Summary
Ring-shaped AAA+ ATPases control a variety of cellular processes by substrate unfolding and remodeling of macromolecular structures. However, how ATP hydrolysis within AAA+ rings is regulated and coupled to mechanical work is poorly understood. Here, we demonstrate coordinated ATP hydrolysis within m-AAA protease ring complexes, conserved AAA+ machines in the inner membrane of mitochondria. ATP binding to one AAA subunit inhibits ATP hydrolysis by the neighboring subunit leading to coordinated rather than stochastic ATP hydrolysis within the AAA ring. Unbiased genetic screens define an intersubunit signaling pathway involving conserved AAA motifs and reveal an intimate coupling of ATPase activities to central AAA pore loops. Coordinated ATP hydrolysis between adjacent subunits is required for membrane dislocation of substrates but not for substrate processing. These findings provide new insight how AAA+ proteins convert energy derived from ATP hydrolysis into mechanical work.
Introduction
Important cellular processes depend on AAA+ molecular machines which utilize the energy derived from ATP binding and hydrolysis for protein unfolding and remodeling of macromolecular assemblies (Hanson and Whiteheart, 2005; Erzberger and Berger, 2006). AAA+ proteins usually form ring structures and share a structurally conserved ATPase domain. The latter contains Walker A and B motifs of P-loop ATPases but is defined as a distinct family by a number of additional conserved structural elements (Hanson and Whiteheart, 2005; Erzberger and Berger, 2006). A characteristic feature of AAA+ ring complexes is the formation of ATP binding pockets at intersubunit interfaces. Most importantly, a conserved arginine residue of one AAA+ domain contributes to the ATP binding pocket of the neighboring subunit by contacting the γ-phosphate of bound ATP (Ogura et al., 2004).
A number of operational modes have been described for AAA+ proteins, which suggest different degrees of intersubunit coupling and include sequential as well as stochastic hydrolysis events within ring complexes (Martin et al., 2005; Enemark and Joshua-Tor, 2006; Moreau et al., 2007; Werbeck et al., 2008). The derived energy is thought to trigger conformational changes exerting mechanical force on substrates. This is perhaps best understood for energy-dependent proteases (Baker and Sauer, 2006). These multisubunit AAA+ machines unfold their substrates and thread unfolded polypeptides through a central pore allowing their degradation in a protected proteolytic chamber. Current evidence suggests that conserved loop regions protruding into the central channel of diverse AAA+ rings play a crucial role in these processes (Song et al., 2000; Yamada-Inagawa et al., 2003; Weibezahn et al., 2004; Hinnerwisch et al., 2005; DeLaBarre et al., 2006; Tatsuta et al., 2007; Martin et al., 2008). Crosslinking studies suggest nucleotide-dependent interactions of substrates with the conserved pore loop-1 at the top and the less conserved pore loop-2 at the bottom of the AAA+ channel (Weibezahn et al., 2004; Hinnerwisch et al., 2005; Martin et al., 2008).
AAA proteases comprise a distinct family of membrane-bound AAA+ unfoldases, in which AAA+ domains are covalently attached to a peptidase domain (Koppen and Langer, 2007). m-AAA proteases are localized in the inner membrane of mitochondria where they form multimeric complexes exposing their catalytic sites to the matrix space. While the yeast orthologue is hetero-oligomeric and consists of homologous Yta10 (Afg3) and Yta12 (Rca1) subunits (Arlt et al., 1996), two isoenzymes exist in human mitochondria: a homo-oligomeric enzyme composed of AFG3L2 subunits and a hetero-oligomeric enzyme containing paraplegin and AFG3L2 subunits (Koppen et al., 2007). Inactivation of mammalian m-AAA proteases leads to axonal degeneration and is associated with various neurodegenerative disorders (Ferreirinha et al., 2004; Maltecca et al., 2008). Complementation studies revealed functional conservation of mammalian and yeast m-AAA proteases (Koppen et al., 2007) which possess versatile activities essential for mitochondrial biogenesis including the processing of key regulatory proteins and quality control surveillance (Esser et al., 2002; Nolden et al., 2005; Koppen and Langer, 2007).
The hetero-oligomeric assembly of the yeast m-AAA protease allows an independent mutagenesis of subunits and thus offers a unique opportunity to define mechanisms regulating ATP hydrolysis by AAA+ machines. Here, we demonstrate coordinated ATP hydrolysis by m-AAA proteases and define conserved sequence motifs within AAA+ domains as crucial components of the underlying intersubunit signaling pathway.
Results
Coordinated ATP hydrolysis within m-AAA protease complexes
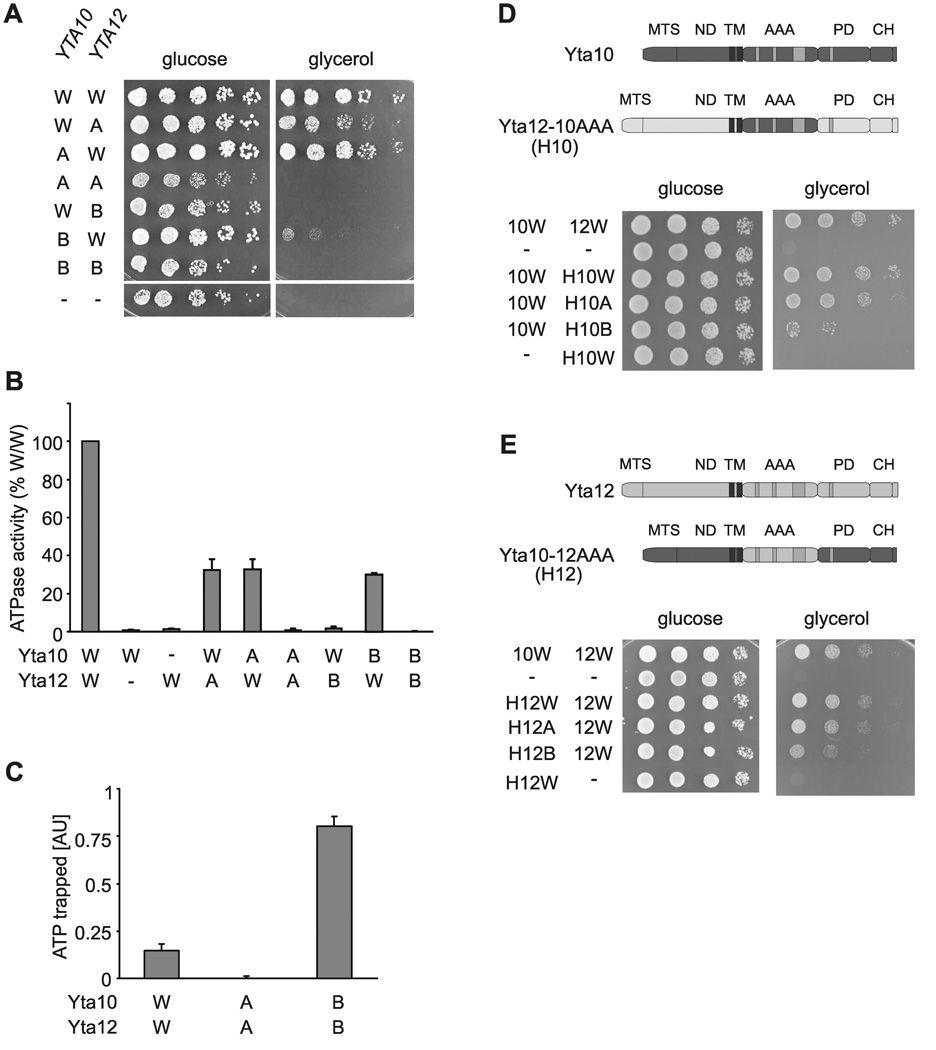
To characterize the regulation of ATP hydrolysis by m-AAA proteases, we introduced point mutations into the Walker A motifs of the conserved AAA domains of Yta10 and Yta12 (Yta10K334A, Yta12K394A). While Δyta10Δyta12 cells expressing Yta10 and Yta12 were able to grow under respiratory conditions, coexpression of Yta10K334A and Yta12K394A did not restore respiratory growth ((Tatsuta et al., 2007); Fig. 1A). In contrast, expression of either the Yta10 or the Yta12 mutant variant in combination with the wild type form of the respective other subunit promoted respiratory growth of Δyta10Δyta12 cells, suggesting that the activity of the m-AAA protease is maintained if only one type of subunit is active (Fig. 1A). Mutations in Walker B motifs of Yta10 or Yta12 (Yta10E388Q, Yta12E448Q) had a strikingly different effect. While mutations in both subunits inhibited respiratory growth as was observed for the Walker A mutants (Tatsuta et al., 2007), coexpression of either Walker B mutant variant with the wild type form of the respective other subunit did not or only weakly restore growth on non-fermentable carbon sources (Fig. 1A).
Figure 1. Coordinated ATP hydrolysis within yeast m-AAA protease complexes.
(A) Growth of Δyta10Δyta12 cells on fermentable (glucose) and non-fermentable (glycerol) carbon sources upon expression of wild type and mutant variants of Yta10 and Yta12. W/W, Yta10-6His/Yta12; W/A, Yta10-6His/Yta12K394A; A/W, Yta10K334A-6His/Yta12; A/A, Yta10K334A-6His/Yta12K394A; W/B, Yta10-6His/Yta12E448Q; B/W, Yta10E388Q-6His/Yta12; B/B, Yta10E388Q-6His/Yta12E448Q; −/−, vector control.
(B) ATPase activities of wild type and mutant m-AAA proteases in vitro. W/−, Yta10-6His/empty vector; −/W, empty vector/Yta12-6His.
(C) Detection of trapped ATP co-purifying with m-AAA protease variants by a luciferase-based assay. Data represent the mean +/− standard deviation of three independent experiments. AU, arbitrary units.
(D) Upper panel, schematic representation of Yta10 and an Yta12-derived chimeric subunit harboring the AAA domain of Yta10 (H10). Lower panel, growth of Δyta10Δyta12 cells expressing Yta10 (10W) and Yta12 (12W) or variants thereof as indicated. MTS, mitochondrial targeting sequence; ND, N-terminal domain; AAA, AAA domain; PD, proteolytic domain; CH, C-terminal helices.
(E) Upper panel, schematic representation of Yta12 and an Yta10-derived chimeric subunit harboring the AAA domain of Yta12 (H12). Lower panel, growth of Δyta10Δyta12 cells expressing Yta12 (12W) and Yta10 (10W) or variants thereof as indicated.
We purified m-AAA protease complexes to define the effect of these mutations on the ATPase activity ((Tatsuta et al., 2007); Fig. S1). Regardless of the relative expression of Yta10 and Yta12, both subunits were present at equimolar concentrations in purified complexes (Arlt et al., 1996) that form hexameric rings (Fig. S1 and unpublished observations) similar to most other AAA+ proteins (Hanson and Whiteheart, 2005; Erzberger and Berger, 2006). Genetic and biochemical evidence suggests an alternating rather than a stochastic subunit arrangement within m-AAA protease complexes (Fig. S2). Mutations in the Walker A or B motifs of Yta10 and Yta12 did not affect assembly of m-AAA complexes but abolished ATP hydrolysis completely when present in both subunits (Fig. 1B; Fig. S1). Similar to other AAA+ proteins, replacement of the conserved lysine residue within the Walker A motif impaired ATP binding, whereas ATP is trapped if the conserved glutamate residue within the Walker B motif is mutated (Fig. 1C; (Weibezahn et al., 2003; Dalal et al., 2004)).
m-AAA proteases with mutant Walker A motifs in one subunit exhibited ATPase activities of ~30% when compared to wild type, irrespective of which subunit carried the mutation (Fig. 1B). Thus, both subunits can hydrolyze ATP independently. The reduced ATPase activity of the mutant m-AAA proteases is still sufficient to maintain respiratory growth (Fig. 1A). Mutations in the Walker B motif of Yta10 reduced the ATPase activity of m-AAA complexes to ~30% (Fig. 1B), whereas the corresponding mutation in Yta12 almost completely inhibited the ATPase activity of the mutant m-AAA protease despite the presence of wild type Yta10 in these complexes (Fig. 1B). Yta10 still binds ATP under these conditions as suggested by the almost unaltered Km for ATP of these complexes (Fig. S3). We therefore conclude that ATP binding to Yta12 blocks ATP hydrolysis by Yta10, pointing to a regulatory mechanism which coordinates ATP hydrolysis by m-AAA protease subunits.
To investigate whether coordinated ATP hydrolysis requires two different AAA domains or can also occur in homo-oligomeric AAA rings, we replaced AAA domains of Yta10 or Yta12 by the AAA domains of the respective other subunit. Expression of the Yta12 hybrid subunit containing the AAA domain of Yta10 (termed H10; Fig. 1D) supported respiratory growth of Δyta10Δyta12 cells only when coexpressed with Yta10 (Fig. 1D) indicating formation of functional complexes (Fig. S4). Similar results were obtained when the AAA domain of Yta10 was replaced by that of Yta12 (Fig. 1E; Fig. S4). Thus, m-AAA protease complexes containing six identical AAA domains are functionally active in vivo. Mutations in the Walker A motifs of H10 or H12 affected respiratory growth only slightly (Fig. 1D, E). Mutations in Walker B motifs of H10 and H12, however, strongly inhibited respiratory growth, ATPase and the proteolytic activities of the assembled proteases in vivo (Fig. 1D, E and Fig. S5), suggesting that ATP binding impairs ATP hydrolysis in homo-oligomeric AAA rings.
ATP binding coordinates ATP hydrolysis in human m-AAA complexes
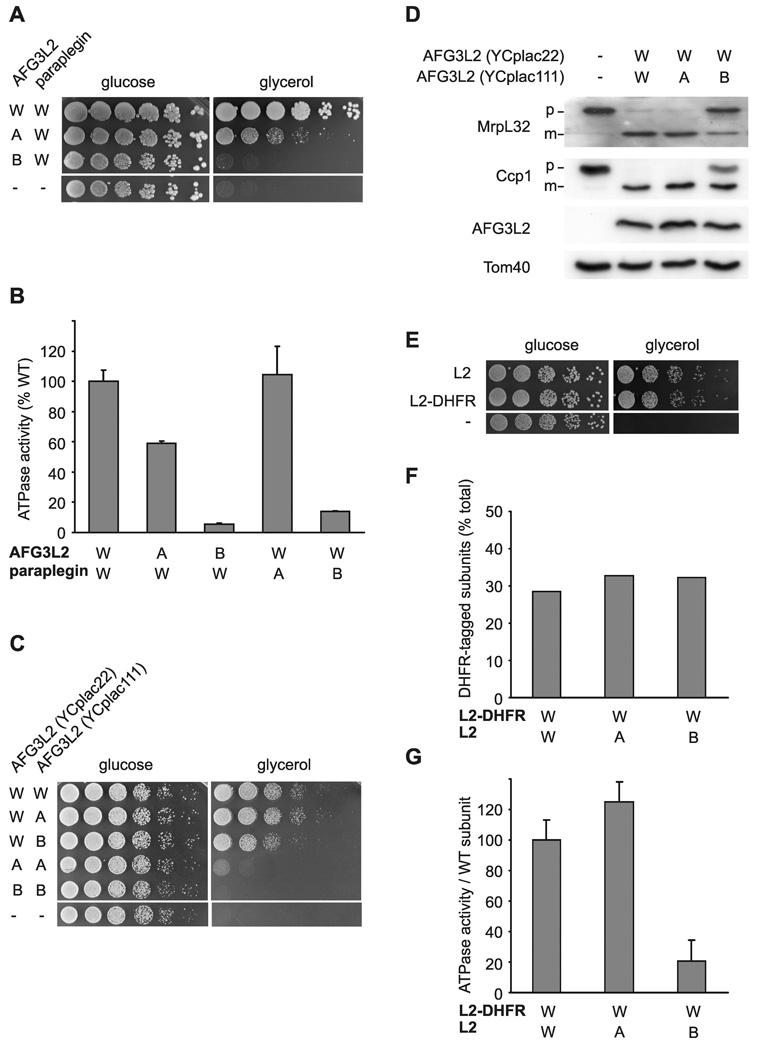
To examine whether coordinated ATP hydrolysis is conserved in human m-AAA proteases, we expressed paraplegin and AFG3L2 or variants thereof harboring mutations in the AAA domain in Δyta10Δyta12 cells. In this experimental setting, respiratory growth depends on paraplegin subunits within the hetero-oligomeric m-AAA isoenzyme when inactive AFG3L2 variants are expressed (Koppen et al., 2007). This allowed us to monitor effects of specific mutations within the AAA domain of AFG3L2 on paraplegin activity in vivo. Coexpression of C-terminally hexahistadine tagged paraplegin with the Walker A AFG3L2 mutant partially restored respiratory growth of Δyta10Δyta12 cells (Fig. 2A). In contrast, respiratory growth was completely abolished when paraplegin was coexpressed with AFG3L2 harboring a mutation in the Walker B motif (Fig. 2A). Mutations in Walker B but not Walker A motifs of AFG3L2 inhibited almost completely the ATPase activity of purified hetero-oligomeric m-AAA complexes in vitro (Fig. 2B). Similar effects were observed when m-AAA proteases with analogous mutant subunits of paraplegin were analyzed (Fig. 2B). Thus, only mutations in the Walker B motif inhibited ATP hydrolysis in hetero-oligomeric human protease complexes, consistent with an inhibitory effect of ATP binding on adjacent subunits.
Figure 2. Dominant negative effects of mutations in the Walker B motif on homo- and hetero-oligomeric human m-AAA proteases.
(A) Growth of Δyta10Δyta12 cells on fermentable (glucose) and non-fermentable (glycerol) carbon sources upon co-expression of wild type paraplegin with AFG3L2 or its mutant variants. W/W, AFG3L2/paraplegin-6His; A/W, AFG3L2K354A/paraplegin-6His; B/W, AFG3L2E408Q/paraplegin-6His −/−, vector control (YCplac22/YEplac181).
(B) In vitro ATPase activities of hetero-oligomeric human m-AAA proteases harboring mutations in Walker A or B motifs. Activities were normalized to the number of assembled complexes. W/A, AFG3L2/parapleginK355A-6His; W/B, AFG3L2/ parapleginE409Q-6His.
(C) Growth of Δyta10Δyta12 cells upon co-expression of wild type and mutant variants of AFG3L2 harboring mutations within Walker A or B motifs. W/W, AFG3L2/AFG3L2; W/A, AFG3L2/AFG3L2K354A; W/B, AFG3L2/AFG3L2E408Q; A/A, AFG3L2K354A/AFG3L2K354A; B/B, AFG3L2 E408Q/AFG3L2E408Q; −/−, vector control.
(D) Processing of MrpL32 and Ccp1 in Δyta10Δyta12 cells expressing AFG3L2 and mutant variants thereof. Total proteins were extracted by alkaline lysis following separation by SDS-PAGE and immune blotting using antisera directed against Ccp1, MrpL32, AFG3L2 and Tom40. p, precursor; m, mature.
(E) Respiratory competence of Δyta10Δyta12 cells expressing AFG3L2 (L2) or DHFR-tagged AFG3L2 (L2-DHFR).
(F) Subunit stoichiometry in homo-oligomeric human m-AAA proteases composed of DHFR- and his-tagged subunits harboring mutations when indicated. After SDS-PAGE and coomassie staining analysis of purified complexes, bands were densitometorically quantified. The proportion of DHFR-tagged subunits is shown. Since m-AAA proteases were purified by metal chelating chromatography from the pool of stochastically formed complexes, his-tagged subunits are overrepresented in the purified fraction.
(G) In vitro ATPase activities of human AFG3L2 complexes composed of L2-DHFR and AFG3L2 subunits carrying mutations in Walker A or B motifs when indicated. Activities were normalized to the number of assembled wild type subunits in the complexes. Data in (B) and (G) represent the mean +/− standard deviation of three independent experiments.
Next, we assessed whether homo-oligomeric AFG3L2 complexes are regulated similarly. Δyta10Δyta12 cells expressing AFG3L2 variants concomitantly from two plasmids were able to grow on non-fermentable carbon sources, while mutations in Walker A- and B motifs introduced into both Afg3l2 genes abolished respiratory growth (Fig. 2C). Interestingly, a Walker A variant of AFG3L2 (AFG3L2K354A) maintained respiratory growth more efficiently than a Walker B variant (AFG3L2E408Q) upon coexpression with AFG3L2 (Fig. 2C). This dominant negative effect was substantiated by monitoring in vivo processing of cytochrome c peroxidase (Ccp1) and of the ribosomal protein MrpL32, two known substrates of the m-AAA protease in yeast mitochondria ((Esser et al., 2002; Nolden et al., 2005); Fig. 2D).
To unambiguously demonstrate assembly of mutant and wild type AFG3L2 subunits, we coexpressed AFG3L2 chimera containing a C-terminal DHFR domain together with AFG3L2 variants which harbor a C-terminal hexahistidine tag and mutations in Walker A-or Walker B-motifs. The fusion of the DHFR domain to the C-terminus of AFG3L2 did not impair respiratory competence (Fig. 2E), but allowed assessment of the relative amount of wild type subunits within assembled complexes. DHFR- and his-tagged AFG3L2 assembled stochastically regardless of the presence of mutations in his-tagged subunits (Fig. 2F). We determined ATPase activities of purified AFG3L2 complexes and normalized them to the number of (DHFR-tagged) wild type subunits (Fig. 2G). These experiments revealed that AFG3L2 subunits carrying a mutation in the Walker A motif did not affect the ATPase activity of wild type subunits in a complex (Fig. 2G). Conversely, subunits harboring a mutation in their Walker B motif exert a dominant negative effect and inhibit ATP hydrolysis by homo-oligomeric AFG3L2 complexes (Fig. 2G).
We therefore conclude that coordinated ATP hydrolysis by AAA domains of m-AAA protease subunits is conserved from yeast to man, and applies to both homo- and hetero-oligomeric m-AAA complexes. While individual m-AAA subunits can hydrolyze ATP independent of each other, ATP hydrolysis is inhibited when neighboring m-AAA subunits are in the ATP-bound state.
Defining intersubunit signaling within m-AAA ring complexes
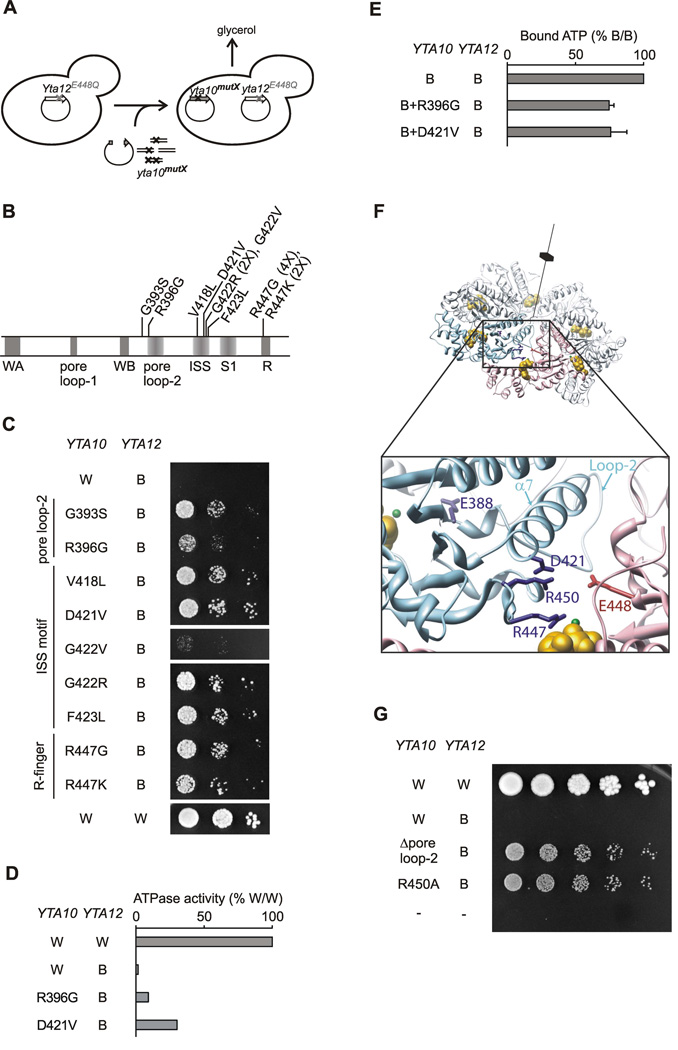
Coordinated ATP hydrolysis within m-AAA rings requires sensing of nucleotide binding by neighboring subunits. To identify amino acid residues involved in intersubunit communication, an unbiased genetic screen was performed to identify suppressor mutations in Yta10, which would alleviate the ATPase activity block of Yta10 imposed upon ATP binding to Yta12 (Fig. 3A). We reasoned that an Yta10 variant, which carries a mutation in an amino acid residue involved in sensing the nucleotide-bound state of Yta12, would be able to hydrolyze ATP and support respiratory growth, even if Yta12 harbors a mutation in the Walker B motif. Notably, mutations of conserved amino acid residues abolishing ATPase activity would not be isolated using such a gain-of-function screening procedure.
Figure 3. Identification of amino acid residues in Yta10 involved in intersubunit signaling.
(A) Screen for suppressor mutations in Yta10 allowing respiratory growth in the presence of a mutation in the Walker B motif of Yta12.
(B) Suppressor mutations in Yta10. Conserved motifs are indicated by grey bars. The number of identified clones harboring the specific mutation is given in brackets. WA, Walker A motif; WB, Walker B motif; S1, sensor-1 motif; R, arginine finger; ISS, intersubunit signaling motif.
(C) Respiratory growth of Δyta10Δyta12 cells co-expressing dominant negative Yta12E448Q (B) and suppressing alleles of Yta10.
(D) ATPase activities of mutant m-AAA protease complexes harboring Yta12E448Q and suppressor alleles of Yta10.
(E) ATP binding to mutant m-AAA protease complexes composed of Yta12E448Q subunits and Yta10E388Q subunits harboring suppressor alleles. ATP co-purifying with m-AAA protease variants was detected using a luciferase-based assay. Data represent the mean +/− standard deviation of three independent experiments.
(F) Localization of suppressors in Yta10 based on an atomic model of the yeast m-AAA protease hexamer. Yta10 is shown in cyan and Yta12 in pink. For clarity, only one pair of Yta10 and Yta12 is colored. Identified suppressor mutations and conserved glutamates in the Walker B motifs are shown as stick model. Bound nucleotide and magnesium ions are shown as CPK model and colored gold and green, respectively. The position of the 6-fold symmetry axis is indicated. The inset depicts a close-up view of the Yta10/Yta12 interface and shows the position of pore loop-2 of Yta10.
(G) Deletion of the pore loop-2 or a mutation of the arginine residue R450 in Yta10 also suppresses the respiratory incompetence caused by the mutation in the Walker B motif of Yta12.
Nine individual mutations in Yta10 were identified, which suppress the inhibitory effect of the Walker B Yta12 mutant (Fig. 3B, C). These suppressor mutations are located in three defined regions within the AAA domain of Yta10 (Fig. 3B; Fig. S6). Mutations replacing the conserved arginine finger residue (Yta10R447) were isolated most frequently. Other mutations cluster to a region (residues 418–423) at the end of the helix α7 and preceding sensor-1 of Yta10. This region, which we termed the ISS (intersubunit signaling) motif, is conserved within the classical AAA clade of proteins (Fig. S6). The third set of suppressor mutations affects a flexible loop after the Walker B motif of Yta10. This loop is highly charged, protrudes into the central pore of the AAA ring and corresponds structurally to the pore loop-2 present in many AAA+ ATPases ((Hinnerwisch et al., 2005; Martin et al., 2008); Fig. S6). Consistent with a role for intersubunit signaling, suppressor mutations did not significantly affect ATP binding by purified m-AAA proteases but did restore, at least partially, the intrinsic ATPase activities of complexes harboring Yta12E448Q subunits upon assembly with Yta10 (Fig. 3D, E; Fig. S7).
To examine the spatial relationship of the affected amino acid residues, we generated an atomic model of the hetero-oligomeric yeast m-AAA protease from the crystal structure of a soluble domain of FtsH (Bieniossek et al., 2006; Suno et al., 2006). In our model, the ISS motif in helix α7 is located close to the arginine finger R447 making possible an interaction of D421 with R450 (Fig. 3F). Moreover, helix α7 directly follows the pore loop-2 and the Walker B motif. This arrangement suggests that a potential interaction between D421 and R450 couples movements of the arginine finger to amino acid residues in the ATP binding pocket of Yta10. This hypothesis was substantiated by mutations introduced at both sites. The growth defect caused by Yta12E448Q was at least partially suppressed by replacing R450 with alanine or a deletion of five amino acids within the pore loop-2 (Fig. 3G).
Taken together, our results are consistent with a signaling cascade from Yta12 to Yta10, which involves sensing of the nucleotide-bound state of Yta12 by the arginine finger R447 of Yta10, and transmission of this information through R450, D421 and helix α7 to the Walker B motif and the ATP binding pocket of Yta10.
Intersubunit signaling within Yta12
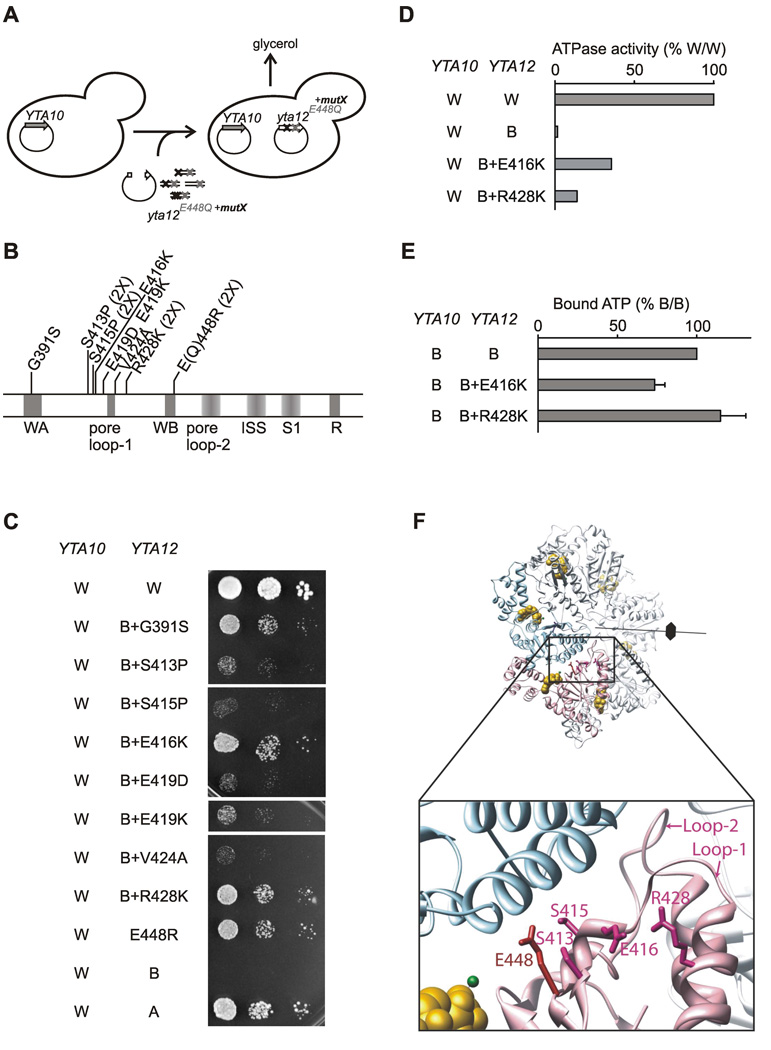
In order to identify amino acid residues in Yta12 required for intersubunit communication, we performed a similar genetic screen to identify intragenic suppressors for the growth defect associated with the Yta12 Walker B mutant (Fig. 4A). Nine suppressor mutations in Yta12E448Q were identified supporting respiratory growth (Fig. 4B). A mutation of the conserved glycine residue within the Walker A motif of Yta12 (G391S) suppresses the inhibitory effect of the Walker B mutation most likely due to impaired nucleotide binding to Yta12 (Fig. 4C). Replacing the originally introduced glutamine in the Walker B motif (E448Q) with arginine also allowed respiratory cell growth (Fig. 4C). The majority of identified suppressor mutations, however, affect the region surrounding the conserved pore loop-1 of Yta12 (Fig. 4C), which is crucial for substrate translocation through the central pore of many AAA+ ATPases (Song et al., 2000; Yamada-Inagawa et al., 2003; Weibezahn et al., 2004; Hinnerwisch et al., 2005; DeLaBarre et al., 2006; Tatsuta et al., 2007; Martin et al., 2008).
Figure 4. Identification of amino acid residues in Yta12 involved in intersubunit signaling.
(A) Screen for intragenic suppressor mutations in Yta12 allowing respiratory growth in the presence of mutations in the Walker B motif of Yta12.
(B) Intragenic suppressor mutations in Yta12. Conserved motifs are indicated as in Fig. 3B.
(C) Respiratory competence of Δyta10Δyta12 cells which co-express wild type Yta10 (W) and Yta12E448Q (B) harboring suppressor mutations.
(D) ATPase activities of mutant m-AAA protease complexes composed of Yta10 subunits and Yta12E448Q variants harboring intragenic suppressor mutations.
(E) ATP binding to mutant m-AAA protease complexes composed of Yta10E388Q subunits and Yta12E448Q subunits harboring suppressor alleles. ATP co-purifying with m-AAA protease variants was detected using a luciferase-based assay. Data represent the mean +/− standard deviation of three independent experiments.
(F) Localization of suppressors in Yta12 based on an atomic model of the yeast m-AAA protease hexamer. Yta10 and Yta12 subunits and other key features are shown as described in Fig. 3F. The inset depicts a close-up view of the Yta10/Yta12 interface and shows the position of pore loop-1 and -2 of Yta12.
The suppressor mutations E416K and R428K restored respiratory growth most efficiently and increased the ATPase activities of purified m-AAA proteases at least to some extent (Figs. 4C, D). Concurrently, they did not inhibit ATP binding to mutant m-AAA complexes and, therefore, could be directly involved in intersubunit signaling (Fig. 4E). In our atomic model of the m-AAA protease, the region preceding the pore loop-1 is close to E448 in the Walker B motif suggesting that many suppressor mutations may affect coordination and positioning of the nucleotide within the ATP binding pocket of Yta12 (Fig. 4F). Interestingly, the corresponding region was recently proposed to regulate the ATPase activity of related AAA+ proteins in response to ligand binding (Joly et al., 2008; Zhang and Wigley, 2008). Consistently, the identification of intragenic suppressors adjacent to the pore loop-1 of Yta12 points to an intimate coupling between coordinated ATP hydrolysis within the AAA+ ring and substrate handling.
Substrate-specific requirement for intersubunit coordination
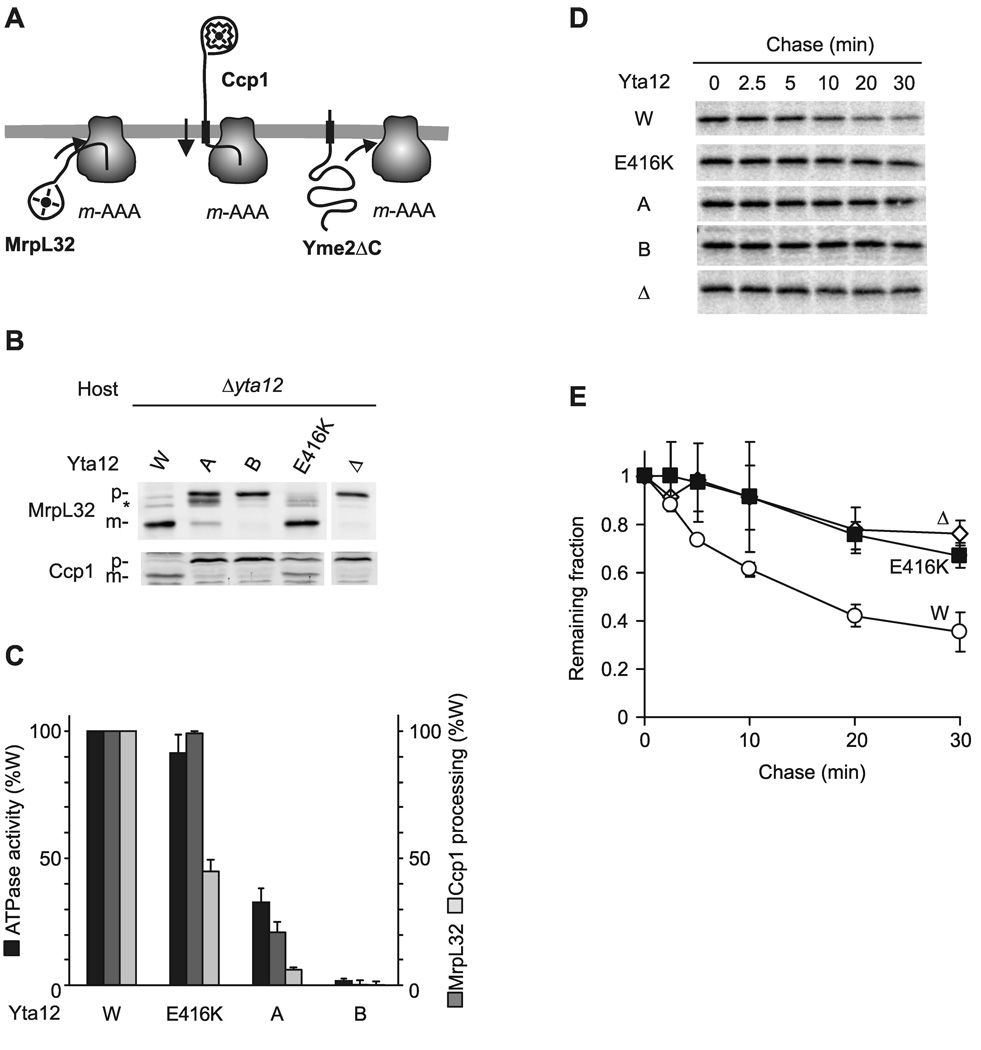
To determine the role of coordinated ATP hydrolysis within the AAA+ ring for substrate handling directly, we analyzed the effect of an impaired intersubunit signaling for various m-AAA protease activities, which differ in their energy requirements (Fig. 5A; (Leonhard et al., 2000; Tatsuta et al., 2007)). For instance, processing of soluble MrpL32 requires only insertion of N-terminal segments of MrpL32 into the proteolytic chamber of the m-AAA protease. It depends on ATP hydrolysis but can be mediated by m-AAA variants with low ATPase activity (Nolden et al., 2005). On the other hand, processing of Ccp1 or turnover of the misfolded inner membrane protein Yme2ΔC requires ATP-dependent membrane dislocation of substrates prior to proteolysis (Leonhard et al., 2000; Tatsuta et al., 2007).
Figure 5. Intersubunit signaling within Yta12 and substrate handling by the m-AAA protease.
(A) Schematic representation of proteolytic substrates of the m-AAA protease. Transmembrane segments are shown as black boxes, folded domains as circles. Ccp1 but not MrpL32 processing depends on ATP-dependent membrane dislocation driven by the m-AAA protease (Tatsuta et al., 2007). Yme2ΔC is extracted from the membrane and degraded processively (Leonhard et al., 2000).
(B) Processing of MrpL32 and Ccp1 in Δyta12 cells (Δ) expressing Yta12 or Yta12 variants harboring mutations in the Walker A- (K394A) or the Walker B- (E448Q) motif or a suppressor mutation preceding the pore loop-1 (E416K). p, precursor form of MrpL32 or Ccp1; m, mature form of MrpL32 or Ccp1; asterisk, intermediate form of MrpL32.
(C) ATPase activities of purified m-AAA protease variants containing mutant Yta12 subunits in vitro and their processing activity (Ccp1, MrpL32) in vivo.
(D) Stability of Yme2ΔC in mitochondria. 35S-labeled Yme2ΔC was imported into mitochondria harbouring Yta10 and variants of Yta12 as indicated. After removal of non-imported precursor proteins by trypsin, samples were incubated at 37°C for the time indicated to allow proteolysis to occur.
(E) 35S-labeled mature Yme2ΔC was quantified at different time points by phosphoimaging. Newly imported Yme2ΔC present in mitochondria before the temperature shift to 37°C was set to 1. Data represent the mean +/− standard deviation of three independent experiments. Open circles, WT (W); open diamonds, Δyta12 (Δ); closed squares, Yta12E416K (E416K).
We expressed Yta12 variants harboring mutations in the Walker A- (K394A) or Walker B- (E448Q) motif, or a suppressor mutation preceding the pore loop-1 (E416K) in Δyta12 cells and monitored their activity upon assembly with Yta10. In agreement with its dominant negative effect on cell growth and ATPase activity of the m-AAA protease (Fig. 1), expression of Yta12E448Q abolished processing of both MrpL32 and Ccp1 as well as turnover of Yme2ΔC (Fig. 5). A mutation in the Walker A motif of Yta12 severely affected the ATPase activity of the m-AAA protease and inhibited Ccp1 processing and turnover of Yme2ΔC (Fig. 5). However, the mutant m-AAA complex allowed respiratory growth (Fig. 1A) and supported at least some processing of MrpL32 (Fig. 5B, C), providing direct evidence for the ability of individual m-AAA subunits to cleave substrates. Strikingly, the suppressor mutation E416K preceding the pore loop-1 did not interfere with processing of MrpL32 but inhibited processing of Ccp1 and degradation of Yme2ΔC strongly (Fig. 5). This is not caused by a deficient ATPase activity of the m-AAA protease, since the ATPase activity was not affected by the Yta12E416K mutation (Fig. 5C).
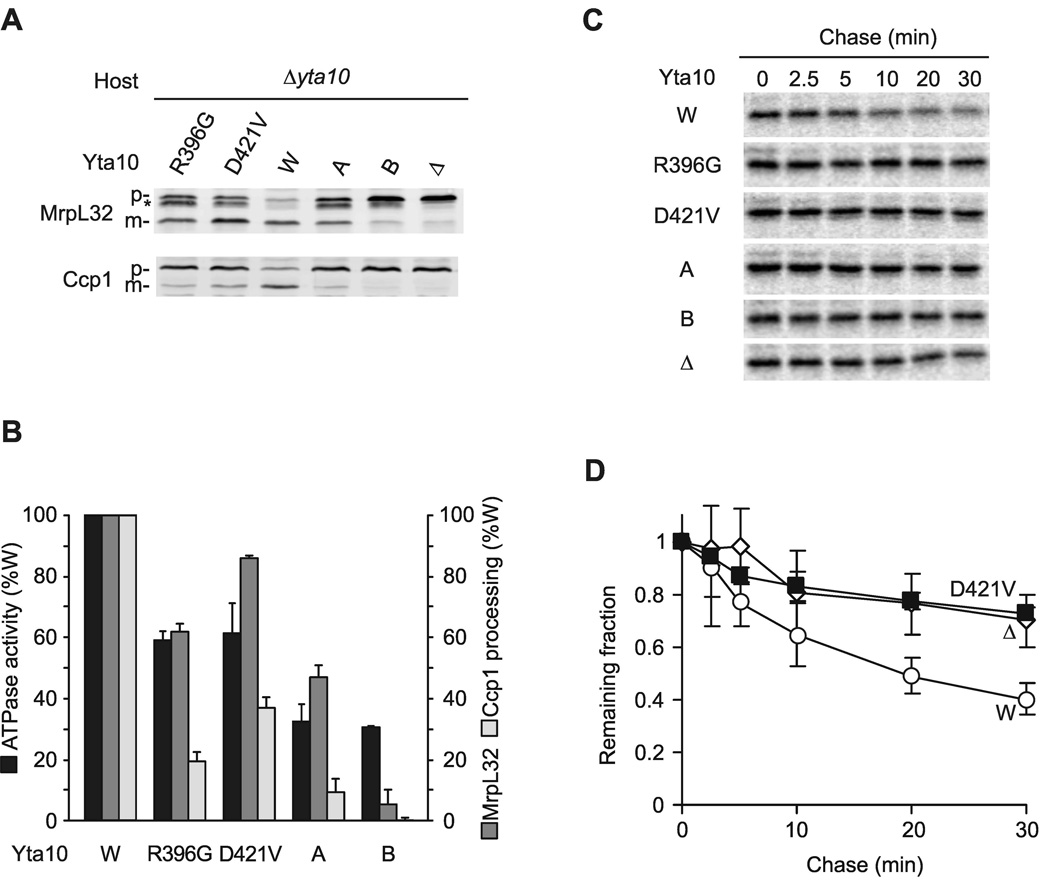
Next, we performed similar experiments with m-AAA protease variants harboring mutations in Yta10 (Fig. 6). Mutations in the Walker A- (K334A) and Walker B- (E388Q) motif of Yta10 reduced significantly the ATPase and proteolytic activities of mutant m-AAA protease complexes (Fig. 6). Ccp1 processing and Yme2ΔC turnover was almost completely inhibited, whereas Yta10K334A allowed MrpL32 processing to an extent sufficient to maintain respiratory growth (Fig. 1A). Mutations in the pore loop-2 (R396G) or the ISS motif (D421V), which prevent intersubunit signaling and suppress the dominant negative effect of Yta12E448Q, affected the ATPase activity to a lesser extent than mutations in either Walker A- or Walker B-motifs (Fig. 6B). Consistently, efficient maturation of MrpL32 occurred in these cells (Fig. 6A, B). However, defective intersubunit signaling impaired severely Ccp1 processing and the degradation of Yme2ΔC (Fig. 6).
Figure 6. Intersubunit signaling in Yta10 and substrate handling by the m-AAA protease.
(A) Processing of MrpL32 and Ccp1 in Δyta10 cells expressing Yta10 or Yta10 variants harboring mutations in the Walker A- (K334A) or Walker B- (E388Q) motif or mutations in pore loop-2 (R396G) or the ISS-motif (D421V) which prevent intersubunit signaling. Membrane fractions (MrpL32) or total cell lysates (Ccp1) were analyzed by SDS-PAGE and immune blotting using antisera directed against Ccp1 and MrpL32. p, precursor form of MrpL32 or Ccp1; m, mature form of MrpL32 or Ccp1; asterisk, intermediate form of MrpL32.
(B) ATPase activities of purified m-AAA proteases containing mutant Yta10 subunits in vitro and their processing activity (Ccp1, MrpL32) in vivo.
(C) Stability of Yme2ΔC in mitochondria. 35S-labeled Yme2ΔC was imported into mitochondria harbouring Yta12 and variants of Yta10 as indicated and the stability was assessed as in Fig. 5D.
(D) 35S-labeled mature Yme2ΔC was quantified at different time points by phosphoimaging as in Fig. 5E. Data represent the mean +/− standard deviation of three independent experiments. Open circles, WT (W); open diamonds, Δyta12 (Δ); closed squares, Yta12D421V (D421V).
Taken together, our experiments suggest that intersubunit signaling and a coordinated ATP hydrolysis within the AAA ring is dispensable for proteolytic activity per se, but essential for the dislocation of substrates by the m-AAA protease.
Coupling of ATPase and substrate processing by pore loop-1
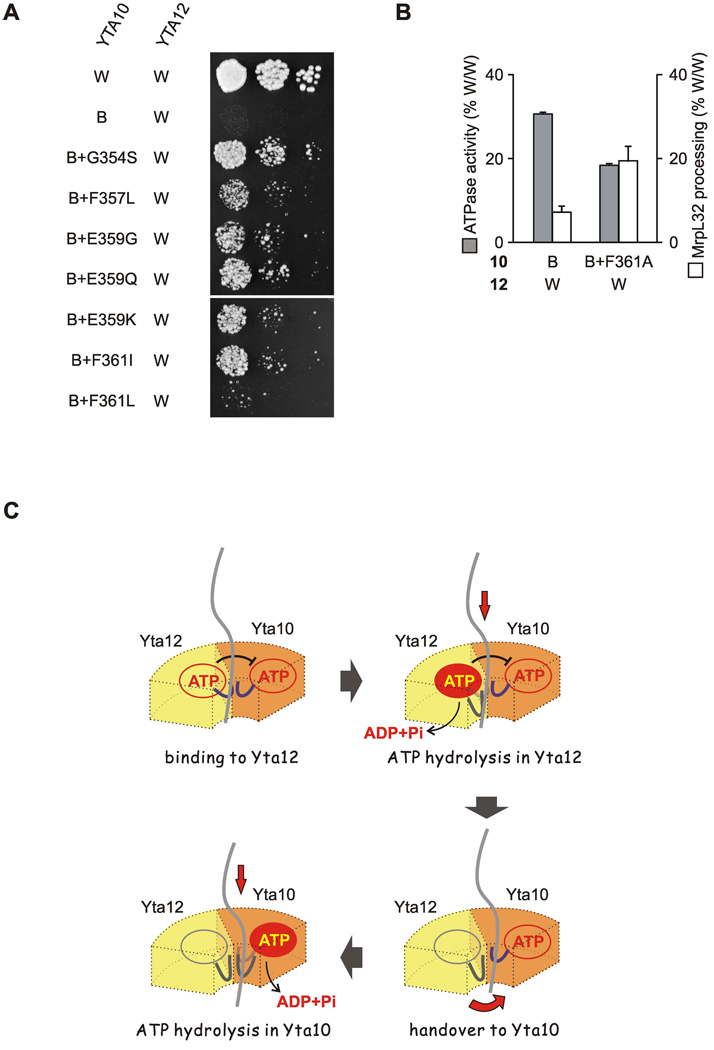
To further dissect the coupling between ATP hydrolysis within the AAA ring and substrate handling, we focused on mutant m-AAA proteases carrying mutations in Walker B motifs of Yta10. These complexes exerted ATPase activity but were barely active in vivo (Fig. 1A, Fig. 6A, B), suggesting an impaired coupling between ATPase and proteolytic activities. We screened for intragenic suppressor mutations in the Walker B mutant of Yta10, which restore the respiratory growth, using a similar strategy as described above. Seven independent suppressor mutations in Yta10E388Q were identified, which cause an amino acid exchange in the pore loop-1 region (Fig. 7A). We purified m-AAA protease complexes composed of wild type Yta12 and mutant Yta10F361A E388Q subunits which harbored mutations in both the Walker B motif and the pore loop-1. Strikingly, ATPase activities of these complexes were slightly reduced when compared to complexes harboring Yta10E388Q subunits, but processing of MrpL32 in vivo was significantly increased (Fig. 7B). These results suggest that ATP binding to Yta10 and pore loop-1 functions in substrate translocation is intimately coupled.
Figure 7. Role of pore loop-1 for coupling of ATPase and proteolytic activities of m-AAA proteases.
(A) Respiratory growth of Δyta10Δyta12 cells expressing Yta12 (W) and Yta10E388Q (B) variants harboring intragenic suppressors. Intragenic suppressor mutations in Yta10E388Q were identified analogous to the gain-of-function screen described in Fig. 4A.
(B) Correlation of MrpL32 processing in vivo with the ATPase activity of a mutant m-AAA protease composed of Yta12 and Yta10E388Q (B) containing a mutation in the pore loop-1 (F361A) when indicated. Data represent the mean +/− standard deviation of three independent experiments.
(C) Coordinated ATP hydrolysis ensures substrate handover between adjacent subunits. See text for details. A heterodimer of Yta10 and Yta12 subunits as the minimal functional unit of the yeast m-AAA protease is shown.
Discussion
Our results reveal coordinated ATP hydrolysis within m-AAA protease ring complexes. ATP binding to one AAA domain inhibits the ATPase activity of the adjacent subunit. ATP hydrolysis abolishes the inhibitory effect and allows subsequent hydrolysis of ATP in the neighboring subunit. Thus, intersubunit signaling within the AAA ring of m-AAA proteases imposes an ordered ATP hydrolysis by at least a subset of subunits.
Dissecting intersubunit signaling: the trans elements
We have identified structural elements involved in intersubunit communication within the AAA ring using a gain-of-function genetic screen and the yeast m-AAA protease as a model. This approach has the advantage that the roles of conserved structural AAA motifs for inter-and intramolecular signaling can be analyzed within a functional AAA complex with ATPase activity. Mutations in conserved structural motifs within Yta10 (trans elements) were found to suppress the inhibitory effect of ATP binding to Yta12: the arginine finger R447, the pore loop-2 and the ISS motif. Our findings assign a central role for signal transmission between AAA domains to the arginine finger, previously demonstrated to be required for ATP hydrolysis in the neighboring subunit (Ogura et al., 2004). The ISS motif is located proximal to the arginine finger and close to R450 at the interprotomer interface, and harbors negatively charged residues conserved within classical AAA proteins. Interestingly, the aspartate residue within the ISS motif of p97 was recently found to interact with an arginine homologous to R450 (Davies et al., 2008). We therefore propose that the salt bridge between the negatively charged residue in the ISS motif and R450 plays an important role in signal transmission between AAA subunits. Although the ISS motif is not conserved in the larger AAA+ superfamily, at least one negatively charged residue is present in the region corresponding to the ISS motif (Iyer et al., 2004). Mutation of this residue abolishes the activity of the DNA helicase RuvB completely (Iwasaki et al., 2000), indicating that the intersubunit signaling mechanism described here may also be conserved within a subset of AAA+ proteins.
The identification of the pore loop-2 of Yta10 as a trans element during intersubunit communication is surprising. In related Clp chaperones, pore loop-2 interacts with substrates and, together with pore loop-1, mediates substrate threading through the pore (Hinnerwisch et al., 2005; Martin et al., 2008). It is therefore conceivable that the nucleotide occupancy in Yta12 affects the conformation of the pore loop-2 in the neighboring Yta10 subunit and thereby controls substrate translocation.
A role of pore loop-1 for intersubunit communication
A coupling of ATP hydrolysis to the conformation of pore loops is also suggested by the observed genetic link between amino acid residues adjacent to pore loop-1 of Yta12 and the nucleotide-binding pocket of Yta12. As suppressor mutations did not abolish ATP binding to Yta12, mutated amino acid residues adjacent to pore loop-1 are likely involved in signaling to Yta10. We favor a model in which conformational changes of the pore loop-1 affect residues in proximity to the Walker B motif, altering the coordination of the bound nucleotide. In agreement with a conservation of this intramolecular pathway within AAA+ proteins, a crucial role for the coupling of ATPase activity and substrate remodeling in the bacterial enhancer binding protein PspF has been assigned to a `glutamate switch´ which include amino acids adjacent to pore loop-1 and the glutamate residue in the Walker B motif (Joly et al., 2008; Zhang and Wigley, 2008).
The pore loop-1 and substrate translocation
The first mechanistic insight how nucleotide occupancy affects the function of pore loop-1 and coordinates substrate translocation between neighboring subunits came from our analysis of m-AAA variants harboring a mutation in the Walker B motif of Yta10. In contrast to corresponding mutations in Yta12, these variants still hydrolyze ATP but are proteolytically inactive, pointing to coupling deficiencies between ATPase and proteolytic activities. Mutations in residues within the pore loop-1 of Yta10, including the conserved hydrophobic residues interacting with substrates in other AAA+ proteins (Song et al., 2000; Yamada-Inagawa et al., 2003; Weibezahn et al., 2004; Hinnerwisch et al., 2005; DeLaBarre et al., 2006; Tatsuta et al., 2007; Martin et al., 2008), alleviate the inhibitory effect and restore proteolysis at least to some extent. Considering that ATP binding increases the substrate affinity of other AAA+ chaperones (Bolon et al., 2004), the presence of ATP-bound Yta10 subunits might trap substrate proteins in the central pore and interfere with substrate translocation by other (active) subunits. Accordingly, suppressor mutations in pore loop-1 of Yta10 may decrease the substrate affinity, allowing release of trapped substrates from mutant subunits and subsequent translocation by active subunits. This model suggests that an ATP-dependent substrate release is required for and directly coupled to substrate translocation by the neighboring subunit.
Coordinated ATP hydrolysis ensures substrate handover between adjacent subunits
Our genetic experiments thus point to a complex regulation of ATP hydrolysis and substrate handling by m-AAA proteases, which is crucial for their functional efficiency. m-AAA protease variants with impaired intersubunit communication exhibit proteolytic activity towards some substrates but cannot handle others, which need to be dislocated from the membrane and therefore require additional energy input (Leonhard et al., 2000; Tatsuta et al., 2007). We propose that coordinated ATP hydrolysis within the AAA ring of m-AAA proteases increases the efficiency of the proteolytic machine by ensuring substrate handover between neighboring subunits (Fig. 7C). Substrates bind to Yta12 subunits in the ATP-bound state. ATP hydrolysis triggers conformational changes of pore loop-1 resulting in substrate translocation and substrate release. Intersubunit coordination of ATP hydrolysis ensures that the adjacent subunit is in the ATP-bound, substrate-acceptor state allowing efficient substrate handover to the neighboring subunit. Notably, evidence for a communication between neighboring subunits in AAA+ rings was previously reported for bacterial ClpX and archaeal MCM (Martin et al., 2005; Moreau et al., 2007). Moreover, substrate translocation by the DNA packaging motor of the bacteriophage φ29, a ring-like ATPase distantly related to AAA+ proteins, depends on successive ATP binding and hydrolysis (Moffitt et al., 2009). Intersubunit coordination of ATP hydrolysis is thus emerging as a common operational principle of ring ATPases.
Operational modes of AAA rings
While a regulated ATP hydrolysis by adjacent subunits is conserved from yeast to human m-AAA proteases, the degree of coordination within the AAA ring remains to be determined and may vary between orthologous enzymes. ATP binding to both human paraplegin and AFG3L2 inhibits ATP hydrolysis in the neighboring subunit but the nucleotide occupancy at a given time will determine how many subunits hydrolyze ATP coordinately. In the yeast m-AAA protease only the ATP-bound state of Yta12, but not that of Yta10, impairs ATP hydrolysis by neighboring subunits, suggesting that dimers of Yta10 and Yta12 are the functional unit of the enzyme. Thus, coordinated ATP hydrolysis does not necessarily involve all six subunits in the AAA ring. Therefore, our findings can also be reconciled with previous observations on the bacterial unfoldase ClpX, which can operate by random ATP hydrolysis within the AAA+ ring (Martin et al., 2005). In view of the observed substrate-specific requirement for intersubunit coordination in m-AAA proteases, it is an attractive possibility that different degrees of coordination may explain differences in the unfolding capacity of AAA+ proteolytic machines (Koodathingal et al., 2009) and reflect adaptations of a conserved regulatory principle to specific biological needs.
Experimental procedures
Yeast strains and growth conditions
Yeast strains used in this study are listed in Table S1 (host strains) and S2 (transformants) in the supplement. Genes were deleted or modified by polymerase chain reaction (PCR)-targeted homologous recombination (Wach et al., 1994). Cells were cultivated at 30°C in YP medium supplemented with 2 % (w/v) glucose or 2 % (w/v) galactose and 0.5 % (w/v) lactate, or synthetic complete (SC) medium supplemented with 2 % (w/v) glucose if not indicated otherwise.
Genetic screening procedures
Identification of trans elements involved in intersubunit signaling
To isolate mutations in Yta10 suppressing the respiratory growth defect imposed by Yta12E448Q, a 2.2 Kb fragment encoding the whole AAA+ domain and flanking regions of Yta10 was amplified from the plasmid YCplac22ADH1-yta10-6His by error-prone PCR with the primers TL 4404 (TTCGCTCGCGCATGCCAGTT) and TL 4405 (TGCCTCAGGCGGTTCGGTAT). YKO200 (Δyta10Δyta12) cells carrying the plasmid YCplac111ADH1-yta12E448Q were transformed with the PCR fragment and a 6.4 Kb fragment resulting from restriction digest of YCplac22ADH1-yta10E388Q-6His by BsmBI and NcoI. Transformants were selected on SC-LEU-TRP plates containing glucose and replica plated on SC-LEU-TRP plates containing glycerol as the sole carbon source for selection. Plasmids were recovered from growing colonies and mutations in Yta10 were identified by DNA sequencing. Single mutations were introduced into YCplac22ADH1-yta10-6His by site-directed mutagenesis and tested for their suppressive activity.
Identification of cis elements involved in intersubunit signaling
To search for intragenic suppressors of Yta12E448Q, a 2.0 Kb fragment encoding the whole AAA+ domain and flanking regions of Yta12 was amplified from YCplac111ADH1-yta12E448Q by error-prone PCR with the primers TL 4406 (ACTATACCGGCTAACTGG) and TL 4407 (CTGTCATGGCATTCTTGG). YKO200 cells carrying the plasmid YCplac22ADH1-yta10-6His were transformed with the PCR fragment and an 8.1 Kb fragment resulting from restriction digest of YCplac111ADH1-yta12E448Q by NcoI and BsaAI. Transformants were selected and suppressor mutations analyzed as described above.
Identification of intragenic suppressors of a Yta10 Walker B mutation
Suppressor mutations were isolated using a similar strategy as described above with minor modifications. The plasmid YCplac22ADH1-yta10E388Q-6His was used as a template for the error-prone PCR and YKO200 cells carrying the plasmid YCplac111ADH1-YTA12 served as a host. Respiratory growth was assessed at 33.5°C.
Detection of ATP bound to the m-AAA protease
ATP co-purifying with m-AAA protease complexes was detected using a commercially available luciferase assay (Biaffin). Protease complexes were dissolved in 50 µl buffer L (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10% [v/v] glycerol, 300 mM imidazole, 0.2 % [v/v] NP-40), at a final concentration of 60 nM (based on an AAA protease hexamer) and incubated for 5 min at 95°C to release cellular ATP bound to the protease. Samples were mixed with 50 µl luciferase reagent (sensitive ATP detection assay, Biaffin), incubated for 10 min at room temperature and finally luminescence was detected at 560 nm (Luminoscan Ascent, Thermo Science).
Miscellaneous
Cloning procedures can be found in the supplement. The purification of the m-AAA protease via a carboxy-terminal hexahistidine tag for in vitro assays and the determination of ATPase activities in vitro have been described previously (Tatsuta et al., 2007). Isolation of mitochondria and protein import and degradation assays in organello were performed as described (Tatsuta and Langer, 2007).
Supplementary Material
Acknowledgements
We are grateful to Dr. U. Baumann (Bern) for sharing unpublished results on the crystal structure of FtsH and Daniela Tils for expert technical assistance. This work was supported by grants of the Deutsche Forschungsgemeinschaft, the European Research Council and the German-Israeli-Project (DIP grant F.5.1.) to T.L., the National Institutes of Health and the Welch Foundation to F.T.F.T., and the American Heart Association to S.L..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental information including eight figures and one table are available with this article online.
References
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12-complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends. Biochem. Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C, Schalch T, Bumann M, Meister M, Meier R, Baumann U. The molecular architecture of the metalloprotease FtsH. Proc. Natl. Acad. Sci. U S A. 2006;103:3066–3071. doi: 10.1073/pnas.0600031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol. Cell. 2004;16:343–350. doi: 10.1016/j.molcel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Dalal S, Rosser MF, Cyr DM, Hanson PI. Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol. Biol. Cell. 2004;15:637–648. doi: 10.1091/mbc.E03-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Mol. Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Esser K, Tursun B, Ingenhoven M, Michaelis G, Pratje E. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J. Mol. Biol. 2002;323:835–843. doi: 10.1016/s0022-2836(02)01000-8. [DOI] [PubMed] [Google Scholar]
- Ferreirinha F, Quattrini A, Priozzi M, Valsecchi V, Dina G, Broccoli V, Auricchio A, Piemonte F, Tozzi G, Gaeta L, Casari G, Ballabio A, Rugarli EI. Axonal degeneration in paraplegin-deficient mice is associated with abnormal mitochondria and impairment of axonal transport. J. Clin. Invest. 2004;113:231–242. doi: 10.1172/JCI20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Han YW, Okamoto T, Ohnishi T, Yoshikawa M, Yamada K, Toh H, Daiyasu H, Ogura T, Shinagawa H. Mutational analysis of the functional motifs of RuvB, an AAA+ class helicase and motor protein for holliday junction branch migration. Mol. Microbiol. 2000;36:528–538. doi: 10.1046/j.1365-2958.2000.01842.x. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Joly N, Burrows PC, Buck M. An intramolecular route for coupling ATPase activity in AAA+ proteins for transcription activation. J. Biol. Chem. 2008;283:13725–13735. doi: 10.1074/jbc.M800801200. [DOI] [PubMed] [Google Scholar]
- Koodathingal P, Jaffe NE, Kraut DA, Prakash S, Fishbain S, Herman C, Matouschek A. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J. Biol. Chem. 2009 doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M, Langer T. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit. Rev. Biochem. Mol. Biol. 2007;42:221–242. doi: 10.1080/10409230701380452. [DOI] [PubMed] [Google Scholar]
- Koppen M, Metodiev MD, Casari G, Rugarli EI, Langer T. Variable and Tissue-Specific Subunit Composition of Mitochondrial m-AAA Protease Complexes Linked to Hereditary Spastic Paraplegia. Mol. Cell. Biol. 2007;27:758–767. doi: 10.1128/MCB.01470-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhard K, Guiard B, Pellechia G, Tzagoloff A, Neupert W, Langer T. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol. Cell. 2000;5:629–638. doi: 10.1016/s1097-2765(00)80242-7. [DOI] [PubMed] [Google Scholar]
- Maltecca F, Aghaie A, Schroeder DG, Cassina L, Taylor BA, Phillips SJ, Malaguti M, Previtali S, Guenet JL, Quattrini A, Cox GA, Casari G. The mitochondrial protease AFG3L2 is essential for axonal development. J. Neurosci. 2008;28:2827–2836. doi: 10.1523/JNEUROSCI.4677-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol. Cell. 2008;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol. Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 2004;146:106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Song HK, Hartmann C, Ramachandran R, Bochtler M, Behrendt R, Moroder L, Huber R. Mutational studies on HslU and its docking mode with HslV. Proc. Natl. Acad. Sci. U S A. 2000;97:14103–14108. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suno R, Niwa H, Tsuchiya D, Zhang X, Yoshida M, Morikawa K. Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH. Mol. Cell. 2006;22:575–585. doi: 10.1016/j.molcel.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Tatsuta T, Augustin S, Nolden M, Friedrichs B, Langer T. m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 2007;26:325–335. doi: 10.1038/sj.emboj.7601514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T, Langer T. Studying proteolysis within mitochondria. Methods Mol. Biol. 2007;372:343–360. doi: 10.1007/978-1-59745-365-3_25. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Poehlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Schlieker C, Bukau B, Mogk A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 2003;278:32608–32617. doi: 10.1074/jbc.M303653200. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan D, Tsai FT, Mogk A, Bukau B. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Werbeck ND, Schlee S, Reinstein J. Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J. Mol. Biol. 2008;378:178–190. doi: 10.1016/j.jmb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Yamada-Inagawa T, Okuno T, Karata K, Yamanaka K, Ogura T. Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J. Biol. Chem. 2003;278:50182–50187. doi: 10.1074/jbc.M308327200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wigley DB. The 'glutamate switch' provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat. Struct. Mol. Biol. 2008;15:1223–1227. doi: 10.1038/nsmb.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.