Summary
TRAIL selectively kills diseased cells in vivo, spurring interest in this death ligand as a potential therapeutic. However, many cancer cells are resistant to TRAIL suggesting the mechanism mediating TRAIL-induced apoptosis is complex. Here we identify PACS-2 as an essential TRAIL effector, required for killing tumor cells in vitro and virally infected hepatocytes in vivo. PACS-2 is phosphorylated at Ser437 in vivo and pharmacologic and genetic studies demonstrate Akt is an in vivo Ser437 kinase. Akt cooperates with 14-3-3 to regulate the homeostatic and apoptotic properties of PACS-2 that mediate TRAIL action. Phosphorylated Ser437 binds 14-3-3 with high affinity, which represses PACS-2 apoptotic activity and is required for PACS-2 to mediate trafficking of membrane cargo. TRAIL triggers dephosphorylation of Ser437, reprogramming PACS-2 to promote apoptosis. Together, these studies identify the phosphorylation state of PACS-2 Ser437 as a molecular switch that integrates cellular homeostasis with TRAIL-induced apoptosis.
The tumor necrosis factor (TNF) superfamily directs signaling pathways that elicit diverse responses ranging from tumor surveillance and inflammation to organogenesis and autoimmunity (Jin and El-Deiry, 2005). The prototypic superfamily ligand, TNFα, is a principal proinflammatory cytokine that mediates NF-κB signaling. However, when NF-κB is repressed, TNFα signals to the cytosolic death domain of the TNFR1 receptor to trigger apoptosis (Ashkenazi and Herbst, 2008). Two additional TNFα superfamily members, FasL and TRAIL, also trigger apoptosis by binding their death-domain containing receptors. The ability of these three TNFα death ligands to eliminate tumors in vivo has been intensively studied (Jin and El-Deiry, 2005). Unfortunately, the severe toxicity elicited by systemic administration of TNFα and FasL prevents their development as cancer therapeutics (Rowinsky, 2005). By contrast, binding of TRAIL to its apoptotic receptors DR4 and DR5 selectively kills cancerous and virally infected cells in vivo without causing harm to healthy cells (Ashkenazi and Herbst, 2008). This ability of TRAIL to selectively kill diseased cells has spurred clinical studies to test the efficacy of TRAIL as a potential therapeutic.
TRAIL is expressed primarily in lymphoid cells and is a multifunctional ligand important for regulation of T-cell homeostasis, innate immunity and tumor surveillance (Grosse-Wilde et al., 2008; Janssen et al., 2005). Virus infection induces DR4 and DR5 expression on the cell surface, which bind TRAIL to trigger the innate immune response leading to the apoptotic destruction of infected cells (Mundt et al., 2005). Similarly, neoplastic transformation of lung, pancreas and colon induce high levels of DR4 and DR5 (Rowinsky, 2005), which in metastatic colorectal cancer correlates with an improved survival rate (Strater et al., 2002). However, approximately 50% of tested tumor cell lines are resistant to TRAIL (Kruyt, 2008), suggesting that the underlying regulation of TRAIL-induced apoptosis is complex. Understanding the molecular basis of TRAIL-induced apoptosis may therefore prove useful in developing new therapeutic strategies.
TRAIL triggers the extrinsic pathway of apoptosis by binding to DR4 and DR5, which directs recruitment of the adaptor FADD and the initiator caspase, procaspase-8, to assemble a death-inducing signaling complex (DISC). Two types of signaling pathways emanate from the DISC to execute the apoptotic program (Jin and El-Deiry, 2005). In type I cells, the DISC/caspase-8 signal is sufficiently robust to directly activate executioner caspases, including caspase-3. Type II cells require an amplification step to activate executioner caspases. This signal is mediated by the proapoptotic Bcl-2 family protein Bid, which links the extrinsic apoptotic pathway to the intrinsic pathway by triggering mitochondria membrane permeabilization and caspase-9 activation, thereby amplifying caspase-3 activation (Aslan and Thomas, 2009; Jin and El-Deiry, 2005). The apoptotic activity of Bid is regulated by proteolysis and by PACS-2, a multi-functional sorting protein that integrates secretory pathway traffic with ER-mitochondrial communication and apoptosis (Atkins et al., 2008; Kottgen et al., 2005; Simmen et al., 2005, Jin and El-Deiry, 2005: Aslan and Thomas, 2009; Youker et al., 2009). Apoptotic inducers switch PACS-2 from a secretory pathway trafficking protein to an apoptotic protein that mediates Bid translocation to mitochondria and Bid cleavage (Simmen et al., 2005). But how apoptotic cues reprogram PACS-2 to become apoptotic is unknown.
Regulators of TRAIL action include a collection of death-inducing and decoy receptors, and several intracellular molecules, including Bcl-2 proteins, c-myc and Akt (Jin and El-Deiry, 2005). The class I PI3K/Akt survival pathway can be tumorigenic (Vivanco and Sawyers, 2002) and constitutive activation of Akt promotes resistance of tumors to apoptotic inducers, including TRAIL (Martelli et al., 2003; Shankar and Srivastava, 2004). Akt also combines with 14-3-3 proteins to repress the action of several proapoptotic proteins, including the Bcl-2 proteins Bad, Bim and the proapoptotic FOXO transcriptional regulators (Manning and Cantley, 2007; Porter et al., 2006).
Unlike Bad and Bim, Bid neither binds 14-3-3 proteins nor is it a known Akt substrate. Yet, elevated expression of Akt prevents TRAIL-induced apoptosis at the level of Bid cleavage (Chen et al., 2001; Kandasamy and Srivastava, 2002; Nesterov et al., 2001). Here we report TRAIL-mediated apoptosis in vivo requires PACS-2 and that Akt phosphorylation of PACS-2 at Ser437 creates a 14-3-3 binding site that regulates PACS-2 apoptotic activity. Our results identify a novel link in the regulation of TRAIL-mediated apoptosis and the molecular mechanism underlying the switch in PACS-2 from a homeostatic to an apoptotic regulator.
Results
TRAIL-mediated apoptosis requires PACS-2 in vivo
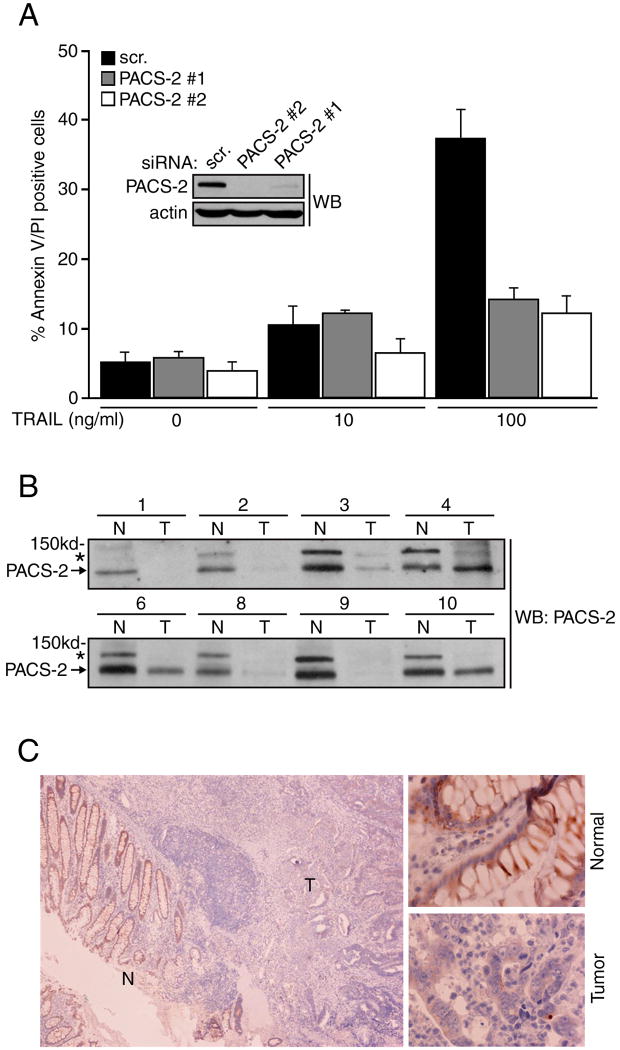
To test whether PACS-2 mediates TRAIL-induced apoptosis, we transfected the type II colon carcinoma cell line HCT116 with a control siRNA or with siRNAs that specifically knocked down PACS-2 (Fig. 1a, inset). The cells were then treated with increasing concentrations of TRAIL for 16 hr and quantification of apoptosis by Annexin V/propidium iodide staining revealed that siRNA knockdown of PACS-2 repressed TRAIL-mediated apoptosis (Fig. 1a). Similar results were obtained using DLD-1 colon carcinoma cells (Fig. S1). The ability of multiple PACS-2 siRNAs to repress apoptosis suggests these results were not due to an off-target effect.
Figure 1. PACS-2 is required for TRAIL-mediated tumor cell death.
(A) HCT116 cells were co-nucleofected with pmaxGFP and either a control siRNA (scr.) or PACS-2 siRNAs (#1, Qiagen, #2, Dharmacon) and then treated with 0, 10 or 100 ng/ml TRAIL for 16 hr. GFP+ cells were analyzed for apoptosis by flow cytometry. Data are represented as mean +/- SEM. Insets: Western blot of PACS-2 in control and PACS-2 knockdown cells. (B) Matched colorectal tumor (T) and adjacent normal (N) colonic tissue from eight volunteers with metastatic (samples 1-3) or stage 3 (samples 4,6 8-10) cancer were homogenized and 100 μg of total protein from each sample was analyzed by western blot. Arrow, PACS-2; *, 150 kDa PACS-2 immunoreactive protein of unknown identity. (C) A paraffin block of colonic tumor was sectioned and analyzed for PACS-2 expression by immunohistochemistry. Left, Normal colonic epithelium (N) and adjacent tumor (T). Right, Magnification of normal and tumor regions.
Consistent with a proapoptotic role for PACS-2 in HCT116 colon carcinoma cells, the PACS-2 locus, located at chromosome 14q32.33, is highly susceptible to loss of heterozygosity in colorectal cancer (Anderson et al., 2001), suggesting that PACS-2 protein expression may be altered during the colon adenoma-carcinoma transition. Therefore, matched cancerous and adjacent normal colon tissue was collected from eight volunteers with metastatic or stage 3 cancers and the expression of PACS-2 in each sample was measured by western blot (Fig. 1b). We found that PACS-2 protein expression was lost in 4 out of 8 cancerous samples. Immunohistochemical staining corroborated these findings, demonstrating PACS-2 was detected in the normal colonic epithelium but absent in the adjacent carcinoma (Fig. 1c). These results suggest PACS-2 expression is frequently lost in colorectal cancer.
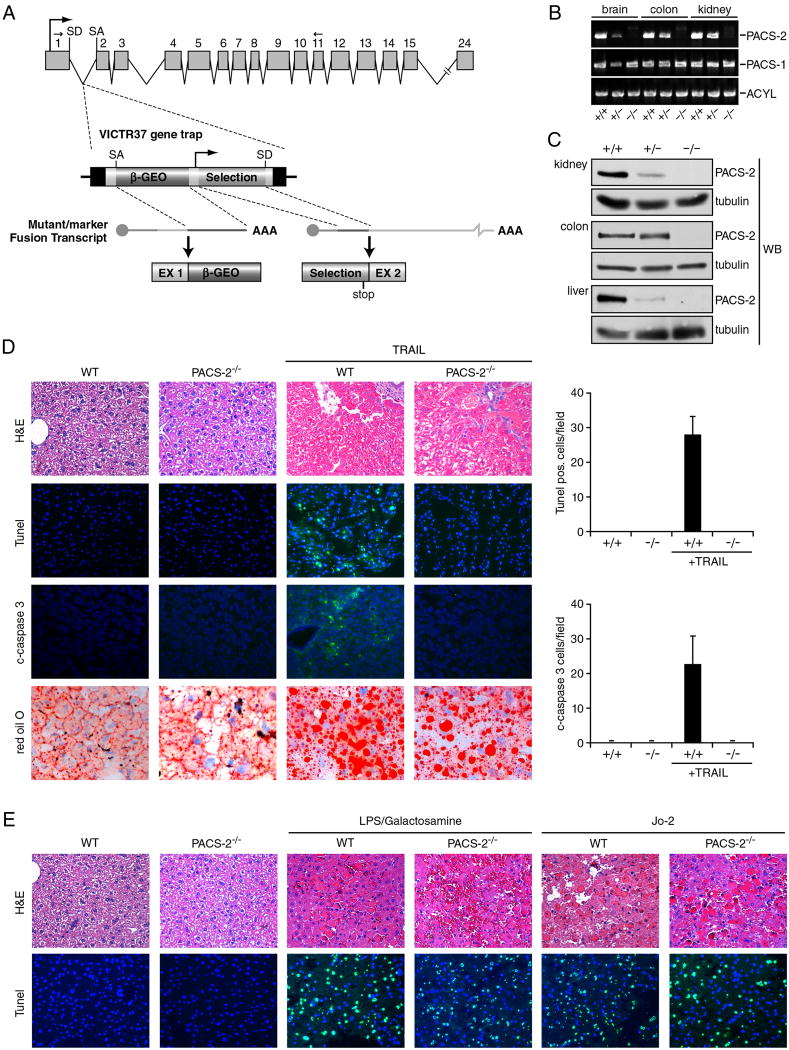
To test the physiological role of PACS-2 as a mediator of TRAIL-dependent apoptosis in vivo, we generated PACS-2-gene trapped mice (Fig. 2a). These mice were viable but lacked detectable 130 kDa (Mr) PACS-2 expression in all organs tested as determined by RT-PCR and western blot (Fig. 2b and c). The mice were subjected to a validated experimental paradigm of TRAIL- and DR5-dependent hepatic apoptosis and steatosis using recombinant adenoviruses that mimic the pathology caused by hepatitis C virus (Mundt et al., 2005). Littermate WT and PACS-2 gene trap mice (hereafter called PACS-2-/- mice) were injected in the tail vein with a recombinant adenovirus expressing GFP for 24 hr to induce DR5 expression (Mundt et al., 2005), followed by injection with a second adenovirus expressing TRAIL, which triggers hepatocyte apoptosis. In agreement with others (Mundt et al., 2005), TRAIL induced significant apoptosis in the infected livers of WT mice as determined by TUNEL staining and formation of cleaved caspase-3 (Fig. 2d). By contrast, TRAIL-induced apoptosis was markedly diminished in livers of PACS-2-/- mice. Surprisingly, the absence of PACS-2 did not prevent TRAIL from eliciting pronounced DR5-dependent hepatic steatosis as indicated by the increased Oil Red O staining of accumulated lipids (Fig. 2d). The TRAIL-induced hepatic apoptosis paradigm was extended to conduct a side-by-side comparison of the role of PACS-2 in mediating apoptosis induced by the other death ligands. We treated WT and PACS-2-/- mice with the anti-Fas antibody Jo2 or with LPS/Galactosamine to trigger TNFα-induced apoptosis (Fig. 2e). The extent of hepatic apoptosis in WT and PACS-2-/- mice treated with these agents was similar, suggesting that PACS-2 is selectively required for TRAIL-induced apoptosis of virally infected hepatocytes.
Figure 2. PACS-2 is required for TRAIL-mediated apoptosis in vivo.
(A) VICTR37 retroviral gene trap inserted into intron 1 of the murine PACS-2 gene. The gene trap (PACS-2GT(OST426086)) produces a truncated protein chimera containing only the first 40 amino acids of the 889 amino acid PACS-2. Arrows above exons 1 and 11 depict PCR primers used in panel B. SD, splice donor site; SA, splice acceptor. (B) RNA was isolated from PACS-2 WT (PACS-2+/+), heterozygote (PACS-2+/GT(OST426086), hereafter called PACS-2+/-), or PACS-2 gene trap ((PACS-2 GT(OST426086)/GT(OST426086), hereafter called PACS-2-/-) littermates and analyzed by RT-PCR to detect PACS-1, PACS-2 and ATP citrate lyase (ACYL, positive control). (C) Tissues from WT, +/- or -/- mice were analyzed by western blot for PACS-2. (D) WT and PACS-2-/- mice (n = 4 in each group) were injected with AdGFP and mouse AdTrail. TUNEL and c-caspase-3 positive cells were counted in liver sections from WT and PACS-2-/- mice were visualized (magnification, 200x) and quantified as the number of TUNEL- and cleaved caspase 3-positive cells per 20X field (graphs). Liver steatosis was detected by Oil Red O staining. (E) WT and PACS-2-/- mice (n = 4 in each group) were injected with the mAb Jo2 to stimulate Fas-mediated apoptosis or with LPS /GalN to stimulate TNFα-mediated apoptosis and TUNEL-positive nuclei were counted (original magnification 200x).
TRAIL requires PACS-2 to trigger cleavage of Bid and executioner caspases
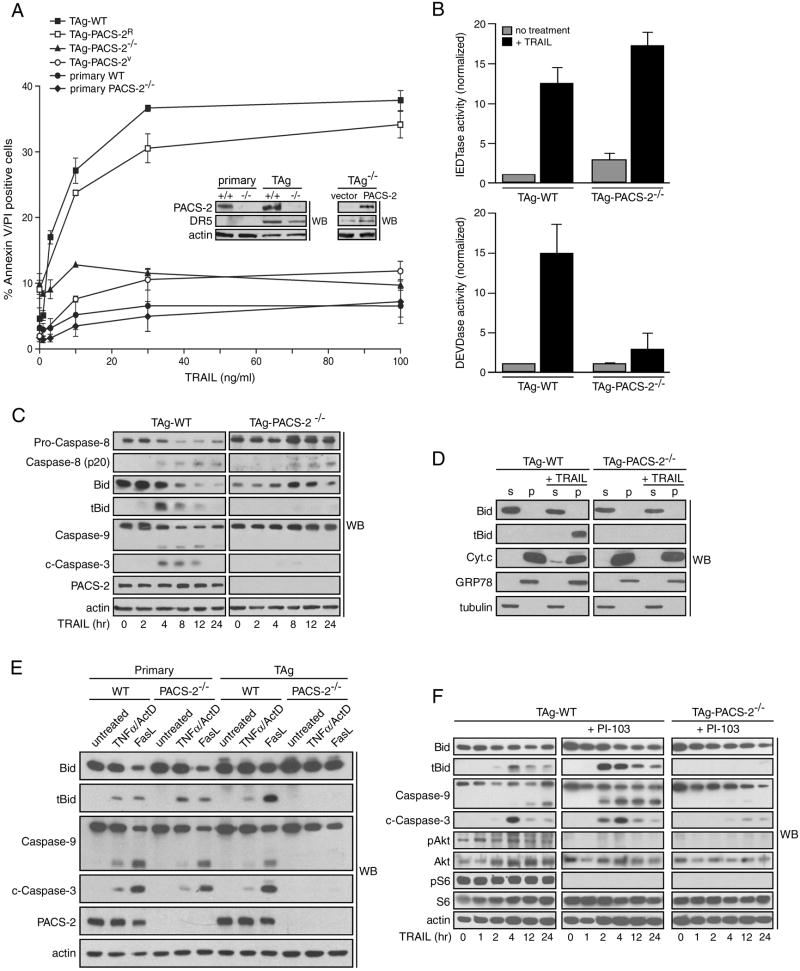
To understand the molecular basis underlying how TRAIL induces PACS-2 to become an apoptotic effector, we examined the ability of TRAIL to trigger apoptosis in embryonic fibroblasts (MEFs) from WT and PACS-2-/- mice. In agreement with others (Wiley et al., 1995), primary WT and PACS-2-/- MEFs were resistant to TRAIL (Fig. 3a). To test whether PACS-2 was required for TRAIL-induced apoptosis, we transformed the primary MEFs with SV40 large T antigen (TAg), which induced expression of the TRAIL receptor DR5 (Fig. 3a, inset). We then treated the TAg-transformed cells with increasing concentrations of TRAIL and measured apoptosis. We found that the TAg-WT MEFs were sensitive to TRAIL. By contrast, the TAg-PACS-2-/- MEFs remained resistant to TRAIL at all concentrations examined. To determine whether loss of PACS-2 was causal to TRAIL resistance, we generated a TAg-PACS-2-/- cell line rescued for PACS-2 expression (TAg-PACS-2R) and found that re-expression of PACS-2 restored sensitivity to TRAIL (Fig. 3a). These findings suggest the resistance of TAg-PACS-2-/- MEFs to TRAIL resulted specifically from loss of PACS-2 and not from an artifact of cell transformation.
Figure 3. PACS-2 is required for TRAIL-induced apoptosis of SV40 large T antigen-transformed MEFs.
(A) Primary WT and PACS-2-/- MEFs, as well as TAg-WT, TAg-PACS-2-/-, TAg-PACS-2V or TAg-PACS-2R MEFs were treated with increasing concentrations of TRAIL for 16 hr and then analyzed for apoptosis by Annexin V/ propidium iodide staining. Inset, western blot of PACS-2 and DR5 expression in the cultures. Data are represented as mean +/- SEM. (B) Lysates from TAg-WT and TAg-PACS-2-/- MEFs treated or not with 25 ng/ml TRAIL for 4 hr were incubated with the caspase-8 substrate Ac-IETD-AFC or the caspase-3 substrate Ac-DEVD-AFC and free AFC fluorescence was quantified. Data are represented as mean +/- SEM. (C) TAg-WT and TAg-PACS-2-/- MEFs were treated with 50 ng/ml TRAIL for the times indicated and the cleavage of each apoptotic factor was determined by western blot. Actin, loading control. (D) TAg-WT and TAg-PACS-2-/- MEFs were treated or not with 25 ng/ml TRAIL for 4 hr. Cells were permeabilized with digitonin and the cytosol supernatant (S) and membrane pellets (P) were separated by centrifugation. GRP78, organelle pellet marker; tubulin, cytosol marker. (E) Primary or TAg MEFs were treated with 100 ng/ml TNFα plus 1 μg/ml Actinomycin D or 250ng/ml FasL for 4 hr and analyzed as described in panel C. (F) TAg-WT and TAg-PACS-2-/- MEFs were pretreated or not with vehicle (DMSO) or 1 μM PI-103 for 1 hr then treated with 50 ng/ml TRAIL for the indicated times and analyzed as described in panel C.
To gain an understanding of the mechanism by which PACS-2 mediates TRAIL-induced apoptosis, we first measured the effect of TRAIL on the enzyme activity of initiator and executioner caspases in TAg-WT and TAg-PACS-2-/- cells. Parallel plates of TAg-WT and TAg-PACS-2-/- cells were treated with TRAIL for 4 hr and the amount of caspase-8 and -3 enzyme activity in cell lysates was measured using fluorogenic substrates (Fig. 3b). Despite the slightly reduced expression of DR5 in TAg-PACS-2-/- cells, TRAIL induced similar amounts of caspase-8 activity in TAg-WT and TAg-PACS-2-/- cells. By contrast, caspase-3 activity was induced only in TAg-WT cells. These results suggest PACS-2 acts subsequent to caspase-8 activation but prior to caspase-3 activation.
Next, to identify the step in TRAIL-induced apoptosis mediated by PACS-2, we measured the cleavage/activation of initiator and executioner caspases as well as Bid cleavage in TAg-WT and TAg-PACS-2-/- cells (Fig. 3c). In TRAIL-treated TAg-WT cells, formation of cleaved caspase-8 coincided with the cleavage of Bid as well cleavage of caspases -9 and -3. In TAg-PACS-2-/- cells, TRAIL induced cleavage of caspase-8. However, formation of tBid, and the subsequent activation of caspases-9 and -3 were inhibited. Interestingly, in TAg-PACS-2-/- cells, TRAIL induced accumulation of 57 kDa procaspase-8. This accumulation together with the lack of caspase-3 activation is consistent with studies suggesting activated caspase-3 mediates a positive feedback loop required for maximal cleavage of procaspase-8 (Lakhani et al., 2006).
The failure of TRAIL to induce caspase-3 activation in TAg-PACS-2-/- MEFs was consistent with our earlier studies that PACS-2 mediates recruitment of Bid to mitochondria leading to membrane permeabilization and cytochrome-c release (Simmen et al., 2005). To determine whether these steps in the apoptotic cascade were blocked in TAg-PACS-2-/- MEFs, we used the plasma membrane selective detergent digitonin to resolve cytosolic from membrane-associated proteins in TRAIL-treated MEFs. We found that TRAIL induced accumulation of tBid on the mitochondria-containing membrane fraction and release of cytochrome c to the cytosol in TAg-WT but not TAg-PACS-2-/- MEFs (Fig. 3d). These results support our prior findings that PACS-2 mediates Bid action downstream of caspase-8 activation (Simmen et al., 2005).
The requirement for Bid, but not PACS-2, for TNFα and FasL-induced apoptosis of healthy livers in vivo (Jin and El-Deiry, 2005 and Fig. 2), raised the possibility that PACS-2 may be selectively required for death ligand-induced apoptosis of diseased cells. To test this possibility directly, we treated parallel plates of primary WT and PACS-2-/- MEFs or TAg-WT and TAg-PACS-2-/- MEFs with TNFα or FasL and then measured apoptosis by western blot (Fig. 3e). We found that TNFα and FasL induced Bid cleavage and caspase-3 activation in primary WT and PACS-2-/- cells as well as in TAg-WT cells but, like TRAIL, not in TAg-PACS-2-/- cells. These results are in agreement with the selective requirement for PACS-2 by TRAIL in vivo in a viral hepatitis model (Fig. 2) and suggest PACS-2 is required by death ligands to induce apoptosis of diseased but not healthy cells.
Akt represses TRAIL action by interfering with Bid cleavage (Chen et al., 2001; Nesterov et al., 2001). We therefore asked whether inhibition of PI3K/Akt would rescue sensitivity of the TAg-PACS-2-/- cells to TRAIL. Parallel plates of TAg-WT and TAg-PACS-2-/- MEFs were treated with TRAIL in the presence or absence of the class I PI3K/mTOR inhibitor PI-103, which blocked Akt activation and phosphorylation of ribosomal protein S6 (Fig. 3f). We found that PI-103 markedly enhanced cleavage of Bid to tBid as well as activation of caspases-9 and -3. In TAg-PACS-2-/- cells, however, PI-103 had little effect on the ability of TRAIL to trigger cleavage of Bid or of procaspases-9 and -3.
Akt phosphorylates PACS-2 at Ser437
Given that inhibition of PI3K/Akt augmented Bid cleavage in TAg-WT but not TAg-PACS-2-/-cells, we asked if Akt may regulate Bid cleavage by acting directly on PACS-2. Inspection of the 889-amino acid PACS-2 protein sequence (Scansite) identified four candidate Akt phosphorylation sites at Ser43, Ser244, Ser435 and Ser437 (Fig. 4a). We therefore tested the ability of Akt to phosphorylate in vitro GST-fusion proteins corresponding to the four segments of PACS-2 (Fig. 4a), and found that Akt phosphorylated only PACS-2MR, which contains Ser244, Ser435 and Ser437 (Fig. 4b). To identify which of these sites were required for phosphorylation, we incubated Akt with PACS-2MR mutants containing Ser→Ala substitutions at Ser244, Ser435 or Ser437 and found that the Ser244Ala and Ser437Ala mutations reduced phosphorylation by ~50%, suggesting Akt phosphorylated both sites in vitro (Fig. 4c). This possibility was confirmed by determining that Akt could not phosphorylate a PACS-2MR mutant containing a Ser244,437Ala double substitution.
Figure 4. Akt phosphorylates PACS-2 Ser437 in vitro and vivo.
(A) Diagram of PACS-2 showing the N-terminal region (NTR), cargo-binding region (FBR), which contains Ser43, the middle region (MR), which contains Ser244, Ser435 and Ser437, and the C-terminal region (CTR). (B) The indicated GST-fusion proteins were incubated with [γ-32P]-ATP and constitutively active HA-Akt (WT) or kinase-dead HA-Akt (KD). Con, immunoisolation from mock-infected cells. Left, autoradiography; Right, input. (C) The indicated GST-fusion proteins were incubated with [γ-32P]-ATP and immunoisolated HA-Akt as described in panel B. Top, autoradiography; Bottom, input. (D) Peptides from in-gel limited trypsin digestion of HA-tagged PACS-2 were evaluated for candidate phosphopeptide by mass spectrometry. A phosphopeptide at [M+2H]=450.7, corresponding to the PACS-2 tryptic peptide RSTpS437LKER with one phosphorylation, was identified by nanoLC-MS/MS. (E) Left; Brain cytosol from WT and PACS-2-/- mice was incubated with affinity purified anti-PACS-2 (Ab 832) and immunoprecipitated proteins were analyzed by western blot using anti-PACS-2 (Ab 193) or mAb 81 to detect pSer437. Right; Liver and small intestine cytosol from WT mice were analyzed as described above. (F) Lysates from WT and Akt1-/- MEFs were analyzed as described in panel E to detect pSer437. (G) HCT116 cells were harvested in proliferative phase (untreated), serum-starved for 16 hr (starved), starved and refed with 20% serum for 3 hr (control), or starved and pre-treated with 1 μM PI-103, 20 nM RAD011, 1 μM BKM-120 or 20 nM BEZ-235 for one hr prior to refeeding with 20% serum for 3 hr. Cells were analyzed by western blot to detect total PACS-2 and PACS-2 pSer437. The pSer437:total PACS-2 ratio was calculated and normalized to the untreated sample. Data are represented as mean +/- SD.
To determine if PACS-2 Ser244 or Ser437 were phosphorylated in vivo, we first performed a mass spectrometry analysis of human PACS-2 purified from cultured cells. Purified PACS-2 trypsinolytic peptides were analyzed by tandem mass spectrometry (Fig. 4d). This analysis yielded the phosphopeptide STpS437LKER, demonstrating PACS-2 Ser437 is a bona fide kinase target in vivo. Identification of the phosphopeptide was confirmed by comparing the fragmentation pattern of the peptide released from PACS-2 in vivo with the synthetic peptide ST(pS)LKER (Fig. S2). The mass spectrometry analysis also identified phosphorylation of PACS-2 at Ser349 and Ser390, suggesting PACS-2 is phosphorylated at multiple sites in vivo (Figs. S3 and S4). The sequence coverage of PACS-2 did not include peptides containing Ser244, precluding phosphorylation analysis of this residue. Second, we identified an Akt substrate phosphospecific antibody, mAb 81, raised against the sequence (RX)RXXpS that recognizes PACS-2 pSer437 (Fig. S5). Western blot of endogenous PACS-2 immunoprecipitated from homogenates of brain, liver and small intestine revealed each tissue from WT mice contained mAb 81-positive PACS-2 (Fig. 4e). These results demonstrate PACS-2 is phosphorylated at Ser437 in multiple tissues.
Our determination that Akt phosphorylated PACS-2 Ser437 in vitro led us to ask whether Akt phosphorylates this site in vivo. We immunoprecipitated endogenous PACS-2 from parallel cultures of WT and Akt1-/- MEFs and then detected pSer437 by western blot (Fig. 4f). We found that PACS-2 was expressed in both WT and Akt1-/- MEFs, but PACS-2 pSer437 was detected only in WT MEFs, suggesting Akt1 is an in vivo PACS-2 Ser437 kinase.
To determine if PI3K/Akt-mediated survival signaling regulates phosphorylation of PACS-2 Ser437, HCT116 cells were serum starved overnight before re-feeding with serum in the absence or presence of either the class I PI3K/mTOR inhibitors PI-103 or BEZ-235, the class I PI3K inhibitor BKM-120 or the mTOR inhibitor RAD001 (Fig. 4g). Serum starvation decreased PACS-2 pSer437 levels 4-fold. Following addition of serum, pSer437 increased 6-fold and this increase was inhibited by PI-103, BEZ-235 or BKM-120 but not RAD001. The weaker inhibition of S6K1 by PI-103 compared to BEZ-235 is consistent with recent studies suggesting PI-103 more potently inhibits class I PI3K relative to mTOR (Thoreen et al., 2009). Interestingly, RAD001 not only blocked activation of S6K1 but also increased the amount of Akt pSer473 and PACS-2 pSer437, which agrees with studies reporting that inhibition of mTOR prevents the S6K1-mediated negative feedback on PI3K/Akt (Um et al., 2006). Together, these results suggest the basal level of phosphorylation at PACS-2 Ser437 is controlled by Akt and not S6K1, despite the ability of both kinases to phosphorylate this site in vitro (Fig. S6).
14-3-3 proteins bind PACS-2 pSer437
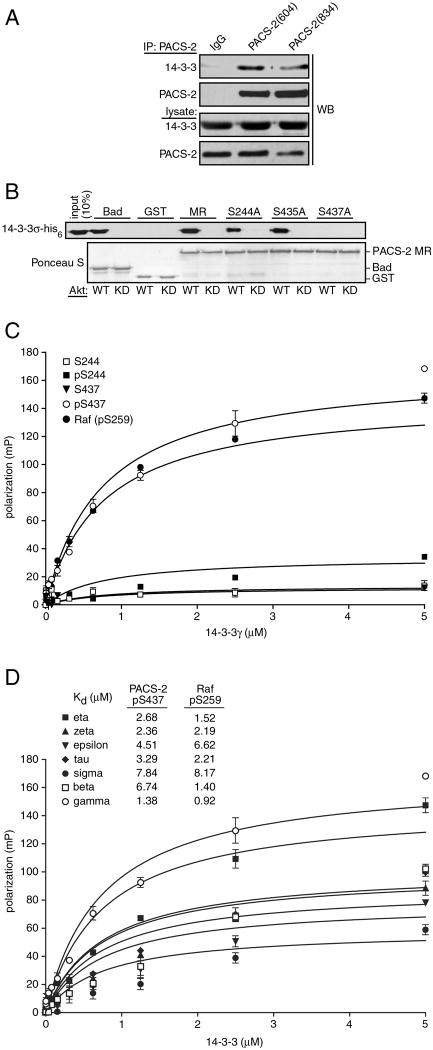
The regulation of proapoptotic activity by binding of 14-3-3 proteins to Akt-phosphorylated serine residues in Bad and FOXO (Porter et al., 2006) led us to ask whether 14-3-3 binds PACS-2 pSer437. We therefore immunoprecipitated PACS-2 from brain cytosol with two different PACS-2 antisera and found that 14-3-3 co-precipitated with PACS-2 (Fig. 5a). Next, GST fusion proteins encoding PACS-2MR or the PACS-2MR mutants containing Ser244Ala, Ser435Ala or Ser437Ala substitutions were pre-incubated with Akt and then mixed with His6-14-3-3 (Fig. 5b). We found that His6-14-3-3 bound to the Akt-phosphorylated PACS-2MR and the Ser244Ala and Ser435Ala mutants but not the Akt-phosphorylated Ser437Ala mutant, suggesting phosphorylation of PACS-2 Ser437 was required for binding 14-3-3 proteins. We then conducted a fluorescence polarization assay to determine quantitatively whether phosphorylated Ser437 was sufficient to bind 14-3-3 proteins. Rhodamine-conjugated, 16-mer peptides corresponding to non-phosphorylated or phosphorylated PACS-2 Ser244 or Ser437 were mixed with increasing concentrations of recombinant 14-3-3γ and the resultant effect on fluorescence polarization was recorded (Fig. 5c). We found only pSer437 peptide bound 14-3-3γ with an affinity similar to the Raf pSer259 positive control peptide. Additionally, we found that PACS-2 pSer437 bound all seven human 14-3-3 isoforms with Kds ranging from 1.34 to 7.84 μM (Fig. 5d), similar to other 14-3-3 client proteins (Du et al., 2006).
Figure 5. 14-3-3 proteins bind PACS-2 phospho-Ser437.
(A) PACS-2 was immunoprecipitated from rat brain cytosol and co-precipitating 14-3-3 was detected by western blot. (B) The indicated GST fusion proteins were incubated with ATP and constitutively active HA-Akt (WT) or kinase-dead HA-Akt (KD), then incubated with 14-3-3σ-his6. Protein complexes were captured with glutathione-sepharose and bound 14-3-3σ-his6 was detected by western blot. (C) (TMR)–labeled 16mer peptides containing pSer259-Raf, PACS-2 Ser244-PACS-2, pSer244-PACS-2, Ser437-PACS-2 or pSer437-PACS-2 (1 nM) were incubated in triplicate with increasing concentrations of GST-14-3-3γ. Fluorescence polarization signals were recorded and used to calculate Kds. (D) 14-3-3 isoforms were incubated with (TMR)-labeled pSer437-PACS-2 or pSer259-Raf peptides and analyzed as described in panel C. Calculated Kd values are shown.
PACS-2 pSer437 binding to 14-3-3 regulates TRAIL-induced apoptosis
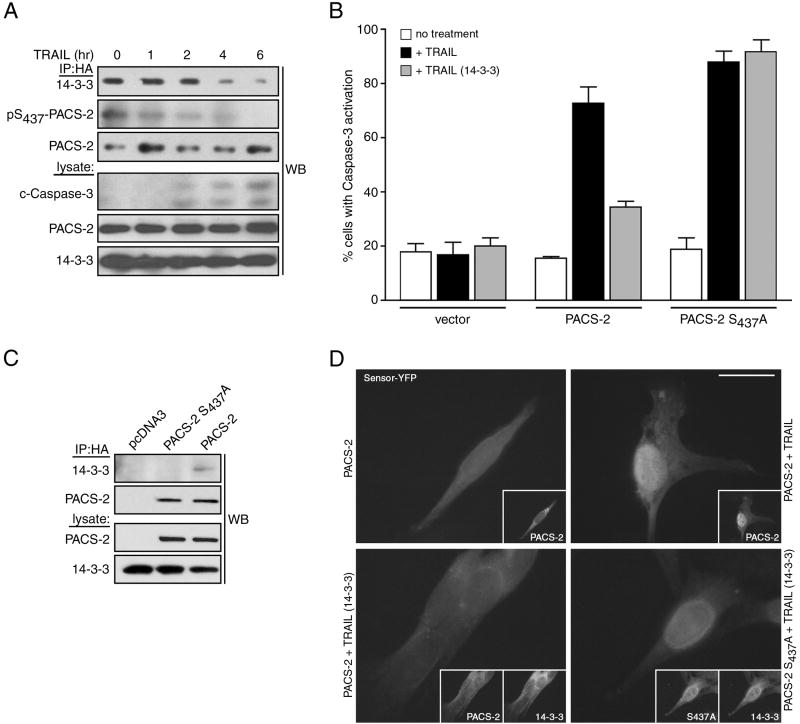
The binding of antiapoptotic 14-3-3 proteins to PACS-2 pSer437 suggested that apoptotic induction would disrupt binding of 14-3-3 to PACS-2, permitting its dephosphorylation. To test this possibility, we treated HCT116 cells co-expressing PACS-2ha and 14-3-3myc with TRAIL to induce apoptosis. PACS-2ha was immunoprecipitated at increasing times after treatment and the status of Ser437 phosphorylation and co-precipitated 14-3-3myc were determined (Fig. 6a). We found that TRAIL induced dephosphorylation of PACS-2 Ser437, which was temporally coupled with the release of bound 14-3-3myc and activation of caspase-3.
Figure 6. 14-3-3 regulates the apoptotic activity of PACS-2 Ser437.
(A) HCT116 cells expressing HA-tagged PACS-2 and myc-tagged 14-3-3ζ were treated with 25 ng/ml TRAIL for the indicated times. PACS-2ha immunoprecipitates were immunoblotted to detect co-precipitating 14-3-3ζ and pSer437. (B) TAg-PACS-2-/- cells were co-transfected with an eYFP-tagged caspase-3 reporter together with pcDNA3.1 (vector) or plasmids expressing HA-tagged PACS-2 or PACS-2S437A alone or with 14-3-3ζ-myc. Top: Cells treated with 10 ng/ml mouse TRAIL for 16 hr were fixed and the number of nuclear eYFP-positive cells indicating activated caspase-3 were analyzed. Data are represented as mean +/- SEM. Bottom: Representative immunofluorescence images of the quantified data. Insets, co-expression of 14-3-3ζ-myc and HA-tagged PACS-2 or PACS-2S437A. Scale bar, 20 μm. (C) TAg-PACS-2-/- cells described in B were lysed, PACS-2 immunoprecipitated and co-precipitating 14-3-3ζ-myc detected by western blot.
To determine if 14-3-3 regulates the apoptotic activity of PACS-2, we used a fluorescent protein-based reporter assay of caspase-3 activation. We co-transfected TAg-PACS-2-/- MEFs cells with plasmids expressing the eYFP-tagged caspase-3 reporter with HA-tagged PACS-2 or PACS-2S437A. The cells were then treated with TRAIL for 16 hr, fixed and the subcellular localization of eYFP was quantified as a marker of caspase-3 activation (Fig. 6b). We found that PACS-2 and PACS-2S437A increased the number of caspase-3-positive cells induced with TRAIL by nearly 5-fold over control cells. Additionally, we found that co-expressed 14-3-3myc repressed caspase-3 activity in cells transfected with PACS-2, but not PACS-2S437A. Consistent with these results, co-immunoprecipitation studies showed that 14-3-3myc interacted with HA-tagged PACS-2 but not PACS-2S437A (Fig. 6c). These results suggest that the binding of 14-3-3 to PACS-2 pSer437 regulates TRAIL-dependent apoptosis.
PACS-2 Ser437 integrates protein traffic with TRAIL-induced apoptosis
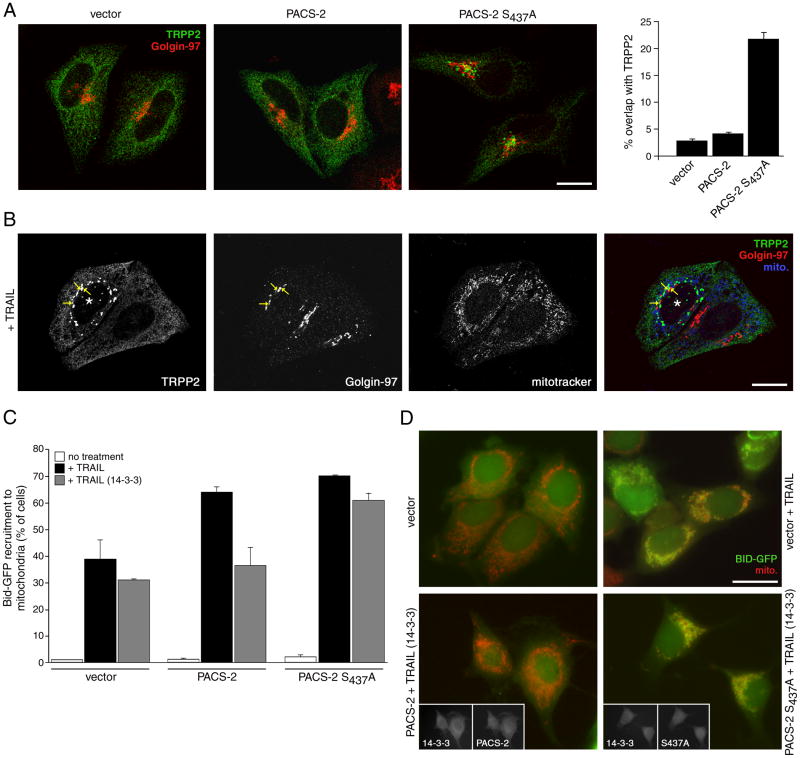
In non-apoptotic cells PACS-2 mediates the localization of cargo proteins to secretory pathway compartments (Youker et al., 2009). We therefore asked whether phosphorylated Ser437 was required for PACS-2 to mediate protein traffic. One PACS-2 cargo protein is the calcium channel polycystin-2 (hereafter called TRPP2), which localizes to the endoplasmic reticulum in a PACS-2-dependent manner (Kottgen et al., 2005). We co-expressed myc-TRPP2 with either a control vector, PACS-2 or PACS-2S437A and monitored its subcellular localization by confocal microscopy (Fig. 7a). In agreement with earlier studies (Kottgen et al., 2005), we found myc-TRPP2 localized to the ER in control cells or cells overexpressing PACS-2. By contrast, PACS-2S437A caused myc-TRPP2 to redistribute from the ER to the paranuclear region where it showed partial overlap with the late Golgi marker Golgin-97. As TRAIL induces PACS-2 Ser437 dephosphorylation (Fig. 6a), we next asked whether the death ligand would also affect the subcellular localization of myc-TRPP2. Similar to the effect of PACS-2S437A, TRAIL caused myc-TRPP2 to redistribute from the ER to a population of paranuclear compartments that showed a partial overlap with Golgin-97 (Fig. 7b). Morphometric analysis showed 78% of TRAIL-treated cells contained punctate myc-TRPP2 and collapsed mitochondria. These results suggest PACS-2 Ser437 dephosphorylation impairs the capacity of PACS-2 to mediate ER trafficking.
Figure 7. PACS-2 Ser437 and 14-3-3 integrate membrane traffic with apoptotic Bid action.
(A) HeLa cells were nucleofected (Amaxa) with myc-TRPP2 together with either empty vector or HA-tagged PACS-2 or PACS-2S437A. After 36 hr, the cells were fixed and processed for confocal microscopy using anti-myc (mAb 9E10, green) and anti-Golgin-97 (red) and visualized using Alexa-conjugated secondary antibodies. Right: Morphometric analysis. Error bars represent mean +/- SEM. Scale bar, 20 μm. (B) HeLa cells expressing myc-TRPP2 were treated with 20 ng/ml TRAIL for 4 hr and processed for confocal microscopy. Mitotracker (pseudocolored blue), anti-myc (green) and anti-Golgin-97 (red) are shown. *, apoptotic cell showing puncate myc-TRPP2 and collapsed mitochondria. Arrows, overlap of myc-TRPP2 and Golgin-97. Scale bar, 20 μm. (C) MCF-7:Bid-GFP cells were nucleofected (Amaxa) with pcDNA3.1 (vector) or plasmids expressing HA-tagged PACS-2 or PACS-2S437A alone or with 14-3-3ζ-myc. The cells were then treated with vehicle or 20 ng/ml TRAIL for 4 hr. Left: The percent of cells containing Bid-GFP translocated to mitochondria (mitotracker, red) in random fields (300 cells minimum) were quantified. Data are represented as mean +/- SD. Right: Immunofluorescence of MCF-7:Bid-GFP expressing the indicated proteins and treated or not with TRAIL. Insets, co-expression of 14-3-3ζ-myc with HA-tagged PACS-2 or PACS-2S437A. Scale bar, 20 μm.
As apoptotic induction triggers PACS-2 to mediate translocation of Bid to mitochondria (Simmen et al., 2005), we asked whether PACS-2 Ser437 dephosphorylation might reprogram PACS-2 from mediating secretory pathway trafficking to promoting Bid translocation. To test this possibility, MCF-7 cells stably expressing GFP-tagged Bid (MCF-7:Bid-GFP) transfected with plasmids expressing PACS-2 or PACS-2S437A or a control vector were treated with TRAIL. After 4 hr the cells were fixed and the translocation of Bid-GFP to mitochondria was quantified by microscopy (Fig. 7c). In agreement with previous studies (Simmen et al., 2005), we found that PACS-2 or PACS-2S437A enhanced the recruitment of Bid-GFP to mitochondria compared to cells expressing empty vector alone (Fig.7c). Next, we repeated the experiment in MCF-7:Bid-GFP co-expressing PACS-2 or PACS-2S437A with 14-3-3myc. We found that 14-3-3myc repressed the ability of PACS-2 but not PACS-2S437A to mediate Bid-GFP translocation to mitochondria. Together, these results suggest the binding of 14-3-3 to pSer437 governs a functional switch of PACS-2 between its roles in membrane traffic and TRAIL-mediated apoptosis.
Discussion
Here we identify PACS-2 as an TRAIL effector, required for killing Type II colon cancer cells in vitro and virally infected hepatocytes in vivo (Figs. 1 and 2). Current models of death ligand-induced apoptosis distinguish target cells as Type I or Type II based upon their requirement for mitochondria membrane permeabilization (Jin and El-Deiry, 2005). Emerging evidence, however, suggests that rather than this strict demarcation, a continuum exists whereby cell transformation or disease can alter cell homeostasis to require a mitochondrial amplification step for inducing cell death (Fehrenbacher et al., 2008). Indeed, our observations that PACS-2 is essential for TRAIL-induced hepatic apoptosis in a model of viral hepatitis but not for TNFα- or FasL-induced apoptosis in healthy liver, together with the ability of TNFα and FasL to efficiently induce apoptosis in primary but not TAg-transformed PACS-2-/- MEFs (Figs. 2 and 3), suggests that signaling pathways leading from death receptor activation to Bid cleavage may fundamentally differ between healthy and diseased (e.g. cancerous, transformed or virally infected) cells. Together, these findings suggest that the Type I/II descriptors warrant additional consideration of healthy versus diseased phenotypes, as diseased cells generally have increased antiapoptotic and survival signaling that may require the action of homeostatic regulators such as PACS-2 to enhance apoptosis through the Type II pathway.
The role of PACS-2 in mediating TRAIL action is not restricted to virally infected cells, as siRNA knockdown of PACS-2 repressed TRAIL-mediated killing of HCT116 and DLD-1 colorectal cancer cells (Figs. 1 and S1). Interestingly, the PACS-2 gene, located at chromosome 14q32.33, is lost in sporadic colorectal cancer (Anderson et al., 2001). In agreement with these genomic studies, our immunohistochemical and western blot analysis revealed that the PACS-2 protein is lost in cancer tissues of ~50% of the patients suffering from colorectal cancer (Fig. 1). These findings raise the possibility that loss of PACS-2 expression may contribute to the colorectal cancer sequence and may serve as a biomarker for TRAIL-resistant cancer. Future studies will investigate the frequency at which loss of heterozygosity at the PACS-2 locus correlates with loss of PACS-2 protein expression and the progression of colorectal cancer.
Although more than 100 putative in vitro Akt substrates have been described in the literature, only about 20 substrates have been rigorously confirmed as bona fide targets in vivo (Manning and Cantley, 2007). PACS-2 Ser437 is embedded within an evolutionally conserved consensus Akt phosphorylation site (Youker et al., 2009), and biochemical, pharmacologic and genetic experiments combine to strongly suggest Akt1 is a PACS-2 Ser437 kinase (Fig. 4). These studies, however, do not exclude the possibility that other kinases, including S6K1 (Fig. S6) may also phosphorylate PACS-2 Ser437. Indeed, in addition to Akt, a mitochondrial pool of S6K1 phosphorylates Bad Ser136 in vivo (Harada et al., 2001). RAD001, however, which blocks activation of S6K1 (Fig. 4), increased the basal phosphorylation of PACS-2 pSer437, presumably through inhibition of an mTOR-dependent negative feedback loop on IRS-1-mediated PI3K/Akt activation (Um et al., 2006). Whether S6K1 may regulate PACS-2 at the mitochondria remains a possibility.
The PI3K/Akt survival pathway increases resistance to chemotherapy, ionizing radiation and TRAIL in solid tumors (Martelli et al., 2003; Shankar and Srivastava, 2004). Correspondingly, inhibition of PI3K/Akt enhances TRAIL-induced apoptosis in tumor cells with weak response to TRAIL (Kandasamy and Srivastava, 2002; Vaculova et al., 2006). The ability of PI-103 to enhance TRAIL-mediated cell death in a PACS-2 dependent manner (Fig. 3), suggests the phosphorylation state of PACS-2 Ser437 represents a key point of regulation in the apoptotic response. Our finding that 14-3-3 can repress the TRAIL-induced apoptotic activity of PACS-2 but not PACS-2S437A (Figs. 6 and 7), supports this possibility. Moreover, the recent demonstration that the class I PI3K inhibitor BEZ-235, which promotes tumor regression in vivo (Engelman et al., 2008), blocks PACS-2 Ser437 phosphorylation (Fig. 4), raised the possibility that new generation PI3K inhibitors may be used to augment the therapeutic activity of TRAIL against PACS-2-expressing tumors.
Genetic and biochemical analyses have revealed that the PACS proteins are metazoan membrane traffic regulators that mediate organ homeostasis and have important roles in diverse pathologies and disease states (Youker et al., 2009). In healthy cells, cytosolic PACS-2 binds membrane cargo proteins containing acidic cluster sorting motifs and mediates their localization to distinct compartments. PACS-2 binds an acidic cluster ER-localization motif in the TRPP2 cytosolic domain and connects this motif to COPI in vitro (Kottgen et al., 2005). Loss of PACS-2 or disruption of the PACS-2—COPI interaction causes TRPP2 to mislocalize from the ER. Our determination that PACS-2S437A or treatment of cells with TRAIL, which induces PACS-2 Ser437 dephosphorylation, disrupts the subcellular localization of TRPP2 (Fig. 7), suggests binding of 14-3-3 to pSer437 may be required for PACS-2 to mediate the trafficking of its client proteins in non-apoptotic cells. How binding of PACS-2 to 14-3-3 and COPI cooperate to mediate cargo traffic in healthy versus apoptotic cells remains to be determined.
Our results suggest the phosphorylation state of PACS-2 Ser437 integrates the separable roles of PACS-2 in mediating membrane traffic in healthy cells with recruitment of Bid to mitochondria in apoptotic cells. Although dephosphorylation of PACS-2 Ser437 and release from 14-3-3 proteins mediates translocation of Bid to mitochondria, this event alone is not sufficient to promote Bid translocation (Fig. 7), indicating additional steps are required for PACS-2 to mediate Bid action. For example, Bid activity can be modulated by CK1 and CK2 phosphorylation proximal to Bid’s caspase cleavage site, which regulates cleavage by caspase-8 and also binding to PACS-2 (Desagher et al., 2001; Simmen et al., 2005 and Fig. 3). While caspase-8 can cleave Bid at Asp59 to form myristoylated tBid that permeabilizes mitochondria, caspase-8 activation is necessary but not sufficient for Bid activity in TRAIL-treated cells (Rokhlin et al., 2002 and see Fig. 3). Thus, the canonical caspase-8/myrisotylation model may not represent the sole mechanism of Bid regulation. Indeed, TRAIL also triggers lysosome permeabilization to release cathepsins, which can cleave Bid at sites other than Asp59 to promote TRAIL-induced cell death (Werneburg et al., 2007). These findings support a model whereby the cooperation of multiple organelles drives the early events leading from death receptor ligation to mitochondria membrane permeabilization (Aslan and Thomas, 2009). As PACS-2 mediates ER-mitochondria communication and endosomal traffic (Atkins et al., 2008; Kottgen et al., 2005; Simmen et al., 2005), our studies raise the possibility that TRAIL triggers dephosphorylated PACS-2 to alter interorganellar communication to drive apoptosis. This model is currently under investigation.
Experimental Procedures
Mice, cancer tissue, reagents, fluorescence polarization and mass spectrometry
Apoptosis assays
In vivo induction of apoptosis in mouse liver
Adenoviruses expressing GFP (Ad-GFP) and membrane-tethered TRAIL (Ad-TRAIL) were prepared as described (Mundt et al., 2003; Mundt et al., 2005). Infection of mice was carried out by administration of 0.25 ml of Ad-GFP virus and of Ad-TRAIL virus solution 24 hours after the first injection into the tail vein with 2×108 pfu/g. 24 hours later mice were sacrificed. To analyze the role of PACS-2 for Fas and TNFα-mediated apoptosis, mice were challenged with intraperitoneal administration of the monoclonal Fas antibody Jo-2 (0.7 μg/g, BD Pharmingen) and of LPS (0.5 μg/g) + D-galactosamine (700 μg/g, Sigma), respectively.
Flow cytometry assays
HCT116 cells, DLD-1 cells or MEFs were treated with recombinant human or mouse TRAIL (R&D Systems #375-TL and #1121-TL, respectively). Apoptosis assays were performed in the OHSU Flow Cytometry Core using Annexin V-FITC (Pharmingen) or Annexin V-APC (Calbiochem) and propidium iodide (Calbiochem) to assay MEFs or on GFP+ cells to assay HCT116 and DLD-1 cells and analyzed with a FACSCalibur (BD) cytometer and CellQuest acquisition/analysis software (BD) as previously described (Simmen et al., 2005).
Caspase activity assays
Cell lysates were mixed with Ac-IETD-AFC (caspase-8) or Ac-DEVD-AFC (caspase-3) fluorometric substrates (AnaSpec) in caspase activity buffer and enzyme reactions were incubated for one hr at 37°C and AFC product was quantified by fluorescence (λex = 400 nm, λex = 505 nm) using a Spectra Max M2/M2e microplate reader (Molecular Devices).
Membrane fractionation assay
Cells were resuspended in digitonin permeabilization buffer (DPB; 250 mM sucrose, 20 mM HEPES pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA and 2 mM DTT) supplemented with protease inhibitors and then incubated with 75 μM digitonin (Calbiochem) on ice for 10 min to permeabilize the plasma membrane. The cells were sedimented (1,500 ×g, 5min) to separate released cytosol from cell husks containing organelles. The fractions were then resuspended in SDS sample buffer and analyzed by western blot.
pSensor reporter assay
TAg-PACS-2-/- MEFs cells were co-transfected with pCaspase3-Sensor (Clontech) and the indicated plasmids, treated or not with mouse TRAIL and processed as indicated in the legend to Fig. 6. Five 20x fields of each sample were visualized Zeiss microscope using Openlab software (Improvision) and scored for eYFP localization in triplicate.
Bid-GFP recruitment assay
MCF-7:Bid-GFP cells were co-nucleofected (Amaxa) with the indicated plasmids, treated or not with human TRAIL and processed as indicated in the legend to Fig. 7 and in (Simmen et al., 2005). Images were captured using a 63x oil immersion objective on a Leica DM-RB microscope and Mamamatsu C4742-95 digital camera and processed with scion image 1.62.
Phosphorylation and binding assays
GST-tagged PACS-2 proteins were prepared as previously described (Simmen et al., 2005). Akt phosphorylated GST proteins (1 μg) were captured on Glutathione Sepharose, washed in binding buffer (20 mM Tris pH 7.4, 1% NP40, 1 mM EDTA), incubated with 10 μg 14-3-3σ-His6 for one hr at RT, washed and then separated by SDS-PAGE. Endogenous PACS-2 was immunoprecipitated from the indicated homogenate by incubating 10 μg affinity purified anti-PACS-2 overnight and then captured with Protein G agarose.
IHC and Confocal Microscopy
Immunohistochemistry
Formalin-fixed, paraffin embedded tissue samples from consenting patients were cut in 3 micron sections, placed on poly-lysine-coated slides, then deparaffinized and antigen was retrieved with heated citric acid (Ventana CC10). The samples were stained with PACS-2 Ab HPA001423 and visualized with diaminobenzamidine (Ventana). The slides were counterstained with hematoxylin and images were captured with a Nikon Eclipse 80i microscope equipped with a Nikon Digital Sight DS-L1 camera.
Confocal microscopy
HeLa cells were nucleofected (Amaxa) with the indicated plasmids. After 16 hr, cells were processed for confocal microscopy and cells were visualized with a 60x oil immersion objective on an Olympus Fluo-View FV300 laser scanning confocal microscope as described (Atkins et al., 2008). Co-localization was quantified morphometrically using Metamorph 7.0 as described (Atkins et al., 2008).
Supplementary Material
Acknowledgments
The authors thank N. Hay, T. Soderling, R. Cone, A. Aitken, P. Rotwein, G. Thomas, S. Somlo, H. Piwnica-Worms, M. Iordanov, A. Guo and B. Vogelstein for reagents, S. Nath and K. Benson for experiments in the early stages of this project and B. Druker and G. Thomas for critical reading of the manuscript. H.F. is Georgia Cancer Coalition Distinguished Cancer Scholar and a Georgia Research Alliance Distinguished Investigator. This work was supported by Deutsche Krebshilfe (AV), and NIH grants T32 AI07472 (JEA), T32 DK007680 (MHB), NRSA DK076343 (RTY), P01CA116676 and P50CA128613 (HF and YD), DK3724, AI49793, P50CA97186 and OCTRI RR0241 (GT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson GR, Brenner BM, Swede H, Chen N, Henry WM, Conroy JM, Karpenko MJ, Issa JP, Bartos JD, Brunelle JK, et al. Intrachromosomal genomic instability in human sporadic colorectal cancer measured by genome-wide allelotyping and inter-(simple sequence repeat) PCR. Cancer Res. 2001;61:8274–8283. [PubMed] [Google Scholar]
- Ashkenazi A, Herbst RS. To kill a tumor cell: the potential of proapoptotic receptor agonists. J Clin Invest. 2008;118:1979–1990. doi: 10.1172/JCI34359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan JE, Thomas G. Death by committee: Organellar Trafficking and Communication in Apoptosis. Traffic. 2009 doi: 10.1111/j.1600-0854.2009.00951.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins KM, Thomas L, Youker RT, Harriff MJ, Pissani F, You H, Thomas G. HIV-1 Nef Binds PACS-2 to Assemble a Multikinase Cascade That Triggers Major Histocompatibility Complex Class I (MHC-I) Down-regulation: ANALYSIS USING SHORT INTERFERING RNA AND KNOCK-OUT MICE. J Biol Chem. 2008;283:11772–11784. doi: 10.1074/jbc.M707572200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Thakkar H, Tyan F, Gim S, Robinson H, Lee C, Pandey SK, Nwokorie C, Onwudiwe N, Srivastava RK. Constitutively active Akt is an important regulator of TRAIL sensitivity in prostate cancer. Oncogene. 2001;20:6073–6083. doi: 10.1038/sj.onc.1204736. [DOI] [PubMed] [Google Scholar]
- Desagher S, Osen-Sand A, Montessuit S, Magnenat E, Vilbois F, Hochmann A, Journot L, Antonsson B, Martinou JC. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol Cell. 2001;8:601–611. doi: 10.1016/s1097-2765(01)00335-5. [DOI] [PubMed] [Google Scholar]
- Du Y, Masters SC, Khuri FR, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay. J Biomol Screen. 2006;11:269–276. doi: 10.1177/1087057105284862. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher N, Bastholm L, Kirkegaard-Sorensen T, Rafn B, Bottzauw T, Nielsen C, Weber E, Shirasawa S, Kallunki T, Jaattela M. Sensitization to the lysosomal cell death pathway by oncogene-induced down-regulation of lysosome-associated membrane proteins 1 and 2. Cancer Res. 2008;68:6623–6633. doi: 10.1158/0008-5472.CAN-08-0463. [DOI] [PubMed] [Google Scholar]
- Grosse-Wilde A, Voloshanenko O, Bailey SL, Longton GM, Schaefer U, Csernok AI, Schutz G, Greiner EF, Kemp CJ, Walczak H. TRAIL-R deficiency in mice enhances lymph node metastasis without affecting primary tumor development. J Clin Invest. 2008;118:100–110. doi: 10.1172/JCI33061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Andersen JS, Mann M, Terada N, Korsmeyer SJ. p70S6 kinase signals cell survival as well as growth, inactivating the pro-apoptotic molecule BAD. Proc Natl Acad Sci U S A. 2001;98:9666–9670. doi: 10.1073/pnas.171301998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen EM, Droin NM, Lemmens EE, Pinkoski MJ, Bensinger SJ, Ehst BD, Griffith TS, Green DR, Schoenberger SP. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature. 2005;434:88–93. doi: 10.1038/nature03337. [DOI] [PubMed] [Google Scholar]
- Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer Biol Ther. 2005;4:139–163. doi: 10.4161/cbt.4.2.1508. [DOI] [PubMed] [Google Scholar]
- Kandasamy K, Srivastava RK. Role of the phosphatidylinositol 3’-kinase/PTEN/Akt kinase pathway in tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in non-small cell lung cancer cells. Cancer Res. 2002;62:4929–4937. [PubMed] [Google Scholar]
- Kottgen M, Benzing T, Simmen T, Tauber R, Buchholz B, Feliciangeli S, Huber TB, Schermer B, Kramer-Zucker A, Hopker K, et al. Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. Embo J. 2005;24:705–716. doi: 10.1038/sj.emboj.7600566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruyt FA. TRAIL and cancer therapy. Cancer Lett. 2008;263:14–25. doi: 10.1016/j.canlet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Lakhani SA, Masud A, Kuida K, Porter GA, Jr, Booth CJ, Mehal WZ, Inayat I, Flavell RA. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli AM, Tazzari PL, Tabellini G, Bortul R, Billi AM, Manzoli L, Ruggeri A, Conte R, Cocco L. A new selective AKT pharmacological inhibitor reduces resistance to chemotherapeutic drugs, TRAIL, all-trans-retinoic acid, and ionizing radiation of human leukemia cells. Leukemia. 2003;17:1794–1805. doi: 10.1038/sj.leu.2403044. [DOI] [PubMed] [Google Scholar]
- Mundt B, Kuhnel F, Zender L, Paul Y, Tillmann H, Trautwein C, Manns MP, Kubicka S. Involvement of TRAIL and its receptors in viral hepatitis. Faseb J. 2003;17:94–96. doi: 10.1096/fj.02-0537fje. [DOI] [PubMed] [Google Scholar]
- Mundt B, Wirth T, Zender L, Waltemathe M, Trautwein C, Manns MP, Kuhnel F, Kubicka S. Tumour necrosis factor related apoptosis inducing ligand (TRAIL) induces hepatic steatosis in viral hepatitis and after alcohol intake. Gut. 2005;54:1590–1596. doi: 10.1136/gut.2004.056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterov A, Lu X, Johnson M, Miller GJ, Ivashchenko Y, Kraft AS. Elevated AKT activity protects the prostate cancer cell line LNCaP from TRAIL-induced apoptosis. J Biol Chem. 2001;276:10767–10774. doi: 10.1074/jbc.M005196200. [DOI] [PubMed] [Google Scholar]
- Porter GW, Khuri FR, Fu H. Dynamic 14-3-3/client protein interactions integrate survival and apoptotic pathways. Semin Cancer Biol. 2006;16:193–202. doi: 10.1016/j.semcancer.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Rokhlin OW, Guseva NV, Tagiyev AF, Glover RA, Cohen MB. Caspase-8 activation is necessary but not sufficient for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis in the prostatic carcinoma cell line LNCaP. Prostate. 2002;52:1–11. doi: 10.1002/pros.10074. [DOI] [PubMed] [Google Scholar]
- Rowinsky EK. Targeted induction of apoptosis in cancer management: the emerging role of tumor necrosis factor-related apoptosis-inducing ligand receptor activating agents. J Clin Oncol. 2005;23:9394–9407. doi: 10.1200/JCO.2005.02.2889. [DOI] [PubMed] [Google Scholar]
- Shankar S, Srivastava RK. Enhancement of therapeutic potential of TRAIL by cancer chemotherapy and irradiation: mechanisms and clinical implications. Drug Resist Updat. 2004;7:139–156. doi: 10.1016/j.drup.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Simmen T, Aslan JE, Blagoveshchenskaya AD, Thomas L, Wan L, Xiang Y, Feliciangeli SF, Hung CH, Crump CM, Thomas G. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. Embo J. 2005;24:717–729. doi: 10.1038/sj.emboj.7600559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strater J, Hinz U, Walczak H, Mechtersheimer G, Koretz K, Herfarth C, Moller P, Lehnert T. Expression of TRAIL and TRAIL receptors in colon carcinoma: TRAIL-R1 is an independent prognostic parameter. Clin Cancer Res. 2002;8:3734–3740. [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mTOR inhibitor reveals rapamycin-insensitive functions of mTORC1. J Biol Chem. 2009 doi: 10.1074/jbc.M900301200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um SH, D’Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Vaculova A, Hofmanova J, Soucek K, Kozubik A. Different modulation of TRAIL-induced apoptosis by inhibition of pro-survival pathways in TRAIL-sensitive and TRAIL-resistant colon cancer cells. FEBS Lett. 2006;580:6565–6569. doi: 10.1016/j.febslet.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Werneburg NW, Guicciardi ME, Bronk SF, Kaufmann SH, Gores GJ. Tumor necrosis factor-related apoptosis-inducing ligand activates a lysosomal pathway of apoptosis that is regulated by Bcl-2 proteins. J Biol Chem. 2007;282:28960–28970. doi: 10.1074/jbc.M705671200. [DOI] [PubMed] [Google Scholar]
- Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- Youker RT, Shinde U, Day R, Thomas G. At the crossroads of homeostasis and disease: roles of the pacs proteins in membrane traffic and apoptosis. Biochem J. 2009 doi: 10.1042/BJ20081016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Hutker S, Mundt B, Waltemathe M, Klein C, Trautwein C, Malek NP, Manns MP, Kuhnel F, Kubicka S. NFkappaB-mediated upregulation of bcl-xl restrains TRAIL-mediated apoptosis in murine viral hepatitis. Hepatology. 2005;41:280–288. doi: 10.1002/hep.20566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.