Abstract
The authors developed a prototype Shared Health Research Information Network (SHRINE) to identify the technical, regulatory, and political challenges of creating a federated query tool for clinical data repositories. Separate Institutional Review Boards (IRBs) at Harvard's three largest affiliated health centers approved use of their data, and the Harvard Medical School IRB approved building a Query Aggregator Interface that can simultaneously send queries to each hospital and display aggregate counts of the number of matching patients. Our experience creating three local repositories using the open source Informatics for Integrating Biology and the Bedside (i2b2) platform can be used as a road map for other institutions. The authors are actively working with the IRBs and regulatory groups to develop procedures that will ultimately allow investigators to obtain identified patient data and biomaterials through SHRINE. This will guide us in creating a future technical architecture that is scalable to a national level, compliant with ethical guidelines, and protective of the interests of the participating hospitals.
Introduction
In May 2008, Harvard received a Clinical and Translation Science Award (CTSA) from the National Institutes of Health to transform patient-oriented research and build an infrastructure that would enable collaboration across the Harvard schools and affiliated hospitals and institutions. Early in the planning stages of designing the structure of Harvard's future Clinical and Translational Science Center (CTSC), we recognized that sharing patient data would be one of our greatest challenges. Therefore, in January 2008, Harvard Medical School began a 6 month project to develop a working prototype of a query tool that can search the clinical data repositories of its three largest affiliated health centers—Beth Israel Deaconess Medical Center (BIDMC), Children's Hospital Boston (CHB), and Partners HealthCare System (PHS). Each of these is an independent financial organization, with its own databases, firewalls, data access policies, and institutional review boards. Each must protect the privacy of their patients, and compete for patient populations, researchers, and grant dollars. As a result, the organizations decided that it would not be desirable to create a combined central data warehouse. Therefore, we chose to use a federated model, where each institution would manage and maintain control over their local databases, but through a standard web service API, distributed queries would still be possible. Admittedly, the system we implemented is truly a prototype, in that it was never designed for widespread use. We plan to replace the system with a scalable peer-to-peer architecture and a streamlined IRB process. However, the technical, regulatory, and political obstacles we faced while creating this prototype will be common themes as institutions across the country increasingly participate in nationwide efforts to share clinical data and participate in other collaborative efforts.
Background
The creation of a federated multi-institution query tool at Harvard was built upon years of experience at each of the hospitals in creating clinical data warehouses and developing processes with the IRBs to allow investigators to access the data for research (▶).
Table 1.
Table 1 The Evolution of a Federated Multi-Institution Query Tool at Harvard. The Table Lists Computer Software and an Innovation that Led to the Creation of SHRINE
| Year | System | Institutions | Innovation |
|---|---|---|---|
| 1989 | ClinQuery | BIH | query a clinical database |
| 1994 | W3-EMRS | BIH, CHB, MGH | federated architecture |
| 1996 | CareWeb/CDR | BIDMC | relational data repository |
| 1998 | DXtractor | CHB | complex queries |
| 1998 | Goldminer | CHB | graphical user interface |
| 1999 | RPDR | BWH, MGH | enterprise roll out |
| 2007 | i2b2 | BWH, MGH | open source code |
| 2007 | SPIN | BIDMC, BWH, CHB, MGH | data sharing policies |
| 2008 | SHRINE | BIDMC, BWH, CHB, MGH | federated query tool |
BIDMC = Beth Israel Deaconess Medical Center; BIH = Beth Israel Hospital; BWH = Brigham and Women's Hospital; CHB = Children's Hospital Boston; MGH = and Massachusetts General Hospital.
Harvard's early work in this area began in 1989 at Beth Israel Hospital with the creation of ClinQuery, a user-friendly computer program that allows health care workers to search for patients in a clinical database without requiring them to write computer code. 1 Doctors, nurses, medical students, and administrators used ClinQuery for a variety of purposes including clinical research, patient care, teaching and education, and hospital administration. In 1994, a collaboration of researchers across Children's Hospital, Beth Israel and Massachusetts General Hospital developed a federated query tool called W3-EMRS that, as a demonstration, queried clinical databases at all three hospitals “on-the-fly” with the results shown dynamically on web pages. 2 In 1996, Beth Israel and Deaconess Hospitals merged to form BIDMC. Part of this merger involved the creation of CareWeb, a secure and retooled version of W3-EMRS to consolidate health records for the two hospitals. 3 In parallel with the development of CareWeb, which uses an HL-7 transactional architecture, BIDMC also created a Clinical Data Repository (CDR), which stores the same information in a relational database for quality improvement reporting. Goldminer, developed by the Children's Hospital Informatics Program, allows authorized investigators to graphically build queries to search the hospital's clinical data repository. 4 Goldminer was based on an earlier application also developed at CHB in 1998 known as DXtractor, which built upon ClinQuery by breaking down the construction of complex queries into combinations of simple “atomic” queries. 5 By combining simple queries with a toolbox of population-based and temporal predicates, 6 users could more easily ask clinically meaningful questions.
Partners HealthCare (PHS) is an integrated health system founded by Brigham and Woman's Hospital (BWH) and Massachusetts General Hospital (MGH). In 1999, PHS, working with the Laboratory of Computer Science at MGH, developed the Research Patient Data Registry (RPDR). 7 The RPDR combines data from multiple clinical systems based at the BWH, the MGH, and four other community and rehabilitation hospitals, and stores it in a relational database. The user interface, which was modeled after Goldminer, includes two components. The web-based Query Tool provides users with a method of identifying patient cohorts defined by an arbitrary Boolean combination of medical concepts. 8 Access to the Query Tool is limited to faculty and their work group members at Partners, but IRB approval is not required since only aggregate counts are returned. 9 The second component of the RPDR is a Data Acquisition Engine, which faculty with an IRB approved protocol can use to retrieve the detailed patient data corresponding to a previously saved cohort defined by the Query Tool. The close relationship between the RPDR development team and the Partner's IRB has made this a highly successful application. To date, over 1,700 investigators have used the RPDR to identify patient populations for clinical studies. 7
Informatics for Integrating Biology and the Bedside (i2b2) is an NIH-funded National Center for Biomedical Computing based at Partners, which seeks to demonstrate the feasibility of using the data accrued during the course of healthcare for discovery research. This has led to the development of the i2b2 Clinical Research Chart (CRC), which is an open source platform and software implementation, which enables a variety of functions required for clinical research including Natural Language Processing (NLP) based querying for phenotype, deidentification that is HIPAA compliant, visualization, access to patient samples, and analytic pipelines. 10 While i2b2 draws upon the functionality of the RPDR, its underlying framework has been rearchitected to enable collaborative software development and to simplify implementation of the software at other institutions. The i2b2 software, known as the “hive”, consists of a collection of independent modules called “cells”, which share a common messaging protocol that allows them to interact using web services and XML messages.
While ClinQuery, CareWeb, DXtractor, Goldminer, RPDR, and i2b2 have successively improved on creating analytic platforms for a single clinical data repository, a separate project at Harvard, known as the Shared Pathology Information Network (SPIN), has tackled the problem of cross-institution data sharing. Initially funded by the National Cancer Institute, SPIN has served as a model of how to share data across a peer-to-peer network in which each participating institution/database maintains autonomy and control of its own data as well as maintaining the privacy of the patients whose data are shared. 11–14
Many commercial products exist for creating distributed database systems, such as Oracle Database Streams, or combining data from multiple sources, such as Microsoft's SQL Server Integration Services. However, a federated query tool for clinical data requires more than out-of-the-box features. It must satisfy the technical restrictions imposed by the hospitals' IRBs and data privacy officers, and it must deal with the specific challenges of working with frequently inaccurate and incomplete medical data.
Design Objectives
Our goal was to build upon the successes of i2b2 and SPIN to create a prototype of a Shared Health Research Information Network (SHRINE) that would enable investigators to search the electronic health records of patients across multiple independent Harvard hospitals. To do this, we would have to bring together the expertise of several informatics research teams, work closely with four different IRBs, and engage senior hospital executives who would need to support the project.
To consider the prototype a success, we had to develop a system that was approved by the IRBs and hospital executives and in real-time could query the clinical data repositories of at least three different health care centers. However, the time frame for the project was extremely short. Harvard Medical School internally sponsored the prototype and provided funding for only 6 months. Thus, we had to be realistic about what we could accomplish and make difficult decisions about what to exclude from the prototype. To protect the developers and ensure they could complete the technical components of the prototype within the limited time frame of the project, there were four significant exclusions from the prototype:
1 The prototype only queried the databases at BIDMC, CHB, and PHS. These are the three largest health centers, and each has experience creating consolidated repositories for research.
2 The prototype was limited to aggregate queries, and only a handful of users were given access to the system. With this, the prototype could be considered a technical proof-of-concept, which poses far fewer concerns for the hospitals and IRBs than a fully functional system open to all researchers.
3 The query interface only allowed users to search patient demographics and diagnoses. Although each of the data repositories has other data types such as laboratory test results, medications, and procedures, there is less consistency in how these concepts are coded than demographics and diagnoses. Since developing a common ontology for all SHRINE databases was a time-consuming part of the prototype implementation, we started with just these two categories.
4 We selected a technical architecture for the prototype that would be the easiest to implement, with an understanding that it would have to be redesigned later to make it scalable to larger numbers of health centers.
Four separate IRBs had to approve the prototype. The IRBs at BIDMC, CHB, and PHS each had to approve the use of their hospital's data, and IRB at Harvard Medical School had to approve the query tool that aggregated the results from the three databases. To protect patient privacy, we included the following additional limitations on the scope of the prototype:
1 There would be no central database. Each hospital would own and manage its data locally and have a local principal investigator responsible for the database.
2 The prototype would only be available for a limited time, after which all data would be destroyed.
3 The local databases at each hospital would include only old data from 2006. After a one-time load, the data would not be refreshed.
4 All patients whose data would be used in the prototype received a HIPAA privacy notice that allows their personal health information to be used for research that has been reviewed and approved by an IRB.
5 The prototype would only allow queries that return aggregate counts of clinical data, such as the total number of patients with diabetes at each health center. No identified data or data collected as part of a research study would be included in this demo.
6 The prototype would obfuscate the aggregate counts by adding a small random number. Thus, the user would see an approximate count of the number of matching patients, not the exact count.9 To make it more difficult for the user to guess the actual number, the prototype would “lock” the user's account if the same query was run multiple times in the same day.
7 If a hospital returned less than ten patients in a query, then “less than 10” would be presented rather than the actual count.
8 An audit of all queries would be logged.
9 In addition to an overall principal investigator for the SHRINE prototype, each hospital would have a local PI who would be responsible for his or her hospital's patient data.
Finally, to protect the hospitals and gain their support, we imposed several other limitations to the prototype, mainly designed to mask the hospitals' identity and minimize risk:
1 Individual hospitals could remove their databases from the prototype at any time.
2 Hospitals would not be identified by name in the demo. Instead, the labels “hospital 1”, “hospital 2”, “hospital 3” would be used.
3 For each query, the aggregate counts would be displayed in a random order so that “hospital 1”, for example, would refer to a different institution each time.
4 The aggregate counts would be multiplied by a scale-factor that is inversely proportional to the number of patients at the hospital. Otherwise, PHS, which includes both BWH and MGH would return aggregate counts that were roughly twice as big on average as the other two hospitals.
5 The counts from the three health centers would be displayed simultaneously instead of one at a time in the order in which they are returned by the hospitals. Otherwise, the speed of a local hospital's database, which is dependent on many factors such as the amount of data and types of servers, could be used to identify the health center from which an aggregate count came.
System Description
Functionality
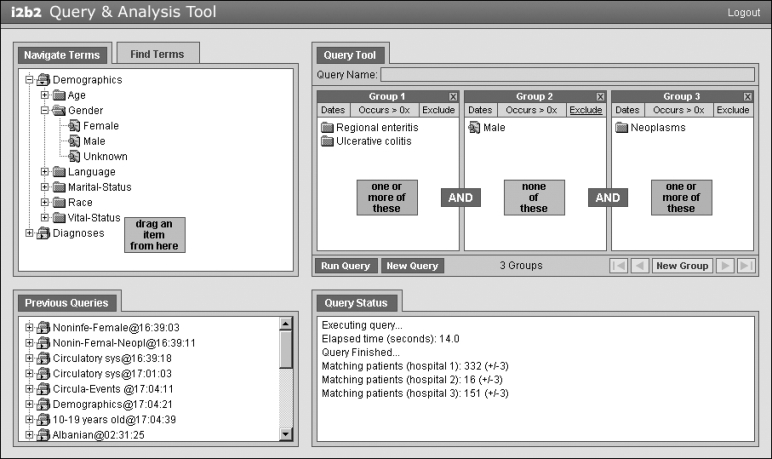
The user interface to SHRINE (▶) is based on the web client developed for i2b2 and RPDR. 8 Users first login to the Web site and then view a “workbench” divided into four modules.
1 The Ontology module contains a list of hierarchical medical concepts organized in an expandable tree. The top two levels in the prototype are demographics and diagnoses. A Find Terms tab lets users search concepts by name or by code, such as ICD-9.
2 Users can drag-and-drop concepts onto “panels” in the Query Tool module to indicate the population of patients that they want to locate. Concepts in the same panel are logically OR'ed, and the panels themselves are AND'ed together. Panels can be negated by clicking an Exclude button. This combination allows the user the flexibility to construct complex Boolean queries. Date ranges can also be placed on a panel. Finally, a minimum number of occurrences can be placed on a panel, which specifies how many times a concept must appear in a patient's medical record. This is useful for increasing the specificity of a search at the expense of decreased sensitivity. Using the full set of search options, users can ask questions such as: How many female patients have at least two diagnoses in their medical record between January 1, 2006, and December 31, 2006? How many of these patients also have a history of neoplasms? How many of these patients were not treated for an intestinal infectious disease?
3 The Query Status module displays the amount of time that a query has been running, and when the query is complete it shows the aggregate counts from each hospital.
4 The Previous Queries module lists the results from all prior queries. Users can drag-and-drop an item from this module onto the Query Tool to view the combination of concepts that were used in the query.
Figure 1.
Interface for SHRINE, which is a modified version of the i2b2 web client. Users drag-and-drop terms from an ontology tree to a query tool that is structured to allow for the creation of Boolean expressions. The Query Status panel indicates the number of matching patients at each hospital, and the Previous Queries panel saves the results of all queries.
Architecture
The prototype SHRINE architecture consists of a Query Aggregator hosted on servers at Harvard Medical School and SHRINE Adapters located at each hospital. The Query Aggregator contains two parts. The first is the web-based interface through which users access the system. The second are web services that broadcast the query to each of the Adapters and receive the counts back from each health center. The web client and back-end services communicate using asynchronous JavaScript and XML (AJAX). The SHRINE Adapters are web services at each hospital that receive queries from the Aggregator and return patient counts. The Adapters are designed so the Aggregator can send the same XML query definition to all health centers. The purpose of the Adapters is to translate the query into a format that is compatible with the institution's source databases, thus hiding the complexity of the local databases from the rest of the SHRINE. Our prototype included three Adapters, though in theory any number of other health centers could be added.
The Aggregator-Adapter model does not define the details of the communication method between the Aggregator and the Adapters. However, because of our prior experience using the i2b2 system and the fact that we had an existing i2b2 web client, we chose to use the i2b2 XML format to encode query definitions and result sets. 13
The SHRINE architecture also places no restrictions on the schemas at the local institutions. Although the Adapters must be designed to accept a standardized XML query definition format, a health center can make any customizations needed in the back-end to connect the Adapter to existing databases. This can be done in one of two ways: the adapter can be modified to work with the local databases, or the local databases can setup to work with the adapter. We chose the latter technique. Separate from the SHRINE prototype, BIDMC, CHB, and PHS were each already planning to implement local i2b2 instances for their researchers. So, each health center created an i2b2 structured database for the SHRINE prototype. This allowed us to design a single Adapter that could be used at each location with only minimal customizations since we could assume that the Adapter would be connecting to an i2b2 database. The i2b2 databases took about a month to set up. This process was facilitated by the fact that the health centers had existing relational clinical repositories and local database administrators worked in close collaboration with members of the i2b2 development team.
Additional details about the i2b2 and SHRINE architecture and downloadable source code can be found at http://www.i2b2.org and http://catalyst.harvard.edu/shrine.
Results
The IRB Approval Process
The IRBs at BIDMC, CHB, PHS, and HMS approved the SHRINE prototype. In addition to the institutions of data origination IRBs, we also needed approval from the Harvard Medical School IRB because we were aggregating the patient data on HMS servers. Approval from the HMS IRB was dependent on approval from all the hospitals, so we submitted our proposal to it last. A faculty member from each institution was designated as the principal investigator on the proposals given to the IRBs, and a separate overall PI was listed on the HMS IRB.
Approval from HMS was straightforward since the three hospitals had already approved the project, and only aggregate counts would be returned to the HMS servers.
Performance
The SHRINE web interface allows users to construct complex Boolean queries. A “typical” query contains two or three concepts, such as “how many patients are female and have a history of both inflammatory bowel disease and cancer”, and takes 10–60 seconds in the prototype. A single concept that matches few or no patients can run in as little as 5 seconds, and a query with many concepts that matches a large percentage of all patients can require up to 2 minutes.
Database Differences
▶ summarizes the types and amounts of data stored in each repository as of summer 2008. To make the counts more comparable, the number of observations from a single year, 2006, are given. ▶ provides a similar summary for the three i2b2 databases, which were generated using only 2006 data from the source repositories. The number of patients in the i2b2 databases represents those who had at least one diagnosis in 2006, and the demographics are only for that subset of the entire patient population. In each repository, ICD-9 codes were used for diagnoses and some procedures; however, there was no common vocabulary for other data types.
Table 2.
Table 2 Summary of the Size of the Source Clinical Data Repository at Each Health Center. Numbers are in Thousands Except Years of Data and Number of Hospitals. Although the PHS Repository Contains Some Data as Far Back as 1986, Significant Amounts do not Start Appearing Until the Early 1990s. Note the Similarities Between BIDMC and CHB Despite Different Patient Populations. Even Adjusted for Number of Patients, the Counts at PHS are Much Higher Because Multiple Data Sources Feed the RPDR, Which Can Result in Duplicate Observations
| Source Repository | BIDMC | CHB | PHS |
|---|---|---|---|
| Year created | 2000 | 2002 | 1999 |
| Years of data | 8 | 6 | 24 |
| Number of hospitals | 1 | 1 | 6 |
| Number of patients | 1,850 | 1,619 | 4,644 |
| Number of providers | 17 | 58 | 74 |
| Observations (2006)—diagnoses | 809 | 1,071 | 12,731 |
| Observations (2006)—EEG Studies | — | 6.4 | 13 |
| Observations (2006)—genomic test results | — | 1.78 | 0.28 |
| Observations (2006)—health maintenance | — | — | 321 |
| Observations (2006)—laboratory values | 11,643 | 6,987 | 50,195 |
| Observations (2006)—medications | 606 | 1,404 | 7,270 |
| Observations (2006)—microbiology studies | 153 | 102 | 515 |
| Observations (2006)—pathology studies | — | 14 | 1,209 |
| Observations (2006)—procedures | 171 | 132 | 13,811 |
| Observations (2006)—radiology studies | 294 | — | 2,265 |
| Observations (2006)—transfusion records | 32 | — | 18 |
| Observations (2006)—vital signs | — | — | 4,909 |
BIDMC = Beth Israel Deaconess Medical Center; CHB = Children's Hospital Boston; PHS = Partners HealthCare System.
Table 3.
Table 3 Summary of the Size of the i2b2 Database at Each Health Center. Numbers are in Thousands Except Years of Data and Number of Hospitals
| i2b2 Database | BIDMC | CHB | PHS |
|---|---|---|---|
| Years of data | 1 | 1 | 1 |
| Hospitals | 1 | 1 | 2 |
| Patients | 147 | 178 | 658 |
| Demographics | 1,029 | 1,243 | 5,264 |
| Diagnoses | 809 | 1,071 | 12,286 |
BIDMC = Beth Israel Deaconess Medical Center; CHB = Children's Hospital Boston; PHS = Partners HealthCare System.
Discussion
It is difficult to overemphasize the significance to the Harvard medical community of what we achieved. Harvard's CTSA award will require its hospitals to collaborate in unprecedented ways to have the transformative effect intended by the grant. After a century of fierce competition among these hospitals, overcoming the regulatory challenges and the mistrust around data sharing is an even more difficult problem than solving the technical hurdles. Creating the SHRINE prototype forced not only the IRBs but also senior executives from different hospitals to work together to reach a common goal. It also required collaboration between multiple research informatics groups to agree on a technical architecture, participation from database and server administrators to set up the i2b2 instances, and help from the security and networking teams to review the systems and open firewall ports. By accomplishing this in a mere 6 months, we have been able to demonstrate to academic Deans and hospital CEOs what is possible, and this has paved the way for future collaborations including jump-starting the conversion of our SHRINE prototype into a scalable enterprise application.
Lessons Learned
Just within Harvard, there is a great deal of heterogeneity among the different data repositories. We avoided some of the challenges of ontology mapping by limiting the prototype to demographics and diagnoses. However, it is already clear that expanding this to more data types and linking this into national ontology efforts will require a great deal of effort.
The use of a SHRINE Adapter greatly simplifies the construction of a federated query tool. The source repositories are too unique to build an Aggregator that includes all the rules necessary to query them directly. The Adapter modularizes the tasks and allows each hospital to solve, independently, the problem of morphing its source databases into a standard SHRINE ontology. It also allows the Aggregator to be developed without concern of the complexities of the local databases.
In the context of clinical data warehouses, discussions about “security and privacy” typically refer to protecting patient data, but we learned that it is just as important to protect the hospitals, which consider their clinical data as one of their most valuable intellectual properties. Over a billion dollars in clinical research is divided among the Harvard affiliated hospitals each year, and having a robust data repository makes an institution more competitive for these funds. The repositories can be used to obtain retrospective data for preliminary results, they simplify the process of identifying patients for trials, and they can assist with collecting data during the course of a study. Each of these can increase the speed of research and result in large cost reductions for a clinical trial. These institutions also compete for patient populations. They fear that the competing hospitals will inappropriately use the data in their repository to generate targeted marketing to urge patients to switch hospitals, or to try to recruit away their best clinicians. These are all issues we had to address when creating the SHRINE prototype, and the sensitivity around them will only heighten as we expand the scope of SHRINE.
Future Directions
Our lessons learned in creating the SHRINE prototype should be used as a road map for other institutions developing a new clinical data repository or a method of integrating multiple existing databases. It should be noted that although we completed our prototype in 6 months, both the technical teams and the IRBs at each of the hospitals had years of experience working with clinical data repositories and using them for research and other applications.
We believe our success in obtaining institutional support for SHRINE was due to the fact that we started with the concerns and issues raised by our local hospital IRBs and senior leadership and designed the technical architecture around that. A different approach would have been to begin with a more generalized vision and develop a standard that could be applied to systems at many institutions. This has been the methodology used by other major efforts to design federated query systems for clinical data, including caGrid, which created the caGrid Query Language (CQL) for sharing objects and object hierarchies; 15 the Biomedical Informatics Research Network (BIRN), which defined an XML schema for data exchange; 16 and the Service-oriented Architecture for NHIN Decision Support (SANDS) system, which enables distributed services to be used together in clinical decision making. 17 Of course, by building SHRINE around local policies, we risk developing a product that only works at one institution. However, sharing clinical data for research requires both technical innovation as well as a cultural shift in attitudes towards collaboration. By addressing the latter first, we pave the way for greater impact and adoption of our software platform. In fact, shortly after the launch of the SHRINE prototype, our local hospitals began discussions about policies for the next iteration of SHRINE, which would be available to a much wider audience and eliminate some of the restrictions such as masking hospital identities.
As we move forward with SHRINE, we face several challenges. We plan to redesign the architecture of the Adapters and Aggregator, most likely based on the scalable model in SPIN. This will allow the SHRINE network to expand and provide a simple mechanism for any number of institutions, including those outside of Harvard, to link in their clinical data repositories. This will require further developing the Adapter so it can use more complex ontologies, and restructuring the Aggregator so multiple Aggregators can exist on the SHRINE network and communicate in a peer-to-peer manner. Finally, we need to continue to work with the IRBs and our CTSC's regulatory committee to develop processes that ultimately allow all investigators to obtain identified patient data and biomaterials through SHRINE. Approaching each of these challenges will guide us in creating a future technical architecture that is scalable to a national level, compliant with ethical guidelines, and protective of the interests of the participating health centers.
Acknowledgments
The authors thank Nicholas A. Benik for his assistance with developing the i2b2/SHRINE web client.
Footnotes
This project was supported by grants 5 U54 LM008748, Informatics for Integrating Biology and the Bedside (i2b2), and 1 UL1 RR025758-01, Harvard Catalyst, The Harvard Clinical and Translational Science Center, from the National Center for Research Resources.
References
- 1.Safran C, Porter D, Lightfoot J, et al. ClinQuery: A system for online searching of data in a teaching hospital Ann Intern Med 1989;111(9):751-756. [DOI] [PubMed] [Google Scholar]
- 2.Kohane IS, Greenspun P, Fackler J, Cimino C, Szolovits P. Building national electronic medical Record systems via the World Wide Web J Am Med Inform Assoc 1996;3(3):191-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halamka JD, Osterland C, Safran C. CareWeb, a web-based medical record for an integrated health care delivery system Int J Med Inform 1999;54(1):1-8. [DOI] [PubMed] [Google Scholar]
- 4.In: Nigrin DJ, Kohane IS, editors. Data mining by clinicians. Orlando, Florida: Hanley and Belfus; 1998. Proceedings, Annual Fall Symposium of the American Medical Informatics Association.
- 5.In: Nigrin DJ, Kohane IS, editors. Scaling a data retrieval and mining application to the enterprise-wide level. 1999. Proceedings, Annual Fall Symposium of the American Medical Informatics Association. [PMC free article] [PubMed]
- 6.Nigrin DJ, Kohane IS. Temporal expressiveness in querying a timestamp-based clinical database J Am Med Inform Assoc 2000;7(2):152-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.In: Nalichowski R, Keogh D, Chueh HC, Murphy SN, editors. Calculating the benefits of a research patient Data Repository. 2006. Proceedings, Annual Fall Symposium of the American Medical Informatics Association. [PMC free article] [PubMed]
- 8.In: Murphy SN, Gainer VS, Chueh H, editors. A visual interface designed for novice users to find research patient cohorts in a large biomedical database. 2003. Proceedings, Annual Fall Symposium of the American Medical Informatics Association. [PMC free article] [PubMed]
- 9.In: Murphy SN, Chueh H, editors. A security architecture for query tools used to access large biomedical databases. 2002. Proceedings, Annual Fall Symposium of the American Medical Informatics Association. [PMC free article] [PubMed]
- 10.In: Murphy SN, Mendis M, Hackett K, et al. editors. Architecture of the open-source clinical research chart from informatics for integrating biology and the bedside. 2007. Proceedings, Annual Fall Symposium of the American Medical Informatics Association. [PMC free article] [PubMed]
- 11.Drake TA, Braun J, Marchevsky A, et al. A system for sharing routine surgical pathology specimens across institutions: The Shared Pathology Informatics Network Hum Pathol 2007;38(8):1212-1225. [DOI] [PubMed] [Google Scholar]
- 12.Holzbach AM, Chueh H, Porter AJ, Kohane IS, Berkowicz D. A query engine for distributed medical databases Medinfo 2004 2004:1519CD.
- 13.McMurry AJ, Gilbert CA, Reis BY, et al. Distributed architecture for public health, research, and clinical care J Am Med Inform Assoc 2007;14(4):527-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Namini AH, Berkowicz DA, Kohane IS, Chueh HC. A submission model for use in the indexing, searching, and retrieval of distributed pathology case and tissue specimens Medinfo 2004:1264-12672004. [PubMed]
- 15.Oster S, Langella S, Hastings S, Ervin D, Madduri R, Phillips J, et al. caGrid 1.0: an enterprise Grid infrastructure for biomedical research J Am Med Inform Assoc 2008;15(2):138-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keator D, Grethe J, Marcus D, et al. A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN) IEEE Trans Inf Technol Biomed 2008;12(2):162-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright A, Sittig DF. SANDS: A service-oriented architecture for clinical decision support in a National Health Information Network J Biomed Inform 2008;41(6):962-981Epub;2008, Mar 14. [DOI] [PMC free article] [PubMed] [Google Scholar]