Abstract
Objective
Machine learning systems can be an aid to experts performing systematic reviews (SRs) by automatically ranking journal articles for work-prioritization. This work investigates whether a topic-specific automated document ranking system for SRs can be improved using a hybrid approach, combining topic-specific training data with data from other SR topics.
Design
A test collection was built using annotated reference files from 24 systematic drug class reviews. A support vector machine learning algorithm was evaluated with cross-validation, using seven different fractions of topic-specific training data in combination with samples from the other 23 topics. This approach was compared to both a baseline system, which used only topic-specific training data, and to a system using only the nontopic data sampled from the remaining topics.
Measurements
Mean area under the receiver-operating curve (AUC) was used as the measure of comparison.
Results
On average, the hybrid system improved mean AUC over the baseline system by 20%, when topic-specific training data were scarce. The system performed significantly better than the baseline system at all levels of topic-specific training data. In addition, the system performed better than the nontopic system at all but the two smallest fractions of topic specific training data, and no worse than the nontopic system with these smallest amounts of topic specific training data.
Conclusions
Automated literature prioritization could be helpful in assisting experts to organize their time when performing systematic reviews. Future work will focus on extending the algorithm to use additional sources of topic-specific data, and on embedding the algorithm in an interactive system available to systematic reviewers during the literature review process.
Introduction and Background
Systematic reviews (SRs) locate, appraise, and synthesize the best available evidence from clinical studies of diagnosis, treatment, prognosis, or etiology, to provide informative empiric answers to specific research questions. 1 They also provide input to medical recommendations and often form the basis for many Health Technology Reports, formal decision analyses, clinical practice guidelines, and economic analyses. Furthermore, SRs are an essential component of evidence-based medicine (EBM), guiding both practice and policy. 2–4 Several groups coordinate the creation of SRs, such as the Cochrane Collaboration and the Evidence-based Practice Centers (EPCs) of the Agency for Healthcare Research and Quality (AHRQ). 3,5 Many SRs focus on specific classes of pharmacological therapies, or on treatments related to a specific disease.
Systematic Reviews leverage the time and experience of independent experts on SR who use the latest scientific and statistical techniques to construct a summary of the best information and practices in an area of medicine with as little bias as possible. This approach has many strengths, but one particularly motivating aspect of this process is that it frees individual physicians from having to review all the available literature themselves, thus enabling them to focus on administering the best possible care to their patients. In constructing an SR, the review team attempts to synthesize all the available evidence about a class of drugs or medical conditions into a set of conclusions that reflect the highest-quality knowledge and the highest level of care available at the time of the review. Some SRs include a meta-analysis, which is an aggregation of the results obtained in specific individual studies by pooling data from similar studies for a specific outcome and performing subsequent statistical analyses on the pooled data. 6 Meta-analyses can make important contributions to our understanding of the true effectiveness of a medical intervention, improving our confidence in a finding by extending the precision and consistency beyond that of a single study. Selection of appropriately similar trials during the SR literature review is an important first step in constructing a meta-analysis.
The SRs make important contributions to improving the quality of health care and making better informed use of available resources. Because medicine is constantly changing, an SR on a given topic is not simply written, published once, and considered permanently completed. To correctly represent the best available medical knowledge at any given time, reviews must undergo periodic updates—although when and exactly how often this should happen is currently an active topic of research. 7–9 Although one study estimated the median time before a SR review becomes out of date to be 5.5 years, many areas of medicine are prone to frequent change (e.g., cardiovascular disease), and information is thought to become out of date within as little as 2.7 years. 10
The Cochrane Collaboration estimates that, to cover most health care problems, at least 10,000 total SRs are needed. Less than half this number has been published, even after 10 years of concerted effort by the EBM community. 11 New clinical trials are published at a rate in excess of 15,000/yr, therefore the evidence supporting EBM is constantly growing and changing. The SRs must expand to cover this new evidence, as well as other current health care problems and future interventions. The creation of a new SR, or the updating of an existing one, can take several months, making the workload of SR teams grow at an exponential pace. Therefore, new tools and methods of keeping up with this expanding workload are necessary.
In this work we address how the creation and update of SRs can be made more efficient with machine learning (ML) techniques. 12,13 Specifically, we have examined how incorporation of ML-based techniques into the initial literature review step can increase the efficiency of the SR process, allowing the team to focus their time and attention on tasks that will most benefit from their experience and expertise. In accordance with the “5S” hierarchical view of medical knowledge and systems proposed by Haynes, this work investigates automated means of reducing the workload for taking medical knowledge from the raw level of individual studies to the more refined level of synopses of the best available evidence. 14 This is a very time- and expertise-intensive step in the process of medical knowledge distribution.
Once the review topic and key questions are defined, the process of creating or updating an SR begins with a three-stage process in which the biomedical literature relevant to that review is identified. In the first stage, a medical librarian creates a set of literature database queries using, for example, PubMed Clinical Queries or Ovid to search MEDLINE, and other electronic databases such as EMBASE and PsycInfo. In the next step, abstract triage, experts review the abstracts for each of the retrieved articles, identifying those that are most likely to meet the inclusion criteria for full text review. Finally, in the full text triage step, these articles are read in full, and decisions are made on which articles contain sufficient quality of evidence to warrant inclusion in the SR. If the SR includes a meta-analysis, an additional step is included, in which data from groups of studies that are sufficiently similar, of adequate quality, and provide suitable data, are pooled together. The system that we present here aims to increase the efficiency of abstract and full text triage by reducing the number of documents requiring manual human expert review in these steps.
The procedures for creating and updating SRs are similar. One important difference, however, is that an SR update already has a base collection of included/excluded article judgments that are based on previous reviews. Once a review topic has been updated several times, a set of associated included/excluded article judgments accumulates. These judgments can serve as input to an ML algorithm, whether they be the relatively few initial judgments performed during the creation of a first review for a given topic with a small set of applicable literature (e.g., anti-platelet drugs, as of 2008), or a large set of judgments that have accumulated over the second or third update for a topic with a large literature base (e.g., ACE inhibitors). This knowledge, along with other information about the subject domain amenable to knowledge engineering, can be used as a training set for an ML system. Once trained, the system can make predictions on the appropriateness of an article for inclusion in the updated SR. The predictions can be used in multiple ways to improve the process of creating or revising the SR.
In an information-intensive era such as the current one, it is common for humans to need to engage in repetitive, labor-intensive processes, such as reviewing large amounts of text. Text processing and ML can be an aid in these and similar situations, such as the creation and update of SRs. Genomics curators continually review the published literature, annotating the accumulated knowledge about genes and gene-gene interactions into scientifically accessible databases. 15,16 Clinical researchers read hundreds of discharge summaries, trying to determine the smoking status of the individuals in their study population. 17 Specific to the work presented here, experts in evidence-based medicine review hundreds or thousands of articles on specific classes of drugs to synthesize the evidence, leading to recommendations that direct the standard of practice, and continually improve the standard of care and its cost-effectiveness. In all of these tasks, text processing and machine learning can help to identify the most promising documents, reducing the human workload, and allowing more time to be spent on other, more analytic parts of tasks.
Work prioritization is a promising application of automated text processing and machine learning. Rather than treating a problem as a binary classification task, in which the system predicts the documents that the reviewers are likely to include or exclude in the final SR, in work prioritization, the ML algorithm uses past inclusion decisions to prioritize documents based on the likelihood that SR reviewers would judge them as necessary to include in an SR. The inclusion assessments can then be used by reviewers to organize and prioritize their manual review work. This is useful for several reasons. By identifying the most likely to-be-included documents first, human reviewers can obtain this set of full-text documents sooner, move these articles through the review process first, and assign reviewing the less-likely documents a lower priority. The processes of article retrieval, data abstraction, quality assessment and finally qualitative and quantitative synthesis take considerable time and effort, such that prioritizing the set of most likely to be included articles to be managed first could be a distinct advantage in the setting of practical time and budget limitations. In reviews with searches that result in a large number of citations to be screened for retrieval, reviewing the documents in order of their likely importance would be particularly useful. The remainder of the citations could be screened over the following months, perhaps by the members of the team with less experience, while the work of reviewing the includable studies is on-going. Lastly, a system incorporating a trained ML algorithm could be set up to monitor newly published literature, and determine whether it is likely to be included in the review topic. This could serve as an aid to answering the question of when a review topic requires an update.
In this work, we extend our prior work in applying ML and automated document classification to literature screening for SRs. We apply our previously described optimized feature representation, and again use support vector machine (SVM) margin-based techniques to perform rank-prioritization of domain-specific documents across a range of SR topics. 12 We extend simple topic-specific training and classification/ranking by incorporating general and topic-specific SR inclusion judgments into the ML model. In the absence of topic-specific data, we propose a method that creates a model by training on data from a combination of other SR topics. As increasing amounts of topic-specific training data become available, our system preferentially incorporates these data into the model, reducing the influence of data from other topics.
In addition to being able to update existing SR topics, our proposed method allows for a very flexible system that can be used for the creation and updating of SR reports. When an SR topic is first created, no data specific to this topic is available for training the ML algorithm. In this situation, our method can only use general, or nontopic-specific data, and incorporate topic-specific training data as expert judgments become available. Furthermore, since our method is capable of combining topic-specific and general nontopic-specific training data, it can be easily applied to topics in domains having a slow publication rate which may not have a large enough base for building an effective training set. Once sufficient topic-specific training data becomes available, the influence of the nontopic-specific data decreases, thereby not impeding performance, compared to a model trained solely on sufficient topic-specific data. The shift in influence from general to topic-specific training data occurs automatically, without human intervention, and does not require any hard threshold or manual decision to change over from a strictly general to strictly topic-specific model.
Methods
We present our methods in three sections. In the first, we describe the data set used to evaluate our system. In the second, we describe the machine learning rationale and approach used for work prioritization. Finally, in the third, we describe our evaluation process.
Data Corpus
In all the experiments described here, the corpus used is based on SR inclusion/exclusion judgments collected by the expert reviewers of the Oregon Evidence-based Practice Center for the Drug Effectiveness Review Project (DERP), during the process of performing initial SRs, and SR updates, on 24 separate topics. The DERP uses a consistent process for collecting the initial literature set and performing and recording the literature review process. This process results in a set of included articles for a given SR, each of which covers a different drug class or drug therapy for a specific disease. For a given article, reviewers base initial inclusion judgments on abstract-level information alone. Citations that are judged “excluded” after reviewing the abstract do not have the associated full-texts retrieved; these are considered excluded at the full text level as well. Those articles that are retrieved after abstract review are read in full, and subsequently given a final inclusion judgment. This process is described in greater detail in our earlier study. 13
Our previous work collected and normalized expert reviewer data by hand, with the aid of regular expression-based processing scripts. We have since expanded our research program to include an automated system (the Systematic Review Information Automated Collection or “SYRIAC” system) for collecting, normalizing, and storing reviewer judgments. 18 The 24 review topics studied in this work, along with the number of articles included and excluded in each study, are shown in ▶. Over 50,000 inclusion/exclusion judgments are included in the full dataset used in the research reported here. Note that the range of both total number of articles and included article percentage varies greatly between studies. The topic HyperlipidemiaCombinationDrugs has the fewest total samples (299), 6.4% of which are included in the SR. The topic ACEinhibitors has the greatest number of total samples (5,558), 2.5% of which are included in the final review. The percentages of included articles also have a wide range. The topic NSAIDs includes 73% of the total 371 articles, while the Opioids topic includes just 0.2%, or seven articles, of the total 4,602 reviewed. As can be seen in ▶, it is typical for SR topics to vary greatly in the both the amount of literature available, as well as in the proportion and absolute number of articles meeting the inclusion criteria. There is no typical number or proportion for these values.
Table 1.
Table 1 Absolute and Relative Percentages of Included and Excluded Articles Across 24 Separate Review Topics, and Across the Collection as a Whole
| Topic | Included | %Included | Excluded | %Excluded | Total |
|---|---|---|---|---|---|
| AceInhibitors | 137 | 2.5% | 5421 | 97.5% | 5558 |
| ADHD | 264 | 10.4% | 2265 | 89.6% | 2529 |
| Antiemetics | 149 | 6.7% | 2068 | 93.3% | 2217 |
| Antihistamines | 99 | 12.1% | 720 | 87.9% | 819 |
| AtypicalAntipsychotics | 507 | 17.2% | 2439 | 82.8% | 2946 |
| BetaAgonistInhaled | 99 | 2.0% | 4858 | 98.0% | 4957 |
| BetaBlockers | 189 | 4.3% | 4229 | 95.7% | 4418 |
| CalciumChannelBlockers | 220 | 7.4% | 2758 | 92.6% | 2978 |
| Diabetes | 37 | 4.2% | 843 | 95.8% | 880 |
| DiabetesCombinationDrugs | 26 | 5.4% | 454 | 94.6% | 480 |
| HepatiticC | 92 | 13.1% | 610 | 86.9% | 702 |
| HormoneReplacementTherapy | 181 | 34.6% | 342 | 65.4% | 523 |
| HyperlipidemiaCombinationDrugs | 19 | 6.4% | 280 | 93.6% | 299 |
| MSDrugs | 131 | 7.3% | 1672 | 92.7% | 1803 |
| NeuropathicPain | 94 | 19.0% | 401 | 81.0% | 495 |
| NSAIDs | 269 | 72.5% | 102 | 27.5% | 371 |
| Opioids | 7 | 0.2% | 4595 | 99.8% | 4602 |
| OralHypoglycemics | 6 | 0.6% | 943 | 99.4% | 949 |
| OveractiveBladder | 96 | 11.0% | 776 | 89.0% | 872 |
| ProtonPumpInhibitors | 173 | 18.4% | 766 | 81.6% | 939 |
| Sedatives | 133 | 8.0% | 1522 | 92.0% | 1655 |
| Statins | 171 | 2.6% | 6516 | 97.4% | 6687 |
| Thiazolidinediones | 227 | 9.4% | 2179 | 90.6% | 2406 |
| Triptans | 141 | 16.7% | 701 | 83.3% | 842 |
| TOTALS | 3467 | 6.8% | 47460 | 93.2% | 50927 |
ACE = acetylcholinesterase; ADHD = attention deficit hyperactivity disorder; MS = multiple sclerosis; NSAID = nonsteroidal antiinflammatory drug.
The data used in the present experiments represents a snap shot of the SYRIAC database as of March 6, 2008. Automated data collection has continued since that time, and, as of this writing, the system contains over 65,000 inclusion/exclusion judgments. By collecting data on an ongoing basis, the system supports both continued refinement and improvement of our ML approach, as well as future prospective evaluations.
Machine Learning Approach and System
The ML system presented here was motivated by an interesting result observed in our earlier studies on work prioritization for SRs. 12 There, we noticed that using SR topic-specific, rather than a mix of general (termed the nontopic-specific or general training data) and topic-specific training data, led to improved classification performance, as measured by the area under the receiver operating curve (AUC) in 14 out of 15 topics. It turned out that the one topic for which we did not observe this improved performance (SkeletalMuscleRelaxants) had a dataset with unique characteristics, namely that it only included 9 positive (Included, as opposed to Excluded) samples out of 1643 total samples, or 0.5%. This percentage is comparable to the smallest number of positive samples for the topics shown in ▶. With so few positive samples, there is inadequate information on which to base future predictions. Furthermore, many ML techniques, including the ones investigated here, attempt to minimize error, and with very few positive samples, this approach may result in a model very biased towards negative prediction.
With such sparse topic-specific positive training data, an AUC of only 0.73 was achieved, while the mean performance across all topics was 0.86. Furthermore, the system using the nontopic-specific training data on SkeletalMuscleRelaxants achieved an AUC of about 0.84, an improvement of 0.11. In addition, while the system trained on topic-specific data consistently outperformed the nontopic system on the other 14 topics, overall, the performance of the nontopic system was encouraging, averaging about 0.73. Keeping in mind that no topic-specific data were used to train the classifiers to achieve this performance, it is clear that there is value in using the relatively voluminous nontopic training data when insufficient topic-specific training data are available.
Initial Cross-Topic Analysis
To better understand the role that training on nontopic data could have across the 24 topics in our current dataset, we systematically analyzed the utility of using one topic to create a predictive model for another. We did this by selecting a test topic, and then training our algorithm with the data from the 23 remaining topics separately, and evaluating the AUC performance of each trained model on the test topic. Throughout the rest of this work, we refer to the process of using nontopic-specific training data to create a machine learning model for a different specific topic as cross-topic learning.
In the present work, our basic ML system is the same as the best-performing system in our prior work. 12 Briefly, the system is support vector machine-based (SVM), 19 and uses one- and two-token n-grams from article titles and abstracts, along with the associated MeSH terms, as a feature set. We use the SVMLight implementation of the SVM algorithm, with a linear kernel and default settings. 20 No feature selection is performed, and articles are ranked using the signed margin distance—the distance in front of, or behind, the SVM separating hyperplane. Articles having the greatest positive margin distance are placed at the top of the ranking, while articles having the most negative margin distance are placed at the end. As in our prior research, AUC is computed using a varying cutoff parameter which is based on the margin distance. 21
▶ presents the results of these initial cross-topic learning experiments. The topic used for training is shown in columns across the top of ▶; the topic used as the test collection is shown across the rows. For comparison, the 5 × 2-way cross validation performance on individual topics (no cross-topic learning) is shown along the diagonal. This is done because otherwise the test and training collections would be the same for the entries, leading to overfitting and overestimation of the performance. A 5 × 2-way cross validation consists of five repetitions of a stratified, randomly split twofold cross-validation. Performance results of these repetitions are then averaged together.
Table 2.
Table 2 AUCs for Cross-Topic Training and Testing Across All 24 Systematic Review Topics. The Mean AUC on the Far Right Represents How Well Models Built on Other Topics Were Able to Classify a Given Topic. The Mean AUC on the Bottom Row Shows How Well Models Built on that Topic Were Able to Classify Other Topics
| Test Collection | Training Collection |
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AceInhibitors | ADHD | Antiemetics | Antihistamines | AtypicalAntipsychotics | BetaAgonistsInhaled | BetaBlockers | CalciumChannelBlockers | Diabetes | DiabetesCombinationDrugs | HepatiticC | HormoneReplacementTherapy | HyperlipidemiaCombinationDrugs | MSDrugs | NeuropathicPain | NSAIDs | Opioids | OralHypoglycemics | OveractiveBladder | ProtonPumpInhibitors | Sedatives | Statins | Thiazolidinediones | Triptans | MEAN | |
| AceInhibitors | 0.91 | 0.80 | 0.75 | 0.81 | 0.79 | 0.71 | 0.84 | 0.84 | 0.77 | 0.65 | 0.76 | 0.73 | 0.73 | 0.85 | 0.70 | 0.57 | 0.49 | 0.69 | 0.73 | 0.76 | 0.74 | 0.81 | 0.82 | 0.72 | 0.74 |
| ADHD | 0.65 | 0.90 | 0.59 | 0.72 | 0.64 | 0.66 | 0.61 | 0.66 | 0.66 | 0.63 | 0.61 | 0.67 | 0.65 | 0.63 | 0.68 | 0.48 | 0.60 | 0.65 | 0.67 | 0.63 | 0.64 | 0.54 | 0.59 | 0.59 | 0.63 |
| Antiemetics | 0.79 | 0.78 | 0.90 | 0.79 | 0.80 | 0.66 | 0.76 | 0.70 | 0.82 | 0.75 | 0.80 | 0.78 | 0.82 | 0.80 | 0.81 | 0.56 | 0.42 | 0.78 | 0.82 | 0.81 | 0.79 | 0.80 | 0.76 | 0.82 | 0.76 |
| Antihistamines | 0.72 | 0.76 | 0.73 | 0.86 | 0.80 | 0.66 | 0.73 | 0.79 | 0.76 | 0.67 | 0.72 | 0.69 | 0.72 | 0.75 | 0.71 | 0.65 | 0.57 | 0.71 | 0.72 | 0.79 | 0.75 | 0.70 | 0.72 | 0.71 | 0.72 |
| AtypicalAntipsychotics | 0.69 | 0.69 | 0.66 | 0.76 | 0.85 | 0.65 | 0.65 | 0.72 | 0.75 | 0.73 | 0.76 | 0.68 | 0.74 | 0.75 | 0.71 | 0.61 | 0.53 | 0.69 | 0.74 | 0.73 | 0.70 | 0.66 | 0.72 | 0.71 | 0.70 |
| BetaAgonistsInhaled | 0.63 | 0.67 | 0.64 | 0.63 | 0.67 | 0.91 | 0.59 | 0.61 | 0.62 | 0.57 | 0.65 | 0.59 | 0.67 | 0.64 | 0.70 | 0.57 | 0.46 | 0.63 | 0.65 | 0.69 | 0.64 | 0.68 | 0.58 | 0.71 | 0.63 |
| BetaBlockers | 0.77 | 0.69 | 0.62 | 0.70 | 0.63 | 0.65 | 0.89 | 0.71 | 0.64 | 0.52 | 0.60 | 0.71 | 0.54 | 0.68 | 0.65 | 0.58 | 0.44 | 0.54 | 0.65 | 0.65 | 0.71 | 0.66 | 0.67 | 0.70 | 0.64 |
| CalciumChannelBlockers | 0.74 | 0.65 | 0.62 | 0.72 | 0.70 | 0.64 | 0.72 | 0.88 | 0.67 | 0.57 | 0.66 | 0.60 | 0.61 | 0.72 | 0.59 | 0.60 | 0.56 | 0.64 | 0.62 | 0.66 | 0.61 | 0.66 | 0.68 | 0.64 | 0.65 |
| Diabetes | 0.82 | 0.94 | 0.89 | 0.94 | 0.93 | 0.63 | 0.74 | 0.79 | 0.98 | 0.94 | 0.95 | 0.85 | 0.90 | 0.97 | 0.92 | 0.64 | 0.71 | 0.92 | 0.95 | 0.90 | 0.93 | 0.85 | 0.95 | 0.90 | 0.87 |
| DiabetesCombinationDrugs | 0.55 | 0.73 | 0.65 | 0.74 | 0.79 | 0.56 | 0.53 | 0.61 | 0.76 | 0.86 | 0.77 | 0.71 | 0.88 | 0.68 | 0.73 | 0.57 | 0.55 | 0.66 | 0.69 | 0.64 | 0.65 | 0.58 | 0.71 | 0.75 | 0.67 |
| HepatiticC | 0.69 | 0.79 | 0.79 | 0.80 | 0.84 | 0.60 | 0.48 | 0.69 | 0.91 | 0.82 | 0.92 | 0.68 | 0.84 | 0.89 | 0.88 | 0.52 | 0.66 | 0.81 | 0.86 | 0.79 | 0.85 | 0.71 | 0.81 | 0.82 | 0.76 |
| HormoneReplacementTherapy | 0.60 | 0.67 | 0.54 | 0.60 | 0.64 | 0.51 | 0.70 | 0.55 | 0.62 | 0.58 | 0.60 | 0.89 | 0.60 | 0.65 | 0.67 | 0.58 | 0.48 | 0.55 | 0.67 | 0.59 | 0.65 | 0.62 | 0.55 | 0.68 | 0.60 |
| HyperlipidemiaCombinationDrugs | 0.68 | 0.80 | 0.81 | 0.78 | 0.83 | 0.72 | 0.54 | 0.64 | 0.74 | 0.89 | 0.81 | 0.65 | 0.92 | 0.77 | 0.68 | 0.54 | 0.55 | 0.74 | 0.75 | 0.83 | 0.69 | 0.74 | 0.72 | 0.79 | 0.73 |
| MSDrugs | 0.82 | 0.86 | 0.73 | 0.85 | 0.85 | 0.74 | 0.74 | 0.81 | 0.87 | 0.79 | 0.88 | 0.73 | 0.83 | 0.91 | 0.80 | 0.52 | 0.59 | 0.82 | 0.82 | 0.83 | 0.82 | 0.80 | 0.85 | 0.82 | 0.79 |
| NeuropathicPain | 0.76 | 0.88 | 0.73 | 0.80 | 0.82 | 0.73 | 0.83 | 0.76 | 0.81 | 0.80 | 0.86 | 0.85 | 0.72 | 0.82 | 0.92 | 0.62 | 0.57 | 0.80 | 0.89 | 0.87 | 0.88 | 0.82 | 0.73 | 0.88 | 0.79 |
| NSAIDs | 0.55 | 0.61 | 0.49 | 0.69 | 0.73 | 0.59 | 0.63 | 0.64 | 0.63 | 0.66 | 0.67 | 0.58 | 0.65 | 0.65 | 0.63 | 0.82 | 0.38 | 0.65 | 0.59 | 0.52 | 0.67 | 0.67 | 0.63 | 0.69 | 0.62 |
| Opioids | 0.53 | 0.60 | 0.58 | 0.61 | 0.51 | 0.58 | 0.50 | 0.72 | 0.63 | 0.62 | 0.67 | 0.49 | 0.60 | 0.65 | 0.54 | 0.48 | 0.78 | 0.68 | 0.67 | 0.56 | 0.53 | 0.61 | 0.48 | 0.58 | 0.58 |
| OralHypoglycemics | 0.67 | 0.89 | 0.87 | 0.88 | 0.75 | 0.79 | 0.62 | 0.77 | 0.88 | 0.72 | 0.86 | 0.59 | 0.90 | 0.89 | 0.88 | 0.68 | 0.68 | 0.94 | 0.94 | 0.74 | 0.83 | 0.85 | 0.84 | 0.80 | 0.80 |
| OveractiveBladder | 0.77 | 0.83 | 0.80 | 0.79 | 0.82 | 0.65 | 0.77 | 0.76 | 0.83 | 0.71 | 0.84 | 0.79 | 0.76 | 0.83 | 0.86 | 0.67 | 0.60 | 0.84 | 0.88 | 0.82 | 0.82 | 0.73 | 0.75 | 0.82 | 0.78 |
| ProtonPumpInhibitors | 0.71 | 0.78 | 0.75 | 0.73 | 0.79 | 0.63 | 0.72 | 0.77 | 0.73 | 0.64 | 0.75 | 0.73 | 0.75 | 0.79 | 0.79 | 0.63 | 0.55 | 0.69 | 0.81 | 0.89 | 0.77 | 0.75 | 0.65 | 0.82 | 0.73 |
| Sedatives | 0.69 | 0.77 | 0.68 | 0.66 | 0.76 | 0.60 | 0.78 | 0.67 | 0.72 | 0.62 | 0.77 | 0.66 | 0.62 | 0.74 | 0.77 | 0.63 | 0.51 | 0.66 | 0.72 | 0.71 | 0.91 | 0.70 | 0.69 | 0.78 | 0.69 |
| Statins | 0.78 | 0.82 | 0.83 | 0.80 | 0.84 | 0.72 | 0.70 | 0.74 | 0.84 | 0.72 | 0.85 | 0.76 | 0.80 | 0.86 | 0.83 | 0.64 | 0.62 | 0.80 | 0.82 | 0.83 | 0.83 | 0.91 | 0.78 | 0.84 | 0.79 |
| Thiazolidinediones | 0.70 | 0.81 | 0.75 | 0.77 | 0.84 | 0.62 | 0.53 | 0.70 | 0.85 | 0.72 | 0.85 | 0.68 | 0.79 | 0.83 | 0.82 | 0.52 | 0.54 | 0.77 | 0.82 | 0.81 | 0.81 | 0.71 | 0.88 | 0.78 | 0.74 |
| Triptans | 0.79 | 0.83 | 0.85 | 0.75 | 0.81 | 0.64 | 0.86 | 0.70 | 0.83 | 0.79 | 0.84 | 0.85 | 0.85 | 0.85 | 0.86 | 0.56 | 0.54 | 0.77 | 0.86 | 0.88 | 0.86 | 0.85 | 0.75 | 0.91 | 0.79 |
| MEAN | 0.70 | 0.77 | 0.71 | 0.75 | 0.76 | 0.65 | 0.68 | 0.71 | 0.75 | 0.70 | 0.76 | 0.70 | 0.74 | 0.77 | 0.75 | 0.58 | 0.55 | 0.72 | 0.76 | 0.74 | 0.75 | 0.72 | 0.71 | 0.75 | |
ACE = acetylcholinesterase; ADHD = attention deficit hyperactivity disorder; AUC = area under the curve; MS = multiple sclerosis; NSAID = nonsteroidal antiinflammatory drug.
The mean score for using a given topic for training and classifying on all the other topics is shown in the bottom row, and the mean score on a given topic, over using each of the other topics for cross-topic learning is shown in the right-most column.
It is clear from the right-most column of ▶ that some topics are “easier” than others, that is, with a large fraction of the available training topics, cross-topic learning results in decent performance. For example, Diabetes is an “easy” topic—it averages 0.87 in cross-topic learning, and has a range of 0.64–0.96 (NSAIDs and MSDrugs, respectively); the performance at the top of this range is similar to Diabetes' cross-validation performance (0.98). Conversely, Opioids appears to be a difficult topic. Here, cross-topic learning averages about 0.58, with a low of 0.48 using, again, NSAIDs for training, and a high of 0.68 using OralHypoglycemics.
Looking at ▶ across the other axis, the best general topics for cross-topic learning are ADHD and MSDrugs, each of which has a mean AUC of about 0.77 when used as training for other topics. This is interesting, because ADHD is not one of the easier test topics when using the other topics for training–it is not simply that ADHD has a lot of commonality in either vocabulary or domain with the other topics. The features learned from ADHD may apply well to other topics, but the converse is not true. Note that the cross-validation performance of ADHD is about 0.90, so ADHD is readily classifiable when topic-specific training data are available.
Certain topics are consistently bad for cross-topic learning. Both NSAIDs and Opioids do not perform well as training topics for cross-topic learning, as they have low mean AUCs when used as training topics. They are also not easy topics to predict when using the other topics for cross-topic learning. Predicting NSAIDs and Opioids with other topics, are, on average, two of the lowest scoring test topics (0.62 and 0.58, respectively), along with HormoneReplacementTherapy. Therefore, not all topics are equivalent in terms of their potential for cross-topic learning, nor in the ease with which they can be predicted based on data from other topics. There are topics that perform well when used for cross-topic learning for some topics, but not for others. For example, MSDrugs is the best cross-topic for learning when testing on ACEinhibitors, but it performs poorly on HyperlipidemiaCombinationDrugs (AUC = 0.77), whereas DiabetesCombinationDrugs performs much better (AUC = 0.89).
Therefore, while it is clear that cross-topic learning has the potential to aid automated machine learning when inclusion/exclusion data specific to a topic is sparse or lacking, it is not obvious how these data are best used. There is no single topic, nor small set of topics, that is the best overall predictor. Furthermore, in the absence of inclusion/exclusion data for test topics, it is unclear how to select a training topic that is “similar enough” to the test topic to achieve good performance. After experimenting with information gain- and KL-divergence-based measures on the topic-specific feature distributions, and abandoning these approaches due to complexity and discouraging results, we decided to apply the cross-topic training data in a manner independent of matching up test topics with good predictive topics.
Cross-Topic Learning Algorithm
The hybrid algorithm we present here is intended to combine nontopic and topic-specific training data in an automatic and flexible manner. It is an enhancement to the baseline topic-specific trained SVM-based classification algorithm described above. We extend this algorithm with nontopic data using a property of SVM—only samples that lie on the boundary of the hyperplane margin have an effect on the location and orientation of the separating hyperplane. Given an SVM model built from a set of training data, the exact same model would be constructed if only the samples lying on the margin boundary (the support vectors) are used to train a subsequent model.
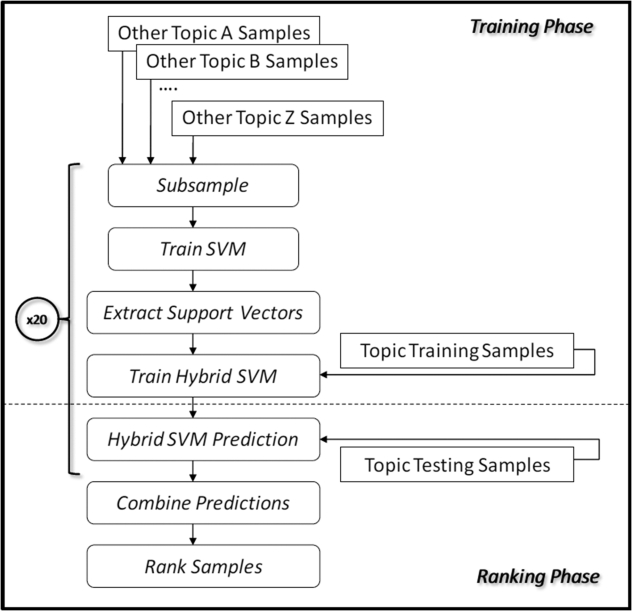
We apply this idea to improving classification with sparse amounts of topic-specific training data in the following manner. First we build a general classification model for a given topic, using the nontopic data (the data samples from the other topics). Next, we extract out the support vectors, and combine them with the available topic-specific training data to build the final predictive model. A diagrammatic representation of the overall process is shown in ▶. The amount of topic-specific training data can range from none, if this topic has never been seen before (i.e., it is a new SR topic), to quite a bit, if this is a review topic's second or third update.
Figure 1.
Process flow model diagram representation of the hybrid topic-specific + nontopic training sample systematic review ranking algorithm.
Supplementing a small amount of topic-specific training data with the support vectors from the general model primes the SVM hyperplane to start off in a reasonable configuration for classifying SR articles. Limiting the nontopic data to the support vectors alone decreases the amount of nontopic data used to train the model, which limits the bias introduced by nontopic influences. The basic idea of priming an SVM using support vectors from related data are taken from the adaptive learning literature, where it is used for tasks which change over time, such as identifying particular kinds of news stories. 22 Instead, here we use it to allow general nontopic training data to compensate somewhat for a lack of topic-specific data.
Several other enhancements are required to make SVM priming work in the present setting. Since we do not have a single topic that changes over time, but instead have a number of partially related topics, we need a method to determine which data to use for creating the priming vectors. As can be seen in ▶, some topics have many more data samples than others. Simply using all the data from the nontopics to create the priming vectors for the model could substantially bias the SVM in the direction of the most common topics. This could subsequently lead to decreased performance, and therefore could be an issue with the reliability of the approach. For example, ACEInhibitors, the topic with the largest number of training samples, creates a poorly performing model for classifying BetaAgonistsInhaled, which is better modeled by several other topics, including Statins and Triptans (see ▶).
To avoid this problem, rather than simply combining all the nontopic data together, we resample it on a topic-by-topic basis, with the goal of including approximately the same number of samples from each topic. This means that the probability of including any individual sample from, say, ACEInhibitors, is much lower than the probability of including any individual sample from HyperlipidemiaCombinationDrugs, the sparsest topic. This, however, raises another issue–since we are resampling over the nontopic data, we are subsequently excluding a large proportion of the total samples. Any given resampling may poorly represent the specific target topic, and adversely affect performance. To address this, we sample the nontopic data, creating several different priming SVM models, and extract the support vectors from each of these models to use as priming vectors. The nontopic data are rejection sampled, that is, sampled without replacement. The probabilities of inclusion for each sample within a given nontopic are adjusted so that approximately the same number of samples from each nontopic is included. This is done using an inverse topic count weighting:
Where c t is the number of samples available for a given nontopic, w t is the unnormalized relative nontopic sampling rate for a given nontopic, and p t is the sample inclusion probability for a given nontopic, with the sum of the p t across all nontopics equal to one.
For each of these sets of priming vectors we train a new hybrid model using the combined priming and topic-specific samples, and then combine the predictions of the individual hybrid models by summing the signed margin distances from each model hyperplane. The final topic rankings are created by ranking the summed signed margin distances. Note that for each of the primed SVM models, the topic-specific training data remains the same—it is only the nontopic data that is sampled. Initial experiments showed that the number of resamplings should be at least 5, and that 20 resamples was sufficient to minimize variability of the results. This number agrees with our previous work using resampling with SVMs. Therefore, we use 20 resamplings for our experiments here. 23
Because the two topics NSAIDs and Opioids both performed so poorly across-the-board when used to cross-train models for SRs, samples from these topics are excluded from the resampling pool. On average, both these topics perform close to random (AUC = ∼0.50) for cross-topic learning; it appears that they have little general information to contribute to the primed SVM model. We do, however, include both topics in our evaluation of this algorithm, presented in the Results section.
Another important issue needing to be addressed is the relative importance of a nontopic priming vector sample, compared to the topic-specific samples in the SVM optimization procedure. Clearly, if the SVM hyperplane can perfectly separate all training samples (nontopic and topic together), it will do so without needing to make a tradeoff between making training errors on topic and nontopic data samples. In general, however, this is not the case, and the optimal separating hyperplane will have to accept training errors on some samples, misclassifying these samples because they fall on the wrong side of the hyperplane. Because, for SR, models trained on topic-specific data perform better than those trained on nontopic data, it is clearly preferable to have the hyperplane make training errors on the nontopic priming samples, and correctly classify the topic-specific ones. The implementation of SVM that we use in this work, SVMLight, 20,24 allows placing a misclassification cost on each training sample individually. This value is used directly in the per-sample loss function optimized by SVMLight. We use this capability to apply a lower cost to misclassifying nontopic training samples, in favor of correctly classifying those that are topic-specific. Our initial experiments showed that the cost factor–the ratio of nontopic priming to topic sample error cost—should be fairly narrow, within the range of 0.20–0.50.
Although the labels that are used to train our system are the set of include/exclude judgments made by expert reviewers after reading the full text of an article, it is important to note that most of the information contained in the full text of the article is not available to our system. Instead, our system uses only the titles, abstracts, and National Library of Medicine assigned MeSH (Medical Subject Headings) terms for a collection of articles to generate a ranking of the likelihood each will be included in an SR after full text review by an expert.
Evaluation Methods
To evaluate our method for automatically combining topic- and nontopic-specific training data, we used a variation of repeated cross-validation, in which all the topic-specific data are split into partitions, some of which are used for training, and the rest for evaluation. The AUC is calculated for each constructed model, and the process is repeated a number of times. All the computed AUCs are averaged together to give a final performance estimate.
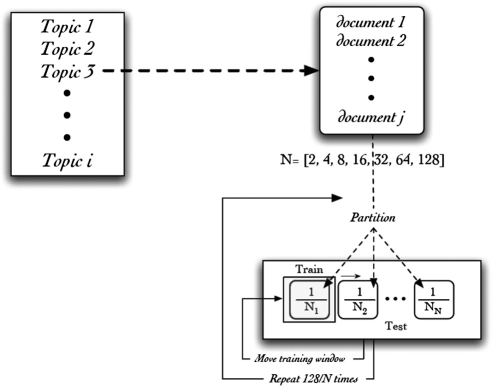
To simulate the effect of having variably reduced amount of training data, we applied N-way cross-validation (with varying N), and, instead of training on most of the data partitions and testing on the remaining partition (as is typically done), we did the opposite—training on a single N-way partition and testing on the combined samples of the remaining partition. By varying N, we controlled the amount of topic-specific training data available to the SVM model, which was equal to 1/N times the total number of topic-specific samples, with (N-1)/N of the samples used to evaluate the model. To have an adequate number of individual AUC measurements to average together, we repeated the process 128/N times. This resulted in a final mean AUC for each topic at each level of N, composed of the average of performance of 128 evaluated models. We used 2, 4, 8, 16, 32, 64, and 128 as the values for N, allowing us to measure the performance of our system using a range of training data—from 1/128th of the available data, up to one half of the available data (corresponding to a standard two-way cross-validation). We call this the reversed cross-valuation method.
Two-way cross-validation tends to produce more accurate predictions of true performance than does leave-one-out cross-validation, or a single 10-way cross-validation, since both of these methods test a given model on a relatively small amount of the data. In two-way cross-validation, each model is tested on a randomly selected half of the data, which leads to more robust estimates, as compared with cross-validation using a higher N. 25 We tried to emulate the positive characteristics of the 2-way cross-validation using our reversed cross-valuation method. For example, at n = 64, the topic-specific data are randomly divided up into 64 partitions, stratified by inclusion/exclusion class to keep the class proportions as constant as possible. One partition (1/64th of the data) was used for training a model which was tested on the remaining 63. This process was repeated for each of the other 63 partitions, and then the entire process was repeated one more time, finally generating 128 AUC measurements that were then averaged together to compute a final mean AUC for that topic at that N-level of topic-specific training data. See ▶ for an illustration of the overall reversed cross-validation procedure.
Figure 2.
Diagrammatic representation of the reversed cross-validation procedure used to sample and estimate classification performance using different fractions of topic-specific training data.
The amount of nontopic-specific priming data are held constant in this process, a characteristic that is accomplished as described in the previous section. In particular, the 20× nontopic data resampling mentioned in the algorithm description above is performed inside each iteration of the cross-validation. This nesting of the resampling inside the iteration made generation of the results presented here somewhat computationally intensive to evaluate. However, this is mostly an artifact of the evaluation process, and not the algorithm itself.
We generated two more sets of results, for the purpose of having a standard of comparison for our algorithm. The first set was simply the average performance of the 20× resampled nontopic model without using any of the topic-specific data for subsequent training. This is equivalent to just summing the signed margin distances from SVMs trained on the nontopic data, without extraction of the priming support vectors or training again with the addition of topic-specific data. Since no topic-specific data are needed to build these models, performance is constant with respect to the cross-validation N used above. In the results presented below, we refer to this as the nontopic performance.
We generated a second set of comparison results by reducing the training set size without using the priming support vectors generated from the nontopic samples. In this case, the resulting AUC shows the effect of a reduced amount of available topic-specific training data. For these tests, we used the same range of N as above, generating mean AUCs for having ½, ¼, etc of the available training data, all the way down to having only 1/128th of the data available for training. In our experimental results we refer to these scores as the baseline performance.
Results
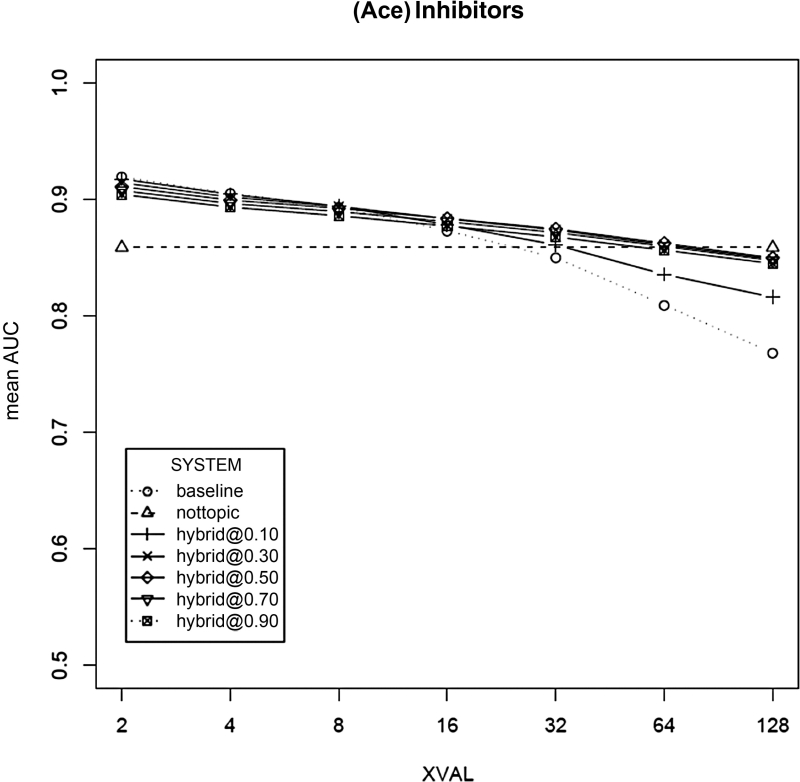
For completeness, we performed the experiments described above for a range of cost factors (0.10–1.0) for each SR topic. The full performance curves across this range of cost factors are shown for ACEInhibitors in ▶, along with the nontopic and baseline scores. The performance curve spread across the cost factors for the other topics is similar. It is clear from these figures that extreme cost factor values (e.g., ∼0.10, and ∼1.0) are not associated with as good a level of performance as are those values in the middle. At cost factors of 0.10, the priming vectors do not appear to have enough influence on the SVM model, leading to performance that is much lower than those at settings where topic-specific training data are sparse (128 and 64 way cross-validation). In the 0.10 cases, performance is still better than baseline, but not as good as can be achieved by allocating more influence to the priming vectors.
Figure 3.
Performance measured by mean area under the receiver operating curve (mean AUC) of the nontopic primed SVM algorithm across seven levels of topic-specific training data for the ACEInhibitors systematic review topic, using a range of cost factors varying between 0.10 and 1.0. The baseline (no nontopic priming) and nontopic only performance is also shown. The x axis numbers represent (N) the partition factor, where higher N means that a smaller amount (1/N of the available topic samples) of topic-specific data were used for training. Here, SYSTEM refers to either the baseline or nontopic system, or the use of the hybrid system where the number listed in the system name is the relative training error cost of a nontopic sample compared to a topic-sample.
At the other end of the curves, when the cost factor is set between 0.50–1.0, the priming vectors have too much influence in the presence of sufficient topic-specific training data (2- and 4-way cross-validation). In these cases, a cost factor of 1.0 leads to the worst performance, while a cost factor of 0.1 results in the highest performance for the system. This makes sense, since, when adequate topic-specific data are available, the nontopic data are more likely to detract from performance, since the nontopic samples contribute less information that is specific to the topic in question.
There is little difference in the performance at cost factors in the middle of the 0.10–0.50 range, and a value somewhere in this range appears to be appropriate. We compared the performance between cost factors using paired Wilcoxon signed rank tests, and found no statistically significant difference among the cost factors between 0.20 and 0.50 (p > 0.05 for all comparisons). Therefore, for our experiments we selected 0.35 as the cost factor in the middle of this range. We use this cost factor for our subsequent results and analysis. We made no attempt to select or identify the optimal performing cost factor for our datasets, as our tests showed that optimization of the cost factor was not necessary, with values in the range of 0.20–0.50 performing equally well.
Figure 4, available as an online data supplement at www.jamia.org, shows the performance curves for all 24 SR topics using a cost factor of 0.35, which is in the middle of the equivalently performing range. As the graphs make clear, our approach gives a good performance boost over the baseline system to almost all topics when topic-specific training data are sparse, and leads to little or no performance penalty when adequate topic-specific training data are available. ▶ shows the AUC in numeric form for all three systems across all levels of reverse cross-validation for each topic.
Table 3.
Table 3 Mean AUC Performance Comparison of the Three Systems Studied on Each Individual Topic, at Each Level of Reverse Cross-Validation
| XVAL |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 |
4 |
8 |
16 |
32 |
64 |
128 |
|||||||||||||||
| Topic | HYBR | BASE | NONT | HYBR | BASE | NONT | HYBR | BASE | NONT | HYBR | BASE | NONT | HYBR | BASE | NONT | HYBR | BASE | NONT | HYBR | BASE | NONT |
| ACE inhibitors | 0.91 | 0.92 | 0.86 | 0.90 | 0.91 | 0.859 | 0.90 | 0.89 | 0.86 | 0.89 | 0.87 | 0.86 | ACEinhibitors | 0.91 | 0.92 | 0.86 | 0.9 | 0.91 | 0.859 | 0.90 | 0.89 |
| ADHD | 0.90 | 0.90 | 0.70 | 0.89 | 0.89 | 0.697 | 0.88 | 0.87 | 0.70 | 0.87 | 0.85 | 0.70 | ADHD | 0.9 | 0.9 | 0.7 | 0.89 | 0.89 | 0.697 | 0.88 | 0.87 |
| Antiemetics | 0.90 | 0.90 | 0.86 | 0.89 | 0.89 | 0.857 | 0.89 | 0.88 | 0.86 | 0.88 | 0.87 | 0.86 | Antiemetics | 0.9 | 0.9 | 0.86 | 0.89 | 0.89 | 0.857 | 0.89 | 0.88 |
| Antihistamines | 0.86 | 0.85 | 0.78 | 0.84 | 0.82 | 0.779 | 0.82 | 0.80 | 0.78 | 0.80 | 0.77 | 0.78 | Antihistamines | 0.86 | 0.85 | 0.78 | 0.84 | 0.82 | 0.779 | 0.82 | 0.80 |
| AtypicalAntipsychotics | 0.85 | 0.85 | 0.76 | 0.84 | 0.83 | 0.762 | 0.82 | 0.82 | 0.76 | 0.81 | 0.80 | 0.76 | AtypicalAntipsychotics | 0.85 | 0.85 | 0.76 | 0.84 | 0.83 | 0.762 | 0.82 | 0.82 |
| BetaAgonistsInhaled | 0.90 | 0.91 | 0.67 | 0.86 | 0.87 | 0.668 | 0.82 | 0.82 | 0.67 | 0.77 | 0.76 | 0.67 | BetaAgonistsInhaled | 0.9 | 0.91 | 0.67 | 0.86 | 0.87 | 0.668 | 0.82 | 0.82 |
| β blockers | 0.90 | 0.90 | 0.76 | 0.88 | 0.87 | 0.758 | 0.86 | 0.85 | 0.76 | 0.84 | 0.81 | 0.76 | β blockers | 0.9 | 0.9 | 0.76 | 0.88 | 0.87 | 0.758 | 0.86 | 0.85 |
| CalciumChannelBlockers | 0.88 | 0.88 | 0.72 | 0.86 | 0.86 | 0.723 | 0.83 | 0.82 | 0.72 | 0.80 | 0.78 | 0.72 | CalciumChannelBlockers | 0.88 | 0.88 | 0.72 | 0.86 | 0.86 | 0.723 | 0.83 | 0.82 |
| Diabetes | 0.99 | 0.98 | 0.97 | 0.99 | 0.98 | 0.967 | 0.98 | 0.98 | 0.97 | 0.98 | 0.97 | 0.97 | Diabetes | 0.99 | 0.98 | 0.97 | 0.99 | 0.98 | 0.967 | 0.98 | 0.98 |
| DiabetesCombinationDrugs | 0.87 | 0.87 | 0.75 | 0.84 | 0.83 | 0.751 | 0.80 | 0.77 | 0.75 | 0.76 | 0.71 | 0.75 | DiabetesCombinationDrugs | 0.87 | 0.87 | 0.75 | 0.84 | 0.83 | 0.751 | 0.80 | 0.77 |
| HepatiticC | 0.93 | 0.93 | 0.90 | 0.92 | 0.92 | 0.895 | 0.91 | 0.91 | 0.90 | 0.90 | 0.89 | 0.90 | HepatiticC | 0.93 | 0.93 | 0.9 | 0.92 | 0.92 | 0.895 | 0.91 | 0.91 |
| Hormone replacement therapy | 0.89 | 0.89 | 0.65 | 0.86 | 0.84 | 0.649 | 0.83 | 0.81 | 0.65 | 0.81 | 0.76 | 0.65 | Hormone replacement therapy | 0.89 | 0.89 | 0.65 | 0.86 | 0.844 | 0.649 | 0.83 | 0.81 |
| HyperlipidemiaCombinationDrugs | 0.91 | 0.90 | 0.84 | 0.87 | 0.84 | 0.842 | 0.84 | 0.78 | 0.84 | 0.83 | 0.71 | 0.84 | HyperlipidemiaCombinationDrugs | 0.91 | 0.9 | 0.84 | 0.87 | 0.84 | 0.842 | 0.84 | 0.78 |
| MSDrugs | 0.91 | 0.90 | 0.89 | 0.89 | 0.89 | 0.89 | 0.88 | 0.88 | 0.89 | 0.86 | 0.86 | 0.89 | MSDrugs | 0.91 | 0.9 | 0.89 | 0.89 | 0.89 | 0.89 | 0.88 | 0.88 |
| NeuropathicPain | 0.93 | 0.93 | 0.87 | 0.91 | 0.91 | 0.871 | 0.89 | 0.90 | 0.87 | 0.87 | 0.88 | 0.87 | NeuropathicPain | 0.93 | 0.93 | 0.87 | 0.91 | 0.911 | 0.871 | 0.89 | 0.90 |
| NSAIDs | 0.85 | 0.82 | 0.64 | 0.82 | 0.79 | 0.643 | 0.80 | 0.76 | 0.64 | 0.78 | 0.73 | 0.64 | NSAIDs | 0.85 | 0.82 | 0.64 | 0.82 | 0.794 | 0.643 | 0.80 | 0.76 |
| Opioids | 0.88 | 0.84 | 0.64 | 0.80 | 0.75 | 0.54 | 0.75 | 0.65 | 0.64 | 0.70 | 0.57 | 0.64 | Opioids | 0.88 | 0.84 | 0.64 | 0.8 | 0.747 | 0.64 | 0.75 | 0.65 |
| OralHypoglycemics | 0.95 | 0.94 | 0.83 | 0.91 | 0.88 | 0.829 | 0.87 | 0.78 | 0.83 | 0.85 | 0.64 | 0.83 | OralHypoglycemics | 0.95 | 0.94 | 0.83 | 0.91 | 0.883 | 0.829 | 0.87 | 0.78 |
| OveractiveBladder | 0.89 | 0.88 | 0.85 | 0.88 | 0.87 | 0.849 | 0.87 | 0.87 | 0.85 | 0.86 | 0.86 | 0.85 | OveractiveBladder | 0.89 | 0.88 | 0.85 | 0.88 | 0.874 | 0.849 | 0.87 | 0.87 |
| Proton pump inhibitors | 0.90 | 0.89 | 0.82 | 0.88 | 0.87 | 0.821 | 0.87 | 0.86 | 0.82 | 0.85 | 0.84 | 0.82 | Proton pump inhibitors | 0.9 | 0.89 | 0.82 | 0.88 | 0.873 | 0.821 | 0.87 | 0.86 |
| Sedatives | 0.91 | 0.91 | 0.76 | 0.90 | 0.88 | 0.763 | 0.88 | 0.85 | 0.76 | 0.87 | 0.83 | 0.76 | Sedatives | 0.91 | 0.91 | 0.76 | 0.9 | 0.882 | 0.763 | 0.88 | 0.85 |
| Statins | 0.91 | 0.91 | 0.86 | 0.90 | 0.90 | 0.862 | 0.88 | 0.88 | 0.86 | 0.87 | 0.86 | 0.86 | Statins | 0.91 | 0.91 | 0.86 | 0.9 | 0.899 | 0.862 | 0.88 | 0.88 |
| Thiazolidinediones | 0.89 | 0.89 | 0.81 | 0.89 | 0.88 | 0.805 | 0.87 | 0.87 | 0.81 | 0.86 | 0.86 | 0.81 | Thiazolidinediones | 0.89 | 0.89 | 0.81 | 0.89 | 0.884 | 0.805 | 0.87 | 0.87 |
| Triptans | 0.91 | 0.91 | 0.89 | 0.91 | 0.90 | 0.891 | 0.90 | 0.90 | 0.89 | 0.90 | 0.89 | 0.89 | Triptans | 0.91 | 0.91 | 0.89 | 0.91 | 0.902 | 0.891 | 0.90 | 0.90 |
ACE = acetylcholinesterase; ADHD = attention deficit hyperactivity disorder; BASE = Baseline System; HYBR = Hybrid System, MS = multiple sclerosis; NONT = Nontopic System; NSAID = nonsteroidal antiinflammatory drugs.
In many cases, the performance boost with very sparse training data (i.e., at 128-way cross-validation) is quite large (▶). On average, the hybrid system we propose here gives almost a 20% boost in performance, with a median of 16.4% and a maximum of 56%. The only negatively performing topic, thiazolidinediones, suffers a small 1.7% decline in AUC. Furthermore, when there is a lot of topic-specific training data, the cost factor of 0.35 does not typically result in a significant performance reduction. For some topics, the performance of our hybrid system continues to be better than that of a system trained only on the baseline topic-specific system. This is the case for antihistamines, NSAIDs, and Opioids. Statins seems to be the only topic where performance on the 2-way cross-validation test decreased a little. For all other topics, the 2-way cross-validation AUC using the hybrid system is about the same as the baseline system trained with topic-specific data.
Table 4.
Table 4 Performance for Proposed Hybrid System Compared to the Baseline System of Training Only on the Available Topic-Specific Training Data. These Results are from the 128-Way Cross-Validation, Where Only 1/128th of the Topic-specific Data are Available for Training
| Topic | AUC |
|||
|---|---|---|---|---|
| Hybrid | Baseline | Delta | %Gain | |
| Ace Inhibitors | 0.86 | 0.77 | 0.09 | 12.30% |
| ADHD | 0.82 | 0.76 | 0.05 | 6.60% |
| Antiemetics | 0.86 | 0.80 | 0.06 | 7.60% |
| Antihistamines | 0.77 | 0.62 | 0.15 | 24.90% |
| AtypicalAntipsychotics | 0.77 | 0.73 | 0.04 | 5.10% |
| BetaAgonistsInhaled | 0.68 | 0.61 | 0.07 | 11.50% |
| BetaBlockers | 0.79 | 0.67 | 0.12 | 18.40% |
| CalciumChannelBlockers | 0.72 | 0.64 | 0.08 | 13.10% |
| Diabetes | 0.96 | 0.62 | 0.33 | 53.20% |
| DiabetesCombinationDrugs | 0.70 | 0.53 | 0.18 | 33.40% |
| HepatiticC | 0.86 | 0.70 | 0.13 | 17.50% |
| HormoneReplacementTherapy | 0.71 | 0.61 | 0.10 | 15.50% |
| HyperlipidemiaCombinationDrugs | 0.78 | 0.52 | 0.26 | 49.80% |
| MSDrugs | 0.85 | 0.77 | 0.09 | 10.10% |
| NeuropathicPain | 0.85 | 0.71 | 0.14 | 19.70% |
| NSAIDs | 0.69 | 0.59 | 0.10 | 17.30% |
| Opioids | 0.63 | 0.51 | 0.12 | 23.10% |
| OralHypoglycemics | 0.81 | 0.52 | 0.29 | 56.20% |
| OveractiveBladder | 0.84 | 0.70 | 0.14 | 19.60% |
| ProtonPumpInhibitors | 0.81 | 0.73 | 0.09 | 12.60% |
| Sedatives | 0.82 | 0.69 | 0.13 | 19.10% |
| Statins | 0.85 | 0.75 | 0.10 | 13.30% |
| Thiazolidinediones | 0.79 | 0.80 | −0.01 | −1.70% |
| Triptans | 0.89 | 0.81 | 0.08 | 9.90% |
| MEAN | 0.80 | 0.67 | 0.12 | 19.50% |
ACE = acetylcholinesterase; ADHD = attention deficit hyperactivity disorder; AUC = area under the curve; NSAID = nonsteroidal antiinflammatory drugs.
▶ shows the results of statistically comparing of our system with the baseline and nontopic systems. The comparisons were performed using nonparametric paired Wilcoxon signed rank tests on the individual topic mean AUC values. The overall average mean AUC across all topics is also shown for each level of cross-validation. As the table makes clear, the performance of the hybrid system is better than the baseline system at all levels of reverse cross-validation, at a statistical significance level of alpha = 0.05. Furthermore, the hybrid system is significantly better than the nontopic system at reverse cross-validation levels of 2 through 32. The hybrid system is no worse, and statistically not different from the nontopic system at reverse cross-validation levels of 64 and 128.
Table 5.
Table 5 Mean AUC Performance Comparison of the Three Systems Studied Averaged Across All Topics. Statistical Significance Shown for Pairs of Systems at the Different Amounts of Training Data Represented by Differing Levels of Reverse Cross-Validation. p Values Computed Using the Nonparametric Paired Wilcoxon Test, Comparing Pairs of System Performance on Each of the 24 SR Topics
| Average Mean AUC Across All Topics |
Topic Pairwise Wilcoxon Test |
Average Mean AUC Across All Topics |
Topic Pairwise Wilcoxon Test |
|||
|---|---|---|---|---|---|---|
| XVAL | Hybrid | Baseline | p-value | Hybrid | Non-Topic | p-value |
| 2 | 0.900 | 0.896 | 0.020 | 0.900 | 0.795 | 0.000 |
| 4 | 0.879 | 0.870 | 0.002 | 0.879 | 0.795 | 0.000 |
| 8 | 0.860 | 0.841 | 0.000 | 0.860 | 0.795 | 0.000 |
| 16 | 0.841 | 0.807 | 0.000 | 0.841 | 0.795 | 0.000 |
| 32 | 0.826 | 0.773 | 0.000 | 0.826 | 0.795 | 0.005 |
| 64 | 0.811 | 0.727 | 0.000 | 0.811 | 0.795 | 0.160 |
| 128 | 0.796 | 0.675 | 0.000 | 0.796 | 0.795 | 0.574 |
AUC = area under the curve; SR = systematic review.
A p-value of 0.0000 Indicates p < 0.00005.
Examining the topic specific graphs in Figure 4, it is clear that, on most individual topics, the performance of our hybrid system with very little topic-specific (128 or 64-way cross validation) training data are equal to, or better than, the nontopic system. As more topic-specific training data becomes available, the performance consistently improves, asymptotically approaching the performance of each of the topics when much topic-specific training data are available. The performance of the hybrid system surpasses that of the nontopic system somewhere between the 128-way and 16-way level of cross-validation for all the 24 topics studied.
With very sparse topic-specific training data, the performance of the nontopic system on individual topics is often better than the baseline system, and is, at times, better than that of our proposed hybrid system. This can be seen in the plots for the topics DiabetesCombinationDrugs, HyperlipidemiaCombinationDrugs, MSDrugs, HepatiticC, NSAIDs, NeuropathicPain, Statins, and Thiazolidinediones. In all these cases, except MSDrugs, the hybrid system improves upon the performance of the nontopic system across the range of N-way cross-validation, as more topic-specific data becomes available. Interestingly, for Thiazolidinediones, the hybrid system is better than the nontopic system throughout most of the range, but the system trained only on the topic-specific data consistently outperforms the other systems; this is the only topic for which this is the case. Nevertheless, at worst, the performance hit is only about 0.02 units of AUC—small, compared to the performance gains on the other topics across the range of available topic-specific training data.
Discussion, Limitations, and Related Work
Overall, the hybrid system significantly outperforms the baseline system when topic-specific training data are sparse. Using 1/128th of the available topic-specific data for training resulted in improved performance for 23 of the 24 topics. There is a performance decline for only one topic, and it is minimal. With increases in the amount of topic-specific training data, the advantage of the hybrid algorithm gradually decreases to the point where, given an adequate amount of topic-specific training data, the performance of the hybrid and baselines systems is almost identical. However, for at least two of the topics, Opioids and NSAIDs, the hybrid system noticeably outperforms the baseline system, even at 2-way cross-validation. This indicates that for these two topics, even at the level of 2-way cross-validation, the topic-specific data are inadequate for producing the best possible model, and the nontopic data still plays a large role in creating an optimal decision surface.
These observations show that there is no significant downside to using the hybrid system with any amount of available topic-specific training data as compared to the baseline system. The hybrid system will either improve performance, sometimes greatly, or not make much difference. This is important because, in a real-world deployment of a system using our hybrid algorithm, users would not have a-priori knowledge of where their data puts them on the topic-specific training data-versus-performance curve. The different topics “saturate” performance at different levels and absolute amounts of training data. Therefore, without performing extensive cross-validation studies as we have done here, users would not know how much topic-specific training data they have, nor would they know how that amount influences performance. This is a great advantage of our proposed approach: it does not rely on the user knowing in advance whether adequate topic-specific training data are already available. As more topic-specific training data becomes available, the user does not need to decide when to stop using the nontopic data, as the system takes care of this automatically, without human intervention.
Although the nontopic system achieves good performance for some topics, it is not nearly as reliable as the hybrid system. The performance of the nontopic system on Diabetes is very good, almost as high as the hybrid system at the level of 2-way cross-validation. For MSDrugs, the performance of the nontopic system surpasses that of the baseline and hybrid systems, up until 4-way cross-validation. However, for many of the topics, the performance of the nontopic system is poor, and incorporating even a little topic-specific data leads to improved performance in the hybrid algorithm (e.g., see the graphs for ADHD, Antiemetics, AtypicalAntipsychotics, BetaAgonistsInhaled, HormoneReplacementTherapy, NSAIDs, and Thiazolidinediones). It is not clear how one would predict the performance characteristics of the topic-specific and nontopic systems on a topic, short of extensive testing such as we have done here. It is unnecessary, with our approach, to try to make this distinction.
Furthermore, the performance of the nontopic system is always surpassed by both the baseline and hybrid system at some level of training data. To use separate nontopic and topic-specific systems, instead of a hybrid approach such as ours, users would have to decide for themselves when to switch over to the baseline (all topic-specific training data) system. As can be seen from the performance curves in Figure 4, this is not always an easy decision. For example, antihistamines has 819 total samples, and the performance of the baseline system surpasses the nontopic system during 8-way cross-validation, or, using approximately 100 training samples, 12 (12%) of which are positive. In contrast, for Sedatives, this point occurs earlier, during 32-way cross-validation, where 50 training samples are being used, only ∼4 (8%) of these samples being positive.
Another interesting aspect of the performance curves is their asymptotic form. For most of the topics, it does not appear that the performance of the system (either hybrid or baseline) has completely leveled off—the ability of the algorithm to improve from additional topic-specific training data has not saturated. Only for Diabetes is the performance curve is very flat; it does not appear that more training data would result in further improvement. Even for topics with high mean AUCs (over 0.90), the slope of the curve is fairly steep between 4-way and 2-way cross-validation (e.g., BetaBlockers, CalciumChannelBlockers, and Opioids). This further substantiates the point that topic-specific data are essential for maximum performance, and that it is necessary to incorporate all topic-specific training data that becomes available during SR creation and updates.
To summarize, the nontopic system performs fairly well with no topic-specific training data, but increasing the amount of topic-specific training data used to build the model results in consistent improvements in performance. It is not clear how one would decide to switch over from the nontopic to the general system, but, with our system, it is not necessary to do so at all, as it does this automatically, and maintains top performance across a wide range of available topic-specific training data.
Why does this technique work? Our explanation for this is based on the idea that every SR topic dataset is composed of a mixture of general-SR and topic-specific SR predictive features. That is, there are some features that are predictive of article inclusion that are specific to the topic of the SR, while there are other features that are more generally predictive of SR inclusion without regard for topic. For example, in our previous work, we noticed that “CI” (an abbreviation for confidence interval) was a strong, positive predictor for some topics. 13 It is likely that this term is general enough that it applies to many SR topics. However, in a small random sample of training data, there may not be enough samples to support the ML algorithm to identify, quantify and model the predictive value of such terms. Combining the nontopic and topic-specific data together, as we have done here, allows the ML algorithm to more readily identify these generally predictive terms.
By contrast, for each topic there are topic-specific predictive terms. These are terms that do not simply imply a high quality of evidence or correct methodology, but instead are terms that predict fitness for the specific inclusion criteria for a given SR. For example, the key questions created by the SRs for ADHD make it clear that studies including both children and adults are necessary, and, in fact, the term “adult” is a strong positive predictive term for this topic. It is not a good predictor for the other topics, however.
There are also features that lie somewhere in the middle, both strongly predictive for one topic, and weakly predictive general (but still useful) features for others. The MeSH term Quality of Life is the top predictive feature (measured by information gain) for CalciumChannelBlockers, however, it is not included in the top 200 predictive features for any other topic. It is at best, a weakly predictive feature for some of the other topics, such as ACEInhibitors and BetaBlockers, where Quality Of Life falls among the top 300 features.
Both topic-specific and general SR features need to be used together to achieve maximum performance with a classification algorithm. With adequate amounts of topic-specific training data, both the topic-specific and general features can be extracted and quantified from the training set. When there is inadequate topic-specific training data, the system can get a performance boost by including nontopic data, which will help to model the general SR features and increase the likelihood of recognizing and including weakly predictive features. This does not help much with the strongly topic-specific features, however, and so performance will increase further if additional topic-specific training samples are made available.
Even though the hybrid system was statistically at least as good as or better than the nontopic system at all levels of reverse cross-validation, nontopic learning can sometimes outperform hybrid learning on individual topics when the topic training sizes are especially small. Determining when to use one of these methods versus the other is not straightforward, and the hybrid method we propose here provides a robust means of avoiding this issue. However, achieving optimal performance may be dependent upon a more topic-specific means of resolving this issue. Investigation of means to predict when one method is preferable to the other, as well as modeling topic-specific features with inadequate training data, is an open area for future exploration.
The practical implications of a SR citation classification system with consistent high performance can be significant. For example, the topic ADHD has had 2,529 articles reviewed to identify the 259 that were included in the final review. The positive prevalence is about 0.10. Our approach produces an AUC of 0.85 at a cross-validation fold of 32, which corresponds to about 80 training samples. Assuming that we have 80 manually classified samples for training leaves 2,449 unclassified samples, including about 245 positives. On the ADHD ROC curve for the classifier with an AUC of 0.85 (data not shown) there is a FPR (false positive rate), TPR (true positive rate) operating point of (0.70, 0.98). Using this as the decision threshold results in 245 × 0.98 = 240 of the positives being correctly identified, missing 5, along with filtering out 660, or about 30%, of the negative articles. Since only 80 training + 240 positive + 1543 remaining negative = 1,863 of the original total 2,529 articles now require manual review, this results in a work savings of about 26%, at a cost of missing about 3% of the positive articles.
There is extensive work in the informatics and ML literature, both in applying ML techniques to literature review for EBM, as well as in using related topic-data for training when inadequate topic-specific data exists. However, none of this work combines the elements in a manner such as we have done here, nor does it improve topic-specific ML performance for SR, as we have here using a combination of topic-specific and nontopic specific training data.
There has been a significant amount of work on optimized clinical queries and automated ML for high quality articles in general for EBM. Haynes et al. and the HEDGES team are among the most published authors in this area. Their optimized clinical queries for EBM are a set of query templates for retrieving a higher proportion of high quality articles from PubMed. This work forms the basis of the Clinical Queries facility in PubMed (http://www.ncbi.nlm.nih.gov/entrez/query/static/clinical.shtml). 26–29 Recently, the HEDGES team has expanded their work to incorporate ML techniques. 30 While this work is certainly useful, it is distinct from the research presented here, as the models are focused on clinical users, rather than on systematic reviewers, and they are not specific to an individual review topic. Similarly, Aphinyanaphongs et al. have studied applying SVM-based and other automated classification ML methods to the identification of high-quality articles for EBM in a nontopic-specific manner, based on using selection for inclusion in the ACP Journal Club as a gold standard. 31–34
Most of the significant previous work in cross-topic learning for text classification appears in the ML literature, where it is also called cross-training. These methods usually focus on pairs of similar topics 35 —they do not create a general model from a group of specific topics and then apply an integrated general + specific model to a new topic, as we have done here. However, Zhai et al. has published work on automatically discovering latent common themes across a set of text collections, which they term comparative text mining. 36 In the bioinformatics literature, Gupta has published work using cross-training to integrate knowledge about protein structure and function to improve the performance of SVM models predicting structure using function annotations and vice-versa. 37 We were unable to find any published work on cross-topic learning or cross-training in the medical informatics literature, except as it relates to cross-language querying and translation. 38,39
There are several limitations to our evaluation. While the data set includes 24 topics and over 50,000 individual citations and expert judgments, by necessity this is a small fraction of the total number of SRs needed, and therefore represents a small fraction of the total amount of literature that might be reviewed in total for all potential SRs. Nevertheless, we think that the range of topics and the size of the data set are large enough to for us to infer that our algorithm will perform well on many additional SR topics.
All the data used in this study was generated by a single SR-producing organization. The DERP uses the most rigorous processes available to maximize quality and consistency. While there may be some differences in the processes used among SR-producing organizations, among groups that produce high quality reviews, by necessity, these processes should be more alike than different. We expect that our system would perform similarly on data from other groups that use rigorous SR methods. 40
All the SR topics studied here relate to classes of pharmacological interventions. Etiology, diagnosis, prognosis, and non-pharmacological therapy are also important areas needing SRs. Evaluation of our approach on these kinds of SRs is an area for further study.
Extensive cross-validation was used to accurately estimate the performance of our method under a large number of conditions. This resampling required ignoring the temporal publication sequence of the individual articles. A deployed system would of course be limited to using prior publications to predict later ones. Whether or not this has implications for the performance of our system depends upon how stable the feature distribution of the literature for a given topic is over time. This topic or conceptual drift can be an important factor in automated Web and news classification systems using on-line learning. 41,42 SRs do on occasion have changes in scope. While some studies have found that, in general, the feature distribution of biomedical literature does change, 43,44 this may or may not be a significant factor in applying ML to individual SR topics. This remains an area for further study.
Finally, we should contrast our approach with another popular ML framework—that of active learning. 45 While active learning also attempts to increase performance using a reduced amount of topic-specific training data, it does so in a very different manner than what we have proposed here. With active learning, the amount of labeled training data that is required is decreased by having the system request labeling for the samples on which it can learn the most, and the rest of the data samples remain unlabeled. Active learning has been proposed for situations in which the definition of a true-positive for a topic changes over time. 46 In these situations, there is usually a continuous flow of data samples, and it is reasonable to expect that experts are available and willing to manually annotate a small number of machine-chosen samples on a regular basis. This is a much more interactive approach than our hybrid classifier, and requires the workflow of the users to be organized around the requirements of the active learning framework. While this is certainly possible for some users, for systematic reviewers at this time, it would negate the usefulness of the system. One of the main benefits of ranking the literature is to allow the reviewers to do work prioritization based on which articles are most likely to be included in the final study. Based on this, the reviewers choose which articles they wish to read, and in which order. But in active learning, the system chooses which articles the reviewers read, at least until the system is fully trained. This reversal of the control over which articles are read first currently makes active learning a less attractive ML approach for SR creation and update than the one that we have proposed here. However, an active learning system that determines which papers to be reviewed based on both its own needs and the needs of the reviewers at a given moment in time is at least possible in principle, and is an interesting area for future research.
Conclusions
We have presented and evaluated a robust and effective method for improving the performance of automated topic-specific ranking on articles for SRs. On average, the method improves performance by about 20%, when the amount of topic-specific training data are scarce. The algorithm works by integrating predictive features from both the available topic-specific training samples and from a large pool of nontopic-specific data sampled from many other topics that, together, result in a large amount of training data. The algorithm maintains effectiveness throughout a wide range in the amount of available topic-specific training data, and therefore no human intervention is necessary to decide when to shift between general nontopic and topic-specific machine learning models. Future work will focus on extending the algorithm to use additional sources of topic-specific data and embedding the algorithm in an interactive system available to systematic reviewers during the literature review process.
Acknowledgments
The authors wish to acknowledge the Drug Evidence Review Project (DERP) of the Oregon EPC for providing the original review inclusion data in the form of Endnote files.
Footnotes
This work was supported by grant 1R01LM009501-01 from the National Library of Medicine.
References
- 1. CRD Report Number 4(2nd edition). York, UK: NHS Centre for Reviews and Dissemination; 2001.
- 2.Haynes RB. What kind of evidence is it that evidence-based medicine advocates want health care providers and consumers to pay attention to? BMC Health Serv Res 2002;2(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helfand M. Incorporating information about cost-effectiveness into evidence-based decision-making: The evidence-based practice center (EPC) modelSuppl Med Care 2005;43(7):33-43. [DOI] [PubMed] [Google Scholar]
- 4.Haynes RB. Of studies, syntheses, synopses, and systems: The “4S” evolution of services for finding current best evidence ACP J Club 2001;134(2):A11-A13. [PubMed] [Google Scholar]
- 5.Grimshaw JM, Santesso N, Cumpston M, Mayhew A, McGowan J. Knowledge for knowledge translation: The role of the Cochrane Collaboration J Contin Educ Health Prof 2006;26(1):55-62. [DOI] [PubMed] [Google Scholar]
- 6.Helfand M. Meta-analysis in deriving summary estimates of test performance Med Decis Mak 1993;13(3):182-183. [PubMed] [Google Scholar]
- 7.Moher D, Tsertsvadze A, Tricco AC, et al. A systematic review identified few methods and strategies describing when and how to update systematic reviews J Clin Epidemiol 2007;60(11):1095. [DOI] [PubMed] [Google Scholar]
- 8.Moher D, Tsertsvadze A, Tricco AC, et al. When and how to update systematic reviews Cochrane Database Syst Rev 2008;1MR000023. [DOI] [PMC free article] [PubMed]
- 9.Sampson M, Shojania KG, McGowan J, et al. Surveillance search techniques identified the need to update systematic reviews J Clin Epidemiol 2008;61(8):755-762. [DOI] [PubMed] [Google Scholar]
- 10.Shojania KG, Sampson M, Ansari MT, et al. How quickly do systematic reviews go out of date?. A survival analysis. Ann Intern Med 2007;147(4):224-233. [DOI] [PubMed] [Google Scholar]
- 11.Mallett S, Clarke M. How many cochrane reviews are needed to cover existing evidence on the effects of health care interventions? ACP J Club 2003;139(1):A11. [PubMed] [Google Scholar]
- 12.Cohen A. Optimizing feature representation for automated systematic review work prioritization AMIA Annu Symp Proc 2008:121-125. [PMC free article] [PubMed]
- 13.Cohen AM, Hersh WR, Peterson K, Yen PY. Reducing workload in systematic review preparation using automated citation classification J Am Med Inform Assoc 2006;13(2):206-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haynes RB. Of studies, syntheses, synopses, summaries, and systems: The “5S” evolution of information services for evidence-based healthcare decisions Evid Based Med 2006;11(6):162-164. [DOI] [PubMed] [Google Scholar]
- 15.Hersh W, Bhupatiraju RT, Ross L, et al. Enhancing access to the bibliome: The TREC 2004 genomics track J Biomed Discov Collab 2006;1(3). [DOI] [PMC free article] [PubMed]
- 16.Donaldson I, Martin J, de Bruijn B, et al. Prebind and Textomy—Mining the biomedical literature for protein–protein interactions using a support vector machine BMC Bioinform 2003;4(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uzuner O, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records J Am Med Inform Assoc 2008;15(1):14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Cohen A, McDonagh MS, SYRIAC The systematic review information automated collection system A data warehouse for facilitating automated biomedical text classification AMIA Annu Symp Proc 2008:825-829. [PMC free article] [PubMed]
- 19.Vapnik VN. The Nature of Statistical Learning Theory2nd edn. New York: Springer; 2000.
- 20.Joachims T. Text categorization with support vector machines: Learning with many relevant features Proceedings of the 10th European Conference on Machine Learning. 1998. pp. 137-142.
- 21.Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms Pattern Recognit 1997;30(7):1145-1159. [Google Scholar]
- 22.Li X, Bilmes J, Malkin J. Maximum margin learning and adaptation of MLP classifiers2005ISCA; 2005.
- 23.Cohen AM. An effective general purpose approach for automated biomedical document classification AMIA Annu Symp Proc 2006:161-165. [PMC free article] [PubMed]
- 24.Joachims T. SVM-light support vector machine, 2004http://svmlight.joachims.org/ 2006. Accessed: May 20, 2009.
- 25.Dietterich TG. Approximate statistical tests for comparing supervised classification learning algorithms Neural Comput 1998;10(7):1895-1924. [DOI] [PubMed] [Google Scholar]
- 26.Haynes RB, Wilczynski N, McKibbon KA, Walker CJ, Sinclair JC. Developing optimal search strategies for detecting clinically sound studies in MEDLINE J Am Med Inform Assoc 1994;1(6):447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilczynski NL, Haynes RB. Developing optimal search strategies for detecting clinically sound causation studies in MEDLINE AMIA Annu Symp Proc 2003:719-723. [PMC free article] [PubMed]
- 28.Wilczynski NL, Haynes RB. EMBASE search strategies for identifying methodologically sound diagnostic studies for use by clinicians and researchers BMC Med 2005;3(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilczynski NL, Morgan D, Haynes RB. An overview of the design and methods for retrieving high-quality studies for clinical care BMC Med Inform Decis Mak 2005;5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kilicoglu H, Demner-Fushman D, Rindflesch TC, Wilczynski NL, Haynes RB. Towards automatic recognition of scientifically rigorous clinical research evidence J Am Med Inform Assoc 2009;16(1):25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aphinyanaphongs Y, Aliferis CF. Text categorization models for retrieval of high quality articles in internal medicine AMIA Annu Symp Proc 2003:31-35. [PMC free article] [PubMed]
- 32.Aphinyanaphongs Y, Aliferis CF. Learning Boolean queries for article quality filtering Medinfo 2004;11(1):263-267. [PubMed] [Google Scholar]
- 33.Aphinyanaphongs Y, Statnikov A, Aliferis CF. A comparison of citation metrics to machine learning filters for the identification of high quality MEDLINE documents J Am Med Inform Assoc 2006;13(4):446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aphinyanaphongs Y, Tsamardinos I, Statnikov A, Hardin D, Aliferis CF. Text categorization models for high-quality article retrieval in internal medicine J Am Med Inform Assoc 2005;12(2):207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarawagi S, Chakrabarti S, Godbole S. Cross-training: Learning probabilistic mappings Between topics2003New York, NY, USA: ACM; 2003. pp. 177-186.
- 36.Zhai CX, Velivelli A, Yu B. A cross-collection mixture model for comparative text mining2004New York, NY, USA: ACM; 2004. pp. 743-748.
- 37.Gupta K, Sehgal V, Levchenko A. A method for probabilistic mapping between protein structure and function taxonomies through cross training BMC Struct Biol 2008;8(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabar N, Krivine S, Jaulent MC. Classification of health webpages as expert and non expert with a reduced set of cross-language features AMIA Annu Symp Proc 2007:284-288. [PMC free article] [PubMed]
- 39.Marko K, Daumke P, Schulz S, Hahn U. Cross-language MeSH indexing using morpho-semantic normalization AMIA Annu Symp Proc 2003:425-429. [PMC free article] [PubMed]
- 40.Helfand M. Using evidence reports: Progress and challenges in evidence-based decision making Health Aff 2005;24(1):123-127. [DOI] [PubMed] [Google Scholar]
- 41.Forman G. Tackling concept drift by temporal inductive transfer Proceedings of the 29th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval 2006:252-259.
- 42.Wang H, Yin J, Pei J, Yu PS, Yu JX. Suppressing model overfitting in mining concept-drifting data streams Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 2006:736-741.
- 43.Cohen AM, Hersh WR, Bhupatiraju RT. Feature generation, feature selection, classifiers, and conceptual drift for biomedical document triage Proceedings of the Thirteeth Text Retrieval Conference—TREC 2004. Gaithersburg, MD. 2004.
- 44.Srinivasan P. Adaptive classifiers, topic drifts and GO annotations AMIA Annu Symp Proc 2007:681-685. [PMC free article] [PubMed]
- 45.Tong S, Koller D. Support vector machine active learning with applications to text classification J Machine Learn Res 2001;2(1):45-66. [Google Scholar]
- 46.Liu Y. Active learning with support vector machine applied to gene expression data for cancer classification J Chem Inf Comput Sci 2004;44(6):1936-1941. [DOI] [PubMed] [Google Scholar]