Abstract
The objective of this work was to identify peptide sequences with high affinity to bone-like mineral (BLM) to provide alternative design methods for functional bone regeneration peptides. Adsorption of preferential binding peptide sequences on four apatite-based substrates [BLM and three sintered apatite disks pressed from powders containing 0% (HA), 5.6% (CA5), 10.5% (CA10)] with varied compositions and morphologies was investigated. A combination of phage display, ELISA, and computational modeling was used to elucidate three 12-mer peptide sequences APWHLSSQYSRT (A), STLPI-PHEFSRE (S), and VTKHLNQISQSY (V), from 243 candidates with preferential adsorption on BLM and HA. Overall, peptides S and V have a significantly higher adsorption to the apatite-based materials in comparison to peptide A (for S vs. A, BLM p = 0.001, CA5 p < 0.001, CA10 p < 0.001, HA p = 0.038; for V vs. A, BLM p = 0.006, CA5 p = 0.033, CA10 p = 0.029). FT-IR analysis displayed carbonate levels in CA5 and CA10 dropped to approximately 1.1–2.2% after sintering, whereas SEM imaging displayed CA5 and CA10 possess distinct morphologies. Adsorption results normalized to surface area indicate that small changes in carbonate percentage at a similar morphological scale did not provide enough carbonate incorporation to show statistical differences in peptide adsorption. Because the identified peptides (S and V) have preferential binding to apatite, their use can now be investigated in bone and dentin tissue engineering, tendon and ligament repair, and enamel formation.
Keywords: Phage display, Bone-like mineral, Peptide adsorption, Carbonated apatite, Apatite
1. Introduction
Understanding biomimetic principles of replicating the formation and structure of tissues can aid in restoring organ function. Repair and restoration of orofacial and long-bone defects caused by trauma or disease could benefit from advances, as current grafting methods have limitations of donor site morbidity, graft rejection, and/or inadequate bone formation and quality [1–3].
Concepts in bone tissue engineering strive to induce bone formation using a combination of scaffolding, biological signals, and/or cells [4,5]. A number of scaffold materials, such as biodegradable polymers [6,7], titanium [8], calcium phosphate-based ceramics [9,10] and glass [11] can elicit the formation of bone tissue from host and/or donor cells. A variety of material choices stem from the requirements that bone implant materials should demonstrate biocompatibility, mechanical integrity, and biodegradability [1,2]. In addition to these design parameters, both the composition and the surface morphology of an implanted material influence protein adsorption and the subsequent cell adhesion [11–14]. Cell adhesion is important for survival, proliferation, and differentiation of multiple cell types, including osteoblasts [15,16].
While both proteins and peptides can promote cell adhesion on biomaterials, peptides have advantages over proteins in that they are smaller, cheaper, less susceptible to degradation, and can be fabricated quickly, all while containing specific target amino acid sequences [17]. Furthermore, peptide fabrication does not involve purification from an animal source, eliminating the chance of an immune response in the recipient. These advantages led to the development of peptide sequences that mimic sections of extra-cellular matrix proteins, including bone sialoprotein [18], osteopontin [19,20], fibronectin [21], statherin [20,22], elastin [23], osteonectin [24], and heparin-binding regions of fibroblast growth factor-2 [25] for improved cell adhesion, proliferation and osteoblastic differentiation. Peptide sequences, other than the common cell adhesion sequence RGD [17], involved in integrin–ligand binding identified from extracellular matrix proteins include PSHRN [26], FHRRIKA [27], and YIGSR [28]. Research investigating the effect of peptide composition and density on cell adhesion to polylactic-co-glycolic-acid (PLGA) polymers, glass, and sintered hydroxyapatite has shown that optimal peptide quantities and conformations are required to elicit a cellular response.
Hydroxyapatite (HA) can fill bone defects and accelerate osseointegration [29]. HA can also be made osteoinductive by incorporation, attachment, or adsorption of biological factors such as growth factors, DNA, proteins, and/or peptides. However, the efficacy of incorporated biological factors is limited by processing conditions (e.g. >1000 °C sintering temperatures). Thus, biomimetic formation of bone-like mineral (BLM) via incubation in supersaturated solutions consisting of selective salt ions at concentrations appropriate for heterogeneous apatite precipitation [30] is an attractive alternative. The benign processing temperature (37 °C) of bone-like mineral permits the retention of biomolecular function. In addition, the biomimetic method of forming BLM on a biodegradable polymeric substrate provides a conductive apatite layer with controlled stoichiometries [31], that provides a template for coprecipitation and adsorption of biological molecules [32,33] and that induces increased osteogenesis in vivo [34].
While BLM can increase osteogenesis in vivo, bone growth is neither uniform nor spatially controlled, suggesting the need for exogenous factors to spatiotemporally influence tissue development. Since the surface of hydroxyapatite-based materials is not easily modified with surface treatments that form functional hydroxyl-, amino-, or carboxyl-groups, such as on polymers and metals, peptides designed to adsorb to these materials are a logical alternative. The long strings of acidic amino acids found in extracellular bone matrix protein sequences (e.g. E7PRGDT derived from bone sialoprotein) increase cell attachment to apatite [35]. These peptides have been derived from bone extracellular matrix proteins [18,36], but have not been designed to preferentially attach to a biomaterial of specific composition. Furthermore, none of these peptides have been investigated on BLM.
The technique of phage display panning is effective in isolating specific peptide sequences attracted to inorganic substrates such as silver, palladium, platinum, titanium, and carbon nanotubes, in addition to plastic [37]. Phage display technology has been mostly used in identifying enzyme substrates and inhibitors, DNA and protein binding peptides, and tissue specific peptides [38]. An objective of this work was to identify peptide sequences with high affinity to apatite to design functional peptides that preferentially adsorb to apatite-based materials. A combinatorial phage display library was used to elucidate specific 12-mer linear peptide sequences that preferentially adsorb to BLM and sintered hydroxyapatite (HA). The preferential affinity of the identified peptides towards apatite-based materials was confirmed with a modified ELISA and synthetic peptide adsorption assays. Another objective of this work was to test the hypothesis that a change in surface morphology and/or the presence of carbonate within apatite will alter the adsorption of the peptides. Surface morphology changes and carbonate incorporation can alter charge distribution on the apatite surface, which could result in altered peptide adsorption. To achieve this second objective, peptide adsorption on four apatite-based materials (BLM, sintered hydroxyapatite disks, and two sintered carbonated apatite disks) was normalized to surface area (obtained via “Brunauer–Emmett–Teller” Method, or BET) to account for changes from substrate composition and/or morphology. Composition and morphology of the apatite-based materials were further characterized using X-ray diffraction (XRD), and Fourier transform Infrared Spectroscopy (FT-IR).
2. Materials and methods
Reagents and solutions were obtained for all experiments from Sigma–Aldrich unless otherwise noted.
2.1. Preparation of biomimetic films and apatite disks
A 5 w/v% 85:15 polylactic-co-glycolic acid (PLGA, Alkermes)–chloroform solution was cast on 15 mm diameter glass slides and dried overnight. The PLGA films were etched in 0.5 M NaOH for 7 min and rinsed in ddH2O. Etched films were soaked in modified simulated body fluid (mSBF) for 5 days at 37 °C [30]. The mSBF was made by dissolving the following reagents in Millipore water (ddH2O) at 25 °C and titrating to pH 6.8 using NaOH: 141 mM NaCl, 4.0 mM KCl, 0.5 mM MgSO4, 1.0 mM MgCl2, 4.2 mM NaHCO3, 5.0 mM CaCl2·2H2O, and 2.0 mM KH2PO4. The mSBF, a supersaturated solution that enables self-assembly of a carbonated apatite layer onto the polymer substrate, was changed daily to maintain supersaturation. Mineralized films were rinsed with ddH2O, dried under a hood, and desiccated until use. Hydroxyapatite disks (10 mm diameter × 4 mm thick) were pressed from powder (Plasma Biotal Ltd. P220) at 1 metric ton for 1 min and sintered at 1350 °C for 1 h (heating rate of 10 °C/min). The sintered disks were sonicated in 10 mM HCl, then in ddH2O. The BLM films and HA disks were placed in 24-well plates and soaked in ddH2O overnight prior to phage panning.
Carbonated apatite disks were made from 5.6% (CA5) and 10.5% (CA10) carbonated apatite powders (generous gift from Dr. Mary Tecklenburg). Carbonated apatite disks (8–10 mm diameter × 2–4 mm thick) were pressed from powders at 0.5 metric ton for 1 min and sintered at 1350 °C for 1 h (heating rate of 10 °C/min). Carbonated disks were also sonicated in 10 mM HCl, then in ddH2O. All sintered disks (CA5, CA10, and HA) were autoclaved prior to use. Macroscopic dimensions of all disks were measured with calipers.
2.2. Identification and sequencing of BLM and HA binding peptides
Peptide sequences with preferential binding to apatite-based materials were identified by screening the Ph.D.12™ Phage Display Library (New England Biolabs, #E8110S), consisting of ~109 different phages with 12-mer amino acid linear peptide inserts, against BLM films and HA sintered disks. A streptavidin control was run in parallel with the two apatite-based materials. The ddH2O was removed and replaced with blocking buffer (Tris–HCl Buffer Solution with 0.1% or 0.5% Tween (TBST) and bovine serum albumin (BSA)) and incubated at 4 °C for 1 h. The blocking buffer was discarded and the substrates were washed 6× with TBST (0.1%). An aliquot of the original Ph.D.12™ library containing 1011 pfu (plaque-forming units) diluted in 1 mL of TBST (0.1%) was introduced to each substrate and gently rocked (LabLine 4625) for 40 min at room temperature. Non-binding phages were discarded and the substrates were washed 10× with TBST (0.1%). The phages bound to the substrates were eluted with 1 mL of Glycine/HCl, pH 2.2, with 1 mg/mL BSA for 10 min at room temperature while being gently rocked. The eluted phages were collected and neutralized with 1 M Tris–HCl, pH 9.1.
A small amount of the phage eluate was titered to determine the number of phages bound to each substrate. The rest of the eluate was amplified with Escherichia coli (ER2738) by culturing the bacteria and phages in 20 mL LB Medium with vigorous shaking for 4.5 h at 37 °C. The culture was centrifuged for 10 min at 10,000 rpm (Sorvall SS-34) at 4 °C. The supernatant was transferred to a fresh tube, respun, and the upper 80% of the final supernatant was placed into a fresh tube and allowed to precipitate with 1/6 the volume of PEG/NaCl (20 w/v% polyethylene glycol-8000 in 2.5 M NaCl) solution. The phages were allowed to precipitate at 4 °C overnight. The PEG precipitation was spun for 15 min at 10,000 rpm at 4 °C, decanted and briefly respun. The pellet was suspended in Tris–HCl Buffered Solution (TBS) and transferred to a microcentrifuge tube, respun in a microcentrifuge for 5 min at 10,000 rpm at 4 °C. The supernatant was precipitated with 1/6 the volume of PEG/NaCl solution for 60 min on ice, recentrifuged for 10 min at 10,000 rpm at 4 °C, and the pellet was resuspended in TBS with 0.02% NaN3. A small amount of the purified phage eluate was titered to determine the number of amplified phage. This amplified and purified phage eluate was used in the next round of panning. Subsequent rounds of panning introduced at least 109 pfu as the input phage concentration. Three to four rounds of panning were performed for each substrate. The entire experiment was repeated 3 times, and a streptavidin control yielding the HPQ consensus sequence was run in parallel each time.
Titering was performed with appropriate serial dilutions for both the eluted phage and amplified phage after each round of panning. Agar plates treated with IPTG (isopropyl-b-D-thiogalactoside) and Xgal (5-bromo-4-chloro-3-indoyl-b-D-thiogalactoside) were able to visually identify correct plaques using blue/white screening. Individual plaques picked from plates having not more than ~100 plaques were amplified by infecting a log phase E. coli culture. Amplified phages from selected phage plaques were purified by PEG/NaCl precipitation. Single stranded DNA was isolated using an iodide buffer extraction, cleaned in ethanol, and resuspended in DNase/RNase free water. The DNA was sequenced via dideoxy chain termination method using a DNA sequencer (Applied Biosystems, 3730XL DNA Analyzer, UM DNA Core) with the NEB-96 gIII sequencing primer provided in the kit.
2.3. Peptide selection
A tri-fold analysis, involving a bioinformatic tool, a biological ELISA, and a computational model, was devised to determine the peptides with greatest ability to adhere to apatite-based materials. The computational model was previously published and is not discussed in the present work [39]. The bioinformatics tool REceptor LIgand Contacts (RELIC) (http://relic.bio.anl.gov/index.aspx) [40,41] was used to analyze the data from the phage display runs. The programs DNA2PRO, AAFREQ, and INFO determined peptide translation sequences from DNA code, amino acid frequency as a function of position, and information numbers for each peptide reflecting the probability of selecting individual phage by chance, respectively. The AAFREQ analysis did not show amino acid specificity by position (data not included). The most informative data were obtained using the INFO program. The INFO program calculates an information measure for each peptide identified that expresses the probability that each peptide occurs by chance (Information measure = –ln[probability]) [40]. For example, a high information measure can be indicative of an uncommon sequence with a low growth rate during amplification; whereas, a low information measure could represent a phage sequence likely to appear based on composition and/or high growth rates. The program INFO was run with subtraction of a random selection of phage from the parent library (12R, 441 random sequences from NEB 12-mer Ph.D. kit).
2.4. Phage binding ELISA
To determine the binding efficiency of the identified phage to the bone-like substrate materials, an ELISA was performed on BLM films, HA disks, PLGA films, and tissue culture polystyrene (TCPS) wells. The substrates were soaked in ddH2O overnight in 24-well plates. The ddH2O was removed and replaced with blocking buffer (TBST w/BSA) and incubated at 4 °C for 1 h. For each plate with samples, a second 24-well plate was blocked and used to prepare serial dilutions for each individual phage. The blocking buffer was discarded and the substrates were washed 6× with TBST (0.1%). In the blocked plate, dilutions of individual phage were prepared in TBST (0.1%) in the concentrations of 106, 107, 108, and 1010 pfu (n = 1 per dilution). Each phage concentration was introduced to the four substrates and incubated at room temperature for 1.5 h with gentle rocking (LabLine 4625). The non-binding phages were discarded and the substrates were washed 6× with TBST (0.1%). HRP-conjugated anti-M13 antibody (GE Healthcare, #27-9421-01) was prepared in 1:5000 ratio in blocking buffer and added to each well. The substrates were washed again 6× with TBST (0.1%). Immediately prior to detection, a 30% H2O2 (Corco, Fairless Hills, PA #1403)–ABTS (2′,2′-azino-bis(3-ethybenz-thiazoline-6-sulphonic acid) diammonium salt) solution in a 0.05 M citric acid solution at pH 4.0, was added to each well and allowed to incubate at room temperature for 50 min. For each sample, an aliquot of each well was read on an UV Spectrophotometer (Biorad SmartSpec 3000) at 410 nm.
2.5. Synthetic peptide adsorption assay
Three high affinity candidate peptides identified from the tri-fold analysis (APWHLSSQYSRT [A], VTKHLNQISQSY [V], and STLPIPHEFSRE [S]) and a positive control peptide (E7PRGDT [E] [18]) were fabricated on a Rainin Symphony synthesizer and determined as >86% pure via HPLC (UM Protein Core). For each peptide, an UV absorbance value vs. absorbance wavelength graph was generated from 200 to 300 nm (BioRad Smartspec 3000). The absorbance wavelength of 225 nm resulted in the highest UV absorbance value for the peptides A, S, and V; whereas, the absorbance wavelength of 220 nm yielded the highest UV absorbance value for the peptide E. A standard curve (peptide concentration vs. UV absorbance (220 or 225 nm)) consisting of the concentrations 0, 1, 5, 10, 25, 50, 75, 100, 250, 500, 750, and 1000 μg/mL was developed for each peptide tested. From duplicate readings per concentration, the error introduced from the detection technique was ~5%. A modified standard curve consisting of values below 100 μg/mL was used for each peptide to calculate the unknown peptide amount that adsorbed onto each material tested. Amino acid analysis was performed to verify peptide concentration for standard curves created with UV absorbance at 205 nm and 220 or 225 nm wavelengths. Standard curves for APWHLSSQYSRT, VTKHLNQISQSY, and STLPIPHEFSRE were also generated using UV Spectroscopy at 280 nm, 274 nm, and 257 nm, respectively, the absorption wavelengths for tryptophan, tyrosine, and phenylalanine. Each peptide was reconstituted in ddH2O and diluted to ~500 μg/mL in 50 mM Trizma buffer (T7818, pH = 7.5). In 24-well tissue culture dishes, BLM films, HA disks, CA5 disks, CA10 disks, and blank TCPS wells were soaked in ddH2O overnight at 4 °C (n = 5). Prior to introducing 1 mL of the peptide or buffer solutions for negative controls, plates were allowed to warm to room temperature and then the overnight solution was removed. The plates were agitated on a Titer Plate Shaker at ~80 rpm for 3 h at room temperature. The films and disks were rinsed with ddH2O and then soaked in a 10 mM HCl solution for 18 h at room temperature on the same shaker. Aliquots of the HCl solution were read on an UV spectrophotometer.
2.6. Statistics
All data are presented as mean values ± one standard deviation. Non-parametric one-way ANOVAs run with the Dunnetts T3 post-hoc test determined the effect of substrate on peptide adsorption (SPSS). The effect of peptide on substrate was also calculated with this statistical test. Statistical significance was defined as p < 0.05.
2.7. Material characterization
2.7.1. Scanning electron microscopy (SEM)
Morphologies of all apatite-based materials were examined using scanning electron microscopy (Philips XL30 FEG Scanning Electron Microscope, UM EMAL). Samples from each group (n = 2) were gold coated and examined at 10–15 kV.
2.7.2. Surface area determination (BET)
Single Point BET surface area at P/P0 = 0.1 was determined for all apatite-based materials on a surface area and porosity analyzer (Micrometrics ASAP 2020). At least 1 g of material was used and data acquisition was performed in duplicate. Peptide adsorption data for all apatite-based substrates were normalized to their respective surface areas using sample dimensions (MacroSA) and BET analysis data (MicroSA). The well area of a 24-well TCPS plate was used for both MacroSA and MicroSA normalizations.
2.7.3. Fourier transform infrared spectroscopy (FT-IR)
Characteristic peaks for apatite were detected using a Fourier Transform Infrared Spectroscopy (Spectrum BX FT-IR, Perkin Elmer). A 300:1 ratio of KBr to apatite sample was used to prepare a pellet and analyze all of the apatite-based materials (n = 3). Spectra were recorded from 400 to 4000 cm−1 and baseline corrected. The weight percentage was determined using FT-IR spectra for each substrate using two established methods [42,43] taking the peak height ratio of 1450 cm−1/569 cm−1 or 1410 cm−1/600 cm−1.
2.7.4. X-ray diffraction analysis (XRD)
XRD spectra were obtained from a representative sample of each composition using a Rigaku Miniflex X-ray Diffractometer with a fixed incidence of 4.2°. A range of 10–90° was scanned using a step size of 0.1° and a scan rate of 1°/min. All spectra were normalized to their respective maximum peaks. Hydroxyapatite from Sigma–Aldrich (#289396) was scanned as a calcium phosphate standard.
3. Results
3.1. Identification of high affinity phage
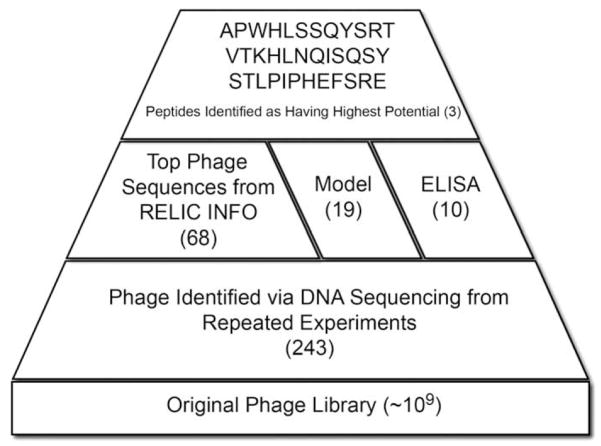
Of the ~109 phages present in the original library, 243 were identified as binders to apatite (Fig. 1). The streptavidin control yielded the HPQ consensus sequence signifying successful panning in all experiments. Within the 243 apatite-binding phages identified, 19 sequences appeared in repeated experiments after DNA sequencing. Of these 19 sequences, 17 were found in more than one experiment on the same substrate, either BLM or HA. Of these 17 phage clones, 7 phages had high affinity towards both BLM and HA (sequences in bold, Table 1). While the frequency data showing 7 sequences bound to both BLM and HA in multiple experiments provided evidence that these sequences were specific to apatite, further validation was required to verify this. Bioinformatic analysis, computational modeling, and an ELISA were therefore used to analyze the entire set or subsets of the original 243 clones identified.
Fig. 1.
Schematic displaying the tri-fold analysis approach used to identify peptides that preferentially adhere to BLM and HA.
Table 1.
Peptide Sequences Identified via Phage Display with Preferential Adsorption Towards Apatite-Based Materials.
| Identified phage sequences | # Clones | Frequency (% of total clones) |
|---|---|---|
| APWHLSSQYSRTb | 53 | 21.8 |
| IDTFYMSTMSHSa,b | 21 | 8.6 |
| ALTLHPQPLDHPb | 12 | 4.9 |
| TALATSSTYDPH | 12 | 4.9 |
| VTKHLNQISQSYa,b | 6 | 2.5 |
| WSSGMTPDTGAPa | 6 | 2.5 |
| ALSSSSNTTTRV | 4 | 1.6 |
| SSLGLTVSSIMYb | 3 | 1.2 |
| NMNTHIHKDRPPa,b | 2 | 0.8 |
| SMRLPLLSSHALa,b | 1 | 0.4 |
| VSPLSFGSPRYPa,b | 1 | 0.4 |
| WSPAPHYIMGTTa,b | 1 | 0.4 |
| STLPIPHEFSREa,b | 1 | 0.4 |
Bold: sequences occurring on both BLM and HA.
Identified as a high information clone via INFO.
Clone ran in modified ELISA.
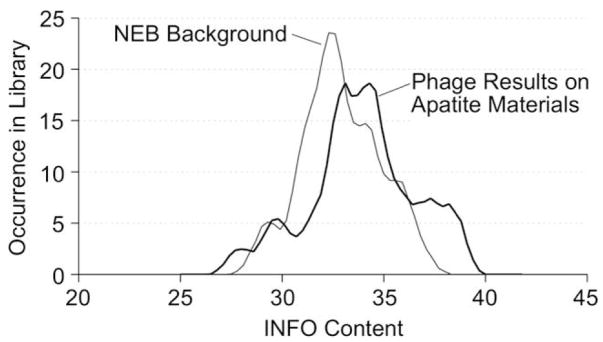
All 243 phage sequences identified as binders to BLM and/or HA were analyzed with the INFO program provided by RELIC. When the phage set identified as adsorbing to the BLM and/or HA was compared to a RELIC provided NEB Background phage set, the BLM/HA data showed a shift towards higher information content (Fig. 2). To isolate this subset of phage valued with high information content, regions of greater occurrence were partitioned from information content ranges 0–28.3, 33.0–35.8, and 36.5–40.0. These ranges were chosen as regions that exhibited higher occurrence levels within the apatite-based material phage set when compared to the NEB background. Sixty-eight sequences were partitioned and deemed “high information clones” (footnote “a” in Table 1).
Fig. 2.
INFO content values calculated in RELIC for NEB background and phage subset identified as high affinity binders to apatite-based materials. High information content correlates to a lower incidence of being selected by chance. The phage subset shows a shift towards a higher information content signifying this subset was selected based on the binding affinity towards HA and BLM.
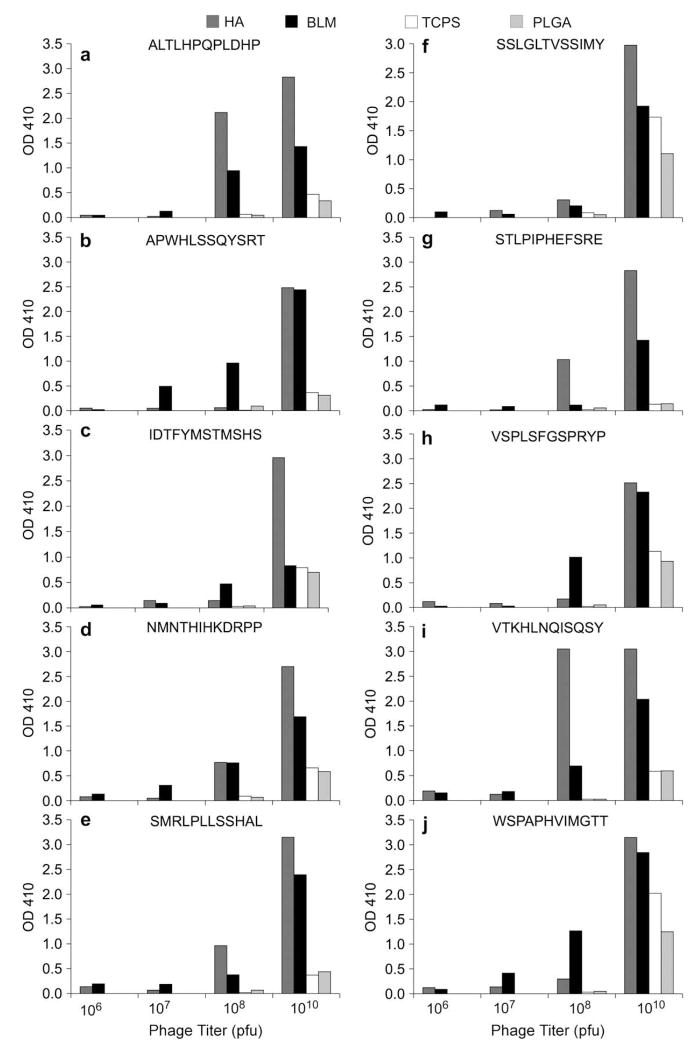
A representative set of 10 clones from the original 243 clones were chosen for ELISA analysis (footnote “b” in Table 1). Of the ten, 6 occurred as repeats, whereas 4 occurred only once. Phage concentrations of 108 and 1010 pfu resulted in positive ELISA readings for most of the peptides, with stronger positive signals for the 1010 pfu (Fig. 3). If the phage absorbance was at least 2× greater than the background absorbance, the ELISA result was considered positive. Apatite-specific phage binding was not observed for VSPLSFGSPRYP and WSPAPHVIMGTT, two of the four single-occurrence phage. Of the 6 repeats tested, four showed positive ELISA results on both apatite-based materials. For the phages with positive ELISA results, TCPS and PLGA films showed consistently low background adsorption for all concentrations tested, illustrating the specificity of the phages identified as binders to BLM and HA.
Fig. 3.
Modified ELISA results for 10 representative peptides on 4 substrates showing a higher absorbance value at OD 410 with a higher concentration of phage. PLGA and TCPS background adsorption of each phage was minimal in 6 of 10 phage sequences tested. An example of a phage with high background is shown for peptide WSPAPHVIMGTT. Positive readings on the modified ELISA support that the phages identified are indeed specific to apatite-based materials.
From the frequency data encompassing all 243 identified clones, the 19 phage sequences that appeared in repeated experiments were also analyzed via computational modeling on a hydroxyapatite lattice (data reported [39], but A, S, and V data are reported (Table 2), so the integrated role of the model along with the bio-informatics and ELISA in peptide selection can be illustrated). Peptides identified as having high peptide adsorption energies, or more negative energies, as neutral or charged peptides were deemed as having greater binding potential. A neutral peptide is assigned an overall charge of zero prior to energy minimization, whereas a charged peptide is assigned a peptide-appropriate overall charge (e.g. VTKHLNQISQSY would be assigned an overall charge of +1). At a pH 7.5, the net charges for A, S, and V were estimated as +1, −1, and +1, respectively. The isoelectric points for A, S, and V were estimated from amino acid contributions of C, D, E, H, K, R, Y, and the amino- and carboxy-termini as 9.9, 5.3, and 9.7, respectively.
Table 2.
Summary of Computational Modeling Data for Peptides A, S, and V on Hydroxyapatite (001) Surface and Step Surface.
| Adsorption on (001) plane |
Adsorption on [010] step in (001) |
||||||
|---|---|---|---|---|---|---|---|
| Peptide | Peptide adsorption energy (kcal/mol) | pI | Peptide | Peptide adsorption energy (kcal/mol) | pI | ||
| Neutral | VTKHLNQISQSY | −336.9 | 9.7 | Neutral | VTKHLNQISQSY | −705.6 | 9.7 |
| APWHLSSQYSRT | −279.6 | 9.9 | APWHLSSQYSRT | −680.8 | 9.9 | ||
| STLPIPHEFSRE | −239.3 | 5.3 | STLPIPHEFSRE | −677.3 | 5.3 | ||
| Charged | VTKHLNQISQSY | −288.2 | 9.7 | Charged | STLPIPHEFSRE | −919.2 | 5.3 |
| STLPIPHEFSRE | −263.9 | 5.3 | VTKHLNQISQSY | −449.6 | 9.7 | ||
| APWHLSSQYSRT | −247.0 | 9.9 | APWHLSSQYSRT | −368.8 | 9.9 | ||
In deciding which peptides to synthetically fabricate for the peptide adsorption experiments, the results from the RELIC analysis, modified ELISA, individual clone frequency over the three phage experiments, and computational modeling were evaluated (Fig. 1). Of the six peptides that exhibited positive ELISA signals, sequences were only considered if they were present in the top 5 ranking of any of the computational models run, eliminating NMNTHIHKDRPP and SMRLPLLSSHAL. From the remaining sequences, only those deemed as high information clones from the INFO analysis were chosen, eliminating ALTLHPQPLDHP. Three phage sequences, APWHLSS-QYSRT [A], STLPIPHEFSRE [S], and VTKHLNQISQSY [V], emerged as having the highest potential for superior binding to apatite.
3.2. Peptide adsorption on the apatite-based materials
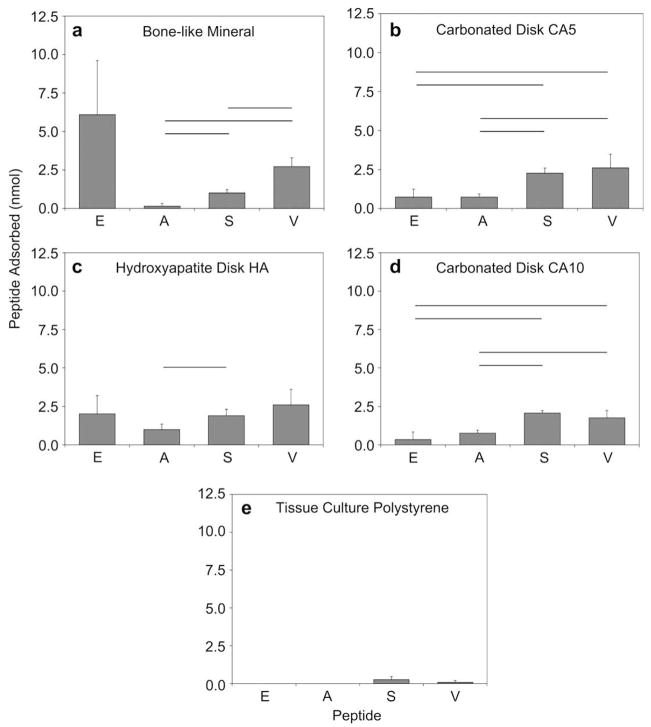
Peptides S and V adsorb in statistically greater amounts on the apatite-based materials in comparison to peptide A, (for S vs. A, BLM p = 0.001, CA5 p < 0.001, CA10 p < 0.001, HA p = 0.038; for V vs. A, BLM p = 0.006, CA5 p = 0.033, CA10 p = 0.029) (Fig. 4). Peptides S and V also have significantly higher adsorption to CA5 and CA10 disks in comparison to peptide E (for S vs. E, CA5 p = 0.005, CA10 p = 0.004; for V vs. E, CA5 p = 0.026, CA10 p = 0.010). Peptide adsorption to HA disks was less specific, as the only significant difference was between peptides A and S, p = 0.038. BLM was the only material which supported a greater adsorption of peptide V when compared to peptide S, p = 0.006. Also on BLM, peptide E exhibited a higher mean adsorption compared to peptides A, S, and V; however, the high standard deviation resulted in no statistical differences against any of the other peptides. Consistently low peptide adsorption to TCPS was found for all peptides, verifying specificity to calcium phosphate substrates. This result suggests that peptide adsorption is not only dependent on substrate composition, but also on the peptide composition.
Fig. 4.
Adsorption of peptides EEEEEEEPRGDT [E], APWHLSSQYSRT [A], STLPIPHEFSRE [S], and VTKHLNQISQSY [V] on (a) bone-like mineral (BLM), (b) sintered disks from 5.6% carbonated apatite powder (CA5), (c) sintered disks from hydroxyapatite powder (HA), (d) sintered disks from 10.5% carbonated apatite powder (CA10), and (e) tissue culture polystyrene (TCPS). Peptides S and V showed significantly increased adherence to all apatite-based materials in comparison to A (for S vs. A, BLM p = 0.001, CA5 p < 0.001, CA10 p < 0.001, HA p = 0.038; for V vs. A, BLM p = 0.006, CA5 p = 0.033, CA10 p = 0.029), and to both carbonated disks CA5 and CA10 in comparison to E (for S vs. E, CA5 p = 0.005, CA10 p = 0.004; for V vs. E, CA5 p = 0.026, CA10 p = 0.010). Differences between S and V were also only seen on bone-like mineral, p = 0.006. These results suggest that peptide adsorption is not only dependent on substrate composition, but also on the peptide composition. Horizontal bars denote statistical differences.
3.3. The effect of substrate composition and morphology on peptide adsorption
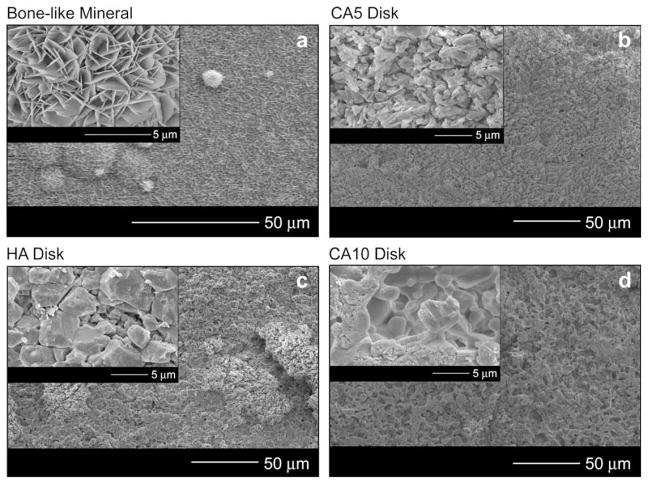
HA, CA5, and CA10 had similar surface feature sizes, whereas the bone-like mineral revealed a smaller plate-like structure with evident spherical areas of nucleation and growth (Fig. 5). Higher magnification images of the surfaces (Fig. 5 insets) illustrate the characteristic plate-like features on the bone-like mineral and more granular surface features on the HA, CA5, and CA10 disks. BET surface areas for BLM, HA, CA5, and CA10 were 121.55, 0.05, 0.11, and 0.19 m2/g, respectively.
Fig. 5.
Representative SEM images showing low and high magnification morphologies of the four apatite-based materials: (a) bone-like mineral films (BLM), (b) sintered disks from 5.6% carbonated apatite powder (CA5), (c) sintered disks from hydroxyapatite powder (HA), and (d) sintered disks from 10.5% carbonated apatite powder (CA10). These higher magnification images show that the bone-like mineral has small surface features that relate to the bone-like mineral having a 100× higher surface area than the other three apatite-based materials. (Original images taken at UM EMAL).
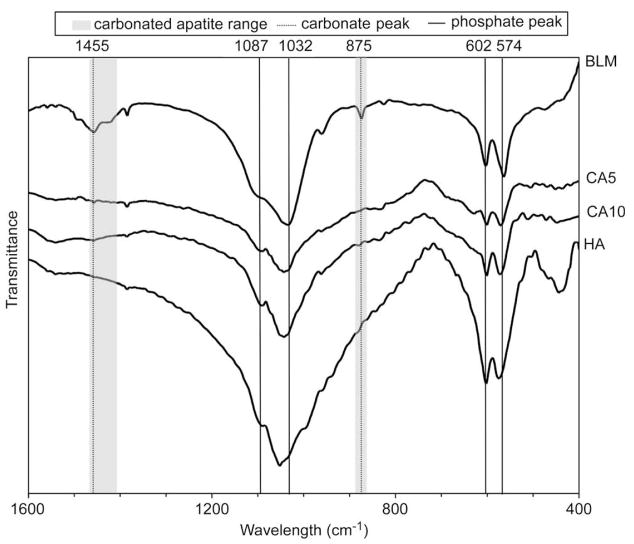
All of the apatite-based materials show phosphate adsorption bands correlating with hydroxyapatite at ca. 1087, 1032, 602, and 574 cm−1 (Fig. 6). Carbonate peaks at ca. 1455 and 875 cm−1 were detected for BLM, CA5, and CA10. The carbonate peaks in BLM indicate that a Type B carbonate substitution is occurring during formation, whereas the small but distinct peak that both CA5 and CA10 exhibited at 1455 cm−1, taken into account with the fabrication temperature of 1350 °C, is indicative of Type A carbonate substitution [44].
Fig. 6.
Representative FT-IR spectra for bone-like mineral (BLM), sintered disks from 5.6% carbonated apatite powder (CA5), sintered disks from 10.5% carbonated apatite powder (CA10), and sintered disks from hydroxyapatite powder (HA) substrates. Phosphate peaks are found for all samples at 1032, 602, and 574 cm−1 and carbonate peaks are found at 875 or 1455 cm−1 for all samples except HA.
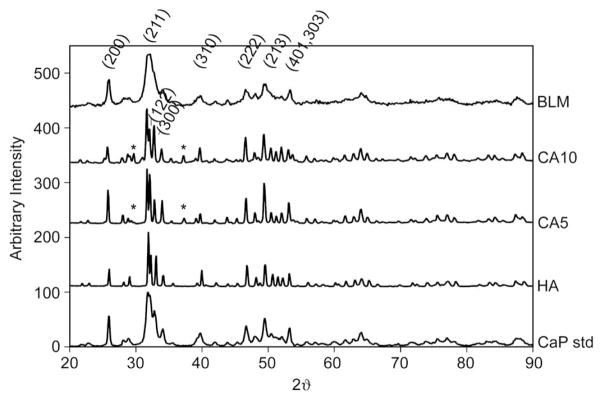
XRD characteristic peaks correlating with hydroxyapatite (PDF 9-432) existed in all of the materials at or around 2θ (crystal family plane) = 25.9° (200), 31.8° (211), 34.0° (202), 39.8° (310), 46.7° (222), 49.4° (213), and 53.1° (401, 303) (Fig. 7). Narrow, distinct peaks were observed in the HA, CA5, and CA10 spectra as a result of the high sintering temperature during disk fabrication. Additional HA peaks were evident for HA, CA5, and CA10 at or ±0.2° for 2θ (crystal family plane) = 32.2° (122) and 32.9° (300). The BLM peaks were broadened in comparison to the sintered disks, revealing that either a less crystalline apatite or a nano-crystalline phase forms over the 5 day soak in the supersaturated ion solution at 37 °C. Small 2θ peaks at 29.7° and 37.3° were found for the sintered disks pressed from carbonated apatite powders, CA5 and CA10. These small peaks are most likely calcium oxide (CaO) and/or α-TCP that did not completely revert back to hydroxyapatite during cooling [44]. These peaks are not present in the HA spectrum. This inconsistency leads to the interpretation that these peaks result from the incorporation of carbonate into the powder prior to sintering the CA5 and CA10 disks. Because the height of this peak is small in relation to the highest apatite peak, the amounts of calcium oxide and/or α-TCP present on the surface are thought to be minimal.
Fig. 7.
X-ray diffraction patterns for bone-like mineral (BLM), sintered disks from 10.5% carbonated apatite powder (CA10), sintered disks from 5.6% carbonated apatite powder (CA5), sintered disks from hydroxyapatite powder (HA), and a calcium phosphate standard (CaP Std). The bone-like mineral was less crystalline than the sintered disks; however, all samples showed characteristic hydroxyapatite peaks (labeled). The disks sintered from carbonate powder, CA10 and CA5, revealed small peaks at 2θ = 29.7° and 37.3° (*), which reflect traces of calcium oxide and/or α-TCP.
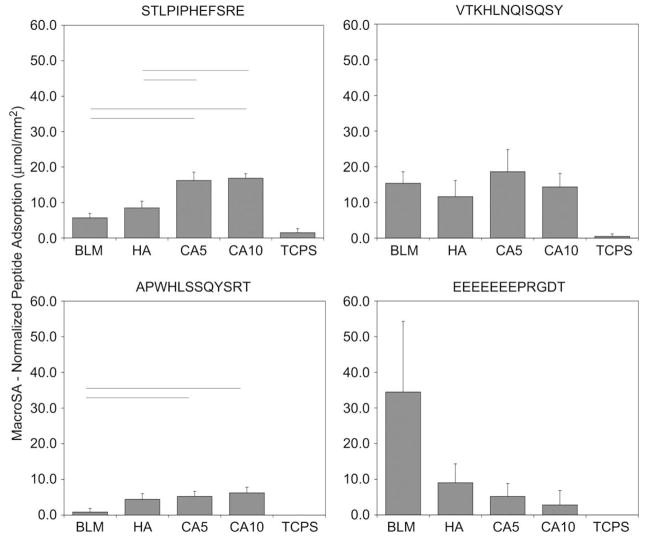
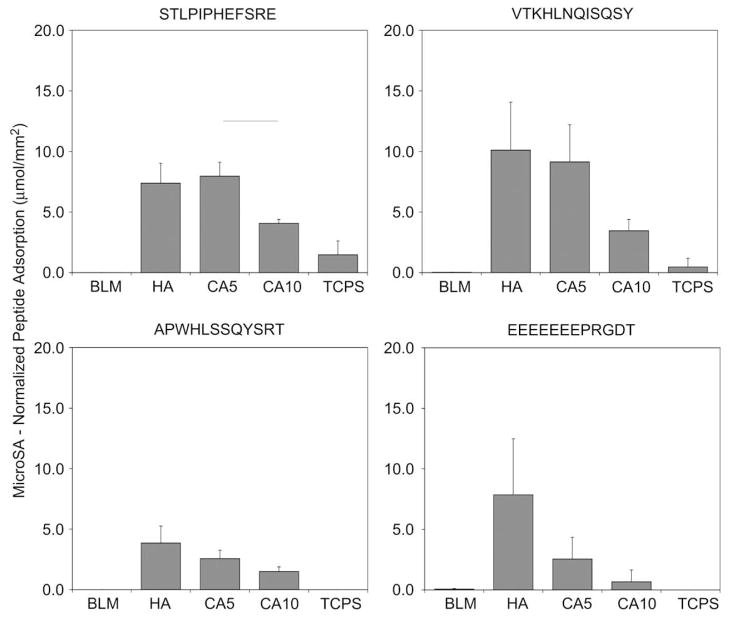
When normalized to MacroSA (Fig. 8) adsorption of A and S was significantly greater on carbonated disks (CA5, CA10) vs. BLM (for A, CA5 p = 0.006, CA10 p = 0.001; for S, CA5 p = 0.001, CA10 p < 0.001). Differences between the carbonated disks and HA disks also existed for peptide S (CA5 p = 0.004, CA10 p = 0.001). Mean normalized adsorption values were consistently higher for peptides S and V when compared to peptide E on CA5 and CA10. The high standard deviation for peptide E on BLM contributes to no significant differences found between E and the other peptides tested. Because the surface area of BLM is three orders of magnitude greater than the sintered disks, the MicroSA normalized peptide adsorption values (Fig. 9) are in the picogram range and are significantly lower than other apatite-based materials, p < 0.050. Despite CA5 and CA10 possessing similar carbonate content, the CA5 disks had a significantly higher ability to adsorb peptide S, p = 0.007. Moreover, a trend of decreased adsorption to CA10 vs. CA5 or HA was seen for A, V, and E. For MacroSA and MicroSA normalizations, all apatite groups were significantly different, p < 0.050, in comparison to TCPS for all four peptides investigated, again indicating the peptides are specific to calcium phosphate substrates.
Fig. 8.
Peptide adsorption data normalized to sample dimensions (MacroSA) show that peptides A and S have increased adsorption on CA5 and CA10 when compared with BLM (for A, CA5 p = 0.006, CA10 p = 0.001; for S, CA5 p = 0.001, CA10 p < 0.001). Differences for peptide S were also evident between HA and the carbonated disks (CA5 p = 0.004, CA10 p = 0.001). All apatite groups were significantly different, p < 0.050, in comparison to TCPS for all four peptides investigated. Horizontal lines represent statistical significance.
Fig. 9.
Peptide adsorption data normalized to BET surface area (MicroSA). Despite CA5 and CA10 possessing similar carbonate content, the CA5 disks had a higher affinity to adsorb peptide S, p = 0.007. Trends of decreased adsorption to CA10 vs. CA5 or HA were seen for peptides A, V, and E. All apatite groups were significantly different, p < 0.050, in comparison to TCPS for all four peptides investigated. Horizontal lines represent statistical significance.
4. Discussion
This work is the first to present detailed methods, to our knowledge, that identify peptide sequences with preferential adsorption towards synthetic bone-like mineral or other apatite substrates. Phage display libraries expressing combinations of linear or cyclic peptide inserts have isolated sequences with high affinity towards inert materials [37,45], cell lines cultured in vitro [46], and cell/organ targets in vivo [47]. Three 7-mer peptide sequences have been reported as specific to hydroxyapatite, but the review presenting this data lacked methods on the preparation of the hydroxyapatite substrate [48]. Through repetitive experimentation and a rigorous, tri-fold validation technique, three peptides APWHLSSQYSRT [A], STLPIPHEFSRE [S], and VTKHLNQISQSY [V] were identified to exhibit preferential adsorption towards apatite-based materials.
Many of the hydroxyapatite binding sequences identified have been from non-collagenous proteins present in the extracellular matrix of bone and/or proteins present in saliva. These proteins include, but are not limited to, bone sialoprotein, osteopontin and osteonectin [4], and a number of these proteins have a high proportion of acidic amino acids, such as glutamic and aspartic acids. Peptide E was included as a positive control to test a known HA binding peptide comprised of a high proportion of glutamic acid against the peptides derived from the phage display. In comparison to the bone sialoprotein derived peptide E, peptides A, S, and V, that did not contain strings of acidic amino acids, adsorbed in comparable, if not increased amounts on all the apatite-based materials with lower variability (Fig. 4). Therefore, phage display derived sequences offer an alternative to peptide identification compared to protein sequence searching and ultimately, contribute to peptide design that can enhance the bioactivity of implanted materials.
Adsorbed or covalently attached peptide sequences mediate cell adhesion, as functionalizing biomaterials with the ubiquitous RGD (Gly-Arg-Asp) sequence increase cell adhesion [17]. Other peptide sequences have been designed to increase cellular adhesion to titanium [8], to contain cyclic RGD sequences [49], or to be modeled from the heparin-binding domain [25,50]. However, these cell adhesive sequences were not designed based on preferential affinity towards a specific material. Weakly attached surface molecules can be redistributed by cells attempting to attach to a surface [51], and softer substrates can cause the disorganization of actin filaments [51]. In both cases, effective formation of focal adhesions will not occur, demonstrating the need to consider both the substrate material and cell source in the peptide design. Furthermore, a cell-influencing sequence can be linked to the substrate-targeting peptides to specify cell behavior, such as attachment.
Not only do the surface peptides need to withstand contractile forces from attached cells, but also the peptide concentration presented on the material surface should be optimized for the specific cellular behavior targeted for control [5]. Adjusting the initial peptide concentration introduced to an apatite-based substrate can provide the specific surface density required for the promotion of cell adhesion. To ensure appropriate surface densities are present for the cells, the amount of peptide adsorbed to the surface must be quantified, as the peptide adsorption can change depending on substrate charge and morphology. The presented work highlights the adsorption capacity through quantification of the adsorption of the high affinity peptides to various apatite-based materials. Combining UV spectrophotometry with amino acid analysis allowed quantification of adsorbed peptide in concentrations >0.5 μg/mL. This range is similar to commercial protein assays, such as Coomassie Blue and MicroBCA, which often depend on the presence of specific amino acids and are more sensitive to presence of buffers and chemicals.
Normalization of peptide adsorbed to MacroSA (Fig. 8) provided insight towards the amount of peptide that would be required to coat apatite-based implants. Slight carbonate incorporation in the CA5 and CA10 groups increased adsorption of peptides S and A compared with HA. The MacroSA normalization data also indicate that a sintered apatite coating on an implant may be more capable of adsorbing S and A peptides compared with a BLM coating. However, the additional factor of peptide surface density must be considered, as extracellular matrix mineralization varies with adhesive peptide surface densities [27].
When normalized to MicroSA (Fig. 9), slight changes in carbonate percentage did not influence peptide adsorption. This result does not rule out that a larger change in carbonate incorporation could affect peptide adsorption. The range of carbonate present in the studied materials did vary; however, at the same morphological scale (CA5 and CA10 vs. HA), the carbonate differences were minimal (~2 wt%). CA5 adsorbed significantly higher amounts of peptide S compared with CA10. A trend of decreased adsorption to CA10 in comparison to CA5 and HA was shown across all peptides, indicating that a morphological difference, not a compositional difference, was evident. The morphology difference between CA5 and CA10 could change the charge distribution on the apatite surface, promoting peptide adsorption. Better control of the surface morphology accompanied by larger differences in carbonate content would determine if carbonate incorporation has an effect on peptide adsorption.
Both material composition and surface morphology influence protein adsorption [12,52] and the resulting cell adhesion and proliferation [53]. Calcium phosphates exhibit the ability to adsorb more protein than other materials, which has led to the suggestion that Ca2+ and present themselves as preferential binding sites for proteins [54]. Changes in surface morphology (e.g. pore size) also affect protein adsorption. Microporosity increases protein adsorption, but decreases cellular differentiation [13]. Taken together, these studies suggest that with a certain composition and morphology, calcium phosphates allow a preferred charge pattern that promotes protein binding. Identifying peptides that preferentially adsorb to apatite-based materials without disrupting surface charge distribution may prove beneficial in vivo by allowing normal protein adsorption to transpire unaffected by the presence of peptide. Despite BLM being classified as a calcium phosphate with a distinct nano-morphology, BLM has not been investigated in biomolecular adsorption studies. The presented data parallel literature discussing factors that affect protein adsorption while offering a new perspective on peptide adsorption to BLM.
Phage display combinatorial libraries offer potential in isolating binding peptide sequences from a vast library, making otherwise unfathomable experiments possible [55]. However, phage libraries can express an unequal representation of each amino acid within the library, are not able to include every peptide combination possible, and oftentimes omit important post-translational modifications such as phosphorylation of serine residues in the parent library. The results from the presented phage display experiments did not preferentially isolate peptide sequences with long strings of acidic amino acids, as the peptides isolated from the phage display had an average overall charge of ±1. This unexpected result could have occurred because poly-acidic peptide sequences were not initially present in the parent library, or because the identified sequences bound with greater affinity than poly-acidic peptide sequences. These data suggest that an overwhelming portion of acidic amino acids need not be present for a peptide to have favorable binding to apatite, but the peptide may only need a small amount of alternating charge that can positively interact with the charge density present on the apatite surface. Nonetheless, the array of peptides presented in these libraries provided the opportunity to identify sequences specific to the apatite-based materials. Furthermore, the A, S, and V peptides identified in this study contain amino acid residues that can be phosphorylated. Phosphorylating the serine, threonine, and/or tyrosine residues on peptides A, S, and V would increase the negative charge on these peptide sequences, possibly enhancing their adsorption to apatite. It is possible with phage display techniques to identify high adsorbing, non-modified sequences that could achieve even higher affinity after synthetic phosphorylation.
While the lack of controlled peptide adsorption was taken into account when analyzing carbonate effect on adsorption by normalizing peptide adsorbed by its respective surface area (micro or macro), we do not report saturation curves, and cannot conclusively say that peptide saturation was attained. However, increased fluorescence intensities with increased concentrations of FITC-labeled E7RGD peptide on HA disks show a strong intensity visible at 100 μg/mL [56]. Since the initial peptide concentrations used in these studies were ~500 μg/mL, it is assumed that saturation was achieved.
The work presented in this manuscript provides a stimulus to explore alternative methods to engineer current tissue repair constructs. For example, apatite-binding peptides can be combined with soft tissue or ligament binding recognition sequences to help re-establish tissue interfaces in the tooth-gum and bone-ligament systems, respectively. A multitude of factors influence not only protein or peptide adsorption, but other biomolecules such as growth factors and DNA as well. Therefore, it is imperative to fully characterize the substrate material to better understand how the material will regulate biomolecular adsorption or incorporation prior to the addition of cells.
5. Conclusions
Three peptides, APWHLSSQYSRT [A], STLPIPHEFSRE [S], and VTKHLNQISQSY [V], were identified as high affinity binders specific to apatite-based materials. The morphology of the apatite-based materials influenced synthetic peptide adsorption more than slight changes (2%) in carbonate content. Because the identified peptides have preferential adsorption to apatite, their use can now be investigated in a variety of bone-based research including bone and dentin tissue engineering, tendon and ligament repair, and enamel formation.
Acknowledgments
This work is supported by the National Institutes of Heath: DE 015411 and T32-DE07057. Thank you to Dr. Udo Becker and Dr. Subhashis Biswas for their work on developing the hydroxyapatite force field and initial validation of the model. We would also like to thank Alisha Diggs for help with disk preparation, Dr. Henriette Remmer and Jen Mahoney for their support that the UM Peptide Core, UM EMAL for their continued support of scanning electron microscopy, and Dr. Xiaoyin Chen for his expertise in the BET analysis. Thanks to Dr. Mary Tecklenberg (Central Michigan University) for supplying the carbonated apatite powders.
References
- 1.Yaszemski MJ, Payne RG, Hayes WC, Langer R, Mikos AG. Evolution of bone transplantation: molecular, cellular and tissue strategies to engineer human bone. Biomaterials. 1996;17:175–85. doi: 10.1016/0142-9612(96)85762-0. [DOI] [PubMed] [Google Scholar]
- 2.Wiesmann HP, Joos U, Meyer U. Biological and biophysical principles in extracorporal bone tissue engineering. Part II. Int J Oral Maxillofac Surg. 2004;33:523–30. doi: 10.1016/j.ijom.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg SE, Aghaloo TL, Cunningham LL., Jr Role of tissue engineering in oral and maxillofacial reconstruction: findings of the 2005 AAOMS research summit. J Oral Maxillofac Surg. 2005;63:1418–25. doi: 10.1016/j.joms.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Shin H, Jo S, Mikos AG. Biomimetic materials for tissue engineering. Biomaterials. 2003;24:4353–64. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]
- 5.Hubbell JA. Bioactive biomaterials. Curr Opin Biotechnol. 1999;10:123–9. doi: 10.1016/s0958-1669(99)80021-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen GP, Ushida T, Tateishi T. A hybrid sponge of poly(DL-lactic-co-glycolic acid), collagen and apatite. Bioceramics. 2000;192:753–6. [Google Scholar]
- 7.Mikos AG, Thorsen AJ, Czerwonka LA, Bao Y, Langer R, Winslow DN, et al. Preparation and characterization of poly(L-lactic acid) foams. Polymer. 1994;35:1068–77. [Google Scholar]
- 8.Ferris DM, Moodie GD, Dimond PM, Giorani CWD, Ehrlich MG, Valentini RF. RGD-coated titanium implants stimulate increased bone formation in vivo. Biomaterials. 1999;20:2323–31. doi: 10.1016/s0142-9612(99)00161-1. [DOI] [PubMed] [Google Scholar]
- 9.Barrere F, van der Valk CM, Dalmeijer RA, van Blitterswijk CA, de Groot K, Layrolle P. In vitro and in vivo degradation of biomimetic octacalcium phosphate and carbonate apatite coatings on titanium implants. J Biomed Mater Res. 2003;64A:378–87. doi: 10.1002/jbm.a.10291. [DOI] [PubMed] [Google Scholar]
- 10.de Groot K, Wolke JG, Jansen JA. Calcium phosphate coatings for medical implants. Proc Inst Mech Eng H. 1998;212:137–47. doi: 10.1243/0954411981533917. [DOI] [PubMed] [Google Scholar]
- 11.Garcia AJ, Ducheyne P, Boettiger D. Effect of surface reaction stage on fibronectin-mediated adhesion of osteoblast-like cells to bioactive glass. J Biomed Mater Res. 1998;40:48–56. doi: 10.1002/(sici)1097-4636(199804)40:1<48::aid-jbm6>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 12.Kilpadi KL, Chang PL, Bellis SL. Hydroxylapatite binds more serum proteins, purified integrins, and osteoblast precursor cells than titanium or steel. J Biomed Mater Res. 2001;57:258–67. doi: 10.1002/1097-4636(200111)57:2<258::aid-jbm1166>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 13.Anselme K, Linez P, Bigerelle M, Le Maguer D, Le Maguer A, Hardouin P, et al. The relative influence of the topography and chemistry of TiAl6V4 surfaces on osteoblastic cell behaviour. Biomaterials. 2000;21:1567–77. doi: 10.1016/s0142-9612(00)00042-9. [DOI] [PubMed] [Google Scholar]
- 14.Redey SA, Razzouk S, Rey C, Bernache-Assollant D, Leroy G, Nardin M, et al. Osteoblast adhesion and activity on synthetic hydroxyapatite, carbonated hydroxyapatite, and natural calcium carbonate: relationship to surface energies. J Biomed Mater Res. 1999;45:140–7. doi: 10.1002/(sici)1097-4636(199905)45:2<140::aid-jbm9>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 15.Menko AS, Boettiger D. Occupation of the extracellular matrix receptor, integrin, is a control point for myogenic differentiation. Cell. 1987;51:51–7. doi: 10.1016/0092-8674(87)90009-2. [DOI] [PubMed] [Google Scholar]
- 16.Damsky CH. Extracellular matrix–integrin interactions in osteoblast function and tissue remodeling. Bone. 1999;25:95–6. doi: 10.1016/s8756-3282(99)00106-4. [DOI] [PubMed] [Google Scholar]
- 17.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24:4385–415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 18.Fujisawa R, Mizuno M, Nodasaka Y, Kuboki Y. Attachment of osteoblastic cells to hydroxyapatite crystals by a synthetic peptide (Glu7-Pro-Arg-Gly-Asp-Thr) containing two functional sequences of bone sialoprotein. Matrix Biol. 1997;16:21–8. doi: 10.1016/s0945-053x(97)90113-x. [DOI] [PubMed] [Google Scholar]
- 19.Shin H, Zygourakis K, Farach-Carson MC, Yaszemski MJ, Mikos AG. Attachment, proliferation, and migration of marrow stromal osteoblasts cultured on biomimetic hydrogels modified with an osteopontin-derived peptide. Biomaterials. 2004;25:895–906. doi: 10.1016/s0142-9612(03)00602-1. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert M, Shaw WJ, Long JR, Nelson K, Drobny GP, Giachelli CM, et al. Chimeric peptides of statherin and osteopontin that bind hydroxyapatite and mediate cell adhesion. J Biol Chem. 2000;275:16213–8. doi: 10.1074/jbc.M001773200. [DOI] [PubMed] [Google Scholar]
- 21.Huebsch JB, Fields GB, Triebes TG, Mooradian DL. Photoreactive analog of peptide FN-C/H-V from the carboxy-terminal heparin-binding domains of fibronectin supports endothelial cell adhesion and spreading on biomaterial surfaces. J Biomed Mater Res. 1996;31:555–67. doi: 10.1002/(SICI)1097-4636(199608)31:4<555::AID-JBM16>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 22.Shaw WJ, Long JR, Dindot JL, Campbell AA, Stayton PS, Drobny GP. Determination of statherin N-terminal peptide conformation on hydroxyapatite crystals. J Am Chem Soc. 2000;122:1709–16. [Google Scholar]
- 23.Simionescu A, Philips K, Vyavahare N. Elastin-derived peptides and TGF-beta1 induce osteogenic responses in smooth muscle cells. Biochem Biophys Res Commun. 2005;334:524–32. doi: 10.1016/j.bbrc.2005.06.119. [DOI] [PubMed] [Google Scholar]
- 24.Fujisawa R, Wada Y, Nodasaka Y, Kuboki Y. Acidic amino acid-rich sequences as binding sites of osteonectin to hydroxyapatite crystals. Biochim Biophys Acta. 1996;1292:53–60. doi: 10.1016/0167-4838(95)00190-5. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Choo J, Choi Y, Lee K, Min D, Pi S, et al. Characterization of the surface immobilized synthetic heparin-binding domain derived from human fibroblast growth factor-2 and its effect on osteoblast differentiation. J Biomed Mater Res Part A. 2007;83A:970–9. doi: 10.1002/jbm.a.31351. [DOI] [PubMed] [Google Scholar]
- 26.Benoit DS, Anseth KS. The effect on osteoblast function of colocalized RGD and PHSRN epitopes on PEG surfaces. Biomaterials. 2005;26:5209–20. doi: 10.1016/j.biomaterials.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 27.Rezania A, Healy KE. Biomimetic peptide surfaces that regulate adhesion, spreading, cytoskeletal organization, and mineralization of the matrix deposited by osteoblast-like cells. Biotechnol Prog. 1999;15:19–32. doi: 10.1021/bp980083b. [DOI] [PubMed] [Google Scholar]
- 28.Massia SP, Hubbell JA. Covalent surface immobilization of Arg-Gly-Asp- and Tyr-Ile-Gly-Ser-Arg-containing peptides to obtain well-defined cell-adhesive substrates. Anal Biochem. 1990;187:292–301. doi: 10.1016/0003-2697(90)90459-m. [DOI] [PubMed] [Google Scholar]
- 29.Geesink RG. Osteoconductive coatings for total joint arthroplasty. Clin Orthop Relat Res. 2002;395:53–65. doi: 10.1097/00003086-200202000-00007. [DOI] [PubMed] [Google Scholar]
- 30.Kokubo T. Apatite formation on organic polymers by a biomimetic process. Eur J Solid State Inorg Chem. 1995;32:819–27. [Google Scholar]
- 31.Shin K, Jayasuriya AC, Kohn DH. Effect of ionic activity products on the structure and composition of mineral self assembled on three-dimensional poly(lactide-co-glycolide) scaffolds. J Biomed Mater Res A. 2007;83:1076–86. doi: 10.1002/jbm.a.31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luong LN, Hong SI, Patel RJ, Outslay ME, Kohn DH. Spatial control of protein within biomimetically nucleated mineral. Biomaterials. 2006;27:1175–86. doi: 10.1016/j.biomaterials.2005.07.043. [DOI] [PubMed] [Google Scholar]
- 33.Segvich S, Smith HC, Luong LN, Kohn DH. Uniform deposition of protein incorporated mineral layer on three-dimensional porous polymer scaffolds. J Biomed Mater Res B Appl Biomater. 2008;84:340–9. doi: 10.1002/jbm.b.30877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohn DH, Shin K, Hong SI, Jayasuriya AC, Leonova EV, Rossello RA, et al. Self-assembled mineral scaffolds as model systems for biomineralization and tissue engineering. Proceedings of the eigth international conference on the chemistry and biology of mineralized tissues; 2005. pp. 216–19. [Google Scholar]
- 35.Itoh D, Yoneda S, Kuroda S, Kondo H, Umezawa A, Ohya K, et al. Enhancement of osteogenesis on hydroxyapatite surface coated with synthetic peptide (EEEEEEEPRGDT) in vitro. J Biomed Mater Res. 2002;62:292–8. doi: 10.1002/jbm.10338. [DOI] [PubMed] [Google Scholar]
- 36.Dettin M, Conconi MT, Gambaretto R, Bagno A, Di Bello C, Menti AM, et al. Effect of synthetic peptides on osteoblast adhesion. Biomaterials. 2005;26:4507–15. doi: 10.1016/j.biomaterials.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 37.Kriplani U, Kay BK. Selecting peptides for use in nanoscale materials using phage-displayed combinatorial peptide libraries. Curr Opin Biotechnol. 2005;16:470–5. doi: 10.1016/j.copbio.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Ladner RC, Sato AK, Gorzelany J, de Souza M. Phage display-derived peptides as therapeutic alternatives to antibodies. Drug Discov Today. 2004;9:525–9. doi: 10.1016/S1359-6446(04)03104-6. [DOI] [PubMed] [Google Scholar]
- 39.Segvich S, Biswas S, Becker U, Kohn D. Identification of peptides with targeted adhesion to bone-like mineral via phage display and computational modeling. Cell Tissue Org. 2008 doi: 10.1159/000151380. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makowski L, Soares A. Estimating the diversity of peptide populations from limited sequence data. Bioinformatics. 2003;19:483–9. doi: 10.1093/bioinformatics/btg013. [DOI] [PubMed] [Google Scholar]
- 41.Mandava S, Makowski L, Devarapalli S, Uzubell J, Rodi DJ. RELIC – a bioinformatics server for combinatorial peptide analysis and identification of protein–ligand interaction sites. Proteomics. 2004;4:1439–60. doi: 10.1002/pmic.200300680. [DOI] [PubMed] [Google Scholar]
- 42.Featherstone JD, Pearson S, LeGeros RZ. An infrared method for quantification of carbonate in carbonated apatites. Caries Res. 1984;18:63–6. doi: 10.1159/000260749. [DOI] [PubMed] [Google Scholar]
- 43.LeGeros RZ. Calcium phosphates in oral biology and medicine. Karger; New York: 1991. p. 201. [PubMed] [Google Scholar]
- 44.Liao CJ, Lin FH, Chen KS, Sun JS. Thermal decomposition and reconstruction of hydroxyapatite in air atmosphere. Biomed Sci Instrum. 1999;35:99–104. [PubMed] [Google Scholar]
- 45.Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature. 2000;405:665–8. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- 46.Samoylova TI, Ahmed BY, Vodyanoy V, Morrison NE, Samoylov AM, Globa LP, et al. Targeting peptides for microglia identified via phage display. J Neuroimmunol. 2002;127:13–21. doi: 10.1016/s0165-5728(02)00071-1. [DOI] [PubMed] [Google Scholar]
- 47.Pasqualini R, Koivunen E, Ruoslahti E. A peptide isolated from phage display libraries is a structural and functional mimic of an RGD-binding site on integrins. J Cell Biol. 1995;130:1189–96. doi: 10.1083/jcb.130.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamerler C, Sarikaya M. Molecular biomimetics: utilizing nature’s molecular ways in practical engineering. Acta Biomater. 2007;3:289–99. doi: 10.1016/j.actbio.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 49.Pallu S, Bareille R, Dard M, Kessler H, Jonczyk A, Vernizeau M, et al. A cyclo peptide activates signaling events and promotes growth and the production of the bone matrix. Peptides. 2003;24:1349–57. doi: 10.1016/j.peptides.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Dee KC, Andersen TT, Bizios R. Design and function of novel osteoblast-adhesive peptides for chemical modification of biomaterials. J Biomed Mater Res. 1998;40:371–7. doi: 10.1002/(sici)1097-4636(19980605)40:3<371::aid-jbm5>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 51.Pelham RJ, Wang YL. Cell locomotion and focal adhesions are regulated by the mechanical properties of the substrate. Biol Bull. 1998;194:348–9. doi: 10.2307/1543109. [DOI] [PubMed] [Google Scholar]
- 52.Rosengren A, Pavlovic E, Oscarsson S, Krajewski A, Ravaglioli A, Piancastelli A. Plasma protein adsorption pattern on characterized ceramic biomaterials. Biomaterials. 2002;23:1237–47. doi: 10.1016/s0142-9612(01)00244-7. [DOI] [PubMed] [Google Scholar]
- 53.McFarland CD, Thomas CH, DeFilippis C, Steele JG, Healy KE. Protein adsorption and cell attachment to patterned surfaces. J Biomed Mater Res. 2000;49:200–10. doi: 10.1002/(sici)1097-4636(200002)49:2<200::aid-jbm7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 54.Zeng HT, Chittur KK, Lacefield WR. Analysis of bovine serum albumin adsorption on calcium phosphate and titanium surfaces. Biomaterials. 1999;20:377–84. doi: 10.1016/s0142-9612(98)00184-7. [DOI] [PubMed] [Google Scholar]
- 55.Rodi DJ, Makowski L. Phage-display technology – finding a needle in a vast molecular haystack. Curr Opin Biotechnol. 1999;10:87–93. doi: 10.1016/s0958-1669(99)80016-0. [DOI] [PubMed] [Google Scholar]
- 56.Sawyer AA, Weeks DM, Kelpke SS, McCracken MS, Bellis SL. The effect of the addition of a polyglutamate motif to RGD on peptide tethering to hydroxyapatite and the promotion of mesenchymal stem cell adhesion. Biomaterials. 2005;26:7046–56. doi: 10.1016/j.biomaterials.2005.05.006. [DOI] [PubMed] [Google Scholar]