Abstract
The homologous series of volatile perfluorinated acids—trifluoroacetic acid (TFA), pentafluoropropionic acid (PFPA) and heptafluorobutyric acid (HFBA)—continue to be excellent anionic ion-pairing reagents for reversed-phase high-performance liquid chromatography (RP-HPLC) after more than two decades since their introduction to this field. It was felt that a thorough, step-by-step re-examination of the effects of anionic ion-pairing reagents over a wide concentration range on RP-HPLC peptide elution behaviour is now due, particularly considering the continuing dominance of such reagents for peptide applications. Thus, RP-HPLC was applied over a range of 1–60 mM phosphoric acid, TFA, PFPA and HFBA to two mixtures of 18-residue synthetic peptides containing either the same net positive charge (+4) or varying positive charge (+1, +2, +3, +4). Peptides with the same charge are resolved very similarly independent of the ion-pairing reagent used, although the overall retention times of the peptides increase with increasing hydrophobicity of the anion: phosphate < TFA− < PFPA− < HFBA−. Peptides of differing charge move at differing rates relative to each other depending on concentration of ion-pairing reagents. All four ion-pairing reagents increased peptide retention time with increasing concentration, albeit to different extents, again based on hydrophobicity of the anion, i.e., the more hydrophobic the anion, the greater the increase in peptide retention time at the same reagent concentration. Interestingly, phosphoric acid produced the best separation of the four-peptide mixture (+1 to +4 net charge). In addition, concentrations above 10 mM HFBA produced a reversal of the elution order of the four peptides (+1 < + 2 < + 3 < + 4) compared to the elution order produced by the other three reagents over the entire concentration range (+4 < + 3 < + 2 < + 1).
Keywords: Volatile perfluorinated acid, RP-HPLC, Peptide
1. Introduction
Since their introduction over two decades ago as anionic ion-pairing reagents for reversed-phase high-performance liquid chromatography (RP-HPLC), the homologous series of volatile perfluorinated acids – trifluoroacetic acid (TFA), pentafluoropropionic acid (PFPA) and heptafluorobutyric acid (HFBA) have proven invaluable for RP-HPLC of peptides [1–9]. TFA, in particular, remains the dominant mobile phase additive for such applications [6,7,9]. In addition, phosphoric acid has seen use as a non-volatile anionic ion-pairing reagent over the same period [1,4–6,10–15]. The negatively charged phosphate, trifluoroacetate (TFA−), pentafluoropropionate (PFPA−) and heptafluorobutyrate (HFBA−) anions will interact (ion-pair) with positively charged peptide residues (arising from the basic side-chains Lys, Arg and His or a free α-amino group). A hydrophilic counterion such as phosphate will neutralize the highly hydrophilic positively charged groups in the peptides, thus decreasing overall peptide hydrophilicity. In contrast, more hydrophobic anions such as the perfluorinated acids will not only neutralize the positively charged groups, thereby decreasing peptide hydrophilicity, but will also increase further the affinity of the peptides for the hydrophobic reversed-phase stationary phase [4], this affinity increasing with increasing hydrophobicity of the anion, i.e., TFA− < PFPA− < HFBA−. Such acidic reagents have generally been employed in a concentration range of 0.05%–0.1% (v/v) for the majority of peptide separations [4–7]. Higher concentrations have generally been avoided in the past due to, among other things, silica-based stationary phase degradation under highly acidic conditions [16,17]. However, the advancement in recent years of reversed-phase, silica-based packings with excellent chemical stability [18–20] has enabled us to revisit the question of the most suitable type and concentration of acidic ion-pairing reagent for separation of peptide mixtures.
In the present study, RP-HPLC was applied over a range of 1–60 mM phosphoric acid, TFA (equivalent to ~0.008%–0.46% TFA), PFPA and HFBA to two mixtures of 18-residue synthetic peptides: (1) a mixture of six peptides with the same net positive charge (+4); and (2) a mixture of four peptides with net positive charges of +1, +2, +3 and +4. From the retention behaviour of these peptides under conditions of increasing concentration and anion hydrophobicity (phosphate < TFA− < PFPA− < HFBA−), insights were gained into optimizing approaches for the separation of sample mixtures containing peptides of varying net charge and hydrophobicity.
2. Experimental
2.1. Materials
Reagent-grade phosphoric acid (H3PO4) was obtained from Caledon Laboratories (Georgetown, Ontario, Canada). TFA was obtained from Hydrocarbon Products (River Edge, NJ, USA); PFPA was obtained from Fluka (Buchs, Switzerland); and HFBA was obtained from Pierce Chemical (Rockford, IL, USA). HPLC-grade water was obtained from EMD Chemicals (Gibbstown, NJ, USA). HPLC-grade acetonitrile was obtained from EM Science (Gibbstown, NJ, USA).
2.2. Column and HPLC conditions
Analytical RP-HPLC runs were carried out on a Zorbax SB300-C8 column (150 mm × 2.1 mm i.d.; 5 μm particle size, 300 Å pore size) from Agilent Technologies (Little Falls, DE, USA), using a linear AB gradient (0.5% acetonitrile/min) at a flow-rate of 0.3 ml/min, where eluent A was 1–60 mM aq. H3PO4, TFA, PFPA or HFBA and eluent B was the corresponding concentration of the respective ion-pairing reagent in acetonitrile; runs were carried out at 25 °C. Peptide bond absorbance was measured by diode array detection at 210 nm. Approximately 1 μmol of each of the peptides in the peptide mixture was injected in a total sample volume of 10 μl.
2.3. Instrumentation
RP-HPLC runs were carried out on an Agilent 1100 Series liquid chromatograph, comprised of a solvent degasser, autosampler, binary pumping system, a thermostatted column compartment and a diode array and multiwavelength detection system.
Peptide synthesis was carried out on an Applied Biosystems Peptide Synthesizer Model 430A (Foster City, CA, USA).
2.4. Peptide synthesis and purification
Synthesis of the peptides was carried out by standard solid-phase synthesis methodology using Nα-tert-butyloxycarbonyl (t-Boc) chemistry on MBHA (methylbenzhydrylamine) resin (0.97 mmol/g) as described previously [21]. The crude peptides were purified by preparative RP-HPLC on an Applied Biosystems 400 solvent-delivery system connected to a 783A programmable absorbance detector. Amino acid analyses of purified peptides were carried out on a Beckman Model 6300 amino acid analyzer (Beckman Instruments, Fullerton, CA, USA) and the correct primary ion molecular masses of peptides were confirmed using a Mariner electrospray ionization time of flight mass spectrometer (Applied Biosystems, Foster City, CA, USA).
3. Results and discussion
3.1. Design of synthetic model peptides
Peptide mixtures designed for the present study followed the view that studies attempting to equate peptide elution behaviour during RP-HPLC with varying run parameters is best achieved by studies using defined model peptide systems, the results of which can then be extrapolated to peptides as a whole. Thus, two groups of 18-residue model peptides exhibiting variations in hydrophobicity and/or net positive charge were designed and synthesized. From Table 1, peptides 1–6 represent a group of six peptides with the same net charge (+4; arising from four lysine residues). Within these peptides, hydrophobicity decreases only subtly between each adjacent peptide (peptide 6 < 5 < 4 < 3 < 2 < 1) due to just single substitutions of glutamine in place of glutamic acid, i.e., between each adjacent peptide, there is a single carboxyl group to amide group substitution. Peptides 1, 7–9 represent a group of four peptides varying in net positive charge through the presence of four lysine residues (peptide 1; +4), three lysine residues (peptide 7; +3), two lysine residues (peptide 8; +2 or one lysine residue (peptide 9; +1). The presence of five glycine residues distributed throughout the sequence ensured negligible secondary structure for these peptides [22,23], i.e., they have a “random coil” conformation, to avoid complications in interpretation of data due to selectivity differences in peptide RP-HPLC retention behaviour arising from conformational variations [15,24].
Table 1.
Sequences of the peptides used in this study. Ac denotes Nα-acetyl and amide denotes C α-amide
| Peptides | Sequence | Number of positive charges |
|---|---|---|
| 1 | Ac-KLKKGGLKGELGGEELEE-amide | 4 |
| 2 | Ac-KLKKGGLKGELGGEELEQ-amide | 4 |
| 3 | Ac-KLKKGGLKGELGGEELQQ-amide | 4 |
| 4 | Ac-KLKKGGLKGELGGEQLQQ-amide | 4 |
| 5 | Ac-KLKKGGLKGELGGQQLQQ-amide | 4 |
| 6 | Ac-KLKKGGLKGQLGGQQLQQ-amide | 4 |
| 7 | Ac-KLKKGGLAGELGGEELEE-amide | 3 |
| 8 | Ac-KLKAGGLAGELGGEELEE-amide | 2 |
| 9 | Ac-KLAAGGLAGELGGEELEE-amide | 1 |
3.2. RP-HPLC stationary phase
The Zorbax SB-300C8 (“SB” denoting “Stable Bond”) is prepared from monofunctional n-octylsilane based on protecting the siloxane bond between the silica and the C8 alkyl chain with bulky side groups (two isopropyl groups, in this case) [18–20]. This packing was originally designed to protect the siloxane bond from acid hydrolysis and has shown excellent chemical and thermal stability at low pH [9,24–29]. It should be noted that the pH values of the aqueous eluents in the present study ranged from ~1.5 to 2.9, depending on the nature and concentration of the ion-pairing reagent, and this pH range is generally referred to as pH 2. Note that even the highest pH value (pH 2.9) is far enough below the pKa values of the positively charged groups in the peptides so as not to affect the net positive charge on the peptides; in addition, if any underivatized silanol groups (pKa ~ 4.0) were on the StableBond packing, they also would remain protonated (i.e., neutral) under the RP-HPLC conditions used in the present study, thus preventing any potential undesirable electrostatic interactions between the positively charged peptides and the hydrophobic stationary phase. Peptide standards have been developed to monitor the presence of non-specific electrostatic interactions with the reversed-phase matrix [30].
3.3. Effect of counterion hydrophobicity on elution behaviour of model peptide mixtures
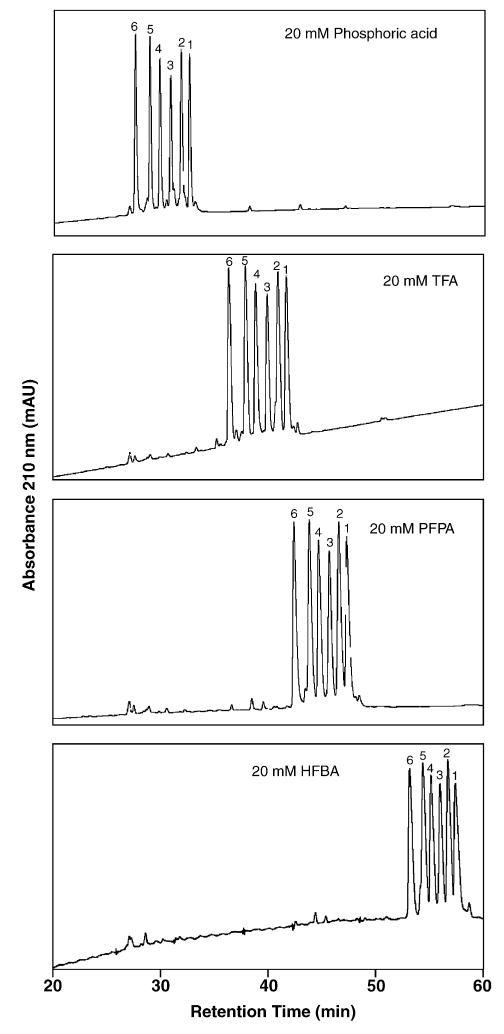
Fig. 1 illustrates the effect of increasing counterion hydrophobicity on the elution behaviour of peptides 1–6 (all +4 net charge) at a constant concentration (20 mM) of each ion-pairing reagent. Ion-pairing reagent concentrations were expressed in mM versus percentage in order to be able to make a direct comparison of different reagents. From Fig. 1, all six peptides are well resolved in the presence of all four reagents, despite the only subtle change in hydrophobicity between adjacent peptides. As expected [1–4,12], the retention times of all six peptides increases with increasing hydrophobicity of the counterion (phosphate < TFA− < PFPA− < HFBA−). A hydrophilic counterion such as phosphate will neutralize the highly hydrophilic positively charged groups in the peptides, thus decreasing overall peptide hydrophilicity. In contrast, more hydrophobic anions such as the perfluorinated acids will not only neutralize the positively charged groups, thereby decreasing peptide hydrophilicity, but will also increase further the affinity of the peptides for the hydrophobic reversed-phase stationary phase, this affinity increasing with increasing hydrophobicity of the anion, i.e., TFA− < PFPA− < HFBA−. Note that the elution range of all six peptides in Fig. 1 remains very similar (mean 4.78 ± 0.54 min) despite the large increase in overall peptide retention time between the 20 mM H3PO4 and 20 mM HFBA systems (e.g., the increase in retention time for peptide 6 is 25.6 min). Such a result would be expected considering that the negatively charged counterions affect peptide retention behaviour through interactions with just the positively charged residues and peptides 1–6 contain an identical number of positively charge lysine residues (Table 1).
Fig. 1.
Effect of ion-pairing reagent hydrophobicity on RP-HPLC behaviour of positively charged model peptides. Conditions: linear AB gradient (0.5% acetonitrile/min) at a flow-rate of 0.3 ml/min, where eluent A is 20 mM aq. H3PO4, TFA, PFPA or HFBA and eluent B is 20 mM of the corresponding ion-pairing reagent in acetonitrile. The sequences of the peptides are shown in Table 1.
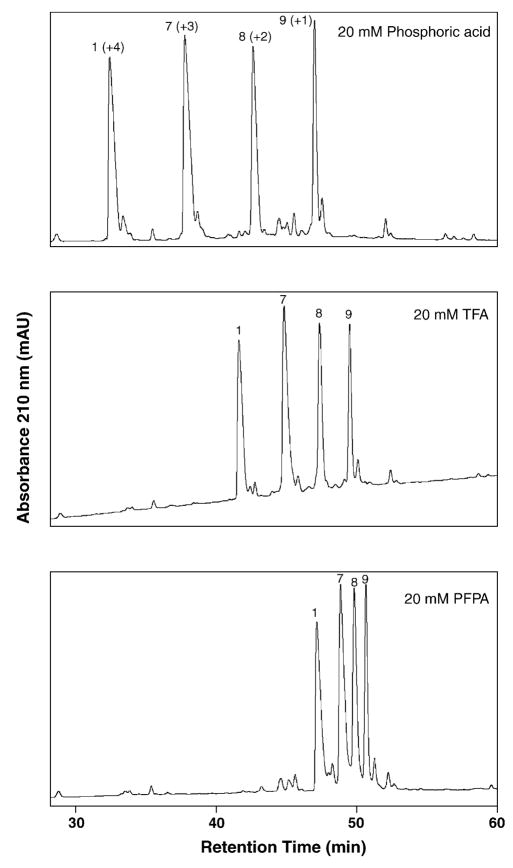
Fig. 2 and Fig. 3 (bottom panel) now illustrate the effect of increasing counterion hydrophobicity on the retention behaviour of peptides 1, 7, 8 and 9 (+4, +3, +2 and +1 net charge, respectively). In a similar manner to the results shown in Fig. 1, the four peptides are again increasing in retention time with increasing counterion hydrophobicity (phosphate < TFA− < PFPA− < HFBA−) albeit peptides of differing charge move at different rates relative to each other. Thus, the greater the net charge on the peptide (+4 > + 3 > + 2 > + 1; peptides 1, 7, 8 and 9, respectively), the greater the effect on its retention time with increasing counterion hydrophobicity (Figs. 2 and 3 (bottom panel)). Thus, at 20 mM concentration of ion-pairing reagent, phosphoric acid produces the best separation of this four-peptide mixture. Interestingly, due to the disproportionate effect of counterion hydrophobicity on peptide retention time, the elution order of the four peptides is completely reversed in 20 mM HFBA (the most hydrophobic reagent) i.e., they are now eluted in the order peptide 9 < 8 < 7 < 1 (Fig. 3, bottom panel) compared to the elution order of 1 < 7 < 8 < 9 achieved with the less hydrophobic H3PO4, TFA and PFPA (Fig. 2). Finally, also due to the disproportionate effect of counterion hydrophobicity on retention times of peptides with different numbers of positively charged groups, the best separation of these four peptides was achieved with phosphoric acid, highlighting the potential value of varying counterion hydrophobicity for specific peptide separations.
Fig. 2.
Effect of ion-pairing reagent hydrophobicity on RP-HPLC behaviour of positively charged model peptides. Conditions: linear AB gradient (0.5% acetonitrile/min) at a flow-rate of 0.3 ml/min, where eluent A is 20 mM aq. H3PO4, TFA or PFPA and eluent B is 20 mM of the corresponding ion-pairing reagent in acetonitrile. The sequences of the peptides are shown in Table 1.
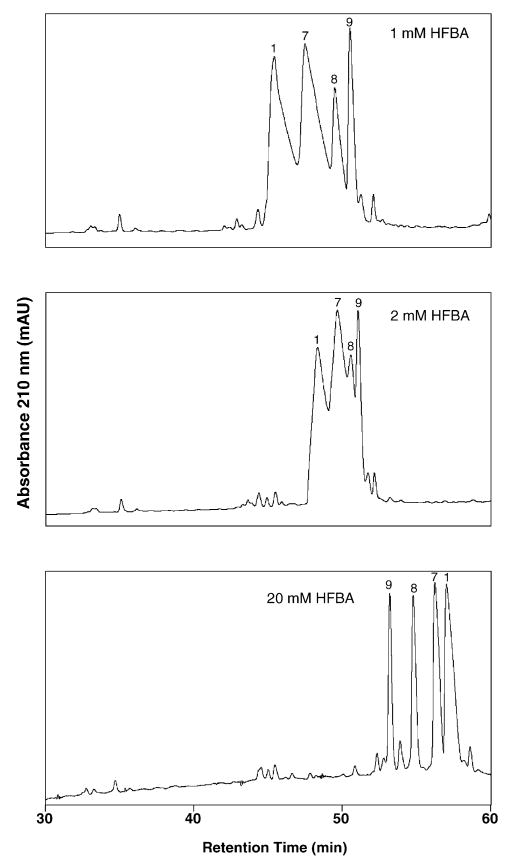
Fig. 3.
Effect of HFBA concentration on RP-HPLC behaviour of positively charged peptides. Conditions: linear AB gradient (0.5% acetonitrile/min) at a flow-rate of 0.3 ml/min, where eluent A is 1, 2 or 20 mM aq. HFBA and eluent B is the corresponding concentration of HFBA in acetonitrile. The sequences of the peptides are shown in Table 1.
3.4. Effect of concentration of ion-pairing reagent on elution behaviour of model peptide mixtures
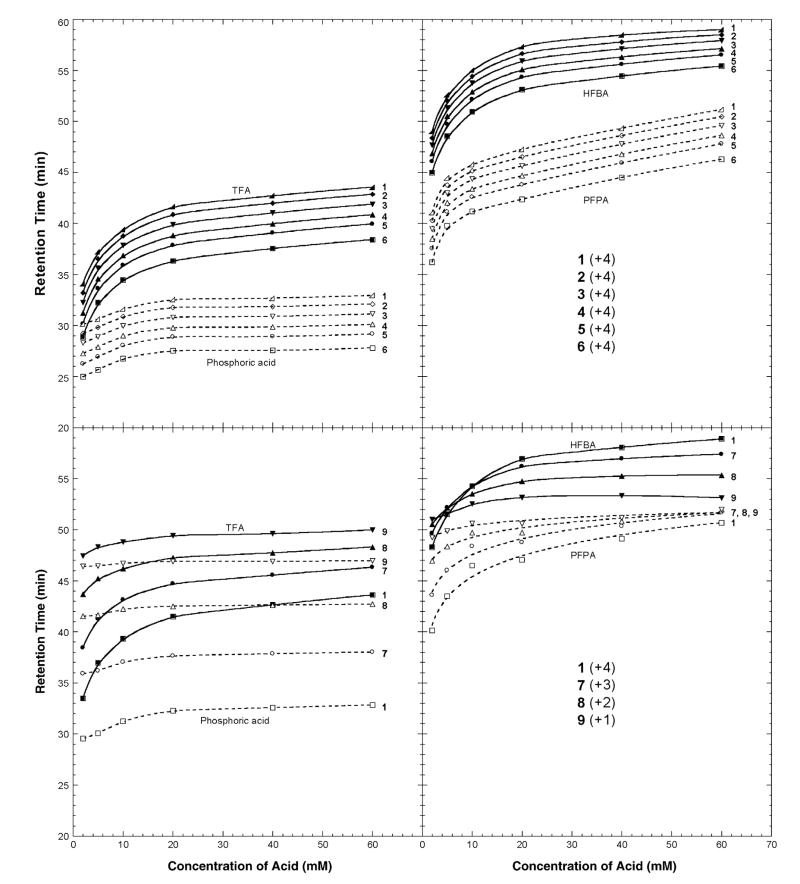
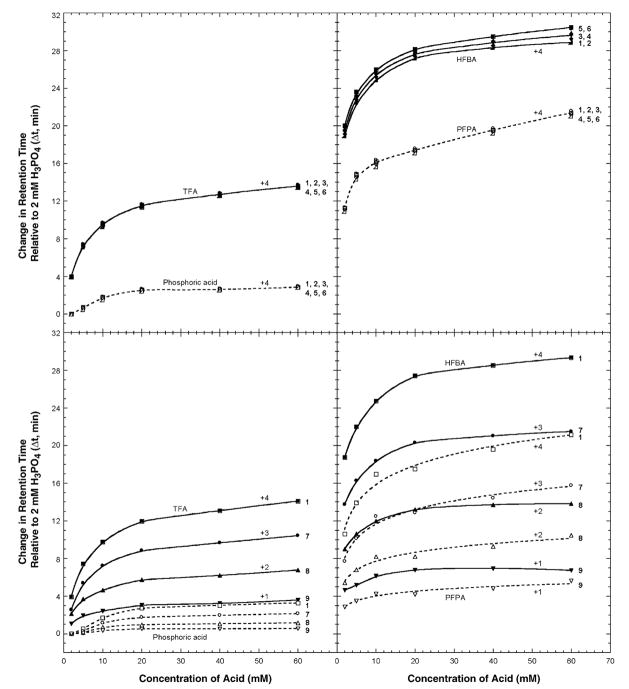
Fig. 4 presents a graphical representation of the effect of increasing counterion concentration (2–60 mM) on elution behaviour of peptides 1–6 (+4 net charge; top panels) and peptides 1, 7, 8 and 9 (+4, +3, +2 and +1 net charge, respectively; bottom panels). From Fig. 4 (top panels), peptides 1–6 all exhibited increasing retention time with increasing counterion concentration for all four ion-pairing reagents, albeit to different extents depending on the counterion hydrophobicity. Thus, although peptides with the same charge are resolved very similarly independent of the ion-pairing reagent used, the effect on peptide retention time is generally more marked in order of phosphate < TFA < PFPA < HFBA. This is particularly noticeable with the relative rapidity of retention time increase at low concentrations of ion-pairing reagent (ca. 2–10 mM), followed by the general leveling off of the profiles at higher concentrations (phosphoric acid, for instance, exhibits an essentially flat profile after 20 mM, whilst the remaining profiles continue to rise, albeit far less steeply than the initial rapid rise), presumably due to saturation of the charged groups at high reagent concentrations. Note that these results suggest that varying the counterion concentration has no advantage for overall separation of peptides, with the same net positive charge. However, as discussed below, such concentration variations may have a profound effect on peptide peak shape and, thus, on resolution.
Fig. 4.
Effect of ion-pairing reagent concentration on RP-HPLC retention times of positively charged model peptides. The sequences of the peptides are shown in Table 1. For RP-HPLC conditions, see Section 2.2.
From Fig. 4 (bottom panels), in a similar manner to the effect of increasing counterion hydrophobicity at the same reagent concentration (20 mM; Fig. 2), peptides of different charge (+4, +3, +2 and +1 for peptides 1, 7, 8 and 9, respectively) increase retention times at different rates depending on both the net charge on the peptide (+4 > + 3 > + 2 > + 1) and the hydrophobicity of the anion (phosphate < TFA− < PFPA− < HFBA−). Hence, the profiles for these four peptides of varying net charge, unlike peptides 1–6 (all +4 net charge), are not parallel, allowing potentially useful selectivity variations with changes in ion-pairing reagent concentration, as well as the nature of the reagent. Thus, under the range of concentrations studied, optimum overall separation of these four peptides was achieved at either low concentrations of phosphoric acid (Figs. 2 and 4) or at the highest (60 mM) concentration of HFBA (Fig. 4), with a concomitant reversal of peptide elution order in HFBA compared to phosphoric acid, TFA and PFPA. For this particular peptide mixture, PFPA is clearly not the reagent of choice for optimum resolution. Indeed, the separation worsens considerably with increasing concentration resulting in co-elution of peptides 7–9 at a 60 mM concentration of PFPA (Fig. 4). It is interesting to note that the aforementioned reversal of peptide elution order was achieved at just 20 mM HFBA (Fig. 4) but was not achieved by any of the other three reagents over the entire concentration range studied, in an analogous manner to the reversal in peptide elution order seen in 20 mM HFBA compared to 20 mM of the less hydrophobic phosphoric acid, TFA and PFPA reagents (Figs. 2 and 3, bottom panel). This again appears to be due to the disproportionate effect of counterion hydrophobicity as the concentration of the reagent is increased, i.e., an increase in concentration of the counterion with the greatest hydrophobicity (HFBA−) has a significantly greater effect on peptide retention behaviour compared to phosphate, TFA− and PFPA−.
Fig. 3 demonstrates the reversal of elution order of peptides 1, 7, 8 and 9 as HFBA concentration is raised from 1 mM (top panel) to 2 mM (middle panel) and 20 mM HFBA (bottom panel). The latter concentration was the lowest employed where all four peptides were essentially resolved to baseline. From Fig. 3, increasing HFBA concentration also clearly results in improved peak shape as well as increasing peptide retention time. This effect is more marked the greater the positive charge on the peptides, i.e., + 4 > + 3 > + 2 > + 1 for peptides 1, 7, 8 and 9, respectively. Similar profiles were obtained for the other three reagents and are discussed below.
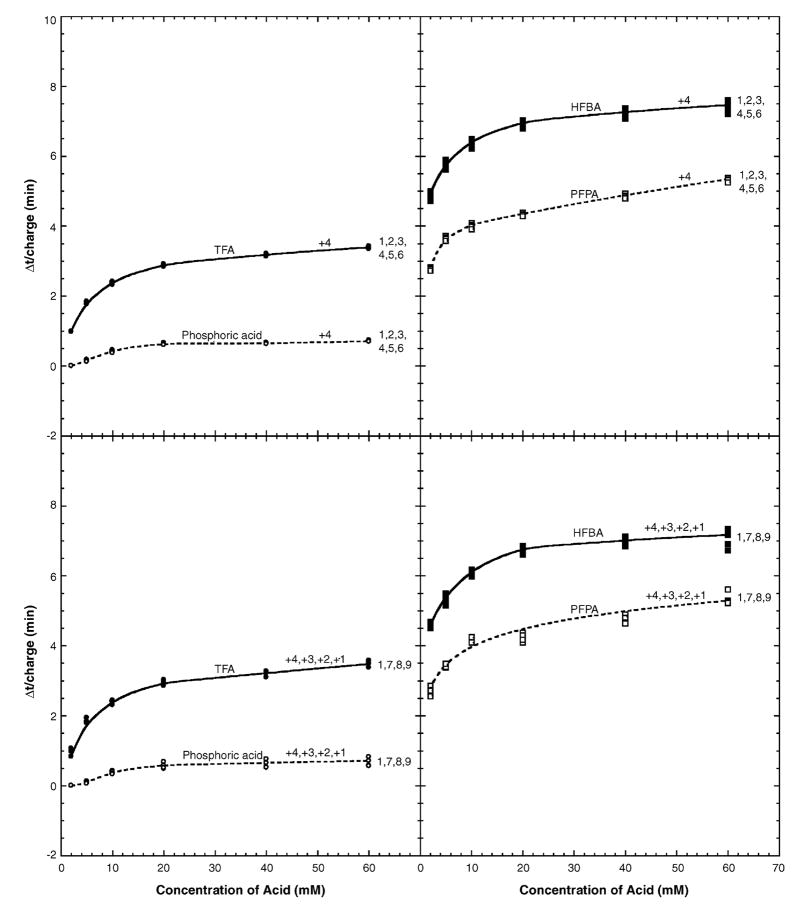
Fig. 5 offers an alternate graphical representation of the effect of counterion concentration on peptide retention behaviour, which plots the increase in retention time of the peptides (Δt) at each reagent concentration over that obtained when using 2 mM phosphoric acid. In effect, this is a normalization of the retention data of the peptides to their behaviour at this initial starting concentration of this ion-pairing reagent. The average Δt values for peptides 1–6 (all +4 net charge) were very similar for the phosphoric acid, TFA and PFPA systems over the entire concentration. The profiles shown for these three systems in Fig. 5 represent their average Δt values.
Fig. 5.
Effect of ion-pairing reagent concentration on RP-HPLC retention times of positively charged model peptides relative to retention time in 2 mM H3PO4 (Δt). The sequences of the peptides are shown in Table 1. For RP-HPLC conditions, see Section 2.2.
Although there is somewhat more of a divergence in Δt values in the HFBA system for peptides 1–6 as the concentration of this reagent increases (Fig. 5, top right panel), this divergence is still relatively small considering the range of movement of the peptides in HFBA over that of 2 mM H3PO4. Such results indicate that, for peptides of the same net positive charge, the effect of increasing ion-pairing reagent concentration on their retention behaviour is the same for most practical purposes.
From Fig. 5 (bottom panels), the variation in Δt values within a group of peptides of varying net charge (peptides 1, 7, 8, 9; +4, +3, +2 and +1 net charge, respectively) as ion-pairing reagent concentration increases is quite clear. These profiles highlight once more the dependence of the response of peptide elution behaviour under varying concentrations of anionic ion-pairing reagent to the number of positive charges they contain.
Finally, Fig. 6 shows the change in retention times of the peptides in the four-ion-pairing reagents relative to those obtained in 2 mM H3PO4 per net positive charge (Δt/N). Overall, the results shown in Fig. 6 indicate that whether peptides have the same net positive charge (peptides 1–6) or varying net positive charge (peptides 1, 7–9), there is an essentially equal effect of counterion concentration on each positively charged residue. Previous work demonstrated accurate prediction of peptide retention behaviour between different ion-pairing reagent systems through the use of internal peptide standards [4]. The above results (Fig. 6) also suggest that prediction of the effect of varying ion-pairing reagent concentration on peptide retention times is also possible and may be of value during development of separation protocols for peptide mixtures.
Fig. 6.
Effect of ion-pairing reagent concentration on RP-HPLC retention times of positively charged model peptides relative to retention time in 2 mM H3PO4 per net positive charge (Δt/charge). The sequences of the peptides are shown in Table 1. For RP-HPLC conditions, see Section 2.2.
A point worth noting from the results presented in Fig. 1 is the relative hydrophobicities of the residues within peptides 1–6 in the presence of different ion-pairing reagents. Data obtained from retention behaviour of peptides during RP-HPLC have frequently been employed to derive relative hydrophilicity/hydrophobicity values or “coefficients” of amino acid side-chains [13,31–42]. Values obtained in this manner may then be used for such purposes as prediction of peptide retention behaviour, e.g., for narrowing down the position of a peptide of interest following RP-HPLC of a complex peptide mixture such as a protein digest. From Fig. 1, it is clear that the observed overall hydrophobicities of the peptides are increasing with increasing counterion concentration. Since the counterions affect peptide retention behaviour through interaction with the positively charged residues (the remaining residues are all neutral, including glutamic acid at pH 2), these residues are responsible for increased peptide hydrophobicity with increasing counterion hydrophobicity. Thus, although such residues would be viewed as inherently hydrophilic due to their charged character, such a definition may not be valid in terms of RP-HPLC, i.e., positively charged residues (lysine in this case) vary in hydrophilicity/hydrophobicity depending on the RP-HPLC conditions under which they are monitored. Indeed, from Fig. 1, it could be viewed that lysine is becoming increasingly hydrophobic with increasing counterion hydrophobicity (phosphate < TFA− < PFPA− < HFBA−). These observations are of importance when attempting to extrapolate peptide retention behaviour during RP-HPLC, in the form of generated hydrophilicity/hydrophobicity coefficients, to alternate applications.
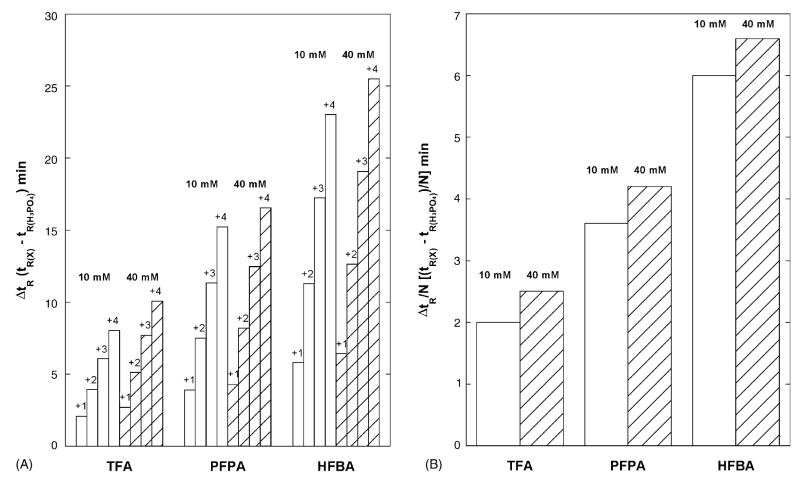
Fig. 7, Panel A shows the change in retention time of the four peptides varying in net charge (+1, +2, +3 and +4) relative to phosphoric acid. It is obvious that the counterion hydrophobicity has a dramatic effect on the retention behavior in the order TFA < PFPA < HFBA. This effect increases with increasing net positive charge on the peptide. For example, the change in retention time for the +1 peptide at 40 mM acid is 2.7, 4.3 and 6.4 min for TFA, PFPA and HFBA, respectively. In contrast, the values for +4 peptide at 40 mM acid are 10.1, 16.6, and 25.5 min for TFA, PFPA and HFBA, respectively. The increase in retention time by varying the acid concentration from 10 to 40 mM is small compared to the effect of counterion hydrophobicity (Fig. 7, Panel A).
Fig. 7.
Panel A is the change in retention time for the group of four peptides (+1, +2, +3 and +4) relative to phosphoric acid as the counterion hydrophobicity increases TFA < PFPA < HFBA and the counterion concentration varies (10 and 40 mM). ΔtR is the retention time, tR of the peptide at a particular acid concentration minus tR of the peptide in H3PO4 at the same concentration. Panel B is the change in hydrophilicity/hydrophobicity of the lysine residues relative to phosphoric acid as the counterion hydrophobicity increases TFA < PFPA < HFBA and the counterion concentration varies (10 and 40 mM) using the group of peptides with the same net charge (+4) and using the average value of ΔtR/N where N is the number of positively charged residues in the peptide. The ΔtR/N values from the group of four peptides of varying charge were similar.
To demonstrate the variation in hydrophobicity of lysine in TFA, PFPA and HFBA, the hydrophobicity coefficient of lysine relative to phosphoric acid was determined at 10 and 40 mM acid concentrations (Fig. 7, Panel B). The hydrophobicity of Lys increases dramatically as the counterion hydrophobicity increases from TFA, PFPA and HFBA relative to phosphoric acid (2.0, 3.6 and 6.0 min, respectively at 10 mM acid concentration). The change in hydrophobicity with acid concentration is small relative to the change in hydrophobicity of the counterion. In going from 10 to 40 mM acid the hydrophobicity of Lys changes from 2.0 to 2.5 min in TFA; 3.6 to 4.2 min in PFPA and 6.0 to 6.6 min in HFBA. Interestingly, the increase in hydrophobicity with acid concentration is independent of the acid (~0.6 min in changing acid concentration from 10 to 40 mM for TFA, PFPA and HFBA (Fig. 7, Panel B).
4. Conclusions
The present study has investigated the effect of varying H3PO4, TFA, PFPA and HFBA concentration on RP-HPLC of two groups of synthetic model peptides containing either one (+4) or multiple (+1, +2, +3, +4) positively charged groups. Overall retention times of the peptides increase with increasing hydrophobicity of the anion: phosphate < TFA− < PFPA− < HFBA−, with peptides of differing charge moving at different rates depending on the hydrophobicity and/or concentration of the ion-pairing reagent. Phosphoric acid produced the best separation of the peptide mixture with differently charged peptides, with concentrations above 10 mM HFBA producing a reversal of the elution order of these peptides compared to the other three reagents over the entire concentration range. We believe that such results will aid in the rational design of peptide separation protocols, including proteomics applications.
Acknowledgments
This work was supported by an NIH grant (RO1 GM61855) to Robert S. Hodges.
References
- 1.Bennett HPJ, Browne CA, Solomon S. J Liq Chromatogr. 1980;3:1353. [Google Scholar]
- 2.Schaaper WMM, Voskamp D, Olieman C. J Chromatogr. 1980;195:181. [Google Scholar]
- 3.Harding DRK, Bishop CA, Tarttellin MF, Hancock WS. Int J Peptide Protein Res. 1981;18:314. doi: 10.1111/j.1399-3011.1981.tb02060.x. [DOI] [PubMed] [Google Scholar]
- 4.Guo D, Mant CT, Hodges RS. J Chromatogr. 1987;386:205. doi: 10.1016/s0021-9673(01)94598-4. [DOI] [PubMed] [Google Scholar]
- 5.Mant CT, Hodges RS, editors. High-Performance Liquid Chromatography of Peptides and Proteins: Separation, Analysis and Conformation. CRC Press; Boca Raton, FL: 1991. [Google Scholar]
- 6.Cunico RL, Gooding KM, Wehr T, editors. HPLC and CE of Biomolecules. Bay Bioanalytical Laboratory; Richmond, VA: 1998. [Google Scholar]
- 7.Mant CT, Hodges RS. In: HPLC of Biological Macromolecules. Gooding KM, Regnier FE, editors. Marcel Dekker; New York, NY: 2002. p. 433. [Google Scholar]
- 8.Popa TV, Mant CT, Hodges RS. Electrophoresis. 2004;25:1219. doi: 10.1002/elps.200305889. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Mehok AR, Mant CT, Hodges RS. J Chromatogr A. 2004;1043:9. doi: 10.1016/j.chroma.2004.03.070. [DOI] [PubMed] [Google Scholar]
- 10.Hancock WS, Bishop CA, Prestidge RL, Harding DRK, Hearn MTW. J Chromatogr. 1978;153:391. [Google Scholar]
- 11.Nice EC, O’Hare MJ. J Chromatogr. 1979;162:401. doi: 10.1016/s0378-4347(00)81527-3. [DOI] [PubMed] [Google Scholar]
- 12.Starratt AN, Stevens ME. J Chromatogr. 1980;194:421. [Google Scholar]
- 13.Gaertner H, Puigserver A. J Chromatogr. 1985;350:279. [Google Scholar]
- 14.Sereda TJ, Mant CT, Quinn AM, Hodges RS. J Chromatogr. 1993;646:17. doi: 10.1016/s0021-9673(99)87003-4. [DOI] [PubMed] [Google Scholar]
- 15.Sereda TJ, Mant CT, Hodges RS. J Chromatogr A. 1995;695:205. doi: 10.1016/0021-9673(94)01147-7. [DOI] [PubMed] [Google Scholar]
- 16.Guo D, Mant CT, Taneja AK, Parker JMR, Hodges RS. J Chromatogr. 1986;359:519. [Google Scholar]
- 17.Glajch JL, Kirkland JJ, Kohler J. J Chromatogr. 1987;384:81. doi: 10.1016/s0021-9673(01)94661-8. [DOI] [PubMed] [Google Scholar]
- 18.Kirkland JJ, Glajch JL, Farlee RD. Anal Chem. 1989;61:2. [Google Scholar]
- 19.Glajch JL, Kirkland JJ. LC GC. 1990;8:140. [Google Scholar]
- 20.Boyes BE, Kirkland JJ. Pept Res. 1993;6:249. [PubMed] [Google Scholar]
- 21.Chen Y, Mant CT, Hodges RS. J Pept Res. 2002;59:18. doi: 10.1046/j.1397-002x.2001.10994.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhou NE, Monera OD, Kay CM, Hodges RS. Protein Pept Lett. 1994;1:114. [Google Scholar]
- 23.Monera OD, Sereda TJ, Zhou NE, Kay CM, Hodges RS. J Pept Sci. 1995;1:319. doi: 10.1002/psc.310010507. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Mant CT, Hodges RS. J Chromatogr A. 2003;1010:45. doi: 10.1016/s0021-9673(03)00877-x. [DOI] [PubMed] [Google Scholar]
- 25.Hancock WS, Chloupek RC, Kirkland JJ, Snyder LR. J Chromatogr A . :686. doi: 10.1016/0021-9673(94)00077-8. [DOI] [PubMed] [Google Scholar]
- 26.Chloupek RC, Hancock WS, Marchylo BA, Kirkland JJ, Boyes BE, Snyder LR. J Chromatogr A. 1994;686:45. doi: 10.1016/S0021-9673(94)89009-9. [DOI] [PubMed] [Google Scholar]
- 27.Mant CT, Kondejewski LH, Cachia PJ, Monera OD, Hodges RS. Methods Enzymol. 1997;289:426. doi: 10.1016/s0076-6879(97)89058-1. [DOI] [PubMed] [Google Scholar]
- 28.Mant CT, Chen Y, Hodges RS. J Chromatogr A. 2003;1009:29. doi: 10.1016/s0021-9673(03)00621-6. [DOI] [PubMed] [Google Scholar]
- 29.Mant CT, Tripet B, Hodges RS. J Chromatogr A. 2003;1009:45. doi: 10.1016/s0021-9673(03)00919-1. [DOI] [PubMed] [Google Scholar]
- 30.Mant CT, Hodges RS. Chromatographia. 1987;24:805. [Google Scholar]
- 31.Meek JL. Proc Natl Acad Sci USA. 1980;77:1632. doi: 10.1073/pnas.77.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meek JL, Rossetti ZL. J Chromatogr. 1981;211:15. [Google Scholar]
- 33.Su SJ, Grego B, Niven B, Hearn MTW. J Liq Chromatogr. 1981;4:1745. [Google Scholar]
- 34.Mabuchi H, Nakahashi H. J Chromatogr. 1981;213:275. doi: 10.1016/s0021-9673(00)81910-x. [DOI] [PubMed] [Google Scholar]
- 35.Wilson KJ, Honegger A, Stötzel RP, Hughes GJ. Biochem J. 1981;199:31. doi: 10.1042/bj1990031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Browne CA, Bennett HPJ, Solomon S. Biochem J. 1982;124:201. doi: 10.1016/0003-2697(82)90238-x. [DOI] [PubMed] [Google Scholar]
- 37.Sasagawa T, Okuyama T, Teller DC. J Chromatogr. 1982;240:329. [Google Scholar]
- 38.Guo D, Mant CT, Taneja AK, Parker JMR, Hodges RS. J Chromatogr. 1986;359:499. [Google Scholar]
- 39.Guo D, Mant CT, Taneja AK, Hodges RS. J Chromatogr. 1986;359:519. [Google Scholar]
- 40.Eng J, Huang CG, Pan YC, Hulmes JD, Yalow RS. Peptides. 1987;8:165. doi: 10.1016/0196-9781(87)90181-1. [DOI] [PubMed] [Google Scholar]
- 41.Sakamoto Y, Kawakami N, Sasagawa T. J Chromtogr. 1988;442:69. doi: 10.1016/s0021-9673(00)94457-1. [DOI] [PubMed] [Google Scholar]
- 42.Jinno K, Tankgawa E. Chromatographia. 1988;25:613. [Google Scholar]