer but some patients develop resistance. It has been found [293] that docetaxel-resistant DRHEp2 cells developed from human laryngeal cancer HEp2 cells had greatly increased mtDNA content whereas reduction of mtDNA content in DRHEp2 cells by ethidium bromide treatment reduced the resistance, suggesting a role for mtDNA-coded MRC proteins in resistant mechanisms. Oligomycin A (a F0–F1 ATPase inhibitor) but not inhibitors of other MRC complexes eliminated docetaxel resistance in these cells, indicating a role for mtDNA-coded ATPase subunits in resistant mechanisms. Docetaxel induced ROS generation in HEp2 cells but not in DRHEp2 cells. Antioxidant pyrrolidine dithiocarbamate eliminated docetaxel-induced cytotoxicity. Furthermore, inhibition of F0–F1 ATPase by oligomycin A induced docetaxel-mediated ROS generation in DRHEp2 cells. Taken together, DRHEp2 cells acquired docetaxel resistance through increasing F0-ATPase, which diminished docetaxel-induced ROS generation and subsequent inhibition of cell death. The mtDNA plays an important role in development of docetaxel resistance through the reduction of ROS generation by regulating F0-ATPase [293].
Persistent alterations of MRC biogenesis and functions induced by E2 and ERs may cause changes in the responsiveness of cells to anticancer drugs that target mitochondria. Thus, regulation of E2/ER-mediated mitochondrial pathway may represent a promising way to overcoming anti-cancer drug resistance.
7.3. Implications in neuro-protective effects
The human brain is only 2% of the body weight, but it consumes about 20% of the total energy in the body at rest. The human brain is the most energy hungry organ in the body thereby increasing its vulnerability. Sufficient supply of mitochondrially-derived energy is essential for the proper functions of the brain. It has been known that if the energy supply is cut off for 10 minutes, there is permanent brain damage. No other organs are as sensitive as brain in respond to the changes in its energy supply. Defective mitochondrial structure and MRC dysfunctions have been observed in patients with neurodegenerative diseases such as Alzheimer’s diseases (AD) and Parkinson’s disease (PD)[294–297]. Epidemiological studies have indicated that estrogen therapy reduced the risk of developing AD and PD in women. Clinical and experimental studies have shown that estrogens are potent modulators of the brain’s functions and have beneficial effects in mental and neurodegenerative diseases. For example, Xu et al. [298] observed significant alterations in mitochondrial ultrastructure and significant reduction in in ATP levels in hippocampal neurons of Ovx rats. These alterations caused by Ovx were reversed by treatment with estrogen and phytoestrogens (e.g. genistein and ipriflavone), revealing a critical role of estrogens in preservation of mitochondrial structure and functions in brain neurons. Comsistent with this observation, a transient alteration of COX activity and mitochondrial ATP content in hippocampi of young OVX rats and a prolonged lowering of COX activity and mitochondrial ATP content were seen in hippocampi of middle-aged OVX rats. Furthermore, treatment of these rats with estradiol benzoate (EB) or genistein reversed this estrogen withdrawal-induced mitochondrial dysfunction in both young and middle-aged rats [299, 300]. The general effects of estrogens on several aspects of mitochondrial functions of brain neurons in relation to estrogen’s neuroprotection on neurodegenrative diseases have been described elsewhere [301–305]. The potential implications of E2/ER-mediated MRC biogenesis and functions in protection against AD and PD are described as the follows.
7.3.1. Implications in Neuro-Protection in Alzheimer’s Disease
Alzheimer’s disease (AD) is by far the most common neurodegenerative dementia among the elderly. Patients with AD initially show memory loss, and as the disease progresses they develop impaired executive function, confusion and personality change and eventually death [306]. The incidence of AD is lower in young women than in men but is increased among postmenopausal women. These observations indicate that estrogens have important protective effects against the development of AD.
There are several pathological hallmarks for AD. One of which is the abnormal aggregation of a protein called Tau, a microtubule-associated protein, within nerve cells. The Taubearing lesions appear initially in the entorhinal cortex and hippocampus, then progress to neocortex of the frontal and temporal poles, and finally involve much of the temporal, frontal, and parietal cortices. Tau associates with microtubules to form Tau aggregation, which appears as neurofibrillary tangles, neuritic plaques, and neuronal threads. Nerve cells in which Tau protein aggregated could not grow and function properly [307]. Tau is closely related to mitochondrial function. Transgenic mice over-expressing the P301L mutant human tau protein exhibited an accumulation of hyperphosphorylated Tau and develop neurofibrillary tangles. Proteomic and functional analyses [308] revealed a mitochondrial dysfunction in P301L tau transgenic mice. Significantly, the reduction in MRC complex V levels, COX I activity and, with age, impaired mitochondrial respiration and ATP synthesis were observed in the P301L tau mutant mice. Mitochondrial dysfunctions were associated with high levels of ROS in aged transgenic mice. Increased Tau pathology seen in aged homozygous P301L Tau mice was accompanied by modified lipid peroxidation levels and up-regulation of antioxidant enzymes in response to oxidative stress. A recent study [309] revealed that a peptide containing residues 26–44 of Tau protein targeted COX and ANT and impaired MRC oxidative phosphorylation at the level of ANT, conferring deleterious effects on cellular availability of mitochondrial ATP.
The second important hallmark of AD is the amyloid plaques comprised of the amyloid A-beta peptides, which cause toxic effects on nerve cells by inducing mitochondrial dysfunction, including altered calcium homeostasis and Bcl-2 expression, associated with increased neuronal apoptosis [310]. It has been shown that soluble beta-amyloid caused mitochondrial defects in amyloid precursor protein and Tau transgenic mice as evidenced by mitochondrial damage such as reduced mitochondrial membrane potential and ATP levels in the brains from these mice before the onset of plaques [311]. A study [312] reported that the amyloid beta-peptide was imported into mitochondria via the Tom import machinery and localized to mitochondrial cristae. Both oligomeric and fibrillar species but not disaggregated (mainly monomeric) form of Abeta42 have been also shown to impair mitochondrial function in P301L Tau transgenic mice. Aging specifically increased the sensitivity of mitochondria to oligomeric Abeta42 damage [313]. The amyloid peptide exposure provoked down regulation of a key anti-apoptotic protein, Bcl-2, and resulted in mitochondrial translocation of Bax, a protein known to promote cell death, and increases mitochondrial peroxide production, nitration and oxidation of proteins and subsequent release of cytochrome c [310].
Mitochondrial dysfunction has been identified in AD [306]. Since both Tau and Abeta act on mitochondria, there could be a synergistic action of Tau and Abeta on the mitochondria. Indeed, P301L tau mitochondria displayed increased vulnerability toward Abeta peptide insult. There may be direct impact of abnormally phosphorylated tau and Abeta on proteins/enzymes involved in metabolism, MRC function and cellular detoxification[314]. However, Tau pathology involves a mitochondrial and oxidative stress disorder which may be distinct from that caused by Abeta.
E2 has been shown to be neuroprotective in a number of animal models of AD. It has multifunctional effects, causing a reduction in the development of AP by preventing or reducting the Tau aggregation and formation of amyloid plaques.
Abnormal Tau hyperphosphorylation by protein kinases such as protein kinase A (PKA) plays an important role in PD pathogenesis [315]. It has been reported[316] that E2 effectively attenuated forskolin-induced over-activation of PKA and elevation of cAMP, and thus prevented Tau from hyperphosphorylation. Serine/threonine protein phosphatases(PP) such as protein phosphatases PP1, PP2A, and calcineurin are involved in hyperphosphorylation of Tau [317]. Using specific inhibitors for PP1, PP2A and calcineurin, Yi and Simpkins [317] have demonstrated that these proteins played a role in estrogen-mediated neuroprotection. Zhang et al. [318] revealed a role of Dickkopf-1 (Dkk1) in attenuation of Tau phosphorylation and E2-induced neuroprotection. DKK1 is an antagonist of the Wnt/beta-catenin signaling pathway. This pathway is a principal mediator of neurodegeneration in cerebral ischemia and AD. E2 at low physiological levels protected the hippocampus CA1 against global cerebral ischemia by preventing Dkk1 elevation. This effect correlated with a reduction of phospho-beta-catenin and elevation of nuclear beta-catenin levels. E2 inhibition of Dkk1 is a critical mechanism underlying its neuroprotective and phospho-tau regulatory effects after cerebral ischemia. Thus, hyperphosphorylation of Tau could be a promising therapeutic target for AD [315].
In addition to attenuating Tau hyperphosphorylation, E2 has preventive effects against amyloid beta-mediated neurotoxicity. E2 exposure prior to neurotoxic insult of hippocampal neurons promoted neuronal defense and survival against amyloid beta. It has been observed [310] that treatment of cultured rat hippocampal neurons with E2 prior to amyloid beta exposure significantly reduced the number of apoptotic neurons and the associated rise in resting intracellular calcium levels. E2 pretreatment inhibited the amyloid beta-induced apoptosis. These anti-apoptotic effects of E2 could be attributed to: 1) activation of antioxidant defense systems scavenging ROS and limiting mitochondrial protein damage [303]. It was observed that E2 induced expression of antioxidative and antiapoptotic thioredoxin (Trx) and MnSOD expression following the induction of NOS1 in human brain-derived SH-SY5Y cells [319]; and 2) improvement of MRC electron transport chain activity and reducing mitochondrial DNA damage as well as increaseing the activity of complex IV of the electron transport chain, improving mitochondrial respiration and ATP production under normal and stressful conditions. In fact, E2 treatment enhanced the expression of genes encoding MRC proteins in rat hippocampus [70] and pituitary tumor cells [71]. In vivo treatment of rats with E2 up-regulated brain mitochondrial proteome, COX IV activity and MRC functions[75, 77].
Inhibition of the assembly of amyloid beta-peptide and the destabilization of preformed betaamyloid fibrils (fAbeta), prevention and/or inhibition of Tau hyperphosphorylation in the central nervous system could represent valuable therapeutic approach for patients with AD. Estrogen has anti-amyloidogenic effects on Alzheimer's fAbeta in vitro [320]. E2 also attenuates Tau hyperphosphorylation. On the other hand, stimulation of the MRC biogenesis and functions in neurons of brain by E2/ERs offsets the damage on mitochondrial functions caused by Tau and fAbeta and thus, are implicated in protection against AD.
Based on the pathological features of AD, it was proposed [321] that drug discovery for AD should include: (i) supplementation therapy, (ii) anti-apoptotic compounds, (iii) substances with a mitochondrial impact, (iv) anti-amyloid substances, (v) anti-protein aggregation and (vi) lipid-lowering drugs. Estrogen replacement therapy for AD appears to fully fill (ii), (iii), (iv) and (v). However, because of the potential carcinogenic effects of E2 in breast cancer (section 7.1) and the highly oxidative cellular environment present brain that may be favorable for oxidative metabolism of estrogens during neurodegeneration, E2 itself is a poor agent for treatment of existing AD [303]. In this regard, in cultured cells and animal models of AD, other estrogenic chemicals, some with less hormonal effects, have been shown to be neuroprotective, including 17α-estradiol [322–324], selective agonists for ERα(PPT) and ERβ (DPN) [229], phytoestrogens [325, 326], and a hybrid structure of E2 and vitamin E [327]
7.3.2. Implications in Neuro-protective Effects against Parkinson’s Disease
Parkinson’s disease (PD), the second most common neurodegenerative disorder, is characterized by the degeneration of dopaminergic neurons in the substantia nigra. The most vulnerable neurons reside in the substantia nigra zona compacta (SNC), whereas the DA neurons in the ventral tegmental area (VTA) and interfascicular (IF) nucleus are less vulnerable to degeneration (for review see [328]). There are two forms of PD, the rare familial form and the more common sporadic form. Both familial and sporadic PD patients have defects in mitochondrial respiration. Exposure to environmental mitochondrial toxins leads to PD-like pathology.
PD is more frequent in men than in women, and is more prevalent in women with short reproductive life [329]. Estrogens are considered neurotrophic for dopamine neurons and are neuroprotective against neurotoxic agents for dopamine neurons in vivo and in vitro[330]. The neuroprotection of E2/ERs on PD could be attributed, in part, to their effects on MRC biogenesis/functions and its potential effects on PD-related proteins that are involved in the control of mitochondrial functions. Several mitochondrial proteins including PTEN-induced kinase 1(PINK1), Parkin, DJ1, α-synuclein and POLG are known to be involved in mediation of both familial and sporadic forms of PD. Recent studies on these genes have revealed the central importance of mitochondrial dysfunction and oxidative stress in both forms of PD [328, 331–333].
Pink1 is a putative serine/threonine kinase localized in mitochondria. It is highly conserved from Drosophila to human. It has been shown that removal of Drosophila Pink1 homologue function resulted in defects in mitochondrial morphology (e.g. fragmented mitochondrial cristae). These effects were associated with male sterility, apoptotic muscle degeneration, and increased sensitivity to multiple stresses including oxidative stress whereas expression of human Pink1 in the Drosophila restored these defects in a portion of pink1 mutants [331]. It has been reported[334] that the germline deletion of the Pink1 gene in mice significantly impaired mitochondrial functions in the striatum but not in the cerebral cortex at 3–4 months of age. Decreased MRC activities in the cerebral cortex were evident in Pink1(−/−) mice at 2 years, indicating that aging can exacerbate MRC dysfunction in these mice. Moreover, MRC defects were induced in the cerebral cortex of Pink1(−/−) mice by cellular stress. Mitochondrial aconitase activity associated with the Krebs cycle was also reduced in the striatum of Pink1(−/−) mice. Because mitochonrdial aconitase contains an iron sulfur core that is oxidized by ROS, the activity of this enzyme is a useful fingerprint of mitochondrial ROS. The reduced activity of this enzyme in the striatum of Pink1(−/−) mice reflects an increased mitochondrial ROS, indicating a key role of Pink1 in reguation of oxidative stress. Furthermore, mutations in human Pink1 caused hereditary early-onset of PD [335]. Together, these findings demonstrate an important role of mammalian Pink1 for mitochondrial function, which provides critical protection against both intrinsic and environmental stress. Thus, loss of Pink1 may lead to nigrostriatal degeneration in PD.
Parkin is a ring protein located in the outer mitochondrial membrane of proliferating cells and is involved in mitochondrial biogenesis. Parkin is functionally linked to Pink1 in that its mitochondrial translocation is controlled by pink1 kinase activity through direct phosphorylation in its ring region [336]. Interestingly, loss of Drosophila parkin showed phenotypes similar to those caused by the loss of pink1 function. Over-expression of parkin rescued the male sterility and mitochondrial morphology defects of pink1 mutants, whereas double mutants removing both pink1 and parkin function showed muscle phenotypes identical to those observed in either mutant alone. These observations suggest that pink1 and parkin function, at least in part, in the same pathway, with pink1 functioning upstream of parkin. The role of the pink1-parkin pathway in regulation of mitochondrial function underscores the importance of mitochondrial dysfunction as a central mechanism of PD pathogenesis. In proliferating cells, transcription and replication of mtDNA are enhanced by parkin over-expression and attenuated by parkin suppression with siRNA. Parkin is associated with Tfam, enhancing Tfam-mediated mtDNA transcription. These results indicate that parkin is involved in the regulation of mitochondrial transcription/replication in these cells [237, 337]. It has been shown [237, 337] that wild-type parkin attenuated ROS production whereas mutant parkin enhanced ROS production in SH-SY5Y and L6 cells and that over-expression of parkin reduced ROS. Parkin prevented apoptosis and enhanced mitochondrial membrane potential in SH-SY5Y and L6 cells but not in COS-1 cells. Expressions and enzymatic activity of MRC complex I but not other MRC complexes were selectively enhanced by Parkin. These results suggest that parkin possesses anti-apoptotic function in neuronal cells.
DJ-1 has a role in protection against oxidative stress, but its direct role in mitochondrial function has not been fully established. Mutations (e.g. homozygous L166P missense mutation) have been identified in DJ-1 gene that cause autosomal recessive early onset of PD. It has been observed [338] that the L166P mutation markedly reduced stability to DJ-1 protein in vivo, resulting from enhanced degradation by the proteasome and that the L166P mutant protein exhibited an impaired ability to self-interact to form homo-oligomers. Wild type DJ-1 protein existed in a homo-dimeric form in vitro, whereas the L166P mutant existed only as a monomer and specifically but differentially associated with parkin. These mutants exhibited impairments in homo-dimer formation, suggesting that parkin may bind to monomeric DJ-1. Parkin failed to specifically ubiquitinate and enhance the degradation of L166P and M26I mutant DJ-1 protein, but instead promoted their stability in cultured cells[339]. The interaction between parkin and L166P DJ-1 may involve a larger protein complex that contains CHIP and Hsp70, perhaps accounting for the lack of parkin-mediated ubiquitination. Oxidative stress also promoted an interaction between DJ-1 and parkin, but this did not result in the ubiquitination or degradation of DJ-1. Parkin-mediated alterations in DJ-1 protein stability may be pathogenically relevant as DJ-1 levels were dramatically increased in the detergent-insoluble fraction from sporadic PD/DLB brains, but were reduced in the insoluble fraction from parkin-linked autosomal recessive juvenile-onset PD brains.
The α–synuclein is an abundant protein found particularly in axonal termini and a component of Lewy bodies, a pathological hallmark of PD. It contains an amino-terminal mitochondrial targeting sequence and is imported into mitochondria induced by acidification of the cytosol or by its over-expression. The import and accumulation of α-synuclein has been shown to impair complex I in human dopaminergic neuronal cultures and PD brain [340]. Defective MRC function or inhibition of MRC with specific MRC inhibitors led to formation of α-synuclein aggregates in cells associated with reduction in ATP levels [341]. Exposure of human embryonic kidney cells over-expressing α-synuclein to rotenone caused increased susceptibility to cell death and lower levels of ATP than control cells. The brainstem neurons of mice that over-expressed the human Ala53Thr form of α-synuclein contained degenerating and dysmorphic mitochondria. The α-synuclein protein also induced oxidative damage and cytochrome c release. Mice with a knockout of the Snca gene exhibited increased resistance to MPTP through modulation of complex I activity, whereas administration of MPTP to mice that overexpressed α-synuclein led to swollen, morphologically abnormal mitochondria. These observations suggest a reciprocal relationship between the activity of this protein and mitochondrial function [328].
The data described above indicate a potential link among Pink1, parkin, DJ-1 and α–synuclein in a common molecular pathway at multiple levels with parkin and α–synuclein being directly involved in the regulation of MRC functions. Understanding the regulation of this Pink1-parkin-DJ-1-α–synuclein pathway may have important implications for understanding the pathogenesis of inherited and sporadic PD. A few studies have suggested that Pink1 [342], parkin [330] and synucleins [343, 344] participate in estrogen response pathways. Thus, E2 and ERs may be involved in the regulation of Pink1-parkin-DJ-1-α–synuclein pathway in neurons. It is likely that the regulation of Pink1-parkin-DJ-1-α–synuclein pathway and MRC biogenesis and functions in neurons by E2 and ERs confer protection for mitochondrial functions, protecting neurons from apoptosis induced by ROS and mitochondrial toxins. Deficience in these E2/ER-mediated mitochondrial effects may be causally related to pathogenesis of PD.
7.4. Implications in Cardiovascular Protective Effects
Cardiovascular diseases (CVDs) are the major cause of morbidity and mortality among both men and women. There is a significant gender difference in incidence of CVDs (e.g. atherosclerotic diseases), in part because of differences in risk factors and hormones [345]. The incidence of CVDs is uncommon in pre-menopausal women, sharply rises in postmenopausal women, and is reduced to premenopausal levels in postmenopausal women who receive estrogen therapy, suggesting that estrogens confer protective effects on the cardiovascular system [346, 347].
E2 has both indirect and direct effects on the cardiovascular system. The indirect effects of E2 include its influences on serum lipoprotein and triglyceride profiles, and the expression of coagulant and fibrinolytic proteins. The direct effects of estrogen occur through both rapid non-genomic and longer-term genomic pathways that are mediated by the ERα and ERβ. The rapid effects of estrogen result in the activation of endothelial nitric oxide synthase, leading to arterial vasodilation, whereas longer-term effects involve changes in gene and protein expression, modulating the response to injury and atherosclerosis [346]. It has been shown [348] that the protective effects of E2 against vascular injury are mediated via ERα.
Mitochondria in the adult mammalian heart have a tremendous capacity for oxidative metabolism, and the conversion of energy by these pathways is critical for proper cardiac function[349]. Several inherited and acquired disorders of mitochondria including defects in MRC proteins, disruption of oxidative phosphorylation and increased ROS oxidative stress, lead to defects in cardiac functions as reflected in exercise intolerance, arrhythmias, dilated cardiomyopathy and heart failure [349–353]. Myocardial ischemia-reperfusion injury also causes mitochondrial dysfunctions, resulting in low ATP levels, malfunctional ion pumps/altered ion homeostasis, ruptured plasma membrane and cell death [186]. These observations suggest that alterations in cardiac mitochondrial structure and functions play a central role in the development of the overt heart failure and heart diseases [354].
Recent studies examining the effects of estrogen on mitochondria of vascular systems have revealed a new paradigm for estrogen’s vascular protection, i.e. the effects of E2/ER on cardiac mitochondrial structure and function contribute, at least in part, to the E2’s cardiac protective effects [355]. E2 has been shown to profoundly affect mitochondrial functions in cerebral blood vessels of rat brain, enhancing efficiency of energy production and suppressing mitochondrial oxidative stress by up-regulating a number of genes encoding MRC proteins and anti-oxidant enzymes such as MnSOD [217, 355, 356]. E2/ERβ pathways mediate down-regulation of genes for nDNA-encoded subunits of all MRC complexes whereas ERα is essential for most of the estrogen-mediated increase in gene expression including MRC proteins and proteins involved in defense against oxidative stress in wild-type rat aortas [116]. In contrast, Hsieh et al. [76] observed up-regulation of the expression and activity of complex IV by E2/ERβ in the heart of male rats that underwent T-H and that these effects were required for E2-mediated inhibition of apoptosis induced by T-H. E2/ERα–[357, 358] and E2/ERβ-pathways [359] were involved in protection of myocardial ischemia-reperfusion injury probably via their effects on preservation and maintenance of mitochondrial structure and functions [186].
7.5. Implications in Beneficial Effects of Estrogens/ERs on Lens Physiology
Cataract formation is a multifactorial eye disease and a leading cause of blindness worldwide. While surgical procedures correct vision loss, this presents a substantial financial burden on national health care systems, mandating the search for pharmaceutical agents that can prevent or delay the onset of cataract. Epidemiological studies indicate an increased frequency of cataract formation in postmenopausal women similar to men of the same age in contrast with lower incidence in premenopausal women, suggesting that the absence of estrogens in postmenopausal women may contribute to their increased risk[360]. Several epidemiological studies have implied that hormone replacement therapy may play some positive role in delaying or preventing cataract. The Beaver Dam Eye Study [361] and the Salisbury Eye Study [362] both supported some degree of protection associated with estrogen and the risk of cataract development. Aina et al. [363], using a large population based study of women with reported cataract and age-matched controls, concluded that, “estrogen and estrogen-progesterone hormone replacement therapies are associated with a reduced risk of cataract.”
Work in the Cammarata laboratory has been aimed at elucidating the biochemical and molecular pathways by which estrogens, which behave as potent biologically active and selective mitochondrial protective compounds, may influence mitochondrial function. But is there a generally established connection between damaged mitochondria and cataractogenesis? “Mitochondriopathies show a chronic, slowly progressive course and present with multiorgan involvement. Although several proteins with signaling, assembling, transport and enzymatic function can be impaired, most frequently the activity of MRC complexes is primarily affected, leading to impaired oxygen utilization and reduced energy production [364]”. Among the most frequently affected systems are eyes and in particular, the lens and the emergence of cataract. Therefore, cataract is not an exceptional occurrence in mitochondrial myopathies and should be included within its multisystem associations [365].
The means by which estrogen and estrogen-progesterone hormone replacement therapies might restrict or delay the onset of cataract is unknown. The primary focus of the Cammarata laboratory has been to demonstrate that E2 protects human lens epithelial cells against oxidative stress by preserving mitochondrial function, in part, via the non-genomic rapid activation of prosurvival signal transduction pathways[59, 217, 366]. Newly obtained information, derived from these recent studies, necessitates a reevaluation of the past interpretation of the cytoprotective scheme. To that end, this section of the review will briefly discuss recent data indicating the coupled duality to E2’s cytoprotective action against acute oxidative stress and offer an interpretation that includes the possibility of genomic, as well as non-genomic interventions, as inclusive aspects of the protective mechanism.
E2 administration to the immortalized human lens epithelial cells, HLE-B3, initiates a rapid increase in intracellular (mitochondrial) ROS. This, in turn, promptly and transiently increases mitochondrial-associated MnSOD activity, without alteration in its mRNA expression or its protein expression profile. The outcome of increasing MnSOD activity is to immediately lower the mitochondrial ROS concentration before oxidative damage might ensue [217]. In agreement, estrogen has previously been reported to increase MnSOD activity levels without affecting the protein levels of MnSOD in mitochondria of rat pheochromocytoma cells (PC-12 cells) [367] and MCF-7 breast cancer cells [54]. Within the same timeframe that estradiol increases MnSOD activity, estradiol also initiates a signaling cascade culminating in the activation of the Ras/Raf/ERK (MAPK) and PKA pathways in cultured HLE-B3 cells [366].
E2 leads to activation (i.e. phosphorylation) of ERK 1/2 (via MAPK) and Bad (via PKA). Transfection of HLE-B3 cells with ERK2-specific siRNA enhanced mitochondrial membrane depolarization subsequent to peroxide insult whereas Bad siRNA-treated and mock-transfected cells did not. The implication is that ERK2, but not Bad, plays a pivotal role in the regulation of mitochondrial membrane potential in human lens epithelial cells[366]. Estrogen treatment does, however, continue to offer some protection against mitochondrial membrane depolarization, despite ERK2 suppression. In agreement with this, an earlier study, using the MEK inhibitor, UO126, also showed that peroxide insult in the presence of estrogen and UO126 retained a considerable level of protection against depolarization [368]. That is, if pERK alone were the sole component driving the protection mechanism, one might reasonably expect that the loss mitochondrial membrane potential be equivalent in the presence of UO126 irrespective of whether estrogen were present or not. The fact that estrogen retains the ability to partially prevent depolarization in the absence of ERK activation suggests at least two parallel protective mechanisms at play. ERK2, and more specifically pERK2, represents one pathway of the estrogen-mediated mitochondrial protection pathway. Alternatively, estrogen may, in part, supersede the necessity for ERK activation and provide a measure of protection via an ERK-independent route[366].
ERβ1 associates with both the nucleus and mitochondria while the ERβ isoform, ERβ2, localizes exclusively with the nucleus [58, 59]. ERβ5 is found primarily in the cytosol [58, 59]. It has been established that mitochondrial-associated ERβ1 is a necessary prerequisite for E2–mediated cytoprotection against H2O2 toxicity as cells transfected with siRNA-wtERβ1 lost the capability to prevent mitochondrial depolarization by estrogen intervention [59]. But in the course of completing these experiments, an interesting fact was uncovered in that suppression of ERβ1, in turn diminished the relative levels of the ERβ isoforms, ERβ2 and ERβ5. As such, the working hypothesis was expanded to include the likelihood that, in conjunction with the non-genomic rapid activation of prosurvival signal transduction pathways, estrogen-mediated cytoprotection may include the possibility that nuclear ERβ2 may provide genomic contribution to the mitochondrial mechanism of estrogen and estrogen receptors in mitochondrial protection, possibly via the upregulation of protective nuclear-encoded mitochondrial proteins.
Clearly, considerably more work is needed to fully comprehend the molecular mechanism(s) of E2 and ERs in mitochondrial protection. The current conceptual framework regarding the protective mechanism(s) activated by E2 is based on the premise that mitochondrial cytoprotection will likely prove to be complex and multi-faceted with contributions from both genomic and non-genomic aspects simultaneously providing a combinatorial defense against mitochondrial permeability transition.
8. Conclusions and Prospective
8.1. Conclusions
A proposed model that summarizes the effects of E2 and ERs on the regulation of several aspects of MRC biogenesis and their physiological, pathological and pharmacological implications is presented in Figure 4. In this fascinating pathway, the transcription factors NRF-1, NRF-2, PGC-lα and Sp1 play master roles in their regulation of almost all aspects of MRC protein biogenesis, whereas E2 and ERs serve as the key “directors” for the entire pathway. This pathway is of physiological importance for the proper functions of a number of cell types and organs. However, persistent over-stimulation or deficiency of E2/ER-mediated MRC biogenesis and functions are implicated in the pathogenesis of a number of diseases in different organs/tissues. Persistent over-induction of this pathway in breast cells may contribute to estrogen carcinogenesis and thus, proper inhibition of this pathway may be valuable for prevention of breast cancer. In contrast, stimulation of this pathway could be highly beneficial in brain, heart and the eye, whereas deficiency of this pathway in these organs/tissues could be causally related to the pathogenesis of neurodegenerative, cardiovascular and the eye diseases.
Fig. 4. Proposed Pathway for Regulation of MRC Biogenesis by E2/ERs and Physiological & Pathological Implications.
This proposed pathway is simultaneously initiated and performed within nuclei and mitochondria. Inside the nuclei, the E2-activated ERα and/or ERβ bind to the EREs and other relevant regulatory elements present in the promoter regions of several key transcription factors including NRF-1, NRF-2 and PGC-lα and activates their transcription. The increased expression of these transcriptional factors, in turn, stimulate the cooperate expression of nuclear genes encoding: A) a number of proteins factors involved in the replication, transcription and translations of mtDNA; B) the MRC proteins and MRC complex assembly proteins and C) several proteins of mitochondrial protein import machinery. These proteins are synthesized on cytosolic ribosomes, and imported into mitochondria via the mitochondrial protein import machinery. The enhanced expression of proteins of mitochondrial import machinery facilitates the mitochondrial import of proteins involved in the replication- transcription-translations of mtDNA, the nDNA-encoded MRC proteins and MRC complex assembly proteins, along with other mitochondrially localized proteins. Within mitochondria, the protein factors involved in mtDNA replication stimulate mtDNA replication and maintain/protection of mtDNA integrity. On the other hand, ERα and/or ERβ are imported into mitochondria. Once inside the mitochondria, E2 enhances their binding to the mtEREs within the regulatory region (i.e. D-loop) of mtDNA, to form the active transcription complexes in association with the transcriptional factors (e.g. Tfam, TFBIM and TFBIIM) and stimulating mtDNA transcription. The enhanced levels of protein factors involved in translation of mitochondrial mRNAs stimulate the translation of thirteen mtRNAs encoding MRC proteins. Both the newly imported, nDNA-encoded MRC subunits and the newly synthesized, mtDNA-encided MRC protein subunits are increasingly assembled into individual MRC complexes and ATP synthase by the increased MRC complex assembly proteins, resulting in increased MRC biogenesis and functions. The consequences of these processes are the increased production of mitochondrial energy in form of ATP and MRC-derived ROS. NRF-1, NRF-2, PGC-lα and Sp1 are the master players involved in the regulation of the majority of proteins and important factors involved in almost all aspects of MRC protein biogenesis. E2 and ERs act as the key “directors” for the entire pathway.
The normal operation of this pathway is of physiological importance for the proper functions of a number of cell types and organs. Persistent over-stimulation or deficiency of E2/ER-mediated MRC biogenesis and functions can cause alterations in mitochondrial functions, resulting in over-abundance or deficiency of ATP and ROS. The changes in ATP and ROS levels and abnormal mitochondrial functions can also trigger retrograde regulation (e.g. redox-sensitive regulation) of the expression of nuclear genes involved a wide variety of physiological processes including oxidative response signaling pathways, various kinase-mediated pathways, cell cycle-related pathways and cell growth/death-related pathways. The combination of these E2/ER-mediated mitochondrial effects and the resulting retrograde regulation of expression of different types of nuclear genes have a number of physiological implications in different types of cells and organs. Persistent induction of these mitochondrial effects can exist in breast cells due to relatively higher levels of E2 and ERs in these cells, leading to over-abundance of these effects, which may contribute to increased cell proliferation and inhibition of apoptosis as well as resistance to anti-cancer drugs. These effects are relevant to estrogen-mediated carcinogenesis in human breast. On the other hand, stimulation of MRC biogenesis/functions by E2 and ERs are highly beneficial for providing the cells in brain, heart and the eye with sufficient MRC-derived energy for their proper functions. Maintenance of cell survival and inhibition of induced-apoptosis by E2/ER-mdiated MRC biogenesis and functions in neurons in brain, cardiomyocytes in heart and lens cells in the eye confer significant protection against AD and PD, heart diseases and the eye diseases (e.g. cataract formation).
8.2. Prospective
It should be pointed out that several key aspects of this important and newly emerging pathway are still far from being understood. Many new studies are needed to address the following aspects.
8.2.1. Understanding the Molecular Mechanisms Underlying the E2/ER-mediated Coordinate Regulation of MRC Biogenesis and Functions
Several lines of evidence presented above indicate that E2 and ERs are involved in the coordinate regulation of almost all aspects of MRC biogenesis, including: 1) nDNA-encoded MRC proteins and MRC complex assembly proteins; 2) nDNA-encoded protein factors involved in the replication, transcription and translation of mtDNA; and 3) nDNA-encoded proteins of mitochondrial protein import machinery and ANT. What is known, to date, is that E2 and ERs are involved in the regulation of the proteins involved in these processes most likely via their stimulation of the expression of several transcription factors, particularly NRF-1, NRF-2, and Sp1. What is unknown is whether E2 and ERs simultaneously up-regulate all of the proteins or whether these factors selectively regulate only specific sets of proteins involved MRC biogenesis. In addition, the nature of the precise molecular mechanisms underlying these effects remains to be determined. Thus, a comprehensive investigation on the global role of E2 and ERs on all the aspects of MRC biogenesis, and many detailed studies on the underlying molecular mechanisms are required. Such studies will allow new insight into the role of E2 and ERs in the regulation of MRC energy metabolism and MRC functions.
8.2.2. Determining whether E2 and ERs Are Directly Involved in the Regulation of MRC Complex Assembly Proteins and the Molecular Mechanisms Involved
The proper assembly of the nDNA- and mtDNA-encoded MRC proteins into functionally correct individual MRC complexes relies on several sets of assembly proteins specific for the MRC complexes. To date, little is known about whether the expression and functions of these assembly proteins are directly regulated by E2 and ERs, though the binding sites for NRFs have been identified in the promoters of a few genes encoding assembly proteins. Because of their crucial role in MRC biogenesis, the expression of these proteins likely parallels the E2/ER-induced changes in both nDNA- and mtDNA-encoded protein subunits. Further investigation on the E2/ER-mediated regulation of these assembly proteins is essential for better insight into this pathway. More importantly, because mutations in assembly proteins are causally related to a number of lethal mitochondrial diseases (see section 2), new knowledge gained about the regulation of these proteins by E2/ERs will allow the identification of the therapeutic targets for these diseases that can be treated with E2 and E2-related agents.
8.2.3. Determining how the Mitochondrial Import of nDNA-encoded Proteins Involved in MRC Biogenesis Is Regulated by E2 and ERs
The nDNA-encoded proteins involved in MRC biogenesis are imported into mitochondria through mitochondrial protein import machinery. The presence of NRF-1 and NRF-2 binding sites in the promoters of the genes for several Tom proteins that are components of mitochondrial protein import machinery strongly points to the possibility that these proteins are regulated by E2 and ERs via NRFs. However, more work is needed to determine whether E2/ERs are directly involved in the regulation of mitochondrial protein import machinery and whether the enhanced expression of these proteins leads to increased mitochondrial import of proteins required for MRC biogenesis. To date, at least four different pathways for the sorting and assembly of nuclear-encoded mitochondrial proteins have been identified [369]. Thus, one needs to determine which pathway(s) are employed for the import of these proteins into mitochondria. Because mutations that disrupt the proper functioning of the mitochondrial import machinery are related to several diseases [370], elucidation of how E2 and ERs regulate the import of these proteins may be valuable for finding ways to control the mitochondrial import of these proteins and may lead to development of diagnostic tools to aid in the identification of mitochondrial diseases caused by defects in the mitochondrial protein import machinery.
8.2.4. Understanding how E2 and ERs Regulate the Expression of mtDNA-encoded MRC Proteins within Mitochondria
A number of studies presented above have demonstrated that the mitochondrial ERα and ERβ are directly involved in the regulation of the expression of mtDNA-encoded MRC proteins within mitochondria. However, the underlying molecular mechanisms are only partially understood. Other mitochondrial transcription factors, particularly Tfam, TFBIM and TFBIIM and several nuclear factors, such as AP-1, Sp1, p53, NF-κB and high mobility group proteins present with in mitochondria, might be also involved. Further studies are needed to fully elucidate the molecular mechanisms by which E2 and ERs in association with these mentioned proteins in regulation of the transcription of mtDNA within mitochondria. In addition, little is known about the role of E2 and ERs in regulation of mitochondrial protein translation and MRC functions. These deficiencies merit future studies.
8.2.5. Understanding the Physiological, Pathological and Pharmacological Implications of E2/ER-mediated Pathway
One of the most important aspects in this area is to delineate the precise physiological and pathological implications of over-stimulation or deficiency of this pathway in breast cancer, neurodegenerative and cardiovascular diseases. Further studies are needed to determine whether over-stimulation of this pathway in human breast cells is causally related to the development of breast cancer. Specifically, studies are needed to investigate the E2/ER-induced changes in this pathway during initiation, promotion and progression of cancinogenesis, invasion, tumoregenesis and metastasis of breast cancer. Such studies would be valuable for development of new intervention approaches to the treatment of breast cancers that would target this pathway. Further studies to investigate the role of this pathway in detoxification of Tau and β-amyloid toxicity in AD and in regulation of the Pink-1-parkin-DJ-1-α-synuclein pathway in AD could also be valuable and highly beneficial for the development of therapeutic treatment for these neuro-degenerative diseases that utilize these target pathways.
Gaining a more complete understanding of E2/ER-meatdied pathways will lead to new insights into the underlying molecular mechanisms. Targeting these pathways characterizes a fresh conceptual approach that will contribute to innovative regimens for prevention or treatment of a wide variety of medical complications.
Table 3.
nDNA-Encoded TOMs Involved Mitochondrial Protein Import Machinery
Acknpwledgements
JQC was graceful to Mrs. Ping He of Johns Hopkins School of Medicine for her personal support and encouragement in writing this work. JDY acknowledged NIH grants CA77550 and CA36701
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hewitt SC, Korach KS. Estrogen receptors: structure, mechanisms and function. Rev Endocr Metab Disord. 2002;3:193–200. doi: 10.1023/a:1020068224909. [DOI] [PubMed] [Google Scholar]
- 2.Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- 3.Levin ER. Rapid signaling by steroid receptors. Am J Physiol Regul Integr Comp Physiol. 2008;295:R1425–R1430. doi: 10.1152/ajpregu.90605.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levin ER, Pietras RJ. Estrogen receptors outside the nucleus in breast cancer. Breast Cancer Res Treat. 2008;108:351–361. doi: 10.1007/s10549-007-9618-4. [DOI] [PubMed] [Google Scholar]
- 5.Song RX, Fan P, Yue W, Chen Y, Santen RJ. Role of receptor complexes in the extranuclear actions of estrogen receptor alpha in breast cancer. Endocr Relat Cancer. 2006;13 Suppl 1:S3–S13. doi: 10.1677/erc.1.01322. [DOI] [PubMed] [Google Scholar]
- 6.Song RX, Santen RJ. Membrane initiated estrogen signaling in breast cancer. Biol Reprod. 2006;75:9–16. doi: 10.1095/biolreprod.105.050070. [DOI] [PubMed] [Google Scholar]
- 7.Fu XD, Simoncini T. Extra-nuclear signaling of estrogen receptors. IUBMB Life. 2008;60:502–510. doi: 10.1002/iub.80. [DOI] [PubMed] [Google Scholar]
- 8.Levin ER. Integration of the extranuclear and nuclear actions of estrogen. Mol Endocrinol. 2005;19:1951–1959. doi: 10.1210/me.2004-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belogrudov G, Hatefi Y. Catalytic sector of complex I (NADH:ubiquinone oxidoreductase): subunit stoichiometry and substrate-induced conformation changes. Biochemistry. 1994;33:4571–4576. doi: 10.1021/bi00181a018. [DOI] [PubMed] [Google Scholar]
- 10.Belogrudov GI, Hatefi Y. Intersubunit interactions in the bovine mitochondrial complex I as revealed by ligand blotting. Biochem Biophys Res Commun. 1996;227:135–139. doi: 10.1006/bbrc.1996.1479. [DOI] [PubMed] [Google Scholar]
- 11.Kowalczyk JE, Zablocka B. Protein kinases in mitochondria. Postepy Biochem. 2008;54:209–216. [PubMed] [Google Scholar]
- 12.Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol Cell Biol. 2007;27:4228–4237. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai Y, Attardi G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 1998;17:4848–4858. doi: 10.1093/emboj/17.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunning CJ, McKenzie M, Sugiana C, Lazarou M, Silke J, Connelly A, Fletcher JM, Kirby DM, Thorburn DR, Ryan MT. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 2007;26:3227–3237. doi: 10.1038/sj.emboj.7601748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogel RO, Janssen RJ, Ugalde C, Grovenstein M, Huijbens RJ, Visch HJ, van den Heuvel LP, Willems PH, Zeviani M, Smeitink JA, Nijtmans LG. Human mitochondrial complex I assembly is mediated by NDUFAF1. FEBS J. 2005;272:5317–5326. doi: 10.1111/j.1742-4658.2005.04928.x. [DOI] [PubMed] [Google Scholar]
- 16.Sugiana C, Pagliarini DJ, McKenzie M, Kirby DM, Salemi R, Abu-Amero KK, Dahl HH, Hutchison WM, Vascotto KA, Smith SM, Newbold RF, Christodoulou J, Calvo S, Mootha VK, Ryan MT, Thorburn DR. Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am J Hum Genet. 2008;83:468–478. doi: 10.1016/j.ajhg.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saada A, Edvardson S, Rapoport M, Shaag A, Amry K, Miller C, Lorberboum-Galski H, Elpeleg O. C6ORF66 is an assembly factor of mitochondrial complex I. Am J Hum Genet. 2008;82:32–38. doi: 10.1016/j.ajhg.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbehti-Green A, Au HC, Mascarello JT, Ream-Robinson D, Scheffler IE. Characterization of the human SDHC gene encoding of the integral membrane proteins of succinate-quinone oxidoreductase in mitochondria. Gene. 1998;213:133–140. doi: 10.1016/s0378-1119(98)00186-3. [DOI] [PubMed] [Google Scholar]
- 19.Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT. Loss of the SdhB, but Not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol Cell Biol. 2008;28:718–731. doi: 10.1128/MCB.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badenhop RF, Jansen JC, Fagan PA, Lord RS, Wang ZG, Foster WJ, Schofield PR. The prevalence of SDHB, SDHC, and SDHD mutations in patients with head and neck paraganglioma and association of mutations with clinical features. J Med Genet. 2004;41:e99. doi: 10.1136/jmg.2003.011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiavi F, Boedeker CC, Bausch B, Peczkowska M, Gomez CF, Strassburg T, Pawlu C, Buchta M, Salzmann M, Hoffmann MM, Berlis A, Brink I, Cybulla M, Muresan M, Walter MA, Forrer F, Valimaki M, Kawecki A, Szutkowski Z, Schipper J, Walz MK, Pigny P, Bauters C, Willet-Brozick JE, Baysal BE, Januszewicz A, Eng C, Opocher G, Neumann HP. Predictors and prevalence of paraganglioma syndrome associated with mutations of the SDHC gene. JAMA. 2005;294:2057–2063. doi: 10.1001/jama.294.16.2057. [DOI] [PubMed] [Google Scholar]
- 22.Neumann HP, Pawlu C, Peczkowska M, Bausch B, McWhinney SR, Muresan M, Buchta M, Franke G, Klisch J, Bley TA, Hoegerle S, Boedeker CC, Opocher G, Schipper J, Januszewicz A, Eng C. Distinct clinical features of paraganglioma syndromes associated with SDHB and SDHD gene mutations. JAMA. 2004;292:943–951. doi: 10.1001/jama.292.8.943. [DOI] [PubMed] [Google Scholar]
- 23.Zara V, Conte L, Trumpower BL. Biogenesis of the yeast cytochrome bc(1) complex. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbamcr.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Schagger H, de Coo R, Bauer MF, Hofmann S, Godinot C, Brandt U. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J Biol Chem. 2004;279:36349–36353. doi: 10.1074/jbc.M404033200. [DOI] [PubMed] [Google Scholar]
- 25.Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C, Moraes CT, Enriquez JA. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell. 2004;13:805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez-Vizarra E, Bugiani M, Goffrini P, Carrara F, Farina L, Procopio E, Donati A, Uziel G, Ferrero I, Zeviani M. Impaired complex III assembly associated with BCS1L gene mutations in isolated mitochondrial encephalopathy. Hum Mol Genet. 2007;16:1241–1252. doi: 10.1093/hmg/ddm072. [DOI] [PubMed] [Google Scholar]
- 27.Hinson JT, Fantin VR, Schonberger J, Breivik N, Siem G, McDonough B, Sharma P, Keogh I, Godinho R, Santos F, Esparza A, Nicolau Y, Selvaag E, Cohen BH, Hoppel CL, Tranebjaerg L, Eavey RD, Seidman JG, Seidman CE. Missense mutations in the BCS1L gene as a cause of the Bjornstad syndrome. N Engl J Med. 2007;356:809–819. doi: 10.1056/NEJMoa055262. [DOI] [PubMed] [Google Scholar]
- 28.Gil-Borlado MC, Gonzalez-Hoyuela M, Blazquez A, Garcia-Silva MT, Gabaldon T, Manzanares J, Vara J, Martin MA, Seneca S, Arenas J, Ugalde C. Pathogenic mutations in the 5' untranslated region of BCS1L mRNA in mitochondrial complex III deficiency. Mitochondrion. 2009 doi: 10.1016/j.mito.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Fontanesi F, Soto IC, Barrientos A. Cytochrome c oxidase biogenesis: new levels of regulation. IUBMB Life. 2008;60:557–568. doi: 10.1002/iub.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fontanesi F, Soto IC, Horn D, Barrientos A , Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am J Physiol Cell Physiol. 2006;291:C1129–C1147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- 31.Banci L, Bertini I, Ciofi-Baffoni S, Hadjiloi T, Martinelli M, Palumaa P. Mitochondrial copper(I) transfer from Cox17 to Sco1 is coupled to electron transfer. Proc Natl Acad Sci U S A. 2008;105:6803–6808. doi: 10.1073/pnas.0800019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rigby K, Cobine PA, Khalimonchuk O, Winge DR. Mapping the functional interaction of Sco1 and Cox2 in cytochrome oxidase biogenesis. J Biol Chem. 2008;283:15015–15022. doi: 10.1074/jbc.M710072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaksch M, Paret C, Stucka R, Horn N, Muller-Hocker J, Horvath R, Trepesch N, Stecker G, Freisinger P, Thirion C, Muller J, Lunkwitz R, Rodel G, Shoubridge EA, Lochmuller H. Cytochrome c oxidase deficiency due to mutations in SCO2, encoding a mitochondrial copper-binding protein, is rescued by copper in human myoblasts. Hum Mol Genet. 2001;10:3025–3035. doi: 10.1093/hmg/10.26.3025. [DOI] [PubMed] [Google Scholar]
- 34.Jaksch M, Horvath R, Horn N, Auer DP, Macmillan C, Peters J, Gerbitz KD, Kraegeloh-Mann I, Muntau A, Karcagi V, Kalmanchey R, Lochmuller H, Shoubridge EA, Freisinger P. Homozygosity (E140K) in SCO2 causes delayed infantile onset of cardiomyopathy and neuropathy. Neurology. 2001;57:1440–1446. doi: 10.1212/wnl.57.8.1440. [DOI] [PubMed] [Google Scholar]
- 35.Tay SK, Shanske S, Kaplan P, DiMauro S. Association of mutations in SCO2, a cytochrome c oxidase assembly gene, with early fetal lethality. Arch Neurol. 2004;61:950–952. doi: 10.1001/archneur.61.6.950. [DOI] [PubMed] [Google Scholar]
- 36.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 37.Smith D, Gray J, Mitchell L, Antholine WE, Hosler JP. Assembly of cytochrome-c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. J Biol Chem. 2005;280:17652–17656. doi: 10.1074/jbc.C500061200. [DOI] [PubMed] [Google Scholar]
- 38.Bundschuh FA, Hoffmeier K, Ludwig B. Two variants of the assembly factor Surf1 target specific terminal oxidases in Paracoccus denitrificans. Biochim Biophys Acta. 2008;1777:1336–1343. doi: 10.1016/j.bbabio.2008.05.448. [DOI] [PubMed] [Google Scholar]
- 39.Piekutowska-Abramczuk D, Popowska E, Pronicki M, Karczmarewicz E, Tylek-Lemanska D, Sykut-Cegielska J, Szymanska-Dembinska T, Bielecka L, Krajewska-Walasek M, Pronicka E. High prevalence of SURF1 c.845_846delCT mutation in Polish Leigh patients. Eur J Paediatr Neurol. 2008 doi: 10.1016/j.ejpn.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 40.Bonnefoy N, Fiumera HL, Dujardin G, Fox TD. Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim Biophys Acta. 2009;1793:60–70. doi: 10.1016/j.bbamcr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Meirleir L, Seneca S, Lissens W, De Clercq I, Eyskens F, Gerlo E, Smet J, Van Coster R. Respiratory chain complex V deficiency due to a mutation in the assembly gene ATP12. J Med Genet. 2004;41:120–124. doi: 10.1136/jmg.2003.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bisetto E, Picotti P, Giorgio V, Alverdi V, Mavelli I, Lippe G. Functional and stoichiometric analysis of subunit e in bovine heart mitochondrial F(0)F (1)ATP synthase. J Bioenerg Biomembr. 2008 doi: 10.1007/s10863-008-9183-5. [DOI] [PubMed] [Google Scholar]
- 43.Cizkova A, Stranecky V, Mayr JA, Tesarova M, Havlickova V, Paul J, Ivanek R, Kuss AW, Hansikova H, Kaplanova V, Vrbacky M, Hartmannova H, Noskova L, Honzik T, Drahota Z, Magner M, Hejzlarova K, Sperl W, Zeman J, Houstek J, Kmoch S. TMEM70 mutations cause isolated ATP synthase deficiency and neonatal mitochondrial encephalocardiomyopathy. Nat Genet. 2008;40:1288–1290. doi: 10.1038/ng.246. [DOI] [PubMed] [Google Scholar]
- 44.Scheller K, Sekeris CE. The effects of steroid hormones on the transcription of genes encoding enzymes of oxidative phosphorylation. Exp Physiol. 2003;88:129–140. doi: 10.1113/eph8802507. [DOI] [PubMed] [Google Scholar]
- 45.Psarra AM, Solakidi S, Sekeris CE. The mitochondrion as a primary site of action of steroid and thyroid hormones: presence and action of steroid and thyroid hormone receptors in mitochondria of animal cells. Mol Cell Endocrinol. 2006;246:21–33. doi: 10.1016/j.mce.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 46.Chen JQ, Yager JD, Russo J. Regulation of mitochondrial respiratory chain structure and function by estrogens/estrogen receptors and potential physiological/pathophysiological implications. Biochim Biophys Acta. 2005;1746:1–17. doi: 10.1016/j.bbamcr.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Psarra AM, Sekeris CE. Steroid and thyroid hormone receptors in mitochondria. IUBMB Life. 2008;60:210–223. doi: 10.1002/iub.37. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Gokhale M, Li Y, Trush MA, Yager JD. Enhanced levels of several mitochondrial mRNA transcripts and mitochondrial superoxide production during ethinyl estradiol-induced hepatocarcinogenesis and after estrogen treatment of HepG2 cells. Carcinogenesis. 1998;19:2187–2193. doi: 10.1093/carcin/19.12.2187. [DOI] [PubMed] [Google Scholar]
- 49.Chen J, Li Y, Lavigne JA, Trush MA, Yager JD. Increased mitochondrial superoxide production in rat liver mitochondria, rat hepatocytes, and HepG2 cells following ethinyl estradiol treatment. Toxicol Sci. 1999;51:224–235. doi: 10.1093/toxsci/51.2.224. [DOI] [PubMed] [Google Scholar]
- 50.Monje P, Boland R. Subcellular distribution of native estrogen receptor alpha and beta isoforms in rabbit uterus and ovary. J Cell Biochem. 2001;82:467–479. doi: 10.1002/jcb.1182. [DOI] [PubMed] [Google Scholar]
- 51.Chen JQ, Delannoy M, Cooke C, Yager JD. Mitochondrial localization of ERalpha and ERbeta in human MCF7 cells. Am J Physiol Endocrinol Metab. 2004;286:E1011–E1022. doi: 10.1152/ajpendo.00508.2003. [DOI] [PubMed] [Google Scholar]
- 52.Chen JQ, Eshete M, Alworth WL, Yager JD. Binding of MCF-7 cell mitochondrial proteins and recombinant human estrogen receptors alpha and beta to human mitochondrial DNA estrogen response elements. J Cell Biochem. 2004;93:358–373. doi: 10.1002/jcb.20178. [DOI] [PubMed] [Google Scholar]
- 53.Chen JQ, Yager JD. Estrogen's effects on mitochondrial gene expression: mechanisms and potential contributions to estrogen carcinogenesis. Ann N Y Acad Sci. 2004;1028:258–272. doi: 10.1196/annals.1322.030. [DOI] [PubMed] [Google Scholar]
- 54.Pedram A, Razandi M, Wallace DC, Levin ER. Functional estrogen receptors in the mitochondria of breast cancer cells. Mol Biol Cell. 2006;17:2125–2137. doi: 10.1091/mbc.E05-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen JQ, Russo PA, Cooke C, Russo IH, Russo J. ERbeta shifts from mitochondria to nucleus during estrogen-induced neoplastic transformation of human breast epithelial cells and is involved in estrogen-induced synthesis of mitochondrial respiratory chain proteins. Biochim Biophys Acta. 2007;1773:1732–1746. doi: 10.1016/j.bbamcr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 56.Yang SH, Liu R, Perez EJ, Wen Y, Stevens SM, Jr, Valencia T, Brun-Zinkernagel AM, Prokai L, Will Y, Dykens J, Koulen P, Simpkins JW. Mitochondrial localization of estrogen receptor beta. Proc Natl Acad Sci U S A. 2004;101:4130–4135. doi: 10.1073/pnas.0306948101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cammarata PR, Chu S, Moor A, Wang Z, Yang SH, Simpkins JW. Subcellular distribution of native estrogen receptor alpha and beta subtypes in cultured human lens epithelial cells. Exp Eye Res. 2004;78:861–871. doi: 10.1016/j.exer.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 58.Cammarata PR, Flynn J, Gottipati S, Chu S, Dimitrijevich S, Younes M, Skliris G, Murphy LC. Differential expression and comparative subcellular localization of estrogen receptor beta isoforms in virally transformed and normal cultured human lens epithelial cells. Exp Eye Res. 2005;81:165–175. doi: 10.1016/j.exer.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 59.Flynn JM, Dimitrijevich SD, Younes M, Skliris G, Murphy LC, Cammarata PR. Role of wild-type estrogen receptor-beta in mitochondrial cytoprotection of cultured normal male and female human lens epithelial cells. Am J Physiol Endocrinol Metab. 2008;295:E637–E647. doi: 10.1152/ajpendo.90407.2008. [DOI] [PubMed] [Google Scholar]
- 60.Solakidi S, Psarra AM, Sekeris CE. Differential subcellular distribution of estrogen receptor isoforms: localization of ERalpha in the nucleoli and ERbeta in the mitochondria of human osteosarcoma SaOS-2 and hepatocarcinoma HepG2 cell lines. Biochim Biophys Acta. 2005;1745:382–392. doi: 10.1016/j.bbamcr.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 61.Solakidi S, Psarra AM, Sekeris CE. Differential distribution of glucocorticoid and estrogen receptor isoforms: localization of GRbeta and ERalpha in nucleoli and GRalpha and ERbeta in the mitochondria of human osteosarcoma SaOS-2 and hepatocarcinoma HepG2 cell lines. J Musculoskelet Neuronal Interact. 2007;7:240–245. [PubMed] [Google Scholar]
- 62.Solakidi S, Psarra AM, Nikolaropoulos S, Sekeris CE. Estrogen receptors alpha and beta (ERalpha and ERbeta) and androgen receptor (AR) in human sperm: localization of ERbeta and AR in mitochondria of the midpiece. Hum Reprod. 2005;20:3481–3487. doi: 10.1093/humrep/dei267. [DOI] [PubMed] [Google Scholar]
- 63.Jonsson D, Nilsson J, Odenlund M, Bratthall G, Broman J, Ekblad E, Lydrup ML, Nilsson BO. Demonstration of mitochondrial oestrogen receptor beta and oestrogen-induced attenuation of cytochrome c oxidase subunit I expression in human periodontal ligament cells. Arch Oral Biol. 2007;52:669–676. doi: 10.1016/j.archoralbio.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 64.Milner TA, Ayoola K, Drake CT, Herrick SP, Tabori NE, McEwen BS, Warrier S, Alves SE. Ultrastructural localization of estrogen receptor beta immunoreactivity in the rat hippocampal formation. J Comp Neurol. 2005;491:81–95. doi: 10.1002/cne.20724. [DOI] [PubMed] [Google Scholar]
- 65.Milner TA, Lubbers LS, Alves SE, McEwen BS. Nuclear and extranuclear estrogen binding sites in the rat forebrain and autonomic medullary areas. Endocrinology. 2008;149:3306–3312. doi: 10.1210/en.2008-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simpkins JW, Yang SH, Sarkar SN, Pearce V. Estrogen actions on mitochondria--physiological and pathological implications. Mol Cell Endocrinol. 2008;290:51–59. doi: 10.1016/j.mce.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stirone C, Duckles SP, Krause DN, Procaccio V. Estrogen increases mitochondrial efficiency and reduces oxidative stress in cerebral blood vessels. Mol Pharmacol. 2005;68:959–965. doi: 10.1124/mol.105.014662. [DOI] [PubMed] [Google Scholar]
- 68.Monje P, Zanello S, Holick M, Boland R. Differential cellular localization of estrogen receptor alpha in uterine and mammary cells. Mol Cell Endocrinol. 2001;181:117–129. doi: 10.1016/s0303-7207(01)00526-3. [DOI] [PubMed] [Google Scholar]
- 69.Milanesi L, Russo de Boland A, Boland R. Expression and localization of estrogen receptor alpha in the C2C12 murine skeletal muscle cell line. J Cell Biochem. 2008;104:1254–1273. doi: 10.1002/jcb.21706. [DOI] [PubMed] [Google Scholar]
- 70.Bettini E, Maggi A. Estrogen induction of cytochrome c oxidase subunit III in rat hippocampus. J Neurochem. 1992;58:1923–1929. doi: 10.1111/j.1471-4159.1992.tb10070.x. [DOI] [PubMed] [Google Scholar]
- 71.Van Itallie CM, Dannies PS. Estrogen induces accumulation of the mitochondrial ribonucleic acid for subunit II of cytochrome oxidase in pituitary tumor cells. Mol Endocrinol. 1988;2:332–337. doi: 10.1210/mend-2-4-332. [DOI] [PubMed] [Google Scholar]
- 72.Chen J, Delannoy M, Odwin S, He P, Trush MA, Yager JD. Enhanced mitochondrial gene transcript, ATP, bcl-2 protein levels, and altered glutathione distribution in ethinyl estradiol-treated cultured female rat hepatocytes. Toxicol Sci. 2003;75:271–278. doi: 10.1093/toxsci/kfg183. [DOI] [PubMed] [Google Scholar]
- 73.Felty Q, Roy D. Estrogen, mitochondria, and growth of cancer and non-cancer cells. J Carcinog. 2005;4:1. doi: 10.1186/1477-3163-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen JQ, Brown TR, Russo J. Regulation of energy metabolism pathways by estrogens and estrogenic chemicals and potential implications in obesity associated with increased exposure to endocrine disruptors. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbamcr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Irwin RW, Yao J, Hamilton RT, Cadenas E, Brinton RD, Nilsen J. Progesterone and estrogen regulate oxidative metabolism in brain mitochondria. Endocrinology. 2008;149:3167–3175. doi: 10.1210/en.2007-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsieh YC, Yu HP, Suzuki T, Choudhry MA, Schwacha MG, Bland KI, Chaudry IH. Upregulation of mitochondrial respiratory complex IV by estrogen receptor-beta is critical for inhibiting mitochondrial apoptotic signaling and restoring cardiac functions following trauma-hemorrhage. J Mol Cell Cardiol. 2006;41:511–521. doi: 10.1016/j.yjmcc.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 77.Nilsen J, Irwin RW, Gallaher TK, Brinton RD. Estradiol in vivo regulation of brain mitochondrial proteome. J Neurosci. 2007;27:14069–14077. doi: 10.1523/JNEUROSCI.4391-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hsieh YC, Yang S, Choudhry MA, Yu HP, Rue LW, 3rd, Bland KI, Chaudry IH. PGC-1 upregulation via estrogen receptors: a common mechanism of salutary effects of estrogen and flutamide on heart function after trauma-hemorrhage. Am J Physiol Heart Circ Physiol. 2005;289:H2665–H2672. doi: 10.1152/ajpheart.00682.2005. [DOI] [PubMed] [Google Scholar]
- 79.Mattingly KA, Ivanova MM, Riggs KA, Wickramasinghe NS, Barch MJ, Klinge CM. Estradiol stimulates transcription of nuclear respiratory factor-1 and increases mitochondrial biogenesis. Mol Endocrinol. 2008;22:609–622. doi: 10.1210/me.2007-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kang D, Kim SH, Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion. 2007;7:39–44. doi: 10.1016/j.mito.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 82.Ikeuchi M, Matsusaka H, Kang D, Matsushima S, Ide T, Kubota T, Fujiwara T, Hamasaki N, Takeshita A, Sunagawa K, Tsutsui H. Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 2005;112:683–690. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- 83.Wang J, Silva JP, Gustafsson CM, Rustin P, Larsson NG. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc Natl Acad Sci U S A. 2001;98:4038–4043. doi: 10.1073/pnas.061038798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoshida Y, Izumi H, Ise T, Uramoto H, Torigoe T, Ishiguchi H, Murakami T, Tanabe M, Nakayama Y, Itoh H, Kasai H, Kohno K. Human mitochondrial transcription factor A binds preferentially to oxidatively damaged DNA. Biochem Biophys Res Commun. 2002;295:945–951. doi: 10.1016/s0006-291x(02)00757-x. [DOI] [PubMed] [Google Scholar]
- 86.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 87.Maniura-Weber K, Goffart S, Garstka HL, Montoya J, Wiesner RJ. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gerard MA, Krol A, Carbon P. Transcription factor hStaf/ZNF143 is required for expression of the human TFAM gene. Gene. 2007;401:145–153. doi: 10.1016/j.gene.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 89.Choi YS, Lee HK, Pak YK. Characterization of the 5'flanking region of the rat gene for mitochondrial transcription factor A (Tfam) Biochim Biophys Acta. 2002;1574:200–204. doi: 10.1016/s0167-4781(01)00361-x. [DOI] [PubMed] [Google Scholar]
- 90.Dong X, Ghoshal K, Majumder S, Yadav SP, Jacob ST. Mitochondrial transcription factor A and its downstream targets are up-regulated in a rat hepatoma. J Biol Chem. 2002;277:43309–43318. doi: 10.1074/jbc.M206958200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Hsieh YC, Frink M, Thobe BM, Hsu JT, Choudhry MA, Schwacha MG, Bland KI, Chaudry IH. 17Beta-estradiol downregulates Kupffer cell TLR4-dependent p38 MAPK pathway and normalizes inflammatory cytokine production following trauma-hemorrhage. Mol Immunol. 2007;44:2165–2172. doi: 10.1016/j.molimm.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rodriguez-Cuenca S, Monjo M, Gianotti M, Proenza AM, Roca P. Expression of mitochondrial biogenesis-signaling factors in brown adipocytes is influenced specifically by 17beta-estradiol, testosterone, and progesterone. Am J Physiol Endocrinol Metab. 2007;292:E340–E346. doi: 10.1152/ajpendo.00175.2006. [DOI] [PubMed] [Google Scholar]
- 93.Wu F, Ivanov I, Xu R, Safe S. Role of Sp Transcription Factors in Hormone-Dependent Modulation of Genes in MCF-7 Breast Cancer Cells: Microarray and RNA Interference Studies. J Mol Endocrinol. 2008 doi: 10.1677/JME-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Safe S, Kim K. Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. J Mol Endocrinol. 2008;41:263–275. doi: 10.1677/JME-08-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee DY, Clayton DA. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J Biol Chem. 1998;273:30614–30621. doi: 10.1074/jbc.273.46.30614. [DOI] [PubMed] [Google Scholar]
- 96.Wanrooij S, Fuste JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc Natl Acad Sci U S A. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martin AN, Li Y. RNase MRP RNA and human genetic diseases. Cell Res. 2007;17:219–226. doi: 10.1038/sj.cr.7310120. [DOI] [PubMed] [Google Scholar]
- 98.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fish J, Raule N, Attardi G. Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science. 2004;306:2098–2101. doi: 10.1126/science.1102077. [DOI] [PubMed] [Google Scholar]
- 100.Evans MJ, Scarpulla RC. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990;4:1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- 101.Champagne AM, Dufresne C, Viney L, Gueride M. Cloning, sequencing and expression of the two genes encoding the mitochondrial single-stranded DNA-binding protein in Xenopus laevis. Gene. 1997;184:65–71. doi: 10.1016/s0378-1119(96)00574-4. [DOI] [PubMed] [Google Scholar]
- 102.Minczuk M, Lilpop J, Boros J, Stepien PP. The 5' region of the human hSUV3 gene encoding mitochondrial DNA and RNA helicase: promoter characterization and alternative pre-mRNA splicing. Biochim Biophys Acta. 2005;1729:81–87. doi: 10.1016/j.bbaexp.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 103.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 104.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Asin-Cayuela J, Gustafsson CM. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem Sci. 2007;32:111–117. doi: 10.1016/j.tibs.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 106.Nozaki Y, Matsunaga N, Ishizawa T, Ueda T, Takeuchi N. HMRF1L is a human mitochondrial translation release factor involved in the decoding of the termination codons UAA and UAG. Genes Cells. 2008;13:429–438. doi: 10.1111/j.1365-2443.2008.01181.x. [DOI] [PubMed] [Google Scholar]
- 107.Ishizawa T, Nozaki Y, Ueda T, Takeuchi N. The human mitochondrial translation release factor HMRF1L is methylated in the GGQ motif by the methyltransferase HMPrmC. Biochem Biophys Res Commun. 2008;373:99–103. doi: 10.1016/j.bbrc.2008.05.176. [DOI] [PubMed] [Google Scholar]
- 108.Wiley DJ, Catanuto P, Fontanesi F, Rios C, Sanchez N, Barrientos A, Verde F. Bot1p is required for mitochondrial translation, respiratory function, and normal cell morphology in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2008;7:619–629. doi: 10.1128/EC.00048-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Overman RG, Jr, Enderle PJ, Farrow JM, 3rd, Wiley JE, Farwell MA. The human mitochondrial translation initiation factor 2 gene (MTIF2): transcriptional analysis and identification of a pseudogene. Biochim Biophys Acta. 2003;1628:195–205. doi: 10.1016/s0167-4781(03)00144-1. [DOI] [PubMed] [Google Scholar]
- 110.Hayashi R, Ueda T, Farwell MA, Takeuchi N. Nuclear respiratory factor 2 activates transcription of human mitochondrial translation initiation factor 2 gene. Mitochondrion. 2007;7:195–203. doi: 10.1016/j.mito.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 111.Sekeris CE. The mitochondrial genome: a possible primary site of action of steroid hormones. In Vivo. 1990;4:317–320. [PubMed] [Google Scholar]
- 112.Demonacos C, Djordjevic-Markovic R, Tsawdaroglou N, Sekeris CE. The mitochondrion as a primary site of action of glucocorticoids: the interaction of the glucocorticoid receptor with mitochondrial DNA sequences showing partial similarity to the nuclear glucocorticoid responsive elements. J Steroid Biochem Mol Biol. 1995;55:43–55. doi: 10.1016/0960-0760(95)00159-w. [DOI] [PubMed] [Google Scholar]
- 113.Chen JQ, Brown TR, Yager JD. Mechanisms of hormone carcinogenesis: evolution of views, role of mitochondria. Adv Exp Med Biol. 2008;630:1–18. [PubMed] [Google Scholar]
- 114.Chen J, Schwartz DA, Young TA, Norris JS, Yager JD. Identification of genes whose expression is altered during mitosuppression in livers of ethinyl estradiol-treated female rats. Carcinogenesis. 1996;17:2783–2786. doi: 10.1093/carcin/17.12.2783. [DOI] [PubMed] [Google Scholar]
- 115.Thompson CJ, Tam NN, Joyce JM, Leav I, Ho SM. Gene expression profiling of testosterone and estradiol-17 beta-induced prostatic dysplasia in Noble rats and response to the antiestrogen ICI 182,780. Endocrinology. 2002;143:2093–2105. doi: 10.1210/endo.143.6.8846. [DOI] [PubMed] [Google Scholar]
- 116.O'Lone R, Knorr K, Jaffe IZ, Schaffer ME, Martini PG, Karas RH, Bienkowska J, Mendelsohn ME, Hansen U. Estrogen receptors alpha and beta mediate distinct pathways of vascular gene expression, including genes involved in mitochondrial electron transport and generation of reactive oxygen species. Mol Endocrinol. 2007;21:1281–1296. doi: 10.1210/me.2006-0497. [DOI] [PubMed] [Google Scholar]
- 117.Araujo GW, Beyer C, Arnold S. Oestrogen influences on mitochondrial gene expression and respiratory chain activity in cortical and mesencephalic astrocytes. J Neuroendocrinol. 2008;20:930–941. doi: 10.1111/j.1365-2826.2008.01747.x. [DOI] [PubMed] [Google Scholar]
- 118.Razmara A, Sunday L, Stirone C, Wang XB, Krause DN, Duckles SP, Procaccio V. Mitochondrial effects of estrogen are mediated by estrogen receptor alpha in brain endothelial cells. J Pharmacol Exp Ther. 2008;325:782–790. doi: 10.1124/jpet.107.134072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Klinge CM. Estrogenic control of mitochondrial function and biogenesis. J Cell Biochem. 2008 doi: 10.1002/jcb.21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Au HC, Scheffler IE. Promoter analysis of the human succinate dehydrogenase iron-protein gene--both nuclear respiratory factors NRF-1 and NRF-2 are required. Eur J Biochem. 1998;251:164–174. doi: 10.1046/j.1432-1327.1998.2510164.x. [DOI] [PubMed] [Google Scholar]
- 121.Hirawake H, Taniwaki M, Tamura A, Amino H, Tomitsuka E, Kita K. Characterization of the human SDHD gene encoding the small subunit of cytochrome b (cybS) in mitochondrial succinate-ubiquinone oxidoreductase. Biochim Biophys Acta. 1999;1412:295–300. doi: 10.1016/s0005-2728(99)00071-7. [DOI] [PubMed] [Google Scholar]
- 122.Piantadosi CA, Suliman HB. Transcriptional Regulation of SDHa flavoprotein by nuclear respiratory factor-1 prevents pseudo-hypoxia in aerobic cardiac cells. J Biol Chem. 2008;283:10967–10977. doi: 10.1074/jbc.M709741200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Virbasius JV, Scarpulla RC. Transcriptional activation through ETS domain binding sites in the cytochrome c oxidase subunit IV gene. Mol Cell Biol. 1991;11:5631–5638. doi: 10.1128/mcb.11.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wong-Riley M, Guo A, Bachman NJ, Lomax MI. Human COX6A1 gene: promoter analysis, cDNA isolation and expression in the monkey brain. Gene. 2000;247:63–75. doi: 10.1016/s0378-1119(00)00121-9. [DOI] [PubMed] [Google Scholar]
- 125.Smith EO, Lomax MI. Structural organization of the bovine gene for the heart/muscle isoform of cytochrome c oxidase subunit VIa. Biochim Biophys Acta. 1993;1174:63–71. doi: 10.1016/0167-4781(93)90092-r. [DOI] [PubMed] [Google Scholar]
- 126.Bachman NJ, Yang TL, Dasen JS, Ernst RE, Lomax MI. Phylogenetic footprinting of the human cytochrome c oxidase subunit VB promoter. Arch Biochem Biophys. 1996;333:152–162. doi: 10.1006/abbi.1996.0376. [DOI] [PubMed] [Google Scholar]
- 127.Basu A, Park K, Atchison ML, Carter RS, Avadhani NG. Identification of a transcriptional initiator element in the cytochrome c oxidase subunit Vb promoter which binds to transcription factors NF-E1 (YY-1, delta) and Sp1. J Biol Chem. 1993;268:4188–4196. [PubMed] [Google Scholar]
- 128.Takahashi Y, Kako K, Arai H, Ohishi T, Inada Y, Takehara A, Fukamizu A, Munekata E. Characterization and identification of promoter elements in the mouse COX17 gene. Biochim Biophys Acta. 2002;1574:359–364. doi: 10.1016/s0167-4781(01)00374-8. [DOI] [PubMed] [Google Scholar]
- 129.Mick DU, Wagner K, van der Laan M, Frazier AE, Perschil I, Pawlas M, Meyer HE, Warscheid B, Rehling P. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 2007;26:4347–4358. doi: 10.1038/sj.emboj.7601862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 131.Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 132.Scarpulla RC. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene. 2002;286:81–89. doi: 10.1016/s0378-1119(01)00809-5. [DOI] [PubMed] [Google Scholar]
- 133.Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. J Cell Biochem. 2006;97:673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- 134.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 135.Ongwijitwat S, Wong-Riley MT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 136.Dhar SS, Ongwijitwat S, Wong-Riley MT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ramachandran B, Yu G, Gulick T. Nuclear respiratory factor 1 controls myocyte enhancer factor 2A transcription to provide a mechanism for coordinate expression of respiratory chain subunits. J Biol Chem. 2008;283:11935–11946. doi: 10.1074/jbc.M707389200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ramachandran B, Yu G, Li S, Zhu B, Gulick T. Myocyte enhancer factor 2A is transcriptionally autoregulated. J Biol Chem. 2008;283:10318–10329. doi: 10.1074/jbc.M707623200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vercauteren K, Gleyzer N, Scarpulla RC. PGC-1-related coactivator complexes with HCF-1 and NRF-2beta in mediating NRF-2(GABP)-dependent respiratory gene expression. J Biol Chem. 2008;283:12102–12111. doi: 10.1074/jbc.M710150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ben-Shachar D, Karry R. Sp1 expression is disrupted in schizophrenia; a possible mechanism for the abnormal expression of mitochondrial complex I genes, NDUFV1 and NDUFV2. PLoS ONE. 2007;2:e817. doi: 10.1371/journal.pone.0000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.de Sury R, Martinez P, Procaccio V, Lunardi J, Issartel JP. Genomic structure of the human NDUFS8 gene coding for the iron-sulfur TYKY subunit of the mitochondrial NADH:ubiquinone oxidoreductase. Gene. 1998;215:1–10. doi: 10.1016/s0378-1119(98)00275-3. [DOI] [PubMed] [Google Scholar]
- 142.Lescuyer P, Martinez P, Lunardi J. YY1 and Sp1 activate transcription of the human NDUFS8 gene encoding the mitochondrial complex I TYKY subunit. Biochim Biophys Acta. 2002;1574:164–174. doi: 10.1016/s0167-4781(01)00377-3. [DOI] [PubMed] [Google Scholar]
- 143.Schuelke M, Loeffen J, Mariman E, Smeitink J, van den Heuvel L. Cloning of the human mitochondrial 51 kDa subunit (NDUFV1) reveals a 100% antisense homology of its 3'UTR with the 5'UTR of the gamma-interferon inducible protein (IP-30) precursor: is this a link between mitochondrial myopathy and inflammation? Biochem Biophys Res Commun. 1998;245:599–606. doi: 10.1006/bbrc.1998.8486. [DOI] [PubMed] [Google Scholar]
- 144.Chau CM, Evans MJ, Scarpulla RC. Nuclear respiratory factor 1 activation sites in genes encoding the gamma-subunit of ATP synthase, eukaryotic initiation factor 2 alpha, and tyrosine aminotransferase. Specific interaction of purified NRF-1 with multiple target genes. J Biol Chem. 1992;267:6999–7006. [PubMed] [Google Scholar]
- 145.Pfanner N, Geissler A. Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol. 2001;2:339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- 146.Brix J, Dietmeier K, Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J Biol Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- 147.Yano M, Terada K, Mori M. Mitochondrial import receptors Tom20 and Tom22 have chaperone-like activity. J Biol Chem. 2004;279:10808–10813. doi: 10.1074/jbc.M311710200. [DOI] [PubMed] [Google Scholar]
- 148.Yamano K, Yatsukawa Y, Esaki M, Hobbs AE, Jensen RE, Endo T. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J Biol Chem. 2008;283:3799–3807. doi: 10.1074/jbc.M708339200. [DOI] [PubMed] [Google Scholar]
- 149.Blesa JR, Prieto-Ruiz JA, Hernandez JM, Hernandez-Yago J. NRF-2 transcription factor is required for human TOMM20 gene expression. Gene. 2007;391:198–208. doi: 10.1016/j.gene.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 150.Blesa JR, Hernandez JM, Hernandez-Yago J. NRF-2 transcription factor is essential in promoting human Tomm70 gene expression. Mitochondrion. 2004;3:251–259. doi: 10.1016/j.mito.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 151.Blesa JR, Hernandez-Yago J. Distinct functional contributions of 2 GABP-NRF-2 recognition sites within the context of the human TOMM70 promoter. Biochem Cell Biol. 2006;84:813–822. doi: 10.1139/o06-064. [DOI] [PubMed] [Google Scholar]
- 152.Blesa JR, Hegde AA, Hernandez-Yago J. In vitro methylation of nuclear respiratory factor-2 binding sites suppresses the promoter activity of the human TOMM70 gene. Gene. 2008;427:58–64. doi: 10.1016/j.gene.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 153.Blesa JR, Prieto-Ruiz JA, Abraham BA, Harrison BL, Hegde AA, Hernandez-Yago J. NRF-1 is the major transcription factor regulating the expression of the human TOMM34 gene. Biochem Cell Biol. 2008;86:46–56. doi: 10.1139/o07-151. [DOI] [PubMed] [Google Scholar]
- 154.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 155.Kwong JQ, Henning MS, Starkov AA, Manfredi G. The mitochondrial respiratory chain is a modulator of apoptosis. J Cell Biol. 2007;179:1163–1177. doi: 10.1083/jcb.200704059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kandasamy K, Srinivasula SM, Alnemri ES, Thompson CB, Korsmeyer SJ, Bryant JL, Srivastava RK. Involvement of proapoptotic molecules Bax and Bak in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced mitochondrial disruption and apoptosis: differential regulation of cytochrome c and Smac/DIABLO release. Cancer Res. 2003;63:1712–1721. [PubMed] [Google Scholar]
- 157.Shankar S, Srivastava RK. Bax and Bak genes are essential for maximum apoptotic response by curcumin, a polyphenolic compound and cancer chemopreventive agent derived from turmeric. Curcuma longa, Carcinogenesis. 2007;28:1277–1286. doi: 10.1093/carcin/bgm024. [DOI] [PubMed] [Google Scholar]
- 158.Choi S, Singh SV. Bax and Bak are required for apoptosis induction by sulforaphane, a cruciferous vegetable-derived cancer chemopreventive agent. Cancer Res. 2005;65:2035–2043. doi: 10.1158/0008-5472.CAN-04-3616. [DOI] [PubMed] [Google Scholar]
- 159.Eun SY, Woo IS, Jang HS, Jin H, Kim MY, Kim HJ, Lee JH, Chang KC, Kim JH, Seo HG. Identification of cytochrome c oxidase subunit 6A1 as a suppressor of Bax-induced cell death by yeast-based functional screening. Biochem Biophys Res Commun. 2008;373:58–63. doi: 10.1016/j.bbrc.2008.05.178. [DOI] [PubMed] [Google Scholar]
- 160.Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, Robinson JP. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem. 2003;278:8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- 161.Wolvetang EJ, Johnson KL, Krauer K, Ralph SJ, Linnane AW. Mitochondrial respiratory chain inhibitors induce apoptosis. FEBS Lett. 1994;339:40–44. doi: 10.1016/0014-5793(94)80380-3. [DOI] [PubMed] [Google Scholar]
- 162.Pelicano H, Feng L, Zhou Y, Carew JS, Hileman EO, Plunkett W, Keating MJ, Huang P. Inhibition of mitochondrial respiration: a novel strategy to enhance drug-induced apoptosis in human leukemia cells by a reactive oxygen species-mediated mechanism. J Biol Chem. 2003;278:37832–37839. doi: 10.1074/jbc.M301546200. [DOI] [PubMed] [Google Scholar]
- 163.Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci. 2007;120:4155–4166. doi: 10.1242/jcs.011163. [DOI] [PubMed] [Google Scholar]
- 164.Nutt LK, Gogvadze V, Uthaisang W, Mirnikjoo B, McConkey DJ, Orrenius S. Indirect effects of Bax and Bak initiate the mitochondrial alterations that lead to cytochrome c release during arsenic trioxide-induced apoptosis. Cancer Biol Ther. 2005;4:459–467. doi: 10.4161/cbt.4.4.1652. [DOI] [PubMed] [Google Scholar]
- 165.Higuchi M, Proske RJ, Yeh ET. Inhibition of mitochondrial respiratory chain complex I by TNF results in cytochrome c release, membrane permeability transition, and apoptosis. Oncogene. 1998;17:2515–2524. doi: 10.1038/sj.onc.1202485. [DOI] [PubMed] [Google Scholar]
- 166.Garcia-Ruiz C, Colell A, Mari M, Morales A, Fernandez-Checa JC. Direct effect of ceramide on the mitochondrial electron transport chain leads to generation of reactive oxygen species. Role of mitochondrial glutathione. J Biol Chem. 1997;272:11369–11377. doi: 10.1074/jbc.272.17.11369. [DOI] [PubMed] [Google Scholar]
- 167.Napolitano M, Zei D, Centonze D, Palermo R, Bernardi G, Vacca A, Calabresi P, Gulino A. NF-kB/NOS cross-talk induced by mitochondrial complex II inhibition: implications for Huntington's disease. Neurosci Lett. 2008;434:241–246. doi: 10.1016/j.neulet.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 168.Dong LF, Freeman R, Liu J, Zobalova R, Marin-Hernandez A, Stantic M, Rohlena J, Valis K, Rodriguez-Enriquez S, Butcher B, Goodwin J, Brunk UT, Witting PK, Moreno-Sanchez R, Scheffler IE, Ralph SJ, Neuzil J. Suppression of tumor growth in vivo by the mitocan alpha-tocopheryl succinate requires respiratory complex II. Clin Cancer Res. 2009;15:1593–1600. doi: 10.1158/1078-0432.CCR-08-2439. [DOI] [PubMed] [Google Scholar]
- 169.Dong LF, Low P, Dyason JC, Wang XF, Prochazka L, Witting PK, Freeman R, Swettenham E, Valis K, Liu J, Zobalova R, Turanek J, Spitz DR, Domann FE, Scheffler IE, Ralph SJ, Neuzil J. Alpha-tocopheryl succinate induces apoptosis by targeting ubiquinone-binding sites in mitochondrial respiratory complex II. Oncogene. 2008;27:4324–4335. doi: 10.1038/onc.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dong LF, Swettenham E, Eliasson J, Wang XF, Gold M, Medunic Y, Stantic M, Low P, Prochazka L, Witting PK, Turanek J, Akporiaye ET, Ralph SJ, Neuzil J. Vitamin E analogues inhibit angiogenesis by selective induction of apoptosis in proliferating endothelial cells: the role of oxidative stress. Cancer Res. 2007;67:11906–11913. doi: 10.1158/0008-5472.CAN-07-3034. [DOI] [PubMed] [Google Scholar]
- 171.Neuzil J, Dong LF, Ramanathapuram L, Hahn T, Chladova M, Wang XF, Zobalova R, Prochazka L, Gold M, Freeman R, Turanek J, Akporiaye ET, Dyason JC, Ralph SJ. Vitamin E analogues as a novel group of mitocans: anti-cancer agents that act by targeting mitochondria. Mol Aspects Med. 2007;28:607–645. doi: 10.1016/j.mam.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 172.Neuzil J, Dyason JC, Freeman R, Dong LF, Prochazka L, Wang XF, Scheffler I, Ralph SJ. Mitocans as anti-cancer agents targeting mitochondria: lessons from studies with vitamin E analogues, inhibitors of complex II. J Bioenerg Biomembr. 2007;39:65–72. doi: 10.1007/s10863-006-9060-z. [DOI] [PubMed] [Google Scholar]
- 173.Nakamura Y, Kawakami M, Yoshihiro A, Miyoshi N, Ohigashi H, Kawai K, Osawa T, Uchida K. Involvement of the mitochondrial death pathway in chemopreventive benzyl isothiocyanate-induced apoptosis. J Biol Chem. 2002;277:8492–8499. doi: 10.1074/jbc.M109760200. [DOI] [PubMed] [Google Scholar]
- 174.Xiao D, Vogel V, Singh SV. Benzyl isothiocyanate-induced apoptosis in human breast cancer cells is initiated by reactive oxygen species and regulated by Bax and Bak. Mol Cancer Ther. 2006;5:2931–2945. doi: 10.1158/1535-7163.MCT-06-0396. [DOI] [PubMed] [Google Scholar]
- 175.Xiao D, Powolny AA, Singh SV. Benzyl Isothiocyanate Targets Mitochondrial Respiratory Chain to Trigger Reactive Oxygen Species-dependent Apoptosis in Human Breast Cancer Cells. J Biol Chem. 2008;283:30151–30163. doi: 10.1074/jbc.M802529200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Han YH, Kim SH, Kim SZ, Park WH. Antimycin A as a mitochondrial electron transport inhibitor prevents the growth of human lung cancer A549 cells. Oncol Rep. 2008;20:689–693. [PubMed] [Google Scholar]
- 177.Gudz TI, Tserng KY, Hoppel CL. Direct inhibition of mitochondrial respiratory chain complex III by cell-permeable ceramide. J Biol Chem. 1997;272:24154–24158. doi: 10.1074/jbc.272.39.24154. [DOI] [PubMed] [Google Scholar]
- 178.Zhang TH, Liu JF, Zhang Y, Li YL, Lu HT, Murata NM, Yamakawa T. Ceramide induces apoptosis in human lung adenocarcinoma A549 cells through mitogen-activated protein kinases. Acta Pharmacol Sin. 2007;28:439–445. doi: 10.1111/j.1745-7254.2007.00505.x. [DOI] [PubMed] [Google Scholar]
- 179.Gholami A, Kassis R, Real E, Delmas O, Guadagnini S, Larrous F, Obach D, Prevost MC, Jacob Y, Bourhy H. Mitochondrial dysfunction in lyssavirus-induced apoptosis. J Virol. 2008;82:4774–4784. doi: 10.1128/JVI.02651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Suter M, Reme C, Grimm C, Wenzel A, Jaattela M, Esser P, Kociok N, Leist M, Richter C. Age-related macular degeneration. The lipofusion component N-retinyl-N-retinylidene ethanolamine detaches proapoptotic proteins from mitochondria and induces apoptosis in mammalian retinal pigment epithelial cells. J Biol Chem. 2000;275:39625–39630. doi: 10.1074/jbc.M007049200. [DOI] [PubMed] [Google Scholar]
- 181.Shaban H, Gazzotti P, Richter C. Cytochrome c oxidase inhibition by N-retinyl-N-retinylidene ethanolamine, a compound suspected to cause age-related macula degeneration. Arch Biochem Biophys. 2001;394:111–116. doi: 10.1006/abbi.2001.2535. [DOI] [PubMed] [Google Scholar]
- 182.Santamaria G, Martinez-Diez M, Fabregat I, Cuezva JM. Efficient execution of cell death in non-glycolytic cells requires the generation of ROS controlled by the activity of mitochondrial H+-ATP synthase. Carcinogenesis. 2006;27:925–935. doi: 10.1093/carcin/bgi315. [DOI] [PubMed] [Google Scholar]
- 183.Roy A, Ganguly A, BoseDasgupta S, Das BB, Pal C, Jaisankar P, Majumder HK. Mitochondria-dependent reactive oxygen species-mediated programmed cell death induced by 3,3'-diindolylmethane through inhibition of F0F1-ATP synthase in unicellular protozoan parasite Leishmania donovani. Mol Pharmacol. 2008;74:1292–1307. doi: 10.1124/mol.108.050161. [DOI] [PubMed] [Google Scholar]
- 184.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 185.Bernardi P, Krauskopf A, Basso E, Petronilli V, Blachly-Dyson E, Di Lisa F, Forte MA. The mitochondrial permeability transition from in vitro artifact to disease target. FEBS J. 2006;273:2077–2099. doi: 10.1111/j.1742-4658.2006.05213.x. [DOI] [PubMed] [Google Scholar]
- 186.Murphy E, Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Leung AW, Halestrap AP. Recent progress in elucidating the molecular mechanism of the mitochondrial permeability transition pore. Biochim Biophys Acta. 2008;1777:946–952. doi: 10.1016/j.bbabio.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 188.Crow MT, Mani K, Nam YJ, Kitsis RN. The mitochondrial death pathway and cardiac myocyte apoptosis. Circ Res. 2004;95:957–970. doi: 10.1161/01.RES.0000148632.35500.d9. [DOI] [PubMed] [Google Scholar]
- 189.Forte M, Bernardi P. The permeability transition and BCL-2 family proteins in apoptosis: co-conspirators or independent agents? Cell Death Differ. 2006;13:1287–1290. doi: 10.1038/sj.cdd.4401957. [DOI] [PubMed] [Google Scholar]
- 190.Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, Robbins J, Molkentin JD. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 191.Clarke SJ, McStay GP, Halestrap AP. Sanglifehrin A acts as a potent inhibitor of the mitochondrial permeability transition and reperfusion injury of the heart by binding to cyclophilin-D at a different site from cyclosporin A. J Biol Chem. 2002;277:34793–34799. doi: 10.1074/jbc.M202191200. [DOI] [PubMed] [Google Scholar]
- 192.Muramatsu Y, Furuichi Y, Tojo N, Moriguchi A, Maemoto T, Nakada H, Hino M, Matsuoka N. Neuroprotective efficacy of FR901459, a novel derivative of cyclosporin A, in in vitro mitochondrial damage and in vivo transient cerebral ischemia models. Brain Res. 2007;1149:181–190. doi: 10.1016/j.brainres.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 193.Kim JS, Qian T, Lemasters JJ. Mitochondrial permeability transition in the switch from necrotic to apoptotic cell death in ischemic rat hepatocytes. Gastroenterology. 2003;124:494–503. doi: 10.1053/gast.2003.50059. [DOI] [PubMed] [Google Scholar]
- 194.Nakayama H, Chen X, Baines CP, Klevitsky R, Zhang X, Zhang H, Jaleel N, Chua BH, Hewett TE, Robbins J, Houser SR, Molkentin JD. Ca2+- and mitochondrial-dependent cardiomyocyte necrosis as a primary mediator of heart failure. J Clin Invest. 2007;117:2431–2444. doi: 10.1172/JCI31060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Oliveira PJ, Seica R, Coxito PM, Rolo AP, Palmeira CM, Santos MS, Moreno AJ. Enhanced permeability transition explains the reduced calcium uptake in cardiac mitochondria from streptozotocin-induced diabetic rats. FEBS Lett. 2003;554:511–514. doi: 10.1016/s0014-5793(03)01233-x. [DOI] [PubMed] [Google Scholar]
- 196.Millay DP, Sargent MA, Osinska H, Baines CP, Barton ER, Vuagniaux G, Sweeney HL, Robbins J, Molkentin JD. Genetic and pharmacologic inhibition of mitochondrial-dependent necrosis attenuates muscular dystrophy. Nat Med. 2008;14:442–447. doi: 10.1038/nm1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Merlini L, Angelin A, Tiepolo T, Braghetta P, Sabatelli P, Zamparelli A, Ferlini A, Maraldi NM, Bonaldo P, Bernardi P. Cyclosporin A corrects mitochondrial dysfunction and muscle apoptosis in patients with collagen VI myopathies. Proc Natl Acad Sci U S A. 2008;105:5225–5229. doi: 10.1073/pnas.0800962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kerkela R, Grazette L, Yacobi R, Iliescu C, Patten R, Beahm C, Walters B, Shevtsov S, Pesant S, Clubb FJ, Rosenzweig A, Salomon RN, Van Etten RA, Alroy J, Durand JB, Force T. Cardiotoxicity of the cancer therapeutic agent imatinib mesylate. Nat Med. 2006;12:908–916. doi: 10.1038/nm1446. [DOI] [PubMed] [Google Scholar]
- 199.Forte M, Gold BG, Marracci G, Chaudhary P, Basso E, Johnsen D, Yu X, Fowlkes J, Rahder M, Stem K, Bernardi P, Bourdette D. Cyclophilin D inactivation protects axons in experimental autoimmune encephalomyelitis, an animal model of multiple sclerosis. Proc Natl Acad Sci U S A. 2007;104:7558–7563. doi: 10.1073/pnas.0702228104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Du H, Guo L, Fang F, Chen D, Sosunov AA, McKhann GM, Yan Y, Wang C, Zhang H, Molkentin JD, Gunn-Moore FJ, Vonsattel JP, Arancio O, Chen JX, Yan SD. Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer's disease. Nat Med. 2008;14:1097–1105. doi: 10.1038/nm.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang H, Guan Y, Wang X, Smith K, Cormier K, Zhu S, Stavrovskaya IG, Huo C, Ferrante RJ, Kristal BS, Friedlander RM. Nortriptyline delays disease onset in models of chronic neurodegeneration. Eur J Neurosci. 2007;26:633–641. doi: 10.1111/j.1460-9568.2007.05663.x. [DOI] [PubMed] [Google Scholar]
- 202.Crompton M, Virji S, Ward JM. Cyclophilin-D binds strongly to complexes of the voltage-dependent anion channel and the adenine nucleotide translocase to form the permeability transition pore. Eur J Biochem. 1998;258:729–735. doi: 10.1046/j.1432-1327.1998.2580729.x. [DOI] [PubMed] [Google Scholar]
- 203.Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- 204.Schinzel AC, Takeuchi O, Huang Z, Fisher JK, Zhou Z, Rubens J, Hetz C, Danial NN, Moskowitz MA, Korsmeyer SJ. Cyclophilin D is a component of mitochondrial permeability transition and mediates neuronal cell death after focal cerebral ischemia. Proc Natl Acad Sci U S A. 2005;102:12005–12010. doi: 10.1073/pnas.0505294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kokoszka JE, Waymire KG, Levy SE, Sligh JE, Cai J, Jones DP, MacGregor GR, Wallace DC. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature. 2004;427:461–465. doi: 10.1038/nature02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Baines CP, Kaiser RA, Sheiko T, Craigen WJ, Molkentin JD. Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat Cell Biol. 2007;9:550–555. doi: 10.1038/ncb1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Morkuniene R, Jekabsone A, Borutaite V. Estrogens prevent calcium-induced release of cytochrome c from heart mitochondria. FEBS Lett. 2002;521:53–56. doi: 10.1016/s0014-5793(02)02820-x. [DOI] [PubMed] [Google Scholar]
- 208.Custodio JB, Moreno AJ, Wallace KB. Tamoxifen inhibits induction of the mitochondrial permeability transition by Ca2+ and inorganic phosphate. Toxicol Appl Pharmacol. 1998;152:10–17. doi: 10.1006/taap.1998.8510. [DOI] [PubMed] [Google Scholar]
- 209.Cardoso CM, Almeida LM, Custodio JB. 4-Hydroxytamoxifen is a potent inhibitor of the mitochondrial permeability transition. Mitochondrion. 2002;1:485–495. doi: 10.1016/s1567-7249(02)00034-x. [DOI] [PubMed] [Google Scholar]
- 210.Moreira PI, Custodio JB, Oliveira CR, Santos MS. Brain mitochondrial injury induced by oxidative stress-related events is prevented by tamoxifen. Neuropharmacology. 2005;48:435–447. doi: 10.1016/j.neuropharm.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 211.Miyaguchi C, Muranaka S, Kanno T, Fujita H, Akiyama J, Yoshioka T, Yasuda T. 17beta-estradiol suppresses ROS-induced apoptosis of CHO cells through inhibition of lipid peroxidation-coupled membrane permeability transition. Physiol Chem Phys Med NMR. 2004;36:21–35. [PubMed] [Google Scholar]
- 212.Hoyt KR, McLaughlin BA, Higgins DS, Jr, Reynolds IJ. Inhibition of glutamate-induced mitochondrial depolarization by tamoxifen in cultured neurons. J Pharmacol Exp Ther. 2000;293:480–486. [PubMed] [Google Scholar]
- 213.Wang Q, Li X, Wang L, Feng YH, Zeng R, Gorodeski G. Antiapoptotic effects of estrogen in normal and cancer human cervical epithelial cells. Endocrinology. 2004;145:5568–5579. doi: 10.1210/en.2004-0807. [DOI] [PubMed] [Google Scholar]
- 214.Fu Y, Li J, Lee AS. GRP78/BiP inhibits endoplasmic reticulum BIK and protects human breast cancer cells against estrogen starvation-induced apoptosis. Cancer Res. 2007;67:3734–3740. doi: 10.1158/0008-5472.CAN-06-4594. [DOI] [PubMed] [Google Scholar]
- 215.Petronilli V, Miotto G, Canton M, Colonna R, Bernardi P, Di Lisa F. Imaging the mitochondrial permeability transition pore in intact cells. Biofactors. 1998;8:263–272. doi: 10.1002/biof.5520080314. [DOI] [PubMed] [Google Scholar]
- 216.Lobaton CD, Vay L, Hernandez-Sanmiguel E, Santodomingo J, Moreno A, Montero M, Alvarez J. Modulation of mitochondrial Ca(2+) uptake by estrogen receptor agonists and antagonists. Br J Pharmacol. 2005;145:862–871. doi: 10.1038/sj.bjp.0706265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Gottipati S, Cammarata PR. Mitochondrial superoxide dismutase activation with 17 beta-estradiol-treated human lens epithelial cells. Mol Vis. 2008;14:898–905. [PMC free article] [PubMed] [Google Scholar]
- 218.Cardoso CM, Moreno AJ, Almeida LM, Custodio JB. Comparison of the changes in adenine nucleotides of rat liver mitochondria induced by tamoxifen and 4-hydroxytamoxifen. Toxicol In Vitro. 2003;17:663–670. doi: 10.1016/s0887-2333(03)00106-1. [DOI] [PubMed] [Google Scholar]
- 219.Belzacq AS, Vieira HL, Kroemer G, Brenner C. The adenine nucleotide translocator in apoptosis. Biochimie. 2002;84:167–176. doi: 10.1016/s0300-9084(02)01366-4. [DOI] [PubMed] [Google Scholar]
- 220.Yang SH, Sarkar SN, Liu R, Perez EJ, Wang X, Wen Y, Yan LJ, Simpkins JW. Estrogen Receptor {beta} as a Mitochondrial Vulnerability Factor. J Biol Chem. 2009;284:9540–9548. doi: 10.1074/jbc.M808246200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Too CK, Giles A, Wilkinson M. Estrogen stimulates expression of adenine nucleotide translocator ANT1 messenger RNA in female rat hearts. Mol Cell Endocrinol. 1999;150:161–167. doi: 10.1016/s0303-7207(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 222.Belzacq AS, Brenner C. The adenine nucleotide translocator: a new potential chemotherapeutic target. Curr Drug Targets. 2003;4:517–524. doi: 10.2174/1389450033490867. [DOI] [PubMed] [Google Scholar]
- 223.Belzacq AS, Vieira HL, Verrier F, Vandecasteele G, Cohen I, Prevost MC, Larquet E, Pariselli F, Petit PX, Kahn A, Rizzuto R, Brenner C, Kroemer G. Bcl-2 and Bax modulate adenine nucleotide translocase activity. Cancer Res. 2003;63:541–546. [PubMed] [Google Scholar]
- 224.Garcia-Segura LM, Cardona-Gomez P, Naftolin F, Chowen JA. Estradiol upregulates Bcl-2 expression in adult brain neurons. Neuroreport. 1998;9:593–597. doi: 10.1097/00001756-199803090-00006. [DOI] [PubMed] [Google Scholar]
- 225.Wang TT, Phang JM. Effects of estrogen on apoptotic pathways in human breast cancer cell line MCF-7. Cancer Res. 1995;55:2487–2489. [PubMed] [Google Scholar]
- 226.Safe S. Transcriptional activation of genes by 17 beta-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm. 2001;62:231–252. doi: 10.1016/s0083-6729(01)62006-5. [DOI] [PubMed] [Google Scholar]
- 227.Alkayed NJ, Goto S, Sugo N, Joh HD, Klaus J, Crain BJ, Bernard O, Traystman RJ, Hurn PD. Estrogen and Bcl-2: gene induction and effect of transgene in experimental stroke. J Neurosci. 2001;21:7543–7550. doi: 10.1523/JNEUROSCI.21-19-07543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Li L, Backer J, Wong AS, Schwanke EL, Stewart BG, Pasdar M. Bcl-2 expression decreases cadherin-mediated cell-cell adhesion. J Cell Sci. 2003;116:3687–3700. doi: 10.1242/jcs.00644. [DOI] [PubMed] [Google Scholar]
- 229.Zhao L, Wu TW, Brinton RD. Estrogen receptor subtypes alpha and beta contribute to neuroprotection and increased Bcl-2 expression in primary hippocampal neurons. Brain Res. 2004;1010:22–34. doi: 10.1016/j.brainres.2004.02.066. [DOI] [PubMed] [Google Scholar]
- 230.Murphy E, Imahashi K, Steenbergen C. Bcl-2 regulation of mitochondrial energetics. Trends Cardiovasc Med. 2005;15:283–290. doi: 10.1016/j.tcm.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 231.Mandal S, Guptan P, Owusu-Ansah E, Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell. 2005;9:843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 232.Schauen M, Spitkovsky D, Schubert J, Fischer JH, Hayashi J, Wiesner RJ. Respiratory chain deficiency slows down cell-cycle progression via reduced ROS generation and is associated with a reduction of p21CIP1/WAF1. J Cell Physiol. 2006;209:103–112. doi: 10.1002/jcp.20711. [DOI] [PubMed] [Google Scholar]
- 233.Stockl P, Hutter E, Zwerschke W, Jansen-Durr P. Sustained inhibition of oxidative phosphorylation impairs cell proliferation and induces premature senescence in human fibroblasts. Exp Gerontol. 2006;41:674–682. doi: 10.1016/j.exger.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 234.Stockl P, Zankl C, Hutter E, Unterluggauer H, Laun P, Heeren G, Bogengruber E, Herndler-Brandstetter D, Breitenbach M, Jansen-Durr P. Partial uncoupling of oxidative phosphorylation induces premature senescence in human fibroblasts and yeast mother cells. Free Radic Biol Med. 2007;43:947–958. doi: 10.1016/j.freeradbiomed.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 235.Han YH, Kim SW, Kim SH, Kim SZ, Park WH. 2,4-dinitrophenol induces G1 phase arrest and apoptosis in human pulmonary adenocarcinoma Calu-6 cells. Toxicol In Vitro. 2008;22:659–670. doi: 10.1016/j.tiv.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 236.Haugen DR, Fluge O, Reigstad LJ, Varhaug JE, Lillehaug JR. Increased expression of genes encoding mitochondrial proteins in papillary thyroid carcinomas. Thyroid. 2003;13:613–620. doi: 10.1089/105072503322239943. [DOI] [PubMed] [Google Scholar]
- 237.Kuroda Y, Mitsui T, Kunishige M, Shono M, Akaike M, Azuma H, Matsumoto T. Parkin enhances mitochondrial biogenesis in proliferating cells. Hum Mol Genet. 2006;15:883–895. doi: 10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- 238.Chang HJ, Lee MR, Hong SH, Yoo BC, Shin YK, Jeong JY, Lim SB, Choi HS, Jeong SY, Park JG. Identification of mitochondrial FoF1-ATP synthase involved in liver metastasis of colorectal cancer. Cancer Sci. 2007;98:1184–1191. doi: 10.1111/j.1349-7006.2007.00527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wright SC, Zhong J, Larrick JW. Inhibition of apoptosis as a mechanism of tumor promotion. FASEB J. 1994;8:654–660. doi: 10.1096/fasebj.8.9.8005393. [DOI] [PubMed] [Google Scholar]
- 240.Fernando RI, Wimalasena J. Estradiol abrogates apoptosis in breast cancer cells through inactivation of BAD: Ras-dependent nongenomic pathways requiring signaling through ERK and Akt. Mol Biol Cell. 2004;15:3266–3284. doi: 10.1091/mbc.E03-11-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zheng A, Kallio A, Harkonen P. Tamoxifen-induced rapid death of MCF-7 breast cancer cells is mediated via extracellularly signal-regulated kinase signaling and can be abrogated by estrogen. Endocrinology. 2007;148:2764–2777. doi: 10.1210/en.2006-1269. [DOI] [PubMed] [Google Scholar]
- 242.Sawada H, Ibi M, Kihara T, Urushitani M, Honda K, Nakanishi M, Akaike A, Shimohama S. Mechanisms of antiapoptotic effects of estrogens in nigral dopaminergic neurons. FASEB J. 2000;14:1202–1214. doi: 10.1096/fasebj.14.9.1202. [DOI] [PubMed] [Google Scholar]
- 243.Zhang Y, Bhavnani BR. Glutamate-induced apoptosis in primary cortical neurons is inhibited by equine estrogens via down-regulation of caspase-3 and prevention of mitochondrial cytochrome c release. BMC Neurosci. 2005;6:13. doi: 10.1186/1471-2202-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Spyridopoulos I, Sullivan AB, Kearney M, Isner JM, Losordo DW. Estrogen-receptor-mediated inhibition of human endothelial cell apoptosis. Estradiol as a survival factor. Circulation. 1997;95:1505–1514. doi: 10.1161/01.cir.95.6.1505. [DOI] [PubMed] [Google Scholar]
- 245.Kim JK, Pedram A, Razandi M, Levin ER. Estrogen prevents cardiomyocyte apoptosis through inhibition of reactive oxygen species and differential regulation of p38 kinase isoforms. J Biol Chem. 2006;281:6760–6767. doi: 10.1074/jbc.M511024200. [DOI] [PubMed] [Google Scholar]
- 246.Boland R, Vasconsuelo A, Milanesi L, Ronda AC, de Boland AR. 17beta-estradiol signaling in skeletal muscle cells and its relationship to apoptosis. Steroids. 2008;73:859–863. doi: 10.1016/j.steroids.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 247.Wang X, Simpkins JW, Dykens JA, Cammarata PR. Oxidative damage to human lens epithelial cells in culture: estrogen protection of mitochondrial potential, ATP, and cell viability. Invest Ophthalmol Vis Sci. 2003;44:2067–2075. doi: 10.1167/iovs.02-0841. [DOI] [PubMed] [Google Scholar]
- 248.Chen J, Gokhale M, Schofield B, Odwin S, Yager JD. Inhibition of TGF-beta-induced apoptosis by ethinyl estradiol in cultured, precision cut rat liver slices and hepatocytes. Carcinogenesis. 2000;21:1205–1211. [PubMed] [Google Scholar]
- 249.Yin Y, Huang WW, Lin C, Chen H, MacKenzie A, Ma L. Estrogen suppresses uterine epithelial apoptosis by inducing birc1 expression. Mol Endocrinol. 2008;22:113–125. doi: 10.1210/me.2007-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Gorodeski GI. Estrogen attenuates P2X7-R-mediated apoptosis of uterine cervical cells by blocking calcium influx. Nucleosides Nucleotides Nucleic Acids. 2004;23:1287–1293. doi: 10.1081/NCN-200027549. [DOI] [PubMed] [Google Scholar]
- 251.Alexaki VI, Charalampopoulos I, Kampa M, Vassalou H, Theodoropoulos P, Stathopoulos EN, Hatzoglou A, Gravanis A, Castanas E. Estrogen exerts neuroprotective effects via membrane estrogen receptors and rapid Akt/NOS activation. FASEB J. 2004;18:1594–1596. doi: 10.1096/fj.04-1495fje. [DOI] [PubMed] [Google Scholar]
- 252.Razandi M, Pedram A, Levin ER. Plasma membrane estrogen receptors signal to antiapoptosis in breast cancer. Mol Endocrinol. 2000;14:1434–1447. doi: 10.1210/mend.14.9.0526. [DOI] [PubMed] [Google Scholar]
- 253.Yager JD, Chen JQ. Mitochondrial estrogen receptors--new insights into specific functions. Trends Endocrinol Metab. 2007;18:89–91. doi: 10.1016/j.tem.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 254.Yune TY, Park HG, Lee JY, Oh TH. Estrogen-induced Bcl-2 expression after spinal cord injury is mediated through phosphoinositide-3-kinase/Akt-dependent CREB activation. J Neurotrauma. 2008;25:1121–1131. doi: 10.1089/neu.2008.0544. [DOI] [PubMed] [Google Scholar]
- 255.Bynoe MS, Grimaldi CM, Diamond B. Estrogen up-regulates Bcl-2 and blocks tolerance induction of naive B cells. Proc Natl Acad Sci U S A. 2000;97:2703–2708. doi: 10.1073/pnas.040577497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Minczuk M, Mroczek S, Pawlak SD, Stepien PP. Human ATP-dependent RNA/DNA helicase hSuv3p interacts with the cofactor of survivin HBXIP. FEBS J. 2005;272:5008–5019. doi: 10.1111/j.1742-4658.2005.04910.x. [DOI] [PubMed] [Google Scholar]
- 257.Miki Y, Suzuki T, Tazawa C, Yamaguchi Y, Kitada K, Honma S, Moriya T, Hirakawa H, Evans DB, Hayashi S, Ohuchi N, Sasano H. Aromatase localization in human breast cancer tissues: possible interactions between intratumoral stromal and parenchymal cells. Cancer Res. 2007;67:3945–3954. doi: 10.1158/0008-5472.CAN-06-3105. [DOI] [PubMed] [Google Scholar]
- 258.Bulun SE, Lin Z, Imir G, Amin S, Demura M, Yilmaz B, Martin R, Utsunomiya H, Thung S, Gurates B, Tamura M, Langoi D, Deb S. Regulation of aromatase expression in estrogen-responsive breast and uterine disease: from bench to treatment. Pharmacol Rev. 2005;57:359–383. doi: 10.1124/pr.57.3.6. [DOI] [PubMed] [Google Scholar]
- 259.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 260.Putignani L, Raffa S, Pescosolido R, Aimati L, Signore F, Torrisi MR, Grammatico P. Alteration of expression levels of the oxidative phosphorylation system (OXPHOS) in breast cancer cell mitochondria. Breast Cancer Res Treat. 2008;110:439–452. doi: 10.1007/s10549-007-9738-x. [DOI] [PubMed] [Google Scholar]
- 261.Abril J, de Heredia ML, Gonzalez L, Cleries R, Nadal M, Condom E, Aguilo F, Gomez-Zaera M, Nunes V. Altered expression of 12S/MT-RNR1, MT-CO2/COX2, and MT-ATP6 mitochondrial genes in prostate cancer. Prostate. 2008;68:1086–1096. doi: 10.1002/pros.20771. [DOI] [PubMed] [Google Scholar]
- 262.Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, Becker K, Yates JR, 3rd, Felding-Habermann B. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 2007;67:1472–1486. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 263.Hooven LA, Baird WM. Proteomic analysis of MCF-7 cells treated with benzo[a]pyrene, dibenzo[a,l]pyrene, coal tar extract, and diesel exhaust extract. Toxicology. 2008;249:1–10. doi: 10.1016/j.tox.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 264.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 265.Zhu W, Qin W, Bradley P, Wessel A, Puckett CL, Sauter ER. Mitochondrial DNA mutations in breast cancer tissue and in matched nipple aspirate fluid. Carcinogenesis. 2005;26:145–152. doi: 10.1093/carcin/bgh282. [DOI] [PubMed] [Google Scholar]
- 266.Parrella P, Xiao Y, Fliss M, Sanchez-Cespedes M, Mazzarelli P, Rinaldi M, Nicol T, Gabrielson E, Cuomo C, Cohen D, Pandit S, Spencer M, Rabitti C, Fazio VM, Sidransky D. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res. 2001;61:7623–7626. [PubMed] [Google Scholar]
- 267.Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67:4687–4694. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- 268.Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X, Niu R. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life. 2007;59:450–457. doi: 10.1080/15216540701509955. [DOI] [PubMed] [Google Scholar]
- 269.Erol A. Retrograde regulation due to mitochondrial dysfunction may be an important mechanism for carcinogenesis. Med Hypotheses. 2005;65:525–529. doi: 10.1016/j.mehy.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 270.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 271.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 272.Felty Q, Singh KP, Roy D. Estrogen-induced G1/S transition of G0-arrested estrogen-dependent breast cancer cells is regulated by mitochondrial oxidant signaling. Oncogene. 2005;24:4883–4893. doi: 10.1038/sj.onc.1208667. [DOI] [PubMed] [Google Scholar]
- 273.Felty Q, Xiong WC, Sun D, Sarkar S, Singh KP, Parkash J, Roy D. Estrogen-induced mitochondrial reactive oxygen species as signal-transducing messengers. Biochemistry. 2005;44:6900–6909. doi: 10.1021/bi047629p. [DOI] [PubMed] [Google Scholar]
- 274.Delsite R, Kachhap S, Anbazhagan R, Gabrielson E, Singh KK. Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol Cancer. 2002;1:6. doi: 10.1186/1476-4598-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kulawiec M, Arnouk H, Desouki MM, Kazim L, Still I, Singh KK. Proteomic analysis of mitochondria-to-nucleus retrograde response in human cancer. Cancer Biol Ther. 2006;5:967–975. doi: 10.4161/cbt.5.8.2880. [DOI] [PubMed] [Google Scholar]
- 276.Obrero M, Yu DV, Shapiro DJ. Estrogen receptor-dependent and estrogen receptor-independent pathways for tamoxifen and 4-hydroxytamoxifen-induced programmed cell death. J Biol Chem. 2002;277:45695–45703. doi: 10.1074/jbc.M208092200. [DOI] [PubMed] [Google Scholar]
- 277.Cardoso CM, Custodio JB, Almeida LM, Moreno AJ. Mechanisms of the deleterious effects of tamoxifen on mitochondrial respiration rate and phosphorylation efficiency. Toxicol Appl Pharmacol. 2001;176:145–152. doi: 10.1006/taap.2001.9265. [DOI] [PubMed] [Google Scholar]
- 278.Cardoso CM, Moreno AJ, Almeida LM, Custodio JB. 4-Hydroxytamoxifen induces slight uncoupling of mitochondrial oxidative phosphorylation system in relation to the deleterious effects of tamoxifen. Toxicology. 2002;179:221–232. doi: 10.1016/s0300-483x(02)00392-x. [DOI] [PubMed] [Google Scholar]
- 279.Kallio A, Zheng A, Dahllund J, Heiskanen KM, Harkonen P. Role of mitochondria in tamoxifen-induced rapid death of MCF-7 breast cancer cells. Apoptosis. 2005;10:1395–1410. doi: 10.1007/s10495-005-2137-z. [DOI] [PubMed] [Google Scholar]
- 280.Moreira PI, Custodio J, Moreno A, Oliveira CR, Santos MS. Tamoxifen and estradiol interact with the flavin mononucleotide site of complex I leading to mitochondrial failure. J Biol Chem. 2006;281:10143–10152. doi: 10.1074/jbc.M510249200. [DOI] [PubMed] [Google Scholar]
- 281.Nagahara Y, Shiina I, Nakata K, Sasaki A, Miyamoto T, Ikekita M. Induction of mitochondria-involved apoptosis in estrogen receptor-negative cells by a novel tamoxifen derivative, ridaifen-B. Cancer Sci. 2008;99:608–614. doi: 10.1111/j.1349-7006.2007.00709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.McLachlan A, Kekre N, McNulty J, Pandey S. Pancratistatin: a natural anti-cancer compound that targets mitochondria specifically in cancer cells to induce apoptosis. Apoptosis. 2005;10:619–630. doi: 10.1007/s10495-005-1896-x. [DOI] [PubMed] [Google Scholar]
- 283.Siedlakowski P, McLachlan-Burgess A, Griffin C, Tirumalai SS, McNulty J, Pandey S. Synergy of Pancratistatin and Tamoxifen on breast cancer cells in inducing apoptosis by targeting mitochondria. Cancer Biol Ther. 2008;7:376–384. doi: 10.4161/cbt.7.3.5364. [DOI] [PubMed] [Google Scholar]
- 284.Nazarewicz RR, Zenebe WJ, Parihar A, Larson SK, Alidema E, Choi J, Ghafourifar P. Tamoxifen induces oxidative stress and mitochondrial apoptosis via stimulating mitochondrial nitric oxide synthase. Cancer Res. 2007;67:1282–1290. doi: 10.1158/0008-5472.CAN-06-3099. [DOI] [PubMed] [Google Scholar]
- 285.Dietze EC, Caldwell LE, Grupin SL, Mancini M, Seewaldt VL. Tamoxifen but not 4-hydroxytamoxifen initiates apoptosis in p53(−) normal human mammary epithelial cells by inducing mitochondrial depolarization. J Biol Chem. 2001;276:5384–5394. doi: 10.1074/jbc.M007915200. [DOI] [PubMed] [Google Scholar]
- 286.Besada V, Diaz M, Becker M, Ramos Y, Castellanos-Serra L, Fichtner I. Proteomics of xenografted human breast cancer indicates novel targets related to tamoxifen resistance. Proteomics. 2006;6:1038–1048. doi: 10.1002/pmic.200500151. [DOI] [PubMed] [Google Scholar]
- 287.Strong R, Nakanishi T, Ross D, Fenselau C. Alterations in the mitochondrial proteome of adriamycin resistant MCF-7 breast cancer cells. J Proteome Res. 2006;5:2389–2395. doi: 10.1021/pr060207c. [DOI] [PubMed] [Google Scholar]
- 288.Tanaka S, Sakai A, Kimura K, Yoshida H, Fushitani H, Ogata A, Miyamoto A, Fukushima M, Wada A, Tanigawa N. Proteomic analysis of the basic proteins in 5-fluorouracil resistance of human colon cancer cell line using the radical-free and highly reducing method of two-dimensional polyacrylamide gel electrophoresis. Int J Oncol. 2008;33:361–370. [PubMed] [Google Scholar]
- 289.Shin YK, Yoo BC, Chang HJ, Jeon E, Hong SH, Jung MS, Lim SJ, Park JG. Down-regulation of mitochondrial F1F0-ATP synthase in human colon cancer cells with induced 5-fluorouracil resistance. Cancer Res. 2005;65:3162–3170. doi: 10.1158/0008-5472.CAN-04-3300. [DOI] [PubMed] [Google Scholar]
- 290.Kojima K, Ohhashi R, Fujita Y, Hamada N, Akao Y, Nozawa Y, Deguchi T, Ito M. A role for SIRT1 in cell growth and chemoresistance in prostate cancer PC3 and DU145 cells. Biochem Biophys Res Commun. 2008;373:423–428. doi: 10.1016/j.bbrc.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 291.Liang XJ, Finkel T, Shen DW, Yin JJ, Aszalos A, Gottesman MM. SIRT1 contributes in part to cisplatin resistance in cancer cells by altering mitochondrial metabolism. Mol Cancer Res. 2008;6:1499–1506. doi: 10.1158/1541-7786.MCR-07-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Qian W, Nishikawa M, Haque AM, Hirose M, Mashimo M, Sato E, Inoue M. Mitochondrial density determines the cellular sensitivity to cisplatin-induced cell death. Am J Physiol Cell Physiol. 2005;289:C1466–C1475. doi: 10.1152/ajpcell.00265.2005. [DOI] [PubMed] [Google Scholar]
- 293.Mizumachi T, Suzuki S, Naito A, Carcel-Trullols J, Evans TT, Spring PM, Oridate N, Furuta Y, Fukuda S, Higuchi M. Increased mitochondrial DNA induces acquired docetaxel resistance in head and neck cancer cells. Oncogene. 2008;27:831–838. doi: 10.1038/sj.onc.1210681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Castellani R, Hirai K, Aliev G, Drew KL, Nunomura A, Takeda A, Cash AD, Obrenovich ME, Perry G, Smith MA. Role of mitochondrial dysfunction in Alzheimer's disease. J Neurosci Res. 2002;70:357–360. doi: 10.1002/jnr.10389. [DOI] [PubMed] [Google Scholar]
- 295.Beal MF. Mitochondrial dysfunction in neurodegenerative diseases. Biochim Biophys Acta. 1998;1366:211–223. doi: 10.1016/s0005-2728(98)00114-5. [DOI] [PubMed] [Google Scholar]
- 296.Beal MF. Mitochondrial dysfunction and oxidative damage in Alzheimer's and Parkinson's diseases and coenzyme Q10 as a potential treatment. J Bioenerg Biomembr. 2004;36:381–386. doi: 10.1023/B:JOBB.0000041772.74810.92. [DOI] [PubMed] [Google Scholar]
- 297.Ramsey CP, Giasson BI. Role of mitochondrial dysfunction in Parkinson's disease: Implications for treatment. Drugs Aging. 2007;24:95–105. doi: 10.2165/00002512-200724020-00002. [DOI] [PubMed] [Google Scholar]
- 298.Xu XW, Shi C, He ZQ, Ma CM, Chen WH, Shen YP, Guo Q, Shen CJ, Xu J. Effects of phytoestrogen on mitochondrial structure and function of hippocampal CA1 region of ovariectomized rats. Cell Mol Neurobiol. 2008;28:875–886. doi: 10.1007/s10571-008-9265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Shi C, Xu J. Increased vulnerability of brain to estrogen withdrawal-induced mitochondrial dysfunction with aging. J Bioenerg Biomembr. 2008;40:625–630. doi: 10.1007/s10863-008-9195-1. [DOI] [PubMed] [Google Scholar]
- 300.Shi C, Xu XW, Forster EL, Tang LF, Ge Z, Yew DT, Xu J. Possible role of mitochondrial dysfunction in central neurodegeneration of ovariectomized rats. Cell Biochem Funct. 2008;26:172–178. doi: 10.1002/cbf.1423. [DOI] [PubMed] [Google Scholar]
- 301.Simpkins JW, Dykens JA. Mitochondrial mechanisms of estrogen neuroprotection. Brain Res Rev. 2008;57:421–430. doi: 10.1016/j.brainresrev.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 302.Singh M, Dykens JA, Simpkins JW. Novel mechanisms for estrogen-induced neuroprotection. Exp Biol Med (Maywood) 2006;231:514–521. doi: 10.1177/153537020623100505. [DOI] [PubMed] [Google Scholar]
- 303.Nilsen J. Estradiol and neurodegenerative oxidative stress. Front Neuroendocrinol. 2008;29:463–475. doi: 10.1016/j.yfrne.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 304.Brinton RD. The healthy cell bias of estrogen action: mitochondrial bioenergetics and neurological implications. Trends Neurosci. 2008;31:529–537. doi: 10.1016/j.tins.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Brinton RD. Estrogen regulation of glucose metabolism and mitochondrial function: therapeutic implications for prevention of Alzheimer's disease. Adv Drug Deliv Rev. 2008;60:1504–1511. doi: 10.1016/j.addr.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Hauptmann S, Keil U, Scherping I, Bonert A, Eckert A, Muller WE. Mitochondrial dysfunction in sporadic and genetic Alzheimer's disease. Exp Gerontol. 2006;41:668–673. doi: 10.1016/j.exger.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 307.Bandyopadhyay B, Li G, Yin H, Kuret J. Tau aggregation and toxicity in a cell culture model of tauopathy. J Biol Chem. 2007;282:16454–16464. doi: 10.1074/jbc.M700192200. [DOI] [PubMed] [Google Scholar]
- 308.David DC, Hauptmann S, Scherping I, Schuessel K, Keil U, Rizzu P, Ravid R, Drose S, Brandt U, Muller WE, Eckert A, Gotz J. Proteomic and functional analyses reveal a mitochondrial dysfunction in P301L tau transgenic mice. J Biol Chem. 2005;280:23802–23814. doi: 10.1074/jbc.M500356200. [DOI] [PubMed] [Google Scholar]
- 309.Atlante A, Amadoro G, Bobba A, de Bari L, Corsetti V, Pappalardo G, Marra E, Calissano P, Passarella S. A peptide containing residues 26–44 of tau protein impairs mitochondrial oxidative phosphorylation acting at the level of the adenine nucleotide translocator. Biochim Biophys Acta. 2008;1777:1289–1300. doi: 10.1016/j.bbabio.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 310.Nilsen J, Chen S, Irwin RW, Iwamoto S, Brinton RD. Estrogen protects neuronal cells from amyloid beta-induced apoptosis via regulation of mitochondrial proteins and function. BMC Neurosci. 2006;7:74. doi: 10.1186/1471-2202-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Eckert A, Hauptmann S, Scherping I, Rhein V, Muller-Spahn F, Gotz J, Muller WE. Soluble beta-amyloid leads to mitochondrial defects in amyloid precursor protein and tau transgenic mice. Neurodegener Dis. 2008;5:157–159. doi: 10.1159/000113689. [DOI] [PubMed] [Google Scholar]
- 312.Hansson Petersen CA, Alikhani N, Behbahani H, Wiehager B, Pavlov PF, Alafuzoff I, Leinonen V, Ito A, Winblad B, Glaser E, Ankarcrona M. The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc Natl Acad Sci U S A. 2008;105:13145–13150. doi: 10.1073/pnas.0806192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Eckert A, Hauptmann S, Scherping I, Meinhardt J, Rhein V, Drose S, Brandt U, Fandrich M, Muller WE, Gotz J. Oligomeric and fibrillar species of beta-amyloid (Abeta42) both impair mitochondrial function in P301L tau transgenic mice. J Mol Med. 2008;86:1255–1267. doi: 10.1007/s00109-008-0391-6. [DOI] [PubMed] [Google Scholar]
- 314.Rhein V, Eckert A. Effects of Alzheimer's amyloid-beta and tau protein on mitochondrial function -- role of glucose metabolism and insulin signalling. Arch Physiol Biochem. 2007;113:131–141. doi: 10.1080/13813450701572288. [DOI] [PubMed] [Google Scholar]
- 315.Gong CX, Iqbal K. Hyperphosphorylation of microtubule-associated protein tau: a promising therapeutic target for Alzheimer disease. Curr Med Chem. 2008;15:2321–2328. doi: 10.2174/092986708785909111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Liu XA, Zhu LQ, Zhang Q, Shi HR, Wang SH, Wang Q, Wang JZ. Estradiol attenuates tau hyperphosphorylation induced by upregulation of protein kinase-A. Neurochem Res. 2008;33:1811–1820. doi: 10.1007/s11064-008-9638-4. [DOI] [PubMed] [Google Scholar]
- 317.Yi KD, Simpkins JW. Protein phosphatase 1, protein phosphatase 2A, and calcineurin play a role in estrogen-mediated neuroprotection. Endocrinology. 2008;149:5235–5243. doi: 10.1210/en.2008-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zhang QG, Wang R, Khan M, Mahesh V, Brann DW. Role of Dickkopf-1, an antagonist of the Wnt/beta-catenin signaling pathway, in estrogen-induced neuroprotection and attenuation of tau phosphorylation. J Neurosci. 2008;28:8430–8441. doi: 10.1523/JNEUROSCI.2752-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Chiueh C, Lee S, Andoh T, Murphy D. Induction of antioxidative and antiapoptotic thioredoxin supports neuroprotective hypothesis of estrogen. Endocrine. 2003;21:27–31. doi: 10.1385/endo:21:1:27. [DOI] [PubMed] [Google Scholar]
- 320.Morinaga A, Hirohata M, Ono K, Yamada M. Estrogen has anti-amyloidogenic effects on Alzheimer's beta-amyloid fibrils in vitro. Biochem Biophys Res Commun. 2007;359:697–702. doi: 10.1016/j.bbrc.2007.05.158. [DOI] [PubMed] [Google Scholar]
- 321.Allain H, Bentue-Ferrer D, Tribut O, Gauthier S, Michel BF, Drieu-La Rochelle C. Alzheimer's disease: the pharmacological pathway. Fundam Clin Pharmacol. 2003;17:419–428. doi: 10.1046/j.1472-8206.2003.00153.x. [DOI] [PubMed] [Google Scholar]
- 322.Simpkins JW, Yang SH, Liu R, Perez E, Cai ZY, Covey DF, Green PS. Estrogen-like compounds for ischemic neuroprotection. Stroke. 2004;35:2648–2651. doi: 10.1161/01.STR.0000143734.59507.88. [DOI] [PubMed] [Google Scholar]
- 323.Dykens JA, Moos WH, Howell N. Development of 17alpha-estradiol as a neuroprotective therapeutic agent: rationale and results from a phase I clinical study. Ann N Y Acad Sci. 2005;1052:116–135. doi: 10.1196/annals.1347.008. [DOI] [PubMed] [Google Scholar]
- 324.Simpkins JW, Wang J, Wang X, Perez E, Prokai L, Dykens JA. Mitochondria play a central role in estrogen-induced neuroprotection. Curr Drug Targets CNS Neurol Disord. 2005;4:69–83. doi: 10.2174/1568007053005073. [DOI] [PubMed] [Google Scholar]
- 325.Zhao L, Mao Z, Brinton RD. A Select Combination of Clinically Relevant Phytoestrogens Enhances Estrogen Receptor {beta}-Binding Selectivity and Neuroprotective Activities In Vitro and In Vivo. Endocrinology. 2008 doi: 10.1210/en.2008-0715. [DOI] [PubMed] [Google Scholar]
- 326.Zeng H, Chen Q, Zhao B. Genistein ameliorates beta-amyloid peptide (25–35)-induced hippocampal neuronal apoptosis. Free Radic Biol Med. 2004;36:180–188. doi: 10.1016/j.freeradbiomed.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 327.Zhao L, Jin C, Mao Z, Gopinathan MB, Rehder K, Brinton RD. Design, synthesis, and estrogenic activity of a novel estrogen receptor modulator--a hybrid structure of 17beta-estradiol and vitamin E in hippocampal neurons. J Med Chem. 2007;50:4471–4481. doi: 10.1021/jm070546x. [DOI] [PubMed] [Google Scholar]
- 328.Henchcliffe C, Beal MF. Mitochondrial biology and oxidative stress in Parkinson disease pathogenesis. Nat Clin Pract Neurol. 2008;4:600–609. doi: 10.1038/ncpneuro0924. [DOI] [PubMed] [Google Scholar]
- 329.Shulman LM. Gender differences in Parkinson's disease. Gend Med. 2007;4:8–18. doi: 10.1016/s1550-8579(07)80003-9. [DOI] [PubMed] [Google Scholar]
- 330.Rodriguez-Navarro JA, Solano RM, Casarejos MJ, Gomez A, Perucho J, de Yebenes JG, Mena MA. Gender differences and estrogen effects in parkin null mice. J Neurochem. 2008;106:2143–2157. doi: 10.1111/j.1471-4159.2008.05569.x. [DOI] [PubMed] [Google Scholar]
- 331.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 332.Davidzon G, Greene P, Mancuso M, Klos KJ, Ahlskog JE, Hirano M, DiMauro S. Early-onset familial parkinsonism due to POLG mutations. Ann Neurol. 2006;59:859–862. doi: 10.1002/ana.20831. [DOI] [PubMed] [Google Scholar]
- 333.Dodson MW, Guo M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson's disease. Curr Opin Neurobiol. 2007;17:331–337. doi: 10.1016/j.conb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 334.Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 336.Kim Y, Park J, Kim S, Song S, Kwon SK, Lee SH, Kitada T, Kim JM, Chung J. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008 doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 337.Kuroda Y, Mitsui T, Kunishige M, Matsumoto T. Parkin affects mitochondrial function and apoptosis in neuronal and myogenic cells. Biochem Biophys Res Commun. 2006;348:787–793. doi: 10.1016/j.bbrc.2006.06.201. [DOI] [PubMed] [Google Scholar]
- 338.Moore DJ, Zhang L, Dawson TM, Dawson VL. A missense mutation (L166P) in DJ-1, linked to familial Parkinson's disease, confers reduced protein stability and impairs homo-oligomerization. J Neurochem. 2003;87:1558–1567. doi: 10.1111/j.1471-4159.2003.02265.x. [DOI] [PubMed] [Google Scholar]
- 339.Moore DJ, Zhang L, Troncoso J, Lee MK, Hattori N, Mizuno Y, Dawson TM, Dawson VL. Association of DJ-1 and parkin mediated by pathogenic DJ-1 mutations and oxidative stress. Hum Mol Genet. 2005;14:71–84. doi: 10.1093/hmg/ddi007. [DOI] [PubMed] [Google Scholar]
- 340.Devi L, Raghavendran V, Prabhu BM, Avadhani NG, Anandatheerthavarada HK. Mitochondrial import and accumulation of alpha-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J Biol Chem. 2008;283:9089–9100. doi: 10.1074/jbc.M710012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Lee HJ, Shin SY, Choi C, Lee YH, Lee SJ. Formation and removal of alpha-synuclein aggregates in cells exposed to mitochondrial inhibitors. J Biol Chem. 2002;277:5411–5417. doi: 10.1074/jbc.M105326200. [DOI] [PubMed] [Google Scholar]
- 342.Abba MC, Hu Y, Sun H, Drake JA, Gaddis S, Baggerly K, Sahin A, Aldaz CM. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6:37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Jiang Y, Liu YE, Goldberg ID, Shi YE. Gamma synuclein, a novel heat-shock protein-associated chaperone, stimulates ligand-dependent estrogen receptor alpha signaling and mammary tumorigenesis. Cancer Res. 2004;64:4539–4546. doi: 10.1158/0008-5472.CAN-03-3650. [DOI] [PubMed] [Google Scholar]
- 344.Jiang Y, Liu YE, Lu A, Gupta A, Goldberg ID, Liu J, Shi YE. Stimulation of estrogen receptor signaling by gamma synuclein. Cancer Res. 2003;63:3899–3903. [PubMed] [Google Scholar]
- 345.Mendelsohn ME, Karas RH. Molecular and cellular basis of cardiovascular gender differences. Science. 2005;308:1583–1587. doi: 10.1126/science.1112062. [DOI] [PubMed] [Google Scholar]
- 346.Mendelsohn ME. Protective effects of estrogen on the cardiovascular system. Am J Cardiol. 2002;89:12E–17E. doi: 10.1016/s0002-9149(02)02405-0. discussion 17E–18E. [DOI] [PubMed] [Google Scholar]
- 347.Mendelsohn ME, Karas RH. The protective effects of estrogen on the cardiovascular system. N Engl J Med. 1999;340:1801–1811. doi: 10.1056/NEJM199906103402306. [DOI] [PubMed] [Google Scholar]
- 348.Pare G, Krust A, Karas RH, Dupont S, Aronovitz M, Chambon P, Mendelsohn ME. Estrogen receptor-alpha mediates the protective effects of estrogen against vascular injury. Circ Res. 2002;90:1087–1092. doi: 10.1161/01.res.0000021114.92282.fa. [DOI] [PubMed] [Google Scholar]
- 349.Russell LK, Finck BN, Kelly DP. Mouse models of mitochondrial dysfunction and heart failure. J Mol Cell Cardiol. 2005;38:81–91. doi: 10.1016/j.yjmcc.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 350.Casademont J, Miro O. Electron transport chain defects in heart failure. Heart Fail Rev. 2002;7:131–139. doi: 10.1023/a:1015372407647. [DOI] [PubMed] [Google Scholar]
- 351.Sheeran FL, Pepe S. Energy deficiency in the failing heart: linking increased reactive oxygen species and disruption of oxidative phosphorylation rate. Biochim Biophys Acta. 2006;1757:543–552. doi: 10.1016/j.bbabio.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 352.Elas M, Bielanska J, Pustelny K, Plonka PM, Drelicharz L, Skorka T, Tyrankiewicz U, Wozniak M, Heinze-Paluchowska S, Walski M, Wojnar L, Fortin D, Ventura-Clapier R, Chlopicki S. Detection of mitochondrial dysfunction by EPR technique in mouse model of dilated cardiomyopathy. Free Radic Biol Med. 2008;45:321–328. doi: 10.1016/j.freeradbiomed.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 353.Rosenberg P. Mitochondrial dysfunction and heart disease. Mitochondrion. 2004;4:621–628. doi: 10.1016/j.mito.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 354.Marin-Garcia J, Goldenthal MJ. Mitochondrial centrality in heart failure. Heart Fail Rev. 2008;13:137–150. doi: 10.1007/s10741-007-9079-1. [DOI] [PubMed] [Google Scholar]
- 355.Duckles SP, Krause DN, Stirone C, Procaccio V. Estrogen and mitochondria: a new paradigm for vascular protection? Mol Interv. 2006;6:26–35. doi: 10.1124/mi.6.1.6. [DOI] [PubMed] [Google Scholar]
- 356.Stirone C, Boroujerdi A, Duckles SP, Krause DN. Estrogen receptor activation of phosphoinositide-3 kinase, akt, and nitric oxide signaling in cerebral blood vessels: rapid and long-term effects. Mol Pharmacol. 2005;67:105–113. doi: 10.1124/mol.104.004465. [DOI] [PubMed] [Google Scholar]
- 357.Zhai P, Eurell TE, Cooke PS, Lubahn DB, Gross DR. Myocardial ischemia-reperfusion injury in estrogen receptor-alpha knockout and wild-type mice. Am J Physiol Heart Circ Physiol. 2000;278:H1640–H1647. doi: 10.1152/ajpheart.2000.278.5.H1640. [DOI] [PubMed] [Google Scholar]
- 358.Zhai P, Eurell TE, Cotthaus R, Jeffery EH, Bahr JM, Gross DR. Effect of estrogen on global myocardial ischemia-reperfusion injury in female rats. Am J Physiol Heart Circ Physiol. 2000;279:H2766–H2775. doi: 10.1152/ajpheart.2000.279.6.H2766. [DOI] [PubMed] [Google Scholar]
- 359.Gabel SA, Walker VR, London RE, Steenbergen C, Korach KS, Murphy E. Estrogen receptor beta mediates gender differences in ischemia/reperfusion injury. J Mol Cell Cardiol. 2005;38:289–297. doi: 10.1016/j.yjmcc.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 360.Klein BE, Klein R, Linton KL The Beaver Dam Eye Study. Prevalence of age-related lens opacities in a population. Ophthalmology. 1992;99:546–552. doi: 10.1016/s0161-6420(92)31934-7. [DOI] [PubMed] [Google Scholar]
- 361.Klein BE, Klein R, Ritter LL The Beaver Dam Eye Study. Is there evidence of an estrogen effect on age-related lens opacities? Arch Ophthalmol. 1994;112:85–91. doi: 10.1001/archopht.1994.01090130095025. [DOI] [PubMed] [Google Scholar]
- 362.Freeman EE, Munoz B, Schein OD, West SK. Hormone replacement therapy and lens opacities: the Salisbury Eye Evaluation project. Arch Ophthalmol. 2001;119:1687–1692. doi: 10.1001/archopht.119.11.1687. [DOI] [PubMed] [Google Scholar]
- 363.Aina FO, Smeeth L, Hubbard R, Hurt LS, Fletcher AE. Hormone replacement therapy and cataract: a population-based case-control study. Eye. 2006;20:417–422. doi: 10.1038/sj.eye.6701877. [DOI] [PubMed] [Google Scholar]
- 364.Grabowski M, Karpinski G, Rdzanek JFKA, Pietrasik A, Wretowski D, Rudowski R, Opolski G. Diagnostic value of BNP in suspected perimyocarditis--a preliminary report. Kardiol Pol. 2004;61:451–458. discussion 459–460. [PubMed] [Google Scholar]
- 365.Pepin B, Mikol J, Goldstein B, Aron JJ, Lebuisson DA. Familial mitochondrial myopathy with cataract. J Neurol Sci. 1980;45:191–203. doi: 10.1016/0022-510x(80)90165-3. [DOI] [PubMed] [Google Scholar]
- 366.Flynn JM, Lannigan DA, Clark DE, Garner MH, Cammarata PR. RNA suppression of ERK2 leads to collapse of mitochondrial membrane potential with acute oxidative stress in human lens epithelial cells. Am J Physiol Endocrinol Metab. 2008;294:E589–E599. doi: 10.1152/ajpendo.00705.2007. [DOI] [PubMed] [Google Scholar]
- 367.Razmara A, Duckles SP, Krause DN, Procaccio V. Estrogen suppresses brain mitochondrial oxidative stress in female and male rats. Brain Res. 2007;1176:71–81. doi: 10.1016/j.brainres.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Moor AN, Flynn JM, Gottipati S, Giblin FJ, Cammarata PR. 17beta-estradiol stimulates MAPK signaling pathway in human lens epithelial cell cultures preventing collapse of mitochondrial membrane potential during acute oxidative stress. Mitochondrion. 2005;5:235–247. doi: 10.1016/j.mito.2005.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Milenkovic D, Muller J, Stojanovski D, Pfanner N, Chacinska A. Diverse mechanisms and machineries for import of mitochondrial proteins. Biol Chem. 2007;388:891–897. doi: 10.1515/BC.2007.097. [DOI] [PubMed] [Google Scholar]
- 370.MacKenzie JA, Payne RM. Mitochondrial protein import and human health and disease. Biochim Biophys Acta. 2007;1772:509–523. doi: 10.1016/j.bbadis.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]