Abstract
Background
Explicit patient consent requirements in privacy laws can have a negative impact on health research, leading to selection bias and reduced recruitment. Often legislative requirements to obtain consent are waived if the information collected or disclosed is de-identified.
Objective
The authors developed and empirically evaluated a new globally optimal de-identification algorithm that satisfies the k-anonymity criterion and that is suitable for health datasets.
Design
Authors compared OLA (Optimal Lattice Anonymization) empirically to three existing k-anonymity algorithms, Datafly, Samarati, and Incognito, on six public, hospital, and registry datasets for different values of k and suppression limits.
Measurement
Three information loss metrics were used for the comparison: precision, discernability metric, and non-uniform entropy. Each algorithm's performance speed was also evaluated.
Results
The Datafly and Samarati algorithms had higher information loss than OLA and Incognito; OLA was consistently faster than Incognito in finding the globally optimal de-identification solution.
Conclusions
For the de-identification of health datasets, OLA is an improvement on existing k-anonymity algorithms in terms of information loss and performance.
Introduction
There have been strong concerns about the negative impact of consent requirements in privacy legislation on the ability to conduct health research. 1–18 Such concerns are reinforced by the compelling evidence that requiring explicit consent for participation in different forms of health research can negatively impact the process and outcomes of the research itself (see online Appendix A at www.jamia.org for the literature search strategy and summary of articles): (a) recruitment rates decline significantly when individuals are asked to consent (opt-in vs. opt-out consent, or explicit consent vs. implied consent), (b) in the context of explicit consent, those who consent tend to be different from those who decline consent on a number of variables (age, sex, race/ethnicity, marital status, rural vs. urban locations, education level, socioeconomic status and employment, physical and mental functioning, language, religiosity, lifestyle factors, level of social support, and health/disease factors such as diagnosis, disease stage/severity, and mortality) hence potentially introducing bias in the results, 19 (c) consent requirements increase the cost of conducting the research and often these additional costs are not covered, and (d) the research projects take longer to complete (because of the additional time and effort needed to obtain consent, as well as taking longer to reach recruitment targets due to the impact on recruitment rates).
One approach to facilitate health research and alleviate some of the problems documented above is to de-identify data beforehand or at the earliest opportunity. 20,21 Many research ethics boards will waive the consent requirement if the data collected or disclosed is deemed to be de-identified. 22
A commonly used de-identification criterion is k-anonymity, and many k-anonymity algorithms have been developed. 23–32 This criterion stipulates that each record in a dataset is similar to at least another k-1 records on the potentially identifying variables. For example, if k = 5 and the potentially identifying variables are age and gender, then a k-anonymized dataset has at least 5 records for each value combination of age and gender.
In this paper we present a new k-anonymity algorithm, Optimal Lattice Anonymization (OLA), which produces a globally optimal de-identification solution suitable for health datasets. We demonstrate on six datasets that OLA results in less information loss and has faster performance compared to current de-identification algorithms.
Methods
Background
Definitions
Quasi-identifiers
The variables that are going to be de-identified in a dataset are called the quasi-identifiers. 33 Examples of common quasi-identifiers are 34–38 : dates (such as birth, death, admission, discharge, visit, and specimen collection), locations (such as postal codes, hospital names, and regions), race, ethnicity, languages spoken, aboriginal status, and gender.
Equivalence Classes
All the records that have the same values on the quasi-identifiers are called an equivalence class. For example, all the records in a dataset about 17-year-old males admitted on Jan 1, 2008 are an equivalence class. Equivalence class sizes potentially change during de-identification. For example, there may be 3 records for 17-year-old males admitted on Jan 1, 2008. When the age is recoded to a five year interval, then there may be 8 records for males between 16 and 20 years old admitted on Jan 1, 2008.
De-identification Optimality Criterion
A de-identification algorithm balances the probability of re-identification with the amount of distortion to the data (the information loss). For all k-anonymity algorithms, disclosure risk is defined by the 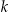 value, which stipulates a maximum probability of re-identification. 39 There are no generally accepted information loss metrics, although some (discussed below) have been proposed and are used in practice. One commonly used criterion to obtain an optimal balance between disclosure risk and information loss is to first find the de-identification solutions that have acceptable disclosure risk, then among these the optimal solution is defined as the one with minimum information loss. 40
value, which stipulates a maximum probability of re-identification. 39 There are no generally accepted information loss metrics, although some (discussed below) have been proposed and are used in practice. One commonly used criterion to obtain an optimal balance between disclosure risk and information loss is to first find the de-identification solutions that have acceptable disclosure risk, then among these the optimal solution is defined as the one with minimum information loss. 40
Requirements for De-identifying Health Data
In this section, we will define four important requirements on a de-identification algorithm to ensure that it is practical for use with health datasets. These requirements are not comprehensive, but represent what we consider a minimal necessary set: if they are not met then the de-identified data may not be practically useful. They are a consensus based on the experiences of the authors de-identifying and analyzing health data. These requirements drove the algorithm we have developed.
Quasi-identifiers are represented as hierarchies
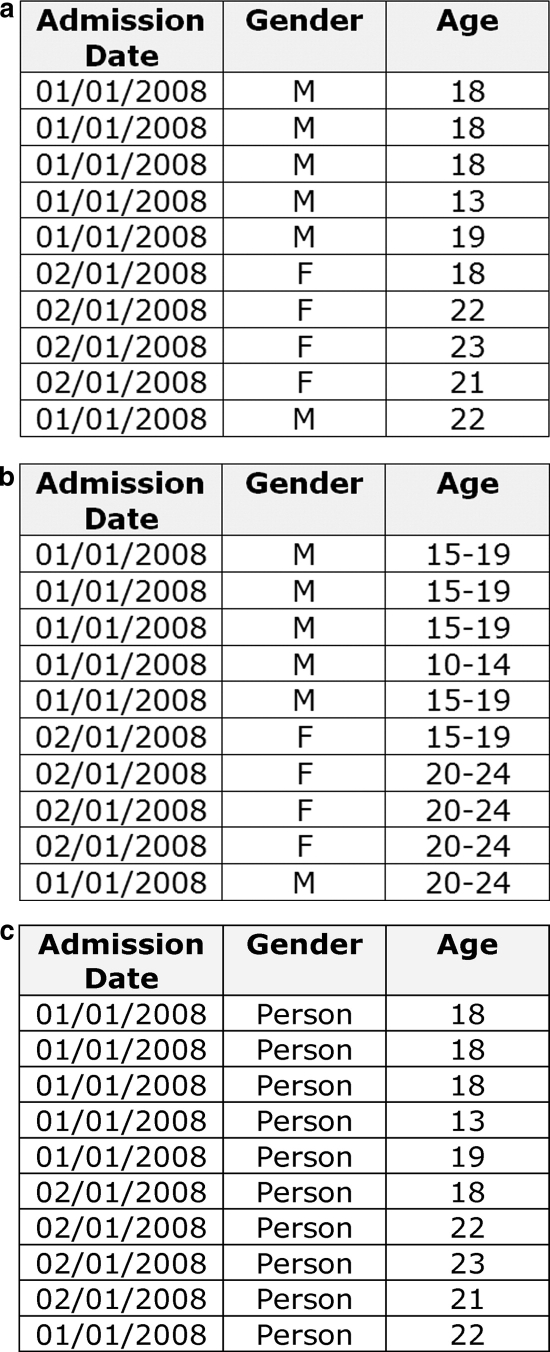
A common way to satisfy the k-anonymity criterion is to generalize values in the quasi-identifiers by reducing their precision. 26 Quasi-identifiers in health data that are used for research, public health, quality improvement, and postmarketing surveillance purposes can be represented as hierarchies. Examples of hierarchies are illustrated in ▶. The precision of the variables is reduced as one moves up the hierarchy. For example, a less precise representation of a postal code “K1H 8L1” would be the first three characters only: “K1H”. Similarly, a date of birth can be represented as a less precise year of birth. Numeric variables can also be represented hierarchically, for instance, age can be converted to a 2-year interval, and then to a 5-year interval. In the context of de-identification, this hierarchical representation is the default approach used in the Canadian Institutes for Health Research privacy guidelines. 41 Therefore, a de-identification algorithm needs to deal with this hierarchical nature of the variables.
Figure 1.
Examples of value generalization hierarchies for three common quasi-identifiers: (a) admission date, (b) gender, and (c) age in years.
Discretization intervals must be definable by the end-user
Some existing k-anonymity algorithms define a total order over all values of a given quasi-identifier, 27 and a quasi-identifier can be recoded to any partition of the values that preserves the order. If this partitioning is performed automatically by a de-identification algorithm, it may produce intervals of unequal sizes (for example, age may be automatically partitioned to intervals such as <0–9> <10–12> <13–25> <26–60>). The unequal interval sizes and the inability to control these in advance by the user make the analysis of such data quite complex and significantly reduce its utility. In practice, the users of the data need to specify the interval sizes that are appropriate for the analysis that they will perform.
Use global recoding instead of local recoding
Several of the k-anonymity algorithms use local recoding. 29–32,42 This means that the generalizations performed on the quasi-identifiers are not consistent across all of the records. For example, if we are considering age, then one record may have a 17 year old recoded to an age interval of <11–19>, and another record with a 17 year old is recoded to the age interval of <16–22>. If the variable was hierarchical, then local recoding may keep one record with the age as 17, and the other record recoded to the <16–20> interval. Such inconsistency in constructing response categories makes the data very difficult to analyze in practice using standard data analysis techniques. Therefore, a more practical approach would be to use global recoding where all the records have the same recoding within each variable.
The de-identification solution must be globally optimal
A globally optimal algorithm satisfies k-anonymity but at the same time minimizes information loss. Some k-anonymity algorithms do work with hierarchical variables but they use heuristics or approximations to the optimal solution, and do not produce a globally optimal solution themselves. 25,28 Excessive information loss can result in the loss of statistical power, inaccurate analysis results, and inefficient use of data that was costly to collect with possible inconvenience to patients. A globally optimal solution mitigates these disadvantages.
Generalization and Suppression
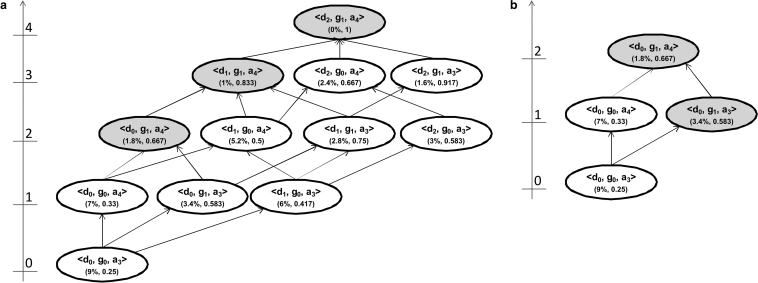
The generalization hierarchies for the three quasi-identifiers in ▶ can be represented as a lattice, as in panel (a) of ▶. The height of each row of nodes is shown on the left hand side, ranging from zero to 7 in this case. The arrows illustrate the possible generalization paths that can be taken through the lattice. A series of connected paths from the bottom node to the top node is a generalization strategy. Panel (b) of ▶ shows two generalization strategies which pass through the node <d0, g1, a2>. Each node in the lattice represents a possible instance of the dataset. One of these nodes is the globally optimal solution and the objective of a k-anonymity algorithm is to find it efficiently.
Figure 2.
(a) An example of a lattice of generalizations. Each node indicates the generalization level for each of the three variables, and in parenthesis the percentage of suppression and the value of the Prec information loss metric. (b) The same lattice showing two generalization strategies through it. The two strategies go through the node <d0, g1, a2>.
All equivalence classes in the dataset that are smaller than  are suppressed. 26 In ▶, 70% of the records were suppressed in the dataset represented by node <d0, g0, a0> because these records were in small equivalence classes. As more generalization is applied, the extent of suppression goes down. For example, node <d0, g0, a1>, with age generalized to 5-year intervals, has only 30% of the records suppressed. Therefore, as we traverse any generalization strategy from the bottom node to the top node, there is a monotonically decreasing level of suppression. 24
are suppressed. 26 In ▶, 70% of the records were suppressed in the dataset represented by node <d0, g0, a0> because these records were in small equivalence classes. As more generalization is applied, the extent of suppression goes down. For example, node <d0, g0, a1>, with age generalized to 5-year intervals, has only 30% of the records suppressed. Therefore, as we traverse any generalization strategy from the bottom node to the top node, there is a monotonically decreasing level of suppression. 24
Suppression is preferable to generalization because the former affects single records whereas generalization affects all the records in the dataset. 24 Therefore, when searching for an optimal solution, a solution that imposes more suppression would be selected instead of one that imposes more generalization.
However, because of the negative impact of missingness on the ability to perform meaningful data analysis, 43 the end-users will want to impose limits on the amount of suppression that is allowed. We will refer to this limit as MaxSup. It is assumed that the data analyst will specify MaxSup such that complete case analysis can be performed or imputation techniques can be used to compensate for the missing data. 44
A node in the lattice is said to be a k-anonymous node if the amount of suppression is less than MaxSup. If we let MaxSup = 5%, then the highlighted nodes in ▶ represent all the possible k-anonymous nodes since they would all satisfy the “suppression <5%” criterion. Once we have identified all the k-anonymous nodes, we need to select the one with the least information loss from among them.
The extent of suppression is not a good measure of information loss because it has counter-intuitive behavior: as we generalize more, suppression decreases (i.e., from a missingness perspective, data utility improves). Whereas information loss is intended to measure the reduction in the utility of the data as it is generalized. 30,45 This is shown in the lattice of ▶, whereby maximum generalization node <d2, g1, a4> has zero suppression, and node <d0, g0, a0> with no generalization has the highest level of suppression at 70% of the records. If we used the extent of suppression as an information loss metric, then node <d2, g1, a4> would be selected as the optimal node because it is k-anonymous and has the lowest suppression. However, this is the node with the maximum possible amount of generalization.
We need to consider other measures of information loss that will allow us to efficiently identify the least generalized node among the k-anonymous nodes.
Information Loss Metrics
Out of the highlighted nodes in the lattice, Samarati 24 proposes that the node with the lowest lattice height should be selected as the optimal solution. In our example of ▶, this would be node <d0, g1, a1>. The assumption being made is that this solution balances the extent of generalization with the extent of suppression.
The lattice height is not considered a good information loss metric because it does not account for the generalization hierarchy depths of the quasi-identifiers. For example, if we generalize “Male” to “Person” then this is given equal weight to generalizing age in years to age in five year intervals. In the former case there is no information left in the gender variable, whereas the five year age interval still conveys a considerable amount of information and there are three more possible generalizations left in the age hierarchy (see ▶).
An information loss metric that takes into account the height of the generalization hierarchy is Precision or Prec. The Prec was introduced by Sweeney 46,47 as an information loss metric that is suitable for hierarchical data. For every variable, the ratio of the number of generalization steps applied to the total number of possible generalization steps (total height of the generalization hierarchy) gives the amount of information loss for that particular variable. For example, in ▶ if age is generalized from age in years to age in five year intervals, then the value is ¼. Overall Prec information loss is the average of the Prec values across all quasi-identifiers in the dataset. As a consequence, the more a variable is generalized, the higher the information loss. Moreover, variables with more generalization steps (i.e., more levels in their generalization hierarchy) tend to have less information loss than ones with shorter hierarchies. Using Prec as the information loss metric, the node <d2, g0, a1> would be the optimal node rather than node <d0, g1, a1> in ▶: the former has a Prec of 0.417 and the latter a Prec of 0.466.
Another commonly used information loss metric is the Discernability Metric or DM. 27,39,48–54 The discernability metric assigns a penalty to every record that is proportional to the number of records that are indistinguishable from it. Following the same reasoning, DM assigns a penalty equal to the whole dataset for every suppressed record (since suppressed records are indistinguishable from all other records). The formula for DM metric appears in online Appendix D at www.jamia.org.
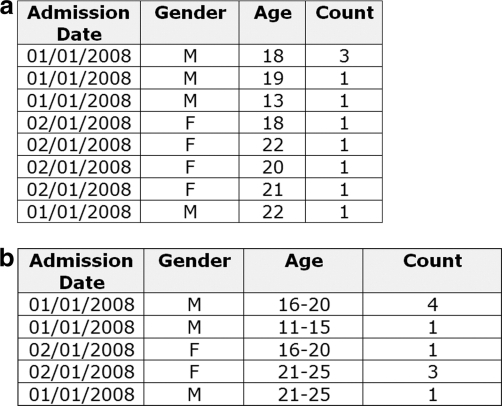
However, DM is not monotonic within a generalization strategy due to the impact of the second term incorporating suppression. The example in ▶ shows two possible datasets for the <d0, g0, a0> and <d0, g0, a1> nodes, where the latter is a direct generalization of the former. We assume that we want to achieve 3-anonymity. For node <d0, g0, a0> seven out of ten records do not achieve 3-anonymity, and therefore the DM value is 79, whereas for node <d0, g0, a1>, the DM value is 55. This reduction in information loss as we generalize means that we would select the k-anonymity solution with the maximum generalization as the best one, which is counter-intuitive. It therefore makes sense not to include the suppression penalty in DM. In other words, we will use a modified version of DM*, as calculated in online Appendix D.
Figure 3.
Three possible datasets representing different nodes in the lattice. Dataset (a) represents node <d0, g0, a0>. Dataset (b) represents node <d0, g0, a1> and is a generalization of (a). Dataset (c) represents node <d0, g1, a0> and is a generalization of (a). We assume that the objective is to achieve 3-anonymity.
The  information loss also solves a weakness with the Prec metric in that Prec does not take into account the size of the equivalence classes. If we generalize gender to “Person” in Table a of ▶ to obtain Table c in ▶, then the Prec for Table (c) would be 0.33 and
information loss also solves a weakness with the Prec metric in that Prec does not take into account the size of the equivalence classes. If we generalize gender to “Person” in Table a of ▶ to obtain Table c in ▶, then the Prec for Table (c) would be 0.33 and 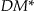 would be 16. However, Table b in ▶ has a Prec of 0.0833 and a
would be 16. However, Table b in ▶ has a Prec of 0.0833 and a 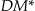 of 28. As can be seen in this case, the higher Prec value had a lower
of 28. As can be seen in this case, the higher Prec value had a lower  value. The reason is that there are more equivalence classes in (b) than in (c), and one of the equivalence classes is larger; the Prec metric does not consider the structure of the data itself.
value. The reason is that there are more equivalence classes in (b) than in (c), and one of the equivalence classes is larger; the Prec metric does not consider the structure of the data itself.
The concept behind DM has been criticized because DM does not measure how much the generalized records approximate the original records. 30,50 For example, suppose we want to achieve 2-anonymity, and we have a single quasi-identifier, age, and six records with the following age values: 9, 11, 13, 40, 42, and 45. The minimal  value is when all of the records are grouped into three pairs: <9,11>, <13,40>, and <42,45>. The criticism is that the second pair has a very wide range and that a more sensible grouping would have only two equivalence classes: <9,11,13> and <40,42,45>. In our case, since we assume that all data are hierarchical and that the end-user would specify the appropriate age grouping in the generalization hierarchy, the end-user may decide that the <13,40> node in a value generalization hierarchy is acceptable. Therefore, this particular criticism is not applicable in the context that we are using
value is when all of the records are grouped into three pairs: <9,11>, <13,40>, and <42,45>. The criticism is that the second pair has a very wide range and that a more sensible grouping would have only two equivalence classes: <9,11,13> and <40,42,45>. In our case, since we assume that all data are hierarchical and that the end-user would specify the appropriate age grouping in the generalization hierarchy, the end-user may decide that the <13,40> node in a value generalization hierarchy is acceptable. Therefore, this particular criticism is not applicable in the context that we are using 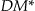 .
.
But the discernability metric has also been criticized because it does not give intuitive results when the distributions of the variables are non-uniform. 55 For example, let's say we have a single quasi-identifier, gender, and two different datasets with 1,000 records. The first has 50 male records and 950 female, and the second has 500 males and 500 females. If gender is generalized to “Person”, then intuitively the information loss for the 950 females in the first dataset should be quite low and the female records dominate the dataset. However, the 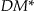 value indicates that the information loss for the nonuniformly distributed dataset is much higher than for the uniformly distributed (second) dataset (905,000 vs. 500,000). One information loss metric that has been proposed based on entropy 56,57 has recently been extended to address the non-uniform distribution problem. 42,58 Formulas for non-uniform entropy calculation appear in online Appendix D.
value indicates that the information loss for the nonuniformly distributed dataset is much higher than for the uniformly distributed (second) dataset (905,000 vs. 500,000). One information loss metric that has been proposed based on entropy 56,57 has recently been extended to address the non-uniform distribution problem. 42,58 Formulas for non-uniform entropy calculation appear in online Appendix D.
Returning to our example, the 50/950 male/female distributed dataset has an entropy of 286 whereas the 500/500 male/female distributed dataset has an entropy of 1,000. Therefore, the information loss in the former dataset is much lower, and this makes more intuitive sense.
The Monotonicity Property
The three information loss metrics that we have presented above (Prec, 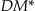 , and non-uniform entropy) are monotonic within any given generalization strategy. This means that as we move up the lattice along any generalization strategy the information loss value will either remain the same or increase.
, and non-uniform entropy) are monotonic within any given generalization strategy. This means that as we move up the lattice along any generalization strategy the information loss value will either remain the same or increase.
This property is important because it means that if we have two k-anonymous nodes in the same generalization strategy, then the one lower in the strategy will always have a lower information loss. We take advantage of this property in our algorithm described below. Furthermore, it has been noted that this monotonicity property is essential to produce de-identified datasets that are more suitable for data analysis. 42
While we have presented three common information loss metrics that have the monotonicity property, this is not intended to be a comprehensive list. There may be other information loss metrics that also have this property.
The OLA Algorithm
In this section, we describe our new algorithm, OLA. We assume that the dataset has more than 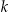 records. The objective of OLA is to find the optimal node in the lattice. The optimal node is k-anonymous and has minimal information loss. For our purposes, information loss can be any one of the three metrics described previously.
records. The objective of OLA is to find the optimal node in the lattice. The optimal node is k-anonymous and has minimal information loss. For our purposes, information loss can be any one of the three metrics described previously.
Main Steps
To find the optimal node, the algorithm proceeds in three steps:
1 For each generalization strategy, conduct a binary search to find all the k-anonymous nodes.
2 For each generalization strategy with k-anonymous nodes, only the k-anonymous node with the lowest height within the strategy is retained. For example, in ▶ both nodes <d0, g1, a1> and <d0, g1, a2> are k-anonymous, but they are both part of the same generalization strategy and <d0, g1, a1> is below <d0, g1, a2> in the lattice. This means that <d0, g1, a1> will have less information loss on all the three metrics we considered. The node <d0, g1, a1> is called a k-minimal node.
3 Now that we have the k-minimal nodes, these are compared in terms of their information loss and the node with the smallest information loss is selected as the globally optimal solution. Because of the monotonicity property, the k-minimal node with the smallest information loss must also have the smallest information loss among all k-anonymous nodes in the lattice.
The most time-consuming computations in OLA are in steps 1 and 3: (a) to find out, for any node, whether it is a k-anonymous node, and (b) to compare the k-minimal nodes. Therefore, to ensure efficiency our algorithm minimizes the number of instances where it needs to determine if a node is k-anonymous by predictively tagging the k-anonymous status of nodes instead of computing it. Predictive tagging is explained further below. OLA also minimizes the number of nodes that need to be compared on information loss (step 3) by ensuring that there are few k-minimal nodes. The complete algorithm itself is described in pseudo code in online Appendix B at www.jamia.org.
Predictive Tagging
Predictive tagging takes advantage of two properties of the lattice. First, if a node N is found to be k-anonymous then all nodes above N on the same generalization strategies that pass through N are also k-anonymous. We therefore tag all of these higher nodes as k-anonymous instead of computing their k-anonymous status. For example, if we are evaluating node <d0, g1, a2> in ▶ and determine that it is k-anonymous, then the following nodes can immediately be tagged as k-anonymous: <d0, g1, a3>, <d0, g1, a4>, <d1, g1, a4>, <d2, g1, a4>, <d1, g1, a2>, <d1, g1, a3>, <d2, g1, a3>, and <d2, g1, a2>. Second, if a node N is found not to be k-anonymous, then all nodes below N on the same generalization strategies that pass through N are not k-anonymous. We therefore tag all of these lower nodes instead of computing their k-anonymous status. For example, if we are evaluating node <d1, g0, a2> in ▶ and determine that it is not k-anonymous, then the following nodes can immediately be tagged as not k-anonymous: <d1, g0, a1>, <d0, g0, a2>, <d0, g0, a1>, <d1, g0, a0>, and <d0, g0, a0>. Predictive tagging results in a significant reduction in the amount of computations that need to be performed, allowing the processing of large lattices efficiently.
Algorithm Walkthrough
The algorithm implements a binary search through the generalization strategies in the lattice. To illustrate our algorithm using the lattice in ▶, we start our search for the globally optimal node through the lattice at the middle height (height equals 3), iterating through the nodes starting from the left. The first node to be examined is <d0, g0, a3>. The extent of suppression is computed and it is determined that this is not a k-anonymous node and is tagged as such. Using the logic of predictive tagging, all nodes below <d0, g0, a3> on all generalization strategies that go through that node are also by definition not k-anonymous nodes: nodes <d0, g0, a0>, <d0, g0, a1>, And <d0, g0, a2> can be tagged as not k-anonymous right away without further computation.
We then focus on the sub-lattice whose bottom node is <d0, g0, a3> and top node is <d2, g1, a4>. This sublattice is illustrated in panel (a) of ▶. The same steps as above are repeated. We go to the middle height in this sub-lattice, which is node <d0, g1, a4>. The extent of suppression is computed for this node, it is determined that this node is k-anonymous, and it is tagged as such. This also means that all other nodes above <d0, g1, a4> on all generalization strategies that go through node <d0, g1, a4> are k-anonymous and can be tagged as such. In this case these are nodes <d1, g1, a4> and <d2, g1, a4>.
Figure 4.
Panel (a) is a sub-lattice of the lattice in ▶. Panel (b) is a sub-lattice of (a). Sub-lattices used in the illustrative example of finding the k-anonymous nodes. The shaded nodes are k-anonymous.
We then focus on the sub-lattice whose top node is <d0, g1, a4> and bottom node is <d0, g0, a3>. This sublattice is illustrated in panel (b) of ▶. The same steps as above are repeated. We go to the middle height in this sub-lattice, which is node <d0, g0, a4>. The extent of suppression is computed for this node, and it is determined that this node is not k-anonymous. This also means that all other nodes below <d0, g0, a4> on all generalization strategies that go through node <d0, g0, a4> are also by definition not k-anonymous. In this case the nodes below it are: <d0, g0, a3>, <d0, g0, a2>, <d0, g0, a1>, and <d0, g0, a0>.
We then proceed to the next node in the most recent sub-lattice, which is node <d0, g1, a3>, which is in panel (b) in ▶. We determine that it is k-anonymous and it is tagged as such. But we can also determine that all other nodes above <d0, g1, a3> on all generalization strategies that go through node <d0, g1, a3> are also by definition k-anonymous solutions and can be tagged. In this case these are nodes <d0, g1, a4>, <d1, g1, a3>, <d1, g1, a4>, <d2, g1, a3>, and <d2, g1, a4>.
Now we go back to the sub-lattice in panel (a) of ▶ and evaluate node <d1, g0, a4>. The suppression is higher than 5% and it is therefore not k-anonymous, and it is tagged as such. We can also determine that all other nodes below <d1, g0, a4> on all generalization strategies that go through node <d1, g0, a4> are also by definition not k-anonymous nodes and are tagged as such.
This search process tags a significant percentage of the nodes without evaluating them.
OLA also maintains a k-minimal solutions list of the k-anonymous nodes that have the lowest height within their generalization strategies. Whenever a node N is tagged as k-minimal, OLA checks if there are other k-minimal nodes above it on the generalization strategies that pass through N. If there are, then these higher nodes are removed from the k-minimal solutions list and node N is added to the list.
The last step in the algorithm is to compare the nodes in the k-minimal solutions list on information loss and select the one with the smallest value as the globally optimal solution.
Empirical Evaluation
We compare OLA to three other existing algorithms: Datafly, Samarati, and Incognito. We include Datafly even though it does not provide a globally optimal solution because it is one of the few k-anonymity algorithms that has been used on actual clinical datasets. 59,60 Samarati 23,24 is an often cited example of a k-anonymity algorithm. 61 Incognito 62 can produce globally optimal results. These three algorithms perform global recoding and can handle hierarchical variables.
Algorithms
Datafly
Datafly uses a heuristic to find a k-anonymous dataset. 25 The quasi-identifier with the most distinct values is selected, and generalized. If the resultant generalized dataset is k-anonymous then the algorithm stops. If not, then the next quasi-identifier with the most unique values is selected and the process repeats as above. We modified the basic Datafly algorithm so the stopping criterion is whether the amount of suppression is less than MaxSup at each iteration.
In any iteration if there is more than one quasi-identifier with the same number of distinct values, then one is selected at random. In our implementation we ran Datafly 100 times and averaged the results to take account of the randomness introduced by the selection of a quasi-identifier when there are ties.
Since Datafly does not explicitly use an information loss metric during its search, it will often produce a solution that is not globally optimal on any chosen information loss metric. In our evaluation we wanted to find out how far Datafly is from the optimal solution.
Samarati
The Samarati algorithm finds the k-anonymous nodes at the lowest height in the lattice through a binary search. 24 The search is based on the observation that if there is no k-anonymous node at height 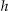 in the lattice then no node at height
in the lattice then no node at height 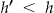 will be k-anonymous.
will be k-anonymous.
If 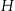 is the total height of the lattice, then it starts by investigating all nodes at
is the total height of the lattice, then it starts by investigating all nodes at  . If a node exists at this height that is k-anonymous, then it will try the nodes at height
. If a node exists at this height that is k-anonymous, then it will try the nodes at height 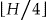 , otherwise it will try the nodes at height
, otherwise it will try the nodes at height 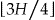 . This process repeats until it reaches the height at which there is at least one node that is k-anonymous, and there are no lower heights with k-anonymous nodes. If there are multiple k-anonymous nodes at the lowest height, one of them can be selected at random or specific preference criteria can be used to select among them. We evaluated the Samarati algorithm with Prec,
. This process repeats until it reaches the height at which there is at least one node that is k-anonymous, and there are no lower heights with k-anonymous nodes. If there are multiple k-anonymous nodes at the lowest height, one of them can be selected at random or specific preference criteria can be used to select among them. We evaluated the Samarati algorithm with Prec, 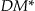 , and non-uniform entropy as the preference criterion. However, because the information loss comparison is only performed on the k-anonymous nodes at the lowest height, this does not guarantee that the selected node will be globally optimal on information loss.
, and non-uniform entropy as the preference criterion. However, because the information loss comparison is only performed on the k-anonymous nodes at the lowest height, this does not guarantee that the selected node will be globally optimal on information loss.
Incognito
Incognito starts by considering all possible subsets of the quasi-identifiers. From our example in ▶, it would consider each lattice with a single quasi-identifier: <Admission Date>, <Gender>, and <Age> separately, then lattices with all combinations of two quasi-identifiers: <Admission Date, Gender>, <Admission Date, Age>, and <Gender, Age>, and finally lattices with all three quasi-identifiers: <Admission Date, Gender, Age>.
Incognito takes advantage of two optimizations. First, when evaluating nodes in each one of these lattices, nodes that are above k-anonymous nodes in the same generalization strategies are tagged as k-anonymous. Second, if a node is not k-anonymous in a smaller quasi-identifier subset, then it will by definition not be k-anonymous in a larger subset of the quasi-identifiers. This means that the lattices for larger subsets of quasi-identifiers can be pruned. In our example, we found that all the nodes in the single quasi-identifier lattices are k-anonymous: <Admission Date>, <Gender>, and <Age>. Therefore, none of these nodes are eliminated. However, when considering lattices with pairs of quasi-identifiers, the following five nodes were not k-anonymous: <d1, g0>, <d0, go>, <d1, a0>, <g0, a0>, and <d0, a0>. Consequently, all nodes containing these generalizations are pruned from the lattice with the three quasi-identifiers.
Incognito would then proceed evaluating the nodes starting from the bottom of the lattice and moving upwards breadth first, tagging the generalizations of k-anonymous nodes that are found. This overall approach results in a significant reduction in the number of nodes that need to be evaluated.
The k-anonymous nodes in the full three quasi-identifier lattice are compared on information loss and the node with the lowest value is selected. This selected node will have the lowest global information loss and is therefore the optimal solution.
There are multiple versions of Incognito. The version we tested is called “Super Root Incognito” as it was shown by the authors to have the best performance. 62
Evaluations
Comparisons
We perform four comparisons of OLA's performance with the above three algorithms on the quality of the results (i.e., information loss) and on its practical utility (i.e., speed and time it takes to produce a result).
In the first evaluation we compare the information loss from using Datafly and Samarati to the globally optimal solution. Since Datafly and Samarati do not guarantee a globally optimal solution on our three information loss metrics, we wanted to find out how far they are from this optimal solution. If these two algorithms produce solutions that are sufficiently close to this optimal solution in practice, then a case can be made for using them given that they are simple, well established and understood.
We do not evaluate Incognito on information loss with respect to proximity to an optimal solution because it already finds the globally optimal solution on the three information loss metrics, and hence gives the same solution as OLA.
We also compare the time it takes for Datafly and Samarati to find a solution in seconds based on the same hardware configuration and datasets. A considerable amount of effort was spent on optimizing the implementation of both algorithms to ensure that the comparison is fair.
The third evaluation compares the theoretical speed performance of OLA with Incognito. Given that Incognito and OLA both produce the same globally optimal solution, the question is whether their performances are different. If OLA performs faster, then a case can be made for using it instead of Incognito.
The final comparison is of the time in seconds it takes OLA and Incognito to produce results. This comparison highlights the practical utility of the algorithms on realistic and large datasets, and re-enforces the results of the theoretical speed performance.
Datasets
The six datasets used are summarized in Table 1 (found in online Appendix C at www.jamia.org). These include datasets that are publicly available as well as hospital and registry datasets. These vary in size, and number and type of quasi-identifiers. The quasi-identifiers are quite typical of what is seen in realistic situations. 35,36,63,64
We included public datasets in our analysis to facilitate the replication of the results by others and because they had quasi-identifiers that are quite typical of those found in health datasets that are often disclosed. The hospital and registry datasets are typical of the types of data that would be de-identified in practice before disclosure: the emergency department data were being disclosed for syndromic surveillance, the pharmacy data were being disclosed to a commercial data aggregator, 65 and the provincial birth registry (Niday) was being disclosed to researchers.
Study Points
In practice, a minimal value of k = 3 is sometimes recommended, 66–69 but more often a value of k = 5 is used. 70–79 To ensure a reasonable amount of variation in our analysis we use values of k between 2 and 15 inclusive. For each dataset the maximum suppression (MaxSup) was set at 1, 5, and 10% of the total number of records.
Measurement
Information Loss
The three information loss metrics described earlier were measured. Since they are unitless, there are no generally accepted absolute benchmarks for their interpretation, and our interest is in determining how far they are from the globally optimal (which is the minimum possible) information loss, we defined relative information loss measures. The baseline for the relative measure was the information loss from the globally optimal solution. The globally optimal solution was generated using a “brute force” approach of evaluating all the nodes in the lattice, and provides the gold standard to compare against. The brute force approach is only useful for evaluation purposes to generate a gold standard as it is extremely slow.
For each metric, we used the gold standard value as the baseline, and the relative information loss values for Datafly and Samarati were represented as a percentage from this baseline. For example, if Datafly has an information loss value of 100% it means that Datafly has the globally optimal (i.e., minimal) information loss value. Alternatively, if it is 200% then that means the information loss for Datafly is twice as large as the globally optimal value. Relative information loss is always greater than 100%.
Performance
The speed of Datafly and Samarati is computed in seconds to produce a result. This is the time as perceived from an end-user perspective, and therefore includes all the preprocessing and reporting time.
We also compare OLA and Incognito in terms of seconds to produce a result. However, such a comparison would be perceived as inherently biased because we would be expected to put more effort optimizing the implementation of OLA. Therefore, we also measure the amount of computations that are inherent to the OLA and Incognito algorithms rather than their implementations. Below we explain how the amount of inherent computation in both algorithms can be measured.
The two most time-consuming activities in all the algorithms that navigate the generalization lattice, such as OLA and Incognito, are (a) evaluating each node to determine whether it is k-anonymous or not, and (b) comparing all the nodes on information loss.
Number of Nodes
An obvious way to compare the algorithms is to count the number of nodes that need to be evaluated as to whether they are k-anonymous or not. The two algorithms use different approaches to minimize the number of nodes to evaluate by predictively tagging nodes and working up from smaller lattices.
The Complexity of Evaluating k-Anonymous Nodes
The complexity of evaluating if a node is k-anonymous is not uniform because the nodes vary in terms of the number of quasi-identifiers being generalized and the number of records being evaluated. A performance measure that takes into account the complexity of each node's evaluation is needed. We propose such a node complexity metric below.
Evaluating whether a node is k-anonymous consists of three tasks:
1 Generalizing the quasi-identifiers.
2 Computing the new equivalence classes on the generalized quasi-identifiers.
3 Summing the number of records in equivalence classes that are smaller than
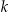 .
.
Our node complexity metric takes into account these three tasks and in practice we have observed that it matches well the actual timing in seconds for both algorithms.
Let each node in a lattice 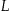 be indexed by its height
be indexed by its height  and position from the left
and position from the left 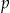 . Each node will have a specific number of quasi-identifiers
. Each node will have a specific number of quasi-identifiers 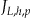 . As noted above, Incognito will create multiple lattices for an increasing number of quasi-identifiers and therefore the nodes across lattices will differ in their
. As noted above, Incognito will create multiple lattices for an increasing number of quasi-identifiers and therefore the nodes across lattices will differ in their 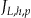 value, but in OLA this value is fixed because we have only one lattice. For example, in our lattice in ▶,
value, but in OLA this value is fixed because we have only one lattice. For example, in our lattice in ▶, 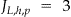 .
.
Not all the quasi-identifiers will be generalized as we move up the lattice. For example, when we want to evaluate node <d0, g1, a0> then we would take node <d0, g0, a0> and only generalize the second quasi-identifier. We let 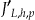 denote the number of quasi-identifiers that need to be generalized. For a given node, it is always the case that
denote the number of quasi-identifiers that need to be generalized. For a given node, it is always the case that 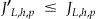 .
.
Each node is associated with a particular instance of the data. The data at each node are represented as a frequency set. 62 This means that we represent the data by the equivalence classes and their counts. For example, ▶ is the frequency set for ▶. The generalization in ▶b has fewer records than (a).
Figure 5.
An example of a frequency set for the datasets shown in ▶. Table (b) is an age generalization of Table (a).
Let the size of the original frequency set be given by  and the size of the generalized frequency set be given by
and the size of the generalized frequency set be given by  . For instance, in ▶ the original frequency set in panel (a) had
. For instance, in ▶ the original frequency set in panel (a) had 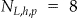 , and the generalized frequency set in panel (b) had
, and the generalized frequency set in panel (b) had 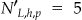 .
.
To generalize we need to go through all the records in the original frequency set. The amount of computation needed to generalize the frequency set is proportional to 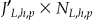 . In the example of ▶, the generalization task is given a score of 1 × 8 = 8. If a given node is involved in more than one generalization strategy, then we would use the smallest frequency set from a node with a lower height in one of those generalization strategies.
. In the example of ▶, the generalization task is given a score of 1 × 8 = 8. If a given node is involved in more than one generalization strategy, then we would use the smallest frequency set from a node with a lower height in one of those generalization strategies.
The second and third tasks are performed on the generalized frequency set, and involve computing the size of the equivalence classes for this frequency set. Doing so requires that we sort the frequency set and then do an additional pass through it to compute the new equivalence class sizes. Formulas for calculating this appear in online Appendix D. 80
We can compare the performance of our algorithm and Incognito by summing this score across all nodes for which we need to compute if it is k-anonymous.
Number of Nodes Compared
Another performance measure to compare OLA with Incognito is the number of nodes for which we evaluate information loss. In the case of OLA these are the k-minimal nodes and in the case of Incognito these are all the k-anonymous nodes that have been found. The information loss metrics take time to compute, and therefore the more nodes for which we need to make this computation the slower the algorithm will be.
Software Verification
To ensure the correctness of the algorithm implementations and accuracy in computing the relative information loss metrics, one programmer developed the k-anonymity programs, and another programmer performed a code review on that code to detect any errors. It is well established in software engineering that peer reviews are one of the best methods for defect detection. 81 In addition, the prospect of having their code being available for scrutiny by their peers motivates programers to be more careful. 82
Additional testing on synthetic datasets and samples from the real datasets was performed to verify correctness. For Datafly and Samarati the correct de-identification results for the test datasets were computed by hand and these were used to verify their implementations. For Incognito and OLA we implemented a brute force search that evaluated all the nodes in the lattice and produced the globally optimal solution, and we checked that the implementations of the two algorithms produced the same result.
The measurement of information loss presented in the results section was performed by a separate program that compared the original and de-identified datasets. The same program was used to evaluate the Datafly and Samarati results. This program was tested by comparing its results to a manually computed gold standard on synthetic datasets and samples of real datasets.
For computing the node evaluation complexity, the results were computed by hand for synthetic datasets and samples of the real datasets, and the computations from the instrumented versions of the algorithms were compared to these to ensure correctness.
Results
In this section, we discuss the results for the 5% suppression limit only. The results for the 1% and 10% suppression limits are provided in online Appendix C.
The information loss results comparing Datafly and Samarati are shown for the 5% suppression limit in Figure 6 (available in online Appendix C at www.jamia.org). The information loss comparisons indicate that both of these algorithms often have information loss values greater than the optimal solution, with Datafly generally having the higher information loss. For the Prec metric, Samarati is closer to the optimal, but the differences are much more pronounced for the 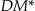 and entropy metrics. These observations hold across different suppression limits as well.
and entropy metrics. These observations hold across different suppression limits as well.
The speed results comparing Datafly and Samarati are shown in Figure 7 (in online Appendix C). It is clear that Datafly is much faster than Samarati. In fact, when compared to other data in Figure 9 (in online Appendix C), Datafly is the fastest k-anonymity algorithm overall. However, as the above results indicate, this comes at the cost of higher information loss.
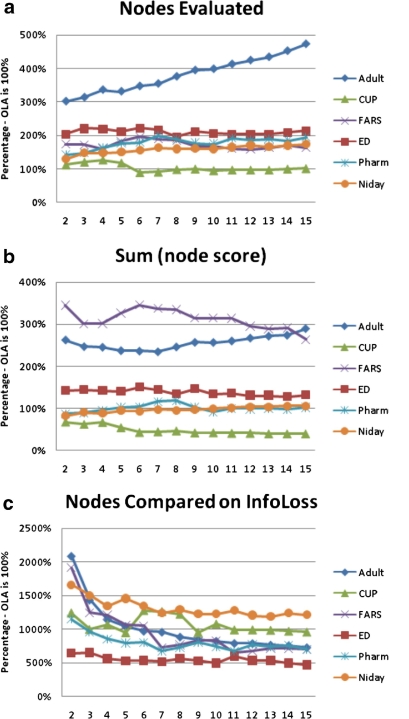
The first comparison of computations performed between OLA and Incognito is in terms of the number of nodes for which we need to evaluate k-anonymity. The results are shown in panel (a) of ▶. OLA almost always evaluates fewer nodes than Incognito across all the datasets.
Figure 8.
The performance metrics comparing our algorithm to Incognito. The results are for the 5% suppression limit. Our algorithm is the 100% value on the y-axis, and if Incognito performs more computations then its value is above 100%, and if it performs less computation then its value is below 100%. The panels show: (a) the total number of nodes for which we need to compute if they are k-anonymous, (b) the node complexity score given by Equation 3, and (c) the number of nodes for which information loss needs to be computed.
The node complexity results are shown in panel (b) of ▶. Incognito is better for the CUP dataset across all values of 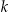 . The amount of computation is more or less the same for the Pharm and Niday datasets, and OLA performs better for the remaining three datasets. In the case of the Adult dataset and FARS, the difference in performance is quite significant in favor of OLA.
. The amount of computation is more or less the same for the Pharm and Niday datasets, and OLA performs better for the remaining three datasets. In the case of the Adult dataset and FARS, the difference in performance is quite significant in favor of OLA.
The third performance comparison is shown in panel (c) of ▶, which plots the relative number of nodes for which information loss needs to be computed. The set of nodes that Incognito produces is quite large and significant resources are consumed evaluating information loss for all of these to perform a comparison and select the node with the lowest information loss.
The overall time in seconds comparing OLA and Incognito is shown in Figure 9 (in online Appendix C). These graphs are dominated by the sum of the computations illustrated in panels (b) and (c) of ▶. The results make clear that OLA consistently, and sometimes quite significantly, finds the optimal solution faster than Incognito. The same results hold across all values of k and suppression limits we tested.
Discussion
Summary
Obtaining consent from patients can be costly and in practice introduces bias in clinical research. The de-identification of datasets at collection or at the earliest opportunity after collection is one alternative to obtaining consent. In this study, we presented a new globally optimal algorithm, OLA, that is suitable for de-identifying health datasets. This new algorithm satisfies the k-anonymity criterion. We empirically compared its performance on six datasets to three other k-anonymity de-identification algorithms: Datafly, Samarati, and Incognito.
Our comparisons showed Datafly and Samarati tended to have higher information loss than the optimal solution for three different common metrics: precision, the discernability metric, and non-uniform entropy. This indicates that there is a non-trivial information loss advantage to using a k-anonymity algorithm which produces a globally optimal result. Comparing the speed of Datafly and Samarati, Datafly was consistently faster, even on quite large datasets. OLA will have the exact information loss as Incognito because both find the same globally optimal solution as defined by the information loss metric in use. OLA's performance is significantly faster than Incognito.
Given the expense and potential inconveniences of collecting health information from patients, it behooves us to minimize the amount of information loss from de-identification. To obtain the result which ensures the minimal possible information loss, the fastest algorithm was shown to be OLA.
Applications in Practice
It is possible that OLA will not find a solution. This would occur if there is no k-anonymous node in the lattice. Under such circumstances the height of the generalization hierarchies would need to be extended to allow for more generalization or the MaxSup value increased and the algorithm run again. Note that if OLA cannot find a solution then no other k-anonymity algorithm which uses generalization and suppression would be able to find a solution either (with the same suppression limit and for the same generalization hierarchies on the quasi-identifiers).
In principle, the number of records flagged for suppression after the application of our algorithm can be as high as the suppression limit provided by the users of the algorithm (MaxSup). However, this does not necessarily mean that these whole records need to be suppressed. A local cell suppression algorithm can be applied to the flagged records and it will only suppress specific values of the quasi-identifiers on the flagged records. 83 Such an approach ensures that the number of cells suppressed from the set of flagged records are minimized.
OLA has already been applied to de-identify a pharmacy dataset being disclosed to a commercial data broker. 65 In this particular case study, the data were being disclosed on a quarterly basis through an automated de-identification and upload process.
Users of OLA will not be able to easily select which of the three information loss metrics to use each time they need to de-identify a dataset. Therefore, in practice it is recommended that non-uniform entropy is used. The reason is that, as can be seen from the review in the Methods Section, it is the one with the least known deficiencies.
There will be many situations where the quasi-identifiers are not equally important to the data recipient. It would be desirable to select a k-anonymity solution which generalizes the most important variables less. Assuming entropy is used as the information loss metric, weighting of quasi-identifiers can be achieved using a weighted non-uniform entropy as indicated in online Appendix D.
A recipient of a dataset may wish to impose constraints on the generalization that is performed. Some common constraints can be accommodated with OLA by limiting the nodes that are included in the lattice. Two specific examples are considered below.
A data recipient may want to impose a maximum allowable generalization. For example, a researcher may say that any generalization of age above a 5-year interval is unacceptable. If we take the age generalization hierarchy in ▶, then a1 would be the highest acceptable height. Such a constraint can be accommodated by creating a lattice where the top node is <d2, g1, a1> instead of <d2, g1, a4>. This ensures that the globally optimal solution will never have an age generalization above 5 years.
Another constraint that is often useful to have is to correlate the generalizations among multiple quasi-identifiers. For example, if a dataset has a date of death and an autopsy date as quasi-identifiers, it would not make sense to generalize the former to the nearest year, and keep the month and year for the latter. In such a case an intruder would be able to infer the month of death quite accurately by knowing the month of the autopsy. Therefore, the additional level of generalization on date of death provides no protection. Consequently we would want to ensure that the generalizations performed on both of these variables match. This can be achieved by not having any nodes in the lattice where these two variables have different levels of generalization. As another example, consider a longitudinal health insurance claims dataset with a patient's residence postal code at the beginning of each year included as a quasi-identifier. For many patients their postal code will be the same from one year to the next. Therefore, it would be prudent to correlate the postal codes for all the years in the dataset to ensure that postal code is generalized to the same height across all years.
Limitations
In principle, OLA requires that the information loss metrics used are monotonic with respect to generalization strategies in the lattice, whereas Incognito does not impose that requirement. We have shown that three different and relatively common information loss metrics that capture different types of generalization costs are monotonic. However, the case has been made that even if an information loss metric is non-monotonic, it rarely exhibits this non-monotonic behavior in practice. 42 To the extent that this empirical observation can be generalized broadly, other nonmonotonic metrics, such as basic entropy or the original discernability metric, may still produce optimal results with OLA.
Instead of generalization and suppression, other de-identification approaches could be used, such as the addition of noise. 57,84 It has been argued that while these approaches may maintain the aggregate properties of the data (such as the mean), they do not preserve the truthfulness of individual records. 24 Furthermore, the optimal type of disturbance will depend on the desired analysis that will be done with the data, which makes it difficult to de-identify datasets for general use. 85 As noted earlier, all variables types can be represented in terms of a generalization hierarchy, and there are very limited perturbation techniques for such hierarchies apart from generalization and suppression.
Acknowledgments
The authors thank Bradley Malin (Vanderbilt University) for reviewing an earlier version of this paper. This work was partially funded by the Canadian Institutes of Health Research and The Natural Sciences and Engineering Research Council of Canada.
References
- 1.Ness R. Influence of the HIPAA privacy rule on health research J Am Med Assoc 2007;298(18):2164-2170. [DOI] [PubMed] [Google Scholar]
- 2.Institute of Medicine Health research and the privacy of health information—The HIPAA privacy rule, 2008http://www.iom.edu/CMS/3740/43729.aspx 2007. Accessed August 4, 2009.
- 3.Institute of Medicine 2006. Effect of the HIPAA privacy rule on health research: Proceedings of a workshop presented to the National Cancer Policy Forum.
- 4.Association of Academic Health Centers HIPAA creating barriers to research and discovery 2008.
- 5.Wilson J. Health insurance portability and accountability Act privacy rule causes ongoing concerns among clinicians and researchers Ann Intern Med 2006;145(4):313-316. [DOI] [PubMed] [Google Scholar]
- 6.Walker D. Impact of the HIPAA Privacy Rule on Health Services Research, Abt Associates, Inc for the Agency for Healthcare Research and Quality 2005.
- 7.Hanna K. The privacy rule and research: Protecting privacy at what cost? Res Pract 2007;8(1):4-11. [Google Scholar]
- 8.Association of American Medical Colleges Testimony on behalf of the Association of American Medical Colleges before the National Committee on Vital and Health Statistics Subcommittee on Privacy 2003.
- 9.Friedman D. HIPAA and research: How have the first two years gone? Am J Opthalmol 2006;141(3):543-546. [DOI] [PubMed] [Google Scholar]
- 10.Nosowsky R, Giordano T. The health insurance portability and accountability Act of 1996 (HIPAA) privacy rule: Implications for clinical research Am Med Rev 2006;57:575-590. [DOI] [PubMed] [Google Scholar]
- 11.Hiatt R. HIPAA: The end of epidemiology, or a new social contract? Epidemiology 2003;14(6):637-639. [DOI] [PubMed] [Google Scholar]
- 12.Erlen J. HIPAA—Implications for research Orthop Nurs 2005;24(2):139-142. [DOI] [PubMed] [Google Scholar]
- 13.Kulynych J, Korn D. The effect of the new federal medical privacy rule on research N Engl J Med 2002;346(3):201-204. [DOI] [PubMed] [Google Scholar]
- 14.O'Herrin J, Fost N, Kudsk K. Health insurance portability and accountability Act (HIPAA) regulations: Effect on medical record research Ann Surg 2004;239(6):772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Cancer Institute. National Cancer Advisory Board The HIPAA Privacy Rule: Feedback from NCI Cancer Centers, Cooperative Groups and Specialized Programs of Research Excellence, SPOREs 2003.
- 16.Shalowitz D. Informed consent for research and authorization under the Health Insurance Portability and Accountability Act Privacy Rule: An integrated approach Ann Intern Med 2006;144(9):685-688. [DOI] [PubMed] [Google Scholar]
- 17.The Academy of Medical Sciences Personal data for public good: using health information in medical research 2006.
- 18.Steinberg M, RUbin E. The HIPAA Privacy Rule: Lacks Patient Benefit, Impedes Patient GrowthAssociation of Academic Health Centers; 2009.
- 19.Harris AL AR, Teschke KE. Personal privacy and public health: Potential impacts of privacy legislation on health research in Canada Can J Pub Health 2008:293-296July–August, 2008. [DOI] [PMC free article] [PubMed]
- 20.Kosseim P, Brady M. Policy by procrastination: Secondary use of electronic health records for health research purposes. McGil. J Law Health 2008:2.
- 21.Lowrance W. Learning from experience: Privacy and the secondary use of data in health research J Health Serv Res Policy 2003;8(S1):2-7. [DOI] [PubMed] [Google Scholar]
- 22.Willison D, Emerson C, Szala-Meneok K, et al. Access to medical records for research purposes: Varying perceptions across Research Ethics Boards J Med Ethics 2008;34:308-314. [DOI] [PubMed] [Google Scholar]
- 23.Samarati P, Sweeney L. Protecting Privacy When Disclosing Information: k-anonymity and its enforcement through generalisation and suppressionSRI International; 1998.
- 24.Samarati P. Protecting respondents' identities in microdata release IEEE Trans Knowl Data Eng 2001;13(6):1010-1027. [Google Scholar]
- 25.Sweeney L. Achieving k-anonymity privacy protection using generalization and suppression Int J Uncertainty, Fuzziness and Knowl-Based Syst 2002;10(5):571-588. [Google Scholar]
- 26.Ciriani V, De Capitani di Vimercati SSF, Samarati P. k-anonymity Secure Data Management in Decentralized Systems. Springer; 2007.
- 27.Bayardo R, Agrawal R. Data privacy through optimal k-anonymization 2005. Proceedings of the 21st International Conference on Data Engineering.
- 28.Iyengar V. Transforming data to satisfy privacy constraints 2002. Proceedings of the ACM SIGKDD International Conference on Data Mining and Knowledge Discovery.
- 29.Du Y, Xia T, Tao Y, Zhang D, Zhu F. 2007. On Multidimensional k-Anonymity with Local Recoding Generalization, IEEE, 23rd International Conference on Data Engineering.
- 30.Xu J, Wang W, Pei J, et al. Utility-based anonymization using local recoding 2006. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining.
- 31.Wong R, Li J, Fu A, Wang K. (Alpa, k)-anonymity: An enhanced k-anonymity model for privacy-preserving data publishing 2006. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining.
- 32.Aggarwal G, Feder T, Kenthapadi K, et al. Approximation algorithms for k-anonymity Journal of Privacy Technology 2005.
- 33.Dalenius T. Finding a needle in a haystack or identifying anonymous census records J Off Stat 1986;2(3):329-336. [Google Scholar]
- 34.El Emam K, Brown A, Abdelmalik P. Evaluating predictors of geographic area population size cutoffs to manage re-identification risk J Am Med Inform Assoc 2009;16(2):256-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El Emam K, Jabbouri S, Sams S, Drouet Y, Power M. Evaluating common de-identification heuristics for personal health information J Med Internet Res 2006;8(4):e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Emam K, Jonker E, Sams S, et al. Pan-Canadian de-identification guidelines for Personal health information (report prepared for the office of the privacy commissioner of Canada), 2007Archived at: http://www.webcitation.org/5Ow1Nko5C]http://www.ehealthinformation.ca/documents/OPCReportv11.pdf 2006. Accessed August 4, 2009.
- 37.Canadian Institutes of Health Research CIHR Best Practices for Protecting Privacy in Health ResearchCanadian Institutes of Health Research; 2005.
- 38. ISO/TS 25,237 Health Informatics: Pseudonymization. 2008.
- 39.El Emam K, Dankar F. Protecting privacy using k-anonymity J Am Med Inform Assoc 2008;15:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trottini M. Assessing Disclosure Risk and Data Utility: A Multiple Objectives Decision Problem Joint, ECE/Eurostat work session on statistical data confidentiality. 2003.
- 41.Canadian Institutes for Health Research Guidelines for protecting privacy and confidentiality in the design, conduct and evaluation of health researchCanadian Institutes for Health Research; 2004.
- 42.Gionis ATassaT. k-Anonymization with minimal loss of information IEEE Trans Knowl Data Eng 2009;21(2):206-219. [Google Scholar]
- 43.Kim J, Curry J. The treatment of missing data in multivariate analysis Soc Methods and Res 1977;6:215-240. [Google Scholar]
- 44.Little R, Rubin D. Statistical Analysis with Missing DataJohn Wiley & Sons; 1987.
- 45.Domingo-Ferrer J, Vicenc T. Risk assessment in statistical microdata protection via advanced record linkage J Stat Comput 2003;13(4).
- 46.Sweeney L. Achieving k-anonymity privacy protection using generalization and suppression. Int. J Uncertainty, Fuzziness and Knowl-Based Syst 2002;10(5):18. [Google Scholar]
- 47.Sweeney L. Computational Disclosure Control: A Primer on Data Privacy ProtectionMassachusetts Institute of Technology; 2001.
- 48.LeFevre K, DeWitt D, Ramakrishnan R. Mondrian multidimensional k-anonymity 2006. Proceedings of the 22nd International Conference on Data Engineering.
- 49.Hore B, Jammalamadaka R, Mehrotra S. Flexible anonymization for privacy preserving data publishing: A systematic search based approach 2007. Proceedings of the SIAM International Conference on Data Mining.
- 50.Xu J, Wang W, Pei J, et al. Utility-based anonymization for privacy preservation with less information loss ACM SIGKDD Explor Newsl 2006;8(2):21-30. [Google Scholar]
- 51.Nergiz M, Clifton C. Thoughts on k-anonymization. Second international workshop on privacy. Data Manag; 2006.
- 52.Polettini S. A Note on the Individual Risk of DisclosureIstituto Nazionale di Statistica; 2003[Italy].
- 53.Machanavajjhala A, Gehrke J, Kifer D, Venkitasubramaniam M. L-diversity: Privacy beyond k-anonymity 2006. Proceedings of the International Conference on Data Engineering.
- 54.Bayardo R, Agrawal R. Data privacy through optimal k-Anonymization 2005. Proceedings of the 21st International Conference on Data Engineering.
- 55.Li T, Li N. Data privacy through optimal k-Anonymization 2006. Proceedings of the Sixth IEEE International Conference on Data Mining.
- 56.de Waal T, Willenborg L. Information loss through global recoding and local suppression Neth Off Statistics 1999;14:17-20. [Google Scholar]
- 57.Willenborg L, de Waal T. Elements of Statistical Disclosure ControlSpringer-Verlag; 2001.
- 58.Domingo-Ferrer J, Vicenc T. Disclosure control methods and information loss for microdataIn: Doyle P, et al. editor. Confidentiality, Disclosure, and Data Access: Theory and Practical Applications for Statistical Agencies. Elsevier; 2001.
- 59.Sweeney L. Computational Disclosure Control for Medical Microdata: The Datafly System. Record Linkage Techniques. National Academy Press; 1997.
- 60.Sweeney L. Guaranteeing anonymity when sharing medical data: The Datafly system 1997. Proceedings of the American Medical Informatics Association Symposium. [PMC free article] [PubMed]
- 61.Ciriani V, De Capitani di Vimercati S, Foresti S, Samarati P. k-Anonymity Secure Data Management in Decentralized Systems. Springer; 2007.
- 62.LeFevre K, DeWitt D, Ramakrishnan R. Incognito: Efficient full domain k-anonymity 2005. International Conference of the ACM Special Interest Group on Management of Data.
- 63.Sweeney L. Uniqueness of Simple Demographics in the US PopulationCarnegie Mellon University, Laboratory for International Data Privacy; 2000.
- 64.Ochoa S, Rasmussen J, Robson C, Salib M. Reidentification of Individuals in Chicago's Homicide Database: A Technical and Legal StudyMassachusetts Institute of Technology; 2001.
- 65.El Emam K, Dankar F, Vaillancourt R, Roffey T, Lysyk M. Evaluating patient re-identification risk from hospital prescription records Can J Hosp Pharm 2009;62(4):307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duncan G, Jabine T, de Wolf S. Private Lives and Public Policies: Confidentiality and accessibility of Government StatisticsNational Academies; 1993.
- 67.de Waal A, Willenborg L. A view on statistical disclosure control for microdata Surv Methodol 1996;22(1):95-103. [Google Scholar]
- 68.Office of the Privacy Commissioner of Quebec (CAI) Chenard v. Ministere de L'Agriculture, des Pecheries et de L'Alimentation 1997. vol 141.
- 69.National Center for Education Statistics NCES Statistical StandardsUS Department of Education; 2003.
- 70. Cancer Care Ontario Data Use and Disclosure Policy. Cancer Care Ont. http://www.cancercare.on.ca/common/pages/UserFile.aspx?fileId=13234 2003. Accessed August 4, 2009.
- 71. Security and confidentiality policies and proceduresHealth Quality Council; 2004.
- 72. Privacy codeHealth Quality Council; 2004.
- 73. Privacy CodeManitoba Center for Health Policy; 2002.
- 74.Subcommittee on Disclosure Limitation Methodology. Federal Committee on Statistical Methodology Working Paper 22, Report on statistical disclosure controlOffice of Management and Budget; 1994.
- 75.Statistics Canada Therapeutic abortion survey, 2007Archived at: http://www.webcitation.org/5VkcHLeQw http://www.statcan.ca/cgi-bin/imdb/p2SV.pl?Function=getSurvey&SDDS=3209&lang=en&db=IMDB&dbg=f&adm=8&dis=2#b9 1994. Accessed August 4, 2008.
- 76.Office of the Information and Privacy Commissioner of British Columbia Order 1998. 261–1998.
- 77.Office of the Information and Privacy Commissioner of Ontario Order P-644http://www.ipc.on.ca/images/Findings/Attached_PDF/P-644.pdf 1994. Accessed August 4, 2009.
- 78.Alexander L, Jabine T. Access to social security microdata files for research and statistical purposes Soc Sec Bull 1978;41(8):3-17. [PubMed] [Google Scholar]
- 79.Ministry of Health And Long term care (Ontario). Corporate Policy 3/1/2021. 1984.
- 80.Goldreich O. Computational ComplexityCambridge University Press; 2008.
- 81.El Emam K. Software Inspection Best Practices, Cutter Consortium 2001(E-Project Management Advisory Service).
- 82.El Emam K. The ROI from Software Quality, CRC Press 2005(Auerbach).
- 83.Aggarwal G, Feder T, Kenthapadi K, et al. Anonymizing tables 2005. Proceedings of the 10th, International Conference on Database Theory (ICDT05).
- 84.Adam N, Wortman J. Security-control methods for statistical databases: A comparative study ACM Comput Surv 1989;21(4):515-556. [Google Scholar]
- 85.Federal Committee on Statistical Methodology Report on Statistical Disclosure Limitation MethodologyOffice of Management and Budget; 2005.