Abstract
Acinetobacter calcoaceticus PHEA-2 exhibited a delayed utilization of phenol in the presence of benzoate. Benzoate supplementation completely inhibited phenol degradation in a benzoate 1,2-dioxygenase knockout mutant. The mphR encoding the transcriptional activator and mphN encoding the largest subunit of multi-component phenol hydroxylase in the benA mutant were significantly downregulated (about 7- and 70-fold) on the basis of mRNA levels when benzoate was added to the medium. The co-transformant assay of E. coli JM109 with mphK::lacZ fusion and the plasmid pETR carrying mphR gene showed that MphR did not activate the mph promoter in the presence of benzoate. These results suggest that catabolite repression of phenol degradation by benzoate in A. calcoaceticus PHEA-2 is mediated by the inhibition of the activator protein MphR.
Introduction
Aromatic hydrocarbons exist ubiquitously in the environment and the most effective and economical way to remove the aromatic pollutants is by means of microbial degradation [1]. The details of the metabolic pathways involved in the degradation of aromatic hydrocarbons at the biochemical and molecular levels have been well described [19]. However, bacteria in the natural environment are often exposed to mixtures of aromatic hydrocarbons. To survive under these complex conditions, bacteria have to quickly adapt to regulate their gene expression to select the best available carbon source for efficient metabolism. In bacteria, the phenomenon of carbon catabolite repression (CCR) is believed to be an energy-saving response whereby readily metabolizable carbohydrates are used in preference to less metabolizable substrates. Several distinct molecular mechanisms of CCR have been identified. In Bacillus subtilis and B. megaterium, glycolysis is involved in signal transduction and mediates activation of the catabolite control protein (CcpA) [8]; In Escherichia coli, the cAMP–cAMP receptor protein complex-mediated activation of genes responsible for degradation of secondary carbon sources does not occur if glucose is available [15]. The catabolite repression control of pseudomonads has been found to be cAMP independent [7]. Moreover, some organic acids repress phenol catabolism in Ralstonia eutropha [2]. Interestingly, in Pseudomonasputida CSV86, aromatic compounds are found to suppress glucose utilization [3]. Up to now, very little information is available on the degradation of mixtures of aromatic compounds or on the degradation of an individual aromatic hydrocarbon when present in a mixture of structurally similar compounds. One interesting catabolite repression-like response is the preferred metabolism of benzoate over 4-hydroxybenzoate (4-HBA) in some gram-negative soil bacteria [6, 13].
Acinetobacter calcoaceticus PHEA-2 can utilize phenol and benzoate as sole carbon and energy sources via the same catechol branch of the β-ketoadipate pathway [20]. The upper pathways involve the mph gene cluster encoding a multicomponent phenol hydroxylase (mphKLMNOP), the transcriptional regulator MphR, ben gene cluster encoding a benzoate 1,2-dioxygenase (benMABCDEKP) and the transcriptional regulator BenM. Here we report that benzoate itself repressed the utilization of phenol when PHEA-2 grows on mixtures of benzoate and phenol as carbon and energy sources. Furthermore, we found that the MphR is the target and that CCR of phenol degradation occurs by disrupting the interaction between the activator protein MphR and phenol. The aim of this study is to clarify the growth behavior and mechanism regulating phenol utilization in the presence of benzoate.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Acinetobacter calcoaceticus PHEA-2 or the benA mutant (PHEA-12, the deletion benA mutant) [20] was cultured in Luria–Bertani (LB) medium (g/l: bacto tryptone 10 g, bacto yeast extract 5 g, NaCl 10 g) or mineral salt (MS) medium (g/l: NaNO3 0.5 g, K2HPO4 0.65 g, KH2PO4 0.17 g, MgSO4 0.10 g) with different carbon sources at 30°C. When necessary, antibiotics were used at the final concentration of 50 μg/ml kanamycin (Km), 10 μg/ml tetracycline (Tc) or 20 or 50 μg/ml ampicillin (Amp).
Phenol and Benzoate Analysis
The quantification of phenol was done using the colorimetric method of Martin [9]. Under alkaline conditions phenol reacts with 4-aminoantipyrine (4-amino-2,3-dimethyl-1-phenyl-3-pyrazolin-5-one) to form a red indophenol dye, which is assayed spectrophotometrically at a wavelength of 510 nm.
For the quantitative analysis of benzoate, cells remaining in the supernatant fractions collected at different times were removed by passage through a low-protein-binding, 0.22-mm pore-size syringe filter (MSI). A 10-ml sample of the filtrate was analyzed on a C18 reversed-phase high-performance liquid chromatography (HPLC) column (Agilent). Elution at a rate of 0.8 ml/min was carried out with 30% acetonitrile and 0.1% phosphoric acid, and the eluent was monitored by UV detection at 254 nm to detect benzoate.
Construction of the mphK::lacZ Transcriptional Fusion Plasmid and β-Galactosidase Assays
A PCR-amplified fragment containing the mph gene promoter sequence (−564 to +3nt from the mphK start codon) was inserted upstream of the lacZ gene of the broad-host range promoter vector pGD926 (promoter probe vector, Tcr) [5]. The complete mphR gene amplified by PCR with primers MPHR-F: 5′-CCGCATATGGCTCGAGTAAAATACGATAC-3′ and MPHR-R: 5′-GCGTCGACACTCAAAGCTGAAATTTTTTAATTC-3′ was ligated to expression vector pET28a (T7 expression vector, Kmr, Novagen) to construct the plasmid pETR. The mphK::lacZ transcriptional fusion was transformed into the wild-type strain PHEA-2 by biparental mating and was also co-transformed into E. coli JM109 containing the plasmid pETR. The resulting strain, PHEA-2(mphK::lacZ) (Ampr and Tcr), grown in MS medium containing 2-mM phenol, 2-mM phenol plus 2-mM or plus 4-mM benzoate with an initial optical density at OD600 of 0.4 was incubated at 30°C and samples were taken at different times. The co-transformant of JM109 (mphK::lacZ and pETR) (Kmr and Tcr) was incubated for 3 h at 37°C in LB medium containing 2-mM phenol, 2-mM benzoate or 2-mM phenol plus 2-mM benzoate with an initial optical density at OD600 of 0.3. The cells were collected and lysed with chloroform and sodium dodecyl sulfate. β-Galactosidase activities were determined according to the methods described by Miller [10].
Reverse Transcription-PCR (RT-PCR)
PHEA-2 cells exposed to 2-mM phenol and 2-mM benzoate were collected at various times and total RNA isolated using an RNeasy total RNA kit (Promega). Where appropriate, a second DNaseI digestion was performed with DNaseI (TaKaRa, Dalian, China) to completely remove the chromosomal DNA. RNA concentration was determined spectrophotometrically and its integrity was assessed by agarose gel electrophoresis. Purified RNA (1 μg) was used to prepare cDNA by using a First Strand cDNA synthesis kit (New England Biolabs, UK). Expressions of benA and mphN were measured by amplification of 108- and 144-bp intergenic regions, respectively. The primers were BenAF(5′-GGTGGCTCGTATGGCTTTGA-3′), BenAR(5′-TTTCGGTGTATTCGTCTGCT-3′); MphNF(5′-AGCCGTTATGGCATTCGT-3′), MphNR(5′-CATTTCTTCGTCGGTTGGA-3′).
Real-Time RT-PCR Assay
Total RNAs were prepared from the benA mutant induced for 3 h with phenol (2 mM) or benzoate (2 mM) plus phenol (2 mM) using the RNeasy total RNA kit (Promega). The concentration and purity of the RNA samples were determined by using a U-3010 spectrophotometer according to the manufacturer’s protocols. RNA samples (1 μg) were transformed into cDNA by using the First Strand cDNA Synthesis Kit (New England Biolabs, UK). The cDNA obtained was stored at −20°C.
The Rotor Gene6000 Real-Time PCR Detection System was used for real-time PCR in a 20-μl volume of PCR mixture containing 10 μl 2 × QuantiTect SYBR Green PCR Master Mix (Qiagen, Germany), 0.5 μl each primer pair and 1 μl cDNA. After 15 min of denaturation at 95°C, 45 cycles were performed. Cycling conditions were 95°C for 15 s, 55°C for 60 s, and 72°C for 30 s (elongation and signal acquisition). The concentration of each gene was calculated by reference to respective standard curves using Corbett Research software and normalized as the ratio of the target. The 16S rRNA was amplified by RT-PCR as an internal control. Each reaction was performed in triplicate. For all isolates, the relative gene expression was reported as the change (n-fold) determined from the mean normalized expression relative to the mean normalized expression of the benA mutant.
Results
The Effect of Benzoate on Phenol Utilization in A. Calcoaceticus PHEA-2
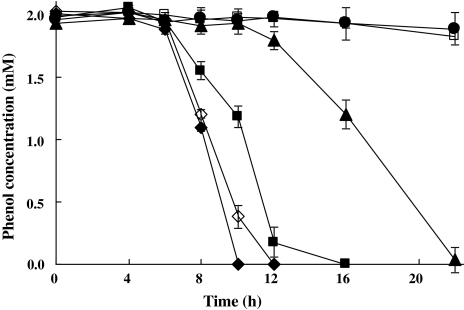
PHEA-2 can utilize phenol and benzoate as the sole carbon and energy sources [20], and exhibits diauxic growth on phenol plus benzoate. To validate the response of PHEA-2 to simultaneously administer aromatic compounds, the effects of benzoate on the metabolism of phenol in PHEA-2 were examined. Phenol consumption was delayed about 2 h in the presence of 2-mM benzoate; however, the latency period was markedly increased when the strain was exposed to higher concentration of benzoate (Fig. 1). The data showed that A. calcoaceticus PHEA-2 preferentially utilizes benzoate over phenol, implying that the presence of benzoate inhibited phenol degradation of PHEA-2.
Fig. 1.
The effects of benzoate on phenol consumption by A. calcoaceticus PHEA-2 and the benA mutant. Filled diamond, filled square, filled triangle, and filled circle—the wild type strain grown in 2-mM phenol plus 0-, 2-, 4-, and 8-mM benzoate, respectively; open diamond and open square—the benA mutant strain grown in 2-mM phenol plus 0- and 2-mM benzoate, respectively. Error bars show standard deviations
Growth of the benA Mutant on Phenol and a Phenol–Benzoate Mixture
In order to better address the role of benzoate in the repression of phenol utilization by A. calcoaceticus PHEA-2, the benA mutant was used. Inactivation of benA (a gene encoding the α-subunit of benzoate 1,2-dioxygenase which catalyzes the first step in the degradation of benzoate), prevented growth of cells on benzoate but did not affect the utilization of phenol [20]. We found that the benA mutant was unable to utilize phenol in the medium supplemented with 2-mM benzoate (Fig. 1). The result indicates that benzoate has the ability to completely inhibit phenol degradation in the A. calcoaceticus PHEA-2 strain.
Role of Benzoate in the Expression of the Phenol Hydroxylase Gene
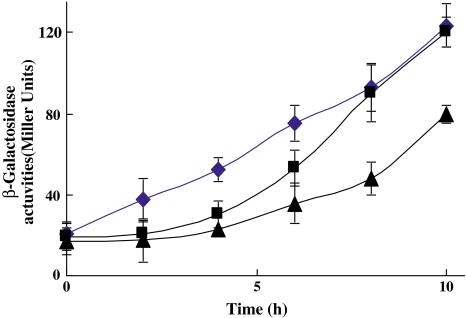
Phenol hydroxylase is a key enzyme that catalyzes the first step in phenol degradation. To investigate the repressing effect of benzoate on the expression of phenol hydroxylase, a transcriptional fusion of the phenol hydroxylase gene (mph) promoter to the promoterless lacZ gene encoding β-galactosidase was conducted. The A. calcoaceticus strain PHEA-2 incorporating the plasmid with this fusion (mphK::lacZ) plasmid was employed. The β-galactosidase activities in cells growing on phenol plus benzoate were lower than those in cells growing on phenol alone, and the higher the concentration of benzoate, the lower the β-galactosidase activities were (Fig. 2). This result demonstrated that benzoate restrained the expression of phenol hydroxylase structural genes (mph) and reduced the activity of phenol hydroxylase when the medium contained benzoate. The higher concentration of benzoate, the lower β-galactosidase activities (Fig. 2). This result demonstrated that benzoate restrained the expression of phenol hydroxylase structural genes (mph) and reduced the activity of phenol hydroxylase when the medium contained benzoate.
Fig. 2.
β-Galactosidase of the mphK::lacZ fusion during growth on various carbon sources. Filled diamond, filled square, and filled triangle—the mphk-lacZ fusion plasmid in Acinetobacter calcoaceticus PHEA-2 is induced by growth on 2-mM phenol, 2-mM phenol -2-mM benzoate, and 2-mM phenol–4-mM benzoate, respectively
Transcriptional Expressions of the mph and ben Gene
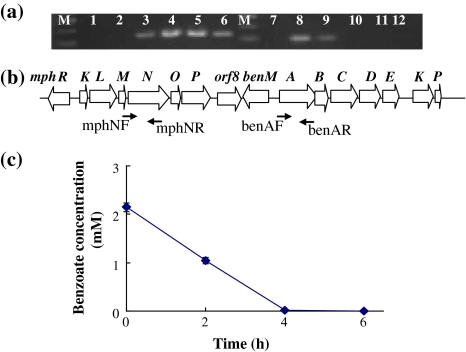
In order to further examine whether benzoate itself repressed the phenol consumption of PHEA-2, the reverse transcription PCR (RT-PCR) analysis was performed with total RNA isolated from the cells exposed to 2-mM phenol plus 2-mM benzoate. As shown in the Fig. 3a and b, benA transcripts were already present at 2 h, and continued to be expressed until 4 h when the benzoate concentration in the culture supernatant approached zero (0.018 ± 0.00013 mM) (Fig. 3c). In contrast, the transcription of the mphN gene, encoding the largest subunit of multi-component phenol hydroxylase, began to appear only at 4 h. These observations indicated that the repression of mphN transcription was specifically released in the presence of benzoate. Moreover, RT-PCR experiments clearly showed a shift in gene expression from benA to mphN between 4 and 6 h. Taken together, the results strongly suggested that benzoate caused transcriptional repression of phenol utilization by transcriptional inhibition of the mph operon in PHEA-2.
Fig. 3.
Preferential utilization of benzoate over phenol by A. calcoaceticus PHEA-2. a Agarose gel electrophoresis of reverse transcription-polymerase chain reaction (RT-PCR) analysis of expressed assays. M 100-bp molecular size markers, lanes from 1 to 6 are the products of mphN at various time points (0, 2, 4, 6, 8, 10 h), lanes from 7 to 12 are the products of benA at the corresponding various time points. b The organization of the mph-ben gene region of PHEA-2. c The concentration of benzoate of PHEA-2 grown on 2-mM phenol–2-mM benzoate
Effect of Benzoate on the Expression of mphR and mphN
MphR is one member of the NtrC family of bacterial transcriptional activators and activates phenol hydroxylase expression in the presence of phenol in A. calcoaceticus PHEA-2. Our data showed that benzoate can repress the expression of the mph operon of PHEA-2 grown on phenol plus benzoate. In P. putida, the inhibitory effect of gene expression was controlled by the negative control factor Crc, and CCR of phenol degradation is mediated by inhibition of the activator protein PhlR [12]. Also, in Rhodopseudomonas palustris, BadM is a transcriptional repressor of benzoylCoA that is the critical enzyme for benzoate degradation under anaerobic conditions [14]. Therefore, there may exist a mechanism whereby the transcriptional repression of mphR mediated by benzoate affects the expression of the mph operon in A. calcoaceticus PHEA-2.
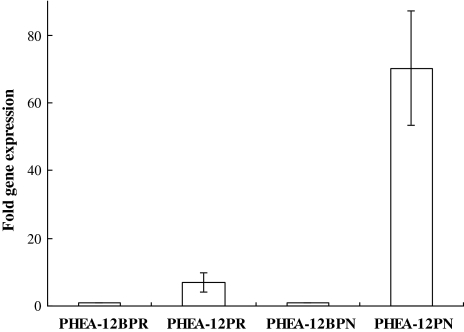
Therefore, to differentiate the effect of MphR on the CCR of phenol degradation, the transcription levels of mphR and mphN from the benA mutant were analyzed by real-time RT-PCR. Compared with phenol as the sole carbon resource, the expression of mphR and mphN were reduced 7- and 70-fold, respectively, in the presence of both phenol and benzoate (Fig. 4). Our data suggested that benzoate repressed the expression of the regulator gene and the mph operon at the transcription levels.
Fig. 4.
Relative abundance of cDNA of mphR and mphN of benA mutant PHEA-12 grown on phenol (PHEA-12PR and PHEA-12PN) or phenol–benzoate (PHEA-12BPR and PHEA-12BPN), respectively, as determined by real-time RT-PCR. Values represent the mean and standard error (n = 3); PHEA-12BPR and PHEA-12BPN are the controls
CR is Mediated by Inhibition of MphR
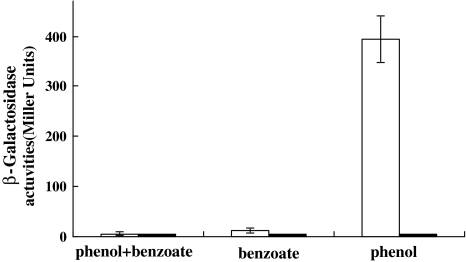
To understand the effect of benzoate on MphR-mediated transcription from the mph promoter, we constructed the co-transformant of E. coli JM109 with mphK::lacZ fusion plasmid and pEMR carrying mphR gene. As shown in Fig. 5, activities of β-galactosidase as high as 394.5 ± 47.1 Miller Units were observed when phenol was used as the sole inducer, whereas only basal β-galactosidase activity was detected in the presence of benzoate, or with the phenol–benzoate combination. These results indicated that benzoate might interfere competitively with the activating capability of the transcription activator MphR, and that the repression of phenol degradation by benzoate was mediated by the inhibition of the activator protein MphR.
Fig. 5.
The mphK::lacZ transcriptional fusion plasmid in E. coli JM109. The mphk-lacZ fusion is induced by the indicated inducers on LB medium in the presence (white bars) or absence (black bars) of the plasmid pEMR in E. coli JM109. Error bars show standard deviations
Discussion
Normally, preferred carbon sources repress the expression of catabolic genes corresponding to the nonpreferred carbon sources that are present. TCA cycle intermediates (e.g., succinate, pyruvate, and citrate), carbohydrates (e.g., glucose and gluconate), and amino acids repress the degradation pathways for aromatic compounds [11, 16, 18]. However in this study, we showed the phenol degradation in Acinetobacter calcoaceticus PHEA-2 is subject to CCR induced by the aromatic compound benzoate. Similarly, benzoate inhibits the catabolism of phenol in Ralstonia eutropha [2], represses 4-hydroxybenzoate degradation genes in P. putida [12], and switches off the degradation pathways for phthalates in Rhodococcus sp. [4] and p-hydroxybenzoate in A. calcoaceticus [6]. The repression control can best be achieved if global regulation is targeted to the peripheral genes, rather than to the central pathways for aromatic compounds.
However, in PHEA-2, phenol and benzoate are independently converted to catechol by phenol hydroxylase and benzoate dioxygenase, respectively, while further degradation proceeds via the same catechol branch of the β-ketoadipate pathway [20]. Therefore, catechol or intermediates of the catechol branch of β-ketoadipate pathway did not act as inhibitors. Furthermore, we observed that a variety of substrates (e.g., lactose, acetate, glucose, pyruvate, succinate, and cis,cis-muconate) did not have the ability to repress the catabolism of phenol in PHEA-2 (data not shown). This phenomenon of PHEA-2 contrasts with observations that benzoate as well as those substrates could diminish the degradation of phenol or other aromatic compounds [2, 6, 13].
A number of underlying mechanisms for the CCR phenomena have been identified. Catabolism of glucose (the preferred carbon source) results in low cAMP levels, which prevent transcription of promoters of the genes for the catabolic pathway of nonpreferred carbon sources [17]. In R. eutropha, benzoate degradation via the catechol branch of the ortho-cleavage pathway blocked the synthesis of phenol hydroxylase, in addition to the synthesis of catechol-2,3-dioxygenase, a key enzyme of phenol degradation through the meta pathway [2]. In P. putida PRS2000, benzoate inhibited expression of the 4-hydroxybenzoate transport gene pcaK which attenuates 4-hydroxybenzoate degradation, and each aromatic compound is degraded via two parallel sequences of reactions that converge at β-ketoadipate [11]. In contrast, in PHEA-2, the inhibitor benzoate and phenol share the same ortho-cleavage branch of the catechol degradation pathway. We hypothesized that benzoate inhibited the activation of the regulator MphR leading to repression of phenol degradation in A. calcoaceticus PHEA-2. MphR is a bacterial transcriptional activator, which interacts with phenol to activate the expression of phenol hydroxylase. Our experiments involving co-transformation of E. coli cells with the mphK::lacZ transcriptional fusion plasmid and the mphR expression plasmid revealed that MphR activated the mph promoter by phenol, but MphR could not activate the mph promoter when benzoate added in the medium. We propose that the high affinity binding of benzoate to MphR results in strong inhibition of the promoter of the phenol hydroxylase genes. The results of this investigation offer new insight into catabolite repression involving binary aromatic mixtures. However, the precise details of the competition between benzoate and phenol for access to MphR await further clarification.
Acknowledgments
This study was supported by National High-Tech Program Nos. 2007AA021305 and 2006AA02Z229) and National Natural Science Foundation of China (Grant Nos. 30470047 and 30770076).
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Wei Zhang, Phone: +86-10-82105038, FAX: +86-10-82106981, Email: zhwmm@caas.net.cn.
Ming Chen, Phone: +86-10-82105038, FAX: +86-10-82106106, Email: chenmingbio@hotmail.com.
References
- 1.Alexander M (1981) Biodegradation of chemicals of environmental concern. Science 211:132–138 [DOI] [PubMed]
- 2.Ampe F, Leonard D, Lindley ND (1998) Repression of phenol catabolism by organic acids in Ralstonia eutropha. Appl Environ Microbiol 64:1–6 [DOI] [PMC free article] [PubMed]
- 3.Basu A, Apte SK, Phale PS (2006) Preferential utilization of aromatic compounds over glucose by Pseudomonas putida CSV86. Appl Environ Microbiol 72:2226–2230 [DOI] [PMC free article] [PubMed]
- 4.Choi KY, Zylstra GJ, Kim E (2007) Benzoate catabolite repression of the phthalate degradation pathway in Rhodococcus sp. strain DK17. Appl Environ Microbiol 73:1370–1374 [DOI] [PMC free article] [PubMed]
- 5.Ditta G, Schmidhauser T, Yakobson E, Lu P, Liang XW, Finlay DR, Guiney D, Helinski DR (1985) Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13:149–153 [DOI] [PubMed]
- 6.Gaines GL III, Smith L, Neidle EL (1996) Novel nuclear magnetic resonance spectroscopy methods demonstrate preferential carbon source utilization by Acinetobacter calcoaceticus. J Bacteriol 178:6833–6841 [DOI] [PMC free article] [PubMed]
- 7.Holtel A, Marques S, Mohler I, Jakubzik U, Timmis KN (1994) Carbon source-dependent inhibition of xyl operon expression of the Pseudomonas putida TOL plasmid. J Bacteriol 176:1773–1776 [DOI] [PMC free article] [PubMed]
- 8.Hueck CJ, Hillen W (1995) Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol Microbiol 15:395–401 [DOI] [PubMed]
- 9.Martin RW (1949) Rapid colorimetric estimation of phenol. Anal Chem 21:1419–1420 [DOI]
- 10.Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor
- 11.Moreno R, Rojo F (2008) The target for the Pseudomonas putida Crc global regulator in the benzoate degradation pathway is the BenR transcriptional regulator. J Bacteriol 190:1539–1545 [DOI] [PMC free article] [PubMed]
- 12.Muller C, Petruschka L, Cuypers H, Burchhardt G, Herrmann H (1996) Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J Bacteriol 178:2030–2036 [DOI] [PMC free article] [PubMed]
- 13.Nichols NN, Harwood CS (1995) Repression of 4-hydroxybenzoate transport and degradation by benzoate: a new layer of regulatory control in the Pseudomonas putida beta-ketoadipate pathway. J Bacteriol 177:7033–7040 [DOI] [PMC free article] [PubMed]
- 14.Peres CM, Harwood CS (2006) BadM is a transcriptional repressor and one of three regulators that control benzoyl coenzyme A reductase gene expression in Rhodopseudomonas palustris. J Bacteriol 188:8662–8665 [DOI] [PMC free article] [PubMed]
- 15.Postma PW, Lengeler JW, Jacobson GR (1993) Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev 57:543–594 [DOI] [PMC free article] [PubMed]
- 16.Putrinš M, Tover A, Tegova R, Saks U, Kivisaar M (2007) Study of factors which negatively affect expression of the phenol degradation operon pheBA in Pseudomonas putida. Microbiology 153:1860–1871 [DOI] [PubMed]
- 17.Stulke J, Hillen W (1999) Carbon catabolite repression in bacteria. Curr Opin Microbiol 2:195–201 [DOI] [PubMed]
- 18.Sze CC, Shingler V (1999) The alarmone (p)ppgpp mediates physiological responsive control at the σ54-dependent Po promoter. Mol Microbiol 31:1217–1228 [DOI] [PubMed]
- 19.Tropel D, van der Meer JR (2004) Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol Mol Biol Rev 68:474–500 [DOI] [PMC free article] [PubMed]
- 20.Zhan Y, Yu H, Yan Y, Chen M, Lu W, Li S, Peng Z, Zhang W, Ping S, Wang J, Lin M (2008) Genes Involved in the Benzoate Catabolic Pathway in Acinetobacter calcoaceticus PHEA-2. Curr Microbiol 57:609–614 [DOI] [PubMed]