SUMMARY
The basic-helix-loop-helix transcription factor HeyL is expressed at high levels by neural crest progenitor cells (NCPs) that give rise to neurons and glia in dorsal root ganglia (DRG). Since HeyL expression was observed in these NCPs during the period of neurogenesis, we generated HeyL null mutants to help examine the factor’s role in ganglion neuronal specification. Homozygous null mutation of HeyL reduced the number of TrkC+ neurons in DRG at birth including the subpopulation that expresses the ETS transcription factor ER81. Conversely, null mutation of the Hey paralog, Hey1, increased the number of TrkC+ neurons. Null mutation of HeyL increased expression of the Hey paralogs Hey1 and Hey2, suggesting that HeyL normally inhibits their expression. Double null mutation of both Hey1 and HeyL rescued TrkC+ neuron numbers to control levels. Thus, the balance between HeyL and Hey1 expression regulates the differentiation of a subpopulation of TrkC+ neurons in the DRG.
Keywords: dorsal root ganglion, TrkC, HeyL, Hey1, differentiation
INTRODUCTION
The basic-helix-loop-helix (bHLH) transcription factors, hairy and Enhancer of split are transcriptional repressors in Drosophila that mediate some of the effects of the Notch signaling pathway on neural stem cell differentiation (Klambt et al., 1989; Lecourtois and Schweisguth, 1995). Related proteins in mammals include the Hes and Hey families of repressors (Akazawa et al., 1992; Leimeister et al., 1999; Nakagawa et al., 1999; Sasai et al., 1992; Thomas and Rathjen, 1992). The Hes family has seven members, Hes1-7, of which Hes1, Hes3, Hes5 and Hes6 are expressed in the developing nervous system (Akazawa et al., 1992; Sakagami et al., 1994; Sasai et al., 1992). In the mammalian central nervous system (CNS) Hes1 and Hes5 act as repressors of proneuronal genes like Mash1 thereby preventing differentiation (Akazawa et al., 1995; Castella et al., 1999; de la Pompa et al., 1997; Ishibashi et al., 1995; Ishibashi et al., 1994; Jarriault et al., 1995; Nakamura et al., 2000). Hes3 has two isoforms, one of which has repressor activity similar to Hes1 while the other lacks repressor activity (Hirata et al., 2000; Hirata et al., 2001). Hes6 represses Hes1 activity and conversely promotes neuronal differentiation (Bae et al., 2000; Gratton et al., 2003; Koyano-Nakagawa et al., 2000). Thus, different Hes proteins play contrasting roles in neural differentiation.
Hey1, Hey2 and HeyL constitute the mammalian Hey (also known as CHF, Hesr, Herp, Hrt) family that, like the Hes proteins, act downstream of the Notch signaling pathway (Iso et al., 2001; Leimeister et al., 1999; Nakagawa et al., 2000; Nakagawa et al., 1999; Steidl et al., 2000). The Hey genes play an important role in the development of the cardiovascular system and loss of Hey function causes defects in vascular specification, septation and valve formation (Donovan et al., 2002; Fischer et al., 2004; Fischer et al., 2007; Gessler et al., 2002; Kokubo et al., 2005; Kokubo et al., 2004; Kokubo et al., 2007; Sakata et al., 2002; Xin et al., 2007). In the developing brain, overexpression of Hey1 and Hey2 prevents neural precursors from differentiating during the neurogenic period and causes glial differentiation by antagonizing neuronal bHLH factors later in development (Sakamoto et al., 2003). HeyL overexpression in the developing retina increases rod differentiation whereas Hey2 increases gliogenesis (Satow et al., 2001). Hey proteins are expressed at high levels in the developing DRG and trigeminal ganglia (Leimeister et al., 1999; Leimeister et al., 2000; Nakagawa et al., 1999) but their roles in sensory ganglion development and neuronal specification are unknown.
Sensory ganglion neurons are divided into three major subpopulations based on their expression of the neurotrophin receptors and the type of sensory information they convey - TrkA+ which convey information from thermoreceptors and nociceptors, TrkB+ that convey mechanosensory information and TrkC+ that are proprioceptive (Snider, 1994). Here, we report that mice carrying a targeted mutation in the HeyL gene have decreased numbers of large TrkC+ neurons in the DRG. Moreover, HeyL in the DRG inhibits Hey1 expression and mice mutant for Hey1 alone have increased numbers of TrkC+ neurons in the DRG suggesting that Hey1 and HeyL play antagonistic roles in TrkC+ neuron differentiation.
MATERIALS AND METHODS
Generation and Maintenance of Mouse lines
The targeting construct was generated from the BAC clone RP23-25K8 (CHORI) containing the HeyL locus by recombineering as previously described (Liu et al., 2003). All vectors and bacterial strains used for creating the targeting vector were a kind gift from Dr. Neal Copeland. In the targeting construct loxP sequences were introduced such that they flanked the exons 2-5 of the HeyL gene and a neomycin resistance cassette (pGK-neo) flanked by FRT sites was introduced downstream of the HeyL gene (Fig. 1A). The targeted region was flanked both upstream and downstream by 3kb homology regions for recombination. The targeting construct was electroporated into HM1 mouse ES cells and the neomycin resistant colonies were screened by southern hybridization with a ∼700b probe downstream of the 3′ homology region. Targeted ES cells were used by the Northwestern University Transgenic and Targeted Mutagenesis Facility to generate chimeras that transmitted the targeted (HeyLfn) allele to their progeny. EIIa-cre mice (Jackson Labs) and ROSAFlp1/Flp1 (Jackson Labs) were bred to HeyLfn/+ and the progeny were backcrossed to WT C57BL6 mice to generate HeyL+/- and HeyLfx/+ mice, respectively. Genotyping was done by polymerase chain reaction (PCR) using a combination of four primers a-5′-GAGCTCTCTCATGCATGTTCTGCG-3′, b-5′-GGCTAGTTTCTCAGAGCTGAAGGATC-3′, c-5′-CCCCCATACACACACCCTGTTATTCTAG-3′ and d-5′-GAAGCTCTAAGAGGAAATGCTGGGG-3′ as shown in Fig. 1C. The Hey1+/- mice were a kind gift from Dr. Manfred Gessler and have been described previously (Fischer et al., 2004). Hey1-/-; HeyL+/- and Hey1+/-; HeyL-/- mice were bred to obtain the control (Hey1+/-; HeyL+/-), Hey1 mutant (Hey1-/-; HeyL+/-), HeyL mutant (Hey1+/-; HeyL-/-) and Hey1/L double mutant (Hey1-/-; HeyL-/-) mice.
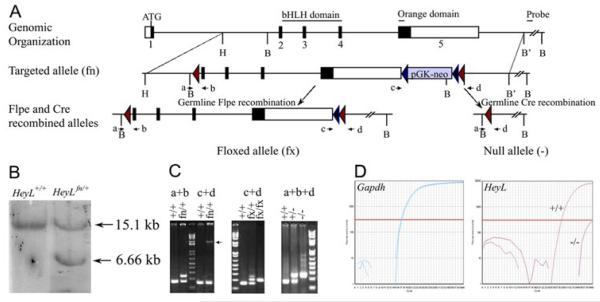
Fig.-1. Targeting and genotyping strategies for HeyL knock out mice.
(A) Exons 2-5 of the HeyL locus that code for the bHLH and Orange domains were flanked by loxP (red triangles) sequences in the targeted HeyLfn allele after homologous recombination. A neomycin resistance cassette flanked by FRT sequences (blue triangles) was also introduced 3′ of the Exon5. The targeted allele in the presence of germline flpe or cre recombinases generated the HeyLfx and HeyL- alleles respectively. (B) Southern hybridization of genomic DNA after BssSI restriction digestion with the 3′ probe in (A) detected a smaller 6.66kb fragment showing the presence of HeyLfn allele. (C) Genotyping primers a-d in (A) were used to distinguish between the different alleles. Note the ∼2kb band (arrow) for the HeyLfn allele when the genomic DNA is amplified with primers c and d. (D) Quantitative real time PCR from trigeminal mRNA with GAPDH as internal control showed the lack of HeyL mRNA in HeyL-/-. B-BssSI, H-HinfI, B’-BslI.
Southern hybridization and Quantitative Real Time PCR
Genomic DNA was extracted from ES cell colonies and 10μg of genomic DNA samples were digested overnight at 37 °C with BssSI (New England Biolabs). The digested DNA was run on a 0.5% agarose gel, denatured using a solution containing 1.5M NaCl and 0.5 N NaOH before being neutralized in 1M Tris (pH 7.4). The DNA fragments were then transferred from the gel to a nitrocellulose membrane overnight by capillary transfer. The membrane was baked at 80 °C for 2 hours to fix the DNA on the filter. DIG labeled probe was generated from genomic DNA by PCR using DIG DNA Labeling and Detection Kit (Roche) and hybridization and detection was done as per manufacturer’s instructions. RNA was extracted from the tissue using RNA Aqueous 4 PCR Kit (Ambion) following manufacturer’s instructions. cDNA was prepared from the RNA samples with OligodT primers using Thermoscript RT (Invitrogen) following manufacturer’s instructions. Real time PCR was done on a Mastercycler ep realplex system (Eppendorf) with SYBR green (Applied Biosystems) PCR mix.
Immunohistochemistry
Mouse embryo, P0 mouse spinal columns with DRGs or P0 mouse thoracic segment with DRGs and supathetic ganglia were fixed in 4% paraformaldehyde for 2hrs and cryoprotected in 30% sucrose solution overnight. The dehydrated tissue samples were frozen in OCT (Tissue Tek) before being cryosectioned on a CM3050S cryostat (Leica) at 10μm thickness for immunohistochemistry. The tissue sections were incubated with primary antibody overnight at 4 °C in a solution containing 1% BSA, 0.25% Triton-X100 and 2% goat serum. For some antibodies antigen retrieval was done by boiling the sections in 10mM Sodium Citrate Solution pH 6 for 10 minutes before incubation with primary antibodies. After washes the tissue sections were incubated with Alexa Fluor conjugated goat secondary antibodies (1:500) (Invitrogen) in a solution containing 1%BSA, 0.25% Triton-X and DAPI nuclear stain (Invitrogen) for 1hr before a second set of washes and being mounted in ProLong Gold antifade reagent (Invitrogen). The primary antibodies used were Rabbit anti-ER81 (1:1000; Covance), Rabbit anti-p75 (1:500; Dr. Louis Reichardt), Rabbit anti-Parvalbumin (1:1000; Swant), Rabbit anti-Sox10 (1: 1000; Abcam), Mouse anti-Substance P (1:500; R&D), Rabbit anti-TrkA (1:500; Dr. Louis Reichardt), Goat anti-TrkB (1:100; R&D), Goat anti-TrkC (1:50; R&D), Mouse anti-Tyrosine hydroxylase (1:500; Sigma).
RNA in situ hybridization
Whole mount RNA in situ hybridization was done as described previously (Avilion et al., 2000). For in situ hybridization on tissue sections 20 μm sections were cut on the cryostat after 4% formaldehyde fixation and 30% sucrose cryoprotection. Plasmid vectors containing cDNA for Hey1, Hey2 and HeyL were a kind gift from Dr. Gessler and have been used previously to generate antisense RNA probes (Leimeister et al., 1999; Leimeister et al., 2000). DIG labeled probes were prepared from the cDNA by PCR using DIG RNA Labeling Kit (Roche) as per manufacturer’s instructions. The sections were hybridized in hybridization solution (50% Formamide, 5X SSC, 1% SDS, 500μg/ml tRNA, 200μg/ml acetylated BSA, 50μg/ml heparin) overnight at 68 °C after which three stringency washes were done for 45 minutes in a wash solution (50% Formamide, 2X SSC, 1% SDS) at 68 °C. The DIG labeling was detected using a peroxidase conjugated anti DIG antibody (1:2500, Roche) and developed in NBT/BCIP solution (Roche).
Cell counts and Statistical Analysis
Sections from six different thoracic DRGs (T1-T6) per animal or the adjacent sympathetic ganglia were immunostained and images were taken using an Axiovert epifluorescence microscope (Zeiss). Cells were counted from four of the largest sections from each DRG, trigeminal and sympathetic ganglia manually using NIH ImageJ software. The area of each DRG, trigeminal and sympathetic ganglia section was also measured using ImageJ and the average number of cells per square millimeter (sq. mm) was calculated for each animal. At least three animals of each genotype were used to get the average number of cells per sq. mm. The examiner was not aware of the genotypes of the specimens while acquiring images and counting the cells. In experiments with only two conditions Student’s t-test assuming unequal variance was used while for multi condition experiments one-way ANOVA followed by Tukey’s post hoc text was used to determine statistical significance. All data is presented as mean±SEM.
RESULTS
Generation of HeyL mutant mice
To generate a HeyL mutant a targeting vector was constructed that contained exons2-5 of the HeyL locus flanked by loxP sequences (Fig.1A). The targeting construct was introduced into mouse ES cells and homologous recombination was identified by southern hybridization with a probe downstream of the targeted region of the HeyL locus after BssSI digestion of the genomic DNA (Fig. 1B). ES cell lines containing the targeted HeyL locus were used to generate chimeras that transmitted the HeyLfn allele to their offspring. Mice carrying the HeyLfn allele were crossed to mice constitutively expressing Flpe recombinase (ROSA-Flpe) and cre recombinase (EIIa-cre) to generate offspring that had the neomycin cassette excised (fx) and the HeyL exons2-5 excised (null) respectively (Fig. 1A). Genotyping primers a,b,c and d in different combinations were used to assess recombination and to distinguish the HeyL WT, fn, fx and null alleles (Fig. 1C). The HeyL transcript was detectable only at background levels in the homozygous null mutant (HeyL-/-) trigeminal ganglion using quantitative RT-PCR (Fig. 1D).
The HeyL-/- animals were viable and were obtained at Mendelian frequencies. Loss of HeyL did not cause any overt effect on the size or weight of the animals and HeyL-/- mice survived to adulthood and were fertile. Since, the HeyL-/- mice were viable, the HeyLfx/fx animals were not used in the analysis and will not be discussed. Thus, functional HeyL is not required for the postnatal survival in mice.
Expression of HeyL in the developing nervous system
In the developing mouse embryo HeyL is expressed in the somites, peripheral nervous system, smooth muscle of all arteries and the mesenchymal tissue surrounding the neural tube (Leimeister et al., 2000; Nakagawa et al., 1999). To investigate which cells in developing peripheral ganglia express HeyL we performed RNA in situ hybridization. In E10.5 embryos HeyL mRNA was expressed in dorsal root, trigeminal, and sympathetic ganglia and their nerve roots (Fig. 2A). This expression pattern was very similar to the expression pattern of the SRY family transcription factor Sox10 that is expressed in migrating neural crest progenitor cells (Britsch et al., 2001; Kuhlbrodt et al., 1998). For closer examination we probed DRGs from adjacent tissue sections for HeyL mRNA and Sox10 protein and found that the HeyL expression (Fig. 2B) was identical to that of Sox10 (Fig. 2C) in the DRG suggesting they are coexpressed. HeyL expression was also detected in the mesenchyme surrounding the neural tube (Fig. 2B) as has been previously described (Nakagawa et al., 1999).
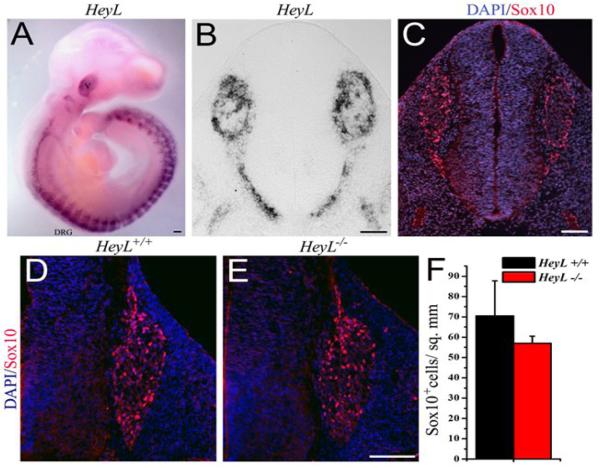
Fig.-2. HeyL is expressed in migrating neural crest cells and is not required for their migration.
(A) Whole mount in situ hybridization for HeyL mRNA on an E10.5 WT embryo showed strong expression in the trigeminal and DRG. (B-C) mRNA in situ hybridization for HeyL (B) and immunohistochemistry for Sox10 (red) and nuclear stain DAPI (blue) (C) on adjacent tissue sections showed identical expression patterns in the DRGs and the spinal nerve roots at E10.5. (D-F) There was no change in the number of Sox10+ cells in the DRGs of E11.5 HeyL-/- (E) compared to WT controls (D). Scale bar, 100 μm
Since, HeyL was expressed in migrating neural crest cells we investigated if loss of HeyL function affected the neural crest progenitor cell population in the embryonic DRG. Comparison of E11.5 HeyL-/- DRGs with their wild type (WT) littermates did not show any change in the number of Sox10 expressing neural crest progenitor cells (Fig. 2D, E and F). We also did not find an increase in cell death in the DRG at E11.5, assessed by cleaved caspase3 immunohistochemistry, in the absence of HeyL (data not shown). Together, these observations suggest that HeyL is expressed in migrating neural crest progenitor cells but is not required for their migration or survival.
Effects of loss of HeyL on Hey family paralogs
The Hey genes are all expressed in the peripheral nervous system during development (Leimeister et al., 1999; Leimeister et al., 2000; Nakagawa et al., 1999; Steidl et al., 2000). To examine the effects of loss of HeyL on the expression pattern of the other family members we performed in situ hybridization with probes for Hey1, Hey2 and HeyL mRNA on DRG and trigeminal ganglia of HeyL-/- and WT littermates at E11.5. As expected HeyL mRNA was not detected in the Hey-/- DRG and trigeminal ganglia (Fig. 3B and D) but could be detected in the WT (Fig. 3A and C).
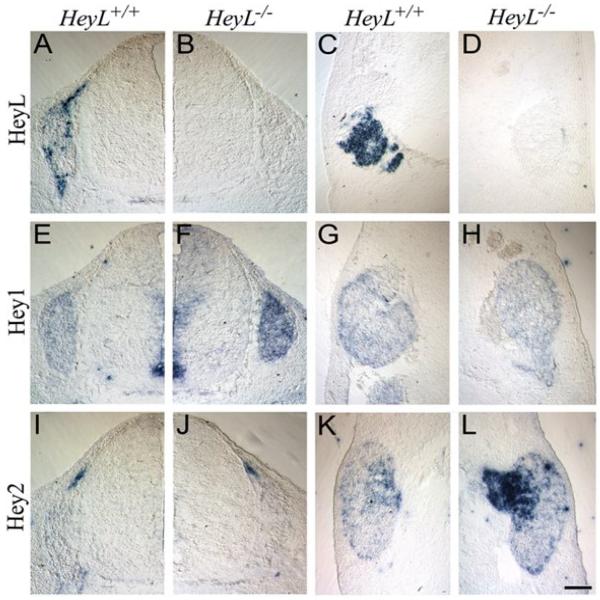
Fig.-3. Loss of HeyL leads to elevated expression of Hey paralogs in the DRG and trigeminal.
(A-D) HeyL expression was detected by in situ hybridization in the E11.5 WT DRG (A) and trigeminal ganglion (C) but not in the HeyL-/- DRG (B) or trigeminal (D). (E-H) Hey1 expression was detected in the WT DRG and spinal cord floor plate (E) and the trigeminal ganglion (G) in the WT. There was an increased level of Hey1 expression in the DRG (F) but not in the trigeminal ganglion (G) in the HeyL-/-. (I-L) Hey2 expression was detected in the boundary cap cells in the WT (I) and was unaffected in the HeyL-/- (J). Hey2 was also detected in the trigeminal ganglion in the WT (K) and an increased level of expression was seen in the trigeminal root of the HeyL-/-(L). Scale bar, 100 μm
In the ventricular zone of the spinal cord and the floor plate cells, regions where HeyL mRNA was not detected (Fig. 3A, B), Hey1 mRNA expression was not affected in HeyL-/- compared to WT mice (Fig. 3E, F). However, in the DRG where Hey1 and HeyL are both expressed, Hey1 mRNA was upregulated compared to the WT (Fig. 3E, F) suggesting that HeyL may inhibit the expression of Hey1 in the DRG. In the trigeminal ganglion Hey1 transcript was detected only at low levels in the WT (Fig. 3G) and the expression was not different in the HeyL-/- mice (Fig. 3H). Interestingly, Hey2 mRNA was detected only in the boundary cap cells (Fig. 3I) that give rise to glia in the spinal nerve roots and some neurons and glia in the DRG (Maro et al., 2004). Loss of HeyL however, did not affect the expression of Hey2 in the boundary cap cells (Fig. 3J). In the trigeminal ganglia there was diffuse expression of Hey2 both in the ganglion and the nerve root (Fig. 3K) and the expression was upregulated in the nerve root in the HeyL-/- mice (Fig. 3L) suggesting that HeyL might be involved in the inhibition of Hey2 in the trigeminal ganglion. Thus, loss of HeyL expression causes an upregulation of Hey paralogs in regions of overlapping expression.
Role of HeyL in differentiation of sensory neurons in the DRG
Soon after neurogenesis begins in the DRG (E11.5) almost all DRG neurons express TrkC and many co-express TrkA and TrkB. At this stage neurotrophin receptor expression does not correspond to the functional subtypes of the DRG neurons (White et al., 1996). Later in development neurotrophin receptor expression becomes specific to the subtype of DRG neuron with the majority expressing TrkA by postnatal day 0 (P0) (Carroll et al., 1992; Mu et al., 1993). Since HeyL is expressed at high levels during the neurogenic period in the DRG we examined if HeyL has a role in the differentiation of different subtypes of neurons.
We immunostained P0 thoracic DRGs with antibodies against TrkA, TrkB, TrkC and the pan neuronal marker NeuN. The total numbers of NeuN+ neurons in the HeyL-/- DRGs were comparable to the WT littermates (Supplementary Fig.1A-B, Fig.4G). On analyzing the specific subtypes, TrkA+ and TrkB+ neuronal populations in the HeyL-/- compared to WT DRG were similar in numbers though the TrkB+ neurons showed a statistically insignificant reduction (Fig. 4A-D and H-I). However, the large TrkC+ neurons showed a significant ∼19% reduction (260.6±7.5 cells/mm2 for WT vs 210.4±11.4 for HeyL-/-; p-value<0.05) in the HeyL-/- DRGs. The TrkA+ neurons are distributed throughout the DRG, while majority of the TrkB+ and TrkC+ neurons are usually found at the periphery (Mu et al., 1993). There was no overall change in the distribution of the TrkA+, TrkB+ or TrkC+ neurons in the DRG of the HeyL-/- animal (Fig. 4A-E).
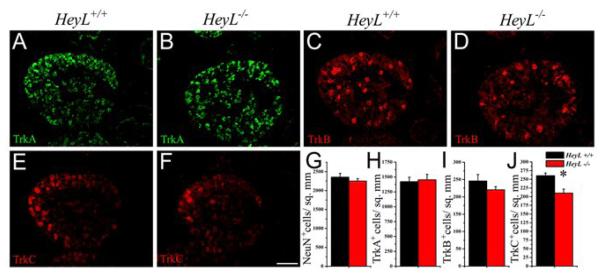
Fig.-4. Loss of HeyL reduces the number of TrkC expressing neurons in the DRG.
(A-D) DRG neuronal populations in WT that immunostained for TrkA (green) (A) and TrkB (red) (C) were not affected by the loss of HeyL (B, D). (E-F) TrkC+ (red) neurons were significantly reduced in the HeyL-/- (F) compared to the WT (E). (G-J) Quantification of the neurons (NeuN) (G), TrkA (H), TrkB (I) and TrkC (J) in WT and HeyL-/-. *p< 0.05 by Student’s t-test. Scale bar 100 μm
Another region of high HeyL expression is the developing trigeminal ganglion (Fig. 2A). We also examined the neuronal subtypes in trigeminal ganglia but did not find any significant changes in the numbers of different sensory neuron subtypes (Supplementary Fig. 1C-N). Hence, loss of HeyL caused a reduction in the number of proprioceptive large TrkC+ neurons specifically in the DRG without affecting the nociceptive and thermoceptive TrkA+ neurons or the mechanosensory TrkB+ neurons numbers or their distribution.
Effects of loss of HeyL on subtypes of TrkC expressing neurons
Since only a small subpopulation of the TrkC+ neurons was lost in the HeyL mutant we investigated the expression of proteins that characterize different subsets of TrkC+ DRG neurons. The calcium binding protein parvalbumin (PV) is expressed in a large subpopulation of the proprioceptive neurons and is related to their functional properties (Antal et al., 1990; Carr et al., 1989a; Carr et al., 1989b; Ringstedt et al., 1997). We immunostained thoracic DRGs from HeyL mutants and WT mice and found a small decrease in the number of PV immunoreactive cells in the mutant (Fig. 5A-C).
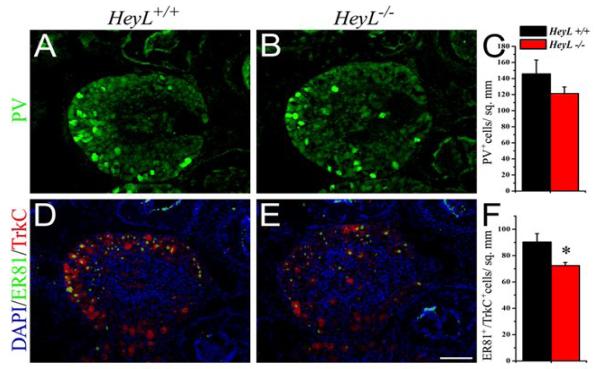
Fig.-5. Mutation in HeyL reduces the ER81 expressing subpopulation of TrkC+ neurons.
(A-C) The PV+ (green) subpopulation of TrkC+ neurons showed a reduction in the HeyL-/- that is not statistically significant (C). (D-F) The TrkC+ (red) neurons in the DRG that expressed ER81 (green) showed a significant reduction in the HeyL-/- (E-F) compared to the WT (D). *p< 0.05 by Student’s t-test Scale bar, 100 μm
A subset of TrkC+ muscle sensory afferent neurons expresses the ETS gene ER81 (Lin et al., 1998). Mice carrying mutations in ER81 exhibit limb ataxia and abnormal flexor-extensor posturing of their limbs and proprioceptive Ia afferents are lost (Arber et al., 2000). Since ER81 is expressed in DRG neurons outside the TrkC+ population (Arber et al., 2000) we immunostained DRGs for both TrkC and ER81 to examine effects on the ER81+ subpopulation. Interestingly, the HeyL mutant DRGs exhibited a significant ∼20% reduction (90.4±6.4 cells/mm2 for WT vs 72.5±2.3 for HeyL-/-; p-value<0.05) in the number of ER81+/TrkC+ neurons compared to the WT (Fig. 5D-F). This is similar to the reduction in the total number of TrkC+ neurons in the DRG of HeyL mutants (Fig. 4J) and suggests that both ER81+ and ER81- subpopulations of TrkC+ neurons are affected in the HeyL mutant. Together, these observations suggest a role for HeyL in the differentiation of multiple subpopulations of TrkC+ neurons in the DRG.
Contrasting roles of Hey1 and HeyL in sensory neuron differentiation
Our previous results demonstrate an upregulation of Hey1 expression and a reduction in the TrkC+ neurons in the DRGs of the HeyL-/- mice. This leads to two contrasting possibilities, either that loss of HeyL expression causes a compensatory increase in the expression of Hey1 that attenuated the reduction in TrkC+ neurons or that HeyL inhibition of Hey1 is required for the differentiation of a subpopulation of TrkC+ neurons. To investigate these possibilities we generated mice carrying mutations in both Hey1 and HeyL and examined neuronal differentiation in the thoracic DRGs. We hypothesized that if the upregulation of Hey1 is compensatory in nature, Hey1 and HeyL should have similar roles in the differentiation process. Hey1 mutants (Hey1-/-; HeyL+/-) would then be expected to have a reduction in the number of TrkC+ neurons, and there might be a more pronounced effect on the differentiation of TrkC+ neurons in Hey1/L double mutant (Hey1-/-; HeyL-/-) mice. Alternatively, if Hey1 repression by HeyL plays a role in the differentiation of TrkC+ neurons in the DRG, then the Hey1 null mice should have an increased number of TrkC+ neurons compared to the controls.
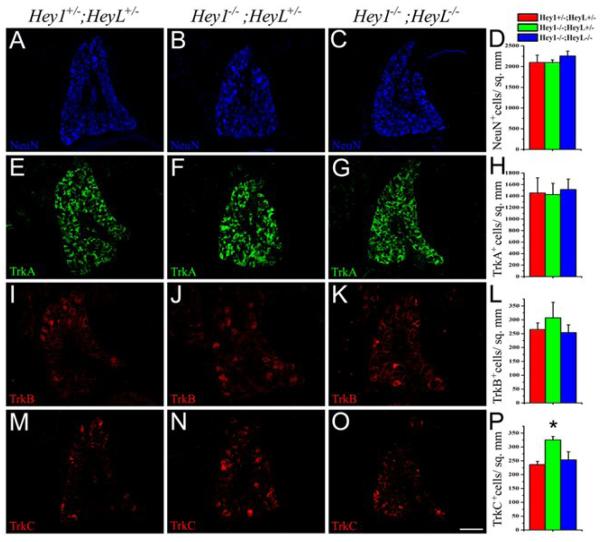
The total numbers of neurons in the DRGs of Hey1 mutant and Hey1/2 double mutant mice assessed by NeuN immunostaining were comparable to control DRGs (Fig. 6A-D). The TrkA+ and TrkB+ neuronal populations in the DRGs of Hey1 or Hey1/L double mutant mice also did not show any changes compared to that in control DRGs (Fig. 6 E-L). Examination of other neuronal subtypes in the DRG based on the expression of the peptide Substance P and the low affinity neurotrophin receptor p75 did not reveal any significant changes in the Hey1, HeyL or Hey1/L double mutants compared to the control animals (Supplementary Fig. 2A-J). Surprisingly, however, the numbers of TrkC+ neurons in the DRG of Hey1 mutants were significantly increased and there were no significant changes in the Hey1/L double (325.3±12.6 for Hey-/- vs 253.41±29.2 for Hey1-/-; HeyL-/- vs 236.5±11.4 for Hey1+/-; HeyL+/-; p-value<0.05) mutants compared to the control DRGs (Fig. 6 M-P). These results together with the observed increase in Hey1 expression in the HeyL-/- ganglia suggest that Hey1 and HeyL have antagonistic roles in the differentiation of TrkC+ neurons in the DRG.
Fig.-6. Loss of Hey1 increases the number of TrkC expressing neurons in the DRG.
(A-C) Thoracic DRGs immunostained for NeuN (blue), TrkA (green) (E-G), TrkB (red) (I-K) and TrkC(red) (M-O) in the Hey1 mutant (Hey1-/-;HeyL+/-) (B, F, J, N), Hey1/L double mutants (Hey1-/-;HeyL-/-) (C, G, K, O) and control (Hey1+/-; HeyL+/-) littermates (A, E, I, M). Note that the TrkC+ subpopulation in the Hey1 mutants (N) is increased and in double mutants (O) it is comparable to that of the control (M) at P0. (D,H,L,P) Quantification of the neurons (NeuN) (D), TrkA (H), TrkB (L) and TrkC (P) immunoreactive cells in the control, Hey1 mutant and Hey1/L double mutant DRGs. *p<0.05 by ANOVA. Scale bar, 100 μm
DISCUSSION
We have created a mouse that carries mutation in the bHLH transcription factor HeyL. The mutation in the HeyL locus deletes sequences that code for all the conserved domains in the HeyL protein including the bHLH, orange and the C-terminal TEIGAF and YHSW motifs and presumably any function performed by the HeyL protein is lost in the mutant animals. The growth, survival and fertility of the mutant mice were not affected by the absence of HeyL function. The only other reported HeyL mutation in mice in which only the bHLH and orange domains were affected (Fischer et al., 2007) also did not affect survival. Thus, the additional loss of the conserved C-terminal motifs in the HeyL mutant reported here did not cause any differential effect on survival.
Comparison of the expression of HeyL and Sox10 indicates that HeyL is expressed by neural crest progenitor cells during the neurogenic period in dorsal root, sympathetic and trigeminal ganglia. Expression of HeyL in neural crest progenitors indicates two contrasting possibilities for the role of HeyL. First, HeyL might be required for the migration and maintenance of the neural crest progenitor population. However the normal number of Sox10+ cells in the DRG indicates that this is not the case. Alternatively, HeyL might play a role in the differentiation of neural crest progenitors into mature cell types. The reduction in the numbers of large TrkC+ neurons in the DRGs of HeyL null mice at P0 provides evidence for the second possibility. The TrkB+ neuronal population also showed a modest reduction in the in the HeyL mutants DRG that was not statistically significant. The loss of HeyL did not change the total number of neurons in the DRG of the mutants though there was a trend towards a reduction. However the TrkB+ and TrkC+ subpopulations of neurons in the DRG together constitute less than 20% of the total neurons (Mu et al., 1993). Thus the changes in the size of these small populations in the HeyL mutant might not be sufficient to detectably change total neuron numbers given the threshold of detection using cell counting methods. The neurons of the sympathetic ganglia, which are derived from the neural crest cells and also expressed HeyL, were not significantly affected in the HeyL mutant mice as assessed by tyrosine hydroxylase immunostaining (Supplementary Fig. 2K-O). Other mature cell types arising from Sox10+ neural crest cells include Schwann cells and satellite glia in the PNS (Britsch et al., 2001). We did not find any gross changes in Schwann cell differentiation after loss of HeyL, but we cannot exclude the possibility of subtle molecular changes in Schwann cells that might indirectly affect the differentiation of DRG neurons.
We find that a null mutation in HeyL increases levels of Hey1 in the DRG and Hey2 in the trigeminal ganglion. Analogy to the Hes family of factors suggests that HeyL directly represses expression of Hey1 and Hey2 by binding to their promoters. In fact direct repression by HeyL of in vitro reporter expression from the Hey2 promoter has been reported (Nakagawa et al., 2000) and our results provide the first evidence of this mechanism in vivo. HeyL shows a weaker response to Notch signaling than Hey1 or Hey2 (Maier and Gessler, 2000; Nakagawa et al., 2000). This suggests that HeyL may also be regulated by other signaling molecules and indeed HeyL expression is upregulated in neural progenitor cells exposed to bone morphogenetic protein4 (BMP4) (Jalali et al., unpublished). Interestingly, expression of Hey1 can be synergistically induced in the presence of Notch and BMP signaling in endothelial cells (Itoh et al., 2004). BMP signaling regulates the expression of TrkC in neurons in the PNS (Chalazonitis, 2004; Chalazonitis et al., 2004; Zhang et al., 1998). The reduction in TrkC+ neurons in the DRG when HeyL function is lost suggests that HeyL might be an effecter of BMP mediated TrkC expression in a subset of TrkC+ neurons.
The Runt family transcription factor Runx3 is expressed in TrkC+ neurons and has been implicated in the cell fate specification in the DRG and projection of sensory afferents (Inoue et al., 2007; Inoue et al., 2002; Kramer et al., 2006; Levanon et al., 2002). However Runx3 does not mediate the expression of TrkC in response to BMPs (Inoue et al., 2007). Recently, it has been shown that there is a late born subpopulation of DRG neurons that expresses TrkC independent of Runx3 and that does not express PV (Nakamura et al., 2008). However the HeyL mutant does not exhibit a significant reduction in PV+ neurons indicating the reduction in TrkC+ neurons is predominantly from the TrkC+/PV- subpopulation. In addition, the HeyL mutant has similar numbers of TrkC+ neurons at an earlier time point DRG at E11.5 (data not shown) and does not exhibit limb ataxia like the Runx3 mutant (Inoue et al., 2002; Levanon et al., 2002). This suggests that HeyL possibly regulates the expression of TrkC in the Runx3 independent subpopulation of TrkC+ neurons.
The neuronal subtypes in the trigeminal ganglion assessed by the expression of different Trk receptors were not affected in the HeyL mutant. The POU domain transcription factor Brn-3a has been implicated in playing a role in the expression of Trk receptors in the trigeminal ganglia (Huang et al., 1999; McEvilly et al., 1996; Xiang et al., 1996). In Brn-3a mutant mice Trk receptor expression and neuronal numbers are reduced in the trigeminal but remain unaffected in the DRGs (Huang et al., 1999; Xiang et al., 1996). Interestingly, though TrkC expression is almost absent in the trigeminal ganglion of the Brn-3a mutants, PV expression appears normal (Huang et al., 1999). The maintenance of PV expression with a reduction in TrkC expression in Brn-3a mutant trigeminal is similar to what we observe in the DRG in HeyL mutants. Hence, both Brn-3a and HeyL may function in a similar manner downstream of signals that regulate the expression of TrkC in the sensory neurons in the trigeminal and DRG respectively.
In the cardiovascular development, Hey2 plays the crucial role with loss of Hey2 alone causing developmental defects (Donovan et al., 2002; Gessler et al., 2002; Kokubo et al., 2004; Sakata et al., 2002) and the HeyL and Hey1 single mutant mice appear normal (Fischer et al., 2004; Fischer et al., 2007). There is functional redundancy between Hey1 and HeyL as the Hey1/L double mutant exhibits cardiac defects and partial perinatal lethality (Fischer et al., 2007). In contrast, our results suggest, Hey1 and HeyL play opposing roles in the differentiation of TrkC+ neurons in the DRG. Moreover, overexpression of HeyL in neural progenitors causes neuronal differentiation (Jalali et al., unpublished) which is antagonistic to that of the known proglial function of Hey1 and Hey2 in the nervous system (Sakamoto et al., 2003; Satow et al., 2001). The functional differences can be attributed partly to the structural differences between HeyL and other Hey proteins. Structurally, HeyL diverges further from the other two Hey proteins in both the orange domain and the YXPW motif (Leimeister et al., 1999; Nakagawa et al., 1999; Steidl et al., 2000), and HeyL lacks the groucho binding domain present in the other two factors. Hes6 which belongs to the closely related Hes family of proteins also diverges in function from other Hes proteins like Hes1, Hes3 or Hes5 in the nervous system (Bae et al., 2000; Koyano-Nakagawa et al., 2000) whereas Hes6 functions similarly to other Hes proteins in non-neural tissues (Cossins et al., 2002). The difference in the functions of Hes6 and other Hes proteins is partly due to a shorter loop in the bHLH sequence (Bae et al., 2000; Koyano-Nakagawa et al., 2000; Pissarra et al., 2000; Vasiliauskas and Stern, 2000). Thus, the divergent structure and function of HeyL in the peripheral nervous system are similar to those of Hes6 in the central nervous system.
Supplementary Material
Acknowledgements
We thank Neal Copeland for providing us the recombineering vectors and bacterial strains, Louis Reichardt for the TrkA and p75 antibodies and Manfred Gessler for providing us the Hey1, Hey2 and HeyL expression constructs and the Hey1 mutant mouse line. This project was supported by NIH grants NS 20778-24and NS 20013-25
Footnotes
Disclosures: There are no conflicts of interest or other relevant issues to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- Akazawa C, Sasai Y, Nakanishi S, Kageyama R. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J Biol Chem. 1992;267:21879–85. [PubMed] [Google Scholar]
- Antal M, Freund TF, Polgar E. Calcium-binding proteins, parvalbumin- and calbindin-D 28k-immunoreactive neurons in the rat spinal cord and dorsal root ganglia: a light and electron microscopic study. J Comp Neurol. 1990;295:467–84. doi: 10.1002/cne.902950310. [DOI] [PubMed] [Google Scholar]
- Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–98. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- Avilion AA, Bell DM, Lovell-Badge R. Micro-capillary tube in situ hybridisation: a novel method for processing small individual samples. Genesis. 2000;27:76–80. [PubMed] [Google Scholar]
- Bae S, Bessho Y, Hojo M, Kageyama R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development. 2000;127:2933–43. doi: 10.1242/dev.127.13.2933. [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr PA, Yamamoto T, Karmy G, Baimbridge KG, Nagy JI. Analysis of parvalbumin and calbindin D28k-immunoreactive neurons in dorsal root ganglia of rat in relation to their cytochrome oxidase and carbonic anhydrase content. Neuroscience. 1989a;33:363–71. doi: 10.1016/0306-4522(89)90216-9. [DOI] [PubMed] [Google Scholar]
- Carr PA, Yamamoto T, Karmy G, Baimbridge KG, Nagy JI. Parvalbumin is highly colocalized with calbindin D28k and rarely with calcitonin gene-related peptide in dorsal root ganglia neurons of rat. Brain Res. 1989b;497:163–70. doi: 10.1016/0006-8993(89)90983-9. [DOI] [PubMed] [Google Scholar]
- Carroll SL, Silos-Santiago I, Frese SE, Ruit KG, Milbrandt J, Snider WD. Dorsal root ganglion neurons expressing trk are selectively sensitive to NGF deprivation in utero. Neuron. 1992;9:779–88. doi: 10.1016/0896-6273(92)90040-k. [DOI] [PubMed] [Google Scholar]
- Castella P, Wagner JA, Caudy M. Regulation of hippocampal neuronal differentiation by the basic helix-loop-helix transcription factors HES-1 and MASH-1. J Neurosci Res. 1999;56:229–40. doi: 10.1002/(SICI)1097-4547(19990501)56:3<229::AID-JNR2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Chalazonitis A. Neurotrophin-3 in the development of the enteric nervous system. Prog Brain Res. 2004;146:243–63. doi: 10.1016/S0079-6123(03)46016-0. [DOI] [PubMed] [Google Scholar]
- Chalazonitis A, D’Autreaux F, Guha U, Pham TD, Faure C, Chen JJ, Roman D, Kan L, Rothman TP, Kessler JA, Gershon MD. Bone morphogenetic protein-2 and -4 limit the number of enteric neurons but promote development of a TrkC-expressing neurotrophin-3-dependent subset. J Neurosci. 2004;24:4266–82. doi: 10.1523/JNEUROSCI.3688-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossins J, Vernon AE, Zhang Y, Philpott A, Jones PH. Hes6 regulates myogenic differentiation. Development. 2002;129:2195–207. doi: 10.1242/dev.129.9.2195. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, Nakano T, Honjo T, Mak TW, Rossant J, Conlon RA. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–48. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- Donovan J, Kordylewska A, Jan YN, Utset MF. Tetralogy of fallot and other congenital heart defects in Hey2 mutant mice. Curr Biol. 2002;12:1605–10. doi: 10.1016/s0960-9822(02)01149-1. [DOI] [PubMed] [Google Scholar]
- Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18:901–11. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Steidl C, Wagner TU, Lang E, Jakob PM, Friedl P, Knobeloch KP, Gessler M. Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ Res. 2007;100:856–63. doi: 10.1161/01.RES.0000260913.95642.3b. [DOI] [PubMed] [Google Scholar]
- Gessler M, Knobeloch KP, Helisch A, Amann K, Schumacher N, Rohde E, Fischer A, Leimeister C. Mouse gridlock: no aortic coarctation or deficiency, but fatal cardiac defects in Hey2 -/- mice. Curr Biol. 2002;12:1601–4. doi: 10.1016/s0960-9822(02)01150-8. [DOI] [PubMed] [Google Scholar]
- Gratton MO, Torban E, Jasmin SB, Theriault FM, German MS, Stifani S. Hes6 promotes cortical neurogenesis and inhibits Hes1 transcription repression activity by multiple mechanisms. Mol Cell Biol. 2003;23:6922–35. doi: 10.1128/MCB.23.19.6922-6935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Ohtsuka T, Bessho Y, Kageyama R. Generation of structurally and functionally distinct factors from the basic helix-loop-helix gene Hes3 by alternative first exons. J Biol Chem. 2000;275:19083–9. doi: 10.1074/jbc.M001075200. [DOI] [PubMed] [Google Scholar]
- Hirata H, Tomita K, Bessho Y, Kageyama R. Hes1 and Hes3 regulate maintenance of the isthmic organizer and development of the mid/hindbrain. EMBO J. 2001;20:4454–66. doi: 10.1093/emboj/20.16.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Zang K, Schmidt A, Saulys A, Xiang M, Reichardt LF. POU domain factor Brn-3a controls the differentiation and survival of trigeminal neurons by regulating Trk receptor expression. Development. 1999;126:2869–82. doi: 10.1242/dev.126.13.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Ito K, Osato M, Lee B, Bae SC, Ito Y. The transcription factor Runx3 represses the neurotrophin receptor TrkB during lineage commitment of dorsal root ganglion neurons. J Biol Chem. 2007;282:24175–84. doi: 10.1074/jbc.M703746200. [DOI] [PubMed] [Google Scholar]
- Inoue K, Ozaki S, Shiga T, Ito K, Masuda T, Okado N, Iseda T, Kawaguchi S, Ogawa M, Bae SC, Yamashita N, Itohara S, Kudo N, Ito Y. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat Neurosci. 2002;5:946–54. doi: 10.1038/nn925. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–48. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes L, Hamamori Y. HERP, a new primary target of Notch regulated by ligand binding. Mol Cell Biol. 2001;21:6071–9. doi: 10.1128/MCB.21.17.6071-6079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh F, Itoh S, Goumans MJ, Valdimarsdottir G, Iso T, Dotto GP, Hamamori Y, Kedes L, Kato M, ten Dijke Pt P. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J. 2004;23:541–51. doi: 10.1038/sj.emboj.7600065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali A, Bassuk AG, Kan L, Israsena N, Mukhopadhyay A, Hu M, Kessler JA. HeyL promotes neuronal differentiation of neural progenitor cells. doi: 10.1002/jnr.22562. unpublished. unpublished. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–8. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Klambt C, Knust E, Tietze K, Campos-Ortega JA. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J. 1989;8:203–10. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo H, Miyagawa-Tomita S, Nakazawa M, Saga Y, Johnson RL. Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev Biol. 2005;278:301–9. doi: 10.1016/j.ydbio.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Kokubo H, Miyagawa-Tomita S, Tomimatsu H, Nakashima Y, Nakazawa M, Saga Y, Johnson RL. Targeted disruption of hesr2 results in atrioventricular valve anomalies that lead to heart dysfunction. Circ Res. 2004;95:540–7. doi: 10.1161/01.RES.0000141136.85194.f0. [DOI] [PubMed] [Google Scholar]
- Kokubo H, Tomita-Miyagawa S, Hamada Y, Saga Y. Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development. 2007;134:747–55. doi: 10.1242/dev.02777. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N, Kim J, Anderson D, Kintner C. Hes6 acts in a positive feedback loop with the neurogenins to promote neuronal differentiation. Development. 2000;127:4203–16. doi: 10.1242/dev.127.19.4203. [DOI] [PubMed] [Google Scholar]
- Kramer I, Sigrist M, de Nooij JC, Taniuchi I, Jessell TM, Arber S. A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron. 2006;49:379–93. doi: 10.1016/j.neuron.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–50. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- Leimeister C, Externbrink A, Klamt B, Gessler M. Hey genes: a novel subfamily of hairy- and Enhancer of split related genes specifically expressed during mouse embryogenesis. Mech Dev. 1999;85:173–7. doi: 10.1016/s0925-4773(99)00080-5. [DOI] [PubMed] [Google Scholar]
- Leimeister C, Schumacher N, Steidl C, Gessler M. Analysis of HeyL expression in wild-type and Notch pathway mutant mouse embryos. Mech Dev. 2000;98:175–8. doi: 10.1016/s0925-4773(00)00459-7. [DOI] [PubMed] [Google Scholar]
- Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, Kremer E, Otto F, Brenner O, Lev-Tov A, Groner Y. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21:3454–63. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Saito T, Anderson DJ, Lance-Jones C, Jessell TM, Arber S. Functionally related motor neuron pool and muscle sensory afferent subtypes defined by coordinate ETS gene expression. Cell. 1998;95:393–407. doi: 10.1016/s0092-8674(00)81770-5. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–84. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier MM, Gessler M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun. 2000;275:652–60. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]
- Maro GS, Vermeren M, Voiculescu O, Melton L, Cohen J, Charnay P, Topilko P. Neural crest boundary cap cells constitute a source of neuronal and glial cells of the PNS. Nat Neurosci. 2004;7:930–8. doi: 10.1038/nn1299. [DOI] [PubMed] [Google Scholar]
- McEvilly RJ, Erkman L, Luo L, Sawchenko PE, Ryan AF, Rosenfeld MG. Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature. 1996;384:574–7. doi: 10.1038/384574a0. [DOI] [PubMed] [Google Scholar]
- Mu X, Silos-Santiago I, Carroll SL, Snider WD. Neurotrophin receptor genes are expressed in distinct patterns in developing dorsal root ganglia. J Neurosci. 1993;13:4029–41. doi: 10.1523/JNEUROSCI.13-09-04029.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa O, McFadden DG, Nakagawa M, Yanagisawa H, Hu T, Srivastava D, Olson EN. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc Natl Acad Sci U S A. 2000;97:13655–60. doi: 10.1073/pnas.250485597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa O, Nakagawa M, Richardson JA, Olson EN, Srivastava D. HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev Biol. 1999;216:72–84. doi: 10.1006/dbio.1999.9454. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Senzaki K, Yoshikawa M, Nishimura M, Inoue K, Ito Y, Ozaki S, Shiga T. Dynamic regulation of the expression of neurotrophin receptors by Runx3. Development. 2008;135:1703–11. doi: 10.1242/dev.015248. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Sakakibara S, Miyata T, Ogawa M, Shimazaki T, Weiss S, Kageyama R, Okano H. The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J Neurosci. 2000;20:283–93. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pissarra L, Henrique D, Duarte A. Expression of hes6, a new member of the Hairy/Enhancer-of-split family, in mouse development. Mech Dev. 2000;95:275–8. doi: 10.1016/s0925-4773(00)00348-8. [DOI] [PubMed] [Google Scholar]
- Ringstedt T, Kucera J, Lendahl U, Ernfors P, Ibanez CF. Limb proprioceptive deficits without neuronal loss in transgenic mice overexpressing neurotrophin-3 in the developing nervous system. Development. 1997;124:2603–13. doi: 10.1242/dev.124.13.2603. [DOI] [PubMed] [Google Scholar]
- Sakagami T, Sakurada K, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure and chromosomal locus of the mouse gene encoding a cerebellar Purkinje cell-specific helix-loop-helix factor Hes-3. Biochem Biophys Res Commun. 1994;203:594–601. doi: 10.1006/bbrc.1994.2224. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Hirata H, Ohtsuka T, Bessho Y, Kageyama R. The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J Biol Chem. 2003;278:44808–15. doi: 10.1074/jbc.M300448200. [DOI] [PubMed] [Google Scholar]
- Sakata Y, Kamei CN, Nakagami H, Bronson R, Liao JK, Chin MT. Ventricular septal defect and cardiomyopathy in mice lacking the transcription factor CHF1/Hey2. Proc Natl Acad Sci U S A. 2002;99:16197–202. doi: 10.1073/pnas.252648999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–34. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Satow T, Bae SK, Inoue T, Inoue C, Miyoshi G, Tomita K, Bessho Y, Hashimoto N, Kageyama R. The basic helix-loop-helix gene hesr2 promotes gliogenesis in mouse retina. J Neurosci. 2001;21:1265–73. doi: 10.1523/JNEUROSCI.21-04-01265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider WD. Functions of the neurotrophins during nervous system development: what the knockouts are teaching us. Cell. 1994;77:627–38. doi: 10.1016/0092-8674(94)90048-5. [DOI] [PubMed] [Google Scholar]
- Steidl C, Leimeister C, Klamt B, Maier M, Nanda I, Dixon M, Clarke R, Schmid M, Gessler M. Characterization of the human and mouse HEY1, HEY2, and HEYL genes: cloning, mapping, and mutation screening of a new bHLH gene family. Genomics. 2000;66:195–203. doi: 10.1006/geno.2000.6200. [DOI] [PubMed] [Google Scholar]
- Thomas PQ, Rathjen PD. HES-1, a novel homeobox gene expressed by murine embryonic stem cells, identifies a new class of homeobox genes. Nucleic Acids Res. 1992;20:5840. doi: 10.1093/nar/20.21.5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliauskas D, Stern CD. Expression of mouse HES-6, a new member of the Hairy/Enhancer of split family of bHLH transcription factors. Mech Dev. 2000;98:133–7. doi: 10.1016/s0925-4773(00)00443-3. [DOI] [PubMed] [Google Scholar]
- White FA, Silos-Santiago I, Molliver DC, Nishimura M, Phillips H, Barbacid M, Snider WD. Synchronous onset of NGF and TrkA survival dependence in developing dorsal root ganglia. J Neurosci. 1996;16:4662–72. doi: 10.1523/JNEUROSCI.16-15-04662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Gan L, Zhou L, Klein WH, Nathans J. Targeted deletion of the mouse POU domain gene Brn-3a causes selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proc Natl Acad Sci U S A. 1996;93:11950–5. doi: 10.1073/pnas.93.21.11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Small EM, van Rooij E, Qi X, Richardson JA, Srivastava D, Nakagawa O, Olson EN. Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function. Proc Natl Acad Sci U S A. 2007;104:7975–80. doi: 10.1073/pnas.0702447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Mehler MF, Song Q, Kessler JA. Development of bone morphogenetic protein receptors in the nervous system and possible roles in regulating trkC expression. J Neurosci. 1998;18:3314–26. doi: 10.1523/JNEUROSCI.18-09-03314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.